The refractory nature of multiple myeloma (MM), as well as the persistence of minimal residual disease (MRD), have long been attributed to the acquisition of drug resistance, in part through the protective role of the microenvironment. Mesenchymal stromal cells (MSC) are one of the major constituents of the bone marrow (BM) microenvironment yet our understanding of their contribution to myelomagenesis remains largely unknown. All attempts to characterize MM-MSC have required in vitro expansion.5-12 While this is understandable given the technical challenge of isolating and characterizing those very few MSC present in BM aspirates, in vitro expansion may alter the biological signature of MSC and preclude correct identification of molecular mechanisms related to aging, malignant transformation and resistance to treatment.7 Hence, more data need to be collected on the properties of non-manipulated MSC from both healthy donors (HD) and MM patients for a better understanding of tumor-microenvironment interactions. 10,13 Here, using freshly isolated MSC we unveil striking new data about the impact of aging on HD-MSC and how tumor cells from patients with active MM may block MSC differentiation and induce an immune suppressive transcriptional program, which may persist even in the presence of various degrees of MRD.
A total of 21 HD, and 56 newly-diagnosed MM patients were studied after providing written informed consent. Multidimensional flow cytometry was used to identify, isolate and characterize BM MSC (Online Supplementary Figure S1A). MSC were sorted by fluorescence activated cell sorting (FACS) according to the phenotype defined by the International Society for Cellular Therapy (CD45–/CD73+/CD105+/CD271+/ CD13+/CD90+), and either lysed for RNA-sequencing or plated for in vitro expansion as previously described.8 Poly-A RNA from MSC was processed for RNA sequencing following the massively parallel single-cell RNA sequencing (MARS) protocol14 optimized for bulk lowcell numbers, and sequenced in an Illumina NextSeq 500. The data pre-processing pipeline included the Cutadapt, STAR and HTSeq bioinformatic packages. Gene expression counts were analyzed with DESeq2 by pairwise contrast across different sample groups. Conservative gene filtering was applied to increase consistency. Gene expression data were normalized with vst (DESeq2) and processed with RemoveBatchEffect functions (limma) for visualization. R suite tools were additionally employed for unsupervised clustering and plot generation. Metascape15 was used to gain insight into the biological meaning of gene expression dynamics. A detailed description of the experimental procedures can be found in the Online Supplementary Materials and Methods.
Quantification of the numbers of MSC in BM aspirates showed that their frequency was significantly higher in patients with active MM [median=0.028%; interquartile range (IQR)=0.045] than in HD (median=0.0038%; IQR=0.012) (Online Supplementary Figure S1A,B). Given the generally low frequencies of MSC in the BM, we first validated, in silico, the identity and purity of MSC sorted by FACS (Online Supplementary Figure S1C, D). In accordance with previous observations,8,9 MM-MSC showed an impaired differentiation capacity compared to that of HD-MSC (Online Supplementary Figure S1E, F), suggesting that a differentiation blockade may be responsible for the accumulation of these cells in the BM of MM patients. More importantly, principal component analysis (PCA) of gene expression data showed that in vitro expansion magnified the differences between HD and MM-MSC, which in turn displayed a transcriptional profile dramatically different from that of their freshly isolated counterparts (Online Supplementary Figure S2A, B). Importantly, in vitro expansion affected MSC from HD or MM patients equally, except for a subset of genes exclusively altered in HDMSC (clusters 1 and 2, Online Supplementary Figure S2B). MM-relevant functions such as immune response activation, cellular growth, tumor necrosis factor-α signaling, or cell substrate adhesion were affected by these changes (Online Supplementary Figure S2C), thereby highlighting the need to study the role of MSC in MM pathogenesis using freshly isolated cells.
