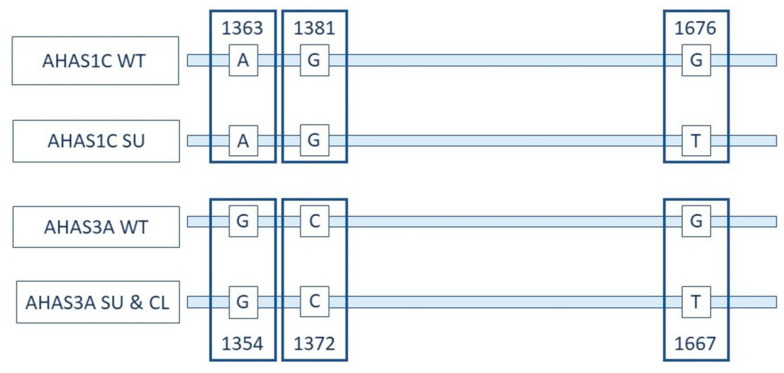
Figure 1.
Relationship between AHAS1C and AHAS3A and positions of a key mutation. Representation of coding strands of the AHAS1C gene and AHAS3A gene in wild-type (WT), genome edited for sulfonylurea and imidazolinone tolerance (SU) and Clearfield (CL) canola varieties. These regions of the AHAS1C and AHAS3A genes are identical except for the bases indicated. Differences in numbering of the two genes is due to differences in sequences upstream of the region depicted. The two SNVs at 1363/1354 and at 1381/1372 differentiate AHAS1C from AHAS3A. The SNV at 1676/1667 differentiates AHAS1C SU, AHAS3A SU and AHAS3A CL from AHAS1C WT and AHAS3A WT. See papers cited in this section for details on structure of the AHAS gene family.