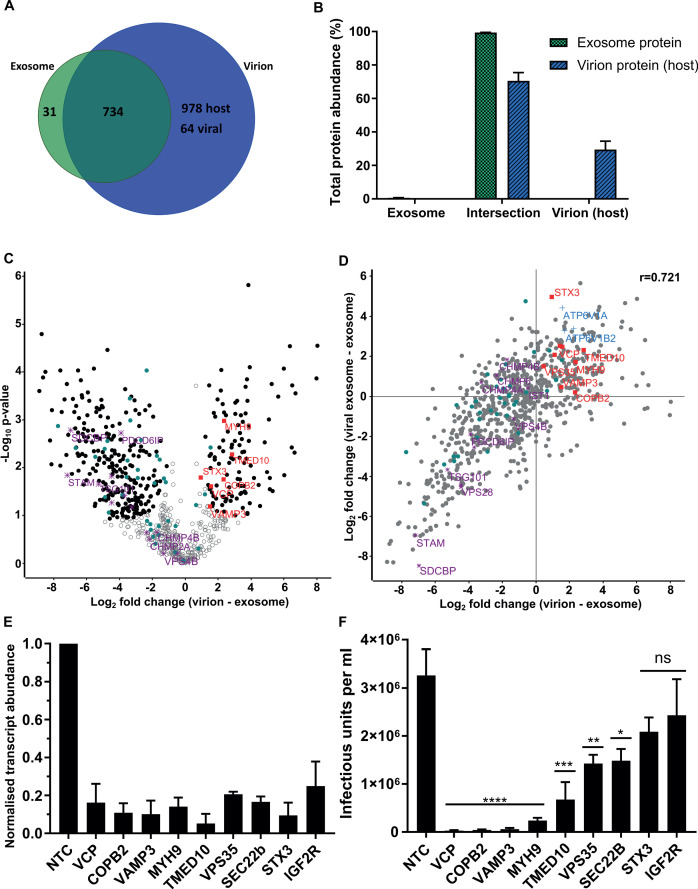
Figure 4. Host proteins in HCMV virions are predominantly exosome constituents.
(A) Venn diagram depicting the total number, and proteins commonly identified in exosomes and virions. (B) Abundance of individual protein groups (based on Venn diagram in panel A), as a proportion of overall total protein abundance, in exosomes or virion host proteins (n = 3, bars = SEM). (C) Volcano plot showing relative enrichment of host proteins in virions (positive fold change), compared to exosomes (negative fold change). Black circles: significant differential expression (n = 3, S0 = 0.4, FDR < 0.05, fold change >2). Red squares: envelopment/egress candidates, Teal circles: classical exosome markers, Purple stars: ESCRT sub-units. (D) Scatter plot correlating relative fold change comparisons of exosome vs viral exosome (Figure 3F), and exosome vs virion (C). Red squares: envelopment/egress candidates, Blue crosses: V-ATPase sub-units, Teal circles: classical exosome markers, Purple stars: ESCRT sub-units, (n = 3). (E) Relative transcript levels of host proteins following siRNA-mediated knock-down and HCMV infection (5 DPI, MOI = 3), normalized to cells treated with non-targeting control siRNA (n = 3, bars = SEM). (F) Quantification of infectious units released from siRNA-treated cells infected with HCMV AD169 5 DPI, MOI = 3. Infectious units in the supernatant were transferred to a reporter plate of uninfected cells, and IE1 assay conducted 24 HPI (n = 3, bars = SEM, ns: not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, one-way ANOVA with Dunnet’s post-test).