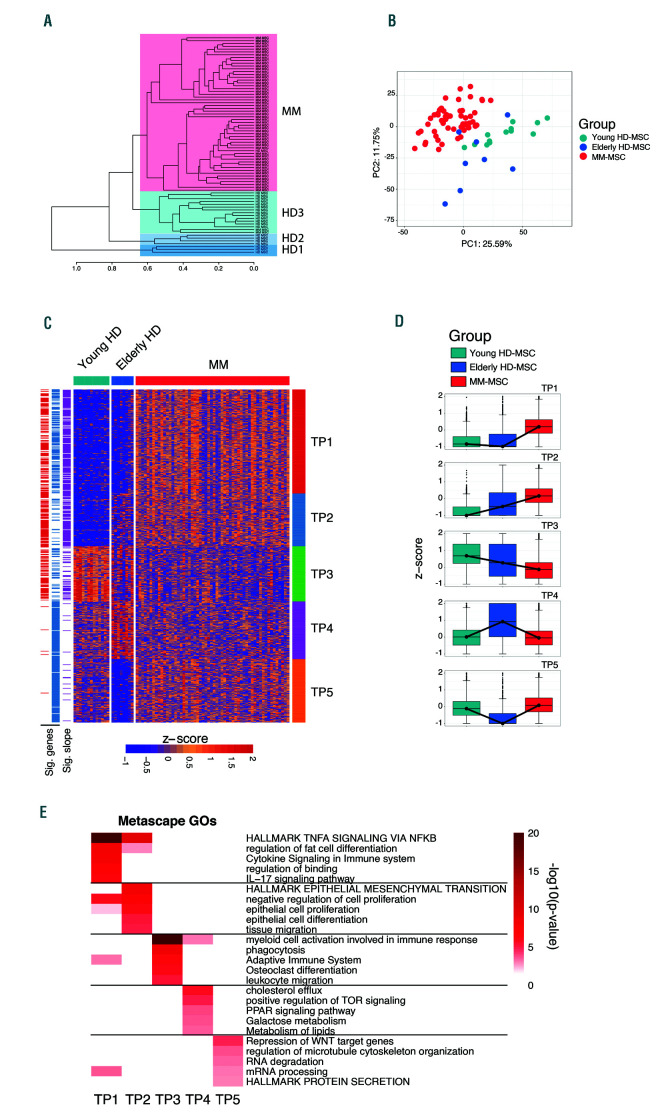
Having shown that in vitro expansion drives transcriptional changes in MSC, we analyzed the transcriptional profile of freshly isolated MSC. Unsupervised hierarchical clustering revealed four main clusters of samples (Figure 1A). One cluster included all MM-MSC, while HD-MSC segregated into three clusters, which showed differences in the ages of the HD (median age HD1: 76, HD2: 66 and HD3: 20 years old). Hence, we classified MSC based on their source and age distribution of the donors into young HD-MSC (median age of donors: 20 years old), elderly HD-MSC (combining both advanced age groups: median age of donors: 69.5 years old), or MM (median age: 72 years old) (Online Supplementary Table S1). PCA further supported the differences between HDMSC from different age groups and those from MM-MSC (Figure 1B). These findings indicate that aging modulates the MSC transcriptional profile, which is further altered by the influence of MM (predominantly a disease of the elderly).
Pairwise comparisons of HD-MSC versus MM-MSC evidenced a total of 606 differentially expressed genes (adjusted P<0.05, |log2FC| >1), distributed in five transcriptional programs (TP) identified by semi-supervised k-means clustering (TP1-5) (Figure 1C, D) and associated with specific functions (Figure 1E). Most MM genes were upregulated when compared to those in HD-MSC and grouped in TP1, which was enriched in functions related to MM pathogenesis such as the interleukin-17 pathway and tumor necrosis factor signaling via nuclear factor-κB, which have been shown to establish an immunosuppressive microenvironment and to induce osteoclastogenic activity via inflammation mediators.3,7,11 Accordingly, the osteoblastogenesis inhibitor SOX9, which blocks MSC differentiation into the osteogenic lineage, was shown to be upregulated in MM-MSC.13,15 TP2 and TP3 included up- and down-regulated genes with significant linear correlation with age and MM onset, revealing previously unrecognized dynamic transcriptional changes associated with age that are further exacerbated by the crosstalk between MSC and tumor cells. MSC proliferation was a hallmark of TP2 based on upregulation of genes implicated in epithelial mesenchymal transition, differentiation and proliferation in MM-MSC. Genes downregulated in TP3 were enriched in functions related to myeloid cell activation, phagocytosis, leukocyte migration and osteoclast differentiation (inhibitory factors), which could help to explain the immune suppressive potential of MMMSC. 