Abstract
GFI1B-related thrombocytopenia (GFI1B-RT) is a rare bleeding disorder mainly caused by the presence of truncated GFI1B proteins with dominant-negative properties. The disease is characterized by low platelet counts, the presence of abnormal platelets, a megakaryocytic expansion, and mild erythroid defects. However, there are no animal models that faithfully reproduce the GFI1B-RT phenotype observed in patients. We had previously generated mice with floxed Gfi1b alleles that can be eliminated by Cre recombinase, but those animals developed a much more severe phenotype than GFI1B-RT patients and were of limited interest in assessing the disease. Using CRISPR/Cas9 technology, we have now established three independent mouse lines that carry mutated Gfi1b alleles producing proteins lacking DNA binding zinc fingers and thereby acting in a dominant negative (DN) manner. Mice heterozygous for these Gfi1b-DN alleles show reduced platelet counts and an expansion of megakaryocytes similar to features of human GFI1B-RT but lacking the distinctively large agranular platelets. In addition, Gfi1b-DN mice exhibit an expansion of erythroid precursors indicative of a mildly abnormal erythropoiesis but without noticeable red blood cell defects. When associated with megakaryocyte-specific ablation of the remaining allele, the Gfi1b-DN alleles triggered erythroid-specific deleterious defects. Gfi1b-DN mice also showed a delayed recovery from platelet depletion, indicating a defect in stress thrombopoiesis. However, injecting Gfi1b-DN mice with romiplostim, a thrombopoietin receptor super agonist, increased platelet numbers even beyond normal levels. Thus, our data support a causal link between DN mutations in GFI1B and thrombocytopenia, and suggest that patients with GFI1B-RT could be treated successfully with thrombopoietin agonists.
Introduction
GFI1B-related thrombocytopenia (GFI1B-RT), also known as bleeding disorder, platelet-type, 17 (BDPLT17) as listed in the Online Mendelian Inheritance in Man (OMIM) database (http://www.omim.org/entry/187900), is a rare dominant congenital platelet disorder caused by mutations in the GFI1B gene, which encodes two protein isoforms of 32 and 37 kDa. These proteins carry an N-terminal SNAG domain that mediates transcriptional repression, an intermediate domain and six C-terminal C2H2-type zinc finger domains.1 In GFI1B-RT, mutations in GFI1B lead to either the production of truncated proteins lacking the last two zinc fingers or to the disruption of the first zinc finger alone, and cause mild to moderate bleeding diathesis, macrothrombocytopenia, an α-granule deficiency in platelets for most but not all cases, and is also often associated with anisocytosis and poikilocytosis. 2-13 GFI1B-RT was first associated with the gray platelet syndrome (GPS), or BDPLT4, caused by mutation in the NBEAL2 gene.14 Although these two forms of thrombocytopenia share a variety of features such as enlarged platelets with reduced α-granule content and abnormal megakaryocytes (MK),6,14,15 they also exhibit significant differences, such as a higher variability in α- granule deficiency in GFI1B-RT, differences in platelet aggregation function, autosomal recessive (NBEAL2) versus autosomal dominant (GFI1B) transmission and GFI1Bspecific red blood cell (RBC) defects.6,7,16,17
Unlike NBEAL2-related GPS, for which mouse models have been described,18-21 no experimental models have been created yet for GFI1B-RT that would faithfully recapitulate the disease phenotype. The rarity of cases makes it difficult to study the cellular mechanisms underlying this thrombocytopenia, and although several Gfi1b-knockout mouse models exist, none of them faithfully reproduce the human disease.22-25 For example, full ablation of Gfi1b is embryonic lethal,22,23 and the use of either inducible Cre or MK-specific Cre to generate homozygous deletions of Gfi1b cause extreme phenotypes such as total abrogation of thrombopoiesis and erythropoiesis that can hardly be correlated with the disease seen in humans.24-26
Here we report results obtained with three Gfi1b mutant mouse lines generated by CRISPR/Cas9 genome editing to alter the region encoding the fifth zinc finger of GFI1B, which mimic similar mutations found in the human GFI1B gene. As in GFI1B-RT patients, the three mutants act in a dominant negative (DN) manner and the mice exhibit a mild to moderate thrombocytopenia with increased sensitivity to thrombopoietic stresses. Importantly, we show that treatment with the clinical drug romiplostim, a thrombopoietin (TPO) analog, could rescue the phenotype and allow the thrombocytopenic mice to significantly increase their platelet count, indicating potential therapeutic avenues for GFI1B-RT patients.
Methods
Mice
Protocols used in this paper were reviewed by the Animal Care Committee (ACC, #2013-04) of the Clinical Research Institute of Montréal and all animals were cared for in compliance with the Canadian Council on Animal Care guidelines (www.ccac.ca).
Hematologic parameters
Circulating platelet counts were measured either on an Advia 120 cell analyzer (Bayer) using the mouse archetype of multispecies software version 2.2.06 or by quantitative flow cytometry using PE-conjugated anti-CD41 (Biolegend), AF647-conjugated anti-CD61 (BioLegend) and 123count eBeads™ counting beads (Invitrogen) as reference. Hematocrit was measured either on the Advia 120 cell analyzer or by centrifugation of microcapillary on a Haematokrit 200 (Hettich).
Other experimental procedures are described in the Online Supplementary Methods.
Results
Introduction of Gfi1b germline mutations similar to those found in GFI1B-RT patients into mice by CRISPR/Cas9
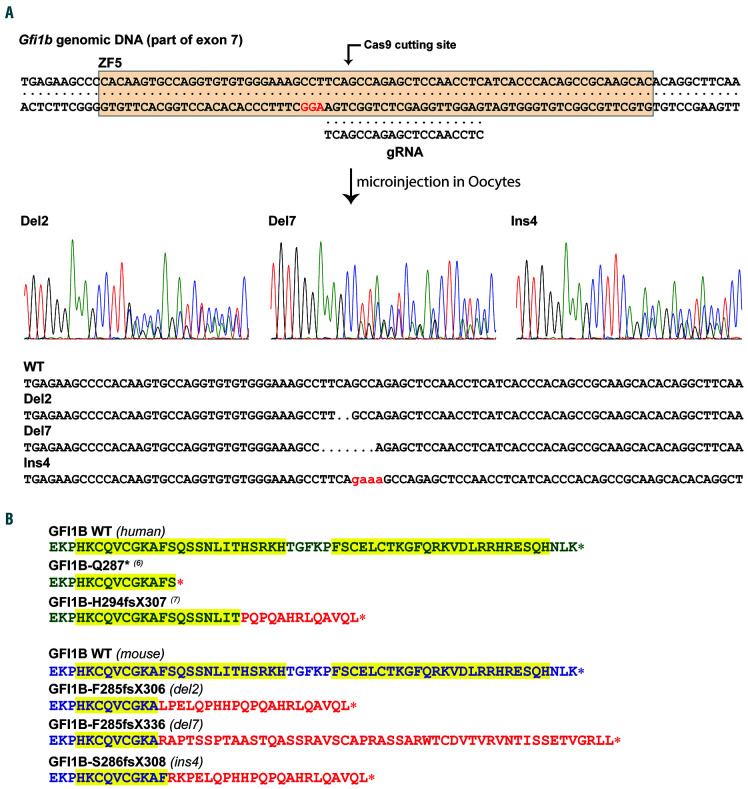
A vector containing both the Cas9 gene and a gRNA targeting the seventh exon of Gfi1b at the position encoding the fifth zinc finger of GFI1B was injected into fertilized murine oocytes. Of 78 pups produced, six carried indel mutations at the target site for a rate of approximately 8%, all of them occurring at the exact Cas9 cutting site. Five founders showed at least some levels of mosaicism, with three founders carrying more than two alleles, indicating that the expression vector remained active for a few cell divisions and that mutations occurred fairly late (Online Supplementary Table S1). Although a total of ten indel alleles were produced, some mutations were identical between several founders. Because of the high level of mosaicism, only two founders were able to transmit the mutant alleles to their progeny, which in turn resulted in three Gfi1b mutant lines carrying alleles modified by deletion of two base pairs (Gfi1bdel2), deletion of seven base pairs (Gfi1bde7), and insertion of four base pairs (Gfi1bins4) (Figure 1A).
