Abstract
Asparagine and glutamine side-chains can form hydrogen-bonded ladders which contribute significantly to the stability of amyloid fibrils. We show, using the example of HET-s(218–289) fibrils, that the primary amide side-chain proton resonances can be detected in cross-polarization based solid-state NMR spectra at fast magic-angle spinning (MAS). J-coupling based experiments offer the possibility to distinguish them from backbone amide groups if the spin-echo lifetimes are long enough, which turned out to be the case for the glutamine side-chains, but not for the asparagine side-chains forming asparagine ladders. We explore the sensitivity of NMR observables to asparagine ladder formation. One of the two possible asparagine ladders in HET-s(218–289), the one comprising N226 and N262, is assigned by proton-detected 3D experiments at fast MAS and significant de-shielding of one of the NH2 proton resonances indicative of hydrogen-bond formation is observed. Small rotating-frame 15N relaxation-rate constants point to rigidified asparagine side-chains in this ladder. The proton resonances are homogeneously broadened which could indicate chemical exchange, but is presently not fully understood. The second asparagine ladder (N243 and N279) in contrast remains more flexible.
Keywords: solid-state NMR, fast MAS, amyloid fibrils, hydrogen bond, asparagine ladder
Introduction
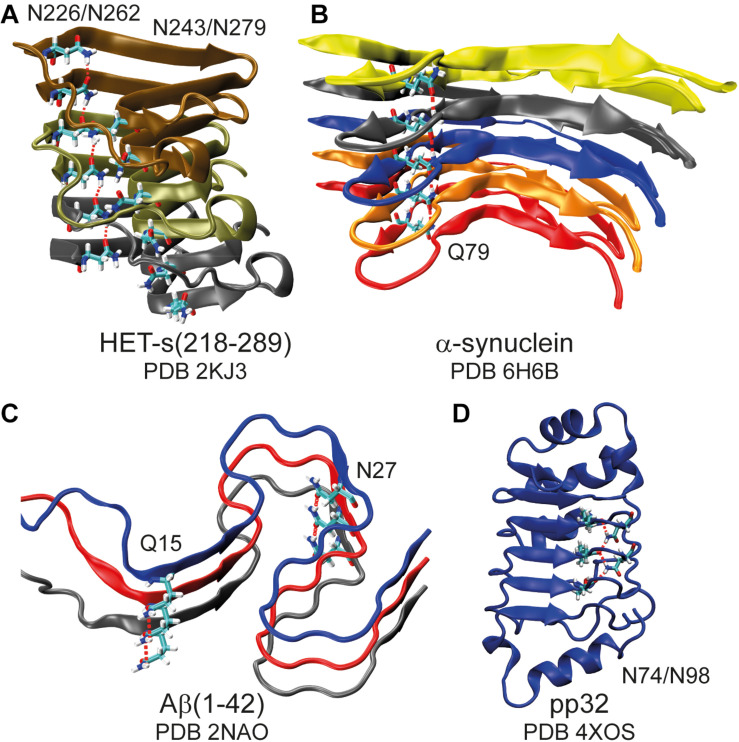
The terminal primary amide group of asparagine and glutamine side-chains can function as both, hydrogen-bond donor via the NH2 group as well as acceptor via the carbonyl group and thus these side-chains are important for protein stability (Vernet et al., 1995; Worth and Blundell, 2010), protein-protein (Van Melckebeke et al., 2010; Talavera et al., 2011; Wälti et al., 2016), protein-ligand (Higman et al., 2004; Raymond et al., 2004) and protein RNA/DNA interactions (Luscombe et al., 2001; Lejeune et al., 2005). In amyloids they can form asparagine/glutamine ladders (Wan and Stubbs, 2014; Riek and Eisenberg, 2016; Eisenberg and Sawaya, 2017; Kurt et al., 2017), e.g., in α-synuclein (Guerrero-Ferreira et al., 2018, 2019), Aβ(1–42) (Colvin et al., 2016; Wälti et al., 2016), the functional amyloid HET-s(218–289) (Wasmer et al., 2008; Van Melckebeke et al., 2010), and the polyglutamine rich Orb2 (Hervas et al., 2020) (for some representative structures see Figure 1 and Supplementary Figure S1). Such side-chains play also important roles in many other amyloids, including the yeast prion Sup35 (Luckgei et al., 2013), huntingtin (Hoop et al., 2016), and prions (Kurt et al., 2017), where these residues form the fibril core, ladders and steric zippers (Sawaya et al., 2007).
FIGURE 1.
Asparagine and glutamine ladders in protein structures. Schematic representation of amyloid fibrils (A–C) highlighting asparagine and glutamine ladders and a leucine-rich repeat protein in which two asparagine side-chains constitute to a second backbone (D). Hydrogen bonds are highlighted by red dashed lines. Note that for (A) structure 16 of the lowest energy structural bundle is shown as it particularly nicely illustrates the ladder motif (for more structures see Supplementary Figure S1).
Solid-state nuclear magnetic resonance (NMR) spectroscopy is a suitable technique to identify asparagine/glutamine ladders in amyloid fibrils: the 1H chemical-shifts are sensitive reporters and less-shielded 1H resonances indicate a participation of the proton in a hydrogen bond (Wagner et al., 1983) whereas relaxation properties can describe dynamical effects. Such spectroscopic information extends the structure information from NMR and cryo-EM (Gremer et al., 2017; Guerrero-Ferreira et al., 2018, 2019; Kollmer et al., 2019; Schmidt et al., 2019; Hervas et al., 2020). Infrared spectroscopy (IR) has also been used recently to identify glutamine ladders in amyloid-forming peptides (Wu et al., 2019) and solution-state NMR has been used to probe asparagine side-chain properties engaged in a dense hydrogen-bond network in the leucine-rich repeat (LRR) proteins pp32 (Klein et al., 2019) (see also Figure 1D). The detection of glutamine and asparagine side-chains by solution-state NMR spectroscopy, e.g., in 15N,1H HSQC-type experiments, has been employed in a variety of biological contexts to probe their structural and dynamic properties (Buck et al., 1995; Vance et al., 1997; Cai et al., 2001; Mulder et al., 2001; Higman et al., 2004; Klein et al., 2019). We show that fast magic-angle spinning (MAS) (Agarwal et al., 2014; Andreas et al., 2015, 2016; Böckmann et al., 2015; Sternberg et al., 2018; Stöppler et al., 2018; Malär et al., 2019; Penzel et al., 2019; Vasa et al., 2019) can detect asparagine and glutamine side-chains in cross-polarization (CP) solid-state NMR proton spectra and, sometimes, also in J-coupling based experiments, in particular INEPT (Morris and Freeman, 1979) experiments, which then can be employed to distinguish the two NH2 side-chain proton resonances from amide NH protons. In amyloid fibrils of HET-s(218–289) of the filamentous fungus Podospora anserina, the asparagine side-chains are visible in CP-based experiments, but not in INEPT experiments at an MAS frequency of 110 kHz. We discuss NMR spectral and relaxation parameters determined for asparagine side-chains forming asparagine ladders, such as high-frequency shifted proton resonances and small rotating-frame 15N relaxation-rate constants.
