Abstract
Background:
To study the specific expression of circular RNA (circRNA) in breast cancer and adjacent normal tissues to identify differentially expressed circRNA in breast cancer patients. To study the correlation between circRNA and clinical data of breast cancer, and to evaluate its potential as a breast cancer biomarker.
Methods:
The differential expression of circRNAs between the breast cancer tissues and adjacent normal tissues was screened by Human CircRNA Microarray. Candidate circRNAs verified by qRT-PCR. CircRNA was analyzed by Agilent GeneSpring 13.0 software. SPSS23.0, GraphPad Prism, and Sigmaplot software were used for statistical analysis. Perform T test, one-way ANOVA, curve regression analysis and ROC curve analysis for evaluating the diagnostic value of circular RNA.
Results:
Among the 2021 differentially expressed circRNAs, 546 were up-regulated and 1475 were down-regulated in breast cancer tissues. The validation study demonstrated that six circRNAs were downregulated. Among them, hsa_circ_0104824 proved high correlation between breast cancer tissues and plasma samples in its expression and diagnostic value.
Conclusion:
Hsa_circ_0104824 may serve as a promising predictive biomarker and therapeutic target for patients with breast cancer.
Keywords: circular RNA (circRNA), breast cancer, microarray, biomarker
Introduction
Breast cancer (BC) is one of the most common malignancy among females.1 Previous research proved that age, abnormal estrogen, late childbirth, and genetic mutation are all associated with breast cancer.2,3 Despite significant advances have been achieved in diagnosis and treatment, the occurrence of metastasis still leads to poor prognosis and low survival of breast cancer.4 Although there are some biomarkers used in clinical diagnostic for BC, like the carbohydrate antigen 153 (CA153), the serum carcinoembryonic antigen (CEA), the carbohydrate antigen 125 (CA125) and the carbohydrate antigen 19-9(CA19-9),5 the sensitivity and specificity of these biomarkers is still not satisfied. Therefore, to achieve an efficient and sensitivity diagnostic marker and development of the targeted therapeutics strategies are urgently needed for early diagnosis and treatment of BC.
CircRNA, together with microRNA and LncRNA, belongs to non-coding RNA family. The circRNAs have a closed and continuous loop structure which is derived from the reverse splicing of exons, introns, or both.6 A number of researches have proved that circRNAs were involved in gene transcription, protein translation, and regulation of miRNA expression through multiple mechanisms. circRNA often performed its function by “sponging” with miRNA. One circRNA could have several miRNA binding sites and one miRNA can bind several circRNAs.7 Previous research revealed that circRNA expressed in cancer tissues and play important roles in cancer cell metastasis by directly bounding (sponge) to its target miRNA.8,9 The circRNAs were also found to be expressed in various cancer tissues and plasma samples of patients.10-12 However, the potential functions and the molecular mechanism of the majority circRNAs in cancer cells are still not clear.
In this study, we investigated the characteristic expression of circRNAs in breast cancer and adjacent normal tissues, analyzed the clinical value of candidate circRNA in breast cancer. Based on our study, we found that hsa_circ_0104824 had lower expression in breast cancer tissues and have high diagnostic value for BC. Therefore, the hsa_circ_0104824 could be a potential biomarker for BC diagnosis and target for treatment.
Materials and Methods
Samples Collection
A total of 83 breast cancer patients peripheral blood samples, 37 pairs of breast cancer tissue samples and adjacent normal tissue samples were obtained from the Department of Tumor Surgery, General Hospital of Ningxia Medical University. All samples in this study were approved by the Ethical Committees of the General Hospital of Ningxia Medical University (Ethics No. 2019—175), and we have obtained written informed consent from all participants involved. Forty nine healthy people (as control) were recruited from Physical Examination Department in the hospital. Detailed clinical, pathological and molecular characterization of these patients were collected accordingly.
Total RNA Isolation
Total RNA was taken from whole blood and tissue samples using an RNA extraction kit (Bioteke, Inc, China) and extracted in strict accordance with the procedure provided with the kit. The purity and concentration of the total RNA were measured by a spectrophotometer Nanodrop 2000 (Thermo Scientific, USA), and the ratio of the OD value of 260/280 was 1.8 to 2.0. 1.3% agarose gel electrophoresis were used for verifying the integrity of total RNA.
Microarray Analysis
Human circRNA Array v2 microarray (Beijing Capital Bio Biotechnology Corporation, China) and GeneSpring 13.0 (Agilent) software were used for circRNA microarray and data analysis. 3 paired breast cancer and adjacent normal tissues were used for microarray analysis. The labeled RNA was hybridized on a microarray containing 162351 human circRNA probes. The original screening criteria for the circRNA microarray results was fold change > 2, P-value <0.05. Based on the criteria, the different expressions of circRNA were separated. Then we raised the screening criteria to fold-change≥4, P-value <0.05 for target circRNA selection and verification.
