Abstract
The emergence of a rapidly spreading and highly infectious coronavirus disease 2019 (COVID-19) outbreak by a novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has caused a global pandemic with unprecedented social and economic dimensions. Therefore, the development of effective strategies is urgent to control the COVID-19 outbreak. According to recent investigations, cell entry of coronaviruses relies on binding of the viral spike glycoprotein to the host cellular receptors. Therefore, the present study aimed to predict immunogenic epitopes in silico by analysing the spike protein. In parallel, by screening the immunogenic SARS-CoV-2 spike-derived epitopes provided in the literature, we chose a set of epitopes that we believed would induce immunogenic response. Next, provided with the epitopes selected by using both approaches, we performed immunoinformatic analysis that mapped identically to the antigen regions and antigenic properties. Finally, after selecting a screened set of epitopes, we designed a novel virus-like particle vaccine optimized to be produced in plants by using molecular farming biotechnology techniques. Our assay may be used as a starting point for guiding experimental efforts towards the development of a vaccine against SARS-CoV-2.
Keywords: COVID-19, epitope prediction, immunoinformatics, severe acute respiratory syndrome coronavirus 2, vaccine, virus-like particles
Introduction
Coronaviruses are positive-sense single-stranded RNA viruses that belong to the family Coronaviridae [1]. They mainly infect animals, including birds and mammals. In humans, they generally cause mild respiratory infections [1]. However, some recent human coronavirus infections have resulted in lethal endemics, which include the severe acute respiratory syndrome (SARS) coronavirus (CoV), Middle East respiratory syndrome (MERS) CoV and the newly emerged severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [2]. On the basis of phylogenetic analysis of the full-length genome, research groups are suggesting that SARS-CoV-2 is quite similar to SARS-CoV [3]. Moreover, they putatively have similar cell entry mechanisms and human cell receptors usage [4]. SARS-CoV-2 has a positive-sense single-stranded RNA ∼30 kb in size, which, like other coronaviruses, encodes for multiple structural and nonstructural proteins [5]. The structural proteins include the spike (S) protein, the envelope (E) protein, the membrane (M) protein and the nucleocapsid (N) protein [6].
We will find therapeutic targets by gaining insight into cell entry mechanisms and viral transmission of SARS-CoV. The spike protein of SARS-CoV and MERS-CoV plays a vital role in receptor binding and membrane fusion [7]. Considering the high genetic similarity of the S protein between SARS-CoV and SARS-CoV-2, we concluded that the S protein would play a vital role in coronavirus disease 2019 (COVID-19) infection. Recent reports have suggested that cell entry of SARS-CoV-2 depends on the binding of the viral S proteins to cellular receptors and on S protein priming by host cell proteases [8]. SARS-CoV-2 binds with a high affinity to human angiotensin-converting enzyme 2 (ACE2) and uses it as an entry receptor to invade target cells [1]. Therefore, the spike protein of SARS-CoV is anticipated to be an important component of candidate vaccines [9] and is anticipated to inhibit spike-mediated entry by SARS-CoV-2. It is vital to consider the spike protein of SARS-CoV-2 as one of the most important targets to develop a vaccine or treatment to cure COVID-19 infection. Recent studies have shown that using the interleukin-6 receptor antagonists and targeting Toll-like receptor 5 may be ways to control COVID-19 [[10], [11], [12]].
Currently, only limited immunologic information, such as probable immunogenic epitopes eliciting antibodies or T-cell responses, are available for SARS-CoV-2. Adaptive immunity mediated by T and B cells is capable of developing a pathogen-specific memory that confers immunologic protection [13]. In particular, B and T cells recognize portions known as epitopes within their cognate antigens. The identification of epitopes in antigens is of great interest for a better understanding of disease aetiology and monitoring the immune system during the infection, which can result in designing diagnostic assays and possibly epitope-based vaccines specific to the occurring infection. Epitope identification is costly and time-consuming, as it requires experimental screening of large arrays of potential epitope candidates. Fortunately, researchers have developed in silico prediction methods that have significantly increased the simplicity of epitope mapping by shortening the list of potential epitope candidates for experimental testing.
The main thing that would help us choose desirable epitopes is to figure out which parts of the SARS-CoV-2 sequence are recognized by human immune responses. Such data and information are unfortunately currently very limited because SARS-CoV-2 is so new. Gaining this information would facilitate evaluating the immunogenicity of vaccine candidates as well as monitoring for the potential consequences of mutational events and epitope escape as the virus is transmitted through human populations [14].