Results and Discussion
Assignment of Asparagine and Glutamine Residues in HET-s(218–289)
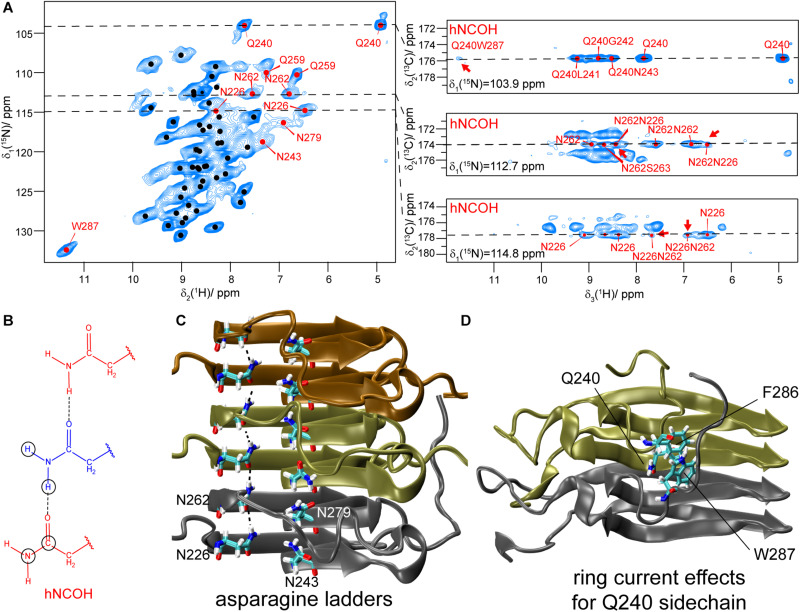
The side-chain amide protons of asparagine (N) and glutamine (Q) side-chains can be observed in proton-detected fast MAS experiments. CP hNH correlation spectra of deuterated and 100% back-exchanged, as well as uniformly 13C and 15N labeled HET-s(218–289) fibrils (abbreviated in the following with DUL), recorded at a MAS frequency of 110 kHz, are shown in Figure 2A. N/Q side-chains resonate in a region somewhat more shielded than backbone amides for both 15N and 1H shifts although the regions overlap. Some of the observed N/Q side-chains (e.g., Q240, N226, N262) lead to two proton correlation peaks with a common 15N frequency indicating no or only a slow rotation around the Cγ -Nδ2 (N) or Cδ -Nε2 (Q) bond as shown in solution-state experiments (Guenneugues et al., 1997). The side-chain assignment is based on a NCOCX 3D experiment performed on a fully protonated HET-s(218–289) sample shown in Supplementary Figure S2 which yields the 15N side-chain resonances. The proton chemical-shift values are extracted from the 2D hNH spectrum (Figure 2A). In total, four out of five asparagine side-chains (N226, N243, N262, N279) and the two glutamine side-chains (Q240, Q259) were identified in the hNH spectrum, only the very C-terminal N289 remains absent probably due to large dynamics (Van Melckebeke et al., 2010). The side-chains of N243, Q259 and N279 have however lower intensity (Supplementary Figure S3 for signal-to-noise ratios extracted from 2D spectra) and remain undetected in proton-detected 3D spectra and thus only one NH2 proton is assigned for N243 and N279. Note that the second NH2 resonance of Q259 is not visible in the CP spectrum, but can be identified in an INEPT-based hNH spectrum (vide infra).
FIGURE 2.
Asparagine and glutamine side-chains in HET-s(218–289) fibrils. (A) 1H,15N CP-hNH spectrum of HET-s fibrils (left) and 2D planes of 3D hNCOH spectra showing spatial proximities for N226, N262 and Q240 (right, the cross peaks mentioned in the text are highlighted by red arrows). The spectra were recorded at a MAS frequency of 110 kHz and an external magnetic field of 20.0 T. The black dots indicate assigned backbone amide correlations based on the BMRB accession code 26913 (Smith et al., 2017). (B) Schematic drawing of an asparagine ladder. Circles indicate connectivities as expected for 3D hNCOH experiments. (C) Asparagine ladders in HET-s highlighted on the PDB structure (PDB accession code 2KJ3). (D) Orientation of the Q240 side-chain toward aromatic residues in HET-s(218–289) highlighted on the PDB structure.
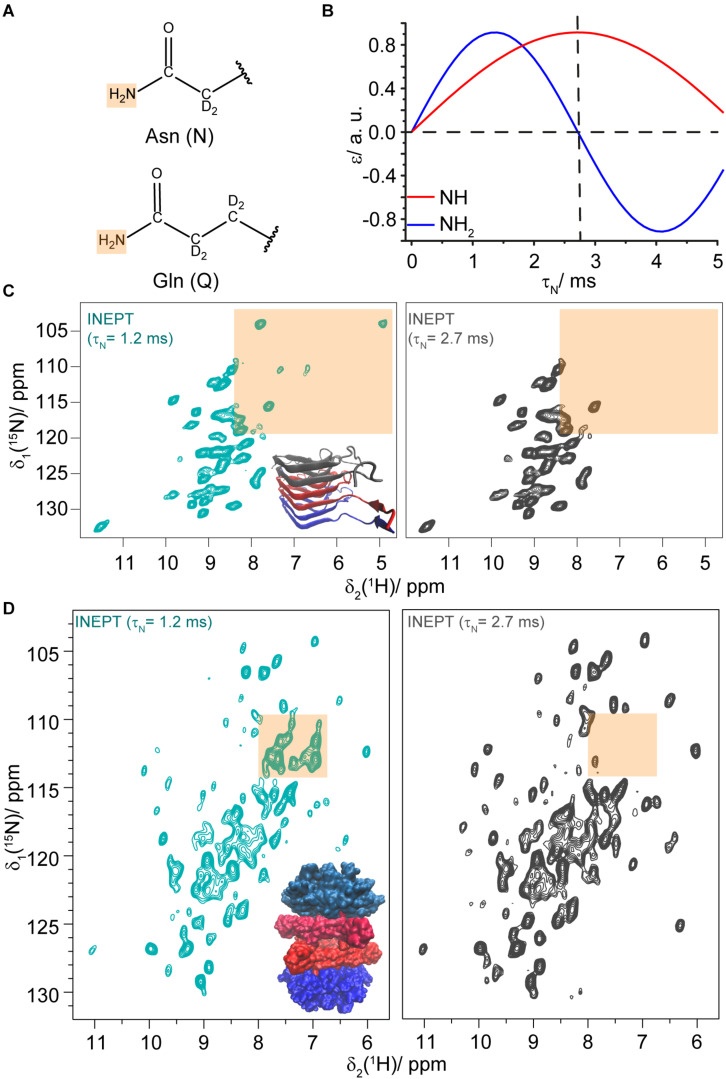
We next address the question whether J-coupling based INEPT-hNH experiments are also suitable to detect N/Q side-chains, similar to 15N,1H HSQC experiments applied in solution-state NMR. The well-known differences in the magnetization-transfer curves of NH and NH2 side-chain resonances in a refocused INEPT-based hNH experiment (Sørensen and Ernst, 1983) could then be used to distinguish the NH2 proton resonances of N/Q side-chains from spectrally overlapping backbone NH resonances (for the chemical structures of N/Q side-chains see Figure 3A). By setting the INEPT delay τN (Supplementary Figure S4) to 2.7 ms (), the NH2 resonances will be strongly suppressed in the INEPT transfer; in contrast, when setting τN close to 1.4 ms, they appear in the spectra (see Figure 3B). For HET-s(218–289) only two of the seven NH2 pairs (assigned to Q240 and Q259) are visible in INEPT experiments (see Figure 3C). None out of the five asparagine residues is detected indicating that proton transverse magnetization lifetimes are too short to detect the HET-s(218–289) asparagine side-chains in INEPT. We attribute this to dynamics as discussed below. Sample deuteration as used herein is crucial for INEPT experiments to prolong proton transverse magnetization lifetimes.