Bioinformatics and Data Analysis
The obtained circRNA microarray data were extracted by Feature Extraction software (CapitalBio). Data standardization, fold change and P value were analyzed by GeneSpring V13.0 (Agilent) software. The heat map was analyzed using the Cluster 3.0 software. The different expression of circRNA and target circRNA being selected based on the GeneSpring V13.0 software. The data was Log2 transformed and median centered by genes using the Adjust Data function of CLUSTER 3.0 software. The circRNA structure was analyzed by circPrimer 1.2 software. The circRNA-miRNA gene network was analyzed by Cytoscape software. The correlation between circRNAs and miRNAs was predicted by circMIR software (miRanda and RNAhybrid database). Targetscan software (http://www.targetscan.org) was used for identifying the binding sites between miRNA and circRNA. KEGG database analysis was used to determine the involvement in circRNA target genes in different biological pathways. The miRNA gene ontology (GO) terms “biological process” analysis was constructed by Cytoscape plug-in ClueGo23.
qRT-PCR
To validate the selected circRNA, 37 pairs of breast cancer tissue and adjacent normal tissue samples, 132 (83 breast cancer patients and 49 healthy women as control) peripheral blood samples were used. Total RNA (500 ng) was reverse transcribed into cDNA using the RevertAid First strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc., USA). The qRT-PCR reaction in a total volume of 20 µL systems, including 0.8 μL/10 μM forward/reverse primers, 10µL TB Green qPCR Mastermix, 2 µL cDNA, and 6.4 µL double-distilled water. The cycle program was 5 mins at 25° C, 60 mins at 42° C, and 5 mins at 70° C. The relative expression of the genes were calculated by the ΔΔCT method.
Statistical Analysis
The data obtained were all statistically analyzed by SPSS 22.0 software (IBM, USA), GraphPad Prism version 6.0 (GraphPad Software, USA) and Sigmaplot version 14.0. Data were presented as mean ± standard deviation (SD). Significance was analyzed by an independent sample t-test or one-way ANOVA. Sensitivity and specificity were determined by ROC curve analysis. The diagnostic value of hsa_circ_0104824 was evaluated using ROC curve and the area under the curve (AUC). If the 2-tailed p <0.05, the difference was considered statistically significant.
Results
Profiling the Expression of circRNAs in BC Patients
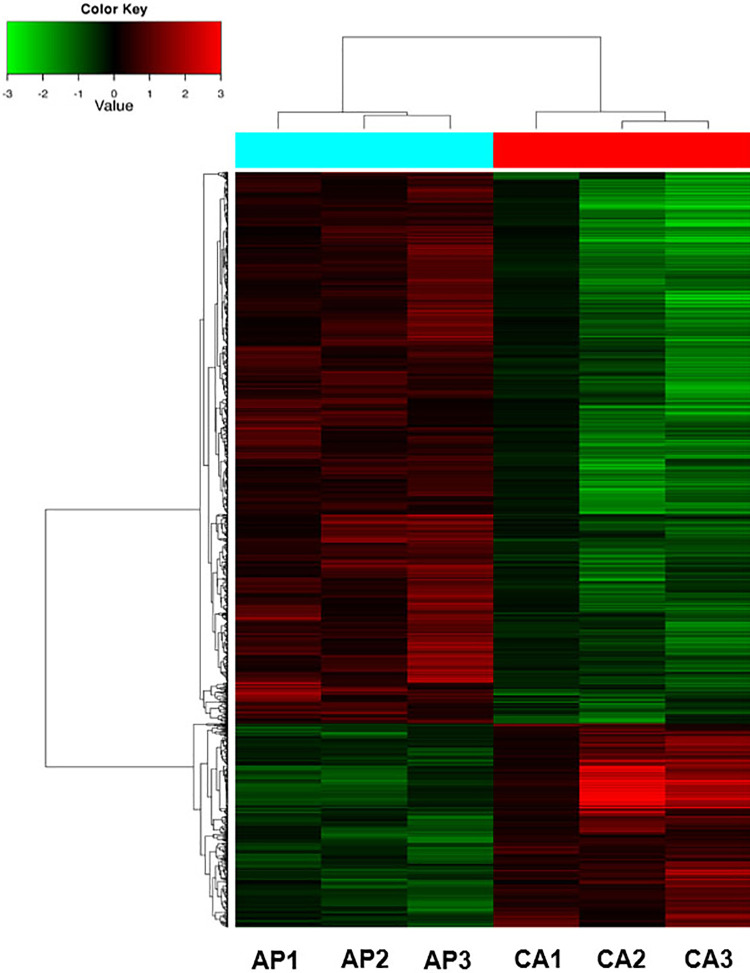
To identify specific circRNAs that are differentially expressed in breast cancer patients, 3 paired breast cancer tissues (CA1-CA3) and adjacent normal tissue (AP1-AP3) samples were subjected for circRNA microarray assay. Differentially expressed circRNAs were detected in the three matched tissue samples. A total of 2021 circRNAs has been identified based on the circRNA microarray. With the filter criteria as fold-change ≥ 2 and P-value <0.05, 546 circRNAs was up-regulated and 1475 circRNAs were down-regulated (Figure 1). The false discovery rate (FDR) method was used to adjust the P values to avert the false positives. After FDR correction and set the filter criteria as fold-change ≥4, P-value <0.05, Fluorescence value ≥100. 6 circRNAs meet these standards, including hsa_circ_0073004, hsa_circ_0054020, hsa_circ_0102618, hsa_circ_0002938, hsa_circ_0073006, and hsa_circ_0104824.