Another approach to map and suggest potential epitopes for designing a vaccine against SARS-CoV-2 is to select them from epitopes provided in previous research. In one recent study, researchers determined the crystal structure of the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein in complex with CR3022, a neutralizing antibody previously isolated from a convalescent SARS patient [15]. A highly conserved epitope is the target of CR3022, which enables cross-reactive binding between SARS-CoV-2 and SARS-CoV. Therefore, among the previously reported anti–SARS-CoV antibodies, we considered CR3022 to be one of the most promising ones. The epitope of CR3022 does not overlap with the ACE2 binding site within SARS-CoV-2 RBD [15].
Virus-like particles (VLPs) have gradually emerged as vaccine delivery agents that are spontaneously assembled from viral structural proteins [16]. These are multimeric structures that can directly stimulate immune cells by mimicking the three-dimensional conformation of native viruses. Moreover, VLPs are devoid of infectious genetic material, which makes them inherently safer than attenuated or inactivated virus preparations [17]. They contain functional viral proteins responsible for cell penetration by the virus, and therefore efficient cell entry (and thus tissue-specific targeting determined by the origin of the virus) is gained, making them a good choice when creating a vaccine. VLP vaccines provide delivery systems that combine good safety profiles with strong immunogenicity and thus are safe alternatives to inactivated infectious viruses. VLPs have excellent adjuvant properties and are capable of inducing innate and adaptive immune responses. They present both high-density B-cell epitopes for antibody production and intracellular T-cell epitopes, thus respectively inducing potent humoral and cellular immune responses [18]. Experience with previously introduced VLP-based vaccines demonstrated that the uptake of VLPs by antigen-presenting cells can trigger efficient immune responses, which result in the control of infection [19]. These features make VLPs a top platform for making a safe and effective vaccine [20]. VLPs are produced using different expression systems, such as cells from bacteria, yeast, mammals, plants or insects. For the present vaccine designed in silico by our research team, we decided to use plants as the expression system. VLPs derived from hepatitis B virus (HBV) are the most studied chimeric VLPs for epitope display [17], and HBV is used to produce different vaccines [21].
By screening for the determined SARS-CoV-2–derived epitopes in the immunogenic structural proteins of SARS-CoV-2 and by using bioinformatic tools, we generated a list of potential epitopes for vaccine design. Our findings will potentially help narrow the quest for powerful targets for an effective peptide-derived SARS-CoV-2 vaccine and help direct development-focused studies to achieve a vaccine for COVID-19 infection as soon as possible.
Materials and methods
Literature review
Recent publications related to SARS-CoV and SARS-CoV-2 epitopes were reviewed, and the most reliable were selected so we could choose epitopes [1,14,22]. Our main focus and concern in choosing epitopes from researchers' suggestions was their immunogenicity. Potential immunogenic peptides from SARS-CoV-2 for vaccine targets were selected by screening the most reliable reports conducted since the COVID-19 outbreak began in late 2019 and early 2020. The epitopes that were mentioned most often and that were most often used in different studies were selected and carefully analysed.
Bioinformatics approaches, data collection and preprocessing
Nucleotide sequence data of whole-genome SARS-CoV-2 isolates accessible until April 2020 were downloaded from the National Center for Biotechnology Information and GISAID database. Data with accession numbers MN95262 (China), MT281530 (Iran), EPIISL415460 (Netherlands), EPIISL417413 (Turkey), EPIISL417444 (Pakistan), EPIISL413515 (South Korea), EPIISL418411 (Finland), MT007544 (Australia), MN997409 (United States) and EPIISL420890 (Japan) for SARS-CoV-2 NC004718 (SARS) and KY417142 (bat SARS-like) were reported from different countries. For comparison of S proteins, nucleotide sequences were aligned using the ClustalW method implemented in Geneious Prime 2019 software.
Identification of epitopes and antigenicity analysis
Antigenic regions of the SARS-CoV-2 S protein were determined using three tools: Geneious Prime 2019 software based bioinformatics tools on the EMBOSS 6.5.7 Tools antigenic; CLC Genomics Workbench 12 with Welling and Kolaskar-Tongaonkar scales; and the Immune Epitope Database (IEDB). Antigenic regions were annotated in S protein amino acid sequences using Geneious Prime 2019 software for better comparison. Then epitopes were analysed for binding to specific major histocompatibility complex (MHC) class I and class II molecules using the IEDB.
