Abstract
This chapter calls out the following contributed articles, and gives a sense of why the tRNA synthetases are an endless frontier for scientific research and the unveiling of a vast world of new biology.
Keywords: Genetic code, Tree of life, Editing, Orthogonal functions, Splice variants, Therapeutics, Lung disease, Covid-19
The genetic code was the greatest discovery of the 20th century. Prior to its discovery, biology was a field of classifications and descriptions of what could be seen by the naked eye or the microscope. Biologists cataloged organisms using specific anatomical features to assign a plant or animal or microbe to one “bin” or another. Darwin saw more deeply the connections between species and the progression, over the eons, to greater and greater complexity. Darwin also understood the selective pressure that forced adaption to a specific environment by culling out the weak from the strong, to capture the traits most needed for survival. These adaptive phenomena were studied in the laboratory in the field of microbial genetics and, outside the laboratory, observers more broadly annotated adaptive phenomena in nature. But, without the benefit of an analytical framework, these efforts, and the knowledge that was gained, were self-limiting. Badly needed was a molecular methodology to interpret the vast complexity of nature with its estimated one trillion species and the apparent cause-effect relationships that could be cataloged by the organization of specific phenotypes into lines of ascent in evolution. The discovery of the genetic code opened the door to a molecular framework that engendered enormous power for understanding the creation of life and its vast diversity (see chapter “The evolution of aminoacyl-tRNA synthetases: From dawn to LUCA” by Ribas de Pouplana).
1. aaRSs establish the genetic code
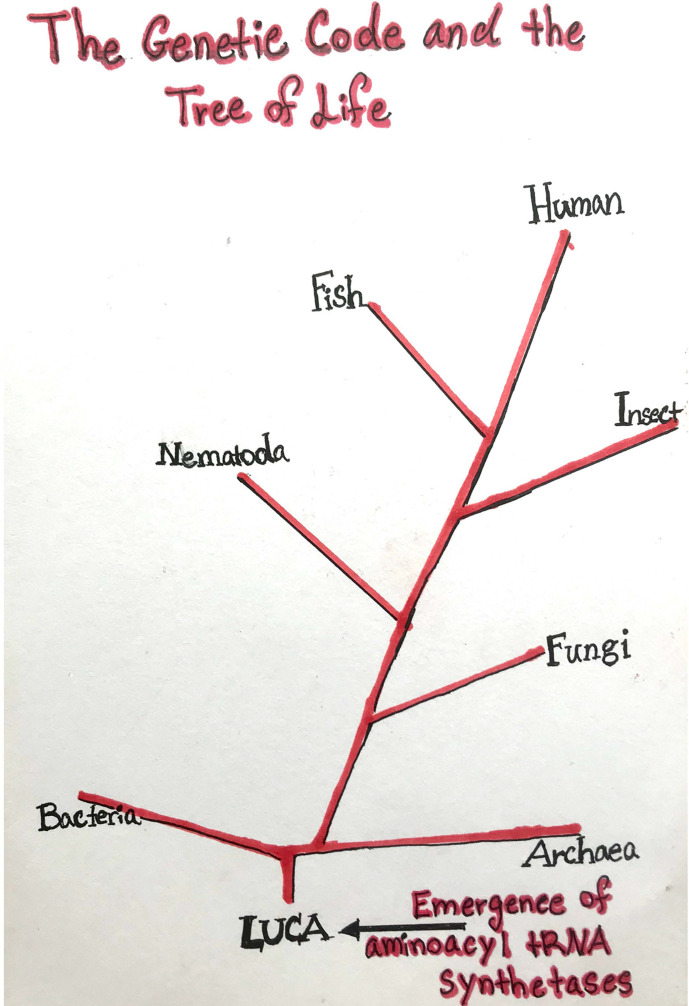
Life is estimated to have appeared less than one billion years after the formation of earth about 4.5 billion years ago. From there a “Tree of Life” eventually emerged with three great kingdoms branching out as major branches designated as Bacteria, Archaea, and Eukarya (Fig. 1 ). At the base of the Tree is a hypothetical organism named “The Last Universal Ancestor,” or LUCA. The genetic code ties together and unites all of the roughly one trillion species that are represented by this Tree.
