Abstract
Seeds are essential for human civilization, so understanding the molecular events underpinning seed development and the zygotic embryo it contains is important. In addition, the approach of somatic embryogenesis is a critical propagation and regeneration strategy to increase desirable genotypes, to develop new genetically modified plants to meet agricultural challenges, and at a basic science level, to test gene function. We briefly review some of the transcription factors (TFs) involved in establishing primary and apical meristems during zygotic embryogenesis, as well as TFs necessary and/or sufficient to drive somatic embryo programs. We focus on the model plant Arabidopsis for which many tools are available, and review as well as speculate about comparisons and contrasts between zygotic and somatic embryo processes.
Keywords: Arabidopsis thaliana, embryogenesis, seed biology, somatic embryo, transcription factor
Introduction
Seeds first appeared a little under four million years ago [1]. Fossil evidence places the origin of angiosperms (flowering plants) during the Cretaceous, but molecular evidence supports an earlier origin [2]. The evolution of the seed was an innovation that led to the predominance of seed plants. Orthodox seeds allow the zygotic embryo to survive in a dry state for long periods, even under adverse conditions [3]. Seeds can perceive environmental cues and complete germination when survival to the next seed set is optimal. The young plant relies on storage products deposited in the seed, generally the endosperm or cotyledons, until the plant becomes autotrophic. Humans directly rely on seeds for ∼70% of our daily calories. Understanding seed development can support work to increase food production and to meet current and future global demands.
Embryogenesis consists of the processes occurring from the one-cell zygote to the mature embryo within the seed. In orthodox seeds, two main phases occur, the first of which is morphogenesis, during which the plant body plan is established. Later in embryogenesis, but overlapping with morphogenesis, maturation programs lead to deposition of storage products such as lipids, carbohydrates, and/or proteins, and accumulation of products for desiccation tolerance in preparation for the loss of water. In addition, dormancy may be established [4].
Early zygotic embryogenesis (ZE) has been difficult to study because the cells involved are few and embedded in several layers of maternal tissues. In addition, forward genetic approaches identifying embryo defective mutants tend to have lesions in essential genes rather than in genes involved in embryogenesis specifically [5]. Similarly, pattern formation mutants tend to be defective in gene products that are needed throughout the life cycle rather than for the establishment of domains in the embryo explicitly [5–7]. A more accessible model for ZE has been somatic embryogenesis (SE, also used as an abbreviation for somatic embryo), whereby somatic cells redifferentiate to follow an embryo program. Whether there is much congruency between molecular programs in ZE and SE remains a question. However, SE is also an important mode of plant propagation and regeneration, allowing genetic engineering to meet agricultural challenges. But, many plants or even particular cultivars of a species are recalcitrant to SE (some recent reviews include [8–10]). Thus, understanding processes promoting SE formation is also relevant. In this review, we focus on key transcription factors (TFs) necessary and/or sufficient for promoting zygotic and somatic embryo programs in the model plant Arabidopsis thaliana (henceforth Arabidopsis).
In both ZE and SE, certain events must occur including the establishment of the body axis (polarity), and of the primary and apical meristems. As such, we briefly discuss key factors involved in the initiation of tissue systems during ZE, and speculate on possible parallels (or not) with SE. Many of the gene products discussed have roles outside of embryogenesis, but we limit discussion to embryo processes.
Overview of morphogenesis
The process of flowering plant seed development begins with double fertilization where one of the sperm nuclei fuses with the egg cell nucleus forming a diploid zygote. In most angiosperms, a second sperm nucleus fuses with two polar nuclei of the central cell forming a triploid endosperm which supports and nourishes the developing embryo [11]. Some recent reviews on double fertilization include [12–14]. The embryo and the endosperm grow covered by maternal integument tissues that later form the seed coat [15]. Thus, the seed consists of three genotypes: the embryo with a 1 : 1 maternal (M) : paternal (P) ratio, the triploid endosperm with a 2M : 1P ratio, and the seed coat with a diploid 2 M genotype.
Upon fertilization, a positive signal from the zygote causes the co-ordinated development of the endosperm [16,17]. There have been various studies showing endosperm development is critical for embryo viability [18–20]. In most angiosperms, the triploid endosperm initially undergoes free nuclear division without cytokinesis forming a syncytium [21]. Later, the endosperm cellularizes [21,22]. The endosperm is largely transient and is mostly consumed by the embryo in some species, such as Arabidopsis, whereas in other plants (e.g. Zea mays), the endosperm is persistent and also supports the young seedling [21]. This review does not address endosperm development. However, for further details on endosperm, the reader may refer to reviews [18,21,23,24]. For some interesting recent literature on endosperm-embryo communication, please see recent reviews [25,26]. Imprinting is also a fascinating phenomenon not covered in this review, but some recent reviews include [27,28].
Embryogenesis is generally considered to occur in two phases: morphogenesis and maturation. Morphogenesis is the phase of embryogenesis during which the plant body is established, starting with the zygote [11,29]. The primary meristems that give rise to the epidermis, vascular system, and ground tissue are established and are called the protoderm, procambium or provascular meristem, and ground meristem, respectively. The shoot and root apical meristems (RAMs) that will perpetuate these primary meristems after completion of germination are also founded. A more detailed description of Arabidopsis morphogenesis and some key TFs involved are outlined below.
When considering developmental processes, it is important to realize that plant cells cannot generally move due to cell walls, so cellular migration, as found in animals, does not contribute to development. However, many plant cells are in symplastic communication via ‘channels’ called plasmodesmata (PD) that are regulated pathways connecting the plasma membrane, endoplasmic reticulum, and cytoplasm, cell-to-cell. PD can selectively allow transport of relatively large molecules including TFs between cells. Some recent reviews include [30,31].
Overlapping with and continuing after morphogenesis, maturation programs are active to accumulate storage reserve materials in the seeds. Additionally, desiccation tolerance is acquired, dormancy programs may be established, and desiccation occurs [32–35]. Excellent reviews for the maturation processes include [36–38].
Morphogenesis in Arabidopsis
Polarity and AC and BC determination
Cellular polarity is an important mechanism involved in many aspects of plant development, including early embryogenesis. In many species, including Arabidopsis, angiosperm egg cells are polarized with most of the cytoplasm and nucleus toward the chalazal (apical) end of the ovule (also a polar structure) and vacuoles at the micropylar (basal) end. Generally, a transient, more symmetrical stage occurs after fusion with the sperm cell, where the nucleus is more centrally located, and the large vacuole divides into many small, more evenly distributed vacuoles within the cell ([39,40] reviewed in [41]). Repolarization of the zygote involves elongation, nuclear migration towards the chalazal end and formation of a large vacuole at the micropylar end. The zygote elongates approximately three-fold in Arabidopsis [42,43].
In almost all flowering plant species the zygote undergoes an asymmetric transverse division [44,45], but there are exceptions (e.g. Triticum aestivum) [45,46]. This division leads to an apical daughter cell (AC) which always provides cells that contribute to the embryo proper (EP; the part of the embryo that upon completion of germination develops into the seedling) and a basal daughter cell (BC) forming the suspensor and at least part of the root (for review [47]). The suspensor is a transient structure that supports the developing embryo. Interestingly, suspensor cells have greater developmental potential and can express EP programs, up to forming viable embryos, when there are defects in the EP (reviewed in [48]).
Many embryogenesis/seed development studies have been focused on the model plant Arabidopsis because of abundant molecular and genetic tools available [49]. In addition, morphogenesis consists of a simple and predictable pattern which is invariant [29,35] and visualization of development is relatively easy [50,51]. Not all dicots show this type of predictable pattern of divisions; for example, Gossypium hirsutum (cotton) has more random early divisions after the first asymmetric division, but organizes a recognizable protoderm by early globular stage [52], as well as the other primary and apical meristems later in morphogenesis.
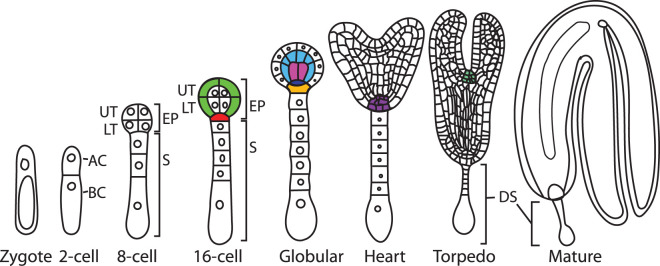
In Arabidopsis, after the first division, the smaller AC generates a spherical pro-embryo by undergoing three rounds of cell division; first two rounds of longitudinal divisions at right angles to form four cells of equal size, followed by one transverse division that gives two tiers, producing an eight-cell stage also called an octant stage embryo (Figure 1) [29,34,41,53,54]. At the octant stage, the apical–basal axis of the embryo can be distinguished as the apical/upper tier embryo domain (UT), which generates the shoot apical meristem (SAM) and most of the cotyledons, and the basal/lower tier embryo domain (LT), which will contribute to the hypocotyl, RAM, and parts of the cotyledons. The BC divides transversely, generating a filament of seven to nine cells by the globular stage of EP development, giving the extraembryonic suspensor (S), with the exception of the uppermost cell that contributes to the root meristem through the formation of the hypophysis (Figure 1). The suspensor connects the pro-embryo to maternal tissue and provides nutrients and growth factors as well as pushing the developing embryo into the nutrient-rich endosperm [35,55,56].
Figure 1. Different stages of embryo development in Arabidopsis (dicot).
Schematic representation of key embryo stages. AC, apical cell; BC, basal cell; UT upper tier cells; LT lower tier cells; EP, embryo proper; S, suspensor; DS degenerating/degenerated suspensor. Establishment of the: protoderm, green; hypophysis, red; ground meristem, light blue; provascular meristem, pink; lens-shaped cell that is the precursor to the quiescent center (QC), dark blue; basal cell from hypophysis that will form part of the root, gold; root apical meristem, purple; and shoot apical meristem, mint. No common scales were used in drawings. Some nuclei are not shown as meristems become established, and from the heart stage on. Cells are approximate from the heart stage on and not shown in the mature embryo. Adapted from [266].
Molecular components of zygote polarity establishment have been identified in Arabidopsis. Several mutants have compromised zygote elongation, defects in the first asymmetric division, and/or problems in basal cell identity and include yoda (yda), gnom (gn), grounded (grd), short suspensor (ssp), wrky2 (named for a protein domain that includes a motif of amino acids WRKY), and brassinosteroid kinase(bsk)1 and bsk2 double mutant.
The YDA pathway is involved in zygote elongation and development of basal cell lineages in Arabidopsis [57,58]. Zygotes homozygous for loss-of-function in yda fail to elongate and show an almost symmetrical first division with a normal-sized AC and a smaller-sized BC. The AC lineage exhibits a normal division pattern up to the octant stage, but the BC shows a random division pattern rather than forming the normal file of cells. YDA encodes a mitogen-activated protein kinase (MAPKKK). MAPKKKs participate in signaling events via a MAP kinase cascade, involving MAPKKs (MKK4 and 5 in the YDA pathway) and MAPKs (MPK3 and MPK6; the double mutant of which also has defects in zygote elongation, division, and lacks a developed suspensor, [59]). Interestingly, a gain-of-function yda mutant that has a deletion in the amino-terminal region, leading to the deregulation of this kinase, results in exaggerated suspensor growth [57].
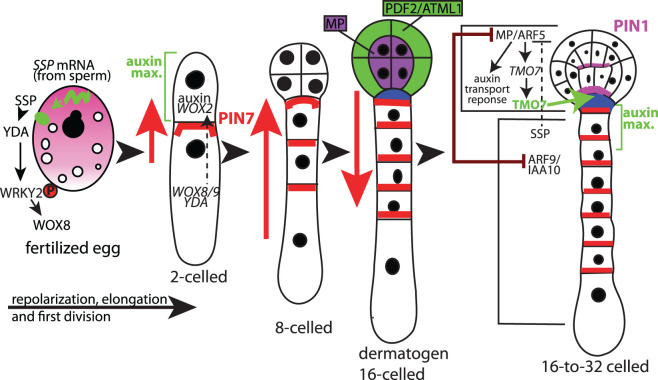
Recently, WRKY DNA-BINDING PROTEIN 2 (WRKY2) was shown to be a target of the YDA/MKK4/5/MPK3/6 kinase cascade [60]. Interestingly, the cascade appears to be initiated upon fertilization, where the sperm contributes SSP mRNA that is then translated in the zygote. SSP is a Brassicaceae specific member of the BSK brassinosteroid signaling kinase family proteins BSK1 and BSK2 that also function upstream of YDA signaling [61]. SSP appears to have lost residues involved in controlling an intramolecular interaction such that the tetratricopeptide (TPR) domain no longer interacts with the kinase domain. Generally, in BSK proteins this interaction is involved in a negative autoregulation that is relieved by phosphorylation. Loss of this interaction in SSP allows it to constitutively activate YDA and subsequently the rest of the kinase cascade, leading to activation of WRKY2 and expression of WUSCHEL-RELATED HOMEOBOX transcriptional regulators WOX8 and WOX9 [61–63].
WOX8 and another family member WOX2, along with WRKY2 are expressed in the egg and zygote, but after the first division, WOX8 is only in the BC along with WOX9, whereas WOX2 is in the AC [62,64,65]. WOX8 and WOX9 are important for BC lineage determination as wox8 wox9 double mutants showed abnormal suspensor development [66]. Defects are also apparent in the EPs that arrest prior to the globular stage or even earlier and develop as finger-like projections [66,67]. Because WOX8 and 9 are initially only expressed in the basal lineage, the EP defects are a non-cell-autonomous effect that functions through WOX8/9 activating WOX2 expression in the apical lineage. WOX2, together with auxin signaling, is involved in the specification of AC fate and are involved in embryonic shoot patterning [66]. Some of these events are diagrammed in Figure 2. Many other factors are also involved, including GRD/RKD4, a RWP-RK transcriptional regulator [58,68], small peptides CLAVATA-Like 8 (CLE8), and EMBRYO SURROUNDING FACTOR's (ESFs) [69,70], and ZYGOTIC ARREST1 (ZAR1) that like SSP, is a receptor-like kinase (RLK)/Pelle kinase [71].
Figure 2. Some key events during early embryogenesis in Arabidopsis.
The mature egg cell is polarized (not shown) but after fertilization undergoes a more symmetric stage. Repolarization of the zygote occurs and involves a signaling pathway that includes SSP kinase (translated in the zygote from SSP mRNA delivered by the sperm). A YDA/MAP kinase signaling cascade ensues, leading to phosphorylation of WRKY2 that then expresses WOX8. WOX2 and WOX8 are present in the zygote, but after the first division, WOX2 is in the AC and WOX8 in the BC. PIN7 efflux auxin carrier moves auxin into the AC/EP, until a reversal at the dermatogen stage, when auxin begins being synthesized in the EP and basipetally distributed. At the dermatogen stage, MP marks the inner cells, and PDF2/ATML1 the protoderm. PIN1, controlled by MP is involved in polar auxin transport. PIN1 and TMO7 are regulated by MP in the EP and TMO7 moves to the top suspensor cell/hypophysis, and along with auxin specifies this cell. ARF9/IAA10 is also important for this process. Not all genes involved, nor all localizations/interactions are shown in order to maintain clarity. For example, PIN1 accumulates earlier than the 32-cell stage, but is more randomly localized.
Auxin and early events
The auxin-dependent pathway is another critical pathway in early embryogenesis. In Arabidopsis, after zygote division, auxin is transported from the basal to the AC mediated by suspensor-specific PIN7 (Figure 2) to form an auxin response maximum that contributes to the pro-embryo specification [72,73]. Auxin biosynthetic pathway components, YUCCA (YUC) family members including YUC3, YUC4, and YUC9 are a source of basal auxin. YUC3/4/9 are initially expressed in the suspensor, but loss-of-function mutants (yuc3/4/9) show abnormalities in apical regions of the embryo [73]. Recently, Robert et al. [74] showed that maternally supplied auxin from synthesis in the integuments is also involved in very early EP development. At the 16-cell EP stage, an apical auxin source is generated by TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1), another key enzyme involved in auxin biosynthesis, and YUC1/YUC4. PIN7 becomes relocalized such that it directs auxin flow basipetally. PIN1, another auxin efflux carrier, contributes to this new auxin maximum in the hypophysis and upper suspensor cells [72,73]. In addition to the PIN transporters, two redundant auxin influx carriers AUX1 and LIKE AUX1 (LAX1) also help to establish the basal auxin accumulation [75].