Figure 1.
Circular RNAs microarray profiles between breast cancer tissues and paired adjacent non-cancerous tissues. Heat map of the circRNA microarray profiles in Group CA VS Group AP, N = 3, CA for breast cancer tissues and AP for adjacent non-cancerous tissues. The expression of circRNAs is hierarchically clustered on the y-axis, tissue samples are hierarchically clustered on the x-axis. The expression levels are presented in red and green, which indicate upregulated and downregulated circRNAs.
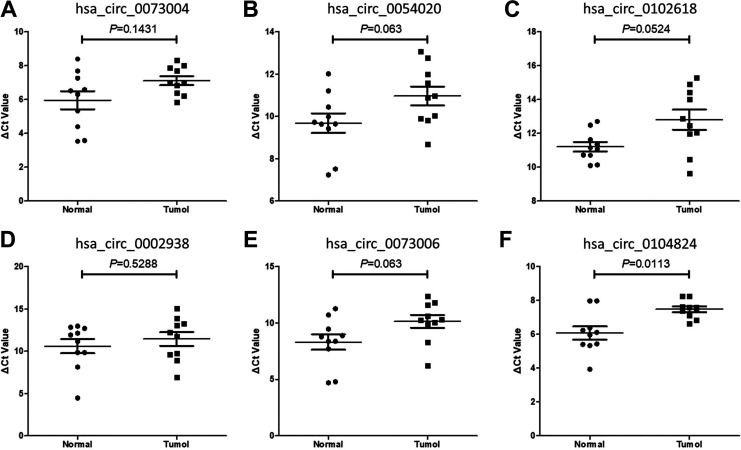
Verify the Candidate circRNAs by qPCR
To verify the six selected candidate circRNAs, qPCR was conducted in another independent cohort consisting of 10 pairs of breast cancer and adjacent normal tissues. The results supported the results from the microarray analysis (Figure 2). Particularly, hsa_circ_0104824 was significantly down-regulated in the tissues (P = 0.0113). Therefore, we then decided to further study the expression of hsa_circ_0104824 as a diagnostic marker for breast cancer.
Figure 2.
The diagnostic capability of 6 candidate circRNAs. (A-F) 6 candidate circRNAs expression between breast carcinoma tissue and para-carcinoma tissue. The ROC analysis results of 6 candidate circRNAs; (A-E) hsa_circ_0073004, hsa_circ_0054020, hsa_circ_0102618, hsa_circ_0002938, hsa_circ_0073006 (P > 0.05), and hsa_circ_0104824 (F) (P < 0.05).
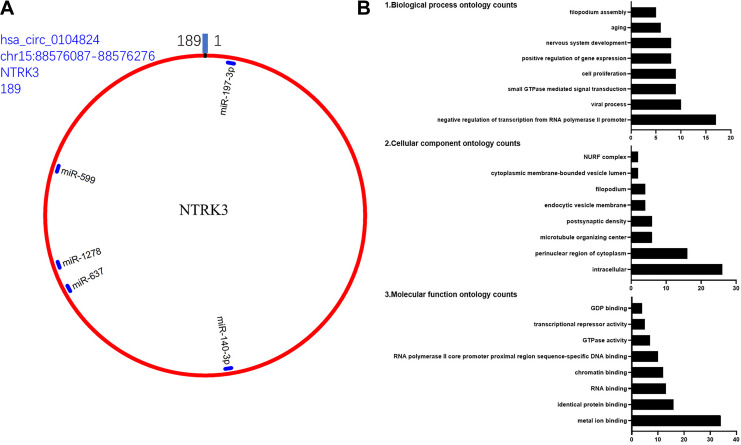
Bioinformatics Analysis of Hsa_circ_0104824
CircRNA can act as a “sponge” for miRNAs through their binding sites and modulate the activity of miRNA. In this study, we predicted the hsa_circ_0104824 binding miRNA by the circMIR software and Targetscan software. As predicted, hsa_circ_0104824 binds with many miRNAs, some of the miRNA have multiple binding sites with hsa_circ_0104824, and some of them only have one binding site. According to the number of binding sites, the top 5 miRNA was selected, listed and shown in Table 1. The specific binding site location and information about the top 5 miRNAs with hsa_circ_0104824 were shown in Figure 3A. The ClueGo analysis of hsa_circ_0104824 target miRNAs demonstrated that hsa_circ_0104824 is involved in enriched GO biological processes such as cancer metastasis (e.g. cell migration and cell–cell adhesion) and proliferation (e.g. regulation of cell cycle and cell differentiation) (Figure 3B).
Table 1.