16-19 Intriguingly, since gene expression in TP3 was linearly correlated with age, these findings also suggest that a pre-existent permissive immune microenvironment may be critical in the development of MM. TP4 and TP5 included genes that changed with age but whose expression levels in MM-MSC resembled that in MSC from young HD. The upregulation of peroxisome proliferator- activated receptor (PPAR) signaling as well as functions such as cholesterol efflux or metabolism of lipids in the elderly (TP4) suggests a reinforced pro-adipogenic microenvironment associated with age, as previously described.20-23 Importantly, this signature diminishes in MM-MSC suggesting that tumor cells modulate the capacity of MSC to differentiate, potentially to support their own survival. TP5 was characterized by enrichment in functions that are compromised in elderly HD-MSC but are inducible by tumor cells, such as secretion of pro tein and RNA metabolism required for microenvironment modulation to promote MM growth. Taken together, our data demonstrate that freshly isolated MM-MSC are transcriptionally different from their normal counterpart and partially influenced by aging, potentially leading to a rewiring of their differentiation capacity and the contribution to a permissive immune microenvironment.
Figure 1.
Transcriptome dynamics associated with age in mesenchymal stromal cells from patients with multiple myeloma and healthy donors. Unsupervised hierarchical clustering of mesenchymal stromal cell (MSC) samples. The dendrogram was cut at 0.65 h resulting in three groups of samples from healthy donors (HD1, HD2, HD3) and a group from patients with multiple myeloma (MM). 1-correlation (Spearman) was used as the distance for complete linkage clustering. (B) Principal component analysis (PCA) from the top most variable genes. Samples are categorized based on clinical status and age range into Young HD-MSC (light blue, median age 20 years), Elderly HD-MSC (dark blue, median age 69.5 years), and MM-MSC (red, median age 72 years). Gene expression dispersion was measured as median absolute deviation (MAD). (C) Semi-supervised k-mean clustering (k=5) of a total of 606 differentially expressed genes (adjusted P<0.05 and |log2FC| >1) between Young HDMSC, Elderly HD-MSC, and MM-MSC. Red and blue lanes (left side) indicate significant differentially expressed genes for Young HD-MSC and Elderly HD-MSC against MM-MSC, respectively. Genes with a P<0.001 for the linear model slopes (i.e., significantly different from slope=0) were considered as lineally correlated with age and MM (purple lane). Gene expression data were z-score standardized by row for visualization purposes. (D) Clusterspecific median gene expression dynamics represented by a boxplot of z-score standardized expression data. (E) Biological function enrichment analysis performed per cluster. Enrichment heatmap of the top five most significant non-redundant biological functions independently selected per cluster. Heatmap colors represent significance value as -log10(P-value).
Figure 2.
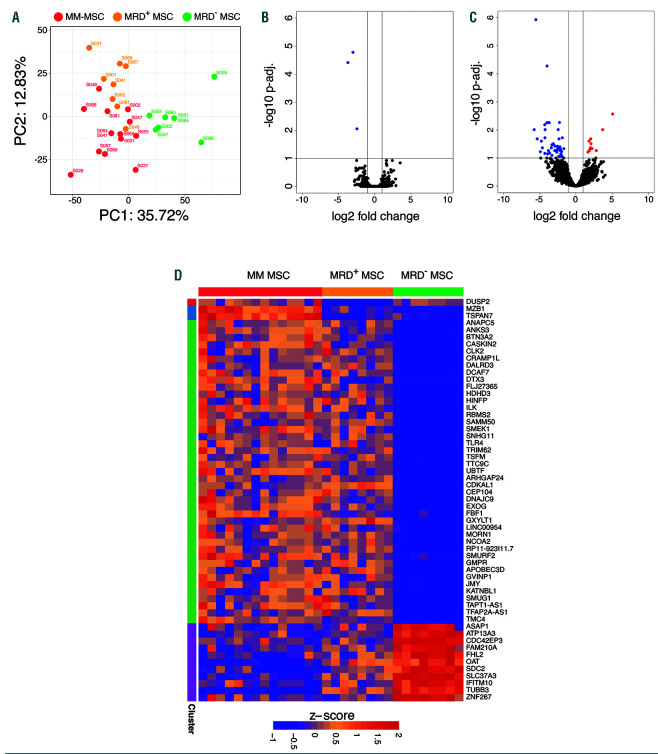