As they all occurred at the cutting site of the Cas9 in the region encoding for the fifth zinc finger of Gfi1b, all mutations produced frameshifts that disrupt the fifth and the sixth zinc fingers. The Gfi1bdel2 and Gfi1bins4 mutations switch to frame 2 and generate truncated proteins similar to the proteins identified in some GFI1B-RT patients,7 whereas the Gfi1bdel7 mutation switches to frame 3 and produces an elongated protein with a different C-terminus (Figure 1B and Online Supplementary Figure S1).
The three Gfi1b mutations act in a dominant negative manner
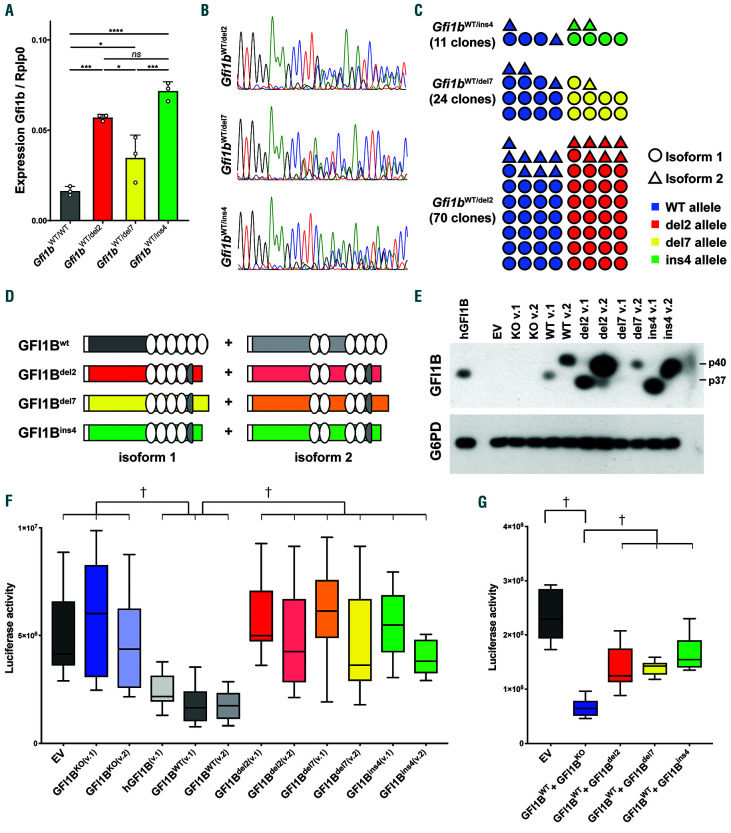
Expression of Gfi1b was first assessed in lineage-depleted bone marrow from Gfi1b mutant heterozygous mice and compared to that from wild-type (WT) animals (Figure 2A). All three Gfi1b mutant lines showed an increased Gfi1b expression with Gfi1bdel2 and Gfi1bins4 being slightly higher than Gfi1bdel7, which is consistent with a loss of GFI1B self-repression.27 Sequencing the cDNA from heterozygous Gfi1bWT/del2, Gfi1bWT/del7, Gfi1bWT/ins4 mice showed mixed chromatograms, demonstrating that the mutant alleles were expressed along with the WT allele (Figure 2B). Moreover, cloning the cDNAs in vectors that were then sequenced, allowed comparison of the ratio of mutant versus WT transcripts of the two isoforms 1 and 2, demonstrating that the mutant mRNAs are present at comparable levels to the transcripts generated by the WT allele, and this, for both isoforms (Figure 2C).
Since it was not possible to distinguish the mutant from the WT GFI1B proteins by western blot in heterozygous mice, we cloned the cDNAs of both isoforms of the three mutants and the WT alleles as well as cDNA prepared from Gfi1bWT/KO mice in an expression vector and transfected 293 cells to detect the proteins (Figure 2D and E and Online Supplementary Figure S2A). As expected, no proteins were detected in cells transfected with either an empty vector or with vectors containing Gfi1bKO cDNAs, which produce mRNAs that do not encode a protein since they lack an open reading frame. Both murine isoforms 1 (37 kDa identified as p37) and 2 (predicted size of 40 kDa and identified as p40) of GFI1BWT and the two mutants GFI1Bdel2 and GFI1Bins4 were readily detectable, whereas the GFI1Bdel7 protein was consistently detected at much lower levels, suggesting possible protein and/or transcript instabilities of this variant. In all cases, the long isoform was expressed at higher levels than the shorter isoform, suggesting that isoform 2 might be more stable than isoform 1.
To assess the functional consequences of the mutated Gfi1b alleles, HEK-293T cells were co-transfected with a luciferase expressing vector under the control of the human GFI1B promoter, which contains GFI1B binding sites,27 along with expression vectors of either isoforms 1 or 2 of GFI1Bdel2, GFI1Bins4, GFI1Bdel7, GFI1BWT or Gfi1bKO, or the isoform 1 of the human GFI1B. As expected, the human and both WT isoforms of the murine GFI1B were efficient in repressing the activity of the GFI1B promoter (Figure 2F). On the contrary, none of the mutants were able to repress the luciferase activity, leading to reads similar to those observed in the absence of GFI1B protein (Figure 2F).
To assess if these mutant alleles exhibit dominant negative (DN) activity as described in human patients,6 the three Gfi1b mutants were transfected along with the WT Gfi1b in the same luciferase assay and we found that, regardless of the isoform, the presence of a mutant allele significantly impaired the capacity of the WT protein to repress the human GFI1B promoter (Figure 2G and Online Supplementary Figure S2B). This indicated that all GFI1B mutants, including GFI1Bdel7, which seemed less stable in vitro than the other two mutants, act in a DN manner independently of the isoform.
Figure 1.
Generation of mice carrying mutant Gfi1b. (A) Mice carrying a mutated allele resembling that of the human mutations causing GFI1B-RT were generated by CRISPR/Cas9 using a gRNA targeting the fifth zinc finger of GFI1B. Three lines capable the mutated allele were produced: one carrying a deletion of two nucleotides (del2), a second one carrying a deletion of seven nucleotides (del7), and one harboring an insertion of four nucleotides (ins4). (B) Resulting C-termini of the proteins produced by two mutants found in human GFI1B-RT patients (green, top) compared to mouse WT gene and the three mutants carried by the Gfi1b mutant mice (blue, bottom). The fifth and sixth zinc fingers are highlighted in yellow and the extraneous peptide sequences generated by the frameshifts are identified in red.
Figure 2.
The three GFI1B mutants are non-functional and as dominant negative. (A) Gfi1b expression analyzed by real-time polymerase chain reaction (RT-PCR) from Gfi1b-mutant lineage-depleted bone marrow. Results are presented as mean±standard deviation of Gfi1b over the housekeeping gene Rplp0. Statistical significance was calculated using a one-way ANOVA (P<0.0001) followed by a Tukey’s multiple comparisons test comparing the mean of each column with the mean of every other column. *Adjusted P<0.05; ***P<0.001; ****P<0.0001. (B) The Gfi1b cDNA derived from erythroblasts of mutant mice was PCR-amplified and sequenced. (C) PCR-amplified cDNAs from (B) were cloned into an expression vector and each clone obtained was sequenced to identify the isoform and the allele. (D) Schematic representation of the two isoforms of GFI1B produced in the mutant mice and cloned in (C). The white rectangle represents the SNAG domain and the white ovals the functional zinc fingers. (E) Detection by western blot of both isoform 1 (v.1; GFI1B-p37) and isoform 2 (v.2; GFI1B-p40) of GFI1B from 293 cells transfected with either the mutants or the wild-type (WT) Gfi1b cDNAs. (F) Box-and-whisker plot of a GFI1B-dependent luciferase assay. Results are presented as luciferase activity (RLU) of six different transfections. EV: empty vector; knockout (KO) is a cDNA cloned from knockout animal and lacking exon 2 to 4; v.1: isoform 1; v.2: isoform 2. (G) Box-and-whisker plot of a GFI1B-dependent luciferase assay measuring the capacity of the GFI1B mutants to inhibit repression by the WT GFI1B. Both isoforms of the three GFI1B mutants were co-transfected along the WT forms and the results are presented as the median of twelve separate experiments. Individual results for the isoforms are presented in Online Supplementary Figure S2B. (F and G) Statistical significance was calculated using a Kruskal-Wallis followed by a Dunn's multiple comparisons comparing the median of each column with the median of all other columns and the full results are presented in Online Supplementary Tables S2 and S3. †Indicates statistical significance.
