Abstract
Klebsiella pneumoniae is a nosocomial pathogen, pointed out by the World Helth Organisation (WHO) as “critical” regarding the highly limited options of treatment. Lipopolysaccharide (LPS, O-antigen) and capsular polysaccharide (K-antigen) are its virulence factors and surface antigens, determining O- and K-serotypes and encoded by O- or K-loci. They are promising targets for antibody-based therapies (vaccines and passive immunization) as an alternative to antibiotics. To make such immunotherapy effective, knowledge about O/K-antigen structures, drift, and distribution among clinical isolates is needed. At present, the structural analysis of O-antigens is efficiently supported by bioinformatics. O- and K-loci-based genotyping by polymerase chain reaction (PCR) or whole genome sequencing WGS has been proposed as a diagnostic tool, including the Kaptive tool available in the public domain. We analyzed discrepancies for O2 serotyping between Kaptive-based predictions (O2 variant 2 serotype) and the actual phenotype (O2 variant 1) for two K. pneumoniae clinical isolates. Identified length discrepancies from the reference O-locus resulted from insertion sequences (ISs) within rfb regions of the O-loci. In silico analysis of 8130 O1 and O2 genomes available in public databases indicated a broader distribution of ISs in rfbs that may influence the actual O-antigen structure. Our results show that current high-throughput genotyping algorithms need to be further refined to consider the effects of ISs on the LPS O-serotype.
Keywords: Klebsiella, O-antigen, lipopolysaccharide, LPS, O-serotype, NMR, WGS, kaptive
1. Introduction
Klebsiella pneumoniae is a Gram-negative bacterium which is part of the human microbiota; however, it is also a frequent cause of nosocomial and community-acquired infections in newborns, the elderly, and immunocompromised patients [1,2,3,4,5,6]. K. pneumoniae belongs to the ESKAPE group of pathogens (ESCAPE is an acronym for Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp.) [2,7] and to the top priority list of “critical” pathogens of the WHO [8], having been indicated as a major target for the development of new prevention and therapeutic strategies.
The global emergence of multidrug-resistant (MDR) strains, especially extended-spectrum β-lactamase (ESBL)- or/and carbapenemase-producing K. pneumoniae (CPKP), has become an ultimate challenge for public health [6,9]. Treatment options against CPKP are sparse and usually limited to only last-line antibiotics, if at all [5,9]. Some of the new therapeutic approaches under development are based on concepts of active and passive immunization against major surface antigens of K. pneumoniae−the capsular polysaccharide (CPS, K-antigen) and lipopolysaccharide (LPS, endotoxin, O-antigen) [5,10]. Several vaccine strategies targeting the most prevalent K-antigens of Klebsiella have been developed, particularly against K1 and K2 characteristics of hypervirulent strains [11,12,13].
Contrary to highly variable K-antigens (more than 80 types) [1,14,15], LPS represents the less variable antigen and is an important virulence factor, triggering the Toll-like receptor 4-dependent immune response. It consists of lipid A, core oligosaccharide, and O-specific polysaccharide (O-PS, O-antigen), with the latter part determining the O-serotype. As only 11 O-serotypes have been identified for K. pneumoniae to date (O1, O2a, O2ac, O2afg, O2aeh (called also O9), O3, O4, O5, O7, O8, and O12) [16,17,18], with two sub-serotypes (O3a, O3b) [1,19,20,21], O-antigens have been suggested as potential targets for active or passive immunization for decades [10].
Promising bactericidal and neutralizing monoclonal antibodies targeting the most common K. pneumoniae O-serotypes (O1, O2, O3, and O5) have recently been developed [19,20,21,22,23]. However, for the success of K/O-antigen-based treatment strategies, comprehensive knowledge on serotype distribution, novel serotypes, their structural diversity, and genotype–phenotype relationships among clinical isolates are mandatory. At present, classical structural analysis based on carbohydrate chemistry is efficiently supported by molecular biology and bioinformatics tools for K- and O-antigen typing [1,24], including Kaptive Web [14]. These tools identify and analyze the specific K- and O-loci encoding individual CPS or O-PS structures, respectively. The O-antigen biosynthesis of K. pneumoniae depends on genes of the rfb locus located between the cps and hisI genes [16,21]. To date, nine O-antigen gene clusters have been defined (for serotypes O1/O2, O3/O5, O4, O8, O12, OL101, OL102, OL103, and OL104) [1]. A survey of genome sequences indicated that the rfb loci of 83% of K. pneumoniae isolates specify the O1, O2, or O3 serotypes [16].
The O1/O2 rfb locus includes the following essential genes: (a) wzm and wzt, encoding transmembrane and nucleotide-binding domains of an ABC transporter; (b) wbbMNO, encoding glycosyltransferases; (c) glf, encoding the UDP-galactopyranose mutase which converts UDP-Galp to UDP-Galf; and (d) kfoC, which is of unknown function [1,16,21,24]. Moreover, the O1/O2 antigen gene cluster occurs in two variants: The O1/O2 variant 1 (v1; O2a, O2ac serotypes), characterized by the presence of D-galactan I, is encoded by the mandatory wzm-wbbO genes [1,21,25]. The second variant (v2; O2afg, O2aeh serotypes) carries an additional three genes, gmlABC (gmlABD, in the case of the O2aeh serotype) [16,20,21]. These genes are in opposite orientation and located downstream of rfb (Figure 1a). The gmlABC products are three putative glycosyltransferases, which modify D-galactan I {→3)-β-D-Galf-(1→3)-α-D-Galp-(1→} (O2a; O2v1) to branched polymers of {→3)-β-D-Galf-(1→3)-[α-D-Galp-(1→4)]-α-D-Galp-(1→} (O2afg; O2v2) or {→3)-β-D-Galf-(1→3)-[α-D-Galp-(1→2)]-α-D-Galp-(1→} (O2aeh) disaccharides, respectively [16,20,21]. O-PS of O1 contains D-galactan I and D-galactan II, built of {→3)-β-D-Galf-(1→3)-α-D-Galp-(1→} and {→3)-α-D-Galp-(1→3)-β-D-Galp-(1→} disaccharide repeating units, respectively. D-galactan II is encoded by a separate operon containing two genes located outside the rfb locus wbbYZ [1,21,24]. The O2 serotype is devoid of D-galactan II.
Figure 1.

Organization and comparative analysis of rfb gene clusters: (a) Organization of the rfb cluster of K. pneumoniae O1/O2 variant 2. The wzm and wzt genes encode transmembrane and nucleotide-binding domains of an ABC transporter. The wbbMNO genes encode glycosyltransferases. The glf gene encodes UDP-galactopyranose mutase. The function of kfoC is unknown. The gml genes (highlighted in grey) encode the structural modification of D-galactan I; and (b) alignment of the gmlB genes of K. pneumoniae BIDMC 7B, ABC152, and two reference strains with gmlABC locus (NTUH-K2044 and 441). Grey areas mark regions of differences in nucleotide sequence. The alignment was performed using the CLC Main Workbench.
