Abstract
Background
Lung cancer has high morbidity and mortality across the globe, and lung adenocarcinoma (LUAD) is the most common histologic subtype. Disordered lipid metabolism is related to the development of cancer. Analysis of lipid-related transcriptome helps shed light on the diagnosis and prognostic biomarkers of LUAD.
Methods
In this study, expression analysis of 1045 lipid metabolism-related genes was performed between LUAD tumors and normal tissues derived from the Cancer Genome Atlas Lung Adenocarcinoma (TCGA-LUAD) cohort. The interaction network of differentially expressed genes (DEGs) was constructed to identify the hub genes. The association between hub genes and overall survival (OS) was evaluated and formed a model to predict the prognosis of LUAD using a nomogram. The model was validated by another cohort, GSE13213.
Results
A total of 217 lipid metabolism-related DEGs were detected in LUAD. Genes were significantly enriched in glycerophospholipid metabolism, fatty acid metabolic process, and eicosanoid signaling. Through network analysis and cytoHubba, 6 hub genes were identified, including INS, LPL, HPGDS, DGAT1, UGT1A6, and CYP2C9. High expression of CYP2C9, UGT1A6, and INS, and low expressions of DGAT1, HPGDS, and LPL, were associated with worse overall survival for 1925 LUAD patients. The model showed that the high-risk score group had a worse OS, and the validated cohort showed the same result.
Conclusions
In this study, a signature of 6 lipid metabolism genes was constructed, which was significantly associated with the diagnosis and prognosis of LUAD patients. Thus, the gene signature can be used as a biomarker for LUAD.
Keywords: Diagnosis, Hub genes, Lipid metabolism, Lung adenocarcinoma, Nomogram, Prognosis, Signature
Background
Lung cancer is the most commonly diagnosed cancer (11.6% of the total cases) and the leading cause of cancer-related death (18.4% of the total cancer-related deaths) in the world [1]. Among lung cancer subtypes, adenocarcinoma is the most common histologic subtype of lung cancer in both men and women [2]. In a study published in 2020 in China, it was reported that in the recent 10 years, the percentage of lung adenocarcinoma (LUAD) increased significantly with a decrease in squamous carcinoma [3]. The increasing incidence of lung adenocarcinoma (LUAD) has also been reported to be associated with air pollution-related factors [4–6]. In previous studies, it was reported that PM2.5 increased the pro-inflammatory lipid metabolism in the lung and was associated with lipid alterations [7, 8]. The importance of alterations related to lipid metabolism is starting to be recognized, and the increase in de novo lipogenesis has considered a new hallmark in many aggressive cancers [9]. Lipid metabolism has been reported to be associated with many types of cancers, including pancreatic, hepatic, and colorectal cancer [10–12]. Cancer cells exhibit strong lipid and cholesterol uptake. Excess lipids and cholesterol in cancer cells are stored in lipid droplets (LDs) [13, 14]. LDs have been found in lung cancer cells [15]. Moreover, in a previous study, it was reported that some lipid-related phenotypic indices were associated with non-small cell lung cancer (NSCLC). Lipid profiles of blood plasma exosomes were used for the early detection of the prevalent NSCLC [16]. Epidemiological data indicated that a certain number of lung cancer patients with high high-density lipoprotein cholesterol (HDL-C) and low-density lipoprotein (LDL) and low-density lipoprotein receptor (LDLR) levels had better survival rates in patients [17, 18]. Compared with healthy subjects, NSCLC patients showed significant increases in levels of phosphatidylcholine (PC) and phosphatidylethanolamine (PE) [19]. Other lipid metabolism indicators associated with LUAD include sphingomyelins, phosphatidylinositols, phosphatidylserines, phosphatidylethanolamine, phospholipids, and phosphatidylcholine [20]. The requirement of cancer cells for metabolic intermediates for macromolecule production is overwhelming. Fatty acid oxidation (FAO) can help generate ATP to support the membranes formation, energy storage, production of signaling molecules by coordinating the activation of lipid anabolic metabolism [21]. Regulation of the lipid metabolism to LUAD is still being explored, and identifying the underlying lipid-related mechanism of the LUAD phenotype will help increase clinical interventions.
To explore the lipid metabolism related to regulatory networks and pathways, an integrated bioinformatic method was used to construct a transcript-wide profile, and a signature of lipid-related genes was analyzed to explore the potential biomarkers for diagnosis and prognostic value of LUAD in terms of lipid metabolism disorder.
Materials and methods
Patients and datasets
From 519 LUAD tissues and 58 normal tissues, mRNA expression data and clinical information were downloaded from The Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) database using the R package TCGAbiolinks [22]. The ensemble ID of TCGA samples was annotated with human genes GRCh38/hg38. To validate the availability of the final prediction model, mRNA expression data and clinical information from 117 LUAD patients were downloaded from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) (GSE13213) using the R package GEOquery [23].
Identification of lipid metabolism-related differentially-expressed genes
After using lipid-specific keywords (fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, sterol lipids, prenol lipids, saccharolipids, and polyketides), 21 lipid metabolism-related pathways and five lipid metabolism-related gene sets were collected from the Kyoto Encyclopedia of Genes and Genomes (KEGG) website (http://www.kegg.jp/blastkoala/) [24] and the Molecular Signatures Database (MisDB) website (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) [25], respectively (Additional file 1). After removing overlapping genes, a total of 1045 lipid metabolism-related genes were obtained. Lipid metabolism-related differentially expressed genes (DEGs) between LUAD tissues and normal tissues were screened through R package edgeR [26]. The parameters set for differential expression analysis were false discovery rate (FDR) < 0.05 and |log2 fold change| (logFC) > 1.