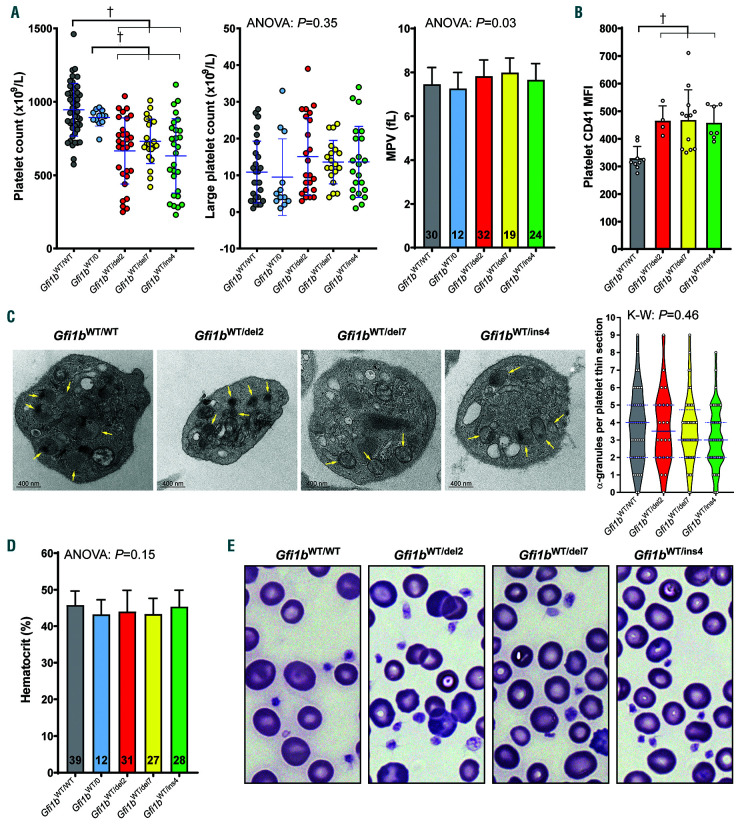
Figure 3.
Moderate thrombocytopenia in Gfi1b-DN mice. (A) Platelet counts, large platelet counts and mean platelet volume (MPV) of Gfi1b-DN and wild-type (WT) mice along hemizygous Gfi1bWT/0 mice. In the scatter dot plots, means±standard deviation are indicated. In the histogram, the numbers given indicate sample size. Statistical significance was calculated using a Brown-Forsythe ANOVA and when significance was found, was followed by a Dunnett's T3 multiple comparisons test comparing the mean of each column with the mean of every other column. (B) Platelet CD41 Mean Fluorescence Intensity (MFI) measured by flow cytometry. Statistical significance was calculated using a one-way ANOVA (P=0.0023) followed by a Holm-Sidak's multiple comparisons test comparing the mean of each column with the mean of every other column. (C) Transmission electron microscopy of thin sections of platelets and violin plot of the quantification of α-granules per platelet thin section. Median and upper/lower quartiles are indicated in blue. In the micrographs, arrows indicate α-granules. Significance was calculated using a Kruskal- Wallis test. (D) Hematocrit of Gfi1b-DN and WT mice along hemizygous Gfi1bWT/0 mice. (E) May-Grünwald Giemsa staining of blood smears showing normal platelets and erythrocytes in both controls and Gfi1b-DN mice. The full results of the Dunnett's T3 (A) and the Holm-Sidak's (B) tests are presented in Online Supplementary Tables S4 and S5, respectively. †Indicates statistical significance.
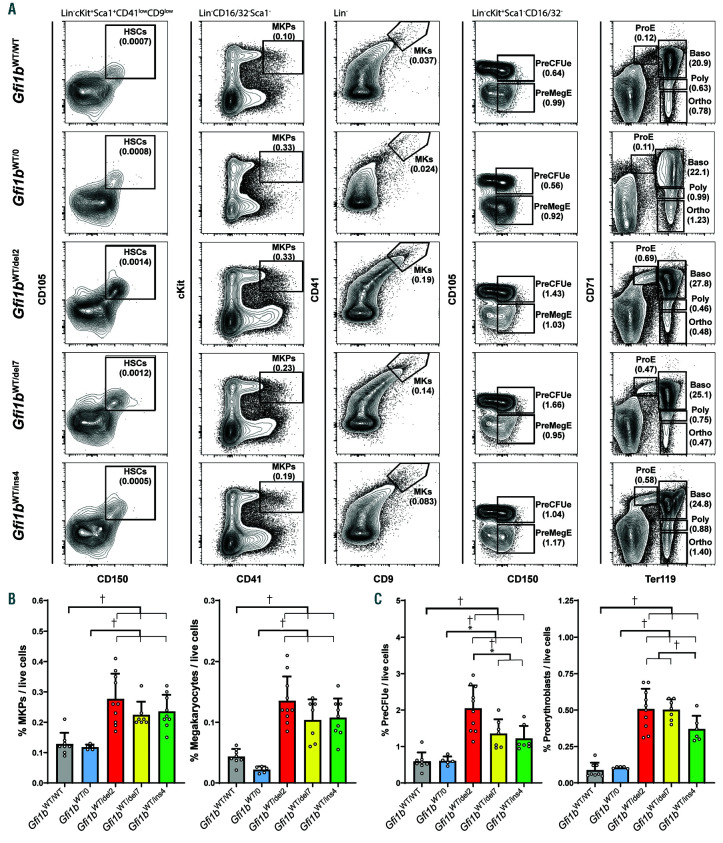
Figure 4.
Megakaryopoiesis and erythropoiesis in Gfi1b-DN mice. (A) Representative FACS analysis of the bone marrow of wild-type (WT) (Gfi1bWT/WT), hemizygotes (Gfi1bWT/0) and Gfi1b-DN (Gfi1bWT/del2, Gfi1bWT/del7 and Gfi1bWT/ins4) mice at steady state. A summary of the gating strategy for all cells is presented in Online Supplementary Figure S3. (B) Quantification of bone marrow megakaryocyte progenitors (MKP) and megakaryocytes (MK). (C) Quantification of bone marrow preerythroid colony forming unit (PreCFUe) and proerythroblasts. All results are reported as mean±standard deviation and significance was calculated by one-way ANOVA tests (P<0.0001 for all graphs) followed by Holm-Sidak’s multiple comparisons tests comparing the mean of each column with the mean of all other columns individually. Full results of the Holm-Sidak’s tests are presented in the Online Supplementary Tables S6 and S7. †Indicates statistical significance.
Figure 5.