Proper localization of the PIN auxin efflux carriers is key to morphogenesis. The gnom mutant was originally isolated as a pattern formation mutant [6,76]. Defects in this gene show a range of phenotypes, but one associated phenotype results in ball-shaped seedlings that lack the normal apical–basal patterning where the first zygotic division is symmetrical. GNOM encodes a GDP/GTP exchange factor for small G proteins of the ARF class (ARF-GEF) and is involved in proper localization of PIN1 to move auxin basipetally via endosomal recycling [77,78].
Interestingly, both PIN1 (auxin efflux) and AUX1/LAX2 (auxin influx) are regulated by the AUXIN RESPONSE FACTOR5/MONOPTEROS (ARF5/MP) in the inner embryonic cells and this is important for hypophysis/root development [75]. ARF's bind to auxin response elements (cis elements, AREs), but have AUX/IAA proteins associated that block ability to regulate target genes. In the case of MP, the AUX/IAA protein is BODENLOS/INDOLE-3-ACETIC ACID INDUCIBLE12 (BDL/IAA12). When auxin is present, it binds to an auxin receptor (TRANSPORT INHIBITOR RESPONSE1, TIR1), activating a complex that will ubiquitinate BDL, marking this protein for degradation by the proteasome and thereby freeing MP to regulate gene expression (for review, [79]). The mp loss-of-function and the bdl gain-of-function (that cannot be degraded via ubiquitination) mutants lack roots [6,80].
Besides impacting on the accumulation of auxin transporters, MP directly regulates expression of TARGET OF MP7 (TMO7), a basic helix-loop-helix (bHLH) TF. MP is present in the inner cells of the very young globular embryo, but TMO7 moves from this site of synthesis to the hypophysis via PD, and is involved in root development [81–83]. Interestingly, an upstream regulator of MP expression involves SSP [84]. In the ssp mutant, MP is ectopically expressed and the ssp mp double mutant partially restores suspensor development compared with the short suspensor found in ssp. Thus, SSP signaling is important to restrict MP expression in suspensor cells.
Another ARF/IAA pair (ARF9/IAA10 and potentially redundant suspensor-expressed factors) have an antagonistic relationship with MP and are important for suspensor and hypophysis development [84]. Defects in auxin response in the suspensor lead to unusual cell divisions and activation of genes normally expressed in the EP. Thus, auxin has different effects in different cells possibly mediated by different contingents of ARF and associated IAA gene expression. In the uppermost suspensor cell, auxin is needed for hypophysis specification and division; however, in the suspensor cells it restricts cell division and EP programs. Some ARFs are activators of gene expression while others repress gene expression, and MP and ARF9 belong to these different groups as measured by a synthetic auxin-responsive promoter driving expression of a reporter gene [85].
The suspensor has a broader developmental potential that is restricted by the EP. Many studies where the EP is ablated, either physically or by a genetic defect, found that cells in the suspensor can take on EP characteristics up to forming viable embryos as in the twin mutants (reviewed in [48]). More recently, Liu et al. [86], used in vivo living cell laser ablation to remove the EP from the S within the developing seed and found that if performed at the globular stage or earlier, the top-most suspensor cell accumulated auxin and formed a new embryo. Recently, transcriptome analysis to examine the suspensor to embryo transition has revealed a role for bHLH TFs in mediating this process [87].
Primary meristem establishment
The upper and lower-tier cells undergo a tangential (also called periclinal) division, giving rise to 16 cells with eight inner cells and eight outer cells. This stage is called the dermatogen stage because from this point on, the outer cells will divide anticlinally, giving rise to the protoderm, while the inner cells are the founder cells of the provasculature and the ground meristem (Figure 1). The WOX TFs, together with auxin signaling, have been linked to these tangential divisions, which is disturbed in wox2 single and wox2 wox8 double mutants [66]. This phenotype is enhanced in combinations of wox2 wox8 double or wox1 wox2 wox3 triple-mutant combinations with mutations in mp/arf5 [66]. An ectopically accumulating, non-degradable version of BODENLOS (BDL) that would inhibit MP/ARF5 gene regulation even in the presence of auxin, shows defects at the transition to dermatogen stage. However, this dominant, gain-of-function bdl mutant later forms a protodermal layer, suggesting that while auxin response is important at the eight to 16 cell transition for protoderm formation, it is not essential for epidermis formation [84].
The differentiation of the outer layer and inner cells accords with the gene expression pattern of two homeodomain leucine zipper class IV (HD-ZIP IV) TFs, ARABIDOPSIS THALIANA MERISTEM LAYER 1 (ATML1) [88], and PROTODERMAL FACTOR 2 (PDF2) [89]. Transcript accumulation corresponding to these two TFs is initially detected throughout the early EP, but immediately after the tangential divisions, expression becomes preferential to the protodermal layer [90–92]. The atml1 pdf2 double mutant shows severe defects in epidermal cell specification leading to embryo lethality [89,93,94]. ATML1 is also sufficient for protodermal identity [95,96].
Further divisions form the globular stages (32–64 cells) where the ground meristem and provascular/procambial meristems are specified that give rise to the ground and vascular tissues, establishing the radial axis [35,53,55,97] (Figure 1). Also, during this stage, the hypophysis, derived from the top-most cell of the basal cell lineage, divides asymmetrically to form the precursor of the quiescent center (QC), which is a smaller lens-shaped cell, and another larger basal cell which is a precursor of the distal stem cells of the root meristem, the columella [35,53,55,97].
Little is known about the initiation of the ground meristem compared with subsequent maintenance and patterning. MP, that ‘marks’ the inner cells at the dermatogen stage, has a role in ground tissue initiation [98], as well as provascular meristem development (discussed below). The ground tissue daughter cells will then divide to give the endodermis (most interior layer of the cortex; roles include regulation of transport of water and ions into the vascular tissue) and the cortex (exterior layer) cell lineages. Furthermore, auxin activity was found to be highest in vascular tissue, lowest in the protoderm, and intermediate in the ground tissue in globular stage embryos. Studies on vasculature formation have largely focused on MP. During early embryogenesis, mp mutants show defects in the characteristic divisions to generate the vascular tissues and fail to develop an embryonic root [80,99–101]. Later, transcript profiling studies revealed numerous direct MP target TFs, which act downstream of MP in root initiation, including bHLH TFs TMO5 and TMO5-LIKE1 (T5L1) [81]. TMO5 marks the provascular initial cells in the early globular stage embryo, indicating that it mediates MP functions in the pro-embryo. The tmo5/t5l1 double mutants show a reduced vascular bundle [81,102]. These defects are also observed in a bHLH146 mutant, also called lonesome highway (lhw) [103]. A double mutation of lhw and its close homolog lonesome highway-like 3 (lhl3) shows severe phenotypic defects in vascular tissue formation [104]. TMO5 and LHW interact to form a heterodimeric complex [102,103], and constitutive co-expression of TMO5 and LHW activates periclinal divisions in all cell types, indicating that the TMO5-LHW dimer is necessary and sufficient to regulate the periclinal divisions in the establishment and for the maintenance of the vasculature in the post-embryonic root [102]. LONELY GUY 4 (LOG4), an enzyme involved in cytokinin (CK, a plant hormone) biosynthesis, was identified as a direct target of the TMO5-LHW complex [105]. The single log4 mutants do not have any phenotype, but higher-order mutants with orthologs show severe defects in embryonic vascular tissue development and patterning [105]. Another interesting study identified LOG3, LOG4, and ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6 (AHP6) as direct targets of the LHW-T5L1 complex [106]. Some additional reading on the initiation of vascular development include reviews [107,108].
Shoot and root apical meristem establishment
At the end of the late globular stage, cells at the top peripheral regions begin dividing more rapidly to form the two cotyledons. Because this is a transition from a radially symmetrical globular stage embryo to a bilaterally symmetric heart stage embryo, this stage is sometimes referred to as ‘transition or triangular stage’ (not shown in Figure 1). The embryo greens during this stage and vascular differentiation (xylem and phloem) becomes apparent. Continued cell division and expansion generate later heart and then torpedo stages, and the RAM is organized. The SAM becomes anatomically apparent during torpedo stages as a population of smaller cells. The suspensor degenerates during later morphogenesis [109] and further expansion causes the embryo to bend to fit within the seed coat (bent cotyledon and mature stage; Figure 1).
Small populations of cells within the SAM and RAM divide slowly to provide the surrounding stem cells, and these regions are called the organizing centers [110]. The organizing centers maintain the stem cell identity of adjacent cells [111]. While the primary meristems are established in the embryo, they are perpetuated after completion of germination by the apical meristems.
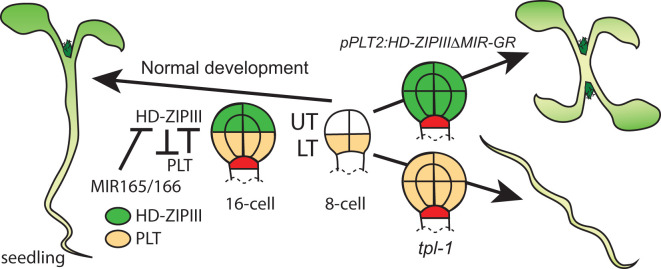
Two families of TFs are involved in shoot versus root identity, specifically the class III HD-ZIP family [112] and the AP2-domain PLETHORA (PLT) family [113,114], respectively. The PLT genes (PLT1, 2, 3, and 4; PLT3 is also called ANINTEGUMENTA-LIKE6, AIL6; PLT4 is also called BABY BOOM, BBM) are expressed in the LT cells at the eight-cell stage of EP development. The HD-ZIP III family includes genes encoding the TFs PHABULOSA (PHB), PHAVOLUTA (PHV), REVOLUTA (REV), ARABIDOPSIS THALIANA HOMEOBOX 8 (ATHB8), and ATHB15 (also called ICU4), that are expressed by the 16-cell stage in the upper tier cells at the globular stage of embryogenesis and are redundant in function for post-embryonic shoot maintenance [112]. HD-ZIP III family TFs regulate the formation of the SAM, the boundary between the SAM and the cotyledons, the central portion of the embryo, and the dorsal/ventral pattern of leaves during post-embryonic development [115]. The HD-ZIPIIIs are post-transcriptionally regulated by microRNA 165/166 (MIR165/166) family members [116]. Constitutive expression of PLT1 or PLT2 induces the hypocotyl, root, and root stem cell niche from the basal region of the embryo, indicating a central role for PLT1 and PLT2 in basal cell fate determination. TOPLESS (TPL) encodes a transcriptional co-repressor that regulates expression of PLT1 and PLT2 [117,118]. In the tpl-1 mutant, both PLT1 and PLT2 are constitutively expressed at both ends of the embryo, resulting in a range of phenotypes with the most severe phenotype being the transformation of the shoot to a root [117]. TPL associates with the PLT promoters [118] so this may be a result of direct regulation, although via interaction with another protein as TPL is not a DNA-binding factor. TPL has been shown to interact with BDL [119] as well as many other TFs [120]. PLTs are induced by auxin, although they do not appear to represent direct ARF targets [113]. A mutation in PHB that eliminates the ability of MIR165/166 to regulate the transcript results in suppression of the double-root phenotype. On the other hand, ectopic expression of forms of HD-ZIPIIIs that cannot be regulated by MIR165/166 using the PLT2 promoter converts basal cells into a second SAM and produces a double-headed seedling. These studies demonstrate antagonistic roles for the HD-ZIP IIIs and PLTs in determining apical and basal cell fate. However, it is not clear whether the HD-ZIP III and PLT directly control each other's expression and how they interact to maintain the boundary between apical and basal cells. These interactions are diagrammed in Figure 3.
Figure 3. PLT and HD-ZIPIII have an antagonistic relationship to determine shoot versus root identity.
PLTs are expressed in the LT cells by the 8-cell stage, while HD-ZIPIII begin to be expressed in the UT cells at the 16-cell stage. MicroRNAs (165/166) are involved in post-transcriptional regulation of the HD-ZIPIIIs. In the tpl-1 mutant, the PLT genes become misregulated and are expressed at both poles of the embryo. The most severe phenotype is the transformation of the shoot into a root. Conversely, if forms of the HD-ZIPIII genes that cannot be regulated by MIR165/166 are expressed in the LT cells, using the PLT2 promoter sequences, the root is transformed into a shoot. Data presented from [117,118].
A comprehensive discussion of all interesting genes involved in SAM and RAM development is beyond the scope of this review. Some of the other TFs (e.g. MP, TMO7) that are involved in establishing the QC of the root are described above, but many other important factors have been reported that are not discussed here. The interested reader is referred to recent reviews for further reading that include [121,122]. Several molecular pathways have been identified that regulate the organization of the SAM. WUSCHEL (WUS) is involved in specifying the OC in the SAM and is involved in feedback regulation with CLV3 [123,124]. Besides the well-known factors, including KNOTTED-1 homeodomain protein homolog SHOOT MERISTEMLESS (STM) [125] and class III HD-ZIP factors [115], more genes have been shown to regulate WUS expression including the class II HD-ZIP genes: ATHB2, ATHB4, HOMEOBOX ARABIDOPSIS THALIANA 1 (HAT1), HAT2, and HAT3 [126–128]. Several class II HD-ZIP genes (e.g. HAT2, HAT3, and ATHB4) are directly regulated by the class III HD-ZIP gene REV, indicating that the HD-ZIP III genes might regulate development by activation of the HD-ZIP II genes [129,130]. For more information, some recent reviews include [131–134].
Overview of somatic embryogenesis
SE is a process by with somatic cells redifferentiate to form embryos. These somatic embryos may form directly on the explant (direct SE) or there may be an intervening embryogenic callus phase (indirect SE). Somatic embryos are bipolar structures that do not have vascular connection with the explant material. SE can be divided into inductive and developmental phases, where changes in differentiation status and acquisition of embryogenic competence occurs during the former phase, and differentiation into SEs during the latter phase [135].
Induction of SE typically involves the use of plant growth regulators (PGRs), often with a stress treatment. Most commonly, treatment with auxin is involved, usually the synthetic auxin 2,4-D, sometimes in combination with CKs (reviewed in [136]). The synthetic auxin 2,4-D may act as an auxin, and/or as a stressing agent, and may induce synthesis of endogenous auxin. The PGR/stress treatment can lead to redifferentiation of cells that are competent for SE, via de- or trans-differentiation to a status with greater potency and establishment of embryo programs by embryo induction. These stages are followed by embryo development, which often requires removal of the exogenously added auxin. Although SE is typically thought of as an in vitro phenomenon, Kalanchoë (commonly known as ‘mother of thousands’) forms somatic embryos that convert into plantlets at the leaf margins [137].
A loss of symplastic communication is also a feature of reprogramming of cells (reviewed in [138]). Recently Godel-Jedrychowska et al. [139], documented that symplastic isolation of embryogenic cells is required for redifferentiation during SE. The cells competent for SE express WOX2 (a marker of AC), but while auxin is common for induction, cells that form embryos show reduced auxin response (as measured by a synthetic auxin promoter driving a reporter gene). A reduction in callose biosynthesis, which would show deficiencies in symplastic isolation, repressed SE. These findings applied to both SE induced from WT immature zygotic embryos (IZE) as well as SE induced by overexpression of BABY BOOM (BBM, discussed below).
Changes in epigenetic state are involved in redifferentiation processes, and involve DNA methylation as well as histone modifications (reviewed in [138]). As discussed below, many Arabidopsis genes when mutated or compromised in expression, result in SE development and these tend to encode factors involved in epigenetic regulation at the developmental transition from embryo to seedling.
Somatic embryos have been used as a more easily accessible model for zygotic processes that occur embedded in multiple layers of maternal tissues, but there are some obvious differences. SE lack communication with endosperm/maternal tissues as they develop without these tissues, and there are differences in maturation processes, specifically lack of desiccation and dormancy. SE's may have a reduced or even lack an obvious suspensor, but morphological stages in the EP are similar to ZE. Often, the initial division of embryogenic cells in SE are asymmetric, similar to ZE (reviewed in [140]). However, for microspore embryogenesis, the asymmetric division necessary for the development of the male gametophyte is replaced by a symmetric division, producing a haploid SE [141].