Bioinformatic analysis
The R package clusterProfiler was used to further explore the biological significance of lipid metabolism-related DEGs [27]. In GO and KEGG analysis, FDR < 0.05 was considered a significant enrichment. Next, DEGs containing gene identifiers and corresponding FDR values and logFC values were uploaded into the IPA software (Qiagen, Redwood City, CA, USA). The “core analysis” function included in the software was used to interpret the DEGs.
Interaction network generation and hub genes analysis
An interaction network of differentially-expressed lipid metabolism-related genes was built using the Search Tool for the Retrieval of Interacting Genes (STRING, http://string-db.org/) database [28]. The combined score of ≥0.4 was the cut-off value. Cytoscape software (version 3.6.0) was used to visualize networks [29]. According to 12 ranking methods in cytoHubba [30], an APP in Cytoscape, the top ten genes of each method were selected for analysis of overlapping genes, and genes with the highest number of overlaps were used as hub genes and potential biomarkers.
The expression level analysis of hub genes
The differences in mRNA expression of hub genes between LUAD tissues and normal tissues were verified using the Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/index.html) [31] and ONCOMINE (http://www.oncomine.org) websites. Gene correlation analysis for hub genes was performed using GEPIA.
Survival analysis
OS analysis of hub genes was employed by Kaplan-Meier Plotter (http://kmplot.com/analysis/), and included clinical data and gene expression information from 719 LUAD patients from GEO and TCGA database [32]. Subsequently, information on the number of cases along with median values of mRNA expression levels, the hazard ratio (HR) with a 95% confidence intervals (CI), and log-rank P-values were extracted from the Kaplan-Meier Plotter webpage. Log-rank P-values < 0.05 were considered statistically significant.
Prediction model
Based on the selected hub genes, the nomogram package of R (“rms”) [33] was used to develop a model to evaluate the prognosis of TCGA-LUAD patients. Using the formula of the nomogram, the prognosis score was calculated for each patient. Based on this score, and using the median classification method, patients were divided into a low-risk score group and a high-risk score group. The prognosis score was validated by the patients’ actual prognosis outcome. To investigate whether the expressions of six genes and prognosis score could be independent factors for OS, multivariate Cox regression analysis was performed, including gender, tumor stage, age, and smoking status. Next, expression data of hub genes and clinical information of 117 LUAD patients were downloaded from a different data set (GSE13213), and calculated the prognosis score of each patient by using the formula of the nomogram. Next, patients were divided into two groups, and survival analysis was performed to validate the availability of this model.
Results
Identification and functional analysis of lipid metabolism-related DEGs
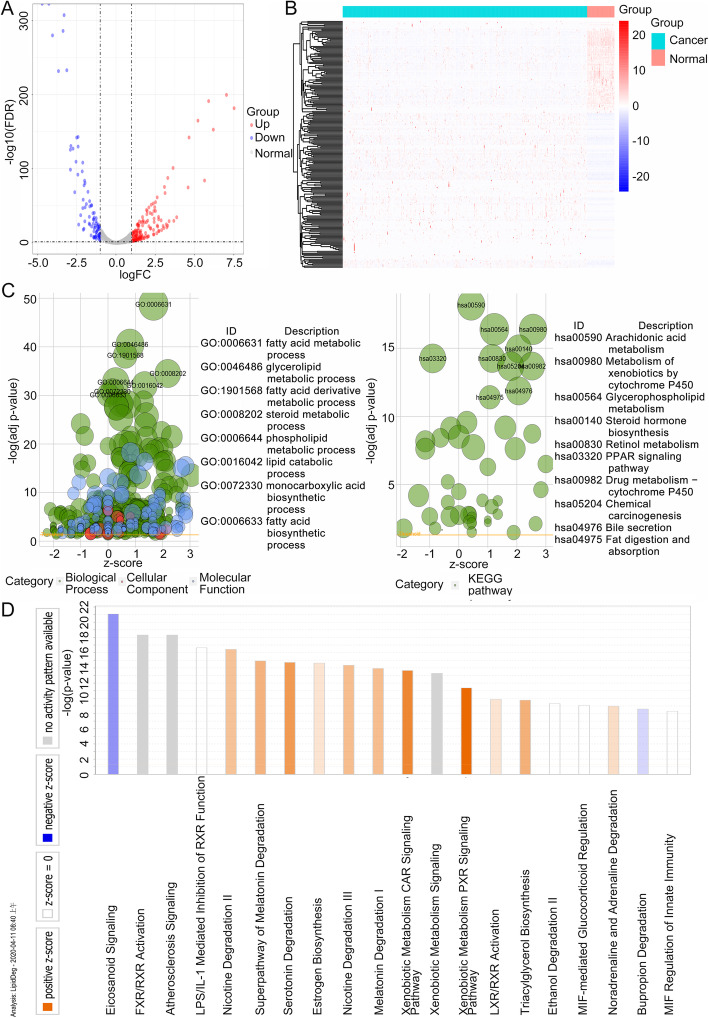
A total of 217 lipid metabolism-related DEGs were identified from the TCGA-LUAD cohort. A volcano plot was constructed to reveal significant DEGs (Fig. 1a), and a heatmap was created to show the hierarchical clustering analysis of the DEGs (Fig. 1b). For the overall understanding of 217 lipid metabolism-related DEGs, GO terms and KEGG pathway enrichment analysis were conducted using the clusterProfiler package, while canonical pathways analysis was performed by IPA. The results of KEGG pathway enrichment showed that DEGs were significantly enriched in arachidonic acid metabolism, metabolism of xenobiotics by cytochrome P450, glycerophospholipid metabolism, and steroid hormone biosynthesis. In contrast, GO terms analysis showed that genes were significantly enriched in the fatty acid metabolic process, glycerolipid metabolic process, fatty acid derivative metabolic process, and steroid metabolic process (Fig. 1c). The genes in each KEGG pathway and GO term are presented in Additional file 2. IPA identified significant canonical networks associated with the DEGs. IPA showed that the top canonical pathways associated with common DEGs including eicosanoid signaling, FXR/RXR activation, and atherosclerosis signaling (Fig. 1d). Combining the results of the three functional analyses showed that DEGs mainly overlapped in glycerophospholipid and steroid metabolism. Furthermore, non-overlapping pathways provided additional information indicating further exploration of the role of lipid metabolism in LUAD.