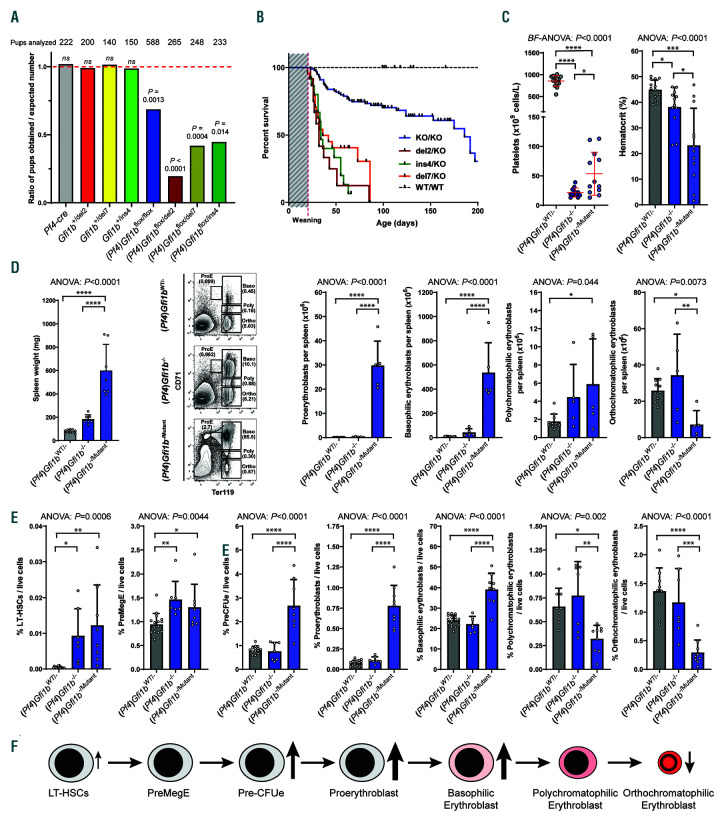
Characterization of Gfi1b-DN mice in the presence of Gfi1b knockout alleles. (A) Ratio of the mice obtained carrying the proper genotype on the expected number (1 representing a perfect Mendelian transmission). The numbers on top indicate the number of pups that survived at least until weaning. Statistical significance between the number obtained compared to the number expected was calculated using a Fisher's exact test. (B) Kaplan-Meier plot of survival of mice with different DN alleles along a megakaryocyte (MK)-specific knockout allele and compared to wild-type (WT) and Pf4-cre:Gfi1bflox/flox mice. Median survival: Pf4-cre:Gfi1bflox/flox (KO/KO) = 186 days; Pf4-cre:Gfi1bflox/del2 (del2/KO) = 32 days; Pf4-cre:Gfi1bflox/del7 (del7/KO) = 36 days; Pf4-cre:Gfi1bflox/Ins4 (ins4/KO) = 34 days. Log-rank Mantel-Cox tests show no significant differences between the three Gfi1bflox/mutant lines (P=0.22) and a significant difference (P<0.0001) between any Gfi1bflox/mutant lines and either Gfi1bWT/WT or Pf4-cre:Gfi1bflox/flox mice. (C) Platelet counts and hematocrits measured from Pf4-cre:Gfi1bflox/mutant (grouping all Pf4-cre:Gfi1bflox/del2, Pf4-cre:Gfi1bflox/del7 and Pf4-cre:Gfi1bflox/Ins4 mice analyzed) to Pf4-cre:Gfi1bflox/flox and Pf4-cre:Gfi1bflox/wt. (D) Spleen weight and quantification by flow cytometry of erythroid precursors from 6- week old female mice with genotypes as in (C). (E) Quantification by flow cytometry of bone marrow of erythroid precursors and progenitors from the same mice as in (C). (C and D) All results are reported as mean±standard deviation and significance was calculated by either a Brown-Forsythe ANOVA (BF-ANOVA) test or a oneway ANOVA (ANOVA) test followed by Dunnett's T3 or Holm-Sidak’s multiple comparisons tests respectively to compare each column with all other columns individually. The statistical significance measured by the post tests are as follow: *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.
Figure 6.
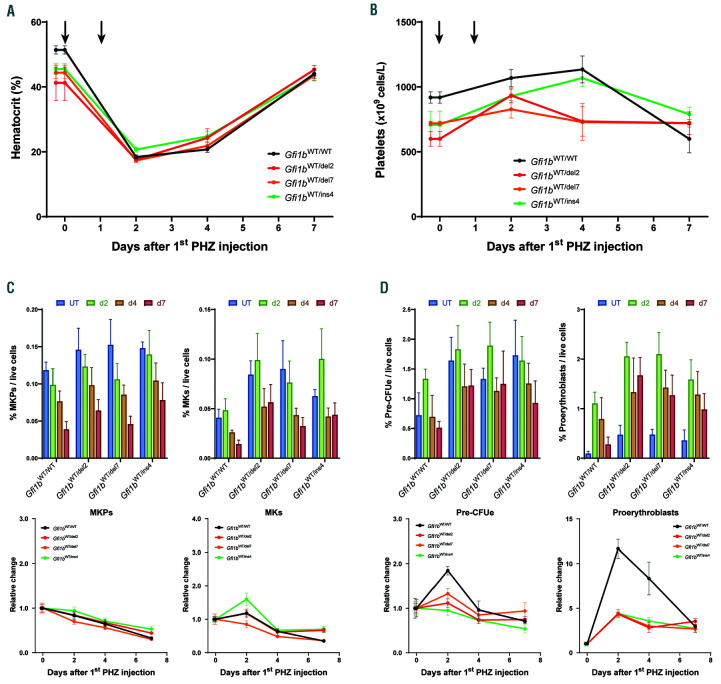
Gfi1b-DN mice can respond adequately to an acute erythropoietic stress. Time course analysis of the response to Phenylhydrazine (PHZ)-induced hemolytic anemia. 4-5 mice for each time point were injected twice with PHZ and sacrificed 2, 4 or 7 days after the first injection for analysis. (A) Measurement of the hematocrit given as the percentage of red blood cell (RBC) volume/total blood volume (with the time of injections indicated by the arrows). (B) Circulating platelet levels. (C and D) Quantification by flow cytometry of bone marrow megakaryocyte progenitors (MKP) and megakaryocytes (MK) (C) or pre-erythroid colony forming unit (PreCFUe) and proerythroblasts (D) and reported as mean±standard deviation of % of cells per live cells (top) or as change relative to day 0 (bottom).
Gfi1b-DN mice exhibit a moderate thrombocytopenic phenotype
To test whether our Gfi1b-DN mice recapitulate the phenotype observed in GFI1B-RT patients, we measured their hematologic parameters and observed that all three lines exhibited lower levels of circulating platelets than WT littermates or hemizygous Gfi1bWT/0 mice that have been produced by crossing Gfi1bflox mice with a Credeleter EIIa-Cre, but only a fraction were thrombocytopenic, i.e., had >50% platelet reduction (Figure 3A). However, unlike GFI1B-RT patients, none of the mice showed signs of spontaneous bleeding; the Gfi1b-DN mice did not show a global increase of large circulating platelets and there was no significant difference in mean platelet volume (MPV) compared to the controls (Figure 3A). Interestingly, Gfi1b-DN mice exhibit higher levels of integrin IIb (CD41) at the surface of platelets than WT mice (Figure 3B), but the number of α-granules in platelets was not significantly decreased as in GFI1B-RT patients (Figure 3C). Finally, none of the Gfi1b-DN mice showed overt erythropoietic defects as revealed by normal hematocrit and RBC shape and size (Figure 3D and E).
Figure 7.