The Binding Sites Between miRNA and Hsa_circ_0104824 by Targetscan.
| CircRNA | CircRNA (Top)—miRNA (Bottom) pairing | Site Type | CircRNA Start | CircRNA End |
|---|---|---|---|---|
| Mirbase ID | ||||
| hsa_circ_0104824 (5′…3′) | GUCAUUGAGAACCCCCAGUACUU | 7mer-m8 | 147 | 153 |
| ||||||| | ||||
| hsa-miR-1278 (3′…5′) | UAUCUACUAUACGUGUCAUGAU | |||
| hsa_circ_0104824 (5′…3′) | AUGCCGGGCCCGACACUGUGGUC | 7mer-m8 | 103 | 109 |
| ||||||| | ||||
| hsa-miR-140-3p (3′…5′) | GGCACCAAGAUGGGACACCAU | |||
| hsa_circ_0104824 (5′…3′) | CCGUGGCUGUCAUCAGUGGUGAG | 7mer-m8 | 19 | 25 |
| ||||||| | ||||
| hsa-miR-197 (3′…5′) | CGACCCACCUCUUCCACCACUU | |||
| hsa_circ_0104824 (5′…3′) | AGUACUUCCGUCAGGGACACAAC | 7mer-m8 | 163 | 169 |
| ||||||| | ||||
| hsa-miR-599 (3′…5′) | CAAACUAUUUGACUGUGUUG | |||
| hsa_circ_0104824 (5′…3′) | CCCUGUCAUUGAGAACCCCCAGU | 7mer-m8 | 143 | 149 |
| ||||||| | ||||
| hsa-miR-637 (3′…5′) | UGCGUCUCGGGCUUUCGGGGGUCA |
Figure 3.
A, The hsa_circ_0104824 structure and miRNA binding sites, including origin gene NTRK3, this gene from Chromosome 15, location between 88576087 and 88576276, the molecule is 189 bp and binding miRNA top 5 (Table 1). B, The ClueGo analysis of its targeted miRNAs revealed hsa_circ_0104824 was involved in enriched GO biological processes such as cancer metastasis (e.g. cell migration and cell–cell adhesion) and proliferation (e.g. regulation of cell cycle and cell differentiation).
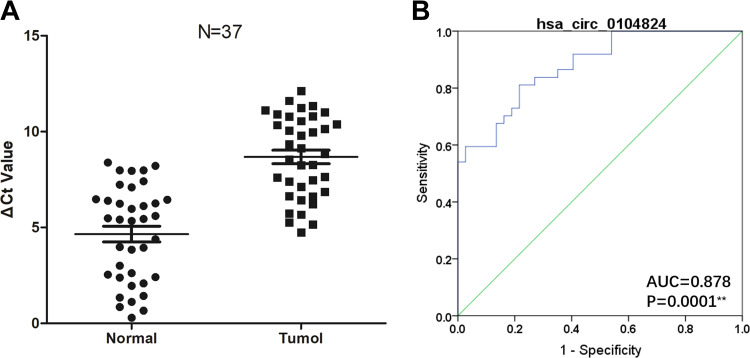
Expression of Hsa_Circ_0104824 in Breast Cancer Tissues
Given the enormous diagnostic and therapeutic role of circRNA in breast cancer, we explored the clinical value of hsa_circ_0104824 by detecting its expression in 37 pairs breast cancer samples. The results demonstrated that hsa_circ_0104824 has significantly lower expression in breast cancer tissues compared to normal controls (Figure 4A). Based on ROC curve analysis, hsa_circ_0104824 has an AUC of 0.823, the sensitivity was 81.1% and specificity was 78.4% (Figure 4B), indicating that hsa_circ_0104824 may be a tissue-specific circRNA in breast cancer.
Figure 4.
Hsa_circ_0104824 is a diagnostic ability in breast cancer. Breast cancer tissue and paracancerous tissues, n = 37; (A) expression level of hsa_circ_0104824 quantified by qPCR; (B) ROC analysis of hsa_circ_0104824, AUC values are given on the graph; * P <0.05, ** P <0.01. CT indicates cycle threshold. Scatter plot drawing with △CT value between CRC and adjacent tissues in Graphpad Software.
Expression of Hsa_Circ_0104824 in Peripheral Blood
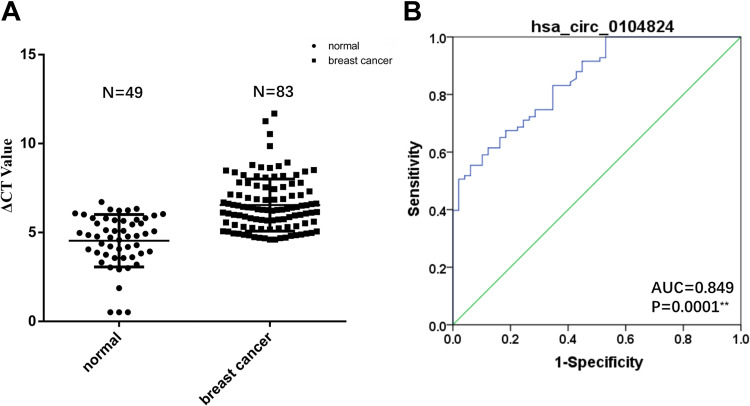
The results of the expressions of hsa_circ_0104824 in the peripheral blood of 83 breast cancer patients and 49 healthy persons showed that hsa_circ_0104824 was significantly decreased in peripheral blood of breast cancer patients (Figure 5A). Based on ROC curve analysis, hsa_circ_0104824 has an AUC of 0.849, a sensitivity of 71.1%, and a specificity of 75.5% (Figure 5B). The results supported that hsa_circ_0104824 could be a blood-specific circRNA in breast cancer.