In this paper, we describe two clinical isolates of K. pneumoniae (strains BIDMC 7B and ABC152), in which Kaptive-based O-serotype prediction and O-antigen structural analysis reveal different O-serotypes. Molecular characterization was performed to explain the genotype–phenotype discrepancies as a result of insertion sequences (ISs) within their rfb regions. Further, large-scale in silico analysis of 8130 K. pneumoniae genomes available in public databases was performed, in order to assess the prevalence of such insertions in rfb of K. pneumoniae O1 and O2 genomes.
2. Results
2.1. O-Antigen Structures of the BIDMC 7B and ABC152 Strains Represent the O2 Variant 1 O-Serotype
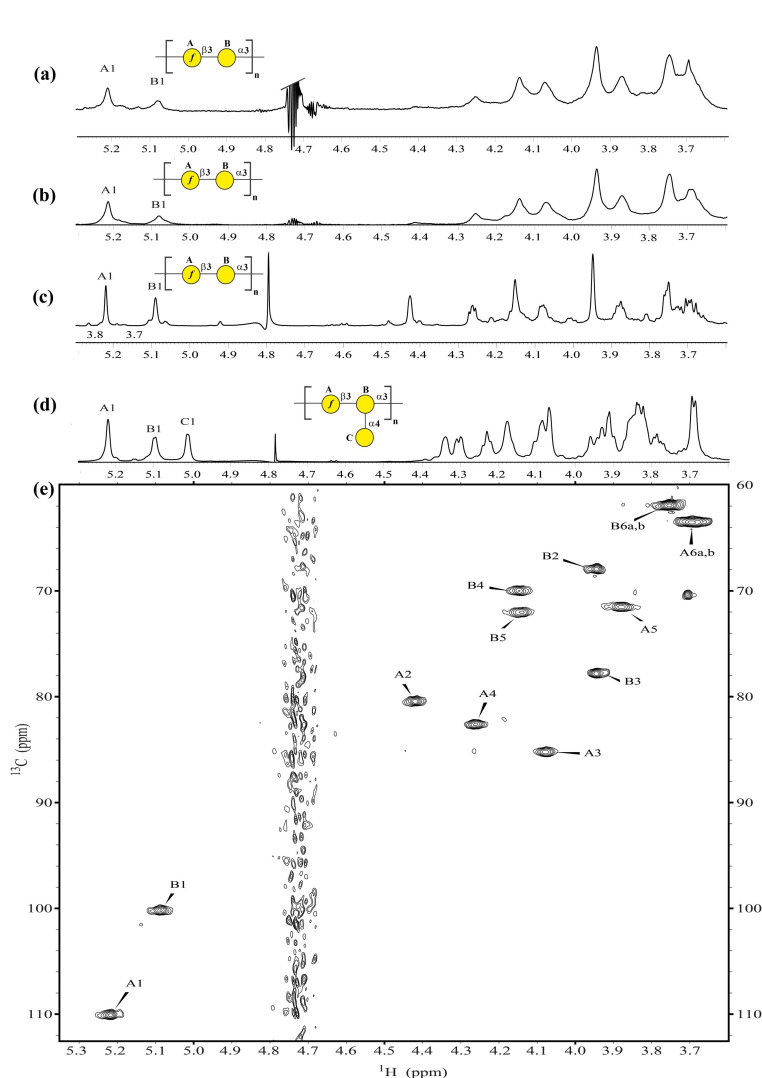
The O-antigen chemical structures of the K. pneumoniae BIDMC 7B and ABC152 clinical isolates were characterized by nuclear magnetic resonance (NMR) spectroscopy on the native isolated LPS. LPS was extracted from bacteria with yields of 0.32% and 0.70% for BIDMC 7B and ABC152, respectively, and then analyzed by the high-resolution magic angle spinning (HR-MAS) 1H, 13C NMR spectroscopy (Figure 2a,b,e).
Figure 2.
Comparative NMR analysis of K. pneumoniae O2v2 and O2v1 O-antigens. 1H HR-MAS NMR spectra of the lipopolysaccharides from (a) K. pneumoniae BIDMC 7B and (b) ABC152; and (c) the O-specific polysaccharides of K. pneumoniae Kp26 strain (O2v1) and (d) the PCM-27 strain (O2v2); (e) 1H,13C HSQC-DEPT NMR spectrum of the BIDMC 7B strain (O2v1). The capital letters refer to protons and/or carbons of O-PS carbohydrate residues, as shown in Table 1. The symbol Nomenclature for Graphical Representation of Glycans is used for O-PS visual representation:  Galactose;
Galactose;  Galactofuranose to show O-specific polysaccharide repeating units: →3)-β-D-Galf-(1→3)-α-D-Galp-(1→ (D-galactan I, O2 variant 1) and →3)-β-D-Galf-(1→3)-[α-D-Galp-(1→4)]-α-D-Galp-(1→ (O2v2) [26].
Galactofuranose to show O-specific polysaccharide repeating units: →3)-β-D-Galf-(1→3)-α-D-Galp-(1→ (D-galactan I, O2 variant 1) and →3)-β-D-Galf-(1→3)-[α-D-Galp-(1→4)]-α-D-Galp-(1→ (O2v2) [26].
Proton and carbon signals of the O-PS region of LPS prevailed in the NMR spectra. The complete assignments of 1H and 13C resonances for both O-PS (Table 1) were performed by interpretation of the NMR spectra, including comparison of the 1H,13C HSQC-DEPT spectrum (Figure 2e) with spectra for the reference O-PS structures of K. pneumoniae Kp26 (O2v1) and PCM-27 (O2v2) isolates (Figure 2c,d) [21].
Table 1.
1H and 13C NMR chemical shifts of O-specific polysaccharides from K. pneumoniae BIDMC 7B and ABC152 LPS.
| Chemical Shift (ppm) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | Residue | Description | H1 C1 |
H2 C2 |
H3 C3 |
H4 C4 |
H5 C5 |
H6a,b C6 |
| BIDMC 7B | A | →3)-β-D-Galƒ-(1→ | 5.22 110.1 |
4.42 80.5 |
4.07 85.2 |
4.26 82.6 |
3.88 71.5 |
3.69nr 63.5 |
| B | →3)-α-D-Galf-(1→ | 5.09 100.2 |
3.94 67.8 |
3.94 77.8 |
4.14 70.1 |
4.14 72.0 |
3.75nr 61.9 |
|
| ABC152 | A | →3)-β-D-Galƒ-(1→ | 5.22 110.0 |
4.42 80.4 |
4.07 85.2 |
4.26 82.6 |
3.88 71.5 |
3.69nr 63.5 |
| B | →3)-α-D-Galf-(1→ | 5.09 100.1 |
3.94 67.9 |
3.94 77.8 |
4.14 70.0 |
4.14 72.0 |
3.75nr 61.9 |
|
nr—not resolved.