Fig. 1.
The genetic code and the tree of life.
Investigations to date, armed with the most advanced tools of informatics and millions of sequences of genes, point to early versions of the aminoacyl tRNA synthetase family of proteins lying at the base of this great Tree (Fig. 1). These enzymes establish the simple rules of the genetic code, whereby codons are matched with amino acids in the catalysis of aminoacylation (see chapter “Putting amino acids onto tRNAs: The aminoacyl-tRNA synthetases as catalysts” by Alexander and Hendrickson). Importantly, although the reaction is simple, it has been imaginatively manipulated and engineered for applications to synthetic biology (see chapter “Engineering aminoacyl-tRNA synthetases for use in synthetic biology” by Krahn et al.). And, nature itself found ways to use the aminoacylation format in creative ways, including the synthesis of key cellular components (see chapter “Noncanonical inputs and outputs of tRNA aminoacylation” by Hemmerle et al.).
Remarkably, the code is just 64 building blocks, perhaps like 64 Legos that have different shapes and colors. From these 64 blocks, roughly one trillion organisms were generated, with an unimaginable diversity of morphologies and abilities to adapt to specific environments. Fig. 2 illustrates a tiny sample of this diversity, from left to right: the tube worms and puffer fish that exist in a deep sea channel running from the artic to the tip of South Africa; The 9-arm pacific octopus; cyanobacteria that can grow at the temperature of boiling water; the exquisite beauty of butterflies and flowers; a huge polar bear searching for fruit; a human of deep intelligence (H. Gobind Khorana, who shared Nobel Prize for discovery of the genetic code); and timber wolves who developed the emotions of affection and bonding. All of this variety, and far more, from the ways that the 64 building blocks of the genetic code are organized and used. Importantly, these building blocks have been separately compartmentalized in the cytoplasm and mitochondria, with mostly distinct single copy nuclear genes for each (see chapter “Mitochondrial aminoacyl-tRNA synthetases” by Chihade).
Fig. 2.
Examples of the vast diversity of living systems that are created by the same genetic code.
2. Problems when simplicity is not quite enough
Not long after the discovery of aaRSs, Pauling pointed out that the accuracy of the genetic code was not possible without another activity that improved the observed specificity of amino acid selection for tRNA matching. This activity had to be separate from the basic aminoacylation aminoacyl adenylate) and of a mischarged tRNA was uncovered. The activity comes from a separate catalytic site for editing, imbedded in the overall architectures of the enzymes. Its role reaction. Some years later, editing the products of amino acid activation (formation of the is to clear away the misactivated complexes and mischarged tRNAs. The field of editing expanded dramatically, with editing activities not only demonstrated for many of the synthetases but, in addition, the discovery of separate trans-acting factors that remove amino acids mis-attached to specific tRNAs (see chapter “Trans-editing by aminoacyl-tRNA synthetase-like editing domains” by Musier-Forsyth and Nagy). Later, interest in the field expanded dramatically with the realization that, in the mouse and fish, pathologies and lethality is associated with even small deficiencies that arise from mutations in the catalytic center for editing. Fig. 3 illustrates an example of warped embryonic development in zebrafish with an editing-defective VARS.
Fig. 3.
Warped embryonic development in zebrafish with an editing-defective VARS.
