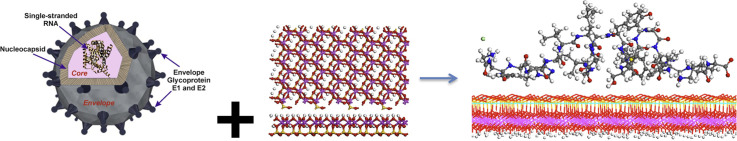
Abstract
This work aimed at studying the potentiality of interactions between kaolinite surfaces and a protein-fragment (350–370 amino acid units) extracted from the glycoprotein E1 in the transmembrane domain (TMD) of hepatitis C virus capsid. A computational work was performed for locating the potential electrostatic interaction sites between kaolinite aluminol and siloxane surfaces and the residues of this protein-fragment ligand, monitoring the possible conformational changes. This hydrated neutralized kaolinite/protein-fragment system was simulated by means of molecular modeling based on atomistic force fields based on empirical interatomic potentials and molecular dynamic (MD) simulations. The MD calculations indicated that the studied protein-fragment interacted with the kaolinite surfaces with an exothermic process and structural distortions were observed, particularly with the hydrophilic aluminol surface by favorable adsorption energy. The viral units isolation or trapping by the adsorption on the kaolinite nanoparticles producing structural distortion of the peptide ligands could lead to the blockage of the entry on the receptor and hence a lack of viral activity would be produced. Therefore, these findings with the proposed insights could be an useful information for the next experimental and development studies in the area of discovering inhibitors of the global challenged hepatitis and other pathogenic viruses based on the phyllosilicate surface activity. These MD studies can be extended to other viruses like the COVID-19 interacting with silicate minerals surfaces.
Keywords: Hepatitis C virus, Kaolinite, Molecular dynamics, Hydration, Adsorption, Virus sorption on minerals, Transmembrane domain, Glycoprotein E1
Graphical abstract
Highlights
-
•
A protein of the Hepatitis C virus is adsorbed favorably on kaolinite surfaces.
-
•
Adsorption on kaolinite provokes changes of Hepatitis C virus.
-
•
Molecular dynamics are useful for exploring virus adsorption on mineral surfaces.
1. Introduction
The human pathogenic viruses have become global health challenges occupying one of the most pressing concerns by the international scientific research community. Hepatitis C virus (HCV) has been estimated at over 185 million infected patients and causes about 1 million deaths per year over the world. The common clinical approaches of the HCV treatments that have been discovered and approved so far include combined dosages of interferon-chemotherapy (pegylated Interferon-α plus ribavirin drug) and herbal compounds (active phytochemical derivatives including polyines, flavonoids, alkaloids, thiophenes and terpenoids) (Foster, 2004; Kamal et al., 2004; Ravikumar et al., 2011). Nowadays, other viruses like COVID-19 coronavirus are generating a global pandemic worldwide with more the 24 million infected people and more than 800.000 of human deaths.
The research gate is accessible always for discovering new strategies and materials to develop antiviral drugs, not only derived from chemical synthesis and biological resources, but also from geological materials (i.e., minerals). Kaolinite (Al2Si2O5(OH)4) is an abundant and inexpensive geomaterial that exhibits hydrated aluminum dioctahedral 1:1 phyllosilicate character widely used in pharmaceutical and biomedicinal applications as excipient and active therapeutic agent (Awad et al., 2017). Previous work of in vitro trial for testing the inhibitory effect of kaolin minerals, as active therapeutic agent against HCV, was attempted, observing a promising antiviral activity with non-cytotoxic effect on the replicon cell line in the use of this mineral for introducing new anti-HCV derivative compounds (Ali et al., 2014). However, the pathway and mechanism of this inhibitory action is still not understood.
In the recent decade, many authors have paid attention to molecular structure and functions of the viral capsid proteins, because most of which are effectively being considered as antiviral targets. Particularly, the transmembrane domains (TMDs) of the structural envelope glycoproteins E1 and E2 play a direct role in heterodimerization that is essential for the formation of the viral envelope, hepatocyte receptor binding, entry and fusion of the virus with the cell. These TMDs consist of two stretches of hydrophobic residues separated by a short segment containing at least one fully conserved charged residue. Hence, it is highly expected that deletion or replacement of these charged residues lead to alter all the functions of the TMDs of E1 and E2 (Selby et al., 1994; Michalak et al., 1997; Beeck et al., 2000; Cocquerel et al., 2000; Voisset and Dubuisson, 2004; Moradpour et al., 2005).
At molecular level, the polypeptides can have charged groups. The negative net charge that appears on the protein molecule arises from carboxylate groups (COO−) of the amino acids (e.g., aspartate ASP and glutamate GLU), while the positive net charge carried by the protein molecule is resulted from the amino groups (NH+ 3) of the basic amino acids (e.g., lysine LYS, arginine ARG and histidine HIS). Accordingly, the positive net charge on the transmembrane domain (TMD) of the HCV glycoprotein E1 molecule is owing to the positive charge basic lysine (LYS) residue (Cocquerel et al., 2000; Ciczora et al., 2005; Bohidar, 2015).
In the layered structure of kaolinite fundamental particle (diameter < 1 μm and thickness < 100 nm), each layer is simply composed of two sheets: a tetrahedral sheet linked by sharing oxygen atoms with an octahedral sheet in the unit cell. Hence, it exhibits two surfaces along the basal plane: siloxane and aluminol (001) surfaces. The charge on these basal planes is predominately polarizable (negative), while the charge on the (110) and (010) edges is pH-dependent amphoteric due to the protonation/deprotonation of hydroxyl groups, where the edges carry a positive charge at low pH and become negative as the pH increases (Armstrong and Clarke, 1971). Recently, kaolinite has been subjected to some useful physical and chemical treatments by modifying its surface and structural properties in a way of enhancing its surface activity and interaction with organics and biomolecules by means of altering charge, porosity and increasing surface area in possible novel active derivatives (Lagaly, 2006; Hu et al., 2013).
Hence, this work sheds light on the application of functionalizing kaolinite and its surface modified derivatives at molecular and nanoscale levels as a novel biophysical antiviral agent. The present study applies methods based on atomistic force fields for molecular modeling and molecular dynamic (MD) simulations to study the potentiality of interaction between protein segment TMD (350–370) of HCV envelope glycoprotein E1 and kaolinite surfaces. Our main aim of this work is to come to predict the behavior of phyllosilicate nanomaterials targeted as anti-HCV agent or kaolinite/protein vaccine, and the extension of this application to other viruses, like COVID-19 coronavirus.
2. Models and computational methodology
The experimental molecular structure of the whole envelope E1 glycoprotein of HCV was not available in the structural databases. Then, only the molecular structure of the TMD fragment (350–370 amino acid units) of this E1 envelope was taken from experimental NMR studies (Beeck et al., 2000) (PDB Code: 1EMZ) (Fig. 1a). This structure was a positively charged peptide. The kaolinite crystal structure was taken from the experimental neutron powder diffraction and Rietveld refinement (Bish, 1993) (Fig. S1 in Supplementary material).
Fig. 1.
Molecular structure of the target segment/protein-fragment 350–370 of HCV glycoprotein E1: (a) experimental structure (Beeck et al., 2000); (b) Model protein-fragment neutralized by chloride anion and optimized with CVFFH; and (c) protein-fragment within a box of an aqueous medium of 626 water molecules optimized.