The third protein structure of the S protein was modelled, and epitopes were validated using SWISS-MODEL Tools and RCSB PDB (http://www.rcsb.org/structure/6VXX). The location of epitopes was determined on the third protein structure of the S protein. Five epitopes from the literature review and five epitopes from bioinformatics tools were selected as the best candidates.
Vaccine construction, modelling and validation
The VLP of HBV core particles (HBc) was used as a platform for exposing SARS-CoV-2 epitopes in the major immunodominant region of HBc. The final production of this vaccine was predicted using SWISS-MODEL Tools.
Analysis of the physicochemical properties of suggested epitopes
In order to characterize the physicochemical parameters of the introduced epitopes, we used the http://web.expasy.org/protparam server. The calculated parameters included molecular weight, extinction coefficient, theoretical pI, aliphatic index and grand average of hydropathicity (GRAVY).
Allergenicity evaluation
The allergenicity of the predicted peptides was computed by the AllerCatPro server (https://allercatpro.bii.a-star.edu.sg/). Accuracy of the AllerCatPro server is 84% compared to other prediction methods, which range from 51% to 73%. At the highest sensitivity needed for conservative assessments, AllerCatPro enhances specificity by 37-fold compared to the Food and Agricultural Organization/World Health Organization rules [23].
Results and discussion
Collection of targeted protein sequence
By spreading SARS-CoV-2 and the resulting COVID-19 epidemic in early 2020, there is an urgent need to design a suitable peptide vaccine component against SARS-CoV-2. Currently there is no approved specific antiviral vaccine targeting SARS-CoV-2, although some drugs are still under investigation. As the first attempt to manage this global concern, scientists compared SARS-CoV-2 to previous coronaviruses. Results of the current investigations indicate that there are several important commonalities between SARS-CoV-2 and SARS-CoV infection, which have led to identifying potential targets for antiviral intervention [7]. According to phylogenetic analysis, a high genetic similarity has been observed between SARS-CoV-2 and SARS-CoV [3]. Also, the complete genomes of different isolates of SARS-CoV-2 have been released online and genomic analyses published [24].
In this study, we report for the first time the comparison of the nucleotide sequences of an Iranian isolate of SARS-CoV-2 with other countries, including China, the Netherlands, Japan, Turkey, Pakistan, South Korea, Finland, Australia and the United States. Our analysis showed that the S protein is conserved between isolates from different countries with 100% and 99% similarity (Table 1). We also compared all of the abovementioned SARS-CoV-2 isolates with bat SARS-like and SARS viruses, which showed about 72% and 75% similarity with SARS and bat SARS-like respectively. The high similarity of the S protein of SARS-CoV-2 in different isolates proves that this protein and suggested epitopes acquired from it are potentially the best targets to use when modelling vaccines against COVID-19. Also, a study showed that the S gene is more conserved regarding variations in the viral population of the patients [25].
Table 1.
Identity matrix of S protein using different isolate of SARS-CoV-2 and bat SARS-like and SARS by country
| China | Iran | Netherlands | Japan | Turkey | Pakistan | South Korea | Finland | Australia | United States | Bat SARS-like | SARS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| China | 100 | 100 | 100 | 99.9 | 100 | 100 | 100 | 100 | 100 | 75.9 | 72.2 | |
| Iran | 100 | 100 | 100 | 99.9 | 100 | 100 | 100 | 100 | 100 | 75.9 | 72.2 | |
| Netherlands | 100 | 100 | 100 | 99.9 | 100 | 100 | 100 | 99.9 | 100 | 75.9 | 72.2 | |
| Japan | 100 | 100 | 100 | 99.9 | 100 | 100 | 100 | 99.9 | 100 | 75.9 | 72.2 | |
| Turkey | 99.9 | 99.9 | 99.9 | 99.9 | 99.9 | 99.9 | 99.9 | 99.9 | 99.9 | 75.8 | 72.2 | |
| Pakistan | 100 | 100 | 100 | 100 | 99.9 | 100 | 100 | 100 | 100 | 75.9 | 72.2 | |
| South Korea | 100 | 100 | 100 | 100 | 99.9 | 100 | 100 | 100 | 100 | 75.9 | 72.2 | |
| Finland | 100 | 100 | 100 | 100 | 99.9 | 100 | 100 | 100 | 100 | 75.9 | 72.2 | |
| Australia | 100 | 100 | 99.9 | 99.9 | 99.9 | 100 | 100 | 100 | 100 | 75.9 | 72.2 | |
| United States | 100 | 100 | 100 | 100 | 99.9 | 100 | 100 | 100 | 100 | 75.9 | 72.2 | |
| Bat SARS-like | 75.9 | 75.9 | 75.9 | 75.9 | 75.8 | 75.9 | 75.9 | 75.9 | 75.9 | 75.9 | 71.8 | |
| SARS | 72.2 | 72.2 | 72.2 | 72.2 | 72.2 | 72.2 | 72.2 | 72.2 | 72.2 | 72.2 | 71.8 |
CoV-2, coronavirus 2; SARS, severe acute respiratory syndrome.