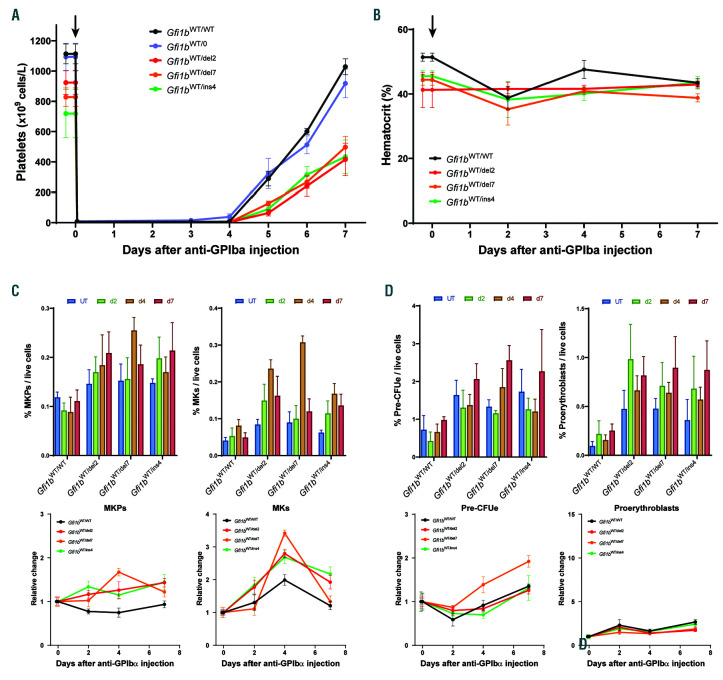
Delay in thrombopoietic stress response in Gfi1b-DN mice. Time course analysis of the response to platelet depletion by intravenous (i.v.) injection of anti- GPIba antibodies. 4-5 mice for each time point received one injection of anti-GPIbα and were either bled daily to measure absolute platelet counts by quantitative flow cytometry on a first experiment, or were sacrificed 2, 4 or 7 days later on a second experiment for analysis of the bone marrow and hematologic parameters. (A) Circulating platelet levels measured daily after the injection (arrows). (B) Measurement of the hematocrit of the mice from the second experiment and given as the percentage of red blood cell (RBC) volume/total blood volume. (C and D) Quantification by flow cytometry of bone marrow megakaryocyte progenitors (MKP) and megakaryocytes (MK) (C) or pre-erythroid colony forming unit (PreCFUe) and proerythroblasts (D) and reported as mean±standard deviation of % of cells per live cells (top) or as change relative to day 0 (bottom).
However, consistent with the phenotype observed in human patients, the three Gfi1b-DN lines presented a moderate megakaryocytic hyperplasia characterized by an increase in both MK and megakaryocyte progenitors (MKP) compared to WT and hemizygous mice (Figure 4A and B). Although no RBC defects were observed in peripheral blood, an analysis of the bone marrow erythroid lineage revealed a strong increase in the pre-erythroid colony-forming unit (PreCFUe) and proerythroblast populations (Figure 4A and C) but no major difference in the earlier pre-megakaryocytic/erythroid progenitor (PreMegE) and hematopoietic stem cell (HSC) populations or in the more differentiated erythroblast subpopulations (Online Supplementary Figure S4). Interestingly, the Gfi1b-DN mice exhibited a mild spleen enlargement compared to the WT and hemizygotes controls (Online Supplementary Figure S5A). However, there was no significant difference in splenic erythroid precursor subpopulations in Gfi1b-DN mice compared to controls (Online Supplementary Figure S5B).
Embryonic lethality and decreased life expectancy of Gfi1b-dominant negative mice in the context of a megakaryocyte-specific Gfi1b knockout
To test whether the Gfi1b-DN mutant proteins retained some residual functions in vivo, we first crossed Gfi1b-DN mice with Gfi1bEGFP animals in which the EGFP gene had been knocked into the Gfi1b locus to replace the coding sequence.23 Gfi1bEGFP/DN mice died in utero around day e14.5 and presented a phenotype similar to that of the full Gfi1b knockout (Online Supplementary Figure S6A).
Figure 8.
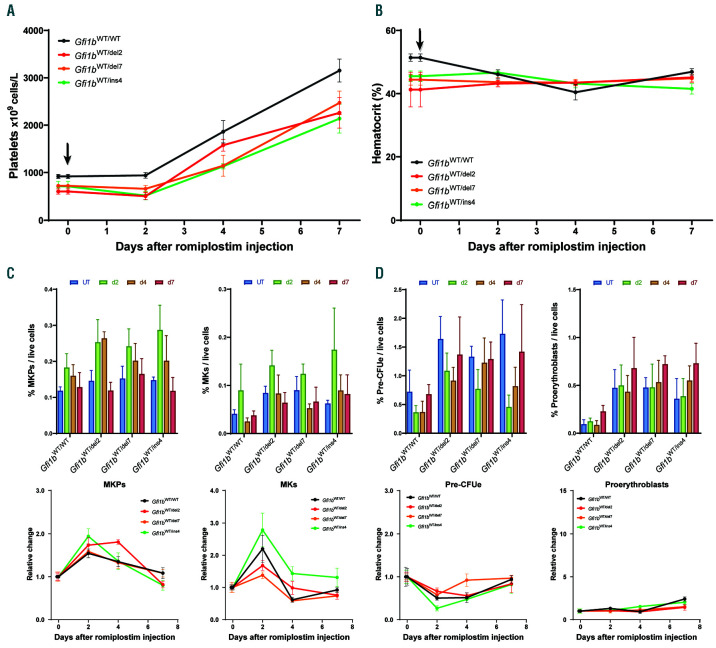
Gfi1b-DN mice respond efficiently to thrombopoietin (TPO) stimulation. Time course analysis of the response to the TPO analog romiplostim-induced thrombocytosis. 4-5 mice for each time point received a single intraperitoneal injection with romiplostim and sacrificed 2, 4 or 7 days later for analysis. (A) Circulating platelet levels (with the time of the injections indicated by the arrows). (B) Measurement of hematocrit given as the percentage of red blood cell (RBC) volume/total blood volume. (C and D) Quantification by flow cytometry of bone marrow megakaryocyte progenitors (MKP) and megakaryocytes (MK) (C) or pre-erythroid colony forming unit (PreCFUe) and proerythroblasts (D) and reported as mean±standard deviation of % of cells per live cells (top) or as change relative to day 0 (bottom).
To measure the effect of the Gfi1b-DN mutants on platelet production in the context of an MK-specific knockout of the remaining Gfi1b allele, we bred Gfi1b- DN mice with Pf4-cre:Gfi1bflox mice that lack GFI1B in MK and exhibit a severe thrombocytopenia (Online Supplementary Figure S6B). Mice with the desired genotype (Pf4-cre:Gfi1bflox/DN) were produced well below the mendelian rate, suggesting either in utero or perinatal lethality (Figure 5A and Online Supplementary Figure S6C). Moreover, the few mice that were born died even faster than Pf4-cre:Gfi1bflox/flox mice, suggesting a deleterious effect mediated by the presence of a Gfi1b-DN allele in this context (Figure 5B).
The few Pf4-cre:Gfi1bflox/DN mice that developed to term, suffered from a severe form of thrombocytopenia similar to that of the Pf4-cre:Gfi1bflox/flox mice although some mice exhibited slightly higher platelet counts than the knockout (Figure 5C). These mice also showed an increase in frequency of bone marrow MKP and MK compared to WT controls and similar to the knockout (Online Supplementary Figure S6D), indicating that, in the megakaryocytic linage, Gfi1b-DN mutants are unable to rescue the knockout phenotype. Interestingly, several Pf4- cre:Gfi1bflox/DN mice had a severely reduced hematocrit suggesting anemia as the probable cause of death (Figure 5C). Indeed, the spleen of the Pf4-cre:Gfi1bflox/DN mice were severely enlarged, and this splenomegaly was associated with a dramatic increase of proerythroblasts and basophilic erythroblasts but with a decrease of splenic orthochromatophilic erythroblasts (Figure 5D and Online Supplementary Figure S6E). An analysis of the bone marrow revealed a similar erythroid pattern with an increase in PreCFUe, proerythroblasts and basophilic erythroblast along with a decrease in orthochromatophilic erythroblasts (Figure 5E). This differentiation block at the basophilic stage suggests an ineffective erythropoiesis (Figure 6F) in the context of a perpetual erythropoietic stress due to recurring spontaneous bleeding.
Gfi1b-dominant negative mice can respond properly to an acute erythropoietic stress
To further explore the possibility that Gfi1b-DN alleles cause defects during erythropoietic stress, we treated WT and heterozygous Gfi1b-DN mice with phenylhydrazine (PHZ) that rapidly destroys circulating RBC. In a first experiment, mice were injected with PHZ, then bled over a period of 3 weeks every 2-4 days for hematocrit measurement to test how fast the RBC pool can be replenished. In this setting, the hematocrit recovered at the same rate in all mice to a full recovery by day 7 (Online Supplementary Figure S7A).
