Abstract
Background
Quinones are reactive to proteins containing cysteine residues and the main protease in Covid-19 contains an active site that includes Cys145. Embelin, a quinone natural product, is known to have antiviral activity against influenza and hepatitis B. Preliminary studies by our group also indicate its ability to inhibit HSV-1 in cultured cells.
Methods
Docking and DFT methods applied to the protease target.
Results
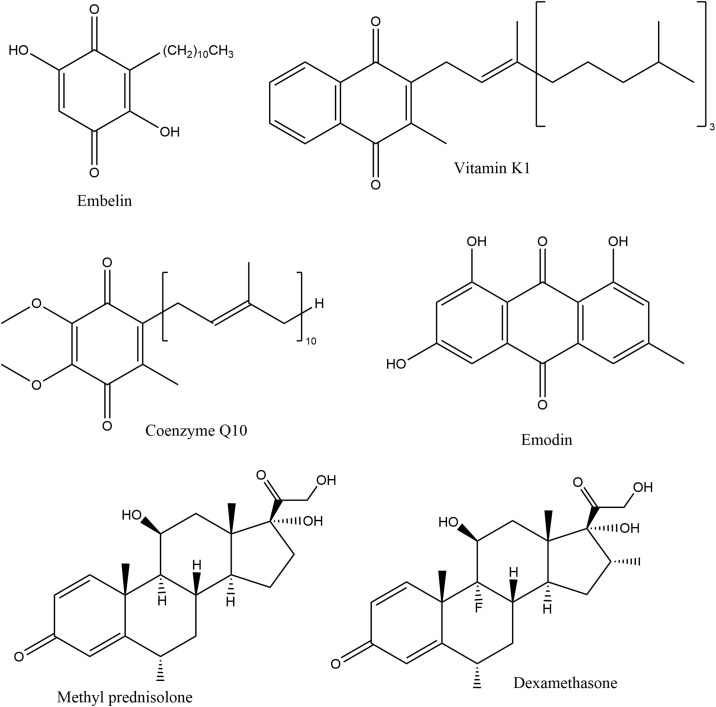
a mechanism for this inhibition of the SARS-CoV-2 Mpro protease is described, specifically due to formation of a covalent bond between S(Cys145) and an embelin C(carbonyl). This is assisted by two protein amino acids (1) N(imidazole-His41) which is able to capture H[S(Cys145)] and (2) HN(His163), which donates a proton to embelin O(carbonyl) forming an OH moiety that results in inhibition of the viral protease. A similar process is also seen with the anti-inflammatory drugs methyl prednisolone and dexamethasone, used for Covid-19 patients. Methyl prednisolone and dexamethasone are methide quinones, and possess only one carbonyl moiety, instead of two for embelin. Additional consideration was given to another natural product, emodin, recently patented against Covid-19, as well as some therapeutic quinones, vitamin K, suspected to be involved in Covid-19 action, and coenzyme Q10. All show structural similarities with embelin, dexamethasone and methyl prednisolone results.
Conclusions
Our data on embelin and related quinones indicate that these natural compounds may represent a feasible, strategic tool against Covid-19.
Keywords: Covid-19, embelin, vitamin K, methyl prednisolone, dexamethasone
Introduction
December 2019 was the beginning of an ongoing global pandemic caused by an unknown coronavirus named Covid-19. This initiated a plethora of infections and deaths around the world [1] and to limit the spread of this pandemic biosafety and hygiene measures are now applied. Unfortunately, there are currently no medications or vaccines proven to be effective for the treatment or prevention of Covid-19. Several antiviral drugs are promising for Covid-19, among them remdesivir is under clinical trial [3]. The antimalarial drug chloroquine has been proposed but its use is controversial in the scientific community [4,5], whereas hydroxychloroquine is a more soluble and less toxic metabolite of chloroquine, which causes fewer side effects [6]. Non-antiviral drugs as methyl prednisolone and dexamethasone show a positive effect for treating Covid patients [7,8].
The recent uncertainties of a future vaccine (for example, AstraZeneca is re-evaluating its clinical trials) suggests the need and urgency for other pharmaceutical options and is also encouraging research into not-yet-approved medicinal compounds. In this context, even if an effective vaccine may become available, effective drugs are currently needed for infected people [9].
A rational based drug design can reduce the time needed in the drug discovery process. Molecular docking can identify lead molecules against the target proteins [10,11]. Plant based compounds have provided the scaffold for many effective pharmaceutical compounds and the natural products are sources of promising antiviral drugs [12]. In fact, many approved antiviral drugs are derived from natural sources [13]. Examples of plant-derived proven antiviral activities include blocking HIV replication [14], inhibiting Dengue virus type-2 (DENV-2) [15] and against Hepatitis C virus (HCV) [16].
The main protease in SARS-CoV-2 is known as 3C-like protease (3CLpro), and it consists of a highly conserved catalytic domain from the SARS virus. It is essential for controlling several functions of Covid-19, hence targeting the 3CLpro would prevent the virus from building its proteins and help in inhibiting viral replication. In fact, its mutation is often lethal to the virus and drugs targeting the enzyme significantly reduce the risk of mutation-mediated drug resistance and display broad-spectrum antiviral activity [17]. The three dimensional X-ray crystal structure of this enzyme in complex with ketoamide inhibitors was recently determined [18], suggesting further investigation for other potential drugs. Due to the rise in the number of infected and death cases from Covid-19 and the lack of effective therapeutic drugs and vaccines, computer-aided drug design may be an important and fast way to find useful drugs.
Embelin, a plant natural product found in Lysimachia punctata (Primulaceae), and Embelia ribes Burm (Myrsinaceae) fruit, possesses interesting biological and pharmacological properties. The fruits contain alkyl-benzoquinones, including embelin [19], embelinol, and embeliol [20]. E. ribes is cardio-protective [21], antibacterial [22] and has anti-inflammatory [23] and antidiabetic activity [24]. Embelin is a cell-permeable, small-molecular weight inhibitor of XIAP and was the first non-peptide XIAP inhibitor [25] which induced additional anti-carcinogenic research [26]. Embelin suppresses paraquat-induced oxidative stress in lung [27], an organ particularly affected by Covid-19. It interacts with several proteins in mammalian cells: as a signal transducer and activator of transcription 3 [28], peroxisome proliferator-activated receptor-γ [29], and plasminogen activator inhibitor-1 [30]. Importantly, embelin is not toxic when given orally to rodents. Daily oral administration of embelin (25 and 50 mg/kg/day) for three weeks causes no side effects in rats [31]. The binding properties of embelin to proteins shows it is bound to human serum albumin through interactions with hydrophobic albumin amino acids [32]. Embelin is a unique chemical species as it includes both quinone and hydroquinone functional groups plus a long hydrophobic tail. Chemical and biological aspects of embelin have been reviewed [33] and recently, embelin was defined as the second solid gold of India, after curcumin [34].
Embelin is the major component of Embelia ribes Burm (Myrsinaceae), as the amount of embelin present in the ethyl acetate extract is 60.7% (w/w). The anti-viral activity of five embelin-containing plant products from Bangladesh were studied against influenza virus A/Puerto Rico/8/34 (H1N1) MDCK infected cells. When treated with increasing concentrations of ethyl acetate extracts of Embelia ribes Burm fruits [35], the highest antiviral activity was IC50 of 0.2 μg/mL. The pure compound, embelin, was isolated and tested against the same virus with an IC50 of 0.3 μM. Embelin also proved to be an inhibitor of hepatitis B virus when tested along with twelve non-cytotoxic natural compounds and was among those showing high inhibition of HBV replication [36]. That quinones show antiviral activity is already known [[37], [38], [39]]. Quinones react with several functional groups of proteins or amino acids. Potential reactive targets for embelin of particular interest are lysine and cysteine, which react through the ε-amino and thiol groups, respectively [40].
The current Covid-19 pandemic has elicited enormous interest in enveloped viruses. Knowing that quinones show antiviral activity on viruses such as influenza, hepatitis B [[35], [36]], together with our preliminary results showing embelin inhibition of the Herpes simplex virus-1 (HSV-1) enveloped DNA virus [41] were stimuli to explore its chemical activity chemical activity on Covid-19. In this study we describe the potential of embelin against Covid-19 virus. We describe our proposed mechanism behind embelin action by using results of DFT computational studies after docking embelin, methyl prednisolone and other related interesting compounds, scheme 1 , at known sites which the viral protease is susceptible to mild quinones.
Scheme 1.
Quinones and quinone derivatives studied with Docking – DFT methods.