Figure 5.
The diagnostic capability of hsa_circ_0104824 in peripheral blood. (Breast cancer peripheral blood, case group, n = 83; healthy person peripheral blood, control group, n = 49; (A) Expression levels of hsa_circ_0104824 quantified by qPCR; (B) the ROC analyses of hsa_circ_0104824. The AUC values are given on the graphs; *P <0.05, **P <0.01. CT indicates cycle threshold.
The Correlation of Hsa_Circ_0104824 With Clinical Characteristics of Breast Cancer
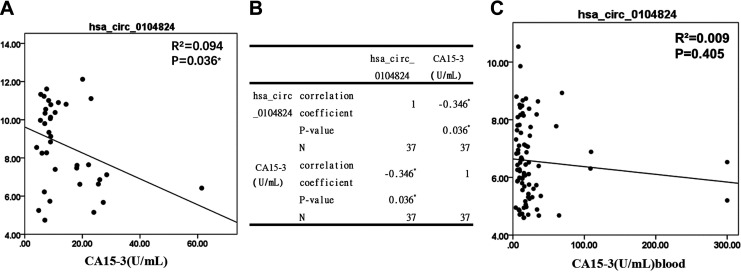
The clinical characteristics of breast cancer patients were analyzed to verify the correlation between hsa_circ_0104824 expression and classification and prognosis of breast cancer. The data demonstrated that hsa_circ_0104824 expression in breast cancer tissues was significantly correlated with CA15-3 (P < 0.05), ER positive (P < 0.05), AR positive (P < 0.05) (Table 2). In peripheral blood of breast cancer, hsa_circ_0104824 expression was significantly correlated with tumor sizes (P < 0.05), TNBC (P < 0.05), ER positive (P < 0.05) and PR positive (P < 0.05) (Table 3, Figure 6). The results suggested that hsa_circ_0104824 expression is specifically and closely related to tumor stage, grading, ER positive and metastasis.
Table 2.
Relationship Between Expression Level of Hsa_Circ_0104824 in Breast Cancer Tissues and Clinicopathological Features of Breast Cancer Patients.
| Parameter | N | Hsa_circ_0104824 level (Mean ± SD) | T-test or One way ANOVA(F) | P |
|---|---|---|---|---|
| Age status | −0.078 | 0.939 | ||
| ≥50 | 21 | 8.65 ± 2.27 | ||
| <50 | 16 | 8.71 ± 2.03 | ||
| CA15-3 status | −5.541 | <0.001** | ||
| CA15-3 positive | 5 | 6.54 ± 0.55 | ||
| CA15-3 negative | 32 | 9.01 ± 2.10 | ||
| CEA status | −0.097 | 0.938 | ||
| CEA positive | 2 | 8.38 ± 4.57 | ||
| CEA negative | 35 | 8.69 ± 2.05 | ||
| Tumor size status (cm) | 0.163 | |||
| ≥2 | 25 | 9.02 ± 2.08 | 1.424 | |
| < 2 | 12 | 7.97 ± 2.17 | ||
| Triple Negative Breast Cancer (TNBC, n) | 0.206 | |||
| Yes | 7 | 9.61 ± 1.83 | 1.288 | |
| No | 30 | 8.46 ± 2.17 | ||
| Her-2 Positive | 0.857 | |||
| Yes | 11 | 8.78 ± 2.55 | 0.181 | |
| No | 26 | 8.64 ± 1.99 | ||
| estrogen receptor (ER, n) | 0.026* | |||
| Positive | 27 | 8.21 ± 2.14 | −2.318 | |
| Negative | 10 | 9.94 ± 1.61 | ||
| progesterone receptor (PR, n) | 0.311 | |||
| Positive | 23 | 8.40 ± 2.12 | −1.027 | |
| Negative | 14 | 9.14 ± 2.16 | ||
| Androgen Receptor (AR, n) | 0.018* | |||
| Positive | 30 | 8.34 ± 2.15 | −2.687 | |
| Negative | 7 | 10.13 ± 1.43 | ||
| Tumor metastasis status | 0.922 | 0.363 | ||
| Yes | 8 | 9.30 ± 1.91 | ||
| No | 29 | 8.51 ± 2.20 | ||
| TNM stage | 0.639 | 0.595 | ||
| I | 11 | 9.11 ± 1.93 | ||
| II | 24 | 8.37 ± 2.26 | ||
| III | 2 | 9.93 ± 1.53 | ||
| IV | 0 | NA | ||
| Edmondson grading | −0.846 | 0.404 | ||
| early stages (I–II) | 35 | 8.61 ± 2.16 | ||
| advanced stages (III–IV) | 2 | 9.93 ± 1.53 |
Abbreviations: TNM, Tumor Node Metastasis; HER-2, human epidermal growth factor receptor-2; CEA, carcino-embryonic antigen; CA15-3, carbohydrate antigen 15-3; NA, NOT applicable.