Fig. 1.
Identification and functional analysis of lipid metabolism-related DEGs. a Volcano plot of lipid metabolism-related genes, b Heatmap analysis of lipid metabolism-related DEGs, c GO and KEGG pathway enrichment analysis by clusterProfiler, d functional and signaling pathway enrichment by IPA. In a and b, red, white, and blue represent higher expression levels, no expression differences, and lower expression levels, respectively
Interaction network construction and cytoHubba analysis
Lipid metabolism-related DEGs were analyzed by the STRING tool. Ultimately, an interaction network with 216 nodes and 1140 edges was established and visualized in Cytoscape (Fig. 2). According to 12 ranked methods in cytoHubba, 6 hub genes were identified by the overlap of the top 10 genes (Additional file 3). Moreover, these genes were related to Insulin (INS), Lipoprotein Lipase (LPL), Hematopoietic Prostaglandin D Synthase (HPGDS), Diacylglycerol O-Acyltransferase 1 (DGAT1), UDP Glucuronosyltransferase Family 1 Member A6 (UGT1A6), and Cytochrome P450 Family 2 Subfamily C Member 9 (CYP2C9).
Fig. 2.
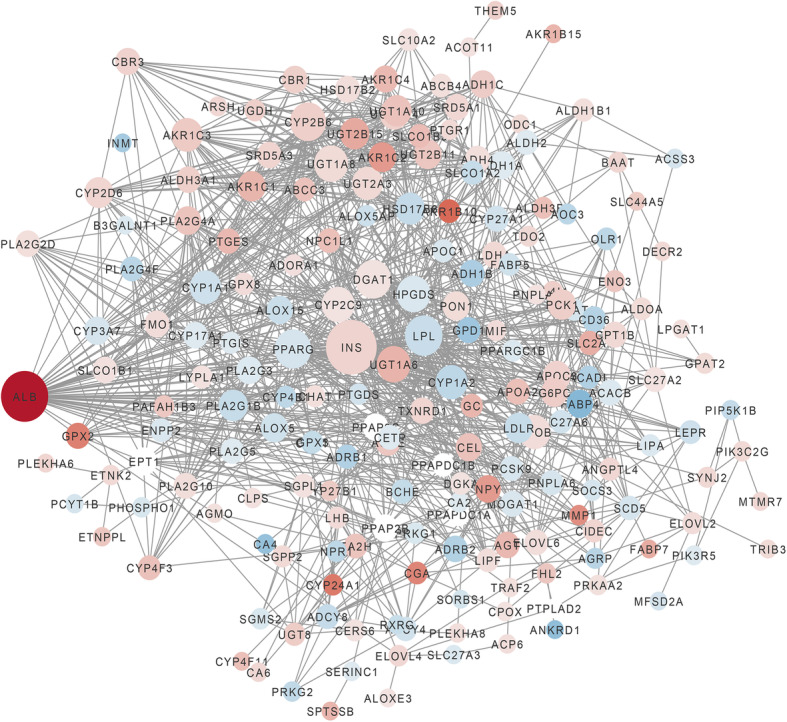
Genes interaction network of lipid metabolism-related DEGs. Red, white, and blue nodes represent upregulated genes, no expression differences genes, and downregulated genes, respectively. The magnitude of the degree is positively correlated with the size of a node
The expression level analysis of hub genes
DEG results of hub genes are presented in Table 1. The data showed that CYP2C9, UGT1A6, INS, and DGAT1 were upregulated, while HPGDS and LPL were downregulated in TCGA-LUAD tissues compared to normal tissues. To verify the expression results of hub genes, GEPIA and ONCOMINE databases were used. In GEPIA databases, HPGDS and LPL were significantly downregulated in LUAD samples (Fig. S1). In addition, correlation analysis showed that LPL and DGAT1 (r = 0.15; P < 0.01), UGT1A6 and HPGDS (r = − 0.11; P = 0.02), and HPGDS and DGAT1 (r = − 0.09; P < 0.05) were significantly correlated (Additional file 4). Meta-analysis of 6 hub genes of lung cancer was performed by ONCOMINE databases, and showed that UGT1A6 and DGAT1 were upregulated, while HPGDS and LPL were downregulated (Fig. S2).