Atomistic calculations based on empirical interatomic potentials forcefields (FF) were performed for the optimizations and total potential energy calculations of the present molecular structures by using Materials Studio™ package (BIOVIA, 2018). Universal FF (UFF) and Interface FF (we named CVFFH in this work), were used with periodic boundary conditions (Heinz et al., 2006; Heinz and Ramezani-Dakhel, 2016). The Ewald summation method was applied for the non-bonding interactions. There are several FF that work well for protein modeling, such as, AMBER, CHARMM, and some others (Tarasova et al., 2017; BIOVIA, 2018). However, it is difficult to find a FF that describes properly mineral and protein structures simultaneously. CVFFH has described well mineral structures and also organic molecules and their interactions with phyllosilicates in previous works (Martos-Villa et al., 2013, Martos-Villa et al., 2014). This is useful for exploring organics/mineral adsorption complexes (Sainz-Díaz et al., 2011). In the present work, molecular dynamics simulations were performed under NVT ensemble at several temperatures (310 K and 400 K) with steps of 1 fs. Several simulations times were used from 100 ps and up to 10 ns. No significant changes were observed after 10 ps of equilibrium time. The average accuracy of these MD simulations is around 0.4%. The use of high temperature was only for enhancing the mobility of the protein and water molecules over the mineral surface to avoid local energy minima. This allowed all peptides of the protein fragment to interact freely with kaolinite surfaces and so, we will be able to study how different peptide residues interact with kaolinite surfaces. Other structural changes, as conformational changes, can occur with static optimization and molecular dynamic conditions. These results were estimated analyzing the radial distribution function (RDF) between O and H atoms.
The protein fragment was positively charged and then was neutralized by adding a chloride anion near of an ammonium residue and successfully optimized within a periodical box of 50 × 50 × 50 Å3. The optimization energies of the kaolinite unit cell and protein fragment structure were calculated after assigning the atomic charges using the QEq method (Rappe and Goddard, 1991). The measured dimensions of the chloride-neutralized protein fragment structure were 12.57 × 30.12 Å2. Then, a (6 × 6 × 1) supercell of kaolinite was used for the adsorption offering an area of 16 nm2 in the ab-plane of 30.43 × 52.41 Å2. This surface was big enough to avoid intermolecular interactions with proteins of vicinal cells during its adsorption in a periodical model. Therefore, the mineral supercell was composed of 1224 atoms distributed within the composition Al144Si144O360(OH)288. A free space of 50 Å along the c axis was applied for representing an external surface without interactions between surfaces. Several orientations of the protein-fragment/kaolinite system were optimized. The interaction energy, E, was calculated by the formula: E = E kaol+prot–[E kaol + E prot], where E kaol+prot is the energy of the adsorption complex of kaolinite with protein-fragment neutralized with chloride; Ekaol is the energy of the optimized kaolinite 6x6x1 supercell; and Eprot is the energy value of the optimized chloride-neutralized protein.
In order to estimate the adsorption affinity and simulate conformational changes of protein/kaolinite interactions in aqueous medium conditions, a cell box of 626 water molecules was generated with a density of 1 g.cm−3. The size of this water box was chosen taking into account the protein dimensions. Then, the chloride-neutralized protein segment was surrounded by 626 water molecules forming a wet protein model with the kaolinite surface (kaol + prot + wat626). The box with the same number of water molecules was optimized separately (wat626). Hence, the interaction energy of the wet system was obtained by the equation: E = E (kaol+prot+wat626) – [E kaol + E prot + E wat626]. The SPC water model was included in the FF, since this model has yielded good results in previous works in protein-water systems (Tarasova et al., 2017) and in the interaction or organics on silicate surfaces (Borrego-Sánchez et al., 2016).
3. Results and discussion
3.1. Molecular structure of kaolinite and protein fragment
The CVFFH reproduces the experimental lattice cell parameters of the crystal structure of kaolinite better than the Universal FF (Table 1 ). Therefore, the CVFFH was chosen to perform the rest of calculations in this work.
Table 1.
Experimental and calculated crystal lattice parameters of kaolinite (distances in Å and angles in °).
| Lattice parameters | EXPa | UFb | CVFFHc |
|---|---|---|---|
| a | 5.15 | 4.61 | 5.18 |
| b | 8.94 | 8.05 | 8.98 |
| C | 7.39 | 7.37 | 7.29 |
| α | 91.9 | 73.3 | 87.2 |
| β | 105.0 | 102.1 | 105.3 |
| ɤ | 89.8 | 89.2 | 90.0 |
EXP: experimental data.
UF: calculated with Universal FF.
CVFFH: calculated with CVFFH.
The molecular structure of the protein fragment was composed of 311 atoms forming 21 amino acids corresponding to the peptide sequence: [LYS-ALA-TRP-ASN-GLY-VAL-MET-SER-PHE-TYR-ALA-ILE-GLY-ALA-LEU-VAL-GLY-TRP-HIS-ALA-GLY]. These amino acids are mainly nonpolar, except histidine (HIS), tyrosine (TYR), serine (SER) and asparagine (ASN) that are polar and neutral. The positively charged residue in this sequence was the N-terminal glycine (GLY) with one ammonium group. In this work, the glycine ammonium group was neutralized with a chloride anion (right part of Fig. 1b) for the present calculations. The carboxylate terminal was negatively charged in a zwitterionic form and belongs to a terminal lysine (LYS) (left side of Fig. 1a).
The protein-segment (Fig. 1a) was taken from the experimental structure determined by NMR technique in aqueous solution (Beeck et al., 2000). The chloride neutralized-protein segment was optimized with CVFFH in aqueous medium (with 626 water molecules) conditions (Fig. 1b) showed conformational changes with respect to the protein-fragment molecular structure optimized in vacuum. A clear volume contraction was observed during the optimization with an inwards folding of the protein (Fig. 1b), whereas the experimental cationic form is more extended (Fig. 1a). Similar effect was observed experimentally in other proteins as Cytochrome C (Castellini et al., 2009). This behavior can be due to the differences in the local environment with the neutralization with chloride. This hydration is favorable with a exothermic hydration energy of the protein-segment of −572.65 kcal/mol. Nevertheless, a certain volume contraction was observed where the protein adopts a zig-zag folding from the positive ammonium terminal group (right zone in Fig. 1c) to the carboxylate terminal group (left hand in Fig. 1c).
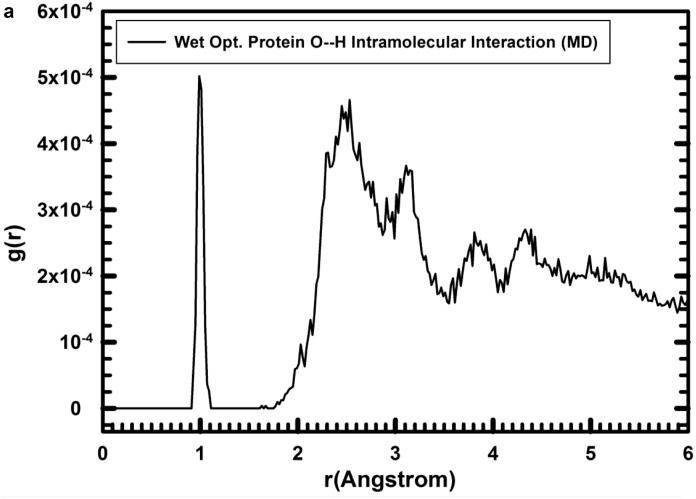
The hydrogen bond and electrostatic interactions between O and H atoms are important for the intramolecular interactions in the protein-fragment. The radial distribution function (RDF) analysis of the intramolecular O…H distances showed a clear peak of the O—H bond at the interval 0.926–0.992 Å in the optimized structure in vacuum, similar to the optimized with water and consistent with the values in the experimental structure (0.948–0.990 Å). The closest intramolecular O…H interactions from the molecular dynamics runs of the protein-fragment/water are close to that of the experimental ones except for the peak at 1.77 Å, supporting the use of this FF for protein (Table 2 ).