Summary of SE systems in Arabidopsis
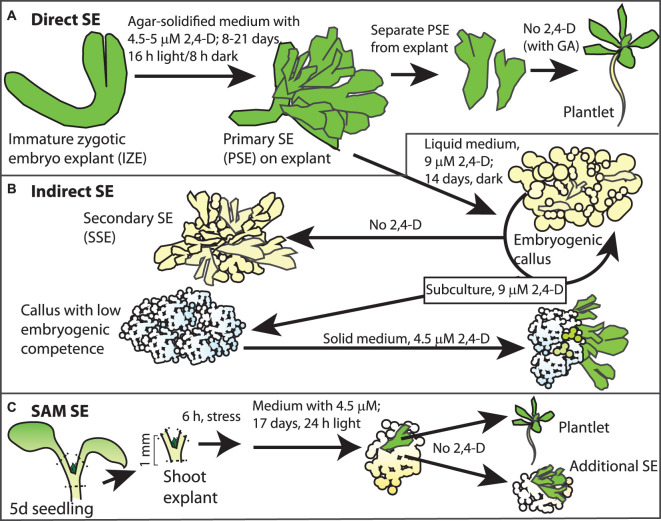
There are both direct and indirect SE systems in Arabidopsis. In almost all cases, embryonic or meristematic tissues serve as the explant source. Gaj [142] used a direct SE system she developed and subsequently documented the requirement of several TF's and proteins involved in hormone synthesis and response [143,144]. Briefly, IZE are isolated from developing seeds and used as the explant. A range of developmental stages were tested, with later-stage corresponding to bent cotyledon embryos (400–700 μm) showing 65–90% SE production for Col and Ws ecotypes [142]. The explants were placed on B5 medium supplemented with 5 μM 2,4-D and sucrose. Within 3 weeks, primary somatic embryos (PSEs) are evident on the explant (Figure 4A). It was possible to move these PSEs to auxin-free medium (in some protocols, also including gibberellic acid (GA)) to allow conversion and recovery of plantlets. The origin of the cells producing the embryos is within the protodermis and subprotodermal layers of the adaxial cotyledons [145]. Formation of PSE requires LEC1, LEC2, and FUS3 (these key embryo TFs are discussed below), with loss-of-function alleles of these genes showing only 1–4% SE production, and this rare SE formation is via an indirect route involving callus [144]. Higher-order mutants of these genes only produce callus. Gaj [146] also reported that younger stage IZE explants more often produce embryogenic callus.
Figure 4. Somatic embryo systems in Arabidopsis.
Diagram showing an example of direct (A), indirect (B), and SAM (C) SE in Arabidopsis. Please see the text for more details.
Su et al. [147], used a modified system from Ikeda-Iwai et al. [148], that involves an indirect route of SE production. As described above, IZEs are cultured to produce PSE that are then moved to a liquid medium with higher concentrations of 2,4-D (9 μM) to generate embryogenic callus. This can be cycled in this higher auxin medium or moved to auxin-free medium to induce secondary SE (SSE). If cultured too long in the high 2,4-D medium, the callus loses embryogenic capacity, but this can be reestablished by culture on solid B5 medium with lower 2,4-D (Figure 4B).
Ikeda-Iwai et al. [149] also developed an Arabidopsis SE system from vegetative tissues (Figure 4C). This system required use of meristematic tissues (SAM, floral buds, or axillary buds; SAM was most efficient) and subjected the explants to osmotic stress before moving to B5 medium with 4.5 μM 2,4-D. After 10–21 days, callus forms at the shoot apical region followed by SE development. If the explants are moved to 2,4-D free medium, additional SE forms on the callus. The best conditions for the Col ecotype involves stress treatment with mannitol for 6 h. The developmental stage of the SAM is important with shoot apical explants from 5 day seedlings giving the most efficient SE at 29%. Younger explants die, whereas explants from older seedlings only generate callus.
Other Arabidopsis SE systems appear to develop at or near SAMs [150–152] either directly or via embryogenic callus, or from IZE explants [153]. Few systems in Arabidopsis are from other tissues. Mathur et al. [154] reported SE-like structures from protoplasts derived from auxin conditioned roots, and Luo and Koop [155] were able to obtain apparent globular stage embryos with a suspensor-like structure from leaf protoplasts, but only early stages of development occurred.
Arabidopsis transcription factors that promote embryo identity
TFs necessary and sufficient for embryogenesis
The LAFLs
While forward genetic screens to identify embryo defective mutants generally result in the identification of non-redundant genes essential for viability rather than for embryo development or designation of domains within the embryo specifically [5,53], some embryo defective mutants have lesions in genes that are more specific for seed development. These include genes encoding four central regulators LEAFY COTYLEDON1 (LEC1), ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEAFY COTYLEDON2 (LEC2), otherwise referred to as the LAFL factors, that are key to embryo development. These genes are necessary and sufficient for embryo processes, and show the highest levels of transcript accumulation in the developing embryo and endosperm ([156] and Arabidopsis eFP Browser [157]), although they have also been reported to have developmental roles after completion of germination [158–163].
To be necessary, a loss-of-function of the factor would result in an embryo defective phenotype (whether specific to embryogenesis or not). Sufficiency is tested by ectopic expression of the factor and assessment of whether it can drive embryo-specific programs outside of the embryo-context. The LAFL factors are among those necessary and sufficient (to different extents; discussed below) to drive embryo processes. Other TFs may be sufficient to drive embryo programs, but not necessary, perhaps due to redundancy. Others may show embryo deficient phenotypes, indicating necessity, but are not sufficient to drive embryo programs, often because they are needed for cell viability rather than embryo specification, but also possibly due to interacting factors missing in the ectopically expressed domains.
While putative orthologs of LEC1 and ABI3, and perhaps LEC2, appear to have arisen prior to the ‘invention’ of the seed, FUS3 orthologs have only been found in seed plants to date ([164] and reviewed in [165]). Roles for putative orthologs in non-seed plants include desiccation tolerance, and many aspects of LAFL loss-of-function phenotypes indicate that they may have been recruited to control processes during the maturation phase and establish the quiescent state of the mature seed.
LEAFY COTYLEDON 1 (LEC1, At1g21970) is a NF-YB/HAP3 subunit of a CCAAT box-binding factor (CBF) [166]. NF-YBs form complexes with NF-YA/HAP2 and NF-YC/HAP5 subunits, with the NF-YA subunit primarily responsible for recognition of the DNA cis element (the CCAAT box) (reviewed in [167]). NF-YB subunits include a broadly conserved histone-like domain (the central B domain) that encompasses residues necessary for complex formation as well as residues contributing to binding DNA [168]. The loss-of-function lec1 shows defects in embryo morphogenesis and embryonic maturation, including decreased accumulation of storage products and loss of acquisition of desiccation tolerance. When developing embryos are rescued prior to desiccation, the resulting lec1 homozygous seedlings show leaf-like traits in the cotyledons, including the development of trichomes (in Arabidopsis trichomes are a leaf, but not a cotyledon feature) [34,169,170]. LEC1 is preferentially expressed in the seeds, beginning early in embryo development and declining during maturation as shown in Figure 5A [166,171–174]. Early defects include abnormal cell divisions in the suspensor [166]. The loss-of-function mutant also shows precocious activation of the SAM during embryogenesis. When LEC1 is constitutively expressed using a semi-constitutive 35S viral promoter [175], the genetically engineered plants produced somatic embryos (SE) on the seedlings, which indicates that LEC1 is sufficient to promote embryo identity [166]. However, this sufficiency has a timeframe. When the 35S : LEC1 transgene included a glucocorticoid receptor (GR) domain that allows the TF to only be transported into the nucleus upon hormone treatment with the synthetic steroid dexamethasone, only early timepoints during or shortly after completion of germination resulted in embryo/embryo-like tissue. By 4 days after germination, the seedlings did not produce this tissue and instead appeared wild type [159]. The central B domain of the LEC1 protein was confirmed to be required and sufficient for embryogenesis. Moreover, an amino acid residue (aspartic acid at position 55), which specifically exists in LEC1-type B domain proteins (LEC1 and L1L), but not in non-LEC1-type NF-YB subunits that have a lysine in this position, was verified to be necessary for embryogenesis [168]. LEC1 interacts with other proteins such as bZIP67, LEC2, and others to control various programs during seed development [167,176]. The propagules that arise on Kalanchoë, by what appears to be a combination of organogenesis and SE, requires an altered form of KdLEC1 [137,177].
Figure 5. Interactions between key regulators of embryogenesis, SE and the transition to seedling development.
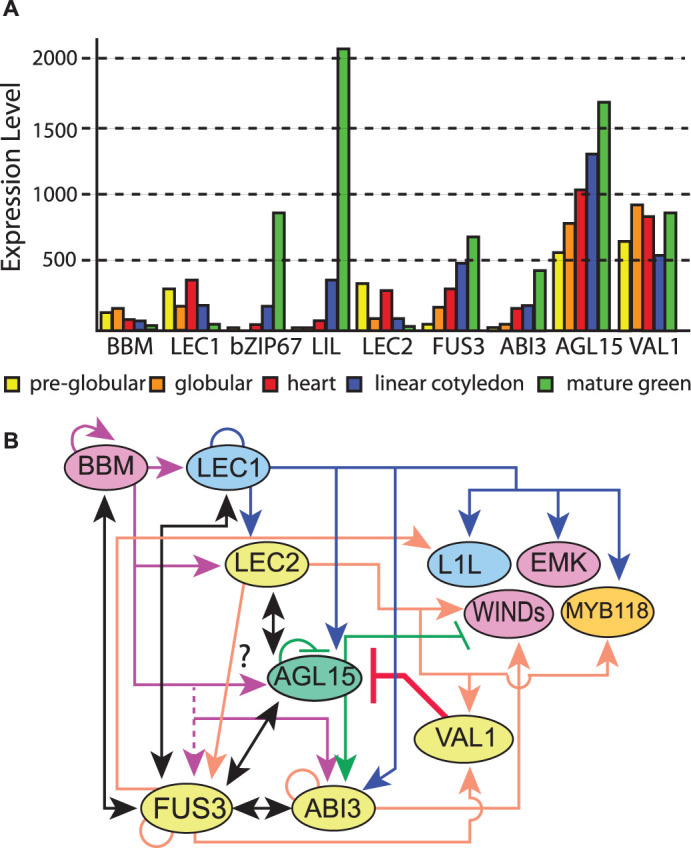
(A) Transcript accumulation in embryos. Data is from the Arabidopsis eFP browser [157] from data provided by Goldberg and Harada. (B) Working model summarizing interaction between select key regulators. Black solid lines with double arrows indicate a positive regulation loop. Other direct induced targets are indicated by arrows; direct repressed by bars. Circles indicate potential autoregulation. Direct responsive targets are from: LEC1 [176]; LEC2 [180,234]; FUS3 [229,232]; ABI3 [230,231]; AGL15 [201]; BBM [209]; VAL1/HSI2 [224]. For BBM targets, while high-throughput analysis indicated FUS3 was a direct expressed target, verification of direct interaction was not found in follow-up studies. Conversely, AGL15 regulatory regions are bound by BBM, but transcript accumulation may not respond. Yellow, B3 domain; pink, AP2 domain; mint, MADS domain; blue, NF-YB/HAP3; orange, MYB domain. Not all interesting interactions or proteins are shown, or relevant references cited in the interest of clarity.
LEAFY COTYLEDON1-LIKE (L1L, At5g47670), is the closest related gene to LEC1, encoding a product with 83% sequence identity that produces a LEC-1 type NF-YB subunit. L1L is primarily expressed in developing seeds and transcript accumulation is highest during maturation (Figure 5A). The l1l RNAi suppression lines showed an incompletely penetrant and range of phenotypes varying from arrest at the globular stage with abnormal suspensor divisions to underdeveloped cotyledons. Although somatic embryos were not produced on vegetative tissues in 35S : L1L, the seedlings did express embryo-specific programs (e.g. accumulation of storage products) in leaves that had cotyledon characteristics. In addition, the overexpression line could suppress the desiccation intolerant phenotype of the lec1 mutant as could LIL driven by the LEC1 regulatory sequences [178].
LEAFY COTYLEDON 2 (LEC2, At1g28300) contains a B3 domain and is another major TF involved in embryogenesis in plants. Loss-of-function lec2 shows a similar phenotype to lec1 in that both have trichomes on their cotyledons upon embryo rescue, and defects in embryo shape [170]. Ectopic LEC2 expression in vegetative cells could induce a somatic embryo-like seedling phenotype demonstrating sufficiency to drive embryo programs [179]. Like LEC1, there is a timeframe for sufficiency. When a 35S : LEC2 transgene is expressed, the ‘seedlings’ produce masses of SE [179]. But when an inducible form (35S : LEC2-GR) is used, while seedlings treated with dexamethasone activate embryo programs and leaves take on embryo-like features, they do not produce somatic embryos [180–182].
FUSCA3 (FUS3, At3g26790) is a B3 domain TF. Loss-of-function of fus3 seeds are desiccation intolerant, but like lec1 and lec2, can be rescued into culture prior to drying, upon which the cotyledons show ‘leafy’ traits (trichomes), but these trichomes were much less prevalent compared with lec1 or lec2 mutant plants and the embryos have a more normal bent cotyledon shape [170,183]. Unlike LEC1 and LEC2 overexpression that produces somatic embryos on the seedlings, the FUS3 ectopic phenotype was milder and produced cotyledon-like leaves [184]. FUS3 transcript is primarily expressed in seeds but is also detected at low levels in tissues after completion of germination [185]. FUS3 transcript accumulation begins by the eight-cell embryo stage, and continues to be expressed relatively late during embryo development compared with LEC1/LEC2 (Figure 5A and [171–174,186–190]). FUS3 is positively regulated by the plant hormone abscisic acid (ABA) and negatively regulated by GA [191,192].
ABSCISIC ACID-INDEPENDENT3 (ABI3, At3g24650) is the third B3 domain TF of the LAFL genes. Like lec1, lec2, and fus3 mutants, abi3 seeds are desiccation intolerant but homozygous plants could be generated by rescue before the seed dries. Unlike other LAFL family members, abi3 did not show abnormal leafy cotyledons, but did show reduced seed storage protein accumulation. ABI3 is also involved in ABA perception and response [193,194]. Ectopic expression of ABI3 did not produce embryos on vegetative tissues, but some seed-specific storage protein mRNA was found in seedlings indicating the ability to ectopically activate some embryo programs [195]. ABI3 is expressed at highest levels during the late stages of embryo development (maturation phase) [171–174,188,195].
Genes that are sufficient for embryo identity/somatic embryogenesis
In addition to the key LAFL factors described above, many other genes were found to be sufficient to confer embryo identity when ectopically expressed, but were not necessary for normal zygotic embryo development. This may be due to redundant gene function(s) in the embryo [196]. Some of these genes, can, like LEC1 and LEC2 promote SE on various parts of the seedlings. Others only promote SE on particular tissues (e.g. roots; for example, WUS) or in particular culture conditions (e.g. AGL15, AGL18).
AGAMOUS-Like15 (AGL15, At5g13790) and AGAMOUS-Like18 (AGL18, At3g57390) encode MADS domain TFs that are preferentially, but not exclusively, expressed during seed development [197,198] Overexpression of these factors dramatically promoted secondary embryo production from zygotic embryo explants in the absence of exogenous hormones [199,200]. Subculturing allowed continued somatic embryo development in the absence of any exogenously added hormones for a long period time with the oldest 35S : AGL15 cultures being nearly 24 years old [199–201]. Additionally, both AGL15 and AGL18 can promote SE development from the SAM when mature seeds are allowed to complete germination in liquid medium containing the synthetic auxin 2,4-D ([199] and Paul and Perry, unpublished). These proteins can interact [202].
While SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 (AtSERK1, At1g71830) does not encode a TF, but rather encodes a leucine-rich repeat (LRR) transmembrane RLK which is broadly expressed including in developing seeds, it can also produce SE when ectopically expressed on auxin containing media [151]. It has been reported to form a complex that includes AGL15 [203]. Phosphorylation of AGL15 has been reported and is important for function [204,205].