FIGURE 3.
Asparagine and glutamine side-chains identified in INEPT experiments. (A) Chemical structures of asparagine and glutamine side-chains. (B) Dependence of the INEPT enhancement ε on the τN mixing time for NH (blue) and NH2 (red) spin pairs. τH was set to 2 ms, a 15N-1H J coupling constant of 92 Hz was assumed and relaxation effects were neglected. (C) 15N,1H INEPT-hNH spectra of DUL HET-s(218–289) fibrils recorded with different τN mixing times (τH was set to 2 ms). The N/Q side-chain region is highlighted in orange. (D) 15N,1H INEPT-hNH spectra of DUL DnaB with different τN mixing times. The N/Q spectral region is highlighted in orange. The spectra were recorded at a MAS frequency of 110 kHz and an external magnetic field of 20.0 T.
We envision that such INEPT-filtering experiments might also be helpful for large proteins to identify and further characterize the corresponding N/Q side-chain region in crowded NMR spectra. To test the feasibility of such an approach, we recorded INEPT-hNH spectra for the hexameric 354 kDa protein DnaB (Wiegand et al., 2019; Wiegand, 2020) (complexed with ADP:AlFx– and DNA). Figure 3D shows a comparison of the spectra with τN = 1.2 ms (cyan) and τN = 2.7 ms (gray, for a difference spectrum see Supplementary Figure S5 and for further τN-times see Supplementary Figures S6, S7) and indeed several N/Q side-chains are identified at 110 kHz MAS. We note that the INEPT spectra of this 488 residue protein (with 24 N and 23 Q residues) are again rather sparse, also for the backbone amides, as the INEPT step filters out resonances with short transverse proton magnetization lifetimes that are on the other hand visible in CP-hNH spectra (Wiegand et al., 2020).
Asparagine Ladders
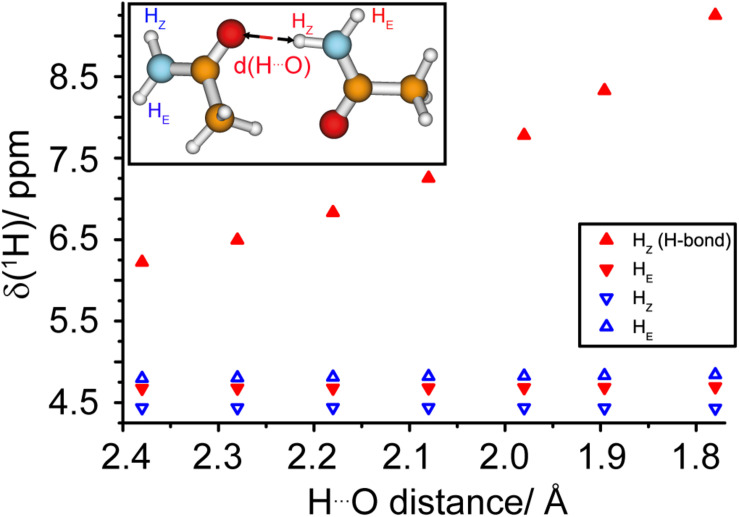
In the amyloid fibrils of HET-s(218–289), two asparagine ladders (one comprising N226 and N262 and the other N243 and N279) have been postulated from the structure obtained by solid-state NMR. N226 and N262 point to the inside of the fibril and are located next to the hydrophobic core, whereas N243 and N279 are outside of the core (see Figure 2C; Wasmer et al., 2008). The NMR structure shows a well-formed ladder for N226/N262 in most structures of the structural bundle whereas N243/N279 do not form a regular ladder. It is, at this point not a priori clear if this difference is a consequence of the limited precision of the NMR structure or if it reflects real structural differences. Such small, but potentially important structural features are not easily addressed by cryo-EM or NMR structures. However, spectroscopic features may be exploited: the proton chemical shift itself is highly sensitive to changes in the environment and can be used as an indicator for hydrogen bonding (Cordier and Grzesiek, 1999) characteristic for ladders (see Figure 2). As illustrated in Density Functional Theory (DFT) calculations on a model consisting of two asparagine side-chains engaged in a hydrogen bond, see Figure 4 and Supplementary Figure S8, a significant de-shielding of the HZ proton (see insert of Figure 4 for the nomenclature) participating in a hydrogen bond is observed. In one calculation, two asparagine side-chain fragments were moved relative to each other along the N-H…O distance vector (Figure 4), while in the second calculation only the HZ proton was moved along that vector and all other atoms of the two fragments were kept fixed (Supplementary Figure S8). Both cases reveal that the shorter the hydrogen bond, the larger the chemical-shift difference between the HZ and HE protons (Barfield, 2002). We speculate that the scenario described in Figure 4 is more likely reflecting the situation in asparagine ladders, although not only the hydrogen bond length effects the strength of the hydrogen bond and thus the proton chemical-shift values observed experimentally, but also the bond angle certainly has an influence.
FIGURE 4.
The proton chemical shift is a sensitive tool to measure hydrogen bond strengths in asparagine ladders. DFT calculations (B3-LYP/def2-TZVP) of 1H chemical-shift values as a function of the hydrogen bond length in a model representing two asparagine side-chains. The geometries of the two molecules were fixed and the two fragments were moved relative to each other along the N-H… O distance vector, see insert.
Experimentally, for N226 we find a large difference between the two proton resonances resonating at 8.4 and 6.5 ppm. For N262 the less-shielded value is 7.6 ppm compared to 6.8 ppm for the more-shielded one. The two NH2 protons are thus shifted in different directions with respect to their average chemical-shift values over all deposited asparagine residues as extracted from the BMRB database (7.3 and 7.1 ppm)1.
3D hNCOH experiments (Figure 2B) do not only identify the two protons in the same NH2 group, but also probe the spatial proximity between such side-chains. A 9 ms 13CO-1H CP-step was employed to achieve a polarization transfer to both NH2 protons of the neighboring asparagine side-chain within the same ladder. Similar experiments have been described recently to identify backbone hydrogen bonding patterns in SH3 and GB1 (Friedrich et al., 2020). And indeed, the 15N planes at the NH2 side-chain chemical-shift value of N262 (112.7 ppm) and N226 (114.8 ppm) clearly show cross peaks to the proton shifts of the NH2 group of N226 and N262, respectively (see Figure 2A, red arrows) indicating their spatial proximity. In HET-s(218–289) this information corroborates the known register in the β-sheets and in other systems of unknown structure this may be of even more interest. For residues N243/N279 that form the second asparagine ladder no signals are detected in the CP-based 3D hNCOH spectrum.
Homogeneous Proton Linewidths of Asparagine and Glutamine Side-Chains
The visibility of the proton resonances in INEPT-type experiments is related to the homogeneous proton linewidth (Δhomo) which is determined by the geometry of the proton dipolar network (coherent contribution; Zorin et al., 2006; Sternberg et al., 2018; Malär et al., 2019), additional additive contributions resulting from Redfield-type relaxation caused by stochastic molecular motion (incoherent contribution from anisotropic interactions) and chemical-exchange broadening effects (incoherent contributions from isotropic chemical-shift differences) (Sternberg et al., 2018; Malär et al., 2019; Penzel et al., 2019). Unfortunately, the three contributions cannot easily be addressed separately, and only their sum is observed experimentally. Coherent and incoherent contributions are expected to decrease with faster spinning (Lewandowski et al., 2011; Andreas et al., 2015; Böckmann et al., 2015; Sternberg et al., 2018; Malär et al., 2019; Penzel et al., 2019) while chemical exchange is largely MAS-frequency independent. Spin-echo decay experiments allow determination of proton T2′ relaxation times and Δhomo = R2′/π = 1/(πT2′) values. The pulse sequences used to measure Δhomo values are shown in Supplementary Figure S9.