Impact of multiple myeloma on the mesenchymal stromal cell transcriptional program according to minimal residual disease status. (A) Principal component analysis (PCA) of gene expression profiles from mesenchymal stromal cells (MSC) from patients with multiple myeloma (MM) at the time of diagnosis and matched samples during minimal residual disease (MRD). Data for PCA were transformed to account for the paired design. Gene expression dispersion was measured as mean absolute deviation (MAD). (B, C) Volcano plot of differentially expressed genes in MSC during the stage of MRD positivity. Paired differential expression analysis showed that three genes were downregulated in MRD+ MSC compared to their counterpart MSC at diagnosis of MM (B) and 56 were deregulated in MRD–MSC compared to the counterpart MSC at MM diagnosis (C). (D) Semi-supervised k-mean clustering (k=4) of genes significantly up- or down-regulated in (B) and (C). Adjusted P<0.1 and |log2FC| >1.
Since our results demonstrated that MM onset modulates gene expression in MSC, we hypothesized that eradication of tumor cells (i.e., undetectable MRD) may revert the transcriptional profile of MSC to normal. We, therefore, collected paired MSC samples from MM patients at diagnosis and while positive for MRD (n=8) or negative for MRD (n=8) (Online Supplementary Table S2).
Intriguingly, gene expression PCA revealed that MSC from MRD– patients emerged as an independent group while MRD+ MSC remained similar to their corresponding cells at the time of diagnosis since most of the transcriptional variability between samples (PC1, 35.72%) supported the gap between MRD– and the rest of the MM stages (Figure 2A).
In fact, differential expression analysis showed that only three genes (DUSP2, MZB1 and TSPAN7) were significantly altered in MSC isolated from MRD+ patients (Figure 2B) as compared to their diagnostic counterparts. By contrast, 56 genes were significantly deregulated in MRD– MSC compared to the time of diagnosis (adjusted P<0.1; |log2FC|>1) (Figure 2 C,D). These results suggest that even residual numbers of tumor cells in MRD+ patients are capable of maintaining a MM-MSC transcriptional signature. This observation becomes relevant when considering the use of immunotherapies to eradicate persistent MRD, which could have their efficacy diminished by an immune suppressive microenvironment maintained, at least in part, by MSC.
In summary, we present striking new data on the impact of aging on HD-MSC and how tumor cells from patients with active MM may block MSC differentiation and promote their expansion to support a protective microenvironment. Moreover, these data indicate that MSC would have the capacity to modulate the effect of conventional therapy and immunotherapy, which should be investigated in both pre-clinical and clinical studies.