The phenotype analysis demonstrated that the O-PS of the BIDMC 7B LPS had an O2v1 serotype structure, characterized by the [→3)-β-D-Galf-(1→3)-α-D-Galp-(1→] disaccharide as the non-modified D-galactan I repeating unit (Figure 2a,e; Table 1). The same D-galactan I structure was determined for the ABC152 LPS (Figure 2b; Table 1). The lack of a terminal α-D-Galp residue, characteristic for the O2v2 serotype (Figure 2d, C1 signal), was confirmed for both isolates. Their NMR spectra were comparable to those recorded for the K. pneumoniae Kp26 O-PS (O2v1 serotype). The comparison of 1H, 13C HSQC-DEPT spectra of BIDMC 7B (Figure 2e) and ABC152 showed a complete overlay, confirming the identity of these O-PSs (Table 1).
2.2. Disruption of the gmlB Gene by IS Affects the O-Antigen Phenotype in BIDMC 7B and ABC152
Kaptive-based O-serotyping was performed with whole-genome sequences of both strains [14]. Contrary to the structural analysis, the O-serotypes were predicted to be O2v2 with high match confidence, according to the Kaptive measures of match quality. However, the BIDMC 7B and ABC152 rfb clusters demonstrated an increased size (by 777 bp) when compared to the reference sequences in the Klebsiella O-locus primary reference database in Kaptive. The gmlABC genes showed 100%, 90.79%, and 97.33% identity, respectively, to those in the Kaptive reference database.
Molecular analysis of BIDMC 7B and ABC152 was performed to explain the discrepancies observed between the O-antigen phenotype and the Kaptive-predicted O-serotype. The alignment of the gmlB genes from BIDMC 7B, ABC152, and from the O1/O2v2 reference strains K. pneumoniae NTUH-K2044 and 441 are shown in Figure 1b. This comparison shows the disruption of gmlBs in both strains by an identical IS element, ISR1, whereas other genes in the rfb locus were intact, in comparison to the reference strains. These results indicated that the ISR1 disruption completely inactivated the GmlB glycosyltransferase gene, resulting in biosynthesis of the O2v1 instead of O2v2 structure, and thus, being the likely reason for the discrepancy between the actual O-antigen phenotype and Kaptive-based predictions.
2.3. ISs Occur in O2v2 and O1v2 K. pneumoniae Isolates—in Silico Study
In order to assess the occurrence of IS elements in rfb loci of K. pneumoniae, 8130 genome sequences available in the public domain were analyzed (Supplementary Material Table S1). Based on the Kaptive results of the O-serotyping (Supplementary Material Table S1, column B), 2281 isolates (≈ 28%) were predicted to be O2v2, and 839 isolates (≈ 10%) to be O1v2. For O2v2, 55 genomes (≈ 2.40%) revealed a significant difference in length of the rfb region (≥ 400 bp), of which 49 genomes were of sufficient quality for further analysis (Supplementary Material Table S2). The presence of different ISs (e.g., ISR1, IS903B, ISKpn14, or ISKpn26) were identified in several genes of these loci; namely, gmlBC, kfoC, wbbMNO, glf, wzm, and wzt (Table 2). In several isolates, the same or two different ISs interrupted two genes; namely, gmlB or gmlC and wbbO, or wbbM and wbbO.
Table 2.
Sequence type and location of IS elements in the rfb clusters of K. pneumoniae isolates selected on the basis of Kaptive-based O2v2 predictions a.
| Isolate | Assembly Accession Number | Sequence Type | Insertion Sequence | Gene | Position of the IS Element (From the First Nucleotide of CDS) |
|---|---|---|---|---|---|
| 27097_7_178-2 | GCF_900776535.1_27097_7_178-2_genomic | ST11 | IS903 | wbbO | 228 bp |
| kpneu028 | GCF_900607955.1_kpneu028_genomic | ST11 | ISR1 | wbbM | 47 bp |
| ASM893134 | GCF_008931345.1_ASM893134v1_genomic | ST12 | IS903B | kfoC | 267 bp |
| UCI 7 | GCF_000492535.1_Kleb_pneu_UCI_7_V1_genomic | ST17 | ISKpn26 | wbbO | 1014 bp |
| ASM966157 | GCF_009661575.1_ASM966157v1_genomic | ST23 | ISR1 | wbbM | 1420 bp |
| ASM170423 | GCF_001704235.1_ASM170423v1_genomic | ST34 | IS903B | gmlC | 7956 bp in rfb (CDS gmlC from 7996 bp) |
| ASM307130 | GCF_003071305.1_ASM307130v1_genomic | ST34 | IS903B | wbbO | 130 bp |
| IS903B | gmlC | 7956 bp in rfb (CDS gmlC from 7996 bp) | |||
| ASM366018 | GCF_003660185.1_ASM366018v1_genomic | ST105 | IS903B | wbbO | 182 bp |
| BIDMC 55 | GCF_000692955.1_Kleb_pneu_BIDMC_55_V1_genomic | ST105 | IS903B | glf | 105 bp |
| ABC152 | GCA_014433645.1 | ST147 | ISR1 | gmlB | 818 bp |
| BIDMC 7B | GCF_000567425.1_Kleb_pneu_BIDMC_7B_V2_genomic | ST258 | ISR1 | gmlB | 818 bp |
| UCI 33 | GCF_000566865.1_Kleb_pneu_UCI_33_V1_genomic | ST258 | ISR1 | kfoC | 656 bp |
| CHS 139 | GCF_001031785.1_Kleb_pneu_CHS139_V1_genomic | ST258 | ISR1 | wbbM | 1881 bp |
| CHS 91 | GCF_001030945.1_Kleb_pneu_CHS91_V1_genomic | ST258 | ISR1 | wbbO | 165 bp |
| CHS 57 | GCF_000694075.1_Kleb_pneu_CHS_57_V1_genomic | ST258 | ISKpn26 | gmlB | 4 bp |
| UCI 38 | GCF_000566805.1_Kleb_pneu_UCI_38_V1_genomic | ST258 | ISKpn26 | gmlB | 453 bp |
| IS1294 | wbbO | 900 bp | |||
| BIDMC 13 | GCF_000567345.1_Kleb_pneu_BIDMC_13_V1_genomic | ST258 | ISKpn26 | gmlB | 453 bp |
| ISKpn26 | wbbO | 490 bp | |||
| CHS 71 | GCF_000694295.1_Kleb_pneu_CHS_71_V1_genomic | ST258 | ISKpn26 | wbbO | 1080 bp |
| CHS 235 | GCF_001033335.1_Kleb_pneu_CHS235_V1_genomic | ST258 | ISKpn26 | wbbO | 641 bp |
| ASM147162 | GCF_001471625.1_ASM147162v2_genomic | ST258 | ISKpn26 | wbbO | 1014 bp |
| ISKpn26 | wbbM | 96 bp | |||
| CHS 105 | GCF_001031225.1_Kleb_pneu_CHS105_V1_genomic | ST258 | ISKpn26 | wbbM | 1548 bp |
| ASM386117 | GCF_003861175.1_ASM386117v1_genomic | ST258 | ISKpn26 | wbbM | 45 bp |
| ISKpn26 | 2156 bp | ||||
| CHS 165 | GCF_001032265.1_Kleb_pneu_CHS165_V1_genomic | ST258 | ISKpn26 | kfoC | 158 bp |
| MGH 51 | GCF_000694435.1_Kleb_pneu_MGH_51_V1_genomic | ST258 | ISKpn26 | wzm | 315 bp |
| CHS 99 | GCF_001031105.1_Kleb_pneu_CHS99_V1_genomic | ST258 | ISKpn26 | wzt | 470 bp |
| ASM205647 | GCF_002056475.1_ASM205647v1_genomic | ST258 | ISKpn14 | kfoC | 782 bp |
| ASM80749 | GCF_000807495.1_ASM80749v2_genomic | ST258 | ISKpn14 | kfoC | 440 bp |
| 18174_7_5 | GCF_900515885.1_18174_7_5_genomic | ST258 | ISKpn14 | wbbO | 377 bp |
| BIDMC 54 | GCF_000692935.1_Kleb_pneu_BIDMC_54_V1_genomic | ST258 | IS1294 | kfoC | 1186 bp |
| CHS 46 | GCF_000693875.1_Kleb_pneu_CHS_46_V1_genomic | ST258 | ISF1 | wbbM | 1138 bp |
| ASM195291 | GCF_001952915.1_ASM195291v1_genomic | ST258 | ISVsa5 | wbbN | 133 bp |
| ASM303004 | GCF_003030045.1_ASM303004v1_genomic | ST512 | ISKpn26 | wbbO | 1014 bp |
| UCI 8 | GCF_000492515.1_Kleb_pneu_UCI_8_V1_genomic | ST1198 | IS9033 | wbbM | 1667 bp |
| IS39 | GCF_000529425.1_IS39v1_genomic | unknown | IS102 | gmlC | 7933 bp in rfb (CDS gmlC from 7996 bp) |
a: O-serotype predictions were performed using the Kaptive Web tool [14].