Table 1.
DEG results of hub genes
| logFC | P | FDR | |
|---|---|---|---|
| CYP2C9 | 1.027946 | 0.001961 | 0.004128 |
| UGT1A6 | 3.382161 | 4.80E-31 | 6.74E-30 |
| INS | 1.773607 | 2.42E-07 | 8.55E-07 |
| DGAT1 | 1.041643 | 4.93E-14 | 3.24E-13 |
| HPGDS | −1.19395 | 6.18E-18 | 5.18E-17 |
| LPL | −1.96376 | 5.62E-83 | 2.01E-81 |
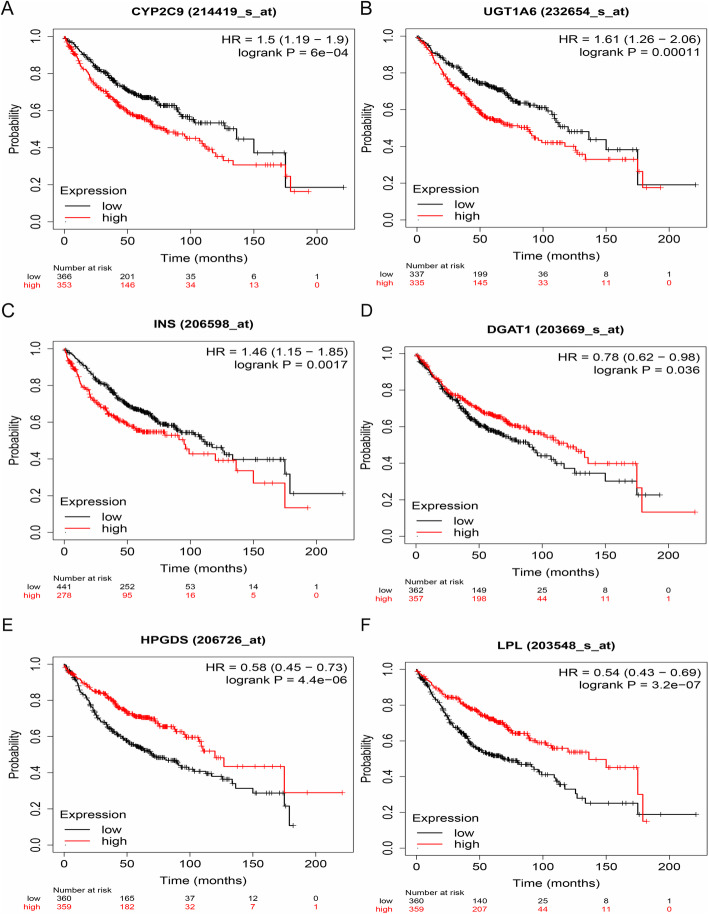
Survival analysis of hub genes
In this study, the relationship between mRNA expression of hub genes and clinical outcome was examined using the Kaplan-Meier plotter. The results showed that high expression of CYP2C9 [HR = 1.50 (1.19–1.90), P < 0.01], UGT1A6 [HR = 1.61 (1.26–2.06), P < 0.01], and INS [HR = 1.46 (1.15–1.85), P < 0.01], and low expression of DGAT1 [HR = 0.78 (0.62–0.98), P = 0.04], HPGDS [HR = 0.58 (0.45–0.73), P < 0.01], and LPL [HR = 0.54 (0.43–0.69), P < 0.01], were associated with a worse OS for 719 LUAD patients (Fig. 3).
Fig. 3.