Materials and Methods
Calculations were performed using programs from Biovia (San Diego, CA, USA). Density functional theory (DFT) code DMol3 was applied to calculate energy, geometry, and frequencies implemented in Materials Studio 7.0 [42]. We employed the double numerical polarized (DNP) basis set that included all the occupied atomic orbitals plus a second set of valence atomic orbitals, and polarized d-valence orbitals [43]; the correlation generalized gradient approximation (GGA) was applied including Becke exchange [44], plus Perdew correlation (PBE) [45]. All electrons were treated explicitly and the real space cutoff of 5 Å was imposed for numerical integration of the Hamiltonian matrix elements. The self-consistent field convergence criterion was set to the root mean square change in the electronic density to be less than 10−6 electron/Å3. The convergence criteria applied during geometry optimization were 2.72 × 10−4 eV for energy and 0.054 eV/Å for force. All calculations did not include solvent effect. Docking studies were performed with the CDOCKER package in Discovery Studio 2020 version [46].
Results
Docking procedure
The importance of the two catalytic residues His41 and Cys145 for the design of α-ketoamide inhibitors against SARS-CoV-2 Mpro has been recently described [18]. In addition, structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease has been suggested [47]. In a recent preliminary report, several anthraquinones were placed using docking procedures into the same important protease protein, one of the most critical drug targets for the blockage of viral replication [48]. Antiviral activity is known to be an important property of anthraquinones [[49], [50]]. The receptor [51] was PDB 6Y84 and comparison was done with remdesivir the first approved drug for Covid-19. Ranking immediately after remdesivir, the quinone emodin’s ΔG value differed from the drug only by 0.51 kcal/ mol.
We have recently described the antioxidant activity of emodin, another quinone plant natural product [52]. Emodin and embelin possess similar redox properties, and their capture of an electron by the quinone ring are closely related [53]. The existence of the Cys145 residue at the protease attracted our attention because of a potential strong bond between Cys and the quinone ring. Therefore, we envisioned embelin as a feasible inhibitor of this important viral enzyme and describe docking and DFT procedures for embelin into the same target protein.