*P < 0.05.
*P < 0.01.
Table 3.
Association Between Hsa_circ_0104824 Expression Level and Clinicopathological Features of Breast Cancer Patients in Breast Cancer Plasma.
| Parameter | N | Hsa_circ_0104824 level (Mean ± SD) | T-test or One way ANOVA(F) | P |
|---|---|---|---|---|
| Age status | 0.522 | 0.603 | ||
| ≥50 | 43 | 6.64 ± 1.36 | ||
| <50 | 40 | 6.49 ± 1.37 | ||
| CA15-3 status | −0.82 | 0.415 | ||
| CA15-3 positive | 16 | 6.32 ± 1.41 | ||
| CA15-3 negative | 67 | 6.63 ± 1.35 | ||
| CEA status | 1.101 | 0.296 | ||
| CEA positive | 10 | 7.12 ± 1.73 | ||
| CEA negative | 73 | 6.49 ± 1.3 | ||
| Tumor size status(cm) | 0.043* | |||
| ≥2 | 56 | 6.35 ± 1.3 | −2.074 | |
| <2 | 27 | 7.02 ± 1.39 | ||
| Tumor grading (G, n) | ||||
| G1 | 8 | 7.30 ± 1.36 | 0.948 | 0.422 |
| G2 | 55 | 6.51 ± 1.15 | ||
| G3 | 18 | 6.39 ± 1.83 | ||
| G4 | 2 | 6.87 ± 2.28 | ||
| Triple Negative Breast Cancer (TNBC, n) | 0.032* | |||
| Yes | 7 | 5.66 ± 0.94 | 0.561 | |
| No | 76 | 6.65 ± 1.37 | ||
| Her-2 Positive | 0.418 | |||
| Yes | 44 | 6.68 ± 1.40 | −0.813 | |
| No | 39 | 6.44 ± 1.32 | ||
| estrogen receptor (ER, n) | 0.028* | |||
| Positive | 62 | 6.75 ± 1.38 | −2.281 | |
| Negative | 21 | 6.04 ± 1.18 | ||
| progesterone receptor(PR, n) | 0.019* | |||
| Positive | 50 | 6.84 ± 1.40 | −2.392 | |
| Negative | 33 | 6.15 ± 1.20 | ||
| Tumor metastasis status | 0.827 | 0.411 | ||
| Yes | 59 | 6.49 ± 1.42 | ||
| No | 24 | 6.76 ± 1.22 | ||
| TNM stage | 0.755 | 0.523 | ||
| I | 12 | 7.08 ± 1.16 | ||
| II | 35 | 6.39 ± 1.17 | ||
| III | 26 | 6.58 ± 1.66 | ||
| IV | 10 | 6.55 ± 1.40 | ||
| Edmondson grading | 0.000 | 1.000 | ||
| early stages (I–II) | 47 | 6.57 ± 1.19 | ||
| advanced stages (III–IV) | 36 | 6.57 ± 1.57 |
Abbreviations: TNM, Tumor Node Metastasis; HER-2, human epidermal growth factor receptor-2; CEA, carcino-embryonic antigen.
* P < 0.05.
** P < 0.01.
Figure 6.
The correlation of hsa_circ_0104824 with breast cancer clinical characteristics. A, Curve regression analysis of CA15-3 and hsa_circ_0104824 in breast cancer tissues. B, Spearman correlation analysis of CA15-3 and hsa_circ_0104824 in breast cancer tissues. C, Curve regression analysis of CA15-3 and hsa_circ_0104824 in breast cancer tissues of breast cancer.
The Diagnostic Value of Hsa_Circ_0104824 for Breast Cancer
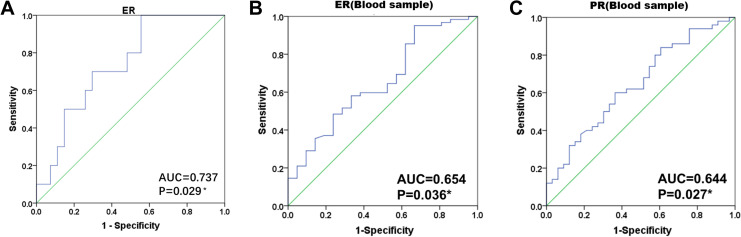
We then analyzed the diagnostic values of hsa_circ_0104824 for breast cancer. In breast cancer tissues, the AUC for hsa_circ_0104824 expression in ER-positive group was 0.737, the cutoff value was 9.86 and the sensitivity and specificity was 70.0% (Figure 7A). In breast cancer blood, the AUC for hsa_circ_0104824 expression in ER-positive group was 0.654, the cutoff value was 6.29, the sensitivity and specificity were 58.1% and 66.7% (Figure 7B). The AUC for hsa_circ_0104824 expression in PR-positive group was 0.644, the cutoff value was 6.35, the sensitivity was 60.0%, and the specificity was 63.6% (Figure 7C). These results indicated that hsa_circ_0104824 has high diagnostic value in consistent with ER, PR positive breast cancer patients.