Table 2.
Main peaks of the protein RDF profiles for the nearest non-bonding intramolecular (O—H) and protein-segment/water intermolecular distances (in Å).
| Intramolecular |
Intermolecular |
||
|---|---|---|---|
| d(O…H)a | d(O…H)b | d(Hprot…Ow)b | d(Oprot…Hw)b |
| 1.77 | 1.83 | 1.71 | 1.55 |
| 2.17 | 2.13 | 1.83 | 1.69 |
| 2.55 | 2.53 | 1.89 | – |
| 2.65 | 2.64 | 1.97 | 2.05 |
| 3.09 | 3.11 | – | 2.39 |
| 3.80 | 3.81 | 2.3–3.0 | 2.63 |
| 4.34 | 4.35 | 3.91 | 2.90–3.60 |
Measured from the experimental structure (Beeck et al., 2000).
Integrated values from the MD simulation of hydrated protein.
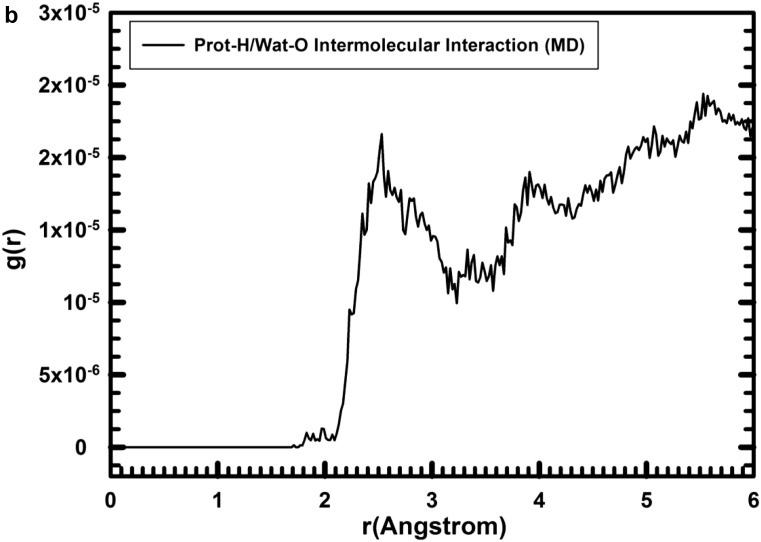
The complex of protein-fragment with water molecules hydrating the protein was equilibrated with MD simulations. The range of the d(O—H) bond length in the integration profile of the MD simulations (100 ps) of the hydrated protein was, d(O—H) = 0.907–1.111 Å (Fig. 2a, first narrow peak). The integrated RDF profile for the non-bonding intramolecular O…H distances (Fig. 2a) shows broad peaks with a qualitatively similar pattern to the equilibrated models and experimental one with hydrogen bonds at 1.77 and 1.83 Å in the experimental and calculated structures (Table 2), respectively, and intense peaks around 2.45–2.8 Å and 3.1 Å. The main peaks of the RDF profiles for the intramolecular non-bonding O···H distances (Fig. 2a) of the hydrated protein were similar to those from the experimental structure (Table 2, first column). This is because the experimental structure has been extracted from NMR analysis in aqueous solution in a similar media that in the MD simulations (Beeck et al., 2000). On the other hand, the intermolecular interactions between the water H atoms and the protein-fragment O atoms in the hydrated structure were shorter than the intramolecular interactions (Fig. 2). In the protein/water intermolecular interactions, the RDF profiles (Fig. 2b,c) showed that the distance between the closest protein-H and water-O atoms, d(Hprot…Ow), were represented by peaks at 1.71, and 1.83 Å (Table 2), but the proportion was the lowest (Fig. 2b). The most intense peaks were in the zone of 2.2–3.1 Å. This profile indicated that the shortest distances below 2.0 Å have very low population being less probable in consequence. This is consistent with the profile of the initial optimized structure, but with few strong interactions with distances smaller than 2.0 Å. On the other hand, in the intermolecular non-bonding distances between the protein-fragment O atoms and the water H atoms, d(Oprot…Hw) (Fig. 2c), short distances were detected after MD simulations at 1.55, 1.69, and 2.05 Å (Fig. 2c). The d(Oprot…Hw) distances were shorter than the d(Hprot…Ow) ones, indicating that the protein is more likely to be an acceptor of hydrogen bonds from water molecules.
Fig. 2.
Radial distribution function (RDF) profiles calculated by integration of the MD simulations of the protein-fragment in water solved form. (a) O…H intramolecular interactions in the protein-fragment; (b) for the O…H intermolecular interactions of protein-fragment H atoms with water O atoms; and (c) protein-fragment O atoms with water H atoms.
3.2. Kaolinite/protein interactions
Significant and potential fragments of HCV capsid proteins could be isolated and loaded on phyllosilicate surface through an albuminous medium for such trial testing as vaccines and for enhancing the immune response. For the adsorption of protein on the kaolinite surface, the most stable surface was considered, the (001) basal one. Two possible (001) surfaces of kaolinite were considered: i) the surface covered with hydroxyl groups of the aluminol groups; and ii) the siloxane surface. In addition, at least two possible orientations of the protein with respect to the mineral surface can be considered for the adsorption process: i) the prot-A, upon which some alkyl groups, some heterocycles, and the terminal ammonium group of glycine neutralized with chloride oriented to the mineral surface; and ii) prot-B, upon which the terminal carboxylate group of the lysine residue, the thioether and phenyl groups oriented to mineral surface (Fig. 1). The protein-fragment was previously stabilized with the presence of 626 water molecules and after placed on the surfaces of the supercell of kaolinite. The whole model has 2161 atoms each.
All possible combinations of these surfaces and orientations of the protein-fragment were optimized as adsorption complexes before and after the MD simulations. In all cases the adsorption processes were exothermic (Table 3 ). After these optimizations and before the MD simulation some differences in the interaction energy between orientations prot-A and prot-B of the protein-fragment were observed, being more exothermic after the MD simulations. This happened in both kaolinite surfaces. Besides, the adsorption was much more favorable with the aluminol surface than in the siloxane surface, as it was expected owing to the interactions between hydroxyl groups of kaolinite and the polar groups of protein-fragment. The most exothermic adsorption was on the aluminol surface and the prot-B orientation of the protein-fragment (Table 3).
Table 3.