Protein PDB 6Y84 crystal structure contains a conserved non-canonical His41-Cys145 dyad located within the cleft between domains I and II [54]. Using Discovery Studio version 2020 we followed the standard procedure of “preparing” the protein, which includes assignment of H atoms and force field (CHARMm), and selected the Cys145-His41 area for a docking sphere, 9.5 Å radius, to perform the docking of embelin, Fig. 1. Of 10 possible poses, numbers 6 and 9 showed a potential sulfur quinone-centroid approach, resembling embelin interaction with the superoxide anion [ 53]. CDocker interaction energy is -36.0 kcal/mol.
Fig. 1.
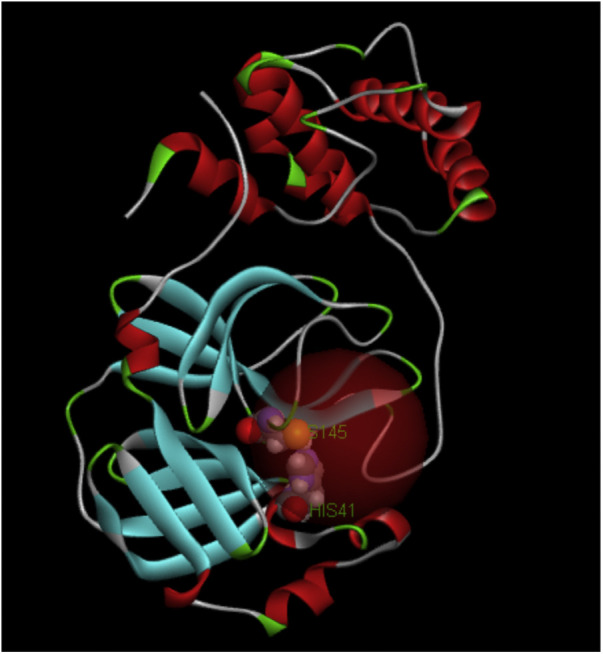
Protein 6Y84, the active site involves Cis145 and His41 CPK style. Docking sphere radius =9.5 Å.
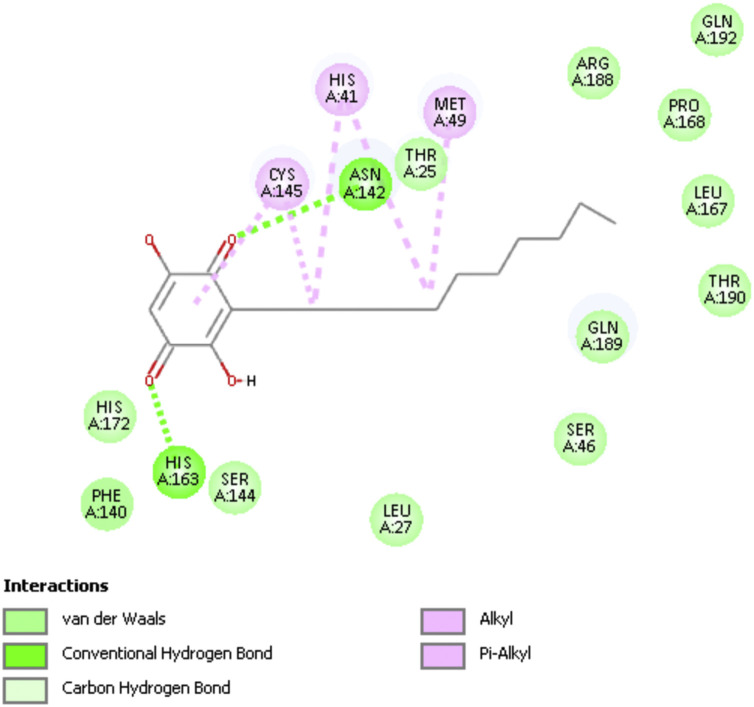
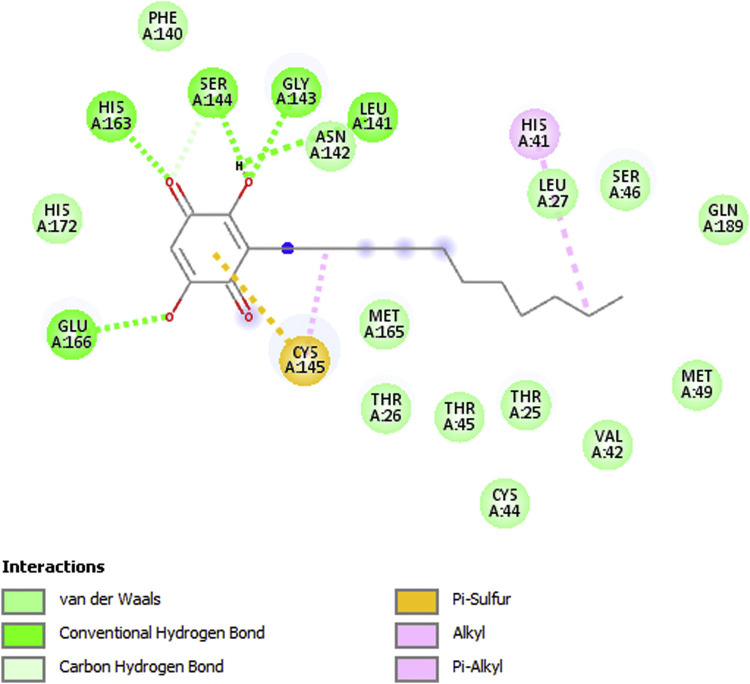
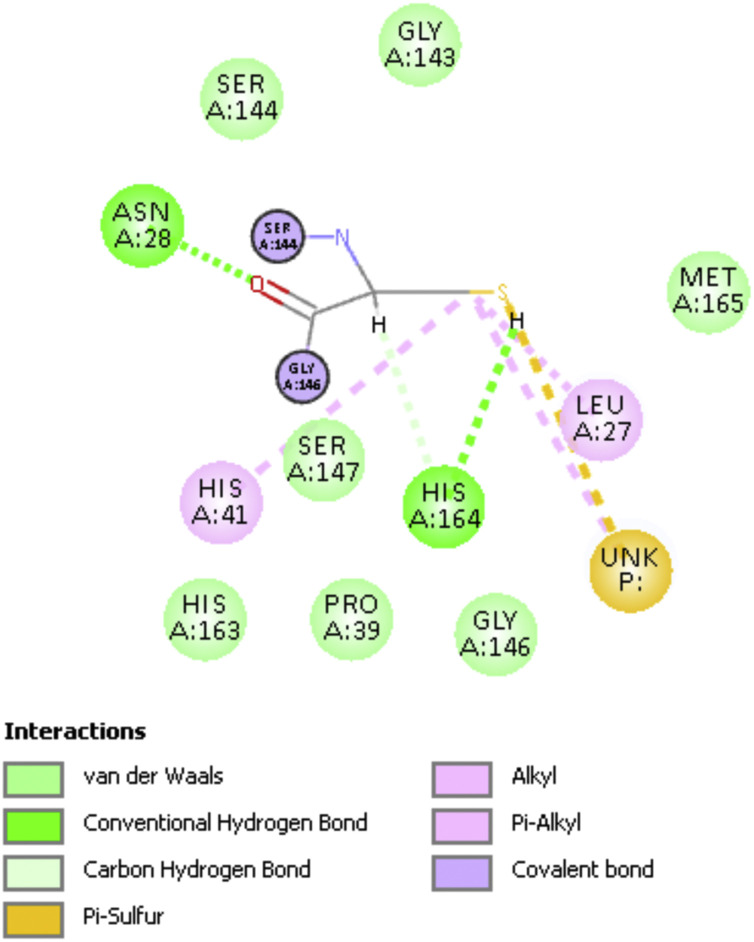
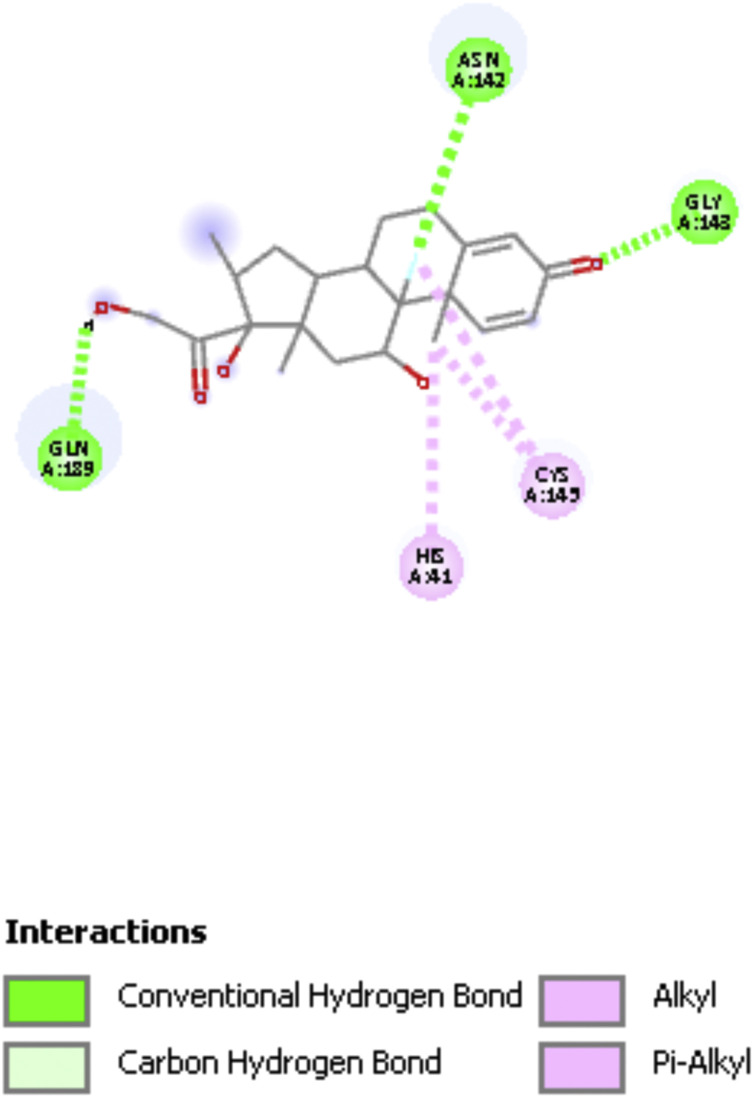
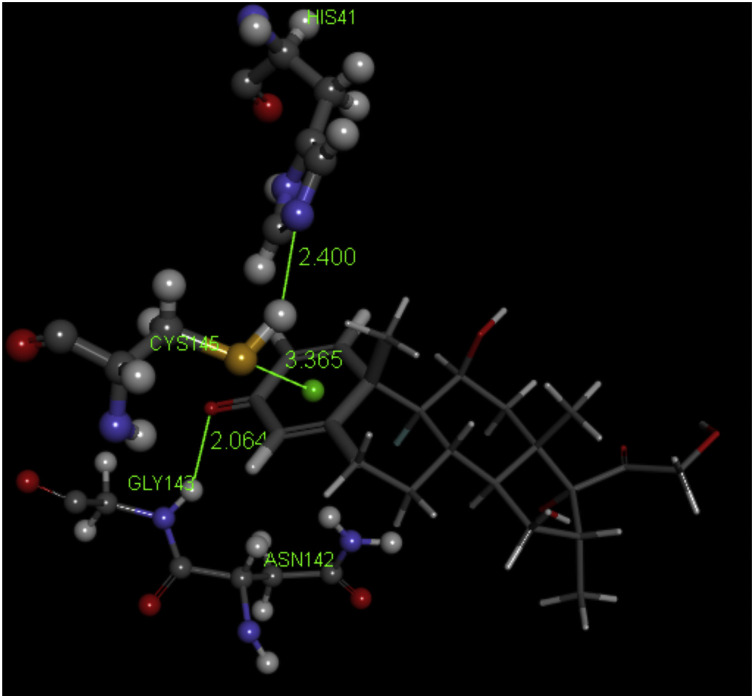
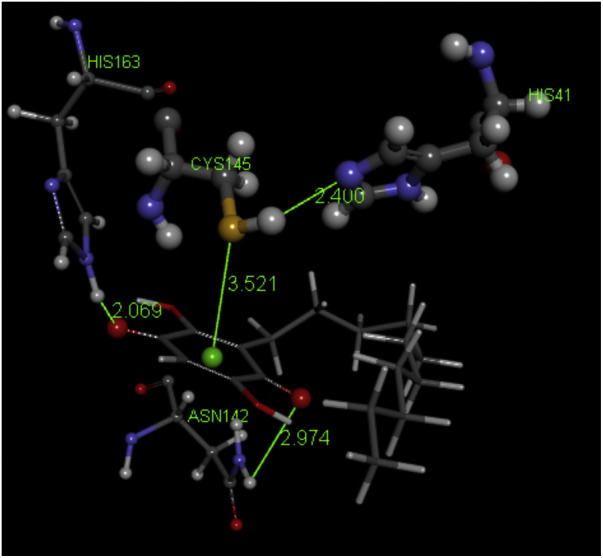
Fig. 2 shows a 2D display of interactions for embelin pose 9, and Fig. 3, Fig. 4 are 2D display of residual interaction with embelin and Cys145, respectively for docked Pose 9 after dynamic cascade. A selection of interacting residues was made to submit a DFT study and Fig. 5 shows the initial set of molecules, N(imidazole-His41) has a H-bond to the H(Cys145), 2.40 Å, and was explored as an inducer of S-H cleavage. A related H-bond was also established by N(His164) with the same H(Cys145) through its O(carbonyl) peptide chain, omitted in Fig. 5 for clarity. Interestingly, after dynamic cascade of pose 9 there is a H-bond between O(carbonyl-His164) and H(Cys145), while H(imidazole-His163) establishes a H-bond with one O(carbonyl) quinone, 1.859 Å, Fig. 6 .
Fig. 2.
Pose 9 docking showing embelin interactions, 6Y84.
Fig. 3.
2D interactions of embelin after dynamic cascade of pose 9 structure, 6Y84.
Fig. 4.
Pose 9 dynamic cascade Cys145 interactions, His41 is now not forming H-bond with Cys145, though His164 is confirmed.
Fig. 5.
3D embelin docked pose 9: H(Cys145) interacts with N(His41), 2.400 Å, and S(Cys145) points to the quinone centroid, 3.521 Å, whereas H(Asn142) is located 2.974 Å from one O(carbonyl) quinone, and the other O(carbonyl) quinone has a H-bond with H(His163), 2.069 Å. The essential amino acids involved are also depicted in Fig. 7.
Fig. 6.
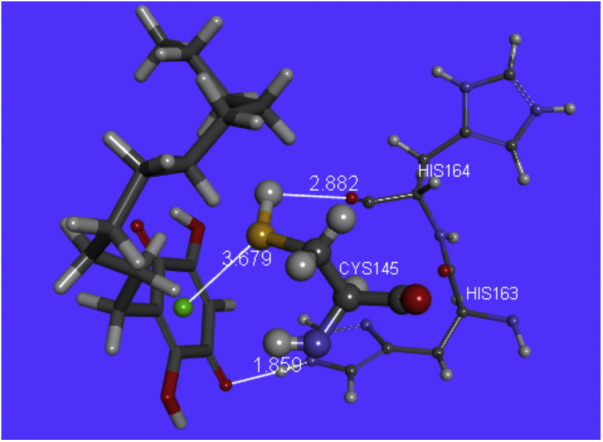
3D Pose 9 cascade of embelin. H(Cys145) interacts with O(carbonyl-His164), 2.882 Å, S(Cys145) points to the quinone centroid, 3.679 Å, whereas H(imidazole-His163) establishes a H-bond with one O(carbonyl) quinone, 1.859 Å.