In a second experimental setting, mice were grouped in cohorts that were sacrificed at given time points following PHZ injections to permit assessment of their hematologic parameters and bone marrow progenitors (Figure 6). Again, the Gfi1b-DN mice did not show any delay in hematocrit recovery compared to the controls (Figure 6A); similarly, platelets remained largely unaffected in all of the mouse treated with PHZ (Figure 6B). Analysis of both erythroid and megakaryocytic lineages in the bone marrow revealed that the Gfi1b-DN mutant mice were able to achieve a normal response to the PHZ-induced hemolytic anemia with the sole exception being the proerythroblasts (and to a lesser extent the PreCFUe) that showed a significantly stronger response in the WT controls than in the Gfi1b-DN mice (Figure 6C and D and Online Supplementary Figure S7B and C).
Gfi1b-dominant negative mice are less efficient at recovering from severe acute thrombocytopenia than controls
Next, we induced an acute thrombocytopenic state in the three Gfi1b-DN lines by intravenous (i.v.) injection of anti-GPIbα (CD42b) antibodies that lead to a rapid and complete platelet depletion. In a first experiment, mice from the three Gfi1b-DN lines as well as WT and hemizygote controls were injected and minimally bled 1 hour after the injection, then every day for a week, for quantitative platelet counts by FACS. The three Gfi1b-DN lines exhibited a slower platelet recovery compared to controls (Figure 7A). To analyze hematopoietic precursors, a new cohort of mice was injected but this time, the animals were sacrificed at day 2, 4 and 7 after platelet depletion, and blood and bone marrow samples were analyzed. The hematocrit remained normal (Figure 7B), but a stronger megakaryopoietic response was seen in Gfi1b-DN mutant mice, which exhibited a stronger increase in both MKP and MK than WT controls (Figure 7C). The erythroid lineage was not affected by the loss of platelets except for a short burst in the orthochromatophilic erythroblast compartment seen in all mice including the controls (Figure 7D and Online Supplementary Figure S8).
Gfi1b-dominant negative megakaryocytes can respond normally to thrombopoietin stimulation
Because the Gfi1b-DN mice exhibited mildly dysregulated thrombopoiesis, we tested their response to TPO stimulation. Mice were given romiplostim, a fusion peptibody analog to TPO with a greater bioavailability than recombinant TPO28 and were analyzed at day 2, 4 or 7 after injection (Figure 8 and Online Supplementary Figure S9). Although romiplostim had no impact on the erythroid lineage except for a short burst in the orthochromatophilic erythroblast population, all mice (controls and Gfi1b-DN) showed a strong thrombopoiesis response as reflected by their high platelet counts associated with an increase in MKP and MK. Although the rate of platelet increase was similar between control and Gfi1b-DN mice, the platelet level remained consistently lower in Gfi1b-DN mice than WT controls (Figure 8A), indicating that MK in Gfi1b-DN mice are less efficient to produce platelets, but are still able to respond to thrombopoietic stress and TPO stimulation. In contrast to Gfi1b- DN mice, Pf4-cre:Gfi1bflox/flox animals treated with romiplostim did not respond at all even after 7 days (Online Supplementary Figure S10).
Discussion
Several autosomal dominant mutations have been identified in the gene GFI1B that are linked to the bleeding disorder type 17 (BDPLT17), three of which cause the production of a truncated protein lacking the two last zinc fingers, one causes the disruption of the last three zinc fingers and the others being missense mutations disrupting non-DNAbinding zinc fingers.6-11,29 Because of the variety of mutations that produce truncated proteins with DN properties, we decided to use CRISPR/Cas9 technology to generate indel mutations in the fifth zinc finger to mimic those found in humans. Injecting the CRISPR vectors directly into oocytes proved to be fairly efficient, although most of the founders produced showed some level of mosaicism that made transmission to F1 progeny more difficult. Interestingly, although ten mutant alleles were generated, only six different alleles were generated as some mutations were obtained multiple times independently, suggesting that the generation of indels might not be entirely random. This result is consistent with other reports suggesting that Cas9- mediated insertions and/or deletions may be context specific. 30-32
When modeling human mutations, including those that produce DN effects, the phenotypic consequences of those alleles may differ when translated into the mouse.33,34 Here, we demonstrate that forms of GFI1B truncated at the fifth zinc finger had a DN effect in vitro similar to the analog human GFI1B mutations found in patients.6 We also demonstrate that both murine short isoform 1 (GFI1B-p37) and long isoform 2 (GFI1B-p40) are not only able to repress target gene expression with an equivalent efficiency, but that they also can both act as DN alleles in a reporter gene assay when carrying the DN mutations. In humans, splice variants 1 (GFI1B-p37) and 2 (GFI1B-p32) exhibit distinct functions, with the long isoform 1 being important for megakaryocytic differentiation and the short isoform 2 being required for proper erythropoiesis.11,13,35,36 Whether the two murine isoforms have a similar lineage-specific functionality remains to be investigated. As both the major human long isoform 1 and the murine short isoform 1 are identical, they are expected to play similar roles. However, the minor isoforms 2 are quite different between mouse and human, with the human version lacking the two first non-DNA-binding zinc fingers and the mouse protein still carrying these domains separated from the four last zinc fingers by a spacer peptide. It is, therefore, difficult to predict if the isoforms 2 behave similarly in mouse and human even if they are expressed at similarly low levels relative to isoform 1.36,37
The reduction in platelet numbers measured in Gfi1b-DN mice was highly variable but never strong enough to cause overt bleedings. This was expected as it has been shown that mice are resistant to bleeding until platelet numbers are decreased by more than 95%.38 The GFI1B-RT patients show variations in severity ranging from severe lifelong bleeding histories to rare manifestations of bleeding events.4-6 Thus, the discrepancy in severity with the Gfi1b-DN mice might be due to the fundamental difference in normal number of platelets between these two organisms, with mice having normally around three times the amount of platelets per volume of blood than humans.39,40 Indeed, a modest decrease in platelet number in humans may easily bring their count under the critical level of 150x109/L, whereas it would take a more drastic decrease to generate a thrombocytopenic state in mice.
We noted that platelets in Gfi1b-DN mice were not significantly larger than controls, which is at variance with the macrothrombocytopenia seen in patients or in MK-specific Gfi1b knockout mice.6,10,25 Similarly, platelets produced in Gfi1b-DN heterozygous mice have normal amounts of α-granules unlike the Gray platelet syndrome reported in GFI1B-RT patients, indicating another difference between this model and the human disease,6,8,9,13,24,41,42 except in the cases of mutations affecting only non-DNA binding zinc fingers of GFI1B and that do not exhibit α-granule deficiency.10-12 Moreover, platelets from Gfi1b-DN mice exhibit increased expression of the fibrinogen receptor integrin αIIbβ3 (CD41/CD61) which also contrasts with GFI1B-RT patients who have normal CD41 expression, but is consistent with MK-specific Gfi1b knockout animals.6,25 On the other hand, all three Gfi1b-DN lines show a megakaryocytic hyperplasia that translates into an increase in MKP and mature MK, which is an important characteristic for GFI1B-RT patients.6
Some studies have reported that mutations in GFI1B lead to defects in erythroid differentiation, albeit infrequently,7,9 but other studies did not observe such a phenotype.6 We could not detect any anisocytosis or poikilocytosis in Gfi1b- DN mice, but all three lines developed a consistent and important expansion of late erythroid progenitors (preCFUe) and early precursor (proerythroblasts) that cannot be explained by a consequence of the thrombocytopenic state of these mice, since such a correlation was not observed in Pf4-cre:Gfi1bflox/flox mice that have an even stronger thrombocytopenia. It has been shown that the loss of GFI1B in mice leads to a complete block of the definitive erythroid lineage, occurring mainly at an erythroid progenitor stage between the PreMegE and the PreCFUe,22,24,43,44 suggesting that these stages of differentiation are particularly dependent on GFI1B.