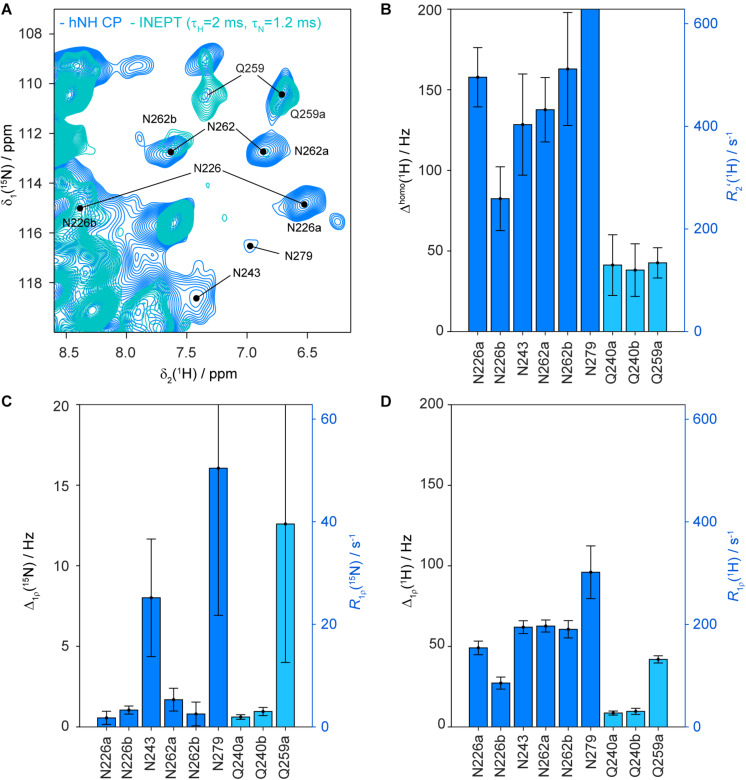
For HET-s(218–289) fibrils, Δhomo values between 80 and 160 Hz for the N226, N243 and N262 side-chain protons are observed (see Figures 5A,B) which explains why these resonances remain absent in INEPT experiments (a Δhomo of 160 Hz, T2′∼2 ms, would lead to an attenuation of ∼88% in the refocused INEPT efficiency with INEPT delays of τH = 2.0 ms and τN = 1.2 ms). Δhomo for N279 is even larger (543 ± 140 Hz). All these values are significantly larger than the corresponding values of the backbone amide protons, which have been reported before, showing on average 22 ± 1 Hz at 100 kHz MAS for DUL HET-s(218–289) (Smith-Penzel, 2019). The glutamine side-chains which are easily observed in INEPT-hNH have, as expected, a smaller Δhomo value (<50 Hz). As an additional reference we also measured the homogeneous proton linewidths for asparagine and glutamine side-chains in DnaB. In accordance with the experimental observation of glutamine side-chains in INEPT spectra, the Δhomo are indeed smaller and amount on average to 31 ± 5 Hz (Supplementary Figure S10).
FIGURE 5.
Homogeneous proton linewidths and 15N and 1H R1ρ rate constants of asparagine and glutamine side-chains in HET-s(218–289) fibrils. Zoom into INEPT-hNH (cyan) and CP-hNH (blue) spectra of HET-s(218–289) fibrils (A) showing the asparagine and glutamine side-chain region and some resonance assignments. Homogeneous proton linewidths for asparagine and glutamine side-chains of HET-s(218–289) fibrils are given in (B). Nitrogen spin-lock relaxation rate constants of asparagine and glutamine side-chains are shown in (C), proton spin-lock relaxation times in (D). All error bars shown correspond to two standard deviations. The relaxation rate constants given on the vertical axis are related according to Δ = R/π. The relaxation times were determined at a MAS frequency of 105 kHz and an external magnetic field of 20.0 T.
To investigate whether the larger-than-expected Δhomo values observed for the asparagine side-chains are related to coherent broadening mechanisms, we have numerically simulated the 1H solid-state NMR spectrum using the geometry of the six NH2 proton spins of N262-N226-N262 taken from the PDB file (Figure 1, PDB accession number 2KJ3, the chosen geometry is taken from structure 16 in which the asparagine ladder N226/N262 is most clearly visible, see also Supplementary Figure S11A). Those simulations yield Δhomo values of less than 5 Hz (see Supplementary Figure S11B) which illustrates that the main contribution to Δhomo is not related to coherent effects resulting from the geometry within an asparagine ladder even if the contribution from a close-by backbone amide is taken into account (Supplementary Figure S12). The numerically simulated spin-echo decay profile using the same six-spin system are shown in Supplementary Figure S11C and also give a negligible Δhomo value of 2 Hz. Cross terms between heteronuclear 15N-1H dipolar and/or J-couplings and the proton homonuclear dipolar couplings were also not found to make an appreciable contribution (Supplementary Figure S11C).
15N Rotating-Frame Relaxation Rates of Asparagine and Glutamine Side-Chains: Rigid Side-Chains in Asparagine Ladders
15N rotating-frame relaxation rate constants, characterized by R1ρ, may give further information about the dynamics of asparagine and glutamine side-chains (for the pulse sequences see Supplementary Figure S13 and for the site-specific decay curves Supplementary Figures S14, S15). Under the experimental conditions used (110 kHz MAS, 13 kHz rf-field amplitude for the spin lock) the experiment is sensitive to dynamics with correlation times in the hundreds of nanoseconds to low microsecond range (Lakomek et al., 2017). Figure 5C shows the 15N R1ρ experimental relaxation-rate constants for HET-s(218–289) fibrils. Typical rigid backbone nitrogen atoms possess R1ρ values of around 2 s−1 (Schanda and Ernst, 2016; Lakomek et al., 2017). The values for the side-chain NH2 nitrogen of N226 and N262 are also in the order of 2 s–1 indicating the absence of dynamics on this timescale for side-chain nitrogen (for details on relaxation properties of NH2 spin systems see Bull (1992). Interestingly, N243 and N279 show significantly larger 15N R1ρ (>25 s–1) pointing to a higher degree of flexibility of these side-chains. Based on the measured R1ρ values, the side-chain of Q240 is similarly rigid as N226 and N262, whereas Q259 is more flexible and shows weaker correlations in 13C-detected 3D spectra (see Supplementary Figure S2). In DnaB, which does not contain N/Q ladders, the side-chain nitrogen R1ρ relaxation-rate constants are ranging between 10 and 32 s–1 (see Supplementary Figure S10C) and are, thus, larger than for the rather rigid N226/N262 side-chains of HET-s(218–289) fibrils.
The higher rigidity of the ladder N226/N262 compared to N243/N279 might be a consequence of a higher water accessibility of N243/N279 in agreement with their location outside of the hydrophobic core. All asparagine residues influence the fibril stability in the presence of guanidine as a chemical denaturant as shown by site-directed mutagenisis (Wan and Stubbs, 2014). However, mutation of N226 and N262 yields more destabilization than mutation of N243 and especially N279, in the latter mutant a similar chemical stability than in wild-type HET-s(218–289) fibrils was observed (Wan and Stubbs, 2014).