Several APETALA2 (AP2) domain TFs including BABY BOOM/PLETHORA4 (BBM/PLT4 At5g17430), EMBRYOMAKER/AINTEGUMENTA-LIKE5/PLT5 (EMK, At5g57390), PLETHORA2 (PLT2, At1g51190), and some of the WOUND INDUCED DEDIFFERENTIATION (WIND) factors promote SE from seedlings in Arabidopsis when ectopically expressed [113,147,206–208]. Other members of the PLT family (PLT1, 2, 3, and 7) can also induce SE when ectopically expressed [209].
Additional TFs can promote SE on roots when ectopically and/or overexpressed. These TFs include WUSCHEL (WUS, At2g17950) [210–212], PLANT GROWTH ACTIVATOR37 (PGA37/MYB118, At3g27785) that encodes MYB transcription factor, and MYB115 (AT5G40360), a paralogous gene of PGA37 [213].
The Arabidopsis genome contains five RWP-RK DOMAIN-CONTAINING (RKD) genes, and expression of AtRKD1 (At1g18790) and AtRKD2 (At1g74480) were detected in reproductive organs, mainly expressed in the egg cell. Single or double mutants of these two genes did not show any phenotype or embryo morphological differences compared with wild type. But overexpression of AtRKD1 or AtRKD2 individually led to the proliferation of tissue expressing egg cell markers [214]. AtRKD4 (At5g53040) is another member of this family that is preferentially expressed in the early EP and suspensor. As discussed above, deficiencies in RKD4 (also called GROUNDED) causes abnormal zygotic cell elongation and subsequent division. Increased ectopic expression of GRD/RKD4 produced embryo-like structures on roots [68].
Loss-of-function mutants that produce ectopic embryos
Many other genes promote SE in Arabidopsis when present as loss-of-function alleles and these include pkl [215,216], val1 val2 [217,218] (VAL1 is also called HSI2; and VAL2 is also called HSL1), clf swn [219], hda6 hda19 RNAi [220], and a double knock-out/down of AtBMI A/B [221] or AtRING1a/b [222]. These mutants/RNAi plants produce embryos/embryonic calli on aberrant seedlings and may be considered as having defects in the transition from embryo to post-germinative development. Interestingly, this group of genes encode proteins that include domains shown or predicted to be involved in epigenetic control of gene expression and in particular involving repression of gene expression. Where it has been investigated, expression of LAFL and other genes mentioned above are derepressed in the seedlings, and in the case of AGL15, VAL1 directly represses gene expression [218,220–224]. VAL1 forms protein interactions with HDA6, while VAL2/HSL1 interacts with HDA19 [225,226]. VAL1 and VAL2 have also been reported to interact [225,227]. Other studies found evidence for direct repression of LAFL genes by VAL–HDA complexes, but these studies used 35S promotor for ChIP and a different stage of seedling development [225,226], rather than the native promoter as used in Chen et al. [224], where the association of VAL1 with LAFL regulatory regions was not found. In addition, CLF and SWN that are components of the Polycomb Repressive Complex2 (PRC2) involved in depositing trimethylation on lysine-27 of histone 3 (H2K27me3 — an epigenetic mark associated with repression of expression), may directly regulate LEC2, FUS3, and ABI3 [228].
Interaction network among these transcription factors
Genes responsive to the accumulation of many of the factors mentioned above have been identified, and in some cases, those that are bound by the TF determined (so-called direct responsive targets) [159,176,180,201,209,224–226,229–234]. Extensive interaction exists among these genes and the products that they encode, and some direct regulatory interactions are shown in Figure 5B. At the bent cotyledon (B-COT) stage during ZE development, LEC1 could directly bind and regulate LEC2, FUS3, ABI3, AGL15, ATSERK1, L1L, PGA37, and EMK, as well as potentially autoregulating itself [176]. Some of these genes are also directly induced targets of LEC1 during SE (FUS3, ABI3, LIL, and EMK). LEC2 directly regulates AGL15 and FUS3 [180,234] and indirectly regulates ABI3 [188]. Recently, WIND1, VAL1/HSI1, and MYB118 were also found to be directly responsive LEC2 targets [234]. Interestingly, LEC2 interacted with FUS3 in vitro by using pull-down method and in vivo by using BiFC assays [162]. LEC1 and LEC2 can be coimmunoprecipitated in the presence of NF-YC2 and the OLE1 promoter (a target DNA sequence) in pull-down experiments [235]. FUS3 direct target genes include LEC1, AGL15, ABI3, BBM, L1L, as well as autoregulating itself [232]. ABI3 could regulate itself and FUS3 [188,231]. BBM regulates several of the LAFL genes [209].
WUS did not bind to any of the genes mentioned above in shoot apices of Arabidopsis [233] or interact with any of them, which suggests that WUS may participate in embryo development through a different pathway or it acted at a relatively down-stream position in the pathway, or the context of shoot apex did not permit binding to these genes. However, several genes that regulate WUS are directly induced targets of LEC1, LEC2, and/or AGL15 in ZE and SE (see below). Regulatory regions of WUS were associated with BBM [236], although the response of transcript accumulation was not documented [237]. Using microarrays to compare transcriptomes in a MYB118 overexpression transgenic line with wild type control showed increased FUS3 transcript accumulation in response to increased MYB118 [238]. In addition, microRNAs are involved in SE. As an example, PLT1 and PLT2 were found to be regulated by miRNA396 via GROWTH REGULATING FACTOR (GRF) TFs [239,240].
Is there congruency between gene regulatory programs during SE and ZE?
At a molecular level, there have been reports of both similarities of transcriptomes in SE and ZE and differences. A study in cotton indicated a high degree of similarity between SE and ZE transcriptomes, with the main difference being transcript accumulation from many stress-associated genes in SE [241]. However, in Arabidopsis, Hofmann et al. [242] found few similarities between ZE and SEs, with the SE datasets that they evaluated appearing more similar to shoot and root tissues, and germinating seeds. In another study, Magnani et al. [135] separated cells that would undergo SE from callus by using an INTACT where the SE competent cells were labeled by driving the chimeric protein used for INTACT using a LEC2 promoter. They found increased transcript accumulation of WIND1 and BBM in the SE compared with callus cells, but not LEC1 or AGL15. They proposed that WIND1 and BBM, along with SERK1 confer embryogenic competence. They also proposed that LEC1 and AGL15 transcripts were absent because the stage sampled was too early for LEC2 to have led to the expression of these genes. Additionally, very early fate markers WOX2 and WOX9 transcripts were not detected possibly due to differences in very early patterning between SE and ZE. As a whole, they found the transcriptome of early embryogenic cells resembled octant stage zygotic embryos and meristematic cells.
The study by Kadokura et al. [243] that used the SE system of Ikeda-Iwai et al. [149] where the embryos formed from the proximity of stress-treated young seedling SAMs (Figure 4C) found key embryo TFs expressed in the SE-forming explants compared with non-SE-forming explants. They determined that embryonic commitment occurs by day 3 of culture at which time they could see the LAFL transcripts preferentially in SE producing explants. As a whole, the SE-forming explants had a mixed embryo, SAM, and root identity.
Wickramasuriya and Dunwell [244] looked at a direct SE system where SE is derived from IZE (from Gaj [142]; Figure 4A), sampling for RNA-seq after 5, 10 and 15 days of culture, comparing these stages to leaf tissue. Many embryo markers, including the LAFLs, L1L, BBM, and AGL15 were expressed at early time points as were many genes encoding TFs involved in polarity and pattern formation (WOX9, WUS, MP, PLT1/2, and STM). Gaj demonstrated that functional LAFL genes are essential for SE in this system [144].
A recent study [234] provided an explanation why use of IZE's are important for SE development in Arabidopsis. They assessed chromatin architecture and transcriptomes in response to culturing bent cotyledon IZE or 3 day seedlings on medium with 2,4-D. The former could produce SE but not the latter. They found that the ‘embryo’ developmental stage of the explants is important for auxin-induced chromatin changes to reprogram cells. Seedlings on 2,4-D had lost the open chromatin architecture at key TFs, including FUS3 and LEC2, that was not able to be reset to an open form in seedlings. While SAM tissue (or nearby tissue) can produce SE's, this only works for young seedlings, or seeds allowed to complete germination under conditions to promote SE [150,243]. It is possible that these very young SAMs are still at least partly ‘embryo’ in nature. For instance, while AGL15 accumulates to its highest level in embryos, one context where similar nuclear accumulation is seen is in the SAM of young seedlings (e.g. 4 days). This is transient, with loss of clear nuclear immunolocalization by 6 days [199].
How do embryo TFs promote (somatic) embryogenesis?
As discussed above, key TF's are necessary and/or sufficient for embryogenesis and there are both similarities and differences in ZE and SE, both morphologically and molecularly. However, in both cases, after fertilization or induction of SE, respectively, the body plan must be established, including initiation of the primary meristems as well as the development of the shoot and root meristems.
Specific roles for some of the TFs discussed above in zygotic processes have been reported based on phenotypic analysis of loss and/or gain-of-function, and genes directly controlled by these factors. For example, depending on the interacting partner, LEC1 is involved in morphogenesis, maturation processes, and photosynthesis [167,176]. LEC2 has roles in auxin production and signaling, and in maturation processes [180,181]. FUS3 and ABI3 both are involved in maturation processes and response to abiotic stress [230–232]. However, LEC1 transcript is already detectable in the egg cell. FUS3 transcript is initially in the basal lineage, while LEC2, AGL15/18, and BBM transcripts are present in the 24 h zygote [245], much earlier than the onset of maturation processes. These factors are also up-regulated early in several SE systems using wild type explants (see the discussion above). Loss-of-function of LEC1 and LEC2 have obvious morphological defects, including abnormal suspensor development. While the other genes do not have obvious morphological defects, the LAFLs show more severe phenotypes with higher-order mutants [188,246]. Likewise, the lack of zygotic phenotypes for the other TFs may reflect redundancy. Because ChIP studies should not be completely impacted by redundant factors, as phenotypes and transcriptomics may be, it is instructive, although perhaps speculative, to investigate genes that may be directly regulated by these key factors that are involved in polarity and meristem development and compare to information from SE systems. Some level of redundancy may result from the fact that many TFs may contribute to the regulation of a particular gene. For example, a 2014 study of genome-wide binding sites of 27 Arabidopsis TFs, identified more than 1000 highly occupied targets (HOTs), defined as genes bound by seven or more of these TFs, with a maximum of 18 of these TFs bound per gene [247]. Target hubs, defined as bound by eight or more of the TFs in the study, were enriched for genes encoding regulatory factors. As an example relevant to factors discussed in this review, several key embryo TFs directly induce expression of HAT's that are in turn involved in WUS regulation. These HATs have redundant functions (reviewed in [134]), and therefore phenotypes in SE (and ZE) SAM development in some mutants may be masked by the regulation of different HATs by diverse embryo TFs.
Previously, based on genes regulated by Arabidopsis AGL15, and its ortholog in soybean (Glycine max), it was reported that AGL15 may impact on early stages in the SE process because many TFs found to be expressed in dedifferentiation processes (also referred to here as ‘cellular reprograming’ that has been proposed as more accurate than dedifferentiation, [248]) are regulated by these genes [249]. Of the 44 ANAC, bZIP and WRKY TFs associated with reprograming [250], half have regulatory regions associated with LEC1 (data of [176]), suggesting that LEC1, like AGL15, may impact on a very early stage of SE. Pelletier et al. [176], looked at the association at early (4 h) and late (8 days) after induction of LEC1 to promote SE, as well as binding of LEC1 in bent cotyledon stage ZE, and the majority of ‘dedifferention’ TF genes showed similar binding between at least one SE stage and ZEs. Nearly half (43% and 41%) have regulatory regions associated with LEC2 and BBM, respectively [180,234,236]. Not all necessarily show a response in terms of transcript accumulation to the level of the potentially controlling TF, but this can be affected by the particular stage/tissue used to assess the transcriptome (for example, ABI3 directly binds regulatory regions for a gene encoding a miRNA, but expression, repression or no response depends on the stage of seed development; [231]). For this reason, we have chosen to report on binding, but those that respond are also shown in Supplementary Table S1 and Supplementary Figure S1. Of the 44 factors, more than three-quarters are bound at least one of the LAFL, AGL15, and/or BBM. Interestingly, of the list of these 44 ‘dedifferentiation’ TFs genes complied by Grafi et al. [250], 84% show transcript accumulation at or before the 32-cell stage of ZE [245] and Supplementary Table S1.
How much potential control of establishment of the primary meristems and apical meristems may these factors have? Interestingly, LEC2 was recently shown to directly up-regulate WOX2 and WOX3 expression during SE, and the products encoded by these genes are necessary but not sufficient for SE [234]. While other factors involved in polarity and fate after the first division may potentially be directly regulated by LAFL/AGL15/BBM during SE, almost no components associated with BC/suspensor development appear to be controlled by these TFs. These include WOX8, YDA, GRD/RDK4, ARF9, and IAA10, as well as some of the components involved in YDA signaling (Figure 2, Supplementary Table S1). Possibly this reflects the fact that an obvious suspensor is not essential for SE development. On the other hand, several genes encoding components involved in AC determination, besides WOX2, are potential direct targets of LAFL/AGL15/BBM during SE including MP, IAA12, and TMO7.
Auxin is an important factor in both SE and ZE. How may biosynthesis and response be controlled by these factors? LEC1, LEC2, and FUS3 have been found to directly induce the YUCCA genes that encode for products involved in auxin biosynthesis. Other genes encoding key TFs are induced in response to auxin, including AGL15, FUS3, LEC1 LEC2, BBM, and WUS [147,184,234,251-253]. As mentioned above, Godel-Jedrychowska et al. [139] showed callose deposition and decreased auxin response (as measured by an artificial auxin response transgene), associated with totipotency. In this regard, the fact that AGL15 appears to limit auxin accumulation and response may be relevant [201,254] although high-throughput data also shows AGL15 directly expresses MP (dataset [201] GSE17742). The regulatory regions of many other genes that encode products involved in auxin biosynthesis, transport, or response are also associated with one or more of the LAFL/AGL15/BBM factors (Supplementary Table S1).
Some of the WOX genes (WOX2, WOX8, WOX1, and WOX3) have roles in the transition from octant to dermatogen stage of ZE. While the lec mutants do not show defects in ZE protoderm development, functional redundancy is possible. WOX2 and WOX3 were shown to be direct LEC2 target genes and necessary for SE [234]. MP, that encodes a TF involved in the development of all three primary meristems, is a direct expressed AGL15 target gene in SE. PDF2, involved in protoderm development, and some of the LOG genes involved in provasculature development are also direct responsive targets of BBM and AGL15, respectively. Jo et al. [167] reported a role for LEC1 in morphogenesis via regulation of PHV, PHB, and SCR. Several factors involved in shoot-root development are also potential LEC1, LEC2, AGL15, and/or ABI3 direct responsive targets including PHB, PHV, ATHB2, ATHB4, HAT2/3, and PLT3. The list of potential directly controlled genes with products involved in meristem formation is longer if only association with regulatory regions is assessed, regardless of response (Supplementary Table S1).
Conclusion
We have highlighted some of the TFs involved in the control of early processes during embryogenesis and in the ability to specify embryo identity. These factors interact in complex networks, by controlling their own and other TFs expression and activity. They also share control of many other directly regulated genes that are not TFs and these targets are what actually ‘builds’ the cell. Transcriptomic studies can now measure and compare transcriptomes in individual cell types [64,65,255]. Some examples include comparison of egg to zygote transcriptomes [256], apical compared with basal cell transcriptomes [257], determination of an embryo epidermal specific transcriptome [258], and other subregions of the seed/embryo [259].
Although this review focuses on Arabidopsis, many of the TFs discussed have orthologs in other species including crop plants and comparison of regulatory networks should reveal interactions conserved and therefore presumably most important for driving developmental processes during seed development. Many orthologs of TFs described can induce SE in other plants and some examples include Nicotiana tabacum and G. max LEC1 [260,261], citrus FUS3 [262], and AGL15 orthologs in G. max and G. hirsutum [263,264]. In addition, computational tools are valuable approaches to determining gene regulatory networks (for a recent review [265]).