One surprising finding was that the combination of a mutant Gfi1b-DN allele with a MK-specific knockout allele led to a higher lethality and a lower transmission rate than a homozygous MK-specific Gfi1b knockout, suggesting that Gfi1b-DN mutant alleles are not only non-functional, but have deleterious effects.25 However, as the Pf4- cre:Gfi1bflox/flox already exhibit an extremely severe thrombocytopenia with spontaneous bleeding,25 a deleterious effect of the Gfi1b-DN mutant alleles on the MK lineage seems unlikely. Rather, the severe splenomegaly associated with an almost complete block at the basophilic erythroblast stage and the severe anemia seen in Pf4-cre:Gfi1bflox/mutant mice is indicative of an ineffective erythropoiesis that could be triggered by excessive spontaneous bleeding caused by the lack of platelets in the MK-specific Gfi1b knockout mice.45,46 Because the Pf4-cre gene is not expressed in erythroid cells, this phenotype, which is not observed in Pf4-cre:Gfi1bflox/flox mice, is therefore very likely a consequence of the action of the GFI1B-DN form in the presence of a functional (non-deleted) floxed allele in erythroid cells. It is thus likely that the deleterious effect of the DN forms in erythroid precursors severely impairs erythropoiesis under chronic stress as for instance in a context of frequent spontaneous hemorrhages.
It was therefore surprising that Gfi1b-DN mice responded just as well as the WT controls to PHZ-induced erythropoietic stress to replenish their RBC pool. This discrepancy might be due to the chronic nature of the erythropoietic stress seen in Pf4-cre:Gfi1bflox/DN mice compared to the acute erythropoietic challenge triggered by the PHZ. However, despite their capacity to replenish their RBC pool, Gfi1b- DN mice were unable to further increase their PreCFUe and proerythroblasts as well as WT animals upon PHZ treatment. This poor response may be explained by the fact that these precursor populations in the heterozygous mutant mice were already at a higher than WT level, suggesting that these mice are already in a “stress response” state, even at steady state.47
Considering that, at steady state, their platelet counts are lower than WT animals, Gfi1b-DN mutant mice were expected to have a poorer thrombopoietic stress response, and indeed, platelet depletion experiments showed that Gfi1b-DN mice had increased response times for platelet recovery despite the fact that the expansion of their MK was even more pronounced than in controls, suggesting an impaired but not entirely dysfunctional platelet production. Treating mice with the TPO analog romiplostim triggered a proper response and did significantly increase platelet counts in these animals, which could be due to the already increased number of MK and MKP that may give the Gfi1b-DN mice a starting advantage. The effect of romiplostim, which is a drug already approved for some patients suffering from chronic idiopathic thrombocytopenia purpura, but was also reported in off-label uses such as treatment of persistent thrombocytopenia associated with stem cell transplantation or congenital amegakaryocytic thrombocytopenia,48-50 confirmed that heterozygous Gfi1b-DN mutant MK are still functional. This finding is important because it suggests that patients suffering from GFI1B-RT could potentially respond to a treatment with thrombopoietic agonists such as romiplostim. Ultimately, the mouse models described here could play a possible future role in drug screening for treatments for GFI1B-RT and other related thrombocytopenias, despite some significant differences compared to the features that are typical for human patients with this disease. A future development of mouse models based on the missense mutations in non-DNA-binding zinc finger seen in some patients could also possibly widen our understanding of the disease.
Supplementary Material
Acknowledgments
The authors would like to thank the animal health technicians of the IRCM animal facility for excellent animal care and skillful technical procedures on animals and Jeannie Mui at the Facility for Electron Microscopy Research of McGill University for help in thin-sections preparation and microscope operation.
Funding Statement
Funding These studies were supported by a Canadian Institutes of Health Research (CIHR) operating grant (MOP-111247), a Canada Research Chair (Tier1) in Hematopoiesis and Immune Cell Differentiation, a grant from the Canadian Hemophilia Society and a CIHR Foundation grant (FDN-148372) to TM; as well as a CIHR Project Grant to GD (PJT-156017).
References
- 1.Tong B, Grimes HL, Yang TY, et al. The Gfi- 1B proto-oncoprotein represses p21WAF1 and inhibits myeloid cell differentiation. Mol Cell Biol. 1998;18(5):2462-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quick AJ, Hussey CV. Hereditary thrombopathic thrombocytopenia. Am J Med Sci. 1963;245:643-653. [PubMed] [Google Scholar]
- 3.Seip MF. Hereditary Hypoplastic Thromobocytopenia. Sangre. 1964;41:382-384. [PubMed] [Google Scholar]
- 4.Kurstjens R, Bolt C, Vossen M, Haanen C. Familial thrombopathic thrombocytopenia. Br J Haematol. 1968;15(3):305-317. [DOI] [PubMed] [Google Scholar]
- 5.Ardlie NG, Coupland WW, Schoefl GI. Hereditary thrombocytopathy: a familial bleeding disorder due to impaired platelet coagulant activity. Aust N Z J Med. 1976; 6(1):37-45. [DOI] [PubMed] [Google Scholar]
- 6.Monteferrario D, Bolar NA, Marneth AE, et al. A dominant-negative GFI1B mutation in the gray platelet syndrome. N Engl J Med. 2014;370(3):245-253. [DOI] [PubMed] [Google Scholar]
- 7.Stevenson WS, Morel-Kopp MC, Chen Q, et al. GFI1B mutation causes a bleeding disorder with abnormal platelet function. J Thromb Haemost. 2013;11(11):2039-2047. [DOI] [PubMed] [Google Scholar]
- 8.Ferreira CR, Chen D, Abraham SM, et al. Combined alpha-delta platelet storage pool deficiency is associated with mutations in GFI1B. Mol Genet Metab. 2017;120(3):288-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kitamura K, Okuno Y, Yoshida K, et al. Functional characterization of a novel GFI1B mutation causing congenital macrothrombocytopenia. J Thromb Haemost. 2016;14(7):1462-1469. [DOI] [PubMed] [Google Scholar]
- 10.Uchiyama Y, Ogawa Y, Kunishima S, et al. A novel GFI1B mutation at the first zinc finger domain causes congenital macrothrombocytopenia. Br J Haematol. 2018;181(6):843-847. [DOI] [PubMed] [Google Scholar]
- 11.Rabbolini DJ, Morel-Kopp MC, Chen Q, et al. Thrombocytopenia and CD34 expression is decoupled from alpha-granule deficiency with mutation of the first growth factor-independent 1B zinc finger. J Thromb Haemost. 2017;15(11):2245-2258. [DOI] [PubMed] [Google Scholar]
- 12.van Oorschot R, Marneth AE, Bergevoet SM, et al. Inherited missense variants that affect GFI1B function do not necessarily cause bleeding diatheses. Haematologica. 2019;104(6):e260-e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulze H, Schlagenhauf A, Manukjan G, et al. Recessive grey platelet-like syndrome with unaffected erythropoiesis in the absence of the splice isoform GFI1B-p37. Haematologica. 2017;102(9):e375-e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahr WH, Hinckley J, Li L, et al. Mutations in NBEAL2, encoding a BEACH protein, cause gray platelet syndrome. Nat Genet. 2011;43(8):738-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larocca LM, Heller PG, Podda G, et al. Megakaryocytic emperipolesis and platelet function abnormalities in five patients with gray platelet syndrome. Platelets. 2015;26(8):751-757. [DOI] [PubMed] [Google Scholar]