While the M and E proteins of SARS-CoV-2 are involved in virus packaging, the spike protein is the prominent intermediate of viral entry [26]. Both the full-length S protein and its antigenic fragments, including the S1 subunit, N-terminal domain (NTD), RBD and S2 subunit, have been reported as potential targets for the development of subunit vaccines [27]. This information, along with the observation of scientists that many epitopes located on the spike protein of SARS-CoV-2 are highly conserved [7] and similar to SARS-CoV, is very important and helpful as we consider this region for designing a VLP-based vaccine against SARS-CoV-2.
Identification of epitopes
Considering the high genetic similarity between SARS-CoV-2 and SARS-CoV, we focused particularly on the epitopes in the S proteins as a result of their previously reported dominant and long-lasting immune response against SARS-CoV [28] and their cell entry role. In recent studies, the S protein of SARS-CoV-2 has been considered as a target for vaccine development and even therapeutic medication development. As a result of the abovementioned importance of the spike glycoprotein, this region of SARS-CoV-2 was analysed for epitope identification in the IEDB. The spike protein's sequence was then also analysed in Geneious 2019 and CLC Genomic Workbench 12 for the identification of the T-cell epitope that can combine with MHC class I and class II molecules (Supplementary Data S1). In order to identify crucial epitopes for designing an effective vaccine, the protein sequence of spike glycoprotein was thoroughly explored using multiple immunoinformatic-based servers and software.
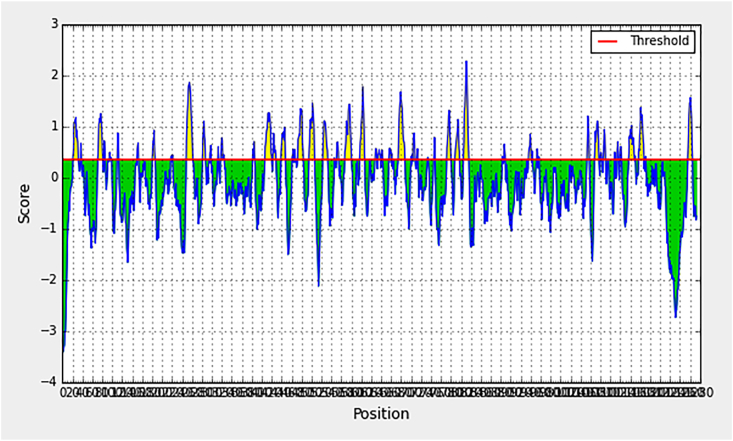
Using immunoinformatics, we could recognize and characterize potential epitopes for the generation of a novel plant-derived VLP-based vaccine [29]. The IEDB is a source of epitope-related information from the scientific literature in the context of infectious disease, allergy and autoimmunity [30]. The IEDB provides bioinformatics tools and algorithms for analysis of epitope data and prediction of potential epitopes from novel sequences (Fig. 1).
Fig. 1.
Antigenic regions of S protein predicted by Immune Epitope Database (IEDB).