Acknowledgements
We apologize to all of our colleagues who have generated excellent data that we could not discuss or cite due to space constraints. We would like to thank our anonymous reviewers for their comments and suggestions to strengthen this review. We are grateful to Ms. Jeanne Hartman for valuable comments on the manuscript, and Ms. Ju-young (Gloria) Yoon for the generation of Figure 1. This work was supported by the National Science Foundation (grant no. IOS-1656380 to S.E.P.) and by the National Institute of Food and Agriculture, U.S. Department of Agriculture, Hatch project (S.E.P.) under accession number 1013409. No conflict of interest is declared.
Abbreviations
- AC
apical daughter cell
- BC
basal daughter cell
- EP
embryo proper
- EP
embryo proper
- IZE
immature zygotic embryos
- PD
plasmodesmata
- PGRs
plant growth regulators
- PSEs
primary somatic embryos
- QC
quiescent center
- RLK
receptor-like kinase
- SE
somatic embryogenesis/somatic embryo
- TFs
transcription factors
- ZE
zygotic embryogenesis/zygotic embryo
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Supplementary Material
References
- 1.Linkies A., Graeber K., Knight C. and Leubner-Metzger G. (2010) The evolution of seeds. New Phytol. 186, 817–831 10.1111/j.1469-8137.2010.03249.x [DOI] [PubMed] [Google Scholar]
- 2.Coiro M., Doyle J.A. and Hilton J. (2019) How deep is the conflict between molecular and fossil evidence on the age of angiosperms? New Phytol. 223, 83–99 10.1111/nph.15708 [DOI] [PubMed] [Google Scholar]
- 3.Roberts E.H. (1973) Predicting the storage life of seeds. Seed Sci. Technol. 1, 499–514 Corpus ID 82377238 [Google Scholar]
- 4.Nonogaki H. (2019) Seed germination and dormancy: the classic story, new puzzles, and evolution. J. Integr. Plant Biol. 61, 541–563 10.1111/jipb.12762 [DOI] [PubMed] [Google Scholar]
- 5.Meinke D.W. (2020) Genome-wide identification of EMBRYO-DEFECTIVE (EMB) genes required for growth and development in Arabidopsis. New Phytol. 226, 306–325 10.1111/nph.16071 [DOI] [PubMed] [Google Scholar]
- 6.Mayer U., Ruiz R.A.T., Berleth T., Misera S. and Jurgens G. (1991) Mutations affecting body organization in the Arabidopsis embryo. Nature 353, 402–407 10.1038/353402a0 [DOI] [Google Scholar]
- 7.Jürgens G., Mayer U., Ruiz R.A.T., Berleth T. and Misera S. (1991) Genetic analysis of pattern formation in the Arabiodpsis embryo. Development 113, 27–38 10.1199/tab.0126 [DOI] [PubMed] [Google Scholar]
- 8.Corredoira E., Merkle S.A., Martinez M.T., Toribio M., Canhoto J.M., Correia S.I. et al. (2019) Non-zygotic embryogenesis in hardwood species. Crit. Rev. Plant Sci. 38, 29–97 10.1080/07352689.2018.1551122 [DOI] [Google Scholar]
- 9.Egertsdotter U., Ahmad I. and Clapham D. (2019) Automation and scale up of somatic embryogenesis for commercial plant production, with emphasis on conifers. Front. Plant Sci. 10, article 109 10.3389/fpls.2019.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yadava P., Abhishek A., Singh R., Singh I., Kaul T., Pattanayak A. et al. (2017) Advances in maize transformation techologies and development of transgenic maize. Front. Plant Sci. 7, article no. 1949 10.3389/fpls.2016.01949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Smet I., Lau S., Mayer U. and Jürgens G. (2010) Embryogenesis–the humble beginnings of plant life. Plant J. 61, 959–970 10.1111/j.1365-313X.2010.04143.x [DOI] [PubMed] [Google Scholar]
- 12.Adhikari P.B., Liu X.Y., Wu X.Y., Zhu S.W. and Kasahara R.D. (2020) Fertilization in flowering plants: an odyssey of sperm cell delivery. Plant Mol. Biol. 103, 9–32 10.1007/s11103-020-00987-z [DOI] [PubMed] [Google Scholar]
- 13.Fatema U., Ali M.F., Hu Z., Clark A.J. and Kawashima T. (2019) Gamete nuclear migration in animals and plants. Front. Plant Sci. 10, article no. 517 10.3389/fpls.2019.00517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprunck S. (2020) Twice the fun, double the trouble: gamete interactions in flowering plants. Curr. Opin. Plant Biol. 53, 106–116 10.1016/j.pbi.2019.11.003 [DOI] [PubMed] [Google Scholar]
- 15.Coen O. and Magnani E. (2018) Seed coat thickness in the evolution of angiosperms. Cell. Mol. Life Sci. 75, 2509–2518 10.1007/s00018-018-2816-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nowack M.K., Grini P.E., Jakoby M.J., Lafos M., Koncz C. and Schnittger A. (2006) A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38, 63–67 10.1038/ng1694 [DOI] [PubMed] [Google Scholar]
- 17.Olsen O.-A. (2004) Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16, S214–S227 10.1105/tpc.017111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lafon-Placette C. and Köhler C. (2014) Embryo and endosperm, partners in seed development. Curr. Opin. Plant. Biol. 17, 64–69 10.1016/j.pbi.2013.11.008 [DOI] [PubMed] [Google Scholar]
- 19.Pignocchi C., Minns G.E., Nesi N., Koumproglou R., Kitsios G., Benning C. et al. (2009) ENDOSPERM DEFECTIVE1 is a novel microtubule-associated protein essential for seed development in Arabidopsis. Plant Cell 21, 90–105 10.1105/tpc.108.061812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhury A.M., Ming L., Miller C., Craig S., Dennis E.S. and Peacock W.J. (1997) Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 94, 4223–4228 10.1073/pnas.94.8.4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J. and Berger F. (2012) Endosperm: food for humankind and fodder for scientific discoveries. New Phytol. 195, 290–305 10.1111/j.1469-8137.2012.04182.x [DOI] [PubMed] [Google Scholar]
- 22.Hehenberger E., Kradolfer D. and Köhler C. (2012) Endosperm cellularization defines an important developmental transition for embryo development. Development 139, 2031–2039 10.1242/dev.077057 [DOI] [PubMed] [Google Scholar]
- 23.Olsen O.-A. (2001) Endosperm development: cellularization and cell fate specification. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 233–267 10.1146/annurev.arplant.52.1.233 [DOI] [PubMed] [Google Scholar]
- 24.Berger F. (2003) Endosperm: the crossroad of seed development. Curr. Opin. Plant Biol. 6, 42–50 10.1016/S1369526602000043 [DOI] [PubMed] [Google Scholar]
- 25.Figueiredo D.D. and Kohler C. (2016) Bridging the generation gap: communication between maternal sporophyte, female gametophyte and fertilization products. Curr. Opin. Plant Biol. 29, 16–20 10.1016/j.pbi.2015.10.008 [DOI] [PubMed] [Google Scholar]
- 26.Robert H.S. (2019) Molecular communication for coordinated seed and fruit development: what can we learn from auxin and sugars? Int. J. Mol. Sci. 20, article no. 936 10.3390/ijms20040936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batista R.A. and Kohler C. (2020) Genomic imprinting in plants-revisiting existing models. Genes Dev. 34, 24–36 10.1101/gad.332924.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han Q., Bartels A., Cheng X., Meyer A., An Y.Q.C., Hsieh T.F. et al. (2019) Epigenetics regulates reproductive development in plants. Plants Basel. 8, article no. 564 10.3390/plants8120564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colette A., Lu K.-J. and Weijers D. (2015) Building a plant: cell fate specification in the early Arabidopsis embryo. Development 142, 420–430 10.1242/dev.111500 [DOI] [PubMed] [Google Scholar]
- 30.Han X., Huang L.J., Feng D., Jiang W.H., Miu W.Z. and Li N. (2019) Plasmodesmata-related structural and functional proteins: the long sought-after secrets of a cytoplasmic channel in plant cell walls. Int. J. Mol. Sci. 20, article no. 2946 10.3390/ijms20122946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otero S., Helariutta Y. and Benitez-Alfonso Y. (2016) Symplastic communication in organ formation and tissue patterning. Curr. Opin. Plant Biol. 29, 21–28 10.1016/j.pbi.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 32.Thomas T.L. (1993) Gene expression during plant embryogenesis and germination: an overview. Plant Cell 5, 1401–1410 10.1105/tpc.5.10.1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laux T. and Jurgens G. (1997) Embryogenesis: a new start in life. Plant Cell 9, 989–1000 10.1105/tpc.9.7.989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.West M. and Harada J.J. (1993) Embryogenesis in higher plants: an overview. Plant Cell 5, 1361–1369 10.2307/3869788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wendrich J.R. and Weijers D. (2013) The Arabidopsis embryo as a miniature morphogenesis model. New Phytol. 199, 14–25 10.1111/nph.12267 [DOI] [PubMed] [Google Scholar]
- 36.Gutierrez L., Wuytswinkel O.V., Castelain M. and Bellini C. (2007) Combined networks regulating seed maturation. Trends Plant Sci. 127, 294–300 10.1016/j.tplants.2007.06.003 [DOI] [PubMed] [Google Scholar]
- 37.Holdsworth M.J., Bentsink L. and Soppe W.J.J. (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179, 33–54 10.1111/j.1469-8137.2008.02437.x [DOI] [PubMed] [Google Scholar]
- 38.Leprince O., Pellizzaro A., Berriri S. and Buitink J. (2017) Late seed maturation: drying without dying. J. Exp. Bot. 68, 827–841 10.1093/jxb/erw363 [DOI] [PubMed] [Google Scholar]
- 39.Faure J.E., Rotman N., Fortune P. and Dumas C. (2002) Fertilization in Arabidopsis thaliana wild type: developmental stages and time course. Plant J. 30, 481–488 10.1046/j.1365-313X.2002.01305.x [DOI] [PubMed] [Google Scholar]
- 40.Kimata Y., Higaki T., Kawashima T., Kurihara D., Sato Y., Yamada T. et al. (2016) Cytoskeleton dynamics control the first asymmetric cell division in Arabidopsis zygote. Proc. Natl. Acad. Sci. U.S.A. 113, 14157–14162 10.1073/pnas.1613979113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palovaara J., de Zeeuw T. and Weijers D. (2016) Tissue and organ initiation in the plant embryo: a first time for everything In Annual Review of Cell and Developmental Biology, vol. 32 (Schekman R., ed.), pp. 47–75 10.1146/annurev-cellbio-111315-124929, Annual Reviews [DOI] [PubMed] [Google Scholar]
- 42.Souter M. and Lindsey K. (2000) Polarity and signalling in plant embryogenesis. J. Exp. Bot. 51, 971–983 10.1093/jexbot/51.347.971 [DOI] [PubMed] [Google Scholar]
- 43.Zhao P., Begcy K., Dresselhaus T. and Sun M.-X. (2017) Does early embryogenesis in eudicots and monocots involve the same mechanism and molecular players? Plant Physiol. 173, 130–142 10.1104/pp.16.01406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mansfield S. and Briarty L. (1991) Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can. J. Bot. 69, 461–476 10.1139/b91-063 [DOI] [Google Scholar]
- 45.Sivaramakrishna D. (1978) Size relationships of apical cell and basal cell in two-celled embryos in angiosperms. Can. J. Bot. 56, 1434–1438 10.1139/b78-166 [DOI] [Google Scholar]
- 46.Kumlehn J., Lorz H. and Kranz E. (1998) Differentiation of isolated wheat zygotes into embryos and normal plants. Planta 205, 327–333 10.1007/s004250050327 [DOI] [Google Scholar]
- 47.Wang K., Chen H.M., Miao Y.J. and Bayer M. (2020) Square one: zygote polarity and early embryogenesis in flowering plants. Curr. Opin. Plant Biol. 53, 128–133 10.1016/j.pbi.2019.10.002 [DOI] [PubMed] [Google Scholar]
- 48.Jacob D. and Brian J. (2020) The short and intricate life of the suspensor. Physiol. Plant. 169, 110–121 10.1111/ppl.13057 [DOI] [PubMed] [Google Scholar]
- 49.Gepstein S. and Horwitz B.A. (1995) The impact of Arabidopsis research on plant biotechnology. Biotechnol. Adv. 13, 403–414 10.1016/0734-9750(95)02003-L [DOI] [PubMed] [Google Scholar]
- 50.Feng J. and Ma L. (2017) A method for characterizing embryogenesis in Arabidopsis. J. Vis. Exp. 126, article no. 55969 10.3791/55969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwartz B.W., Yeung E.C. and Meinke D.W. (1994) Disruption of morphogenesis and transformation of the suspensor in abnormal suspensor mutants of Arabidopsis. Development 120, 3235–3245 PMID: [DOI] [PubMed] [Google Scholar]
- 52.Reeves R.G. (1935) Development of the cotton embryo. J. Agric. Res. 51, 935–944 Accession Number: WOS:000188351000062, ISSN: 0095-9758 [Google Scholar]
- 53.Jürgens G. (2001) Apical–basal pattern formation in Arabidopsis embryogenesis. EMBO J. 20, 3609–3616 10.1093/emboj/20.14.3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshida S., de Reuille P.B., Lane B., Bassel G.W., Prusinkiewicz P., Smith R.S. et al. (2014) Genetic control of plant development by overriding a geometric division rule. Dev. Cell 29, 75–87 10.1016/j.devcel.2014.02.002 [DOI] [PubMed] [Google Scholar]
- 55.Laux T., Würschum T. and Breuninger H. (2004) Genetic regulation of embryonic pattern formation. Plant Cell 16, S190–S202 10.1105/tpc.016014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lau S., Slane D., Herud O., Kong J. and Jürgens G. (2012) Early embryogenesis in flowering plants: setting up the basic body pattern. Annu. Rev. Plant Biol. 63, 483–506 10.1146/annurev-arplant-042811-105507 [DOI] [PubMed] [Google Scholar]
- 57.Lukowitz W., Roeder A., Parmenter D. and Somerville C. (2004) A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116, 109–119 10.1016/S0092-8674(03)01067-5 [DOI] [PubMed] [Google Scholar]
- 58.Jeong S., Palmer T.M. and Lukowitz W. (2011) The RWP-RK factor GROUNDED promotes embryonic polarity by facilitating YODA MAP kinase signaling. Curr. Biol. 21, 1268–1276 10.1016/j.cub.2011.06.049 [DOI] [PubMed] [Google Scholar]
- 59.Zhang M.M., Wu H.J., Su J.B., Wang H.C., Zhu Q.K., Liu Y.D. et al. (2017) Maternal control of embryogenesis by MPK6 and its upstream MKK4/MKK5 in Arabidopsis. Plant J. 92, 1005–1019 10.1111/tpj.13737 [DOI] [PubMed] [Google Scholar]
- 60.Ueda M., Aichinger E., Gong W., Groot E., Verstraeten I., Vu L.D. et al. (2017) Transcriptional integration of paternal and maternal factors in the Arabidopsis zygote. Genes Dev. 31, 617–627 10.1101/gad.292409.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neu A., Eilbert E., Asseck L.Y., Slane D., Henschen A., Wang K. et al. (2019) Constitutive signaling activity of a receptor-associated protein links fertilization with embryonic patterning in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 116, 5795–5804 10.1073/pnas.1815866116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haecker A., Groß-Hardt R., Geiges B., Sarkar A., Breuninger H., Herrmann M. et al. (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131, 657–668 10.1242/dev.00963 [DOI] [PubMed] [Google Scholar]
- 63.Ueda M., Zhang Z. and Laux T. (2011) Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev. Cell 20, 264–270 10.1016/j.devcel.2011.01.009 [DOI] [PubMed] [Google Scholar]
- 64.Palovaara J. and Weijers D. (2019) Adapting INTACT to analyse cell-type-specific transcriptomes and nucleocytoplasmic mRNA dynamics in the Arabidopsis embryo. Plant Reprod. 32, 113–121 10.1007/s00497-018-0347-0 [DOI] [PubMed] [Google Scholar]
- 65.Palovaara J., Saiga S. and Weijers D. (2013) Transcriptomics approaches in the early Arabidopsis embryo. Trends Plant Sci. 18, 514–521 10.1016/j.tplants.2013.04.011 [DOI] [PubMed] [Google Scholar]
- 66.Breuninger H., Rikirsch E., Hermann M., Ueda M. and Laux T. (2008) Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14, 867–876 10.1016/j.devcel.2008.03.008 [DOI] [PubMed] [Google Scholar]