3. Orthogonal functions created from pieces and decorations
The enzymes are ancient and, given their early appearance, had eons to evolve additional functions and characteristics going far beyond the simple aminoacylation reaction. On that point, the discovery of non-catalytic functions of aaRSs was one of the major developments of the last 20 years (see chapters “Structures and functions of multi-tRNA synthetase complexes” by Kim and Kim, “Non-canonical functions of human cytoplasmic tyrosyl-, tryptophanyl- and other aminoacyl-tRNA synthetases” by Wakasugi and Yokosawa, “Aminoacyl-tRNA synthetases in cell signaling” by Yao and Fox, “Human diseases linked to cytoplasmic aminoacyl-tRNA synthetases” by Jiang et al., and “Novel functions of cytoplasmic aminoacyl-tRNA synthetases shaping the hallmarks of cancer” by Wang and Yang). Because these functions are detached from anything even remotely resembling the aminoacylation reactions, we can think of them as analogous to a caterpillar turning into a butterfly (Fig. 4 ).
Fig. 4.
New eyes: Morphogenesis to give a form and function.
These new functions include a vast repertoire of nuclear and extra-cellular activities and are associated with a large number of novel and aaRS-specific interacting partners. Synthetases can be decorated, by phosphorylation or acetylation, by the accretive and progressive (in evolution) addition of new non-catalytic domains, and they can be split into pieces that are reassembled by alternative splicing. More than 200 splice variants have been annotated and these appear in tissue- and developmental-stage specific patterns. Interestingly, most splice variants ablate the catalytic site in part or in whole. These ablations clearly point to a functional redirection.
4. Functions and diseases linked to tRNA synthetases
The cytoplasmic and mitochondrial genes for tRNA synthetases are essential because they are present in nuclear-encoded single copies and because life cannot exist without ensuring the implementation of the code. Thus, homozygous mutations that inactivate the catalytic site are lethal, and such mutation-bearing cells/organisms are discarded. This essentiality gave an obvious opportunity to develop medicines that target pathogen-specific tRNA synthetases, which vary in their active-site architectures just enough to have drugs distinguish them from their human counterparts (see chapter “Aminoacyl-tRNA synthetases as drug targets” by Lukarska and Palencia). However, the non-catalytic elaborations on the basic framework of the core catalytic domain are targets for mutations that will not necessarily be lethal but rather will introduce pathologies (see chapters “Structures and functions of multi-tRNA synthetase complexes” by Kim and Kim, “Non-canonical functions of human cytoplasmic tyrosyl-, tryptophanyl- and other aminoacyl-tRNA synthetases” by Wakasugi and Yokosawa, “Aminoacyl-tRNA synthetases in cell signaling” by Yao and Fox, “Human diseases linked to cytoplasmic aminoacyl-tRNA synthetases” by Jiang et al., and “Novel functions of cytoplasmic aminoacyl-tRNA synthetases shaping the hallmarks of cancer” by Wang and Yang). Indeed, over 100 recessive cytoplasmic synthetase mutations have been linked to diseases in human populations. (Mutations in mitochondrial synthetases are also linked to diseases (see chapter “Mitochondrial aminoacyl-tRNA synthetases” by Chihade).) Others are dominant, such as those that are causally associated with neurological pathologies grouped as one form or another of Charcot-Marie-Tooth Disease (CMTD). These gain-of-function mono-allelic mutations often show no evidence of any change in the catalytic efficiency or specificity of the mutant synthetase. New mutation-induced interacting partners, or enhanced interactions with normal existing partners, can be the root of the cause of the disease phenotype.
Other investigations made associations between aaRSs and cancer, and suggested roles in tumor suppression (see chapter “Novel functions of cytoplasmic aminoacyl-tRNA synthetases shaping the hallmarks of cancer” by Wang and Yang). Immune disorders, and especially the anti-synthetase auto-immune inflammatory condition, led to new insights into immune modulatory roles. Other mutations resulted in widespread system failures, and yet have not always been linked to failures of catalytic activity. These and other effects, across many diseases, are manifestations of alternative functions, such as well documented roles in angiogenesis and hematopoiesis. And, in addition, and perhaps surprisingly, the long-known organization in eukaryotes of some tRNA synthetases into a multi-synthetase cluster(s) is increasingly viewed as being connected to novel functions and their regulation (see chapters “Structures and functions of multi-tRNA synthetase complexes” by Kim and Kim and “Aminoacyl-tRNA synthetases in cell signaling” by Yao and Fox). Here again, disease connections are being made.