Protease DFT studies
Embelin docked pose 9 was treated with DFT methods. The essential molecular components of this pose, Cys145, His41 and model embelin with its C11 tail substituted by an ethyl group, are shown in Fig. 7, containing 3 molecules as found after docking procedure. For simplification, the peptide chain is replaced by 2 H atoms, which were kept fixed during the geometry optimization. This is imposed to conserve the protein environment; Fig. 8 shows the best refined structure after DFT minimization and Fig. 9 its converged structure after adding an H atom to the embelin O(carbonyl) and performing geometry optimization.
Fig. 7.
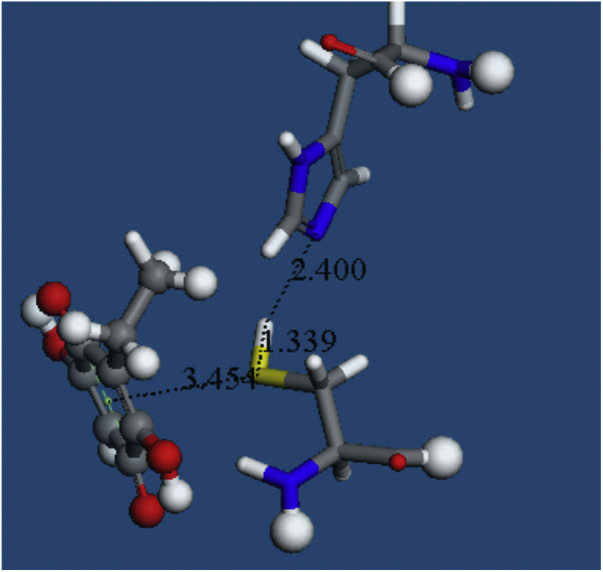
Cys145, His41 and embelin as found in pose 9 of docking. Embelin model has its C11 tail substituted by a C2H5 group, one of these H atoms is stick style, kept fixed during minimization.
Fig. 8.
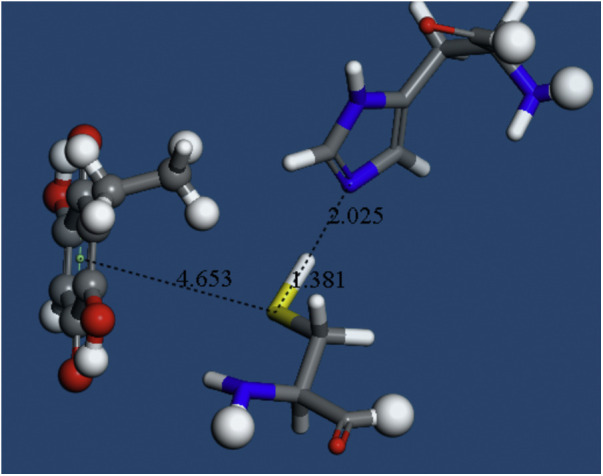
The converged system after geometry optimization of Fig. 7. The S atom points to the quinone ring centroid, 4.653 Å and the N(imidazole) approaches H(Cys145), 2.025 Å.
Fig. 9.
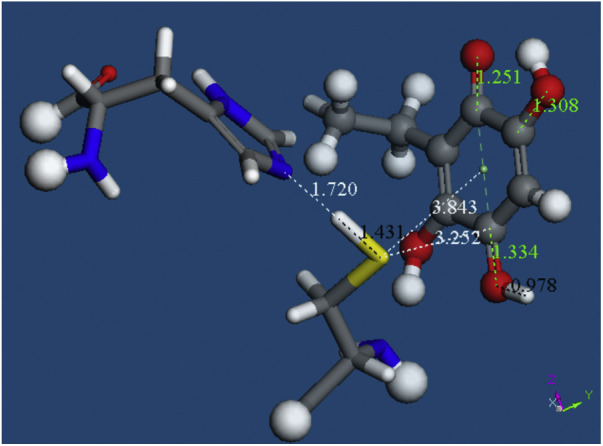
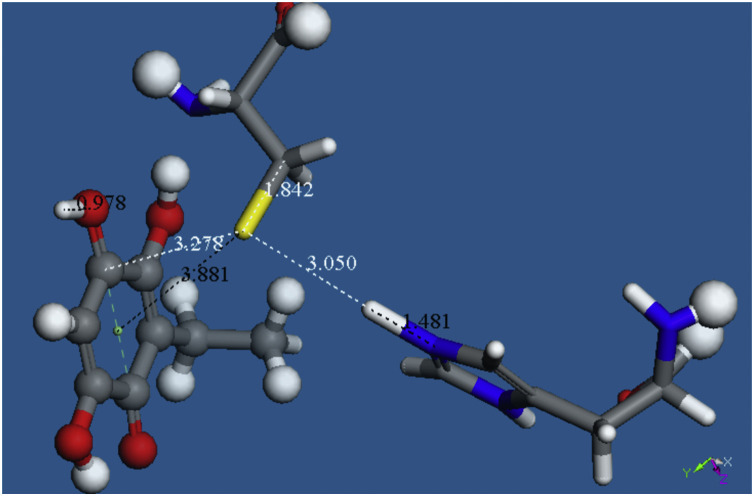
Starting from the system shown in Fig. 8, we added a proton, stick style, separated at van der Waals distance from the embelin O(carbonyl) located farther away from the C2H5 group. After geometry optimization this H binds to the O(carbonyl), 0.978 Å and S(Cys145) points to the C(quinone) associated to the proton approach, 3.252 Å.
We added a proton to the system shown in Fig. 8, separated at van der Waals distance from the embelin O(carbonyl) located farther away from the C2H5 group, indicated as stick style in Fig. 9. The rational of this insertion is based on the H-bond contact shown in Fig. 5, H(His163) – O(carbonyl) =2.069 Å. The result of minimization is shown in Fig. 9 and includes the Cys145 S-H bond now lengthened, 1.431 Å, compared with that of Fig. 8, 1.381 Å.
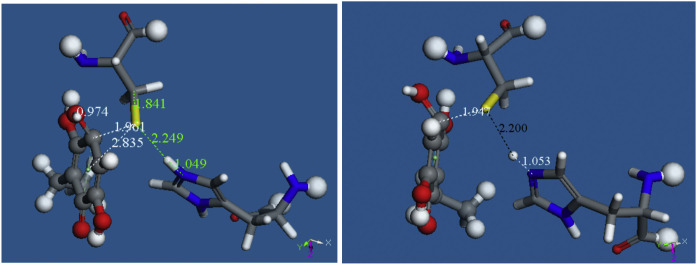
Our next approach was to study the product of this reaction, shown in Fig. 10, Fig. 11. This was obtained after linking the H atom to N(imidazole) at bonding distance, and leaving the remaining Cys145 group at van der Waals separation, d(S---H) =3.05 Å. ΔG of this reaction is -11.7 kcal/mol. H(Cys145) was placed slightly farther from N(imidazole), 1.481 Å, instead of the expected 1 Å, making this calculation more exigent. As a result, H is captured by N(imidazole); Fig. 11 left shows the H(Cys145) retained by N(imidazole-His41), d(N-H) =1.049 Å, while S(Cys145) establishes a chemical bond with the C(quinone), d(S-C) =1.961 Å and this is the same C atom already bound to the proton, d(C-H) =0.974 Å, as seen in Fig. 9.
Fig. 10.
Initial atomic positions to study a potential product of Fig. 8, having H captured by N(imidazole).
Fig. 11.
Left: Final atomic positions obtained after performing a geometry optimization from the system shown in Fig. 10. Right: After minimization of the same system, but not imposing any restraint.
We were interested to see if there is some energy barrier in the process of capturing H(Cys145) by N(imidazole-His41). Since we mimicked the polypeptide role by replacing the amino acid chain linked to Cys145 and His41 with 2 H atoms each, these 4 H were kept fixed to conserve the protein environment. However, fixing atoms both in the reagent and the product does not allow a canonical search for transition state to be performed. Basically, our quantum mechanical program does not permit atoms to be in equivalent positions in both the reagent and the product; in this case the “H” atoms fixed during minimizations that replaced the polypeptide chain. We therefore decided to perform geometrical minimization without any constraints in both systems, corresponding to Figs. 9 (reagent) and 11 left (product). From these geometry optimizations the product showed minimal atomic variation, that is, the product is confirmed. In contrast, for the reagent the obtained structure was just the product, Fig. 11 right, which ensures that no barrier exists, and the system can evolve spontaneously towards the product. We conclude that the docking of pose 9 provides solid elements to establish an important role for embelin, as it is able to inhibit the protease by establishing a strong S(Cys145) bond to the quinone ring.