- 67.Wu X., Chory J. and Weigel D. (2007) Combinations of WOX activities regulate tissue proliferation during Arabidopsis embryonic development. Dev. Biol. 309, 306–316 10.1016/j.ydbio.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waki T., Hiki T., Watanabe R., Hashimoto T. and Nakajima K. (2011) The Arabidopsis RWP-RK protein RKD4 triggers gene expression and pattern formation in early embryogenesis. Curr. Biol. 21, 1277–1281 10.1016/j.cub.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 69.Costa L.M., Marshall E., Tesfaye M., Silverstein K.A., Mori M., Umetsu Y. et al. (2014) Central cell-derived peptides regulate early embryo patterning in flowering plants. Science 344, 168–172 10.1126/science.1243005 [DOI] [PubMed] [Google Scholar]
- 70.Fiume E. and Fletcher J.C. (2012) Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8. Plant Cell 24, 1000–1012 10.1105/tpc.111.094839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu T.Y., Shi D.Q., Jia P.F., Tang J., Li H.J., Liu J. et al. (2016) The Arabidopsis receptor kinase ZAR1 is required for zygote asymmetric division and its daughter cell fate. PLoS Gen. 12, article no. e1005933 10.1371/journal.pgen.1005933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T. et al. (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153 10.1038/nature02085 [DOI] [PubMed] [Google Scholar]
- 73.Robert H.S., Grones P., Stepanova A.N., Robles L.M., Lokerse A.S., Alonso J.M. et al. (2013) Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr. Biol. 23, 2506–2512 10.1016/j.cub.2013.09.039 [DOI] [PubMed] [Google Scholar]
- 74.Robert H.S., Park C., Gutierrez C.L., Wojcikowska B., Pencik A., Novak O. et al. (2018) Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat. Plants 4, 548–553 10.1038/s41477-018-0204-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robert H.S., Grunewald W., Sauer M., Cannoot B., Soriano M., Swarup R. et al. (2015) Plant embryogenesis requires AUX/LAX-mediated auxin influx. Development 142, 702–711 10.1242/dev.115832 [DOI] [PubMed] [Google Scholar]
- 76.Mayer U., Buttner G. and Jurgens G. (1993) Apical-basal pattern formation in the Arabidopsis embryo: studies on the role of the gnom gene. Development 117, 149–162 Accession Number: WOS:A1993KP05500012, ISSN: 0950-1991 [Google Scholar]
- 77.Busch M., Mayer U. and Jurgens G. (1996) Molecular analysis of the Arabidopsis pattern formation gene GNOM: gene structure and intragenic complementation. Mol. Gen. Gen. 250, 681–691 10.1007/BF02172979 [DOI] [PubMed] [Google Scholar]
- 78.Steinmann T., Geldner N., Grebe M., Mangold S., Jackson C.L., Paris S. et al. (1999) Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286, 316–318 10.1126/science.286.5438.316 [DOI] [PubMed] [Google Scholar]
- 79.Roosjen M., Paque S. and Weijers D. (2018) Auxin response factors: output control in auxin biology. J. Exp. Biol. 69, 179–188 10.1093/jxb/erx237 [DOI] [PubMed] [Google Scholar]
- 80.Hamann T., Benkova E., Bäurle I., Kientz M. and Jürgens G. (2002) The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 16, 1610–1615 10.1101/gad.229402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schlereth A., Möller B., Liu W., Kientz M., Flipse J., Rademacher E.H. et al. (2010) MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464, 913–916 10.1038/nature08836 [DOI] [PubMed] [Google Scholar]
- 82.Bayer M., Nawy T., Giglione C., Galli M., Meinnel T. and Lukowitz W. (2009) Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323, 1485–1488 10.1126/science.1167784 [DOI] [PubMed] [Google Scholar]
- 83.Lu K.J., De Rybel B., van Mourik H. and Weijers D. (2018) Regulation of intercellular TARGET OF MONOPTEROS 7 protein transport in the Arabidopsis root. Development 145, dev152892 10.1242/dev.152892 [DOI] [PubMed] [Google Scholar]
- 84.Rademacher E.H., Lokerse A.S., Schlereth A., Llavata-Peris C.I., Bayer M., Kientz M. et al. (2012) Different auxin response machineries control distinct cell fates in the early plant embryo. Dev. Cell 22, 211–222 10.1016/j.devcel.2011.10.026 [DOI] [PubMed] [Google Scholar]
- 85.Ulmasov T., Hagen G. and Guilfoyle T.J. (1999) Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. U.S.A. 96, 5844–5849 10.1073/pnas.96.10.5844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu Y., Li X.B., Zhao J., Tang X.C., Tian S.J., Chen J.Y. et al. (2015) Direct evidence that suspensor cells have embryogenic potential that is suppressed by the embryo proper during normal embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 112, 12432–12437 10.1073/pnas.1508651112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Radoeva T., Lokerse A.S., Llavata-Peris C.I., Wendrich J.R., Xiang D.Q., Liao C.Y. et al. (2019) A robust auxin response network controls embryo and suspensor development through a basic helix loop helix transcriptional module. Plant Cell 31, 52–67 10.1105/tpc.18.00518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mukherjee K., Brocchieri L. and Bürglin T.R. (2009) A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 26, 2775–2794 10.1093/molbev/msp201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Abe M., Katsumata H., Komeda Y. and Takahashi T. (2003) Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development 130, 635–643 10.1242/dev.00292 [DOI] [PubMed] [Google Scholar]
- 90.Lu P., Porat R., Nadeau J.A. and O'Neill S.D. (1996) Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. Plant Cell 8, 2155–2168 10.1105/tpc.8.12.2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sessions A., Weigel D. and Yanofsky M.F. (1999) The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20, 259–263 10.1046/j.1365-313x.1999.00594.x [DOI] [PubMed] [Google Scholar]
- 92.Takada S. and Jürgens G. (2007) Transcriptional regulation of epidermal cell fate in the Arabidopsis embryo. Development 134, 1141–1150 10.1242/dev.02803 [DOI] [PubMed] [Google Scholar]
- 93.Ogawa E., Yamada Y., Sezaki N., Kosaka S., Kondo H., Kamata N. et al. (2015) ATML1 and PDF2 play a redundant and essential role in Arabidopsis embryo development. Plant Cell Physiol. 56, 1183–1192 10.1093/pcp/pcv045 [DOI] [PubMed] [Google Scholar]
- 94.San-Bento R., Farcot E., Galletti R., Creff A. and Ingram G. (2014) Epidermal identity is maintained by cell–cell communication via a universally active feedback loop in Arabidopsis thaliana. Plant J. 77, 46–58 10.1111/tpj.12360 [DOI] [PubMed] [Google Scholar]
- 95.Takada S., Takada N. and Yoshida A. (2013) ATML1 promotes epidermal cell differentiation in Arabidopsis shoots. Development 140, 1919–1923 10.1242/dev.094417 [DOI] [PubMed] [Google Scholar]
- 96.Peterson K.M., Shyu C., Burr C.A., Horst R.J., Kanaoka M.M., Omae M. et al. (2013) Arabidopsis homeodomain-leucine zipper IV proteins promote stomatal development and ectopically induce stomata beyond the epidermis. Development 140, 1924–1935 10.1242/dev.090209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chandler J., Nardmann J. and Werr W. (2008) Plant development revolves around axes. Trends Plant Sci. 13, 78–84 10.1016/j.tplants.2007.11.010 [DOI] [PubMed] [Google Scholar]
- 98.Möller B.K., ten Hove C.A., Xiang D.Q., Williams N., Lopez L.G., Yoshida S. et al. (2017) Auxin response cell-autonomously controls ground tissue initiation in the early Arabidopsis embryo. Proc. Natl. Acad. Sci. U.S.A. 114, E2533–E2539 10.1073/pnas.1616493114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Berleth T. and Jurgens G. (1993) The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118, 575–587 Accession Number: WOS:A1993LH32700024, ISSN: 0950-1991 [Google Scholar]
- 100.Hardtke C.S. and Berleth T. (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411 10.1093/emboj/17.5.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hamann T., Mayer U. and Jurgens G. (1999) The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126, 1387–1395 PMID: [DOI] [PubMed] [Google Scholar]
- 102.De Rybel B., Möller B., Yoshida S., Grabowicz I., de Reuille P.B., Boeren S. et al. (2013) A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev. Cell 24, 426–437 10.1016/j.devcel.2012.12.013 [DOI] [PubMed] [Google Scholar]
- 103.Ohashi-Ito K. and Bergmann D.C. (2007) Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development 134, 2959–2968 10.1242/dev.006296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ohashi-Ito K., Matsukawa M. and Fukuda H. (2013) An atypical bHLH transcription factor regulates early xylem development downstream of auxin. Plant Cell Physiol. 54, 398–405 10.1093/pcp/pct013 [DOI] [PubMed] [Google Scholar]
- 105.De Rybel B., Adibi M., Breda A.S., Wendrich J.R., Smit M.E., Novák O. et al. (2014) Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science 345, article no. 1255215 10.1126/science.1255215 [DOI] [PubMed] [Google Scholar]
- 106.Ohashi-Ito K., Saegusa M., Iwamoto K., Oda Y., Katayama H., Kojima M. et al. (2014) A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr. Biol. 24, 2053–2058 10.1016/j.cub.2014.07.050 [DOI] [PubMed] [Google Scholar]
- 107.De Rybel B., Breda A.S. and Weijers D. (2014) Prenatal plumbing-vascular tissue formation in the plant embryo. Physiol. Plant. 151, 126–133 10.1111/ppl.12091 [DOI] [PubMed] [Google Scholar]
- 108.Ohashi-Ito K. and Fukuda H. (2014) Initiation of vascular development. Physiol. Plant. 151, 142–146 10.1111/ppl.12111 [DOI] [PubMed] [Google Scholar]
- 109.Bozhkov P.V., Filonova L.H. and Suarez M.F. (2005) Programmed cell death in plant embryogenesis. Curr. Top. Dev. Biol. 67, 135–179 10.1016/S0070-2153(05)67004-4 [DOI] [PubMed] [Google Scholar]
- 110.Weigel D. and Jurgens G. (2002) Stem cells that make stems. Nature 415, 751–754 10.1038/415751a [DOI] [PubMed] [Google Scholar]
- 111.Scheres B. (2007) Stem-cell niches: nursery rhymes across kingdoms. Nat. Rev. Mol. Cell Biol. 8, 345–354 10.1038/nrm2164 [DOI] [PubMed] [Google Scholar]
- 112.Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A. et al. (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774 10.1016/j.cub.2003.09.035 [DOI] [PubMed] [Google Scholar]
- 113.Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C. et al. (2004) The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120 10.1016/j.cell.2004.09.018 [DOI] [PubMed] [Google Scholar]
- 114.Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R. et al. (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057 10.1038/nature06206 [DOI] [PubMed] [Google Scholar]
- 115.Prigge M.J., Otsuga D., Alonso J.M., Ecker J.R., Drews G.N. and Clark S.E. (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17, 61–76 10.1105/tpc.104.026161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mallory A.C., Reinhart B.J., Jones-Rhoades M.W., Tang G., Zamore P.D., Barton M.K. et al. (2004) MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 23, 3356–3364 10.1038/sj.emboj.7600340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Long J.A., Ohno C., Smith Z.R. and Meyerowitz E.M. (2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523 10.1126/science.1123841 [DOI] [PubMed] [Google Scholar]
- 118.Smith Z.R. and Long J.A. (2010) Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464, 423–426 10.1038/nature08843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Szemenyei H., Hannon M. and Long J.A. (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386 10.1126/science.1151461 [DOI] [PubMed] [Google Scholar]
- 120.Causier B., Ashworth M., Guo W.J. and Davies B. (2012) The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158, 423–438 10.1104/pp.111.186999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Perini S., Mambro R. and Sabatini S. (2012) Growth and development of the root apical meristem. Curr. Opin. Plant Biol. 15, 17–23 10.1016/j.pbi.2011.10.006 [DOI] [PubMed] [Google Scholar]
- 122.Yoshida S., Saiga S. and Weijers D. (2013) Auxin regulation of embryonic root formation. Plant Cell Physiol. 54, 325–332 10.1093/pcp/pcs170 [DOI] [PubMed] [Google Scholar]
- 123.Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M. and Simon R. (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619 10.1126/science.289.5479.617 [DOI] [PubMed] [Google Scholar]
- 124.Schoof H., Lenhard M., Haecker A., Mayer K.F.X., Jurgens G. and Laux T. (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644 10.1016/S0092-8674(00)80700-X [DOI] [PubMed] [Google Scholar]
- 125.Long J.A., Moan E.I., Medford J.I. and Barton M.K. (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69 10.1038/379066a0 [DOI] [PubMed] [Google Scholar]
- 126.Ruberti I., Sessa G., Ciolfi A., Possenti M., Carabelli M. and Morelli G. (2012) Plant adaptation to dynamically changing environment: the shade avoidance response. Biotechnol. Adv. 30, 1047–1058 10.1016/j.biotechadv.2011.08.014 [DOI] [PubMed] [Google Scholar]
- 127.Bou-Torrent J., Salla-Martret M., Brandt R., Musielak T., Palauqui J.C., Martinez-Garcia J.F. et al. (2012) ATHB4 and HAT3, two class II HD-ZIP transcription factors, control leaf development in Arabidopsis. Plant Signal. Behav. 7, 1382–1387 10.4161/psb.21824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Reymond M.C., Brunoud G., Chauvet A., Martínez-Garcia J.F., Martin-Magniette M.-L., Monéger F. et al. (2012) A light-regulated genetic module was recruited to carpel development in Arabidopsis following a structural change to SPATULA. Plant Cell 24, 2812–2825 10.1105/tpc.112.097915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Brandt R., Salla-Martret M., Bou-Torrent J., Musielak T., Stahl M., Lanz C. et al. (2012) Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 72, 31–42 10.1111/j.1365-313X.2012.05049.x [DOI] [PubMed] [Google Scholar]
- 130.Reinhart B.J., Liu T., Newell N.R., Magnani E., Huang T., Kerstetter R. et al. (2013) Establishing a framework for the ad/abaxial regulatory network of Arabidopsis: ascertaining targets of class III HOMEODOMAIN LEUCINE ZIPPER and KANADI regulation. Plant Cell 25, 3228–3249 10.1105/tpc.113.111518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fuchs M. and Lohmann J.U. (2020) Aiming for the top: non-cell autonomous control of shoot stem cells in Arabidopsis. J. Plant Res. 133, 297–309 10.1007/s10265-020-01174-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Voronina A.S. and Pshennikova E.S. (2020) Plant stem cells. Mol. Biol. 54, 163–177 10.1134/S002689332002017X [DOI] [PubMed] [Google Scholar]
- 133.Somssich M., Je B., Simon R. and Jackson D. (2016) CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143, 3238–3248 10.1242/dev.133645 [DOI] [PubMed] [Google Scholar]
- 134.Roodbarkelari F. and Groot E.P. (2017) Regulatory function of homeodomain-leucine zipper (HD-ZIP) family proteins during embryogenesis. New Phytol. 213, 95–104 10.1111/nph.14132 [DOI] [PubMed] [Google Scholar]
- 135.Magnani E., Jimenez-Gomez J.M., Soubigou-Taconnat L., Lepiniec L. and Fiume E. (2017) Profiling the onset of somatic embryogenesis in Arabidopsis. BMC Genom. 18, 998 10.1186/s12864-017-4391-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wojcik A.M., Wojcikowska B. and Gaj M.D. (2020) Current perspectives on the auxin-mediated genetic network that controls the induction of somatic embryogenesis in plants. Int. J. Mol. Sci. 21, article no. 1333 10.3390/ijms21041333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Garces H.M.P., Champagne C.E.M., Townsley B.T., Park S., Malho R., Pedroso M.C. et al. (2007) Evolution of asexual reproduction in leaves of the genus Kalanchole. Proc. Natl. Acad. Sci. U.S.A. 104, 15578–15583 10.1073/pnas.0704105104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sugimoto K., Temman H., Kadokura S. and Matsunaga S. (2019) To regenerate or not to regenerate: factors that drive plant regeneration. Curr. Opin. Plant Biol. 47, 138–150 10.1016/j.pbi.2018.12.002 [DOI] [PubMed] [Google Scholar]