Since so far coherent and incoherent broadening mechanisms cannot account for the observed proton homogeneous linewidth, we also consider the possibility of chemical-exchange broadening, namely slow to intermediate exchange (below coalescence) on a millisecond timescale to explain the broader-than-expected lines. Such processes have been observed for asparagine side-chains in T4 lysozyme (Mulder et al., 2001) or in coiled-coil homo dimers (Thomas et al., 2017) in solution. The exchange is most likely asymmetric with one dominating conformer which is detected in our spectra. In our model, the conformational states represent asparagine side-chains hydrogen-bonded in a ladder (the most populated state observed experimentally) and an additional so far unobserved state. We performed 15N chemical-exchange saturation transfer (CEST) experiments (Vallurupalli et al., 2012), but were not able to observe this state requiring further experimental work. The NH2 proton relaxation-rate constants in the rotating frame measured for the asparagine side-chains are larger compared to backbone amides possibly as a consequence of a modulation of the proton homonuclear dipolar couplings by the exchange process (see Figure 5D and Supplementary Figure S16). However, the exchange contribution to the measured proton R1ρ values is negligible at a spin-lock field of 13 kHz as used in our experiment (Palmer and Massi, 2006). Whether this observation is a general feature of N/Q ladders has to be investigated on further amyloid fibrils in future work, it is however interesting that the glutamine side-chains in HET-s(218–289) do not show these effects, and their homogeneous linewidths are roughly by a factor of three smaller.
Characterization of Glutamine Side-Chains and the Orientation of Q240
For the two glutamine side-chains of HET-s(218–289), no ladder formation is possible from the structure (Van Melckebeke et al., 2010). Both glutamine side-chain NH2 are observed in the CP and INEPT-spectra (Figures 2A, 3D). For Q240, one proton is shifted to the more shielded region (4.8 ppm), the other one is at 7.8 ppm, slightly less shielded compared to the average chemical shift values taken from the BMRB database (7.22/7.04 ppm). The increased shielding of one of the NH2 proton of Q240 might be associated with ring-current effects, since Q240 points toward a hydrophobic pocket constituted by aromatic residues (F286 and W287, Figure 2D; Van Melckebeke et al., 2010). The cross signal between Q240 and W287 in the 3D hNCOH experiment reveals that the side-chain of Q240 is indeed in spatial proximity to the side-chain NH group of W287 (Figure 2A).
Conclusion
We detected asparagine and glutamine NH2 side-chains proton signals in fast MAS spectra of HET-s(218–289) in its amyloid form. The spectral and relaxation properties are described and the influence of the formation of ladders is investigated. Table 1 summarizes the NMR properties of such side-chains. The following observations were made for the asparagines involved in a well-formed ladder: (i) strong de-shielding of the proton shift of one of the NH2 protons, (ii) a large chemical-shift difference between the two protons, (iii) correlations in 3D through-space hCONH experiments, and (iv) small 15N R1ρ relaxation-rate constants. These properties characterize the N226-N262 asparagine ladder. In addition (v) a large homogeneous proton linewidth and (vi) a large 1H R1ρ relaxation-rate constants were found. 15N rotating-frame relaxation rate constants indicate that the side-chains involved into the N226-N262 asparagine ladder are more rigid than for the N243-N279 ladder which appear more dynamic not forming a strong restraining hydrogen network. The large homogeneous proton linewidths for asparagine side-chains involved in the ladder, not fully understood yet, could be related to millisecond timescale chemical exchange characteristic for protons engaged in an asparagine ladder in HET-s(218–289) fibrils. The glutamine side-chain amide protons of HET-s(218–289) not participating in such interactions are less homogeneously broadened and have linewidths comparable to the backbone NH protons. Fast MAS NMR experiments thus probe protein side-chain hydrogen bonding, in particular in the context of asparagine/glutamine ladder formation. While more systems must be investigated for a full understanding, the results on HET-s(218–289) clearly show the potential of the approach.
TABLE 1.
Overview of asparagine and glutamine side-chain spectral and relaxation properties.
| N226 | N262 | N243 | N279 | Q240 | Q259 | |
| Visible in CP-hNH? | Yes | Yes | Yes | Yes | Yes | Yes |
| Visible in INEPT-hNH? | No | No | No | No | Yes | Yes |
| δ(15Nδ2/15Nε2)/ppm | 114.9 (115.4a) | 112.8 (113.5a) | 118.9 (n.d.a) | 116.5 (115.4a) | 104.0 | 110.2 (110.8a) |
| δ(13Cγ/13Cδ)/ppm | 177.6 (177.4a) | 173.9 (176.5a) | 176.1 (176.1a) | 176.4 (176.3a) | 175.5 (175.7a) | 180.3 (180.3a) |
| δ(1Hδ21/δ22/1Hε21/ε22)/ppm | 8.4/6.5 | 7.6/6.8 | 7.4/n.d. | 7.0 (broad) | 7.8/4.9 | 7.3/6.7 |
| Δ[δ(1H)]/ppm | 1.9 | 0.8 | n.d. | n.d. | 2.9 | 0.6 |
| Δhomo (1H)/Hzc | 157 ± 18 | 138 ± 20 | 128 ± 50 | 543 ± 140 | 41 ± 20 | 43 ± 10 |
| 82 ± 18 | 160 ± 40 | 38 ± 18 | ||||
| R1ρ (15N)/s–1 | 2.6 ± 0.7b | 3.8 ± 1.6b | 25 ± 10 | 50 ± 30 | 2.5 ± 0.5b | 40 ± 30 |
| R1ρ (1H)/s–1 | 120 ± 9b | 196 ± 9b | 190 ± 17 | 302 ± 52 | 29 ± 4b | 132 ± 7 |
aDetermined on protonated HET-s fibrils in 13C-detected MAS experiments (BMRB accession code 11064). Differences to the values derived from 1H-detected experiments are related to temperature differences in the experiments. bThe average value over the relaxation rate constants for both resonances is given. The error is derived by error propagation of the individual experimental errors (a 2σ error is given). cΔhomo is calculated according to R2’/π = 1/(πT2’).
Materials and Methods
Protein Preparations
The preparations of the DUL HpDnaB:ADP:AlF4–:DNA (Wiegand et al., 2019) and DUL HET-s(218–289) (Wasmer et al., 2008) samples were prepared as described previously. In short, the proteins were recombinantly expressed in D2O in presence of deuterated 13C-glucose (2 g/L) and 15N-ammonium chloride (2 g/L) as sole sources of carbon-13 and nitrogen-15. In case of HET-s(218–289), fibrillization was performed in protonated solvent (50 mM citric acid at pH 5 in H2O) for 24 h which yields 100% back-exchanged fibrils (Smith et al., 2017). For DnaB, back-exchange was achieved by purifying the protein in a protonated buffer (2.5 mM sodium phosphate, pH 7.5, 130 mM NaCl). The complex formation with ADP:AlFx− and DNA (a single-stranded DNA molecule with 20 thymine bases was used, purchased from Microsynth) is described in reference Wiegand et al. (2019). UL HET-s(218–289) fibrils were prepared identically, but only using H2O instead of D2O and protonated 13C-glucose (2 g/L) and 15N-ammonium chloride (2 g/L).