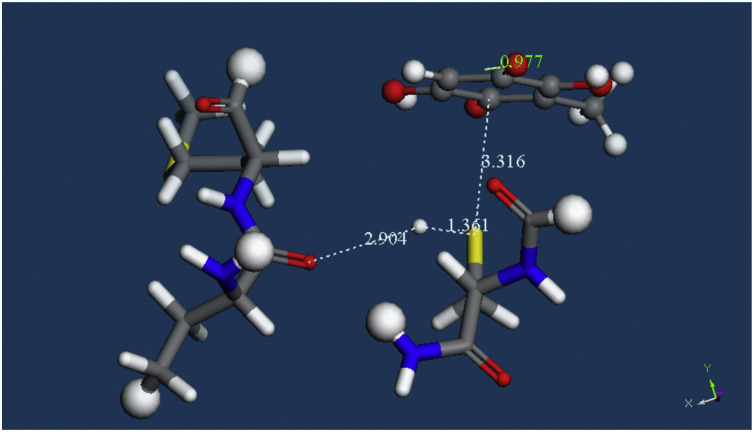
We already mentioned that after docking of embelin into the protease site, pose 9 provided two potential interactions for H(Cys145), one having N(imidazole-His41), explained above. The other was O(carbonyl-His164), at the peptide chain, which was confirmed after a standard dynamic cascade, Fig. 6. We proceeded with this configuration along the same lines shown in Fig. 7, Fig. 8, Fig. 9, Fig. 10, Fig. 11 and the behavior was similar. However, when we tried to minimize the expected product, equivalent to Fig. 11, that is having the H[S(Cys145)] captured by O(His164), this H atom returned to the S-Cys moiety and so there is no capture of H by the O(carbonyl), Fig. 12. In fact, the H atom became bound to the cysteine molecule, d(S-H) =1.361 Å. We conclude that His164 is not useful to assist in cleavage of the S-H bond. Therefore, His41 is better suited than the O(carbonyl-His164) at establishing a H-bond useful for reactivity, so that the S(Cys145) ends up bonding to the C(quinone) located farther away from the C 11 tail. Fig. 5 shows that this process is assisted by an initial approach of a proton towards the same O(carbonyl) quinone to be bound by S(Cis145): a H-bond from H(His163), 2.069 Å. The binding of Cys34 on human serum albumin (HSA) was studied and binding experiments with HSA demonstrated that Cys34 (the only reduced thiol in HSA) is the preferred target for benzoquinone binding and that the bond formed is irreversible and covalent [55]. Moreover, in a very recent communication, the crystal structure of the same protein protease used by us here has been reported along with inhibitors of Covid-19 containing keto-amide groups [18]. It was shown the role of Cys145, reacted with the C(aldehyde) of two inhibitors, at the catalytic site of SARS-CoV-2 Mpro, forming a standard 1.8 Å C–S covalent bond. Using a fluorescence resonance energy transfer (FRET)-based, one of these inhibitors exhibited 100% SARS-CoV-2 Mpro inhibition activity, with IC50 of 0.053 ± 0.005 μM. This result is in agreement with the reactivity of embelin at molecular level using DFT methods we describe here.
Fig. 12.
Embelin: The H atom was bound to the O(Carbonyl) of His164. Upon geometry minimization the O(carbonyl) rejected the H atom, d(O-H) =2.904 Å.
Emodin
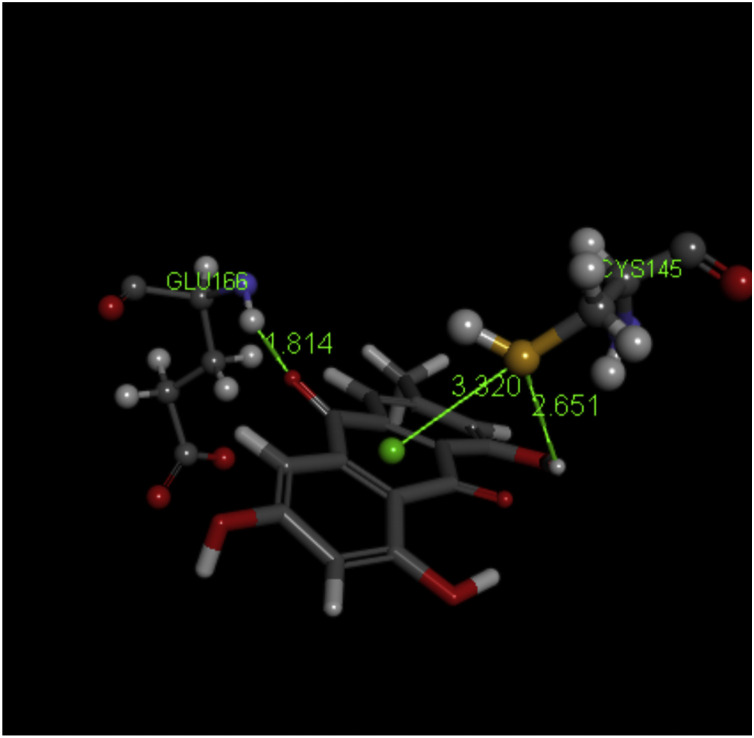
Emodin is another interesting natural quinone found in goji berry (Lycium barbarum and Lycium chinense). Docking on PDB 6Y84 protein never shows Cys145 pointing towards the quinone ring, rather in several poses S is directed to one aromatic ring adjacent to the quinone ring. Glu166 provides a H-bond to a carbonyl, suggesting a similar action than found for embelin [donation of H by HN(His163)] to generate a hydroxyl moiety. Interestingly, Asn142 points its two H atoms (NH2) to the quinone ring, thus suggesting a potential Schiff reaction with the O(carbonyl) not engaged in internal H bonds of emodin. This type of reaction between cysteine and quinone has been described [40,56]. CDocker interaction energy is -29.6 kcal/mol. Therefore, we performed a dynamic cascade on one pose and the S atom became close to the quinone center suggesting a potential role for emodin as an inhibitor of this SARS-CoV-2 3CLpro, also known as Mpro protease, Fig. 13 . We plan to do further work to completely elucidate this reactivity of emodin.
Fig. 13.
Essential amino acids interacting with emodin, after standard dynamic cascade S(Cys145) shifts from the docking position pointing now to the quinone centroid, 3.320 Å. The emodin carbonyl not engaged in internal H-bonds seems to accept a H atom from Glu166, d(O-H) =1.814 Å.
Discussion
The present study shows that for embelin some amino acid residues at the Covid-19 protease active site, including Cys145, His41 and His163, are important factors to establish interaction with the protease. The associated mechanism is indicated in the following scheme 2 .
Scheme 2.
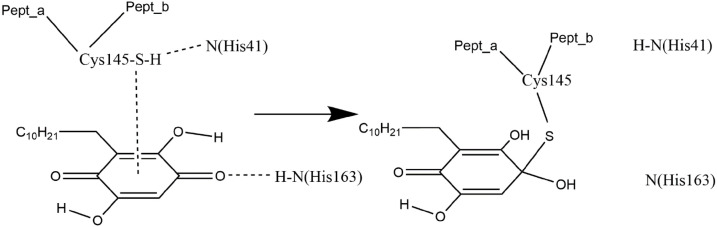
Reactivity of embelin with Covid-19 protease. The S atom points to the quinone centroid, while the N(imidazole-His41) acceptor attacks H[S(Cys145)] and H(His163) donor provides an H to one O(carbonyl). Pept_a and Pept_b represent the polypeptide chain bound to Cys145.
The most important result is the covalent bond formed between Cys145, after losing its H atom, with one carbonyl of embelin, thus inhibiting the protease virus. The other potential aminoacid residue able to establish an interaction similar to His41, [His164 through its O(carbonyl)], is shown to be not effective. Emodin seems to act similarly, and progress in its potential mechanism is underway by our group. Interestingly, it was only after we applied molecular dynamics that the interaction between S(Cys145) and the quinone centroid was established. That is, the docking procedure showed, instead, S(Cys145) interacting with one aromatic ring centroid adjacent to the quinone ring.