- 139.Godel-Jedrychowska K., Kulinska-Lukaszek K., Horstman A., Soriano M., Li M.F., Malota K. et al. (2020) Symplasmic isolation marks cell fate changes during somatic embryogenesis. J. Exp. Bot. 71, 2612–2628 10.1093/jxb/eraa041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Feher A. (2015) Somatic embryogenesis - stress-induced remodeling of plant cell fate. Biochim. Biophys. Acta 1849, 385–402 10.1016/j.bbagrm.2014.07.005 [DOI] [PubMed] [Google Scholar]
- 141.Segui-Simarro J.M. and Nuez F. (2008) Pathways to doubled haploidy: chromosome doubling during androgenesis. Cytogenet. Genome Res. 120, 358–369 10.1159/000121085 [DOI] [PubMed] [Google Scholar]
- 142.Gaj M.D. (2001) Direct somatic embryogenesis as a rapid and efficient system for in vitro regeneration of Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 64, 39–46 10.1023/A:1010679614721 [DOI] [Google Scholar]
- 143.Gaj M.D., Trojanowska A., Ujczak A., Medrek M., Koziol A. and Garbaciak B. (2006) Hormone-response mutants of Arabidopsis thaliana (L.) heynh. impaired in somatic embryogenesis. Plant Growth Regul. 49, 183–197 10.1007/s10725-006-9104-8 [DOI] [Google Scholar]
- 144.Gaj M.D., Zhang S.B., Harada J.J. and Lemaux P.G. (2005) Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222, 977–988 10.1007/s00425-005-0041-y [DOI] [PubMed] [Google Scholar]
- 145.Kurczynska E.U., Gaj M.D., Ujczak A. and Mazur E. (2007) Histological analysis of direct somatic embryogenesis in Arabidopsis thaliana (L.) Heynh. Planta 226, 619–628 10.1007/s00425-007-0510-6 [DOI] [PubMed] [Google Scholar]
- 146.Gaj M.D. (2011) Somatic embryogenesis and plant regeneration in the culture of Arabidopsis thaliana (L.) Heynh. immature zygotic embryos In Plant Embryo Culture: Methods and Protocols (Thorpe T.A. and Yeung E.C., eds.), pp. 257–265 10.1007/978-1-61737-988-8_18 Springer Publishing Company; [DOI] [PubMed] [Google Scholar]
- 147.Su Y.H., Zhao X.Y., Liu Y.B., Zhang C.L., O'Neill S.D. and Zhang X.S. (2009) Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J. 59, 448–460 10.1111/j.1365-313X.2009.03880.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ikeda-Iwai M., Satoh S. and Kamada H. (2002) Establishment of a reproducible tissue culture system for the induction of Arabidopsis somatic embryos. J. Exp. Bot. 53, 1575–1580 10.1093/jxb/erf006 [DOI] [PubMed] [Google Scholar]
- 149.Ikeda-Iwai M., Umehara M., Satoh S. and Kamada H. (2003) Stress-induced somatic embryogenesis in vegetative tissues of Arabidopsis thaliana. Plant J. 34, 107–114 10.1046/j.1365-313X.2003.01702.x [DOI] [PubMed] [Google Scholar]
- 150.Mordhorst A.P., Voerman K.J., Hartog M.V., Meijer E.A., van Went J., Koornneef M. et al. (1998) Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics 149, 549–563 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hecht V., Vielle-Calzada J.P., Hartog M.V., Schmidt E.D.L., Boutilier K., Grossniklaus U. et al. (2001) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127, 803–816 10.1104/pp.010324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.von Recklinghausen I.R., Iwanowska A., Kieft H., Mordhorst A.P., Schel J.H.N. and van Lammeren A.A.M. (2000) Structure and development of somatic embryos formed in Arabidopsis thaliana pt mutant callus cultures derived from seedlings. Protoplasma 211, 217–224 10.1007/BF01304489 [DOI] [Google Scholar]
- 153.Wu Y., Haberland G., Zhou C. and Koop H.U. (1992) Somatic embryogenesis, formation of morphogenetic callus and normal development in zygotic embryos of Arabidopsis thaliana in vitro. Protoplasma 169, 89–96 10.1007/BF01323608 [DOI] [Google Scholar]
- 154.Mathur J., Koncz C. and Szabados L. (1995) A simple method for isolation, liquid culture, transformation and regernation of Arabidopsis thaliana protoplasts. Plant Cell Rep. 14, 221–226 10.1007/BF00233637 [DOI] [PubMed] [Google Scholar]
- 155.Luo Y.K. and Koop H.U. (1997) Somatic embryogenesis in cultured immature zygotic embryos and leaf protoplasts of Arabidopsis thaliana ecotypes. Planta 202, 387–396 10.1007/s004250050141 [DOI] [PubMed] [Google Scholar]
- 156.Le B.H., Cheng C., Bui A.Q., Wagmaister J.A., Henry K.F., Pelletier J. et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. U.S.A. 107, 8063–8070 10.1073/pnas.1003530107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V. and Provart N.J. (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, article no. e718 10.1371/journal.pone.0000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Junker A. and Baumlein H. (2012) Multifunctionality of the LEC1 transcription factor during plant development. Plant Signal. Behav. 7, 1718–1720 10.4161/psb.22365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Junker A., Monke G., Rutten T., Keilwagen J., Seifert M., Tuyet M.N.T. et al. (2012) Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 71, 427–442 10.1111/j.1365-313X.2012.04999.x [DOI] [PubMed] [Google Scholar]
- 160.Huang M.K., Hu Y.L., Liu X., Li Y.G. and Hou X.L. (2015) Arabidopsis LEAFY COTYLEDON1 mediates postembryonic development via interacting with PHYTOCHROME-INTERACTING FACTOR4. Plant Cell 27, 3099–3111 10.1105/tpc.15.00750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chiu R.S., Nahal H., Provart N.J. and Gazzarrini S. (2012) The role of the Arabidopsis FUSCA3 transcription factor during inhibition of seed germination at high temperature. BMC Plant Biol. 12, article no. 15 10.1186/1471-2229-12-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tang L.P., Zhou C., Wang S.S., Yuan J., Zhang X.S. and Su Y.H. (2017) FUSCA3 interacting with LEAFY COTYLEDON2 controls lateral root formation through regulating YUCCA4 gene expression in Arabidopsis thaliana. New Phytol. 213, 1740–1754 10.1111/nph.14313 [DOI] [PubMed] [Google Scholar]
- 163.Bedi S., Sengupta S., Ray A. and Chaudhuri R.N. (2016) ABI3 mediates dehydration stress recovery response in Arabidopsis thaliana by regulating expression of downstream genes. Plant Sci. 250, 125–140 10.1016/j.plantsci.2016.06.006 [DOI] [PubMed] [Google Scholar]
- 164.Han J.D., Li X., Jiang C.K., Wong G.K.S., Rothfels C.J. and Rao G.Y. (2017) Evolutionary analysis of the LAFL genes involved in the land plant seed maturation program. Front. Plant Sci. 8, article no. 439 10.3389/fpls.2017.00439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Boulard C., Fatihi A., Lepiniec L. and Dubreucq B. (2017) Regulation and evolution of the interaction of the seed B3 transcription factors with NF-Y subunits. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 1069–1078 10.1016/j.bbagrm.2017.08.008 [DOI] [PubMed] [Google Scholar]
- 166.Lotan T., Ohto M., Yee K.M., West M.A.L., Lo R., Kwong R.W. et al. (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205 10.1016/S0092-8674(00)81463-4 [DOI] [PubMed] [Google Scholar]
- 167.Jo L., Pelletier J.M. and Harada J.J. (2019) Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J. Integr. Plant Biol. 61, 564–580 10.1111/jipb.12806 [DOI] [PubMed] [Google Scholar]
- 168.Lee H.S., Fischer R.L., Goldberg R.B. and Harada J.J. (2003) Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 100, 2152–2156 10.1073/pnas.0437909100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Meinke D.W. (1992) A homeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258, 1647–1650 10.1126/science.258.5088.1647 [DOI] [PubMed] [Google Scholar]
- 170.Meinke D.W., Franzmann L.H., Nickle T.C. and Yeung E.C. (1994) Leafy cotyledon mutants of Arabidopsis. Plant Cell 6, 1049–1064 10.2307/3869884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Casson S., Spencer M., Walker K. and Lindsey K. (2005) Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. Plant J. 42, 111–123 10.1111/j.1365-313X.2005.02355.x [DOI] [PubMed] [Google Scholar]
- 172.Klepikova A.V., Kasianov A.S., Gerasimov E.S., Logacheva M.D. and Penin A.A. (2016) A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 88, 1058–1070 10.1111/tpj.13312 [DOI] [PubMed] [Google Scholar]
- 173.Nakabayashi K., Okamoto M., Koshiba T., Kamiya Y. and Nambara E. (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J. 41, 697–709 10.1111/j.1365-313X.2005.02337.x [DOI] [PubMed] [Google Scholar]
- 174.Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M. et al. (2005) A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506 10.1038/ng1543 [DOI] [PubMed] [Google Scholar]
- 175.Dey N., Sarkar S., Acharya S. and Maiti I. (2015) Synthetic promoters in planta. Planta 242, 1077–1094 10.1007/s00425-015-2377-2 [DOI] [PubMed] [Google Scholar]
- 176.Pelletier J.M., Kwong R.W., Park S., Le B.H., Baden R., Cagliaria A. et al. (2017) LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. U.S.A. 114, E6710–E6719 10.1073/pnas.1707957114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Garces H.M.P., Koenig D., Townsley B.T., Kim M. and Sinha N.R. (2014) Truncation of LEAFY COTYLEDON1 protein Is required for asexual reproduction in Kalanchoe daigremontiana. Plant Physiol. 165, 196–206 10.1104/pp.114.237222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kwong R.W., Bui A.Q., Lee H., Kwong L.W., Fischer R.L., Goldberg R.B. et al. (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15, 5–18 10.1105/tpc.006973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Stone S.L., Kwong L.W., Yee K.M., Pelletier J., Lepiniec L., Fischer R.L. et al. (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U.S.A. 98, 11806–11811 10.1073/pnas.201413498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Braybrook S.A., Stone S.L., Park S., Bui A.Q., Le B.H., Fischer R.L. et al. (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 3468–3473 10.1073/pnas.0511331103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Stone S.L., Braybrook S.A., Paula S.L., Kwong L.W., Meuser J., Pelletier J. et al. (2008) Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 3151–3156 10.1073/pnas.0712364105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Feeney M., Frigerio L., Cui Y.H. and Menassa R. (2013) Following vegetative to embryonic cellular changes in leaves of Arabidopsis overexpressing LEAFY COTYLEDON2. Plant Physiol. 162, 1881–1896 10.1104/pp.113.220996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Keith K., Kraml M., Dengler N.G. and McCourt P. (1994) FUSCA3 - a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6, 589–600 10.2307/3869865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gazzarrini S., Tsuchiya Y., Lumba S., Okamoto M. and McCourt P. (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 7, 373–385 10.1016/j.devcel.2004.06.017 [DOI] [PubMed] [Google Scholar]
- 185.Luerssen K., Kirik V., Herrmann P. and Misera S. (1998) FUSCA3 encodes a protein with a conserved VP1/ABI3-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 15, 755–764 10.1046/j.1365-313X.1998.00259.x [DOI] [PubMed] [Google Scholar]
- 186.Baumlein H., Misera S., Luerssen H., Kolle K., Horstmann C., Wobus U. et al. (1994) The FUS3 gene of Arabidopsis thaliana is a regulator of gene expression during late embryogenesis. Plant J. 6, 379–387 10.1046/j.1365-313X.1994.06030379.x [DOI] [Google Scholar]
- 187.Roscoe T.J., Vaissayre V., Paszkiewicz G., Clavijo F., Kelemen Z., Michaud C. et al. (2019) Regulation of FUSCA3 expression during seed development in Arabidopsis. Plant Cell Physiol. 60, 476–487 10.1093/pcp/pcy224 [DOI] [PubMed] [Google Scholar]
- 188.To A., Valon C., Savino G., Guilleminot J., Devic M., Giraudat J. et al. (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18, 1642–1651 10.1105/tpc.105.039925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tsuchiya Y., Nambara E., Naito S. and McCourt P. (2004) The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J. 37, 73–81 10.1046/j.1365-313X.2003.01939.x [DOI] [PubMed] [Google Scholar]
- 190.Willmann M.R., Mehalick A.J., Packer R.L. and Jenik P.D. (2011) MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol. 155, 1871–1884 10.1104/pp.110.171355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chiu R.S., Saleh Y. and Gazzarrini S. (2016) Inhibition of FUSCA3 degradation at high temperature is dependent on ABA signaling and is regulated by the ABA/GA ratio. Plant Signal. Behav. 11, article no. e1247137 10.1080/15592324.2016.1247137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lu Q.S., dela Paz J., Pathmanathan A., Chiu R.S., Tsai A.Y.L. and Gazzarrini S. (2010) The C-terminal domain of FUSCA3 negatively regulates mRNA and protein levels, and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis. Plant J. 64, 100–113 10.1111/j.1365-313X.2010.04307.x [DOI] [PubMed] [Google Scholar]
- 193.Giraudat J., Hauge B.M., Valon C., Smalle J., Parcy F. and Goodman H.M. (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4, 1251–1261 10.1105/tpc.4.10.1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Nambara E., Naito S. and McCourt P. (1992) A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new ABI3 allele. Plant J. 2, 435–441 10.1111/j.1365-313X.1992.00435.x [DOI] [Google Scholar]
- 195.Parcy F., Valon C., Raynal M., Gaubiercomella P., Delseny M. and Giraudat J. (1994) Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6, 1567–1582 10.1105/tpc.6.11.1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Cutler S. and McCourt P. (2005) Dude, where's my phenotype? Dealing with redundancy in signaling networks. Plant Physiol. 138, 558–559 10.1104/pp.104.900152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Heck G.R., Perry S.E., Nichols K.W. and Fernandez D.E. (1995) AGL15, a MADS domain protein expressed in developing embryos. Plant Cell 7, 1271–1282 10.1105/tpc.7.8.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Rounsley S.D., Ditta G.S. and Yanofsky M.F. (1995) Diverse roles for MADS box gene in Arabidopsis development. Plant Cell 7, 1259–1269 10.1105/tpc.7.8.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Harding E.W., Tang W.N., Nichols K.W., Fernandez D.E. and Perry S.E. (2003) Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol. 133, 653–663 10.1104/pp.103.023499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Adamczyk B.J., Lehti-Shiu M.D. and Fernandez D.E. (2007) The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 50, 1007–1019 10.1111/j.1365-313X.2007.03105.x [DOI] [PubMed] [Google Scholar]
- 201.Zheng Y.M., Ren N., Wang H., Stromberg A.J. and Perry S.E. (2009) Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21, 2563–2577 10.1105/tpc.109.068890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Serivichyaswat P., Ryu H.S., Kim W., Kim S., Chung K.S., Kim J.J. et al. (2015) Expression of the floral repressor miRNA156 is positively regulated by the AGAMOUS-like proteins AGL15 and AGL18. Mol. Cells 38, 259–266 10.14348/molcells.2015.2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Karlova R., Boeren S., Russinova E., Aker J., Vervoort J. and de Vries S. (2006) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18, 626–638 10.1105/tpc.105.039412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Patharkar O.R., Macken T.A. and Walker J.C. (2016) Serine 231 and 257 of AGAMOUS-Like 15 are phosphorylated in floral receptacles. Plant Signal. Behav. 11, article no. UNSP e1199314 10.1080/15592324.2016.1199314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Patharkar O.R. and Walker J.C. (2015) Floral organ abscission is regulated by a positive feedback loop. Proc. Natl. Acad. Sci. U.S.A. 112, 2906–2911 10.1073/pnas.1423595112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Boutilier K., Offringa R., Sharma V.K., Kieft H., Ouellet T., Zhang L.M. et al. (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14, 1737–1749 10.1105/tpc.001941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tsuwamoto R., Yokoi S. and Takahata Y. (2010) Arabidopsis EMBRYOMAKER encoding an AP2 domain transcription factor plays a key role in developmental change from vegetative to embryonic phase. Plant Mol. Biol. 73, 481–492 10.1007/s11103-010-9634-3 [DOI] [PubMed] [Google Scholar]