Calculated interaction energy values (in kcal/mol) of hydrated protein on kaolinite surfaces optimized before and after MD simulations.
| Protein orientation | Optimization states | Aluminol surface | Siloxane surface |
|---|---|---|---|
| Prot-A | Before MD | −204.60 | −136.25 |
| After MD | −292.80 | −265.73 | |
| Prot-B | Before MD | −192.08 | −153.97 |
| After MD | −372.57 | −306.25 |
The presence of water is important for a more realistic scenario. Therefore, this work is focused on the protein-mineral interaction in wet conditions. Nevertheless, the analysis of the interaction of this protein fragment and kaolinite surfaces along with MD simulations in dry conditions were described and discussed in the Supplementary material section for comparison.
In the aluminol surface, the water molecules were closer to surface than in the siloxane surfaces (Fig. 3 ). The protein configurations were similar on aluminol surface that on siloxane surface. In all cases the protein structures were different to those observed in hydrated state without kaolinite (see later). The protein was not in direct contact with the mineral surface. In general, a layer of water molecules was localized between protein and kaolinite surface. A certain interaction between the indole H atom of tryptophan and the basal tetrahedral O siloxane atoms of kaolinite was observed at 3.75 Å.
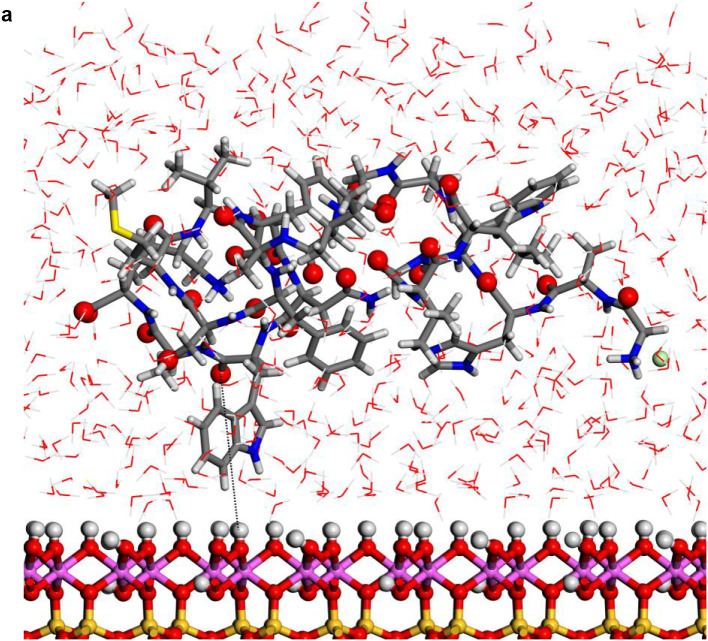
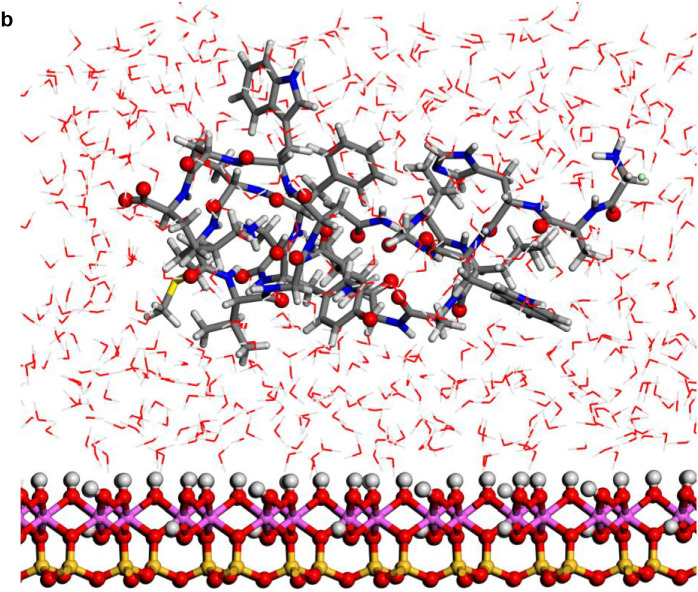
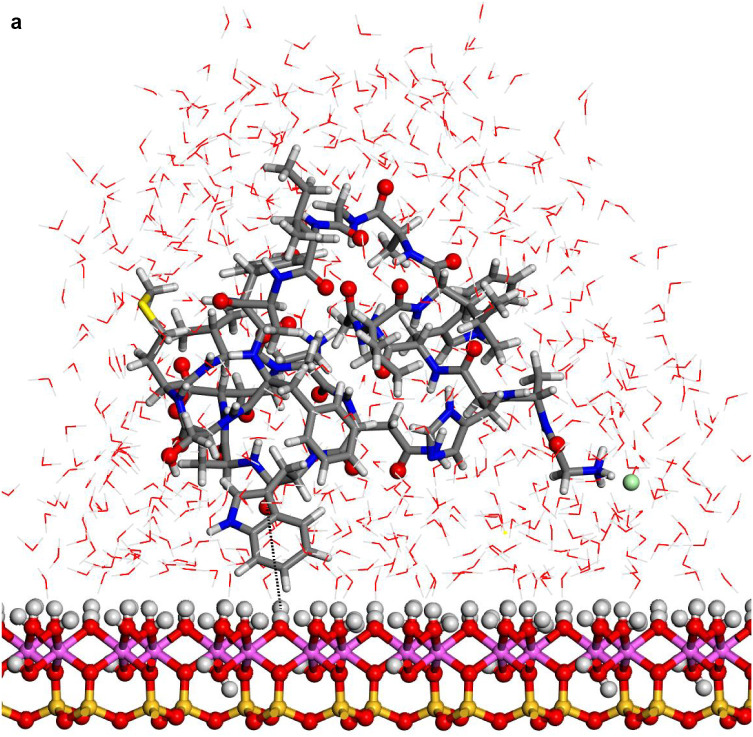
Fig. 3.
Structures optimized of the hydrated protein-fragment adsorbed on kaolinite surfaces: (a) prot-fragment-A, and (b) prot-fragment-B adsorbed on kaolinite aluminol surface; (c) prot-fragment-A, and (d) prot-fragment-B adsorbed on kaolinite siloxane surface.
After the MD simulations, the water molecules were more strongly interacting with the aluminol surface than the siloxane surface. However, water molecules tended to be clustered as a globular form in all surfaces. This effect seems to be more clear in the siloxane surface than in the aluminol surface, due probably to the hydrophobic affinity of the siloxane surface, whereas protein adsorbate is closer to the aluminol surface (Fig. 4 ).
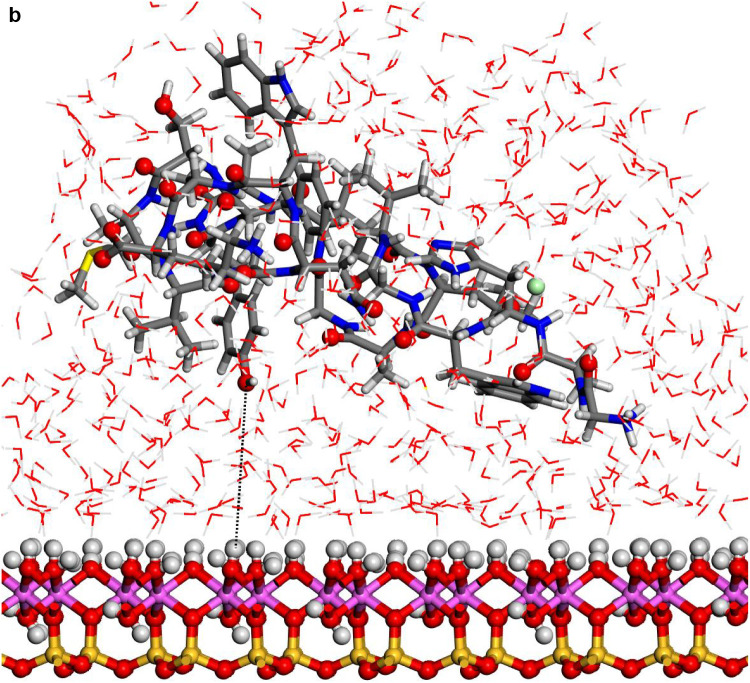
Fig. 4.