Among the 839 Kaptive-identified O1v2 isolates, significant length discrepancies (≥700 bp) occurred in the rfb region of six isolates (≈0.7%), one of which was excluded due to the low quality of reads (Supplementary Material Table S3). Selected rfb genes of these isolates, namely gmlABC, wbbM, and wzm, were interrupted by IS5, IS102, IS903B, or ISKpn14 (Table 3). Two and one O1v2 isolates, two and four genes, respectively, were disrupted simultaneously. In both the O2v2 and O1v2 groups, the same ISs were observed at the same positions of the same genes in several isolates, suggesting their close genetic relatedness. The gmlB:ISR1 (nt 818) disruption of the studied isolates BIDMC 7B and ABC152 was found in four other genomes.
Table 3.
Sequence type and location of IS elements in the rfb clusters of K. pneumoniae isolates selected on the basis of Kaptive-based O1v2 predictions a.
| Isolate | Assembly Accession Number | Sequence Type | Insertion Sequence | Gene | Position of the IS Element (From the First Nucleotide of CDS) |
|---|---|---|---|---|---|
| ASM492431 | GCF_004924315.1_ASM492431v1_genomic | ST23 | IS102 | gmlC | 1217 bp |
| ASM275277 | GCF_002752775.1_ASM275277v1_genomic | ST29 | IS102 | gmlB | 472 bp |
| IS903B | gmlC | 526 bp | |||
| ASM296687 | GCF_002966875.1_ASM296687v1_genomic | ST34 | IS5 | wzm | 315 bp |
| IS903B | wbbM | 465 bp | |||
| IS903B | gmlB | 926 bp | |||
| IS903B | gmlC | 143 bp | |||
| ASM290977 | GCF_002909775.1_ASM290977v2_genomic | ST231 | ISKpn14 | gmlA | 95 bp |
a: O-serotype predictions were performed using the Kaptive Web tool [14].
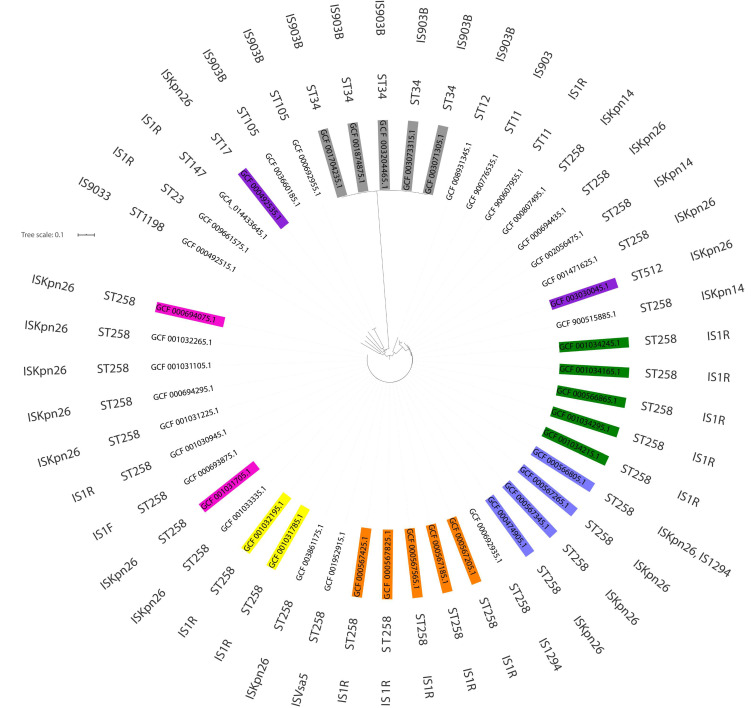
In order to sort out the approximate number of independent IS insertions into the rfb loci of the available K. pneumoniae O2v2 and O2v1 genomes, clonality (MLST) and phylogenetic analyses were performed on the isolates using the ABC152 strain as a reference (Supplementary Material Tables S2 and S3; Figure 3). These confirmed that some individual disruptions within the rfb locus have spread in K. pneumoniae populations clonally with specific lineages, indicating single IS insertion events at their origins. This was demonstrated by clusters of O2v2 ST258 isolates with kfoC:ISR1 (nt 656) or wbbM:ISR1 (nt 1,881) disruptions, ST258 with double gmlB:ISKpn26 (nt 453) plus wbbO:ISKpn26 (nt 490) disruptions, or ST34 with gmlC:IS903B (nt 7,956). In some cases, an additional IS insertion likely marked on-going diversification within a lineage, such as wbbO:IS903B (nt 130) in ST34 with gmlC:IS903B (nt 7,956). An interesting case was the gmlB:ISR1 (nt 818) disruption in the study isolates BIDMC 7B and ABC152, which was observed also in four others. BIDMC 7B plus the four others formed a closely related cluster of ST258 isolates. ABC152 was of a non-related ST147, suggesting a horizontal transfer and recombination event. A similar case was represented by the disruption wbbO:ISKpn26 (nt 1,014), present in two ST258 and ST512 close relatives, as well as a non-related ST17 isolate. Based on these results, it may be assumed that IS disruptions within the rfb loci in K. pneumoniae O2v2 and O2v1 genomes might have occurred at least ≈35 and ≈10 times, respectively (Table 2 and Table 3).