Figure 7.
ROC curve analysis of hsa_circ_0104824 in breast cancer tissues and peripheral blood. A, ROC analysis of hsa_circ_0004585 and ER in 37 pairs of tissues of breast cancer patients. B, C: ROC analysis of ER, PR and hsa_circ_0104824 in 83 blood samples from breast cancer patients. AUC indicates area under curve; ROC, receiver-operating characteristic.
Discussion
As a type of non-coding RNA, circRNA has a closed continuous loop and lack of 3′ and 5′ ends. circRNA is involved in many biological processes by act as miRNA “sponging” to regulate the function of mRNA expression.13 Previous studies indicated that circRNA plays an important role in oncogenesis and influence the cancer cell proliferation, migration and apoptosis.14,15 For example, Chen Yang et al. found that circARHGAP10 promotes GLUT1 expression through sponge with miR-150-5p and inhibits NSCLC progression.16 Liang WC et al. reported that circRNA can regulate the non-canonical function of liver cancer cell growth through Wnt/β-catenin pathway.17 Xue D et al. found that Circ-AKT3 inhibits clear cell renal cell carcinoma (ccRCC) metastasis by competitively binding to miR-296-3p and to promote E-cadherin expression, Circ-AKT3 can serve as a novel therapeutic target for better inhibition of ccRCC metastasis.18 Zhu K et al. showed that circDNMT3B regulating miR-20b-5p and BAMBI to promote vascular dysfunction in diabetic retina.19 And Chen Q et al. reported that circRAPGEF5 plays an important role in inhibiting RCC through the miR-27a-3p/TXNIP pathway and may be a promising prognostic biomarker for RCC patients.20 These evidences supported circRNA with the great potential to be an ideal biomarkers for diagnosis and prognosis of breast cancer.
Breast cancer was one of high morbidity disease in women. Early diagnosis and type differentiation is helpful for the treatment of breast cancer patients. circRNAs had been reported involving breast cancer cells growth and metastasis. For example, Ye F et al. reported that circFBXW7 acts as a sponge to adsorb miR-197-3p and encodes FBXW7-185aa protein, thereby up-regulating the expression of FBXW7, and finally inhibiting the growth and migration of TNBC.21 Yang CY et al. found that circRNA_100876 can inhibit the expression of miRNA-361-3p, thereby promoting the transfer and proliferation of BC cells.22 Liu Y et al reported that Circular RNA hsa_circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axis.23 Those studies have provided new insights into the pathogenicity of breast cancer and supported circRNA as a novel therapeutic target for the treatment of breast cancer.
In this study, we first screened the expression profiling of circRNA in three paired BC and normal adjacent tissues following a microarray screening. With the cluster analysis and GO analysis, a total of 2021 circRNAs being identified. Based on the filter criteria as fold-change ≥2 and P-value <0.05, 546 circRNAs were up-regulated and 1475 circRNAs were down-regulated. We then raised the screening criteria to fold-change ≥4, and selelcted 6 candidate circRNAs. After verifying by RT-qPCR in an independent cohort in BC tissue and patients’ peripheral samples, hsa_circ_0104824 being identified as a candidate target gene for BC.
CircRNA performing its function by acting as a “sponging” to miRNAs. The binding of circRNA with miRNA is quite different. In this study, we screened 5 possible hsa_circ_0104824 targets miRNAs by the circMIR software and Targetscan software. Functional analysis revealed that the biological processes of these 5 miRNAs were all related to cell cycle and cell proliferation, proved that hsa_circ_0104824 related to cell proliferation and differentiation, and may intervene in tumor occurrence and development through binding these miRNAs as a “sponge”.
To further identify the correlation between hsa_circ_0104824 and BC, we analyzed the expression of hsa_circ_0104824 in BC tissues and peripheral blood. ROC curve analysis revealed that hsa_circ_0104824 has the highest diagnostic value for breast cancer tissues (AUC = 0.823). The sensitivity of it was 81.1% and specificity was 78.4%. In the peripheral blood group, the AUC of hsa_circ_0104824 for BC was 0.849. The sensitivity and specificity were 71.1% and 75.5%, indicating that hsa_circ_0104824 is a blood-specific circRNA in breast cancer. Correlation analysis of hsa_circ_0104824 with breast cancer clinical characteristics showed that hsa_circ_0104824 expression was significantly correlated with CA15-3, ER positive, AR positive, tumor size, and TNBC, suggesting that hsa_circ_0104824 expression was specific and closely related to breast cancer tumor stage, grading and metastasis.
In summary, our study indicated that circRNA hsa_circ_0104824 was downregulated both in the BC tissues and patients’ peripheral blood. ROC curve analysis demonstrated that hsa_circ_0104824 had high diagnostic value in BC. Therefore, we hypothesized that hsa_circ_0104824 could be applied as a potential biomarker for BC diagnosis and treatment. However, further research is needed to investigate the mechanism of hsa_circ_0104824 in BC tumorigenesis and metastasis.