Interaction of some important compounds with the same Covid-19 protease
(1) The anti-inflammatory drug methyl prednisolone is being used to treat Covid-19 patients [7]. Our interest was stimulated by its p-methide-quinone moiety, resembling quinone embelin. We used crystal coordinates from the CSD database (refcode MTHPRG) for docking in PDB 6Y84, and see the methide-quinone ring establishes interactions with the receptor in agreement to those shown earlier by embelin. CDocker interaction energy is -35.2 kcal/mol for this pose, Fig. 14 .
Fig. 14.
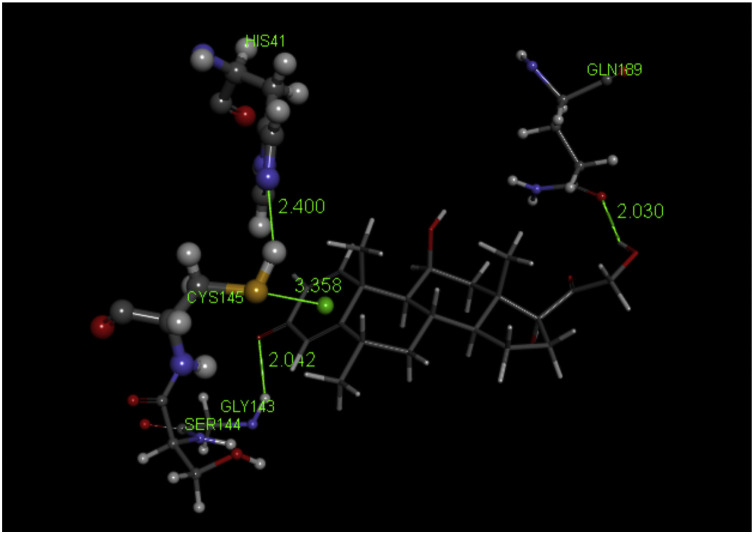
Methyl prednisolone docking pose 2, its O(carbonyl) has a H-bond contact with N(Gly143), 2.042 Å; S is pointing to the ring centroid, 3.358 Å, and N(His41) has a H-bond to H(Cys145), 2.400 Å.
(2) Recently, doctors at the University Medical Center, Maastricht, The Netherlands noted severely reduced vitamin K levels in Covid-19 patients [57], and suggested it was due to a pathological condition. There is marked structural similarity between embelin and vitamin K, including their long alkyl tails. Docking of Vitamin K on the same PDB 6Y84 Covid-19 protease receptor shows CDocker interaction energy of -32.8 kcal/mol. Although the separation between S(Cys145) and the quinone center is slightly longer than in methyl prednisolone, Fig. 15 , the pattern is similar to that of embelin. We conclude that vitamin K decrease in Covid patients may be due to S(Cys145)-vitamin K bond formation.
Fig. 15.
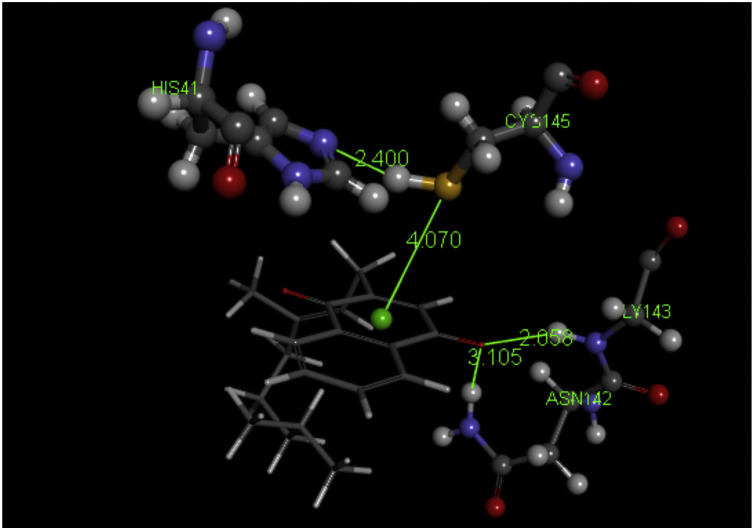
Vitamin K pose 3: The quinone vitamin K ring centroid is 4.070 Å from S(Cys145). A double H-bond is established by Gly143 and Asn142, 2.058 Å and 3.105 Å, respectively, to one O(carbonyl).
Vitamin K, embelin and coenzyme Q10, are all quinones having long alkyl tails that allow insertion in the host cell membrane, a property recently described for embelin [53]. Therefore, we also explored coenzyme Q10 to verify its insertion at the protease active site. Again, correlation with the earlier described embelin mechanism was observed, Fig. 16, and its CDocker interaction energy is -33.2 kcal/mol. We conclude the viral protein PDB 6Y84 protease is an excellent target receptor for quinones such as embelin, vitamin K or coenzyme Q10. This is consistent with the anti-inflammatory drug methyl prednisolone, recently used to treat Covid-19 patients.
Fig. 16.
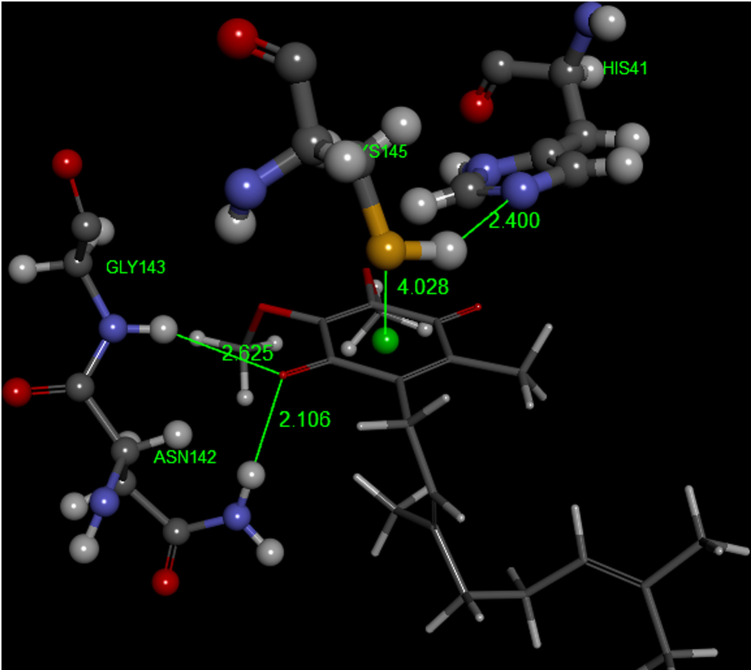
A model of coenzyme Q10 Pose 5 docking on 6Y84 virus protease. As for vitamin K, a double H-bond is established by Gly143 and Asn142. S(Cys145) points to the quinone center, 4.028 Å, and N(His41) is H-bound to H(Cys145), 2.400 Å.
Embelin can inhibit viral infection via a variety of mechanisms and is active against influenza and hepatitis B viruses [[35], [36]]. Our preliminary biological study indicates that it is also an inhibitor for HSV-1 by acting directly on the virions [41], providing elements of investigation related to Covid-19 virus. Recently, we demonstrated that, as an antioxidant scavenger of the superoxide radical, embelin acts in an unusual way. Instead of providing an H atom from one of its hydroxyls to the radical as do most polyphenolic antioxidants, it prefers to capture the superoxide electron (effect a) [53]. This is accompanied by inserting a proton at one O(carbonyl) of embelin, to form a single C-OH bond (effect b). This mechanism seems to be reflected in this study when embelin is docked into the Covid-19 virus protease PDB 6Y84: Cys145 points its S atom to the ring centroid of embelin (effect a), while another amino acid establishes an important H-bond with the embelin O(carbonyl), from H(His163), 2.069 Å, effect b. Meanwhile, N(His41) establishes a H-bond to the H[S(Cys145)] assisting in its cleavage from the S atom. That is, it increases the negative charge on the S atom, inducing it to point to the quinone center, Fig. 5. The final result is bond formation between S(Cys145) and the same carbon also attacked by the proton through the HN(His163) donation: a Michael addition [56].
On 15 June 2020 the World Health Organization (WHO) “welcomed the preliminary results about clinical trials performed by the corticosteroid dexamethasone, used in treating critically ill COVID-19 patients. For patients on ventilators, the treatment was shown to reduce mortality by about one third” [8] and this result was very recently published [58]. Since dexamethasone is also a methide-quinone, we were interested in performing a docking calculation using the crystal structure coordinates from the CSD Database (refcode DEXMET11). We correlate its structural features with other compounds of this study and describe its docking into the same Covid-19 protease PDB 6Y84. The CDocker interaction energy is -29.8 kcal/mol for pose 8 shown in Fig. 17, Fig. 18. H(Gly143) has a H-bond to the O(carbonyl) of the drug methide-quinone ring, 2.064 Å, having the same role of H-N(His163) in Scheme 2. Gly143 exerts the same role in methyl prednisolone, Fig. 14.