Adsorptions of the hydrated protein-fragment molecule orientations on the kaolinite surfaces after MD simulations: (a) prot-A and (b) prot-B (b) adsorbed on kaolinite aluminol surface; (c) prot-fragment-A, and (d) prot-fragment-B adsorbed on kaolinite siloxane surface.
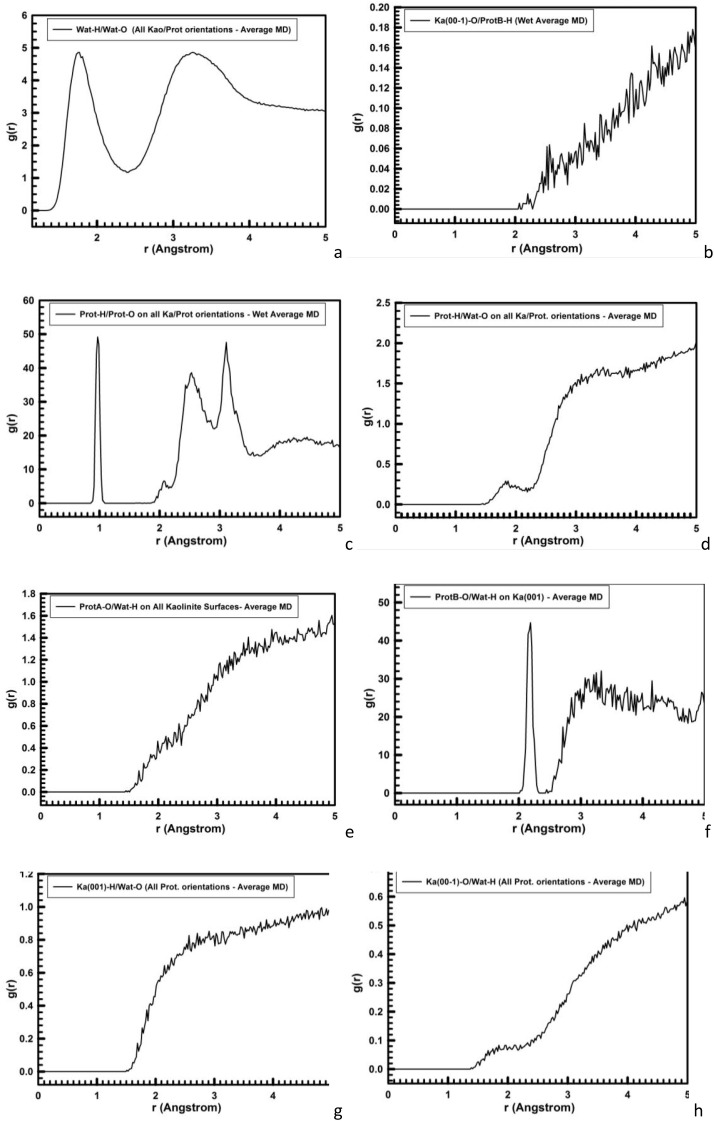
The MD simulations (100 ps) of the hydrated systems showed average peak positions on the RDF profiles of the O···H intramolecular and intermolecular interactions of the calculated 626 water molecules with all the hydrated kaolinite and protein surface orientations (Fig. 5 ). Notice, the average O···H peak positions on RDF profile of the present simulated water molecules (Fig. 5a) were in good agreement with the previously reported liquid water SPC/E model and the experimental data obtained with neutron diffraction (Soper and Phillips, 1986; Mark and Nilsson, 2001). The RDF profile integrated from MD sampling of the intermolecular O···H interactions between the H atoms of prot-B and the kaolinite siloxane surface O atoms in the MD simulations showed peak positions for H···O distances at 2.07, 2.21, 2.44, 2.52, and 2.6 Å (Fig. 5b), the last two peaks being the most intense. This indicated hydrogen bonds between protein and kaolinite siloxane surface. The average closest distances of the hydrated protein-fragment intramolecular O···/—H interactions on the RDF profile (Fig. 5c) exhibited identical peak positions with all the orientations on kaolinite surfaces. The O—H bond lengths are described by the intense peak at 0.98 Å; whereas, intramolecular hydrogen bonds are observed at 2.08, 2.52, 2.84, and 3.11 Å. This RDF profile shows some differences with that observed without kaolinite surface (Fig. 2a) indicating that the configuration of the protein is different. These changes could suggest a certain denaturalization of the protein owing to the surface effect (Yu et al., 2000), however no clear evidence can be concluded with these results and further investigations in this way should be performed.
Fig. 5.
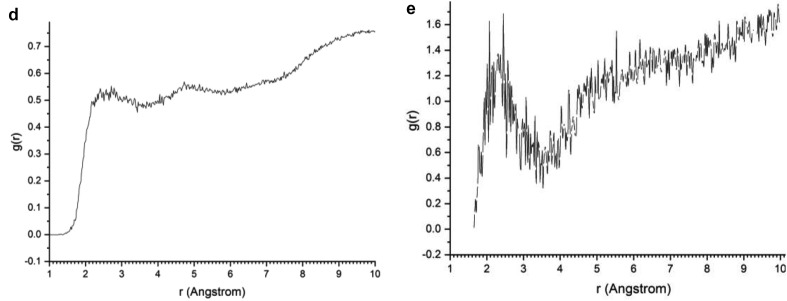
RDF profiles of the MD simulations. (a) non-bonding distances between the H-water and O-water atoms of the wet kaolinite/protein adsorption complexes; (b) protein-fragment-H atoms with the kaolinite siloxane surface O atoms; (c) the H and O atoms of the protein-fragment for all orientations and surfaces; (d) protein-fragment H atoms with water O atoms for all orientations; (e) O atoms of the protein-fragment with water H atoms in the complex adsorbed on the kaolinite siloxane surface; (f) O atoms of the prot-fragment-B with water H atoms on the kaolinite aluminol surface; (g) water O atoms with H atoms of the kaolinite aluminol surface; and (h) water H atoms with O atoms of the kaolinite siloxane surface.
The RDF profiles of the intermolecular interactions between the protein-fragment H-atoms and water O atoms were similar for all protein orientations on kaolinite surfaces. The first peak position was broad and prominent at 1.82 Å (Fig. 5d). However, the maximum peaks appear at longer distances than 2.7 Å, whereas in the MD simulation of wet protein-fragment without kaolinite maximum peaks at shorter distances (2.2–2.6 Å) were observed (Fig. 2b). The water H atoms interacted with the O atoms of the protein-fragment in presence of kaolinite surfaces showing H/O RDF profiles with low populations at short distances (1.6–2.2 Å, Fig. 5e). However, the protein-fragment O atoms interacted with the H atoms of the water molecules adsorbed on the kaolinite aluminol surface exhibiting a very intensive and distinguished H/O RDF peak at 2.18 Å (Fig. 5f).