Figure 3.
Results of clonality (MLST) and phylogenetic analyses of O2v2 K. pneumoniae genomes characterized by ST and insertion sequences distribution. Each color indicates closely related strains characterized by genetic similarity, shown in detail in Supplementary Material Table S2. Separately analyzed isolates are not colored.
3. Discussion
Owing to their universality, reproducibility, varied resolution, and standardized high-throughput protocols, molecular biology methods have become an excellent tool for pathogen characterization, finding wide application in microbiology diagnostics and surveillance. In recent years, these have been revolutionized by WGS, an increasingly common approach used in public health laboratories for the control of antimicrobial resistance or bacterial genotyping. At present, WGS is also successfully used to complement laborious structural chemical analyses, such as those of microbial surface antigens, being key pathogenicity factors as well as critical targets for vaccines and therapeutic strategies [27,28,29].
As the molecular genetics of the K. pneumoniae O- and K-antigen biosynthesis has been well-elucidated, new O-genotyping techniques have been demonstrated to be useful for serotyping. There are several useful examples of tracking O- or K-antigen diversity among K. pneumoniae isolates [1,14,24]. For example, Fang et al. used a PCR-based O-genotyping approach to explore the distribution of the O-antigen genetic determinants in 87 clinical K. pneumoniae strains, showing a high prevalence of O1 (≈ 57%), followed by the O2a, O3, and O5 O-genotypes [24]. Follador et al. analyzed over 500 whole-genome sequences and reached a similar conclusion: that O1, O2, and O3 serotypes were the most common, with approximately 80% of all isolates [1]. Finally, Wick et al. presented the user-friendly Kaptive Web, an online tool for the rapid typing of K. pneumoniae surface polysaccharide loci, and demonstrated its utility using more than 500 K. pneumoniae genomes [14].
Kaptive Web-supported differentiation between K. pneumoniae O1/O2v1 and O1/O2v2 serotypes based on two steps: First, Kaptive recognizes the serotype by searching for the D-galactan-II-encoding genes (wbbY, wbbZ) characteristics of the O1 serotype. Second, the O1 and O2 serotypes are distinguished by the analysis of genes found in the rfb cluster. Finally, these are reported as variant v1 or v2 [14]. As the final result, the tool prediction is accompanied by length discrepancy information, which may indicate the possibility of some rearrangements in the rfb region.
In this study, we presented two cases of genotype–phenotype discrepancies for O-antigens in the K. pneumoniae clinical isolates BIDMC 7B and ABC152, the actual phenotype of which was O2v1, whereas Kaptive predicted O2v2. However, the tool provided an alert about “length disruption” within the rfb region and recommended further analyses. In the case of BIDMC 7B and ABC152 isolates, the structural analysis by the HR-MAS NMR spectroscopy proved the O-antigen structure to be O2v1 (Figure 2). The ISR1 element was identified in the gmlB gene, one of the three responsible for the D-galactan I conversion from v1 to v2. The presence of IS, actual O-antigen structures, and the lack of other obvious differences between the analyzed genomes and O2v1 reference strains indicated that the IS disruption was the reason for the discrepancy between the O-antigen phenotype and the Kaptive-based prediction. The large-scale in silico analysis of publicly available genomes of K. pneumoniae O2v2 and O1v2 clearly showed that various insertions have occurred in several rfb fragments, possibly causing similar divergences between the O-serotype prediction and phenotype. A variety of ISs have been identified, including the common elements ISR1, ISKpn14, ISKpn26, and IS903B.
As only structural verification in each strain could provide definite proof of the O-phenotype, the in silico survey only suggested the influence of IS on the O-antigen chemical structure. By analogy with the BIDMC 7B and ABC152 strains, fourteen O2 strains (e.g., ASM170423, CHS57, and IS39) revealed ISs in the gmlABC region with higher prevalence of gmlB and gmlC disruptions, likely representing similar genotype–phenotype discrepancies. Three O2 strains with an IS in the gmlABC region had additional disruption within the wzm–wbbO region, suggesting failure of the O-antigen biosynthesis and the rough form of LPS, devoid of O-PS (i.e., ASM307130, UCI 38, and BIDMC 13). Other identified cases also suggested O-antigen biosynthesis failure, including O1v2 isolates (Table 2 and Table 3). Regarding the genetic background of O1 and O2 antigen biosynthesis, the presence of an IS in the O-locus may influence the O-antigen phenotype by: (i) O2v2 to O2v1 or O1v2 to O1v1 conversion; or (ii) conversion from smooth to rough LPS. It is noteworthy that the results obtained from Kaptive Web depended on the IS location. In the case of gene disruption or frameshift mutation, the results will indicate the lack of an enzyme specific for the analyzed serotype, which may contribute to the false serotype prediction by the algorithm. For instance, the Kaptive results for the isolate IS39 indicated the absence of the wbbM and gmlABC genes (Table S2). Detailed analysis of the rfb region has shown the presence of these genes with a gmlC IS disruption and point mutations in the other three genes. Although Kaptive suggests the possibility of the presence of IS by reporting differences in length discrepancy, it is worth analyzing the nucleotide sequence of the rfb region more precisely, in order to exclude falsely predicted serotypes based on errors occurring during O-genotyping.
Sequence analysis of isolates from the database confirmed the occurrence of many IS insertions in the rfb region; however, the frequency of such events is hard to evaluate. Although there are no previous data on IS disruptions within the O-locus, in general, these elements are common in K. pneumoniae genomes [30]. The hyperepidemic clone ST258, characterized by the notorious production of KPC-type carbapenemases and extensive drug resistance, has more ISs than an average K. pneumoniae isolate of another ST [30,31]. Several IS types are especially frequent in ST258, such as ISKpn26 [30,32]. In our study, the majority of O2 isolates identified belonged to ST258 (≈70%), and ISKpn26 was commonly found in these (≈50%). These data further emphasized the impact of ISs on the evolution of K. pneumoniae ST258; however, one must also consider the over-representation of ST258 genomes in public databases, resulting from the high clinical and epidemiological relevance of these organisms.
According to Adams et al., 94% of K. pneumoniae strains have at least one IS in their genome, where transposition of these elements within the genome causes rearrangements and may create new genotypes [30]. One consequence of an IS disruption of the rfb cluster genes may be the protection of bacteria against the host immune system. Although the presence of ISs in gmlABC genes may cause the phenotype–genotype discrepancy discussed above, the disruption of the genes determining D-galactan I elements may abolish the O-antigen synthesis, as in the case of the wzm or wzt genes, coding for ABC transporters [1,16,25]. IS elements in the rfb and/or wbbYZ operon can either inhibit the expression of the O-antigen on the surface of the bacterial cell [24] (resulting in the rough phenotype) or cause the switch from one serotype to another. In both cases, the change in phenotype can alter or impair the virulence of the bacteria [33]. Structural large-scale analysis of K. pneumoniae isolates could determine the consequences of ISs in the rfb region and their effects on bacterial antigenicity and host interactions. Such changes can significantly affect the ability of bacteria to survive during antibacterial therapies; for example, by changing the surface antigens and virulence in regard to reactivity with the complement, antibodies, or phage resistance [34]. The antigenic drift of LPS can be a way by which bacteria avoid the immune system. This is the case, for example, in the Salmonella species, the O-antigen composition of which affects the host–pathogen interactions during infection. Strains belonging to one serovar can have a different repertoire of O-antigen-modifying genes. Moreover, their expression is different depending on the phase variations. The gtrABC operon acquired by horizontal gene transfer is such a set of genes for modification of the Salmonella O-antigen. These genes encode proteins showing functional homology to the glycosyltransferases encoding by the gmlABC genes cluster in K. pneumoniae [35,36].