- 208.Ikeuchi M., Iwase A., Rymen B., Harashima H., Shibata M., Ohnuma M. et al. (2015) PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis. Nat. Plants 1, 1–7 10.1038/nplants.2015.89 [DOI] [PubMed] [Google Scholar]
- 209.Horstman A., Li M.F., Heidmann I., Weemen M., Chen B.J., Muino J.M. et al. (2017) The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 175, 848–857 10.1104/pp.17.00232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zuo J.R., Niu Q.W., Frugis G. and Chua N.H. (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 30, 349–359 10.1046/j.1365-313X.2002.01289.x [DOI] [PubMed] [Google Scholar]
- 211.Gallois J.L., Nora F.R., Mizukami Y. and Sablowski R. (2004) WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 18, 375–380 10.1101/gad.291204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Jha P., Ochatt S.J. and Kumar V. (2020) WUSCHEL: a master regulator in plant growth signaling. Plant Cell Rep. 39, 431–444 10.1007/s00299-020-02511-5 [DOI] [PubMed] [Google Scholar]
- 213.Wang X.C., Niu Q.W., Teng C., Li C., Mu J.Y., Chua N.H. et al. (2009) Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in Arabidopsis. Cell Res. 19, 224–235 10.1038/cr.2008.276 [DOI] [PubMed] [Google Scholar]
- 214.Koszegi D., Johnston A.J., Rutten T., Czihal A., Altschmied L., Kumlehn J. et al. (2011) Members of the RKD transcription factor family induce an egg cell-like gene expression program. Plant J. 67, 280–291 10.1111/j.1365-313X.2011.04592.x [DOI] [PubMed] [Google Scholar]
- 215.Ogas J., Cheng J.-C., Sung Z.R. and Somerville C. (1997) Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277, 91–94 10.1126/science.277.5322.91 [DOI] [PubMed] [Google Scholar]
- 216.Ogas J., Kaufmann S., Henderson J. and Somerville C. (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 96, 13839–13844 10.1073/pnas.96.24.13839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Suzuki M. and McCarty D.R. (2008) Functional symmetry of the B3 network controlling seed development. Curr. Opin. Plant Biol. 11, 548–553 10.1016/j.pbi.2008.06.015 [DOI] [PubMed] [Google Scholar]
- 218.Suzuki M., Wang H.H.Y. and McCarty D.R. (2007) Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 143, 902–911 10.1104/pp.106.092320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chanvivattana Y., Bishopp A., Schubert D., Stock C., Moon Y.H., Sung Z.R. et al. (2004) Interaction of polycomb-group proteins controlling flowering in Arabidopsis. Development 131, 5263–5276 10.1242/dev.01400 [DOI] [PubMed] [Google Scholar]
- 220.Tanaka M., Kikuchi A. and Kamada H. (2008) The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146, 149–161 10.1104/pp.107.111674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bratzel F., Lopez-Torrejon G., Koch M., Del Pozo J.C. and Calonje M. (2010) Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 20, 1853–1859 10.1016/j.cub.2010.09.046 [DOI] [PubMed] [Google Scholar]
- 222.Chen D.H., Molitor A., Liu C.L. and Shen W.H. (2010) The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 20, 1332–1344 10.1038/cr.2010.151 [DOI] [PubMed] [Google Scholar]
- 223.Rider S.D., Henderson J.T., Jerome R.E., Edenberg H.J., Romero-Severson J. and Ogas J. (2003) Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 35, 33–43 10.1046/j.1365-313X.2003.01783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Chen N.C., Veerappan V., Abdelmageed H., Kang M. and Allen R.D. (2018) HSI2/VAL1 silences AGL15 to regulate the developmental transition from seed maturation to vegetative growth in Arabidopsis. Plant Cell 30, 600–619 10.1105/tpc.17.00655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Chhun T., Chong S.Y., Park B.S., Wong E.C.C., Yin J.L., Kim M. et al. (2016) HSI2 repressor recruits MED13 and HDA6 to down-regulate seed maturation gene expression directly during Arabidopsis early seedling growth. Plant Cell Physiol. 57, 1689–1706 10.1093/pcp/pcw095 [DOI] [PubMed] [Google Scholar]
- 226.Zhou Y., Tan B., Luo M., Li Y., Liu C., Chen C. et al. (2013) HISTONE DEACETYLASE19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell 25, 134–148 10.1105/tpc.112.096313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Chen N.C., Wang H., Abdelmageed H., Veerappan V., Tadege M. and Allen R.D. (2020) HSI2/VAL1 and HSL1/VAL2 function redundantly to repress DOG1 expression in Arabidopsis seeds and seedlings. New Phytol. 227, 840–856 10.1111/nph.16559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Shu J., Chen C., Thapa R.K., Bian S.M., Nguyen V., Yu K.F. et al. (2019) Genome-wide occupancy of histone H3K27 methyltransferases CURLY LEAF and SWINGER in Arabidopsis seedlings. Plant Direct. 3, article no. Unsp 100 10.1002/pld3.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Yamamoto A., Kagaya Y., Usui H., Hobo T., Takeda S. and Hattori T. (2010) Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell Physiol. 51, 2031–2046 10.1093/pcp/pcq162 [DOI] [PubMed] [Google Scholar]
- 230.Mönke G., Seifert M., Keilwagen J., Mohr M., Grosse I., Hahnel U. et al. (2012) Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res. 40, 8240–8254 10.1093/nar/gks594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Tian R., Wang F., Zheng Q., Niza V., Downie A.B. and Perry S.E. (2020) Direct and indirect targets of the Arabidopsis seed transcription factor ABSCISIC ACID INSENSITIVE3. Plant J. 103, 1679–1694 10.1111/tpj.14854 [DOI] [PubMed] [Google Scholar]
- 232.Wang F.F. and Perry S.E. (2013) Identification of direct targets of FUSCA3, a key regulator of Arabidopsis seed development. Plant Physiol. 161, 1251–1264 10.1104/pp.112.212282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Busch W., Miotk A., Ariel F.D., Zhao Z., Forner J., Daum G. et al. (2010) Transcriptional control of a plant stem cell niche. Dev. Cell 18, 849–861 10.1016/j.devcel.2010.03.012 [DOI] [PubMed] [Google Scholar]
- 234.Wang F.-X., Shang G.-D., Wu L.-Y., Xu Z.-G., Zhao X.-Y. and Wang J.-W. (2020) Chromatin accessibility dynamics and a hierarchical transcriptional regulatory network structure for plant somatic embryogenesis. Dev. Cell 54, 742–757 10.1016/j.devcel.2020.07.003 [DOI] [PubMed] [Google Scholar]
- 235.Baud S., Kelemen Z., Thevenin J., Boulard C., Blanchet S., To A. et al. (2016) Deciphering the molecular mechanisms underpinning the transcriptional control of gene expression by master transcriptional regulators in Arabidopsis seed. Plant Physiol. 171, 1099–1112 10.1104/pp.16.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Horstman A., Fukuoka H., Muino J.M., Nitsch L., Guo C.H., Passarinho P. et al. (2015) AIL and HDG proteins act antagonistically to control cell proliferation. Development 142, 454–464 10.1242/dev.117168 [DOI] [PubMed] [Google Scholar]
- 237.Passarinho P., Ketelaar T., Xing M.Q., van Arkel J., Maliepaard C., Hendriks M.W. et al. (2008) BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol. Biol. 68, 225–237 10.1007/s11103-008-9364-y [DOI] [PubMed] [Google Scholar]
- 238.Zhang Y., Cao G., Qu L.-J. and Gu H.J.P.C.R. (2009) Involvement of an R2R3-MYB transcription factor gene AtMYB118 in embryogenesis in Arabidopsis 28, 337–346 10.1007/s00299-008-0644-4 [DOI] [PubMed] [Google Scholar]
- 239.Szczygiel-Sommer A. and Gaj M.D. (2019) The miR396-GRF regulatory module controls the embryogenic response in Arabidopsis via an auxin-related pathway. Int. J. Mol. Sci. 20, article no. 5221 10.3390/ijms20205221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Szyrajew K., Bielewicz D., Dolata J., Wojcik A.M., Nowak K., Szczygiel-Sommer A. et al. (2017) MicroRNAs are intensively regulated during induction of somatic embryogenesis in Arabidopsis. Front. Plant Sci. 8, article no. 18 10.3389/fpls.2017.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Jin F.Y., Hu L.S., Yuan D.J., Xu J., Gao W.H., He L.R. et al. (2014) Comparative transcriptome analysis between somatic embryos (SEs) and zygotic embryos in cotton: evidence for stress response functions in SE development. Plant Biotechnol. J. 12, 161–173 10.1111/pbi.12123 [DOI] [PubMed] [Google Scholar]
- 242.Hofmann F., Schon M.A. and Nodine M.D. (2019) The embryonic transcriptome of Arabidopsis thaliana. Plant Reprod. 32, 77–91 10.1007/s00497-018-00357-2 [DOI] [PubMed] [Google Scholar]
- 243.Kadokura S., Sugimoto K., Tarr P., Suzuki T. and Matsunaga S. (2018) Characterization of somatic embryogenesis initiated from the Arabidopsis shoot apex. Dev. Biol. 442, 13–27 10.1016/j.ydbio.2018.04.023 [DOI] [PubMed] [Google Scholar]
- 244.Wickramasuriya A.M. and Dunwell J.M. (2015) Global scale transcriptome analysis of Arabidopsis embryogenesis in vitro. BMC Genom. 16, article no. 301 10.1186/s12864-015-1504-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Zhou X., Liu Z., Shen K., Zhao P. and Sun M.-X. (2020) Cell lineage-specific transcriptome analysis for interpreting cell fate specification of proembryos. Nat. Commun. 11, 1366 10.1038/s41467-020-15189-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Roscoe T.T., Guilleminot J., Bessoule J.J., Berger F. and Devic M. (2015) Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in Arabidopsis. Plant Cell Physiol. 56, 1215–1228 10.1093/pcp/pcv049 [DOI] [PubMed] [Google Scholar]
- 247.Heyndrickx K.S., Van de Velde J., Wang C.M., Weigei D. and Vandepoele K. (2014) A functional and evolutionary perspective on transcription factor binding in Arabidopsis thaliana. Plant Cell 26, 3894–3910 10.1105/tpc.114.130591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Feher A. (2019) Callus, dedifferentiation, totipotency, somatic embryogenesis: what these terms mean in the era of molecular plant biology? Front. Plant Sci. 10, 536 10.3389/fpls.2019.00536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Perry S.E., Zheng Q.L. and Zheng Y.M. (2016) Transcriptome analysis indicates that GmAGAMOUS-Like 15 may enhance somatic embryogenesis by promoting a dedifferentiated state. Plant Signal. Behav. 11, article no. UNSP e1197463 10.1080/15592324.2016.1197463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Grafi G., Chalifa-Caspi V., Nagar T., Plaschkes I., Barak S. and Ransbotyn V. (2011) Plant response to stress meets dedifferentiation. Planta 233, 433–438 10.1007/s00425-011-1366-3 [DOI] [PubMed] [Google Scholar]
- 251.Ledwon A. and Gaj M.D. (2009) LEAFY COTYLEDON2 gene expression and auxin treatment in relation to embryogenic capacity of Arabidopsis somatic cells. Plant Cell Rep. 28, 1677–1688 10.1007/s00299-009-0767-2 [DOI] [PubMed] [Google Scholar]
- 252.Ledwon A. and Gaj M.D. (2011) LEAFY COTYLEDON1, FUSCA3 expression and auxin treatment in relation to somatic embryogenesis induction in Arabidopsis. Plant Growth Regul. 65, 157–167 10.1007/s10725-011-9585-y [DOI] [Google Scholar]
- 253.Zhu C. and Perry S.E. (2005) Control of expression and autoregulation of AGL15, a member of the MADS-box family. Plant J. 41, 583–594 10.1111/j.1365-313X.2004.02320.x [DOI] [PubMed] [Google Scholar]
- 254.Zheng Q.L., Zheng Y.M., Ji H.H., Burnie W. and Perry S.E. (2016) Gene regulation by the AGL15 transcription factor reveals hormone interactions in somatic embryogenesis. Plant Physiol. 172, 2374–2387 10.1104/pp.16.00564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Palovaara J., Saiga S., Wendrich J.R., Hofland N.V., van Schayck J.P., Hater F. et al. (2017) Transcriptome dynamics revealed by a gene expression atlas of the early Arabidopsis embryo. Nat. Plants 3, 894–904 10.1038/s41477-017-0035-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Kimata Y. and Ueda M. (2020) Intracellular dynamics and transcriptional regulations in plant zygotes: a case study of Arabidopsis. Plant Reprod. 33, 89–96 10.1007/s00497-020-00389-7 [DOI] [PubMed] [Google Scholar]
- 257.Zhou X.M., Shi C., Zhao P. and Sun M.X. (2019) Isolation of living apical and basal cell lineages of early proembryos for transcriptome analysis. Plant Reprod. 32, 105–111 10.1007/s00497-018-00353-6 [DOI] [PubMed] [Google Scholar]
- 258.Sakai K., Taconnat L., Borrega N., Yansouni J., Brunaud V., Paysant-Le Roux C. et al. (2018) Combining laser-assisted microdissection (LAM) and RNA-seq allows to perform a comprehensive transcriptomic analysis of epidermal cells of Arabidopsis embryo. Plant Methods 14, article number 10 10.1186/s13007-018-0275-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Becker M.G., Hsu S.W., Harada J.J. and Belmonte M.F. (2014) Genomic dissection of the seed. Front. Plant Sci. 5, article no. 464 10.3389/fpls.2014.00464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Guo F.D., Liu C.L., Xia H., Bi Y.P., Zhao C.Z., Zhao S.Z. et al. (2013) Induced expression of AtLEC1 and AtLEC2 differentially promotes somatic embryogenesis in transgenic tobacco plants. PLoS One 8, article no. e71714 10.1371/journal.pone.0071714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Jo L., Pelletier J.M., Hsu S.W., Baden R., Goldberg R.B. and Harada J.J. (2020) Combinatorial interactions of the LEC1 transcription factor specify diverse developmental programs during soybean seed development. Proc. Natl. Acad. Sci. U.S.A. 117, 1223–1232 10.1073/pnas.1918441117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Liu Z., Ge X.X., Qiu W.M., Long J.M., Jia H.H., Yang W. et al. (2018) Overexpression of the CsFUS3 gene encoding a B3 transcription factor promotes somatic embryogenesis in citrus. Plant Sci. 277, 121–131 10.1016/j.plantsci.2018.10.015 [DOI] [PubMed] [Google Scholar]
- 263.Yang Z.R., Li C.F., Wang Y., Zhang C.J., Wu Z.X., Zhang X.Y. et al. (2014) GhAGL15s, preferentially expressed during somatic embryogenesis, promote embryogenic callus formation in cotton (Gossypium hirsutum L.). Mol. Genet. Genom. 289, 873–883 10.1007/s00438-014-0856-y [DOI] [PubMed] [Google Scholar]
- 264.Zheng Q.L. and Perry S.E. (2014) Alterations in the transcriptome of soybean in response to enhanced somatic embryogenesis promoted by orthologs of AGAMOUS-Like15 and AGAMOUS-Like18. Plant Physiol. 164, 1365–1377 10.1104/pp.113.234062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kulkarni S.R. and Vandepoele K. (2020) Inference of plant gene regulatory networks using data-driven methods: a practical overview. Biochim. Biophys. Acta Gene Regul. Mech. 1863, article no. 194447 10.1016/j.bbagrm.2019.194447 [DOI] [PubMed] [Google Scholar]
- 266.Goldberg R.B., De Paiva G. and Yadegari R. (1994) Plant embryogenesis: zygote to seed. Science 266, 605–614 10.1126/science.266.5185.605 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.