- 16.Nurden AT, Nurden P. Should any genetic defect affecting alpha-granules in platelets be classified as gray platelet syndrome? Am J Hematol. 2016;91(7):714-718. [DOI] [PubMed] [Google Scholar]
- 17.Gunay-Aygun M, Falik-Zaccai TC, Vilboux T, et al. NBEAL2 is mutated in gray platelet syndrome and is required for biogenesis of platelet alpha-granules. Nat Genet. 2011; 43(8):732-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomberg K, Khoriaty R, Westrick RJ, et al. Spontaneous 8bp Deletion in Nbeal2 Recapitulates the Gray Platelet Syndrome in Mice. PLoS One. 2016;11(3):e0150852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kahr WH, Lo RW, Li L, et al. Abnormal megakaryocyte development and platelet function in Nbeal2(-/-) mice. Blood. 2013; 122(19):3349-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deppermann C, Cherpokova D, Nurden P, et al. Gray platelet syndrome and defective thrombo-inflammation in Nbeal2-deficient mice. J Clin Invest. 2013;123(8):3331-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerrero JA, Bennett C, van der Weyden L, et al. Gray platelet syndrome: proinflammatory megakaryocytes and alpha-granule loss cause myelofibrosis and confer metastasis resistance in mice. Blood. 2014; 124(24):3624-3635. [DOI] [PubMed] [Google Scholar]
- 22.Saleque S, Cameron S, Orkin SH. The zincfinger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 2002; 16(3):301-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vassen L, Okayama T, Moroy T. Gfi1b:green fluorescent protein knock-in mice reveal a dynamic expression pattern of Gfi1b during hematopoiesis that is largely complementary to Gfi1. Blood. 2007;109(6):2356-2364. [DOI] [PubMed] [Google Scholar]
- 24.Foudi A, Kramer DJ, Qin J, et al. Distinct, strict requirements for Gfi-1b in adult bone marrow red cell and platelet generation. J Exp Med. 2014;211(5):909-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beauchemin H, Shooshtarizadeh P, Vadnais C, Vassen L, Pastore YD, Moroy T. Gfi1b controls integrin signaling-dependent cytoskeleton dynamics and organization in megakaryocytes. Haematologica. 2017;102(3):484-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vassen L, Beauchemin H, Lemsaddek W, Krongold J, Trudel M, Moroy T. Growth factor independence 1b (gfi1b) is important for the maturation of erythroid cells and the regulation of embryonic globin expression. PLoS One. 2014;9(5):e96636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vassen L, Fiolka K, Mahlmann S, Moroy T. Direct transcriptional repression of the genes encoding the zinc-finger proteins Gfi1b and Gfi1 by Gfi1b. Nucleic Acids Res. 2005;33(3):987-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang B, Nichol JL, Sullivan JT. Pharmacodynamics and pharmacokinetics of AMG 531, a novel thrombopoietin receptor ligand. Clin Pharmacol Ther. 2004;76(6):628-638. [DOI] [PubMed] [Google Scholar]
- 29.van Oorschot R, Hansen M, Koornneef JM, et al. Molecular mechanisms of bleeding disorder-associated GFI1BQ287* mutation and its affected pathways in megakaryocytes and platelets. Haematologica. 2019;104(7):1460-1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemos BR, Kaplan AC, Bae JE, et al. CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc Natl Acad Sci U S A. 2018;115(9):E2040-E2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Overbeek M, Capurso D, Carter MM, et al. DNA Repair Profiling Reveals Nonrandom Outcomes at Cas9-Mediated Breaks. Mol Cell. 2016;63(4):633-646. [DOI] [PubMed] [Google Scholar]
- 32.Taheri-Ghahfarokhi A, Taylor BJM, Nitsch R, et al. Decoding non-random mutational signatures at Cas9 targeted sites. Nucleic Acids Res. 2018;46(16):8417-8434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hara S, Takada S. Genome editing for the reproduction and remedy of human diseases in mice. J Hum Genet. 2018;63(2): 107-113. [DOI] [PubMed] [Google Scholar]
- 34.Vandamme TF. Rodent models for human diseases. Eur J Pharmacol. 2015;759:84-89. [DOI] [PubMed] [Google Scholar]
- 35.Polfus LM, Khajuria RK, Schick UM, et al. Whole-Exome Sequencing Identifies Loci Associated with Blood Cell Traits and Reveals a Role for Alternative GFI1B Splice Variants in Human Hematopoiesis. Am J Hum Genet. 2016;99(2):481-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laurent B, Randrianarison-Huetz V, Frisan E, et al. A short Gfi-1B isoform controls erythroid differentiation by recruiting the LSD1-CoREST complex through the dimethylation of its SNAG domain. J Cell Sci. 2012;125(Pt 4):993-1002. [DOI] [PubMed] [Google Scholar]
- 37.Osawa M, Yamaguchi T, Nakamura Y, et al. Erythroid expansion mediated by the Gfi- 1B zinc finger protein: role in normal hematopoiesis. Blood. 2002;100(8):2769-2777. [DOI] [PubMed] [Google Scholar]
- 38.Morowski M, Vogtle T, Kraft P, Kleinschnitz C, Stoll G, Nieswandt B. Only severe thrombocytopenia results in bleeding and defective thrombus formation in mice. Blood. 2013;121(24):4938-4947. [DOI] [PubMed] [Google Scholar]
- 39.Fukuda T, Asou E, Nogi K, Goto K. Evaluation of mouse red blood cell and platelet counting with an automated hematology analyzer. J Vet Med Sci. 2017;79(10):1707-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crowther MA, Cook DJ, Meade MO, et al. Thrombocytopenia in medical-surgical critically ill patients: prevalence, incidence, and risk factors. J Crit Care. 2005;20(4):348-353. [DOI] [PubMed] [Google Scholar]
- 41.Stevenson WS, Morel-Kopp MC, Ward CM. Platelets are not all gray in GFI1B disease. Clin Genet. 2015;87(3):299. [DOI] [PubMed] [Google Scholar]
- 42.Fixter K, Rabbolini DJ, Valecha B, et al. Mean platelet diameter measurements to classify inherited thrombocytopenias. Int J Lab Hematol. 2018;40(2):187-195. [DOI] [PubMed] [Google Scholar]
- 43.Randrianarison-Huetz V, Laurent B, Bardet V, Blobe GC, Huetz F, Dumenil D. Gfi-1B controls human erythroid and megakaryocytic differentiation by regulating TGFbeta signaling at the bipotent erythromegakaryocytic progenitor stage. Blood. 2010;115(14):2784-2795. [DOI] [PubMed] [Google Scholar]
- 44.Garcon L, Lacout C, Svinartchouk F, et al. Gfi-1B plays a critical role in terminal differentiation of normal and transformed erythroid progenitor cells. Blood. 2005;105(4):1448-1455. [DOI] [PubMed] [Google Scholar]
- 45.Sjogren U. Erythroblastic islands and ineffective erythropoiesis in acute myeloid leukaemia. Acta Haematol. 1975;54(1):11-17. [DOI] [PubMed] [Google Scholar]
- 46.Suragani RN, Zachariah RS, Velazquez JG, et al. Heme-regulated eIF2alpha kinase activated Atf4 signaling pathway in oxidative stress and erythropoiesis. Blood. 2012;119(22):5276-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh RP, Grinenko T, Ramasz B, et al. Hematopoietic Stem Cells but Not Multipotent Progenitors Drive Erythropoiesis during Chronic Erythroid Stress in EPO Transgenic Mice. Stem Cell Reports. 2018;10(6):1908-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doobaree IU, Newland A, McDonald V, et al. Primary Immune Thrombocytopenia (ITP) Treated with Romiplostim in Routine Clinical Practice: Retrospective Study from the United Kingdom ITP Registry. Eur J Haematol. 2019;102(5):416-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bento L, Bastida JM, Garcia-Cadenas I, et al. Thrombopoietin Receptor Agonists for Severe Thrombocytopenia after Allogeneic Stem Cell Transplantation: Experience of the Spanish Group of Hematopoietic Stem Cell Transplant. Biol Blood Marrow Transplant. 2019;25(9):1825-1831. [DOI] [PubMed] [Google Scholar]
- 50.Pecci A, Ragab I, Bozzi V, et al. Thrombopoietin mutation in congenital amegakaryocytic thrombocytopenia treatable with romiplostim. EMBO Mol Med. 2018;10(1):63-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.