Fig. 17.
2D interaction of amino acid with dexamethasone docked pose 8.
Fig. 18.
Essential amino acids interactions with dexamethasone. H(Gly143) probably exerts the same role as H-N(His163) in Scheme 2.
When comparing embelin interaction at the 6Y84 protease target with methyl prednisolone, dexamethasone, vitamin K and coenzyme Q10, a consistent trend is observed. The S(Cys145) is ready to interact with the quinone centroid (a quinone methide for methyl prednisolone and dexamethasone), and there is an acceptor amino acid prone to establish a H-bond with H[S(Cys145)] for S-H cleavage, and another amino acid residue which provides H-bond donation to a quinone O(carbonyl), with the final result being S(Cys145) bound to a C(carbonyl) belonging to the quinone (or quinone methide for methyl prednisolone and dexamethasone) and which inhibits the virus protease. Emodin, a quinone present in goji berry, has recently received approval for a patent against Covid-19 [59] and we plan to further explore emodin interaction with Covid-19 protease.
Besides the recent important study of the main protease Covid-19 crystal structure containing α-ketoamide inhibitors [18], the interaction between potential drugs and this active site is subject of current interest and intense studies using molecular mechanics techniques. This is shown in very recent literature [[60], [61], [62], [63]] for several compounds docked at the site. However, to our knowledge, specific detailed chemical mechanisms regarding molecular interaction are not described. In contrast, our work on the action of quinones, as described here, suggests inhibition of the protease by a well-defined chemical mechanism. Thus, there is a specific covalent bond formation between a quinone carbonyl and the Cys(145) thiolate, helped by 2 proton transfers from neighboring residues. One residue, His41, allows cleavage of the H-S(Cys145) bond by capturing the H-(Cys145), and the second residue releases a proton to the same C(carbonyl) quinone engaged with S(Cys145) thiolate. The second amino acid residue may differ when comparing embelin with dexamethasone, methyl prednisolone, coenzyme Q10 and Vitamin K docking into the protease target, but the mechanism appears to be similar.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.https://www.gilead.com/purpose/advancing-global-health/covid-19/remdesivir-clinical-trials.
- 4.https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)31324-6/fulltext.
- 5.Funck-Brentano C., Nguyen L.S., Salem J.-E. Retraction and republication: cardiac toxicity of hydroxychloroquine in COVID-19. Lancet. 2020;39 doi: 10.1016/S0140-6736(20)31528-2. 10245, e2-e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahraei Z., Shabani M., Shokouhi S., Saffaei A. Aminoquinolines against coronavirus disease 2019 (COVID-19): Chloroquine or hydroxychloroquine. Internat J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fadel R., Morrison A.R., Vahia A., Smith Z.R., Chaudhry Z., Bhargava P., Miller J., Kenney R.M., Alangaden G., Ramesh M.S. Early short course corticosteroids in hospitalized patients with COVID-19. Clinic Infect Dis. 2020 doi: 10.1093/cid/ciaa601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.https://www.who.int/news-room/detail/16-06-2020-who-welcomes-preliminary-results-about-dexamethasone-use-in-treating-critically-ill-covid-19-patients.
- 9.Atzrodt C.L., Maknojia I., McCarthy R.D.P., Oldfield T.M., Po J., Ta K.T.L., Stepp H.E., Clements T.P. A guide to COVID-19: a global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS J. 2020 doi: 10.1111/febs.15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng X.-Y., Zhang H.-X., Mezei M., Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. 2011;7:146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sethi A., Joshi K., Sasikala K., Alvala M. Molecular docking in modern drug discovery: principles and recent applications. Drug Discov Dev Adv Intech Open. 2019 doi: 10.5772/intechopen.85991. [DOI] [Google Scholar]
- 12.Liu A.-L., Du G.-H. Antiviral properties of phytochemicals. Diet. Phytochem Microb. 2012:93–126. doi: 10.1007/978-94-007-3926-0_3. [DOI] [Google Scholar]
- 13.Newman D.J., Cragg G.M. Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 14.Notka F., Meier G., Wagner Concerted inhibitory activities of Phyllanthus amarus on HIV replication in vitro and ex vivo. Antivir Res. 2004;64:93–102. doi: 10.1016/j.antiviral.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Parida M.M., Upadhyay C., Pandya G., Jana A.M. Inhibitory potential of neem (Azadirachta indica Juss) leaves on dengue virus type-2 replication. J Ethnopharmacol. 2002;79:273–278. doi: 10.1016/S0378-8741(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 16.Rehman S., Ashfaq U.A., Riaz S., Javed T., Riazuddin S. Antiviral activity of Acacia nilotica against hepatitis C virus in liver infected cells. Virol J. 2011;8:220. doi: 10.1186/1743-422X-8-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goyal B., Goyal D. Targeting the Dimerization of the Main Protease of Coronaviruses: A Potential Broad-Spectrum Therapeutic Strategy. ACS Comb Sci. 2020;22:297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;80 doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chopra R.N., Nayar S.L., Chopra I.C. CSIR, "Glossary of Indian Medicinal Plants” New Delhi. Quart Rev Biol. 1958;33:156. [Google Scholar]
- 20.Haq K., Ali M., Siddiqui A.W. New compounds from the seeds of Embelia ribes Burm. Pharmazie. 2005;60:69–71. [PubMed] [Google Scholar]
- 21.Bhandari U., Ansari M.N., Islam F. Cardioprotective effect of aqueous extract of Embelia ribes Burm fruits against isoproterenol-induced myocardial infarction in albino rats. Indian J Exp Biol. 2008;46:35–40. [PubMed] [Google Scholar]
- 22.Chitra M., Devi C.S., Sukumar E. Antibacterial activity of embelin. Fitoterapia. 2003;74:401–403. doi: 10.1016/s0367-326x(03)00066-2. [DOI] [PubMed] [Google Scholar]
- 23.Ahn K.S., Sethi G., Aggarwal B.B. Embelin, an inhibitor of X chromosome-linked inhibitor-of-apoptosis protein, blocks nuclear factor-κB (NF-κB) signaling pathway leading to suppression of NF-κB-regulated antiapoptotic and metastatic gene products. Mol Pharmacol. 2007;71:209–219. doi: 10.1124/mol.106.028787. [DOI] [PubMed] [Google Scholar]
- 24.Naik S.R., Niture N.T., Ansari A.A., Shah P.D. Anti-diabetic activity of embelin: Involvement of cellular inflammatory mediators, oxidative stress and other biomarkers. Phytomedicine. 2013;20:797–804. doi: 10.1016/j.phymed.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Nikolovska-Coleska Z., Xu L., Hu Z., Tomita Y., Li P., Roller P.P., Wang R., Fang X., Guo R., Zhang M., Lippman M.E. Discovery of embelin as a cell-permeable, small-molecular weight inhibitor of xiap through structure-based computational screening of a traditional herbal medicine three-dimensional structure database. J Med Chem. 2004;47:2430–2440. doi: 10.1021/jm030420+. [DOI] [PubMed] [Google Scholar]
- 26.Allensworth J.L., Aird K.M., Aldrich A.J., Batinic-Haberle I., Devi G.R. XIAP inhibition and generation of reactive oxygen species enhances trail sensitivity in inflammatory Breast Cancer Cells. Mol Cancer Ther. 2012;11:1518–1527. doi: 10.1158/1535-7163. [DOI] [PubMed] [Google Scholar]
- 27.SreeHarsha N. Embelin impact on paraquat-induced lung injury through suppressing oxidative stress, inflammatory cascade, and MAPK/NF-κB signaling pathway. J Biochem Mol Toxicol. 2020;34 doi: 10.1002/jbt.22456. [DOI] [PubMed] [Google Scholar]
- 28.Heo J.Y., Kim H.J., Kim S.M., Park K.R., Park S.Y., Kim S.W. Embelin impact on paraquat-induced lung injury through suppressing oxidative stress, inflammatory cascade, and MAPK/NF-κB signaling pathway. Cancer Lett. 2011;308:71–80. doi: 10.1002/jbt.22456. [DOI] [PubMed] [Google Scholar]
- 29.Dai Y., Qiao L., Chan K.W., Yang M., Ye J., Ma J., Zou B., Gu Q., Wang J., Pang R., Lan H.Y. Peroxisome Proliferator-Activated Receptor-γ Contributes to the Inhibitory Effects of Embelin on Colon Carcinogenesis. Cancer Res. 2009;69:4776–4783. doi: 10.1158/0008-5472. [DOI] [PubMed] [Google Scholar]