A key element in protection against coronaviruses is the neutralizing antibody response. Notably, a previously reported SARS-CoV RBD-specific human neutralizing monoclonal antibody called CR3022 has been recently noted to be able to bind to SARS-CoV-2 RBD with high affinity. Evidently CR3022 recognizes an epitope on the RBD that does not overlap with the ACE2 binding site [22]. Therefore, analysis of the region of the previously introduced CR3022 monoclonal antibody was also performed in the SARS-CoV-2 S protein sequence as a strategy to obtain the maximum reliable epitopes possible. Additionally, we analysed the reported spike protein sequence that binds to ACE2 because RBDs of SARS-CoV-2 spike binding to human ACE2 correlate with the efficient spread of SARS-CoV-2 among humans. This region of 223 aa on the spike protein is believed to play a critical role in the binding of SARS-CoV-2 to human host cells. Overall, by a multistep computational approach, we predicted various antigenic regions within the S protein of CoV-2 that concur with our five identified epitopes (Table 2). In another approach, we identified multiple specific regions in SARS-CoV-2 spike protein that are ideal candidates as epitopes for vaccine design according to recent studies conducted on SARS-CoV-2, which resulted in finding another five specific epitopes (Table 2).
Table 2.
Candidate linear epitopes of S protein for exposing on virus-like particles
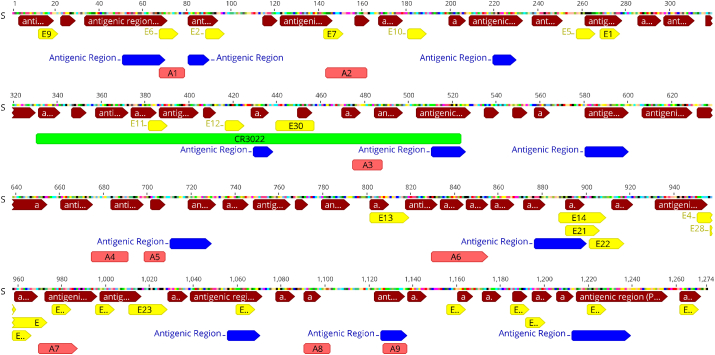
With a high sequence similarity to immunogenic peptides but with a greater predicted immunogenicity score, our predicted epitopes can be used to build a peptide vaccine (Fig. 2). Also, these epitopes can be used as diagnostic tools for identifying SARS-CoV-2. We additionally incorporated the information about the associated MHC alleles to provide a list of epitopes that seek to maximize population coverage globally (Supplementary Data S1).
Fig. 2.
Binding sites and antigenic regions of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein. Yellow indicates epitopes recommended in literature; brown, antigenic regions predicted by Genius 2019 software; blue, antigenic regions predicted by CLC Genomic Workbench 12 software; and pink, predicted epitopes.
Validation of epitopes
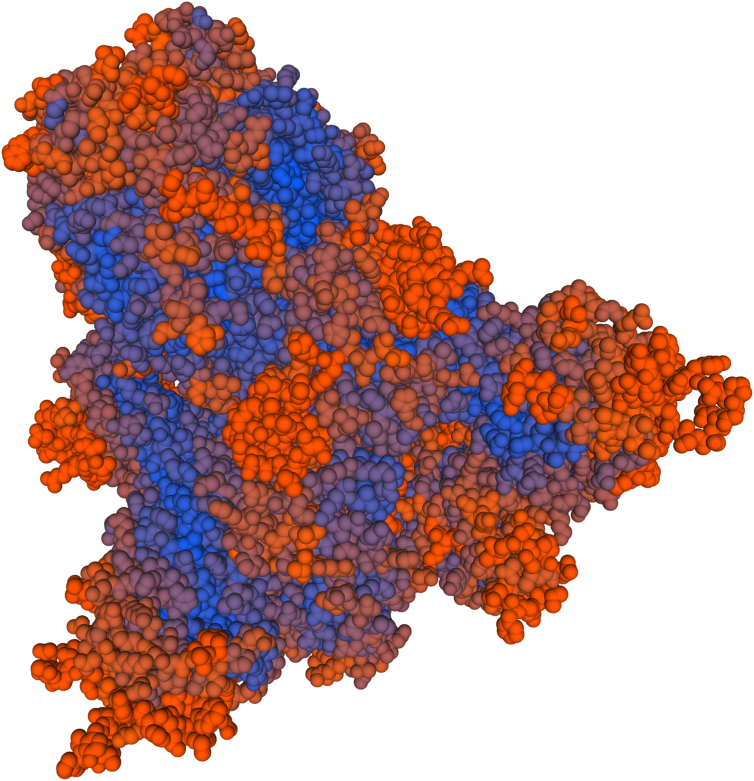
For validation of selected epitopes, we used SWISS-MODEL Tools. The third structure of the S protein was predicted, and the position of epitopes on the protein structure was determined (Fig. 3). This result confirmed that epitopes were selected from positions that can interact with host proteins such as antibodies. In order to be more confident, the S protein model and epitopes were checked using RCSB PDB, with results similar to that of SWISS-MODEL Tools.