Funding Statement
Funding: this study was supported by Instituto de Salud Carlos III (Miguel Servet tipo I: CPII19/00028), Centro de Investigación Biomédica en Red - Área de Oncología - del Instituto de Salud Carlos III (CIBERONC; CB16/12/00369 and CB16/12/00489), formerly named Cooperative Research Thematic Network (grant n. RD12/0036/0058 and RD12/0036/0068) of the Red de Cancer (Cancer Network of Excellence); Asociación Española Contra el Cáncer (GCB120981SAN, AIO16163636SAEZ); the International Myeloma Foundation (IMF) junior award (to PM) and Cancer Research UK, FCAECC and AIRC under the Accelerator Award Program
References
- 1.Meads MB, Gatenby RA, Dalton WS. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat Rev Cancer. 2009;9(9):665-674. [DOI] [PubMed] [Google Scholar]
- 2.Podar K, Chauhan D, Anderson KC. Bone marrow microenvironment and the identification of new targets for myeloma therapy. Leukemia. 2009;23(1):10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reagan MR, Ghobrial IM. Multiple myeloma mesenchymal stem cells: characterization, origin, and tumor-promoting effects. Clin Cancer Res. 2012;18(2):342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119-150. [DOI] [PubMed] [Google Scholar]
- 5.Arnulf B, Lecourt S, Soulier J, et al. Phenotypic and functional characterization of bone marrow mesenchymal stem cells derived from patients with multiple myeloma. Leukemia. 2007;21(1):158-163. [DOI] [PubMed] [Google Scholar]
- 6.Corre J, Mahtouk K, Attal M, et al. Bone marrow mesenchymal stem cells are abnormal in multiple myeloma. Leukemia. 2007;21(5):1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernando RC, Mazzotti DR, Azevedo H, et al. Transcriptome analysis of mesenchymal stem cells from multiple myeloma patients reveals downregulation of genes involved in cell cycle progression, immune response, and bone metabolism. Sci Rep. 2019;9(1):1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garayoa M, Garcia JL, Santamaria C, et al. Mesenchymal stem cells from multiple myeloma patients display distinct genomic profile as compared with those from normal donors. Leukemia. 2009; 23(8):1515-1527. [DOI] [PubMed] [Google Scholar]
- 9.Garderet L, Mazurier C, Chapel A, et al. Mesenchymal stem cell abnormalities in patients with multiple myeloma. Leuk Lymphoma. 2007;48(10):2032-2041. [DOI] [PubMed] [Google Scholar]
- 10.Reagan MR, Mishima Y, Glavey SV, et al. Investigating osteogenic differentiation in multiple myeloma using a novel 3D bone marrow niche model. Blood. 2014;124(22):3250-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schinke C, Qu P, Mehdi SJ, et al. The pattern of mesenchymal stem cell expression is an independent marker of outcome in multiple myeloma. Clin Cancer Res. 2018;24(12):2913-2919. [DOI] [PubMed] [Google Scholar]
- 12.Wallace SR, Oken MM, Lunetta KL, Panoskaltsis-Mortari A, Masellis AM. Abnormalities of bone marrow mesenchymal cells in multiple myeloma patients. Cancer. 2001;91(7):1219-1230. [DOI] [PubMed] [Google Scholar]
- 13.Giuliani N, Mangoni M, Rizzoli V. Osteogenic differentiation of mesenchymal stem cells in multiple myeloma: identification of potential therapeutic targets. Exp Hematol. 2009;37(8):879-886. [DOI] [PubMed] [Google Scholar]
- 14.Jaitin DA, Kenigsberg E, Keren-Shaul H, et al. Massively parallel single- cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343(6172):776-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou G, Zheng Q, Engin F, et al. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc Natl Acad Sci U S A. 2006; 103(50):19004-19009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen MH. The targeting of immunosuppressive mechanisms in hematological malignancies. Leukemia. 2014;28(9):1784-1792. [DOI] [PubMed] [Google Scholar]
- 17.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329-360. [DOI] [PubMed] [Google Scholar]
- 18.Kawano Y, Roccaro AM, Ghobrial IM, Azzi J. Multiple myeloma and the immune microenvironment. Curr Cancer Drug Targets. 2017; 17(9):806-818. [DOI] [PubMed] [Google Scholar]
- 19.Kerkar SP, Restifo NP. Cellular constituents of immune escape within the tumor microenvironment. Cancer Res. 2012;72(13):3125-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caers J, Deleu S, Belaid Z, et al. Neighboring adipocytes participate in the bone marrow microenvironment of multiple myeloma cells. Leukemia. 2007;21(7):1580-1584. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann JN, Birmann BM, Teras LR, et al. Low levels of circulating adiponectin are associated with multiple myeloma risk in overweight and obese individuals. Cancer Res. 2016;76(7):1935-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonald MM, Fairfield H, Falank C, Reagan MR. Adipose, bone, and myeloma: contributions from the microenvironment. Calcif Tissue Int. 2017;100(5):433-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medina EA, Oberheu K, Polusani SR, Ortega V, Velagaleti GV, Oyajobi BO. PKA/AMPK signaling in relation to adiponectin's antiproliferative effect on multiple myeloma cells. Leukemia. 2014; 28(10):2080-2089. [DOI] [PubMed] [Google Scholar]