Survival analysis of hub genes. LUAD patients were subdivided into high/low gene expression groups based on the median expression level of each gene in LUAD tissues. a OS analysis of CYP2C9, b OS analysis of UGT1A6, c OS analysis of INS, d OS analysis of DGAT1, e OS analysis of HPGDS, and f OS analysis of LPL
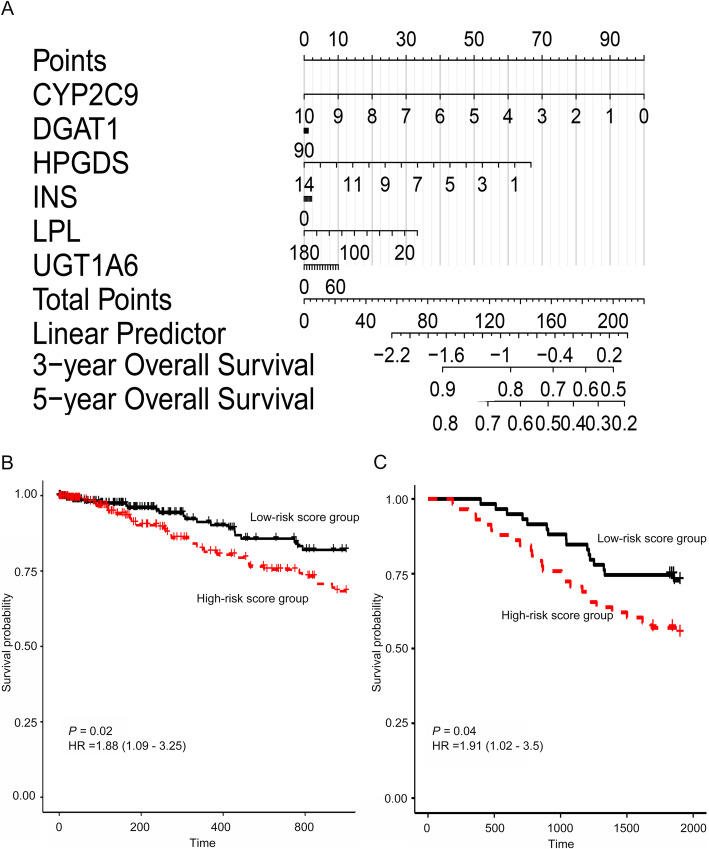
Prediction model based on survival-related hub genes and validation
Based on the Cox regression model, a nomogram was built to predict the prognosis of TCGA-LUAD patients, using mRNA expression of the six survival-related hub genes (CYP2C9, DGAT1, UGT1A6, INS, HPGDS, and LPL) (Fig. 4a). The concordance index of the nomogram was 0.61. Subsequently, the prognosis score of each patient was calculated, which showed that patients in the high-risk score group had a worse OS of 3 years [HR = 1.88 (1.09–3.25), P = 0.02] (Fig. 4b). A total of 486 patients with complete information, including gender, tumor stage, age, and smoking status were included for the multivariate Cox regression analysis. Except for HPGDS and LPL, the HR of CYP2C9, DGAT1, UGT1A6, and INS was not significant. In addition, the risk score calculated from the six-gene signature was an independent prognostic factor (Fig. S3). The model was validated and demonstrated that patients in the high-risk score group had a worse OS [HR = 1.91 (1.02–3.50), P = 0.04] (Fig. 4c).
Fig. 4.
Prediction model based on survival-related hub genes and validation. a Nomogram of 6 survival-related genes, b survival analysis between the high-risk score group and low-risk score group, and c validation of the model
Discussion
Metabolic changes have been widely observed in a variety of cancer cells [34]. Among the metabolisms involved, the lipid metabolism widely participated in the regulation of many cellular processes, including cell growth, proliferation, differentiation, survival, apoptosis, inflammation, motility, membrane homeostasis, chemotherapy response, and drug resistance [35]. In several recent studies, some components of PM2.5 have been reported as risk factors of lung cancer [36–38], because the PM2.5 components promoted pulmonary injury by modifying lipid metabolism [7] and might be involved in the development of lung cancer. However, studies on the association between lipid metabolism and lung cancer regarding transcriptome-wide analysis are limited. In this study, a LUAD cohort was used to generate a transcriptome-wide profile that included 217 lipid-related genes. The enrichment biological pathway found in LUAD patients included fatty acids, glycerolipids, and glycerophospholipids and were the primary driving enrichment biological function reported [39]. Furthermore, arachidonic acid metabolism, PPAR signaling pathway, insulin resistance, eicosanoids signaling, and other pathways have also been reported in cancer [40–44].
The results indicated that LUAD-related lipid metabolism was associated with nicotine, estrogen biosynthesis, melatonin, and atherosclerosis. Similar to PM2.5, nicotine may promote LUAD development by regulating disordered lipid metabolism. The interaction between estrogen biosynthesis and lipid metabolism may be one of the high-risk factors for LUAD, and is consistent with the observation that LUAD incidence is rising in women, and that the incidence rate among women was higher than that among men [2]. Lipid and cancer-related genes have been shown to be enriched in atherosclerosis and cancer [45]. For the long-term survival of LUAD patients, their health management should be managed by oncologists and cardiologists.
The network of genes was constructed and identified six hub genes related to lipid metabolism and LUAD. CYP2C9, a drug target in lung cancer, can inhibit the occurrence of lung cancer by acting on cytochrome P450, thereby regulating tumorigenesis [46, 47]. LUAD patients with a lower expression of CYP2C9 have a better prognosis than those with a higher expression of CYP2C9. UGT1A variants may play a minor role in the risk of other types of lung cancer [48]. LUAD patients with a lower expression of UGT1A6 have a better prognosis than one those with a higher expression of UGT1A6. DGAT1 catalyzes the final step in triglyceride synthesis [49], and LPL is a key lipolytic enzyme that plays a crucial role in the catabolism of triglycerides in triglyceride-rich particles [50]. Both are involved in triglyceride synthesis, and triglycerides combined with HPGDS have been reported to have therapeutic potential in allergic inflammation [51]. Serum triglyceride concentrations were reported to be involved in the pathogenesis of lung cancer [52]. INS encodes insulin and plays a vital role in the regulation of carbohydrate and lipid metabolism. LUAD patients with a lower expression of INS have a better prognosis. The regulation of triglyceride synthesis, insulin, and inflammatory control may be an effective intervention of LUAD patients. Based on those six genes, CYP2C9, DGAT1, UGT1A6, INS, HPGDS, and LPL, a risk model was constructed, including that LUAD patients from two cohorts with a lower risk score had a better prognosis.