The water molecules on the kaolinite surfaces show a different behavior for each mineral surface. The RDF profile of the aluminol surface H atoms with the water O atoms (Fig. 5g) shows short non-bonding distances at 1,6–2.0 Å, corresponding to the H bonds at similar distances that in the first coordination sphere of liquid water (Fig. 5a). However, the RDF profile of the siloxane surface O atoms with the water H atoms shows a low density of H bonds at 1.5–2.2 Å and the main density correspond to longer distances (Fig. 5h). This indicates the hydrophilia of kaolinite aluminol surface with respect to the more hydrophobia of the siloxane surface.
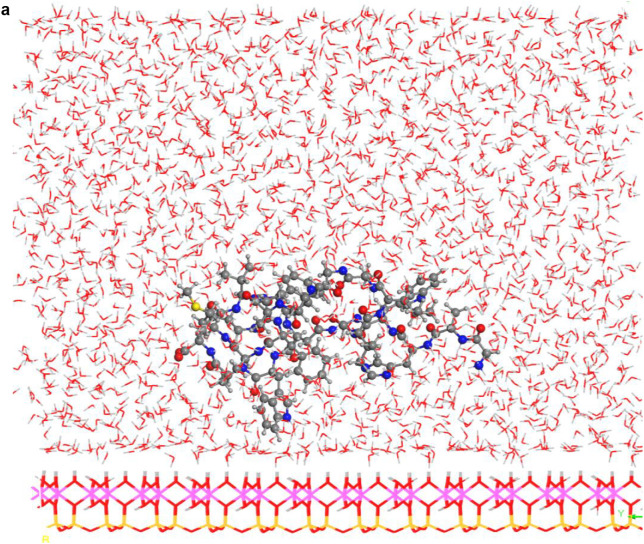
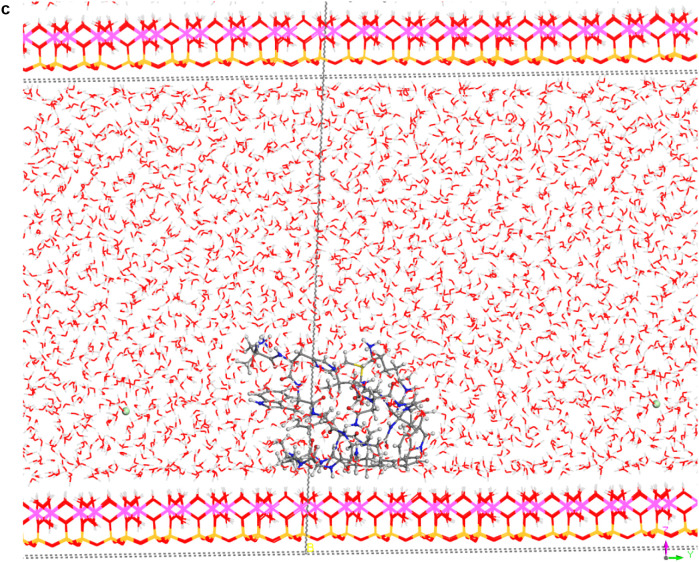
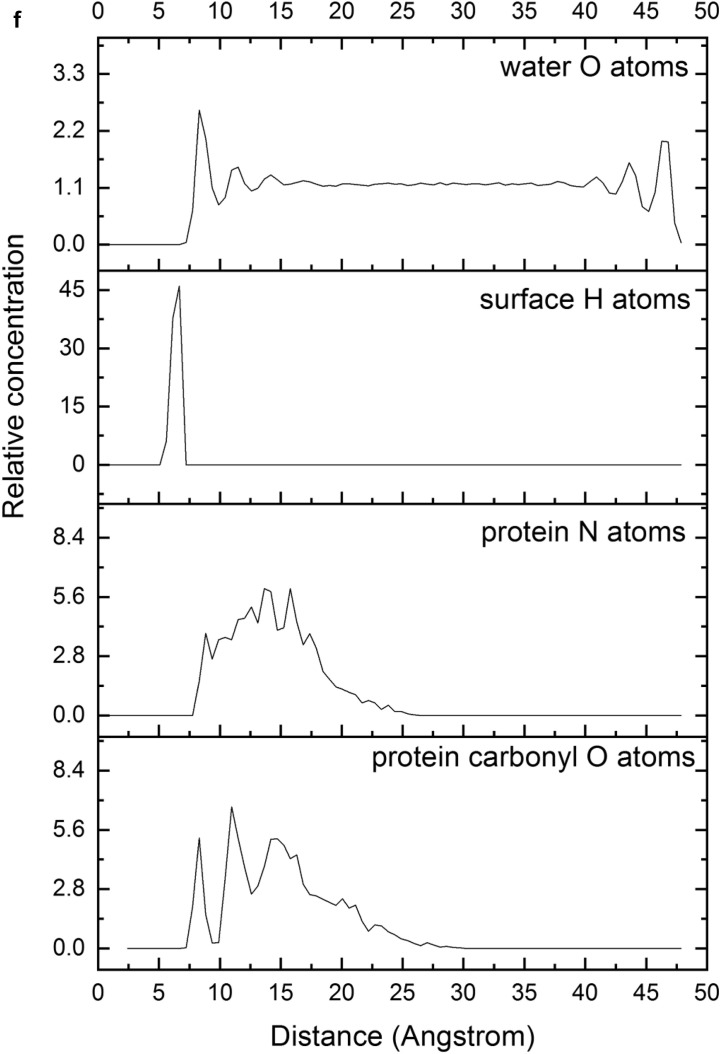
In spite of the whole protein-fragment was completely hydrated and surrounded by water molecules, this model could be considered limited because the amount of water does not cover completely the interlayer space between mineral surfaces, provoking a solid-liquid-air interphase. Hence this model was extended filling the whole interlayer space with disordered water molecules. The total amount of water was 2400 water molecules per protein fragment. This model (8730 atoms) was optimized obtaining a similar structure than above model. Molecular dynamics simulations were performed at 310 K with 1 fs steps during 0.7 ns. The final structure (Fig. 6a) was similar to previous ones with a layer of water between protein and mineral surface (see Supplementary Video 1 in Supplementary material). The RDF between C atoms of protein-fragment and H atoms of the aluminol surface showed low intensity peaks at 2.5–4.5 Å, corresponding mainly to the tryptophan residue (Fig. 6b). Although there is enough water medium space in the upper part of the model, the protein-fragment was maintained close to the aluminol surface, indicating an affinity to the mineral surface. The calculated adsorption energy was −10.31 kcal/mol corroborating this affinity. This is consistent with previous experimental (Lambert, 2008; Docoslis et al., 2001) and theoretical (Patwardhan et al., 2012) works on adsorption of peptides with silicate surfaces. Longer MD simulations were performed with this model up to 10 ns. The structure of the adsorption complex samples was different to the former one after 0.7 ns. The protein conformation changed completely in a more compact form with a certain zone close to the mineral surface. With this long MD simulation, a certain flexibility of the mineral layer is observed (Fig. 6c). The RDF integrated on the sampling frames along the simulations showed different profiles with respect to the previous shorter simulations. The RDF profile of the water O atoms with respect to the aluminol surface H atoms shows strong H bond interactions at 1.8–2.5 Å (Fig. 6d). The RDF profile of the carbonyl O atoms of protein with respect to the surface H atoms shows a certain group close to the surface at H bond distances 1.7–2.8 Å (Fig. 6e). The atomic density profile along the 001 direction on the interlayer space showed that the protein (O and N atoms protein profiles) remains close to one surface not in the middle of the interlayer space and close to the aluminol surface instead of the siloxane surface. Water molecules are more concentrated close to the mineral surfaces, being much more close to the aluminol surface than to the siloxane one. A short-range ordering of the water molecule is observed being more random in the middle of the interlayer space. As observed in the concentration profiles, certain carbonyl groups approach very close to the aluminol surface indicating that some carbonyl groups do not form only collinear H bonds, but several H bonds with surrounding surface H atoms allow being closer to the surface (Fig. 6f).