This study showed that some K. pneumoniae isolates, flagged by Kaptive Web as having length discrepancy within the rfb locus and advised for further analysis, may be basically mis-O-serotyped by the tool. As a methodology for the identification of an actual O-antigen phenotype is not broadly available, we assume that the development of the Kaptive algorithm in that direction would increase its high quality and usefulness, particularly for inexperienced users. Such development might be based on broader studies of isolates with non-clear O-serotyping results or identified O-genotype–phenotype discrepancies, with the use of structural analysis to precisely elucidate the O-genotype–phenotype relationships.
As O- and K-antigens represent target molecules for therapeutic strategies against Klebsiella infections, it is important to broaden our knowledge about genotype–phenotype relationships. Filling all detected gaps will improve serotype predictions based on bioinformatic tools. Exact fast prediction will enable the monitoring of K. pneumoniae antigen drift, which is vulnerable to selective pressure by therapies and vaccines.
4. Materials and Methods
4.1. Bacteria and Growth Conditions
K. pneumoniae BIDMC 7B (urine isolate) was obtained through BEI Resources, NIAID, NIH: “Klebsiella pneumoniae, strain BIDMC 7B, NR-41923”, as a reagent bought as a part of the Klebsicure-Eurostars project (no. E!7563). Strain ABC152 (urine isolate) was recovered from the Abu Dhabi Hospital (UAE) in 2013, kindly provided by Agnes Pal-Sonnevend and Tibor Pal from the United Arab Emirates University. Both strains were selected for a previous large-scale serotyping study (unpublished results), due to the inconsistency between the lack of LPS reactivity with O2v2-specific monoclonal antibody [21] and PCR results showing the presence of gmlABC genes. Bacteria were grown on Trypcase Soy Agar plates. For semi-preparative scale LPS preparation, the strains were cultured in Luria–Bertani (LB) broth in 500 mL flasks with shaking (at 37 °C), inactivated overnight with 3% formalin at 22 °C, then harvested by centrifugation, washed with water, and freeze-dried.
4.2. O-Specific Polysaccharides
K. pneumoniae PCM-27 and Kp26 O-specific polysaccharides were obtained from the Laboratory of Microbial Immunochemistry and Vaccines in the Ludwik Hirszfeld Institute of Immunology and Experimental Therapy, PAS (Wroclaw, Poland) and isolated as previously described [21].
4.3. LPS Preparation
LPS of K. pneumoniae strains BIDMC 7B and ABC152 were isolated by hot phenol/water extraction [37] and purified by dialysis and ultracentrifugation, as described elsewhere [38], followed by freeze-drying.
4.4. NMR Spectroscopy
All NMR spectra were obtained at 298 K using an Avance III 600 MHz (Bruker BioSpin GmbH, Rheinstetten, Germany) spectrometer equipped with a PH HR MAS probe (LPS analysis) or 5 mm QCI cryoprobe with z-gradients (O-PSs analysis). NMR spectra of isolated O-PSs were obtained in 2H2O, processed and analyzed as described previously [39]. For high-resolution magic angle spinning (HR-MAS) NMR spectroscopy, LPS (3–4 mg) was suspended in 2H2O and placed into the ZrO2 rotor. Acetone was used as an internal reference (δH/δC 2.225/31.05 ppm) for both O-PS and LPS spectra [21]. The processed spectra (1H, 13C HSQC-DEPT, 1H, 1H COSY, and TOCSY) were assigned with the use of NMRFAM-SPARKY (v1.2, NMRFAM, Madison, Wisconsin, USA) [40].
4.5. DNA Isolation
Genomic DNA of the BIDMC 7B and ABC152 strains were extracted from overnight cultures with the Genomic Mini kit (A&A Biotechnology, Gdynia, Poland).
4.6. DNA Library Preparation and Sequencing
Libraries were prepared using the Nextera XT DNA Library Preparation Kit (Illumina Inc., San Diego, California, USA) and sequenced on an Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA).
4.7. Sequence Analysis
The obtained reads were trimmed by Trimmomatic v0.39 [41]. The genomes of the BIDMC 7B and ABC152 strains were assembled using SPAdes v3.12 [42]. Assembled genomes of the BIDMC 7B and ABC152 isolates were analyzed by the Kaptive Web algorithm (https://github.com/katholt/Kaptive) for in silico O-serotyping [14]. Sequences of the rfb gene clusters were compared with those of the reference strains K. pneumoniae NTUH-K2044 (serotype O1v2) and K. pneumoniae 441 (O1/O2v2) (GenBank accession numbers, AB117611 and LT174602, respectively) using the CLC Main Workbench version 20 software (https://digitalinsights.qiagen.com; Qiagen, Hilden, Germany). All K. pneumoniae whole-genome sequences available from GenBank (n = 8130, as of 3 December 2019) were downloaded and screened using Kaptive for the O1v2- and O2v2-predicted serotype isolates, and for the rfb region length discrepancies among these (excess of ≥ 700 bp and ≥ 400 bp, respectively). Genomes of all such isolates were typed by 7-loci MLST with Multi-Locus Sequence Typing 2.0 (http://cge.cbs.dtu.dk/services/MLST) [43]. Identification of ISs was performed by ISfinder (http://www-is.biotoul.fr) [44]. Subsequently, all putative O2v2 genomes with rfb length discrepancies were aligned against the strain ABC152, inferring SNP-based phylogeny with Parsnp v.1.2. [45]. The phylogenetic tree was visualized with iTOL (https://itol.embl.de) [46].
4.8. Data Availability
The sequence of the BIDMC 7B strain is available in GenBank under accession number JCNG00000000.1. The sequence of the ABC152 strain was submitted to the GenBank database under accession number JACENF000000000.