Study strengths and limitations
The main strength of the study is the establishment of a lipid metabolic transcriptome-wide profile of LUAD and a gene signature that is significantly associated with the diagnosis and prognosis of LUAD patients. Limitations of this study include the following: 1) The data field information of these two cohorts is limited. Therefore, covariables related to LUAD might be missed and caused bias; 2) The internal mechanism of the six lipid-related genes is not illuminated clearly. In the future, a well-designed study based on the results is warranted.
Conclusions
In summary, a lipid metabolic transcriptome-wide profile of LUAD patients was generated and showed that lipid metabolic pathways were correlated with LUAD. A signature of six lipid metabolic genes was significantly associated with the diagnosis and prognosis of LUAD patients. Taken together, this gene signature can be used as a biomarker for LUAD to guide the prevention of the occurrence of LUAD and improve the prognosis of LUAD patients.
Supplementary information
Additional file 3: Table S1. Hub genes for lipid metabolism-related DEGs ranked in cytoHubba.
Additional file 4: Table S2. Correlation analysis of expression levels of hub genes in LUAD by GEPIA.
Additional file 5: Figure S1. Expression level analysis of 6 hub genes in GEPIA databases. Red and gray represent cancer and normal, respectively. (A) CYP2C9, (B) UGT1A6, (C) INS, (D) DGAT1, (E) HPGDS, and (F) LPL. * P < 0.05.
Additional file 6: Figure S2. Meta-analysis of 6 hub genes of lung cancer in ONCOMINE databases.
Additional file 7: Figure S3. Forrest plot of the multivariate Cox regression analyzis in TCGA-LUAD. (A) Risk score, (B) CYP2C9, (C) UGT1A6, (D) INS, (E) DGAT1, (F) HPGDS, and (G) LPL.
Acknowledgments
Not applicable.
Abbreviations
- LUAD
Lung adenocarcinoma
- TCGA-LUAD
The Cancer Genome Atlas Lung Adenocarcinoma
- HDL-C
High-density lipoprotein cholesterol
- LDL
Low-density lipoprotein
- LDLR
Low-density lipoprotein receptor
- PC
Phosphatidylcholine
- PE
Phosphatidylethanolamine
- FAO
Fatty acid oxidation
- NSCLC
Non-small-cell lung carcinoma
- TCGA
The Cancer Genome Atlas
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MisDB
Molecular Signatures Database
- DEGs
Differentially expressed genes
- FDR
False discovery rate
- logFC
Log2 fold change
- STRING
Search Tool for the Retrieval of Interacting Genes
- OS
Overall survival
- HR
Hazard ratio
- CI
Confidence interval
- INS
Insulin
- LPL
Lipoprotein Lipase
- HPGDS
Hematopoietic Prostaglandin D Synthase
- DGAT1
Diacylglycerol O-Acyltransferase 1
- UGT1A6
UDP Glucuronosyltransferase Family 1 Member A6
- CYP2C9
Cytochrome P450 Family 2 Subfamily C Member 9
- RXRα
Retinoid X receptor alpha
- FXRE
FXR response element
Authors’ contributions
JYL and QL analyzed the data and helped draft the manuscript. FZ and HTM supervised the study and edited and revised the manuscript. ZYS, QS, YZ, TNF, and JYJ prepared figures and contributed to the drafting of the manuscript. The authors read and approved the final version of the manuscript.
Funding
This study was supported by the Jiangsu Digital Innovative Medical Equipment Application Demonstration Research project of National Key Research and Development Program (2017YFC0114303), Youth Project of Wuxi Municipal Health and Family Planning Commission (Q201803), Youth Project of Jiangnan University Public Health Research Center (JUPH201826), Startup Fund for Youngman Research at SJTU (17X100040015), and Shanghai Jiao Tong University Medical and Industrial Cross Project (YG2017QN70).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the TCGA & GEO databases, [https://cancergenome.nih.gov/] & [https://www.ncbi.nlm.nih.gov/geo/].
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jinyou Li and Qiang Li contributed equally to this work.
Contributor Information
Feng Zhang, Email: zf9958@163.com.