Fig. 6.
(a) A snapshot of a 0.7 ns MD simulation of the adsorption complex of protein with 2400 water molecules on kaolinite aluminol surface; (b) RDF profile integrated from 0.7 ns MD sampling between the protein C atoms and H atoms of kaolinite aluminol surface; (c) snapshot of a long 10 ns MD simulation; (d) RDF profile integrated from 10 ns MD sampling of water O atoms and surface H aluminol atoms; (e) RDF profile integrated from 10 ns MD sampling of protein carbonyl O atoms and surface H aluminol atoms; (f) concentration profiles along the 001 direction integrated from 10 ns MD sampling.
Recent experimental results of zeta potential titration of kaolinite have shown dissociation of the kaolinite surface at neutral pH (Jacquet et al., 2018). Then, at neutral pH some OH groups of kaolinite can be dissociated. This situation can be described with a model were some OH groups of the aluminol surface of kaolinite were deprotonated remaining a negatively charged surface. The original form of the protein fragment was protonated. Then, the protein fragment can be adsorbed as a cation in this scenario on the negatively charged mineral surface. These models were optimized as a static model obtaining adsorption energy of −77.18 kcal/mol. Some Lewis acid-base interactions with electrostatic forces can be also be present in these conditions.
On the other hand, the adsorption can be produced by cation exchange with the kaolinite surface negatively charged neutralized with the presence of a Na+ cation and the protein cation with a chloride anion. With both systems, the adsorption complex will have the protein cation adsorbed on the kaolinite anion and the solvated Na+ and Cl− ions out of the adsorption process. These models were also optimized (Fig. 7 ) obtaining higher adsorption energy (−147.56 kcal/mol) than the previous one. Both cases indicate that the effect of pH at neutral aqueous media favors the adsorption of this protein fragment on the mineral surface. Such conditions could be mainly considered experimentally in case of design applications and development in biomedicine where the normal human blood pH is tightly regulated between 7.35 and 7.45 and the plasma could be neutralized in some critical therapeutic conditions by buffering systems, also for possible application in the design of viral-adsorbent agents for treatment of water contaminated by human viruses.
Fig. 7.
Adsorption complexes of protein on a kaolinite aluminol surface dissociated by the pH effect.
4. Conclusions
This work introduces a new modeling and mechanism that can help for the understanding of the experimental behavior and improve the applicability and development of kaolinite as antiviral agent. The present study sheds light on the possible interactions between the viral capsid proteins and phyllosilicate surfaces. This work was focused only on the most critical fragment for the fixation of virus to human cell receptors. Further studies on the whole protein structure and the glycoxy moiety should be done in future. The molecular dynamics simulations and calculations indicated that the presence of kaolinite surfaces changes the conformations of the main protein segment TMD (350–370) of HCV envelope glycoprotein E1. This is likely to be adsorbed on the kaolinite surfaces favorably with an exothermic process. The intermolecular interactions were mainly hydrogen bonds and electrostatic ones. The hydration effect was critical for the conformation of protein and for the adsorption of the protein fragment on kaolinite surfaces. In a system saturated with water, the protein was completely solvated by water and remains attracted to the mineral surface. This adsorption was more favorable to be with the hydrophilic aluminol (001) surface than with the siloxane surface. Longer MD simulations corroborate the strong interactions with H bonds of the protein with the aluminol mineral surface. The adsorption affinity becomes more favorable at neutral pH media, where some OH groups of the kaolinite were partially dissociated. Therefore, targeted kaolinite nanoparticles can interact with the charged residues in the transmembrane domains of hepatitis C virus (HCV) glycoproteins leading to molecular conformational changes and alter dysfunctions of the TMDs causing viral inhibition. This study can predict that the kaolinite can be considered in an exploratory research for a therapeutic treatment against HCV.
The computational results could be considered as initial stage for evaluating limitations and take previous conclusion to go to a long-term project, intending to further trials on antiviral therapeutic, including the COVID-19 coronavirus, or treatment of human virus-contaminated water strategies based on mineral surface properties. Besides, this kind of interactions can be also produced in any prebiotic reaction pathway on the mineral surfaces in early Earth.
The following is the supplementary data related to this article.
Molecular dynamics simulation of the hydrated protein fragment onto the kaolinite aluminol (001) surface.
Supplementary material
Declaration of Competing Interest
None.
Acknowledgements
Authors would like to acknowledge the contribution of the European COST Action CA17120 supported by the EU Framework Program Horizon 2020 and the financial support of the Andalusia Government projects [RNM1897 and RNM363, and CTS-946]; the MINECO and FEDER projects [FIS2016-77692-C2-2-P, PCIN-2017-098]. The first author is especially thankful to Prof. Mahmoud Mohamed El Rahmany (Faculty of Science, Al-Azhar University in Cairo) for encouragement and fruitful discussions.