Acknowledgments
Agnes Sonnevend-Pal and Tibor Pal from United Arab Emirates University are kindly acknowledged for clinical isolate ABC152. The following reagent was obtained through BEI Resources, NIAID, NIH: K. pneumoniae, Strain BIDMC 7B, NR-41923. Alona Tsybulska is acknowledged for technical support in LPS extraction. We also acknowledge the consultation, support, and WGS services provided by Lukasz Laczmanski from the Laboratory of Genomics and Bioinformatics.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/18/6572/s1. Table S1: Kaptive Web analysis results for the 8130 assemblies of K. pneumoniae isolates. Isolates belonging to the O2v2 serotype are marked in green. Isolates belonging to the O1v2 serotype are marked in blue; Table S2: Kaptive analysis results extracted for the 55 K. pneumoniae O2 variant 2 genomes characterized by IS occurrence. Each color indicates closely related strains characterized by genetic similarity. Separately analyzed isolates are not colored; Table S3: Kaptive analysis results extracted for the 5 K. pneumoniae O1 variant 2 genomes characterized by IS occurrence. Isolates belonging to the same strain are marked in blue.
Author Contributions
Conceptualization, J.L., V.S., M.G., D.A., and R.I.; investigation, D.A., J.L., A.M., M.K., R.I., and A.H.; writing—original draft preparation, J.L. and D.A.; writing—review and editing, A.D., R.I., A.M., M.K., A.H., V.S., M.G., and J.L.; funding acquisition, J.L.; project administration, J.L.; supervision, J.L. All authors have read and agreed to the published version of the manuscript. D.A. was supported in part by the Wroclaw Doctoral School of Institutes of Polish Academy of Sciences, and this research forms part of her PhD thesis. D.A. cultured bacteria, elucidated O-PS structures, isolated DNA, assisted in HR-MAS NMR and WGS experiments, and performed most of bioinformatic analyses.
Funding
This research was funded by the National Science Centre, Poland, grant number 2018/31/B/NZ7/04002.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| MDR | Multidrug-Resistant |
| ESBL | Extended-Spectrum Beta-Lactamases |
| COSY | Correlation Spectroscopy |
| CPKP | Carbapenemase-Producing K. pneumoniae |
| CPS | Capsular Polysaccharide; K antigen |
| DEPT | Distortionless Enhancement by Polarization Transfer |
| HR-MAS | High Resolution-Magic Angle Spinning |
| HSQC | Heteronuclear Single Quantum Correlation |
| IS | Insertion Sequence |
| LPS | Lipopolysaccharide |
| MLST | Multi-locus Sequence Typing |
| NMR | Nuclear Magnetic Resonance |
| O-PS | O-specific Polysaccharide |
| PCR | Polymerase Chain Reaction |
| ST | Sequence Type |
| TOCSY | Total Correlation Spectroscopy |
| WGS | Whole-genome Sequencing |
References
- 1.Follador R., Heinz E., Wyres K.L., Ellington M.J., Kowarik M., Holt K.E., Thomson N.R. The diversity of Klebsiella pneumoniae surface polysaccharides. Microb. Genom. 2016;2:e000073. doi: 10.1099/mgen.0.000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt K.E., Wertheim H., Zadoks R.N., Baker S., Whitehouse C.A., Dance D., Jenney A., Connor T.R., Hsu L.Y., Severin J., et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA. 2015;112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin R.M., Bachman M.A. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell. Infect Microbiol. 2018;8:4. doi: 10.3389/fcimb.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navon-Venezia S., Kondratyeva K., Carattoli A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017;41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 5.Paczosa M.K., Mecsas J. Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 2016;80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyres K.L., Lam M.M.C., Holt K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020 doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 7.Boucher H.W., Ambrose P.G., Chambers H.F., Ebright R.H., Jezek A., Murray B.E., Newland J.G., Ostrowsky B., Rex J.H. White paper: Developing antimicrobial drugs for resistant pathogens, narrow-spectrum indications, and unmet needs. J. Infect Dis. 2017;216:228–236. doi: 10.1093/infdis/jix211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tacconelli E., Magrini N. WHO, Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. WHO; Geneva, Switzerland: 2017. [Google Scholar]
- 9.Grundmann H., Glasner C., Albiger B., Aanensen D.M., Tomlinson C.T., Andrasević A.T., Cantón R., Carmeli Y., Friedrich A.W., Giske C.G., et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): A prospective, multinational study. Lancet Infect Dis. 2017;17:153–163. doi: 10.1016/S1473-3099(16)30257-2. [DOI] [PubMed] [Google Scholar]
- 10.Adamo R., Margarit I. Fighting antibiotic-resistant Klebsiella pneumoniae with “sweet” immune targets. mBio. 2018;9 doi: 10.1128/mBio.00874-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edelman R., Taylor D.N., Wasserman S.S., McClain J.B., Cross A.S., Sadoff J.C., Que J.U., Cryz S.J. Phase 1 trial of a 24-valent Klebsiella capsular polysaccharide vaccine and an eight-valent Pseudomonas O-polysaccharide conjugate vaccine administered simultaneously. Vaccine. 1994;12:1288–1294. doi: 10.1016/S0264-410X(94)80054-4. [DOI] [PubMed] [Google Scholar]
- 12.Feldman M.F., Mayer Bridwell A.E., Scott N.E., Vinogradov E., McKee S.R., Chavez S.M., Twentyman J., Stallings C.L., Rosen D.A., Harding C.M. A promising bioconjugate vaccine against hypervirulent Klebsiella pneumoniae. Proc. Natl. Acad. Sci. USA. 2019;116:18655–18663. doi: 10.1073/pnas.1907833116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegerle N., Choi M., Sinclair J., Amin M.N., Ollivault-Shiflett M., Curtis B., Laufer R.S., Shridhar S., Brammer J., Toapanta F.R., et al. Development of a broad spectrum glycoconjugate vaccine to prevent wound and disseminated infections with Klebsiella pneumoniae and Pseudomonas aeruginosa. PLoS ONE. 2018;13:e0203143. doi: 10.1371/journal.pone.0203143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wick R.R., Heinz E., Holt K.E., Wyres K.L. Kaptive Web: User-friendly capsule and lipopolysaccharide serotype prediction for Klebsiella genomes. J. Clin. Microbiol. 2018;56 doi: 10.1128/JCM.00197-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyres K.L., Wick R.R., Gorrie C., Jenney A., Follador R., Thomson N.R., Holt K.E. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2016;2:e000102. doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke B.R., Ovchinnikova O.G., Kelly S.D., Williamson M.L., Butler J.E., Liu B., Wang L., Gou X., Follador R., Lowary T.L., et al. Molecular basis for the structural diversity in serogroup O2-antigen polysaccharides in Klebsiella pneumoniae. J. Biol. Chem. 2018;293:4666–4679. doi: 10.1074/jbc.RA117.000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly S.D., Clarke B.R., Ovchinnikova O.G., Sweeney R.P., Williamson M.L., Lowary T.L., Whitfield C. Klebsiella pneumoniae O1 and O2ac antigens provide prototypes for an unusual strategy for polysaccharide antigen diversification. J. Biol. Chem. 2019 doi: 10.1074/jbc.RA119.008969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen D.S., Mestre F., Albertí S., Hernández-Allés S., Álvarez D., Doménech-Sánchez A., Gil J., Merino S., Tomás J.M., Benedí V.J. Klebsiella pneumoniae lipopolysaccharide O typing: Revision of prototype strains and O-group distribution among cClinical isolates from different sources and countries. J. Clin. Microbiol. 