5. Therapeutics with splice variants
The many splice variants of aaRSs have activities outside of translation and now are being investigated and applied to treat specific disease indications. Fig. 5 illustrates a lung that is badly fibrotic. Polluted air is thought to contribute to this pathology.
Fig. 5.
Loss of homeostasis: A lung at the end-stage of interstitial lung disease.
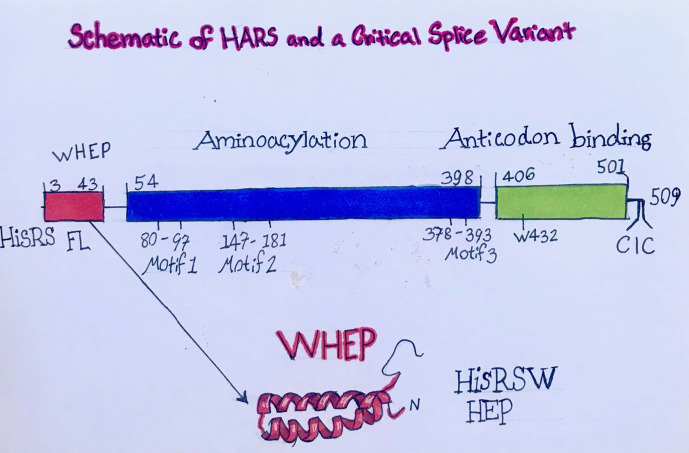
Using an engineered version of a splice variant of HARS Fig. 6 , a bleomycin-induced inflammatory lung condition in the mouse can be dramatically improved by systemic injection of modest amounts of this variant. This improvement can be traced to the ability of the “drug” to tamper down organ-targeted immune responses, such as in the lung. Inflamed lungs are seen commonly in Covid-19 patients. As the inflammation progresses, victims are put on ventilators. Because of the strong homeostatic effects of the HARS splice variant construct, it is now being administered in FDA-approved trials to hospitalized Covid-19. Patients, prior to their being put on a ventilator. These and other studies are unmasking and applying the power of the aaRSs well beyond translation (Fig. 7 ).
Fig. 6.
Schematic of HARS and a critical splice variant.
Fig. 7.
HARS is potent in bleomycin lung injury model.
6. The endless frontier
This volume gives some glimpse of what has now placed tRNA synthetases on “center stage,” where the coming decades will reveal some of the most profound and rich biology that the broad field of life sciences has ever seen. Because of their deep roots at the origin of life, the enzymes will continue to provide a framework and opportunity to understanding their roles and structural adaptations which facilitated the creation of over one trillion species. These structural changes correlate well with the development of ex-translational functions associated with major steps in evolution, such as the development of the closed vascular system of vertebrates. Immuno-modulatory roles are emerging and, given the early appearance of the enzymes, may lead to insights in connections between the innate and adaptive immune systems. Neurological roles are also emerging, and those roles may in part explain diseases of the nervous system that link to tRNA synthetases. The world of secreted and nuclear tRNA synthetases has been established but, except for a few specific examples, what happens in those worlds is barely known. Undoubtedly, knowledge of the role of tRNA synthetases in aminoacyl adenylate transactions, such as protein and lipid modifications, will open another new world. And with the ability to manipulate the activated amino acids attached to tRNAs for creation of new materials, we will see more activity in this area as well. So, the stage is wide and deep, and offers more opportunities and excitement than ever.
Acknowledgements
This work was supported by NIH grant R01 GM125908 and a fellowship from the National Foundation of Cancer Research. All art is original and provided by Heidi Herbawi using material provided by Paul Schimmel.