Solid-State NMR
Solid-state NMR spectra were acquired at 20.0 T static magnetic-field strength using a Bruker 0.7 mm probe. The MAS frequency was set to 110 kHz. The 2D spectra were processed with the software TOPSPIN (version 3.5, Bruker Biospin) with a shifted (3.0) squared cosine apodization function and automated baseline correction in the indirect and direct dimensions. The sample temperature was set to 293 K (Böckmann et al., 2009). All spectra were analyzed with the software CcpNmr (Fogh et al., 2002; Vranken et al., 2005; Stevens et al., 2011) and referenced to 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS). Site-specific relaxation-rate constants were extracted from the 2D hNH relaxation experiments, using the INFOS software (Smith, 2017) (see Supplementary Figures S14, S15 for the mono-exponential fits of all relaxation decay curves for DUL HET-s). Error bars for all relaxation data have been derived by bootstrapping methods, using 500 iterations and are shown as 2σ, where σ denotes the obtained error. For more details see Supplementary Table S1. The NMR spectra can be accessed at https://doi.org/10.3929/ethz-b-000430151.
Numerical Simulations
The numerical simulations of 1H MAS spectra were performed with SIMPSON (Bak et al., 2000) using six proton spins in the geometry extracted from the pdb file 2JK3 (structure 16). The spin-echo decay curves were obtained using the same proton six-spin system with and without three nitrogen spins. The simulations were performed in the GAMMA spin simulations environment (Smith et al., 1994). The program code for the simulations is provided in the Supplementary Material.
DFT Calculations
The DFT calculations were performed with TURBOMOLE (Ahlrichs et al., 1989) version 6.4.0. Geometry optimizations were performed on a B3-LYP (Stephens et al., 1994)/def2-TZVP (Weigend and Ahlrichs, 2005) level employing the D3 dispersion correction (Grimme et al., 2010). In all TURBOMOLE SCF calculations, an energy convergence criterion of 10–7 Eh was used, and in the geometry optimizations, an energy convergence criterion of 10–6 Eh was employed. The integration grid was set to m4. Magnetic shieldings were calculated within the GIAO framework on the same theoretical level and converted into chemical-shift values according to δ = σref-σ with σref being the magnetic shielding of the protons of TMS calculated at the same level of theory (σref = 31.9 ppm). The orientation-dependence of the 1H chemical-shift values was studied by moving the two molecules relative to each other along the N-H… O distance vector (Figure 4) or moving the proton only along that vector (Supplementary Figure S8). No additional geometry optimization was performed.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
RC prepared the samples. TW and AM performed the NMR experiments. TW, AM, ME, AB, and BM analyzed the data. TW and ME performed the simulations. TW, AB, ME, and BM designed and supervised the research. All authors contributed to the writing of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
TW thanks Denis Lacabanne for helpful discussions.
Funding. This work was supported by the ETH Career SEED-69 16-1 (TW) and the ETH Research Grant ETH-43 17-2 (TW), an ERC Advanced Grant (BM, Grant No. 741863, Faster), by the Swiss National Science Foundation (BM, Grant Nos. 200020_159707 and 200020-188711, ME, Grant No. 200020_188988), the French Agence Nationale de Recherche (AB, ANR-14-CE09-0024B), and the LABEX ECOFECT (AB, ANR-11-LABX-0048) within the Université de Lyon program Investissements d’Avenir (AB, ANR-11-IDEX-0007).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2020.582033/full#supplementary-material
References
- Agarwal V., Penzel S., Szekely K., Cadalbert R., Testori E., Oss A., et al. (2014). De Novo 3D structure determination from sub-milligram protein samples by solid-State 100 kHz MAS NMR spectroscopy. Angew. Chem. Int. Ed. 53 12253–12256. 10.1002/anie.201405730 [DOI] [PubMed] [Google Scholar]
- Ahlrichs R., Bär M., Häser M., Horn H., Kölmel C. (1989). Electronic structure calculations on workstation computers: the program system turbomole. Chem. Phys. Lett. 162 165–169. 10.1016/0009-2614(89)85118-8 [DOI] [Google Scholar]
- Andreas L. B., Jaudzems K., Stanek J., Lalli D., Bertarello A., Le Marchand T., et al. (2016). Structure of fully protonated proteins by proton-detected magic-angle spinning NMR. Proc. Natl. Acad. Sci. U.S.A. 113 9187–9192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreas L. B., Le Marchand T., Jaudzems K., Pintacuda G. (2015). High-resolution proton-detected NMR of proteins at very fast MAS. J. Magn. Reson. 253 36–49. 10.1016/j.jmr.2015.01.003 [DOI] [PubMed] [Google Scholar]
- Bak M., Rasmussen J. T., Nielsen N. C. (2000). SIMPSON: a general simulation program for solid-state NMR spectroscopy. J. Magn. Reson. 147 296–330. 10.1006/jmre.2000.2179 [DOI] [PubMed] [Google Scholar]
- Barfield M. (2002). Structural dependencies of interresidue scalar coupling h3JNC‘ and donor 1H chemical shifts in the hydrogen bonding regions of proteins. J. Am. Chem. Soc. 124 4158–4168. 10.1021/ja012674v [DOI] [PubMed] [Google Scholar]
- Böckmann A., Ernst M., Meier B. H. (2015). Spinning proteins, the faster, the better? J. Magn. Reson. 253 71–79. 10.1016/j.jmr.2015.01.012 [DOI] [PubMed] [Google Scholar]
- Böckmann A., Gardiennet C., Verel R., Hunkeler A., Loquet A., Pintacuda G., et al. (2009). Characterization of different water pools in solid-state NMR protein samples. J. Biomol. NMR 45 319–327. 10.1007/s10858-009-9374-3 [DOI] [PubMed] [Google Scholar]
- Buck M., Boyd J., Redfield C., MacKenzie D. A., Jeenes D. J., Archer D. B., et al. (1995). Structural determinants of protein dynamics: analysis of 15N NMR relaxation measurements for main-chain and side-chain nuclei of hen egg white lysozyme. Biochemistry 34 4041–4055. 10.1021/bi00012a023 [DOI] [PubMed] [Google Scholar]
- Bull T. E. (1992). Relaxation in the rotating frame in liquids. Prog. Nucl. Magn. Reson. Spectrosc. 24 377–410. 10.1016/0079-6565(92)80002-w [DOI] [Google Scholar]
- Cai M., Huang Y., Clore G. M. (2001). Accurate orientation of the functional groups of asparagine and glutamine side chains using one- and two-bond dipolar couplings. J. Am. Chem. Soc. 123 8642–8643. 10.1021/ja0164475 [DOI] [PubMed] [Google Scholar]
- Colvin M. T., Silvers R., Ni Q. Z., Can T. V., Sergeyev I., Rosay M., et al. (2016). Atomic resolution structure of monomorphic Aβ42 amyloid fibrils. J. Am. Chem. Soc. 138 9663–9674. 10.1021/jacs.6b05129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordier F., Grzesiek S. (1999). Direct observation of hydrogen bonds in proteins by interresidue 3hJNC‘ scalar couplings. J. Am. Chem. Soc. 121 1601–1602. 10.1021/ja983945d [DOI] [PubMed] [Google Scholar]
- Eisenberg D. S., Sawaya M. R. (2017). Structural studies of amyloid proteins at the molecular level. Annu. Rev. Biochem 86 69–95. 10.1146/annurev-biochem-061516-045104 [DOI] [PubMed] [Google Scholar]