Supplemental Material
Supplemental Material, Figue_S1_Flow_Chart_ for Identification of Hsa_circ_0104824 as a Potential Biomarkers for Breast Cancer by Xiaohan Li, Fang Ma, Ligang Wu, Xu Zhang, Jinhai Tian, Jinping Li, Jia Cao, Yunfei Ma, Liang Zhang and Libin Wang in Technology in Cancer Research & Treatment
Acknowledgments
We thank for the efforts of all the authors.
Authors’ Note: Xiaohan Li and Fang Ma contributed equally. This work has been approved by the ethical committees of the General Hospital of Ningxia Medical University (Ethics Number 2019—175), and we have obtained written informed consent from all participants involved in the study.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article. This study was funded by the Science Research Project of Ningxia Higher Education (Number NGY2018-91). The National Natural Science Foundation of China (Number 81860470). The Ningxia Autonomous Region Key R&D Programs (Number 2019BFH02012). The Natural Science Foundation of Ningxia (Number 2019AAC03209). The NingXia Natural Science Foundation (Number 2018AAC03156).
ORCID iDs: Xiaohan Li  https://orcid.org/0000-0001-6370-4652
https://orcid.org/0000-0001-6370-4652
Libin Wang  https://orcid.org/0000-0002-7861-1560
https://orcid.org/0000-0002-7861-1560
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Spronk I, Schellevis FG, Burgers JS, de Bock GH, Korevaar JC. Incidence of isolated local breast cancer recurrence and contralateral breast cancer: a systematic review. Breast. 2018;39:70–79. [DOI] [PubMed] [Google Scholar]
- 2. Wang L, Shi JF, Zhu J, et al. Health-related quality of life and utility scores of patients with breast neoplasms in China: a multicenter cross-sectional survey. Breast. 2018;39:53–62. [DOI] [PubMed] [Google Scholar]
- 3. Donovan MG, Wren SN, Cenker M, Selmin OI, Romagnolo DF. Dietary fat and obesity as modulators of BC risk: focus on DNA methylation. Br J Pharmacol. 2020;177(6):1331–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sridharan S, Howard CM, Tilley AM, et al. Novel and alternative targets against breast cancer stemness to combat chemoresistance. Front Oncol. 2019;9:1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang W, Xu X, Tian B, et al. The diagnostic value of serum tumor markers CEA, CA19-9, CA125, CA15-3, and TPS in metastatic breast cancer. Clin Chim Acta. 2017;470:51–55. [DOI] [PubMed] [Google Scholar]
- 6. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guan YJ, Ma JY, Song W. Identification of circRNA-miRNA-mRNA regulatory network in gastric cancer by analysis of microarray data. Cancer Cell Int. 2019;19(1):183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. [DOI] [PubMed] [Google Scholar]
- 9. Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. 2017;10:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dou Y, Cha DJ, Franklin JL, et al. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang S, Zeng X, Ding T, et al. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8(1):2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng J, Zhuo H, Xu M, et al. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J Transl Med. 2018;16(1):216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu Q, Lu G, Luo Z, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/β-catenin axis. Biochem Biophys Res Commun. 2018;497(2):626–632. [DOI] [PubMed] [Google Scholar]
- 15. Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9(2):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jin M, Shi C, Yang C, Liu J, Huang G. Upregulated circRNA ARHGAP10 Predicts an unfavorable prognosis in NSCLC through regulation of the miR-150-5p/GLUT-1 Axis. Mol Ther Nucleic Acids. 2019;18:219–231. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Liang WC, Wong CW, Liang PP, et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xue D, Wang H, Chen Y, et al. Circ-AKT3 inhibits clear cell renal cell carcinoma metastasis via altering miR-296-3p/E-cadherin signals. Mol Cancer. 2019;18(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhu K, Hu X, Chen H, et al. Downregulation of circRNA DMNT3B contributes to diabetic retinal vascular dysfunction through targeting miR-20b-5p and BAMBI. EBioMedicine. 2019;49:341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Q, Liu T, Bao Y, et al. CircRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR-27a-3p/TXNIP pathway. Cancer Lett. 2020;469:68–77. [DOI] [PubMed] [Google Scholar]
- 21. Ye F, Gao G, Zou Y, et al. circFBXW7 inhibits malignant progression by sponging miR-197-3p and encoding a 185-aa protein in triple-negative breast cancer. Mol Ther Nucleic Acids. 2019;18:88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang CY, Zhang FX, He JN, Wang SQ. CircRNA_100876 promote proliferation and metastasis of breast cancer cells through adsorbing microRNA-361-3p in a sponge form. Eur Rev Med Pharmacol Sci. 2019;23(16):6962–6970. [DOI] [PubMed] [Google Scholar]
- 23. Liu Y, Lu C, Zhou Y, Zhang Z, Sun L. Circular RNA hsa_circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axis. Biochem Biophys Res Commun. 2018;502(3):358–363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Figue_S1_Flow_Chart_ for Identification of Hsa_circ_0104824 as a Potential Biomarkers for Breast Cancer by Xiaohan Li, Fang Ma, Ligang Wu, Xu Zhang, Jinhai Tian, Jinping Li, Jia Cao, Yunfei Ma, Liang Zhang and Libin Wang in Technology in Cancer Research & Treatment