Haitao Ma, Email: mahaitaoprofessor@163.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12944-020-01390-9.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung Cancer. Ann Glob Health. 2019;85:8. doi: 10.5334/aogh.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zang R, Shi JF, Lerut TE, Wang L, Liu CC, Brunelli A, Petersen RH, Ng CSH, Lim E, Gao S. Ten-year trends of Clinicopathologic features and surgical treatment of lung Cancer in China. Ann Thorac Surg. 2020;109:389–395. doi: 10.1016/j.athoracsur.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 4.Tseng C-H, Tsuang B-J, Chiang C-J, Ku K-C, Tseng J-S, Yang T-Y, Hsu K-H, Chen K-C, Yu S-L, Lee W-C, et al. The relationship between air pollution and lung Cancer in nonsmokers in Taiwan. J Thorac Oncol. 2019;14:784–792. doi: 10.1016/j.jtho.2018.12.033. [DOI] [PubMed] [Google Scholar]
- 5.Groen HJM, Hiltermann TJN. Air pollution and adenocarcinoma in never-smokers. J Thorac Oncol. 2019;14:761–763. doi: 10.1016/j.jtho.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Chen F, Jackson H, Bina WF. Lung adenocarcinoma incidence rates and their relation to motor vehicle density. Cancer Epidemiol Prev Biomarkers. 2009;18:760–764. doi: 10.1158/1055-9965.EPI-08-0741. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S-Y, Shao D, Liu H, Feng J, Feng B, Song X, Zhao Q, Chu M, Jiang C, Huang W, Wang X. Metabolomics analysis reveals that benzo [a] pyrene, a component of PM2.5, promotes pulmonary injury by modifying lipid metabolism in a phospholipase A2-dependent manner in vivo and in vitro. Redox Biol. 2017;13:459–469. doi: 10.1016/j.redox.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ku T, Zhang Y, Ji X, Li G, Sang N. PM2.5-bound metal metabolic distribution and coupled lipid abnormality at different developmental windows. Environ Pollut. 2017;228:354–362. doi: 10.1016/j.envpol.2017.05.040. [DOI] [PubMed] [Google Scholar]
- 9.Salvador MM, de Cedrón MG, Rubio JM, Martínez SF, Martínez RS, Casado E, de Molina AR, Sereno M. Lipid metabolism and lung cancer. Crit Rev Oncol Hematol. 2017;112:31–40. doi: 10.1016/j.critrevonc.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Guillaumond F, Bidaut G, Ouaissi M, Servais S, Gouirand V, Olivares O, Lac S, Borge L, Roques J, Gayet O. Cholesterol uptake disruption, in association with chemotherapy, is a promising combined metabolic therapy for pancreatic adenocarcinoma. Proc Natl Acad Sci. 2015;112:2473–2478. doi: 10.1073/pnas.1421601112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delmas D, Rebe C, Micheau O, Athias A, Gambert P, Grazide S, Laurent G, Latruffe N, Solary E. Redistribution of CD95, DR4 and DR5 in rafts accounts for the synergistic toxicity of resveratrol and death receptor ligands in colon carcinoma cells. Oncogene. 2004;23:8979–8986. doi: 10.1038/sj.onc.1208086. [DOI] [PubMed] [Google Scholar]
- 12.Stratton SA, Barton MC. p53-mediated regulation of hepatic lipid metabolism: forging links between metabolism, atherogenesis, and cancer. J Hepatol. 2012;56:518–519. doi: 10.1016/j.jhep.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Bozza PT, Viola JP. Lipid droplets in inflammation and cancer. Prostaglandins Leukot Essent Fat Acids. 2010;82:243–250. doi: 10.1016/j.plefa.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Yue S, Li J, Lee S-Y, Lee HJ, Shao T, Song B, Cheng L, Masterson TA, Liu X, Ratliff TL. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014;19:393–406. doi: 10.1016/j.cmet.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhury R, Amin MA, Bhattacharyya K. Intermittent fluorescence oscillations in lipid droplets in a live Normal and lung Cancer cell: time-resolved confocal microscopy. J Phys Chem B. 2015:10868–75. [DOI] [PubMed]
- 16.Fan TWM, Zhang X, Wang C, Yang Y, Kang W-Y, Arnold S, Higashi RM, Liu J, Lane AN. Exosomal lipids for classifying early and late stage non-small cell lung cancer. Anal Chim Acta. 2018;1037:256–264. doi: 10.1016/j.aca.2018.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou T, Zhan J, Fang W, Zhao Y, Yang Y, Hou X, Zhang Z, He X, Zhang Y, Huang Y. Serum low-density lipoprotein and low-density lipoprotein expression level at diagnosis are favorable prognostic factors in patients with small-cell lung cancer (SCLC) BMC Cancer. 2017;17:269. doi: 10.1186/s12885-017-3239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pei-Dong C, Wen L, Hao C, et al. High-density lipoprotein cholesterol is a favorable prognostic factor and negatively correlated with Creactive protein level in non-small cell lung carcinoma.[J]. PLoS ONE. 2014;9:e91080. [DOI] [PMC free article] [PubMed]
- 19.Chen Y, Ma Z, Shen X, Li L, Zhong J, Min LS, Xu L, Li H, Zhang J, Dai L. Serum Lipidomics profiling to identify biomarkers for non-small cell lung Cancer. Biomed Res Int. 2018;2018:5276240. doi: 10.1155/2018/5276240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marien E, Meister M, Muley T, Fieuws S, Bordel S, Derua R, Spraggins J, Van de Plas R, Dehairs J, Wouters J. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. Int J Cancer. 2015;137:1539–1548. doi: 10.1002/ijc.29517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo X, Cheng C, Tan Z, Li N, Tang M, Yang L, Cao Y. Emerging roles of lipid metabolism in cancer metastasis. Mol Cancer. 2017;16:76. doi: 10.1186/s12943-017-0646-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis S, Meltzer PS. GEOquery: a bridge between the gene expression omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 24.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–d361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Kattan MW. Drawing Nomograms with R: applications to categorical outcome and survival data. Ann Transl Med. 2017;5:211. doi: 10.21037/atm.2017.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 36.Cao Q, Rui G, Liang Y: Study on PM2.5 pollution and the mortality due to lung cancer in China based on geographic weighted regression model. BMC Public Health 2018, 18:925. [DOI] [PMC free article] [PubMed]