Fig. 3.
Third structure of S protein determined by SWISS-MODEL Tools. Positions of epitopes on protein structure are shown in orange.
Vaccine construction, modelling and validation
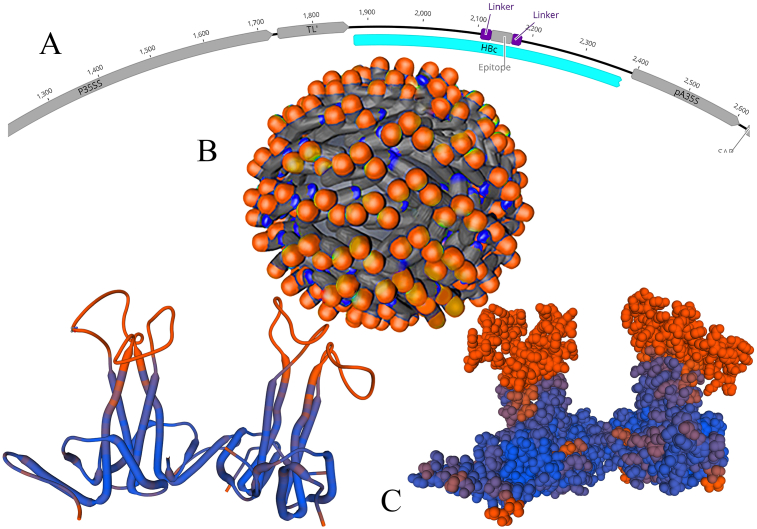
Finally, the vaccine component was modelled using SWISS-MODEL Tools. Our obtained results indicate that the designed vaccine model is of good quality (Fig. 4). We predicted the VLP of HBc-exposing epitopes of SARS-CoV-2 (Fig. 4). HBc is the best choice as a platform for commercial vaccine production, not only against HBV but also against many other viral diseases. HBc showed high immunogenicity and enhanced presentation to the immune system. This VLP was expressed using an open reading frame and has the flexibility to allow a wide variety of foreign insertions without affecting the protein self-assembly and VLP function [31,32]. This result confirmed that inserting the epitope in the HBc major immunodominant region can expose these epitopes on the surface of VLP. The present study indicates the effectiveness of selected epitopes within the spike glycoprotein of SARS-CoV-2. These epitopes can be used to make an immunogenic multiepitopic peptide vaccine against SARS-CoV-2. For better exposing of epitopes on VLP, they need to link by a peptide linker (Linker Database, IBIVU). The (EAAAK)3 linker was suggested, which easily fused with the virus coat protein [33].
Fig. 4.
General design of virus-like particle (VLP) on hepatitis B core virus. (A) Proposed structure based on PVX virus along with VLP of hepatitis B core protein (HBc) virus and epitopes. (B) Final design of hepatitis virus and epitopes (orange on body of virus). (C) Predicted model of third structure of HBc along with epitopes (orange).
Physical and chemical properties
Using the PROTPARAM tool, the physical and chemical properties of epitopes were identified (Table 3). According to the result, the A1 epitope has the highest theoretical pI. Moreover, most peptides seem to remain stable, with an instability index of <40. The stability index is calculated on the basis of N-terminal ubiquitin-mediated degradation. Among all, E30, E24, A9 and A7 showed the highest thermostability due to their aliphatic index. Considering the GRAVY, the negative index indicates that the peptide is hydrophilic and soluble in aqueous solutions [34]. A2, E13, E29 and A1 showed the highest potential to interact with water molecules because they showed the least GRAVY. Overall, we found a number of epitopes that would serve as efficient vaccines against SARS-CoV-2 infection. However, E29 is predicted to be one of the most promising epitopes because it has high thermostability and is strongly hydrophilic.
Table 3.