- 30.Chen F., Zhang G., Hong Z., Lin Z., Lei M., Huang M. Hu Design, synthesis, and SAR of embelin analogues as the inhibitors of PAI-1 (plasminogen activator inhibitor-1) Bioorg Med Chem Lett. 2014;24:2379–2382. doi: 10.1016/j.bmcl.2014.03.045. [DOI] [PubMed] [Google Scholar]
- 31.Omura S., editor. The search for bioactive compounds from microorganisms. Springer; New York: 1992. ISBN 978-1-4612-4412-4417. [Google Scholar]
- 32.Yeggoni D.P., Rachamallu A., Subramanyam R. A comparative binding mechanism between human serum albumin and α-1-acid glycoprotein with corilagin: biophysical and computational approach. J Photochem Photobiol B. 2016;160:248–259. doi: 10.1016/j.jphotobiol.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 33.Shuveksh P.S., Ahmed K., Padhye S., Rainer S.R., Biersack B. Chemical and Biological Aspects of the Natural 1,4-Benzoquinone Embelin and its (semi-)Synthetic Derivatives. Curr Med Chem. 2017;24:1998–2009. doi: 10.2174/0929867324666170116125731. [DOI] [PubMed] [Google Scholar]
- 34.Radhakrishnan N., Gnanamani A. 2,5-dihydroxy-3-undecyl-1,4-benzoquinone (Embelin)-A second solid gold of India—a review. Int J Pharm Pharmaceutical Sci. 2014;6:23–30. [Google Scholar]
- 35.Hossan M.S., Fatima A., Rahmatullah M., Khoo T.J., Nissapatorn V., Galochkina A.V., Slita V., Shtro A.A., Nikolaeva V.V., Zarubaev V.V., Wiart C. Antiviral activity of Embelia ribes Burm. f. against influenza virus in vitro. Arch Virol. 2018;163:2121–2131. doi: 10.1007/s00705-018-3842-6. [DOI] [PubMed] [Google Scholar]
- 36.Parvez M.K., Rehman T., Alam P., Al-Dosari M.S., Alqasoumi S.I., Mohammed F., Alajmi M.F. Plant-derived antiviral drugs as novel hepatitis B virus inhibitors: Cell culture and molecular docking study. Saudi Pharm J. 2019;27:389–400. doi: 10.1016/j.jsps.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hashim N.M., Rahmani M., Sukari M.A., Yahayu M., Oktima W., Ali A.M., Go R. Antiproliferative activity of xanthones isolated from Artocarpus obtusus. J Biomed Biotechnol. 2012;130627:1–9. doi: 10.1155/2012/130627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono K., Nakane H., Shimizu S., Koshimura S. Inhibition of HIV-reverse transcriptase activity by asterriquinone and its analogues. Biochem Biophys Res Commun. 1991;174:56–62. doi: 10.1016/0006-291X(91)90484-O. [DOI] [PubMed] [Google Scholar]
- 39.Bogdanova N.S., Pershin G.N., Nikolaeva I.S., Grinev A.N., Shvedov V.I. Antiviral activity of p-benzoquinone and hydroquinone derivatives. Farmakol Toksikol. 1970;33:488–496. [PubMed] [Google Scholar]
- 40.Bittner S. When quinones meet amino acids: chemical, physical and biological consequences. Amino Acids. 2006;30:205–224. doi: 10.1007/s00726-005-0298-2. [DOI] [PubMed] [Google Scholar]
- 41.Rossi M., Caruso F., Adams S. to be submitted.
- 42.Delley B.J. From molecules to solids with the DMol3 approach. J Chem Phys. 2000;113:7756–7764. doi: 10.1063/1.1316015. [DOI] [Google Scholar]
- 43.Perdew J.P., Chevary J.A., Vosko S.H., Jackson K.A., Pederson M.R., Singh D.J., Fiolhais C. Atoms, molecules, solids, and surfaces: Applications of the generalized gradient approximation for exchange and correlation. Phys Rev B Cond Mat Mater Phys. 1992;46:6671–6687. doi: 10.1103/physrevb.46.6671. [DOI] [PubMed] [Google Scholar]
- 44.Becke A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys Rev A. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 45.Perdew J.P., Wang Y. Accurate and simple analytic representation of the electron-gas correlation energy. Phys Rev B. 1992;45:13244–13249. doi: 10.1103/physrevb.45.13244. [DOI] [PubMed] [Google Scholar]
- 46.Wu G., Robertson D.H., Brooks C.L., Vieth M.J. Detailed analysis of grid-based molecular docking: A case study of CDOCKER-A CHARMm-based MD docking algorithm. J Comput Chem. 2003;24:1549–1562. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- 47.Dai W., Zhang B., Su H., Li J., Zhao Y., Xie X. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das S., Roy A.S. Naturally occurring anthraquinones as potential inhibitors of SARS-CoV-2 main protease: A molecular docking study. ChemRxiv database. 2020 doi.org/10.26434/chemrxiv.12170904.v1. [Google Scholar]
- 49.Kamata M., Wu R.P., An D.S., Saxe J.P., Damoiseaux R., Phelps M.E., Huang J., Chen I.S. Cell-based chemical genetic screen identifies damnacanthal as an inhibitor of HIV-1 Vpr induced cell death. Biochem Biophys Res Commun. 2006;348:1101–1106. doi: 10.1016/j.bbrc.2006.07.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen H., Muhammad I., Zhang Y., Ren Y., Zhang R., Huang X., Diao L., Liu H., Li X., Sun X., Abbas G., Li G.L. Antiviral activity against infectious bronchitis virus and bioactive components of hypericum L. Front Pharmacol. 2019;10:1272. doi: 10.3389/fphar.2019.01272. https://doi-org.libproxy.vassar.edu/10.3389/fphar.2019.01272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Owen CD, Lukacik P, Strain-Damerell CM, Douangamath A, Powell AJ et al. Covid-19 main protease with unliganded active site, PDB.org Code 6Y84.
- 52.Rossi M., Wen K., Caruso F., Belli S. Emodin scavenging of superoxide radical includes π–π interaction. X-ray crystal structure, hydrodynamic voltammetry and theoretical studies. Antioxidants. 2020;9:194. doi: 10.3390/antiox9030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caruso F., Rossi M., Kaur S., Garcia-Villar E., Molasky N., Belli S., Sitek J.D., Gionfra F., Pedersen J.Z., Incerpi S. Antioxidant properties of embelin in cell culture. electrochemistry and theoretical mechanism of scavenging. potential scavenging of superoxide radical through the membrane cell. Antioxidants. 2020;5(9):E382. doi: 10.3390/antiox9050382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Q.-W., Su Y., Sheng J.-T., Gu L.-M., Zhao Y. Anti-influenza A virus activity of rhein through regulating oxidative stress, TLR4, Akt, MAPK, and NF-κB signal pathways. PLoS One. 2018;13 doi: 10.1371/journal.pone.0191793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mbiya W., Chipinda I., Siegel P.D., Mhike M., Simoy R.H. Substituent effects on the reactivity of benzoquinone derivatives with thiols. Chem Res Toxicol. 2013;26:112–123. doi: 10.1021/tx300417z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wehr H.M. 1972. Reactions of protein with phenols and quinones evaluation of amino acid modification and protein digestibility. Thesis.https://ir.library.oregonstate.edu/concern/graduate_thesis_or_dissertations/qb98mh88s [Google Scholar]
- 57.Dofferhoff ASM, Piscaer I, Schurgers LJ, Walk J et al. Reduced vitamin K status as a potentially modifiable prognostic risk factor in Covid-19 Preprints (www.preprints.org) Posted: 29 May 2020.
- 58.Ledford H. Coronavirus breakthrough: dexamethasone is first drug shown to save lives. Nature. 2020;582 doi: 10.1038/d41586-020-01824-5. 469-469. [DOI] [PubMed] [Google Scholar]
- 59.Wen H. Compounds for preventing and treating virus diseases and its application in preparing anti-viral drugs. Patent CN 111320541, Jun 23, 2020. Application number CN 2020-10222005.
- 60.Kumar Y., Singh H.C., Patel C.N. In silico prediction of potential inhibitors for the Main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug. J Infect Public Health. 2020 doi: 10.1016/j.jiph.2020.06.016. published in advance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: Drug repurposing approach. Life Sci. 2020;252 doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu R., Chen L., Lan R., Shen R., Li P. Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Internat J Antimicrob Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peele K.A., Durthi C.P., Srihansa T., Krupanidhi S., Ayyagari V.S., Babu D.J., Indira M., Reddy A.R., Venkateswarulu T.C. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: A computational study. Informat Med Unlock. 2020;19 doi: 10.1016/j.imu.2020.100345. [DOI] [PMC free article] [PubMed] [Google Scholar]