- 37.Lequy E, Siemiatycki J, Zhivin S, Vienneau D, Bentayeb M, Wagner V, Stempfelet M, Le Tertre A, Goldberg M, Zins M, et al. Reducing PM2.5 levels is associated with lower cancer incidence. Environ Epidemiol. 2019;3:178. [Google Scholar]
- 38.Gogna P, Narain TA, O'Sullivan DE, Villeneuve PJ, Demers PA, Hystad P, Brenner DR, Friedenreich CM, King WD. Estimates of the current and future burden of lung cancer attributable to PM2.5 in Canada. Prev Med. 2019;122:91–99. doi: 10.1016/j.ypmed.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Huang C, Freter C. Lipid metabolism, apoptosis and cancer therapy. Int J Mol Sci. 2015;16:924–949. doi: 10.3390/ijms16010924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marks F, Fürstenberger G, Müller-Decker K. Metabolic targets of cancer chemoprevention: interruption of tumor development by inhibitors of arachidonic acid metabolism. Recent Results Cancer Res. 1999;151:45–67. doi: 10.1007/978-3-642-59945-3_4. [DOI] [PubMed] [Google Scholar]
- 41.Go R-E, Hwang K-A, Choi K-C. Cytochrome P450 1 family and cancers. J Steroid Biochem Mol Biol. 2015;147:24–30. doi: 10.1016/j.jsbmb.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 42.Ashton KA, Proietto A, Otton G, Symonds I, McEvoy M, Attia J, Gilbert M, Hamann U, Scott RJ. Polymorphisms in genes of the steroid hormone biosynthesis and metabolism pathways and endometrial cancer risk. Cancer Epidemiol. 2010;34:328–337. doi: 10.1016/j.canep.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 43.Chinetti G, Fruchart J-C, Staels B. Peroxisome proliferator-activated receptors (PPARs): nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm Res. 2000;49:497–505. doi: 10.1007/s000110050622. [DOI] [PubMed] [Google Scholar]
- 44.Michalik L, Wahli W. PPARs mediate lipid signaling in inflammation and Cancer. PPAR Res. 2008;2008:134059. doi: 10.1155/2008/134059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mukai M, Komori K, Oka T. Mechanism and Management of Cancer Chemotherapy-Induced Atherosclerosis. J Atheroscler Thromb. 2018;25:994–1002. doi: 10.5551/jat.RV17027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Bearz A, Kim D, Mamdani H, Bauman J, Chiari R, Ou S, Solomon B, Soo R, Felip E. P1. 01–84 Interaction of Lorlatinib with CYP2B6, CYP2C9, UGT, and P-gp Probe Drugs in Patients with Advanced Non-Small Cell Lung Cancer. J Thorac Oncol. 2019;14:S392–S393. doi: 10.1016/j.jtho.2019.08.799. [DOI] [Google Scholar]
- 47.Sausville LN, Gangadhariah MH, Chiusa M, Mei S, Wei S, Zent R, Luther JM, Shuey MM, Capdevila JH, Falck JR, et al. The cytochrome P450 slow metabolizers CYP2C9*2 and CYP2C9*3 directly regulate tumorigenesis via reduced Epoxyeicosatrienoic acid production. Cancer Res. 2018;78:4865–4877. doi: 10.1158/0008-5472.CAN-17-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schieffer KM, Angstadt AY, Zhu J, Lazarus P, Gallagher CJ. Associations between polymorphisms and haplotypes in the UDP-glucuronosyl transferase 1A gene family with lung cancer risk. Next Generat Sequenc Applic. 2016;3:124. doi: 10.4172/2469-9853.1000124. [DOI] [Google Scholar]
- 49.Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hata A, Robertson M, Emi M, Lalouel JM. Direct detection and automated sequencing of individual alleles after electrophoretic strand separation: identification of a common nonsense mutation in exon 9 of the human lipoprotein lipase gene. Nucleic Acids Res. 1990;18:5407–5411. doi: 10.1093/nar/18.18.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rittchen S, Heinemann A. Therapeutic potential of hematopoietic prostaglandin D2 synthase in allergic inflammation. Cells. 2019;8:619. doi: 10.3390/cells8060619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ulmer H, Borena W, Rapp K, Klenk J, Strasak A, Diem G, Concin H, Nagel G, Vhm, Group PPS Serum triglyceride concentrations and cancer risk in a large cohort study in Austria. Br J Cancer. 2009;101:1202–1206. doi: 10.1038/sj.bjc.6605264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 3: Table S1. Hub genes for lipid metabolism-related DEGs ranked in cytoHubba.
Additional file 4: Table S2. Correlation analysis of expression levels of hub genes in LUAD by GEPIA.
Additional file 5: Figure S1. Expression level analysis of 6 hub genes in GEPIA databases. Red and gray represent cancer and normal, respectively. (A) CYP2C9, (B) UGT1A6, (C) INS, (D) DGAT1, (E) HPGDS, and (F) LPL. * P < 0.05.
Additional file 6: Figure S2. Meta-analysis of 6 hub genes of lung cancer in ONCOMINE databases.
Additional file 7: Figure S3. Forrest plot of the multivariate Cox regression analyzis in TCGA-LUAD. (A) Risk score, (B) CYP2C9, (C) UGT1A6, (D) INS, (E) DGAT1, (F) HPGDS, and (G) LPL.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the TCGA & GEO databases, [https://cancergenome.nih.gov/] & [https://www.ncbi.nlm.nih.gov/geo/].