Physicochemical properties of SARS-CoV-2 epitopes
| Epitope | Molecular weight | Theoretical pI | Extinction coefficient | Instability index | Aliphatic index | GRAVY |
|---|---|---|---|---|---|---|
| A1 | 1316.48 | 11.00 | Not visible by UV spectrophotometry | 7.42 | 56.67 | −0.550 |
| A2 | 2455.69 | 8.39 | 9970 | 86.29 unstable | 15.26 | −1.505 |
| A3 | 1355.46 | 4.00 | 125 | −8.89 | 27.86 | −0.071 |
| A7 | 1947.26 | 5.96 | Not visible by UV spectrophotometry | 27.39 | 146.11 | 0.650 |
| A9 | 1142.32 | 3.80 | Not visible by UV spectrophotometry | −14.06 | 176.36 | 1.291 |
| E13 | 2076.38 | 8.59 | Not visible by UV spectrophotometry | 121.50 unstable | 65.00 | −0.689 |
| E21 | 1815.18 | 8.75 | 1490 | 27.11 | 73.75 | 0.631 |
| E24 | 916.17 | 5.52 | Not visible by UV spectrophotometry | −0.54 | 227.78 | 3.056 |
| E29 | 1056.23 | 8.75 | Not visible by UV spectrophotometry | 8.89 | 130.00 | −0.589 |
| E30 | 1042.24 | 5.81 | Not visible by UV spectrophotometry | 51.69 unstable | 205.56 | 0.867 |
GRAVY, grand average of hydropathicity; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Allergenicity evaluation
Allergen peptides or proteins stimulate an IgE antibody response [35]. The predicted vaccine candidate epitopes should not result in an allergic response to the body. Based on the prediction of the AllerCatPro server, all predicted epitopes are nonallergenic.
Conclusions
We attempted to find various epitopes against SARS-CoV-2 by using immunoinformatic tools because quick identification of these epitopes is crucial to design a vaccine component against COVID-19. The spike protein of SARS-CoV-2, which is currently believed to play the most important role in the binding and entry of these viruses to human cells, was analysed to find epitopes. We chose five optimal epitopes for SARS-CoV-2 from the literature, and our parallel bioinformatics predictions identified five more. These epitopes are ideal candidates to study while formulating a multiepitopic peptide vaccine—not only because of their being selected from the ACE2 binding site and CR3022 antibody epitopic regions of spike protein, but also because their antigenic property was confirmed by bioinformatics tools. On the basis of the selected epitopes, we designed a vaccine that may be generated in a short time in the near future. Our novel platform of a VLP-based vaccine is designed to target the immune response towards these conserved epitope regions, which are potent sites to generate immunity against SARS-CoV-2.
Next we plan to study the significance and effectiveness of our suggested epitopes as ideal vaccine candidates against SARS-CoV-2 in plant-derived VLPs. These immunoinformatic analyses will require several in vitro and in vivo validations before designing the vaccine to resist COVID-19.
The independent identification of the same regions using two approaches reflects the high probability that our suggested epitopes are promising targets for immune recognition of SARS-CoV-2. In conclusion, the use of available information related to SARS-CoV and SARS-CoV-2 epitopes, in conjunction with bioinformatics predictions, points to specific regions of SARS-CoV-2 that have a high likelihood of being recognized by human immune responses.
Conflict of interest
None declared.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nmni.2020.100786.