References
- Ali L., Idrees M., Ali M., Hussain A., Ur Rehman I., Ali A., Iqbal S.A., Kamel E.H. Inhibitory effect of kaolin compounds against hepatitis C virus in Huh-7 cell lines. BMC Res. Notes. 2014;247:1–5. doi: 10.1186/1756-0500-7-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N., Clarke C. The adsorption of crystal violet by kaolin. J. Pharm. Pharmacol. 1971;23:95–100. doi: 10.1111/j.2042-7158.1971.tb08776.x. [DOI] [PubMed] [Google Scholar]
- Awad M.E., López-Galindo A., Setti M., El-Rahmany M.M., Iborra C.V. Kaolinite in pharmaceutics and biomedicine. Int. J. Pharm. 2017;533:34–48. doi: 10.1016/j.ijpharm.2017.09.056. [DOI] [PubMed] [Google Scholar]
- Beeck A., Montserret R., Duvet S., Cocquerel L., Cacan R., Barberot B., Le Maire M., Penin F., Dubuisson J. The transmembrane domains of hepatitis C virus envelope glycoproteins E1 and E2 play a major role in heterodimerization. J. Biol. Chem. 2000;275:31428–31437. doi: 10.1074/jbc.M003003200. [DOI] [PubMed] [Google Scholar]
- BIOVIA . 2018. Materials Studio, Dassault Systèmes. [Google Scholar]
- Bish D.L. Rietveld refinement of the kaolinite structure at 1.5 K. Clay Clay Miner. 1993;41:738–744. [Google Scholar]
- Bohidar H.B. Cambridge University Press; UK: 2015. Fundamentals of Polymer Physics and Molecular Biophysics. [Google Scholar]
- Borrego-Sánchez A., Viseras C., Aguzzi C., Sainz-Díaz C.I. Molecular and crystal structure of praziquantel. Spectroscopic properties and crystal polymorphism. Eur. J. Pharm. Sci. 2016;92:266–275. doi: 10.1016/j.ejps.2016.04.023. [DOI] [PubMed] [Google Scholar]
- Castellini E., Ranieri A., Simari D.A., Di Rocco G. Thermodynamic aspects of the adsorption of cytochrome C and its mutants on kaolinite. Langmuir. 2009;25:6849–6855. doi: 10.1021/la9001016. [DOI] [PubMed] [Google Scholar]
- Ciczora Y., Callens N., Montpellier C., Bartosch B., Cosset F., Beeck A.O.D., Dubuisson J. Contribution of the charged residues of hepatitis C virus glycoprotein E2 transmembrane domain to the functions of the E1E2 heterodimer. J. Gen. Virol. 2005;86:2793–2798. doi: 10.1099/vir.0.81140-0. [DOI] [PubMed] [Google Scholar]
- Cocquerel L., Wychowski C., Minner F., Penin F., Dubuisson J. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a key role in the processing, subcellular localization and assembly of these envelope proteins. J. Virol. 2000;74:3623–3633. doi: 10.1128/jvi.74.8.3623-3633.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docoslis A., Rusinski L.A., Giese R.F., Van Oss C.J. Kinetics and interaction constants of protein adsorption onto mineral microparticles — measurement of the constants at the onset of hysteresis. Colloids Surf. B: Biointerfaces. 2001;22:267–283. [Google Scholar]
- Foster G.R. Pegylated interferons: chemical and clinical differences. Aliment. Pharmacol. Ther. 2004;20:825–830. doi: 10.1111/j.1365-2036.2004.02170.x. [DOI] [PubMed] [Google Scholar]
- Heinz H., Ramezani-Dakhel H. Simulations of inorganic–bioorganic interfaces to discover new materials: insights, comparisons to experiment, challenges, and opportunities. Chem. Rev. 2016;45:412–448. doi: 10.1039/c5cs00890e. [DOI] [PubMed] [Google Scholar]
- Heinz H., Vaia R.A., Farmer B.L. Interaction energy and surface reconstruction between sheets of layered silicates. J. Chem. Phys. 2006;124:224713. doi: 10.1063/1.2202330. [DOI] [PubMed] [Google Scholar]
- Hu Y., Liu L., Min F., Zhang M., Song S. Hydrophobic agglomeration of colloidal kaolinite in aqueous suspensions with dodecylamine. Colloids Surf. A Physicochem. Eng. Asp. 2013;434:281–286. [Google Scholar]
- Jacquet A., Geatches D.L., Clark S.J., Greenwell H.C. Understanding cationic polymer adsorption on mineral surfaces: kaolinite in cement aggregates. Minerals. 2018;8:130. [Google Scholar]
- Kamal S.M., Ismail A., Graham C.S., He Q., Rasenack J.W., Peters T., Tawil A.A., FehrJJKhalifa K.S., Madwar M.M., Koziel M.J. Pegylated interferon alpha therapy in acute hepatitis C: relation to hepatitis C virus-specific T cell response kinetics. Hepatology. 2004;39:1721–1731. doi: 10.1002/hep.20266. [DOI] [PubMed] [Google Scholar]
- Lagaly G. Colloid clay science. In: Bergaya F., Theng B.K.G., Lagaly G., editors. Handbook of Clay Science. Elsevier, Developments of Clay Science; Amsterdam: 2006. pp. 141–247. [Google Scholar]
- Lambert J.-F. Adsorption and polymerization of amino acid on mineral surfaces: a Review. Orig. Life Evol. Biosph. 2008;38:211–242. doi: 10.1007/s11084-008-9128-3. [DOI] [PubMed] [Google Scholar]
- Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A. 2001;105:9954–9960. [Google Scholar]
- Martos-Villa R., Francisco-Márquez M., Mata M.P., Sainz-Díaz C.I. Crystal structure, stability and spectroscopic properties of methane and CO2 hydrates. J. Mol. Graph Model. 2013;44:253–265. doi: 10.1016/j.jmgm.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Martos-Villa R., Mata M.P., Sainz-Díaz C.I. Characterization of CO2 and mixed methane/CO2 hydrates intercalated in smectites by means of atomistic calculations. J. Mol. Graph Model. 2014;49:80–90. doi: 10.1016/j.jmgm.2014.01.008. [DOI] [PubMed] [Google Scholar]
- Michalak J.P., Wychowski C., Choukhi A., Meunier J.C., Ung S., Rice C.M., Dubuisson J. Characterization of truncated forms of hepatitis C virus glycoproteins. J. Gen. Virol. 1997;78:2299–2306. doi: 10.1099/0022-1317-78-9-2299. [DOI] [PubMed] [Google Scholar]
- Moradpour D., Brass V., Penin F. Function follows form: the structure of the N-terminal domain of HCV NS5A. Hepatology. 2005;42:732–735. doi: 10.1002/hep.20851. [DOI] [PubMed] [Google Scholar]
- Patwardhan S.V., Emami F.S., Berry R.J., Jones S.E., Naik R.R., Deschaume O., Heinz H., Perry C.C. Chemistry of aqueous silica nanoparticle surfaces and the mechanism of selective peptide adsorption. J. Am. Chem. Soc. 2012;134:6244–6256. doi: 10.1021/ja211307u. [DOI] [PubMed] [Google Scholar]
- Rappe A.K., Goddard W.A. Charge equilibration for molecular dynamics simulations. J. Phys. Chem. 1991;95:3358–3363. [Google Scholar]
- Ravikumar Y.S., Ray U., Nandhitha M., Perween A., Naikaa H.R., Khanna N., Das S. Inhibition of hepatitis C virus replication by herbal extract: phyllanthusamarus as potent natural source. Virus Res. 2011;158:89–97. doi: 10.1016/j.virusres.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Sainz-Díaz C.I., Francisco-Márquez M., Vivier-Bunge A. Adsorption of Polyaromatic heterocycles on pyrophyllite surface by different theoretical approaches. Environ. Chem. 2011;8:429–440. [Google Scholar]
- Selby M.J., Glazer E., Masiarz F., Houghton M. Complex processing and protein: protein interactions in the E2:NS2 region of HCV. Virol. 1994;204:114–122. doi: 10.1006/viro.1994.1515. [DOI] [PubMed] [Google Scholar]
- Soper A.K., Phillips M.G. A new determination of the structure of water at 25°C. Phys. Chem. 1986;107:47–60. [Google Scholar]
- Tarasova E., Farafonov V., Khayat R., Okimoto N., Komatsu T.S., Taiji M., Nerukh D. All-atom molecular dynamics simulations of entire virus capsid reveal the role of ion distribution in capsid’s stability. J. Phys. Chem. Lett. 2017;8:779–784. doi: 10.1021/acs.jpclett.6b02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisset C., Dubuisson J. Functional hepatitis C virus envelope glycoproteins. Biol. Cell. 2004;9:413–420. doi: 10.1016/j.biolcel.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Yu C.H., Norman M.A., Newton S.Q., Miller D.M., Teppen B.J., Schäfer L. Molecular dynamics simulations of the adsorption of proteins on clay mineral surfaces. J. Mol. Struct. 2000;556:95–103. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Molecular dynamics simulation of the hydrated protein fragment onto the kaolinite aluminol (001) surface.
Supplementary material