1999;37:56–62. doi: 10.1128/JCM.37.1.56-62.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guachalla L.M., Stojkovic K., Hartl K., Kaszowska M., Kumar Y., Wahl B., Paprotka T., Nagy E., Lukasiewicz J., Nagy G., et al. Discovery of monoclonal antibodies cross-reactive to novel subserotypes of K. pneumoniae O3. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-017-06682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stojkovic K., Szijártó V., Kaszowska M., Niedziela T., Hartl K., Nagy G., Lukasiewicz J. Identification of D-Galactan-III as part of the lipopolysaccharide of Klebsiella pneumoniae serotype O1. Front. Microbiol. 2017;8:684. doi: 10.3389/fmicb.2017.00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szijártó V., Guachalla L.M., Hartl K., Varga C., Banerjee P., Stojkovic K., Kaszowska M., Nagy E., Lukasiewicz J., Nagy G. Both clades of the epidemic KPC-producing Klebsiella pneumoniae clone ST258 share a modified galactan O-antigen type. Int. J. Med. Microbiol. 2016;306:89–98. doi: 10.1016/j.ijmm.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Rollenske T., Szijarto V., Lukasiewicz J., Guachalla L.M., Stojkovic K., Hartl K., Stulik L., Kocher S., Lasitschka F., Al-Saeedi M., et al. Cross-specificity of protective human antibodies against Klebsiella pneumoniae LPS O-antigen. Nat. Immunol. 2018;19:617–624. doi: 10.1038/s41590-018-0106-2. [DOI] [PubMed] [Google Scholar]
- 23.Szijártó V., Guachalla L.M., Hartl K., Varga C., Badarau A., Mirkina I., Visram Z.C., Stulik L., Power C.A., Nagy E., et al. Endotoxin neutralization by an O-antigen specific monoclonal antibody: A potential novel therapeutic approach against Klebsiella pneumoniae ST258. Virulence. 2017;8:1203–1215. doi: 10.1080/21505594.2017.1279778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang C.-T., Shih Y.-J., Cheong C.-M., Yi W.-C. Rapid and accurate determination of lipopolysaccharide O-antigen types in Klebsiella pneumoniae with a novel PCR-based O-genotyping method. J. Clin. Microbiol. 2016;54:666–675. doi: 10.1128/JCM.02494-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke B.R., Whitfield C. Molecular cloning of the rfb region of Klebsiella pneumoniae serotype O1:K20: The rfb gene cluster is responsible for synthesis of the D-galactan I O polysaccharide. J. Bacteriol. 1992;174:4614–4621. doi: 10.1128/JB.174.14.4614-4621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varki A., Cummings R.D., Aebi M., Packer N.H., Seeberger P.H., Esko J.D., Stanley P., Hart G., Darvill A., Kinoshita T., et al. Symbol nomenclature for graphical representations of glycans. Glycobiology. 2015;25:1323–1324. doi: 10.1093/glycob/cwv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Artyszuk D., Wołkowicz T. Zastosowanie sekwencjonowania pełnogenomowego do genotypowania bakterii. Post Mikrob. 2018;57:179–193. [Google Scholar]
- 28.Quainoo S., Coolen J.P.M., van Hijum S.A.F.T., Huynen M.A., Melchers W.J.G., van Schaik W., Wertheim H.F.L. Whole-genome sequencing of bacterial pathogens: The future of nosocomial outbreak analysis. Clin. Microbiol. Rev. 2017;30:1015–1063. doi: 10.1128/CMR.00016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wołkowicz T. The utility and perspectives of NGS-based methods in BSL-3 and BSL-4 laboratory–sequencing and analysis strategies. Briefings Funct. Genom. 2018;17:471–476. doi: 10.1093/bfgp/elx033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams M.D., Bishop B., Wright M.S. Quantitative assessment of insertion sequence impact on bacterial genome architecture. Microb. Genom. 2016;2 doi: 10.1099/mgen.0.000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baraniak A., Izdebski R., Żabicka D., Bojarska K., Górska S., Literacka E., Fiett J., Hryniewicz W., Gniadkowski M. Multiregional dissemination of KPC-producing Klebsiella pneumoniae ST258/ST512 genotypes in Poland, 2010–2014. J. Antimicrob. Chemother. 2017;72:1610–1616. doi: 10.1093/jac/dkx054. [DOI] [PubMed] [Google Scholar]
- 32.Lev A.I., Astashkin E.I., Shaikhutdinova R.Z., Platonov M.E., Kartsev N.N., Volozhantsev N.V., Ershova O.N., Svetoch E.A., Fursova N.K. Identification of IS1R and IS10R elements inserted into ompk36 porin gene of two multidrug-resistant Klebsiella pneumoniae hospital strains. FEMS Microbiol. Lett. 2017;364 doi: 10.1093/femsle/fnx072. [DOI] [PubMed] [Google Scholar]
- 33.Lukácová M., Barák I., Kazár J. Role of structural variations of polysaccharide antigens in the pathogenicity of Gram-negative bacteria. Clin. Microbiol. Infect. 2008;14:200–206. doi: 10.1111/j.1469-0691.2007.01876.x. [DOI] [PubMed] [Google Scholar]
- 34.Siguier P., Gourbeyre E., Chandler M. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Microbiol. Rev. 2014;38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kintz E., Heiss C., Black I., Donohue N., Brown N., Davies M.R., Azadi P., Baker S., Kaye P.M., van der Woude M. Salmonella enterica serovar Typhi lipopolysaccharide O-antigen modification impact on serum resistance and antibody recognition. Infect. Immun. 2017;85 doi: 10.1128/IAI.01021-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mann E., Whitfield C. A widespread three-component mechanism for the periplasmic modification of bacterial glycoconjugates. Can. J. Chem. 2016;94:883–893. doi: 10.1139/cjc-2015-0594. [DOI] [Google Scholar]
- 37.Westphal O., Lüderitz O., Bister F. Über die extraktion von bakterien mit phenol/wasser. Z. Nat. B. 1952;7:148–155. doi: 10.1515/znb-1952-0303. [DOI] [Google Scholar]
- 38.Lukasiewicz J., Jachymek W., Niedziela T., Kenne L., Lugowski C. Structural analysis of the lipid a isolated from Hafnia alvei 32 and PCM 1192 lipopolysaccharides. J. Lipid Res. 2010;51:564–574. doi: 10.1194/jlr.M001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szijártó V., Lukasiewicz J., Gozdziewicz T.K., Magyarics Z., Nagy E., Nagy G. Diagnostic potential of monoclonal antibodies specific to the unique O-antigen of multidrug-resistant epidemic Escherichia coli clone ST131-O25b:H4. Clin. Vaccine Immunol. 2014;21:930–939. doi: 10.1128/CVI.00685-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larsen M.V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R.L., Jelsbak L., Sicheritz-Pontén T., Ussery D.W., Aarestrup F.M., et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Treangen T.J., Ondov B.D., Koren S., Phillippy A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letunic I., Bork P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.