Contributor Information
A. Afsharifar, Email: afshari@shirazu.ac.ir.
A. Pormohammad, Email: pormohammadali@yahoo.com.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaki A.M., Van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 3.Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int J Biol Sci. 2020;16:1678. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J., Liu W. Puzzle of highly pathogenic human coronaviruses (2019-nCoV) Protein Cell. 2020;11:235–238. doi: 10.1007/s13238-020-00693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C. A SARS-CoV-2 vaccine candidate: in-silico cloning and validation. Inform Med Unlocked. 2020;20:100394. doi: 10.1016/j.imu.2020.100394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakraborty C., Sharma A.R., Bhattacharya M., Sharma G., Lee S.S., Agoramoorthy G. Consider TLR5 for new therapeutic development against COVID-19. J Med Virol. 2020 doi: 10.1002/jmv.25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakraborty C., Sharma A.R., Bhattacharya M., Sharma G., Lee S.S., Agoramoorthy G. COVID-19: consider IL6 receptor antagonist for the therapy of cytokine storm syndrome in SARS-CoV-2 infected patients. J Med Virol. 2020 doi: 10.1002/jmv.26078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc Natl Acad Sci U S A. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratajczak W., Niedźwiedzka-Rystwej P., Tokarz-Deptuła B., Deptuła W. Immunological memory cells. Cent Eur J Immunol. 2018;43:194. doi: 10.5114/ceji.2018.77390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan M., Wu N.C., Zhu X., Lee C.C.D., So R.T., Lv H. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Q., Lai H. Plant-derived virus-like particles as vaccines. Hum Vaccin Immunother. 2013;9:26–49. doi: 10.4161/hv.22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pushko P., Pumpens P., Grens E. Development of virus-like particle technology from small highly symmetric to large complex virus-like particle structures. Intervirology. 2013;56:141–165. doi: 10.1159/000346773. [DOI] [PubMed] [Google Scholar]
- 19.Chroboczek J., Szurgot I., Szolajska E. Virus-like particles as vaccine. Acta Biochim Pol. 2014;61 [PubMed] [Google Scholar]
- 20.Jain N.K., Sahni N., Kumru O.S., Joshi S.B., Volkin D.B., Middaugh C.R. Formulation and stabilization of recombinant protein based virus-like particle vaccines. Adv Drug Deliv Rev. 2015;93:42–55. doi: 10.1016/j.addr.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 21.Pang E.L., Peyret H., Ramirez A., Loh H.S., Lai K.S., Fang C.M. Epitope presentation of dengue viral envelope glycoprotein domain III on hepatitis B core protein virus-like particles produced in Nicotiana benthamiana. Front Plant Sci. 2019;10:455. doi: 10.3389/fpls.2019.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng M., Song L. Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. Cell Mol Immunol. 2020;17:536–538. doi: 10.1038/s41423-020-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurer-Stroh S., Krutz N.L., Kern P.S., Gunalan V., Nguyen M.N., Limviphuvadh V. AllerCatPro—prediction of protein allergenicity potential from the protein sequence. Bioinformatics. 2019;35:3020–3027. doi: 10.1093/bioinformatics/btz029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian X., Li C., Huang A., Xia S., Lu S., Shi Z. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghorbani A., Samarfard S., Ramezani A., Izadpanah K., Afsharifar A., Eskandari M.H. Quasi-species nature and differential gene expression of severe acute respiratory syndrome coronavirus 2 and phylogenetic analysis of a novel Iranian strain. Infect Genet Evol. 2020;85:104556. doi: 10.1016/j.meegid.2020.104556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020 Sep 6 doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y., Jiang S., Du L. Prospects for a MERS-CoV spike vaccine. Exp Rev Vaccines. 2018;17:677–686. doi: 10.1080/14760584.2018.1506702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Channappanavar R., Zhao J., Perlman S. T cell-mediated immune response to respiratory coronaviruses. Immunol Res. 2014;59:118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosales-Mendoza S. Will plant-made biopharmaceuticals play a role in the fight against COVID-19? Expert Opin Biol Ther. 2020 doi: 10.1080/14712598.2020.1752177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kazaks A., Lu I.N., Farinelle S., Ramirez A., Crescente V., Blaha B. Production and purification of chimeric HBc virus-like particles carrying influenza virus LAH domain as vaccine candidates. BMC Biotechnol. 2017;17:1–11. doi: 10.1186/s12896-017-0396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji M., Xie X.X., Liu D.Q., Lu S., Zhang L.X., Huang Y.R. Engineered hepatitis B core virus-like particle carrier for precise and personalized Alzheimer’s disease vaccine preparation via fixed-point coupling. Appl Mater Today. 2020;19:100575. [Google Scholar]
- 33.Bhattacharya M., Sharma A.R., Patra P., Ghosh P., Sharma G., Patra B.C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J Med Virol. 2020;92:618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vita R., Mahajan S., Overton J.A., Dhanda S.K., Martini S., Cantrell J.R. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res. 2019;47:D339–D343. doi: 10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Droppa-Almeida D., Franceschi E., Padilha F.F. Immune-informatic analysis and design of peptide vaccine from multi-epitopes against Corynebacterium pseudotuberculosis. Bioinform Biol Insights. 2018;12 doi: 10.1177/1177932218755337. 1177932218755337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.