- Fogh R., Ionides J., Ulrich E., Boucher W., Vranken W., Linge J. P., et al. (2002). The CCPN project: an interim report on a data model for the NMR community. Nat. Struct. Mol. Biol. 9 416–418. 10.1038/nsb0602-416 [DOI] [PubMed] [Google Scholar]
- Friedrich D., Perodeau J., Nieuwkoop A. J., Oschkinat H. (2020). MAS NMR detection of hydrogen bonds for protein secondary structure characterization. J. Biomol. NMR 74 247–256. 10.1007/s10858-020-00307-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremer L., Schölzel D., Schenk C., Reinartz E., Labahn J., Ravelli R. B. G., et al. (2017). Fibril structure of amyloid-β(1–42) by cryo–electron microscopy. Science 358 116–119. 10.1126/science.aao2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimme S., Antony J., Ehrlich S., Krieg H. (2010). A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 132:154104 10.1063/1.3382344 [DOI] [PubMed] [Google Scholar]
- Guenneugues M., Drevet P., Pinkasfeld S., Gilquin B., Ménez A., Zinn-Justin S. (1997). Picosecond to hour time scale dynamics of a “three finger” toxin: correlation with its toxic and antigenic properties. Biochemistry 36 16097–16108. 10.1021/bi971293k [DOI] [PubMed] [Google Scholar]
- Guerrero-Ferreira R., Taylor N. M. I., Arteni A.-A., Kumari P., Mona D., Ringler P., et al. (2019). Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy. eLife 8:e48907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Ferreira R., Taylor N. M. I., Mona D., Ringler P., Lauer M. E., Riek R., et al. (2018). Cryo-EM structure of alpha-synuclein fibrils. eLife 7:e36402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervas R., Rau M. J., Park Y., Zhang W., Murzin A. G., Fitzpatrick J. A. J., et al. (2020). Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila. Science 367 1230–1234. 10.1126/science.aba3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higman V. A., Boyd J., Smith L. J., Redfield C. (2004). Asparagine and glutamine side-chain conformation in solution and crystal: a comparison for hen egg-white lysozyme using residual dipolar ouplings. J. Biomol. NMR 30 327–346. 10.1007/s10858-004-3218-y [DOI] [PubMed] [Google Scholar]
- Hoop C. L., Lin H. K., Kar K., Magyarfalvi G., Lamley J. M., Boatz J. C., et al. (2016). Huntingtin exon 1 fibrils feature an interdigitated beta-hairpin-based polyglutamine core. Proc. Natl. Acad. Sci. U.S.A. 113 1546–1551. 10.1073/pnas.1521933113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S. A., Majumdar A., Barrick D. (2019). A second backbone: the contribution of a buried asparagine ladder to the global and local stability of a leucine-rich repeat protein. Biochemistry 58 3480–3493. 10.1021/acs.biochem.9b00355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmer M., Close W., Funk L., Rasmussen J., Bsoul A., Schierhorn A., et al. (2019). Cryo-EM structure and polymorphism of Aβ amyloid fibrils purified from Alzheimer’s brain tissue. Nat. Commun. 10:4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt T. D., Aguilar-Calvo P., Jiang L., Rodriguez J. A., Alderson N., Eisenberg D. S., et al. (2017). Asparagine and glutamine ladders promote cross-species prion conversion. J. Biol. Chem. 292 19076–19086. 10.1074/jbc.m117.794107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakomek N.-A., Penzel S., Lends A., Cadalbert R., Ernst M., Meier B. H. (2017). Microsecond dynamics in ubiquitin probed by solid-state 15N NMR spectroscopy R1ρ relaxation experiments under fast MAS (60–110 kHz). Chem. Eur. J. 23 9425–9433. 10.1002/chem.201701738 [DOI] [PubMed] [Google Scholar]
- Lejeune D., Delsaux N., Charloteaux B., Thomas A., Brasseur R. (2005). Protein–nucleic acid recognition: statistical analysis of atomic interactions and influence of DNA structure. Proteins Struct. Funct. Bioinform. 61 258–271. 10.1002/prot.20607 [DOI] [PubMed] [Google Scholar]
- Lewandowski J. R., Dumez J.-N., Akbey Ü, Lange S., Emsley L., Oschkinat H. (2011). Enhanced resolution and coherence lifetimes in the solid-state NMR spectroscopy of perdeuterated proteins under ultrafast magic-angle spinning. J. Phys. Chem. Lett. 2 2205–2211. 10.1021/jz200844n [DOI] [Google Scholar]
- Luckgei N., Schütz A. K., Bousset L., Habenstein B., Sourigues Y., Gardiennet C., et al. (2013). The conformation of the prion domain of Sup35p in isolation and in the full-length protein. Angew. Chem. Int. Ed. 52 12741–12744. 10.1002/anie.201304699 [DOI] [PubMed] [Google Scholar]
- Luscombe N. M., Laskowski R. A., Thornton J. M. (2001). Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 29 2860–2874. 10.1093/nar/29.13.2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malär A. A., Dong S., Kehr G., Erker G., Meier B. H., Wiegand T. (2019). Characterization of H2 -splitting products of frustrated lewis pairs: benefit of fast magic-angle spinning. ChemPhysChem 20 672–679. 10.1002/cphc.201900006 [DOI] [PubMed] [Google Scholar]
- Malär A. A., Smith-Penzel S., Camenisch G. M., Wiegand T., Samoson A., Bockmann A., et al. (2019). Quantifying proton NMR coherent linewidth in proteins under fast MAS conditions: a second moment approach. Phys. Chem. Chem. Phys. 21 18850–18865. 10.1039/c9cp03414e [DOI] [PubMed] [Google Scholar]
- Morris G. A., Freeman R. (1979). Enhancement of nuclear magnetic resonance signals by polarization transfer. J. Am. Chem. Soc. 101 760–762. 10.1021/ja00497a058 [DOI] [Google Scholar]
- Mulder F. A. A., Skrynnikov N. R., Hon B., Dahlquist F. W., Kay L. E. (2001). Measurement of slow (μs-ms) time scale dynamics in protein side chains by 15N relaxation dispersion NMR spectroscopy: application to Asn and Gln residues in a cavity mutant of T4 lysozyme. J. Am. Chem. Soc. 123 967–975. 10.1021/ja003447g [DOI] [PubMed] [Google Scholar]
- Palmer A. G., Massi F. (2006). Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem. Rev. 106 1700–1719. 10.1021/cr0404287 [DOI] [PubMed] [Google Scholar]
- Penzel S., Oss A., Org M.-L., Samoson A., Böckmann A., Ernst M., et al. (2019). Spinning faster: protein NMR at MAS frequencies up to 126 kHz. J. Biomol. NMR 73 19–29. 10.1007/s10858-018-0219-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond A. C., Rideout M. C., Staker B., Hjerrild K., Burgin A. B. (2004). Analysis of human tyrosyl-DNA phosphodiesterase I catalytic residues. J. Mol. Biol. 338 895–906. 10.1016/j.jmb.2004.03.013 [DOI] [PubMed] [Google Scholar]
- Riek R., Eisenberg D. S. (2016). The activities of amyloids from a structural perspective. Nature 539 227–235. 10.1038/nature20416 [DOI] [PubMed] [Google Scholar]
- Sawaya M. R., Sambashivan S., Nelson R., Ivanova M. I., Sievers S. A., Apostol M. I., et al. (2007). Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature 447 453–457. 10.1038/nature05695 [DOI] [PubMed] [Google Scholar]
- Schanda P., Ernst M. (2016). Studying dynamics by magic-angle spinning solid-state NMR spectroscopy: principles and applications to biomolecules. Prog. Nucl. Magn. Reson. Spectrosc. 96 1–46. 10.1016/j.pnmrs.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Wiese S., Adak V., Engler J., Agarwal S., Fritz G., et al. (2019). Cryo-EM structure of a transthyretin-derived amyloid fibril from a patient with hereditary ATTR amyloidosis. Nat. Commun. 10:5008. 10.1038/s41467-019-13038-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. A. (2017). INFOS: spectrum fitting software for NMR analysis. J. Biomol. NMR 67 77–94. 10.1007/s10858-016-0085-2 [DOI] [PubMed] [Google Scholar]
- Smith A. A., Ravotti F., Testori E., Cadalbert R., Ernst M., Böckmann A., et al. (2017). Partially-deuterated samples of HET-s(218–289) fibrils: assignment and deuterium isotope effect. J. Biomol. NMR 67 109–119. 10.1007/s10858-016-0087-0 [DOI] [PubMed] [Google Scholar]
- Smith S. A., Levante T. O., Meier B. H., Ernst R. R. (1994). Computer simulations in magnetic resonance. An object-oriented programming approach. J. Magn. Reson. Ser. A 106 75–105. 10.1006/jmra.1994.1008 [DOI] [Google Scholar]
- Smith-Penzel S. (2019). Solid-State NMR Under Fast Magic-Angle Spinning (90-150 kHz) for Biological Applications. Zürich: ETH Zürich. [Google Scholar]
- Sørensen O. W., Ernst R. R. (1983). Elimination of spectral distortion in polarization transfer experiments. Improvements and comparison of techniques. J. Magn. Reson. 51 477–489. 10.1016/0022-2364(83)90300-1 [DOI] [Google Scholar]
- Stephens P. J., Devlin F. J., Chabalowski C. F., Frisch M. J. (1994). Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J. Phys. Chem. 98 11623–11627. 10.1021/j100096a001 [DOI] [Google Scholar]
- Sternberg U., Witter R., Kuprov I., Lamley J. M., Oss A., Lewandowski J. R., et al. (2018). 1H line width dependence on MAS speed in solid state NMR – comparison of experiment and simulation. J. Magn. Reson. 291 32–39. 10.1016/j.jmr.2018.04.003 [DOI] [PubMed] [Google Scholar]
- Stevens T., Fogh R., Boucher W., Higman V., Eisenmenger F., Bardiaux B., et al. (2011). A software framework for analysing solid-state MAS NMR data. J. Biomol. NMR 51 437–447. 10.1007/s10858-011-9569-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöppler D., Macpherson A., Smith-Penzel S., Basse N., Lecomte F., Deboves H., et al. (2018). Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR. PLoS Biol. 16:e2006192. 10.1371/journal.pbio.2006192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera D., Robertson D. L., Lovell S. C. (2011). Characterization of protein-protein interaction interfaces from a single species. PLoS One 6:e21053. 10.1371/journal.pone.0021053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas F., Niitsu A., Oregioni A., Bartlett G. J., Woolfson D. N. (2017). Conformational dynamics of asparagine at coiled-coil interfaces. Biochemistry 56 6544–6554. 10.1021/acs.biochem.7b00848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallurupalli P., Bouvignies G., Kay L. E. (2012). Studying “invisible” excited protein states in slow exchange with a major state conformation. J. Am. Chem. Soc. 134 8148–8161. 10.1021/ja3001419 [DOI] [PubMed] [Google Scholar]
- Van Melckebeke H., Wasmer C., Lange A., Ab E., Loquet A., Böckmann A., et al. (2010). Atomic-resolution three-dimensional structure of HET-s(218-289) amyloid fibrils by solid-state NMR spectroscopy. J. Am. Chem. Soc. 132 13765–13775. 10.1021/ja104213j [DOI] [PubMed] [Google Scholar]
- Vance C. K., Kang Y. M., Miller A.-F. (1997). Selective 15N labeling and direct observation by NMR of the active-site glutamine of Fe-containing superoxide dismutase. J. Biomol. NMR 9 201–206. 10.1023/A:1018662421878 [DOI] [PubMed] [Google Scholar]
- Vasa S. K., Singh H., Grohe K., Linser R. (2019). Assessment of a large enzyme-drug complex by proton-detected solid-state NMR spectroscopy without deuteration. Angew. Chem. Int. Ed. Engl. 58 5758–5762. 10.1002/anie.201811714 [DOI] [PubMed] [Google Scholar]
- Vernet T., Tessier D. C., Chatellier J., Plouffe C., Lee T. S., Thomas D. Y., et al. (1995). Structural and functional roles of asparagine 175 in the cysteine protease papain. J. Biol. Chem. 270 16645–16652. 10.1074/jbc.270.28.16645 [DOI] [PubMed] [Google Scholar]
- Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., et al. (2005). The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59 687–696. 10.1002/prot.20449 [DOI] [PubMed] [Google Scholar]
- Wagner G., Pardi A., Wuethrich K. (1983). Hydrogen bond length and proton NMR chemical shifts in proteins. J. Am. Chem. Soc. 105 5948–5949. 10.1021/ja00356a056 [DOI] [Google Scholar]
- Wälti M. A., Ravotti F., Arai H., Glabe C. G., Wall J. S., Böckmann A., et al. (2016). Atomic-resolution structure of a disease-relevant Aβ(1–42) amyloid fibril. Proc. Natl. Acad. Sci. U.S.A. 113 E4976–E4984. 10.1073/pnas.1600749113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W., Stubbs G. (2014). Fungal prion HET-s as a model for structural complexity and self-propagation in prions. Proc. Natl. Acad. Sci. U.S.A. 111 5201–5206. 10.1073/pnas.1322933111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmer C., Lange A., Van Melckebeke H., Siemer A. B., Riek R., Meier B. H. (2008). Amyloid fibrils of the HET-s(218-289) prion form a β solenoid with a triangular hydrophobic core. Science 319 1523–1526. 10.1126/science.1151839 [DOI] [PubMed] [Google Scholar]
- Weigend F., Ahlrichs R. (2005). Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: design and assessment of accuracy. Phys. Chem. Chem. Phys. 7 3297–3305. 10.1039/b508541a [DOI] [PubMed] [Google Scholar]
- Wiegand T. (2020). A solid-state NMR tool box for the investigation of ATP-fueled protein engines. Prog. Nucl. Magn. Reson. Spectrosc. 117 1–32. 10.1016/j.pnmrs.2020.02.001 [DOI] [PubMed] [Google Scholar]
- Wiegand T., Cadalbert R., Lacabanne D., Timmins J., Terradot L., Bockmann A., et al. (2019). The conformational changes coupling ATP hydrolysis and translocation in a bacterial DnaB helicase. Nat. Commun. 10:31. 10.1038/s41467-018-07968-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand T., Schledorn M., Malär A. A., Cadalbert R., Däpp A., Terradot L., et al. (2020). Nucleotide binding modes in a motor protein revealed by 31P- and 1H-detected MAS solid-state NMR. ChemBioChem 21 324–330. 10.1002/cbic.201900439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth C. L., Blundell T. L. (2010). On the evolutionary conservation of hydrogen bonds made by buried polar amino acids: the hidden joists, braces and trusses of protein architecture. BMC Evol. Biol. 10:161. 10.1186/1471-2148-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Saltzberg D. J., Kratochvil H. T., Jo H., Sali A., DeGrado W. F. (2019). Glutamine side chain 13C-18O as a nonperturbative IR probe of amyloid fibril hydration and assembly. J. Am. Chem. Soc. 141 7320–7326. 10.1021/jacs.9b00577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorin V. E., Brown S. P., Hodgkinson P. (2006). Origins of linewidth in H1 magic-angle spinning NMR. J. Chem. Phys. 125:144508 10.1063/1.2357602 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.