Abstract
The novel severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) is the cause of an ongoing pandemic with significant case fatality ratio (CFR) worldwide. Although SARS-CoV-2 primarily causes respiratory infection by binding to ACE2 receptors present on alveolar epithelial cells, studies have been published linking the disease to the small intestine enterocytes and its microbiome. Dysbiosis of microbiome, mainly intestinal and lung, can affect the course of the disease. Environmental factors, such as reduced intake of commensal bacteria from the environment or their products in the diet, play an important role in microbiome formation, which can significantly affect the immune response. In elderly, obese or chronically ill people, the microbiota is often damaged. Therefore, we speculate that a good microbiome may be one of the factors responsible for lower CFR from the coronavirus disease 2019 (COVID-19). An approach using tailored nutrition and supplements known to improve the intestinal microbiota and its immune function might help minimize the impact of the disease at least on people at higher risk from coronavirus.
Introduction to coronavirus biology
The new SARS-CoV-2 virus (severe acute respiratory syndrome Coronavirus 2) belongs to the subfamily Coronavirinae (order Nidovirales, family Coronaviridae), which includes a diverse group of similar viruses of medical and veterinary importance. Coronaviruses (CoVs) are enveloped, positive-sense RNA viruses that contain the largest known RNA genomes with a length of up to 32 kb. This subfamily is divided into four genera: alpha-, beta-, gamma- and delta-coronavirus (α-, β-, γ- and δ-CoV). The first two families include species capable of causing disease in humans. The α-coronavirus class includes two viruses, HCoV-NL63 and HCoV-229E, while the β-coronavirus class includes two other viruses, HCoV-OC43 and HCoV-HKU1. These four common HCoV viruses generally cause mild upper respiratory tract disease and contribute to 15–30% of colds in adults. Severe and life-threatening lower respiratory tract infections such as pneumonia and / or bronchitis caused by α-coronavirus HCoV-NL-63 and β-coronavirus HCoV-HKU1 can sometimes occur in infants, elderly patients or immunocompromised patients. However, the β-coronaviruses group also includes species causing severe disease more often (SARS-CoV, SARS-CoV-2 and MERS-CoV) (Fig. 1). The tropism of different coronaviruses varies between the respiratory tract and the gastrointestinal tract and has a remarkable potential for switch (Hulswit et al. 2016; Walls et al. 2020).
Fig. 1.
Overview of currently known spike protein (S-protein) binding domains for diverse human coronaviruses. Schematic representation of coronavirus spike proteins drawn to scale. Orange boxes indicate signal peptides. Blue boxes indicate the N-terminal regions in α- and β-coronavirus spike proteins, which were mapped based on sequence homology between viruses within the same genus. Green boxes indicate known receptor-binding domains (RBD) in the C-terminal region of S1. Known receptors are indicated in the boxes: APN, aminopeptidase N; ACE2, angiotensin-converting enzyme 2; O-ac Sia, O-acetylated sialic acid; DPP4, dipeptidyl peptidase-4. Red boxes indicate transmembrane domains. Spike proteins are shown for Human CoV NL63 (GB: YP_003767.1), Human CoV 229E strain inf-1 (GB:NP_073551.1), HCoV-OC43 strain Paris (GB: AAT84362), HCoV-HKU1 (GB: AAT98580), SARS-CoV strain Urbani (GB: AAP13441), MERS-CoV strain EMC/2012 (GB: YP_009047204), SARS-CoV-2 strain (GB: AAP13441)
The coronavirus particle comprises at least four canonical structural proteins: E (envelope protein), M (membrane protein), N (nucleocapsid protein) and S (spike protein). Viruses belonging to the β-coronavirus lineage also promote expression of membrane-bound HE (hemagglutinin esterase) protein. The key protein responsible for viral entry into the host cell is glycoprotein S (known as the spike protein). Glycoprotein S has two subunits, the N-terminal subunit of S1 which has a receptor binding domain (RBD) and the C-terminal subunit of S2 which participates in membrane fusion (Hulswit et al. 2016). The SARS-CoV-2 glycoprotein S trimer exists in at least two conformational states, resulting from the opening of the S1 glycoprotein subunit at the top of the trimer. In contrast, only closed glycoprotein S trimers were detected in the four above mentioned human-infecting coronaviruses (HCoV-NL63, HCoV-OC43, HCoV-HKU1 and HCoV-229E).
After the attachment of the virus to angiotensin-converting enzyme 2 (ACE2), the cell surface receptor, the C-terminal subunit is released. ACE2 is present in the lung tissue but also in esophageal and intestinal epithelium (Li et al. 2020). Cellular proteases, such as furin-like enzymes or transmembrane protease serine 2 (TMPRSS2) are responsible for the cleavage of S protein in polybasic motif, also known as “priming” (Braun and Sauter 2019; Coutard et al. 2020; Matsuyama et al. 2020). The strong polybasic motif is exactly the difference factor between SARS-CoV (Leu-Arg) and SARS-CoV-2 (Pro-Arg-Arg-Ala-Arg), allowing the efficient release of the S2 subunit of the glycoprotein (Fig. 2). Conformation changes are crucial during fusion. Some coronaviruses deliver their genomes to the cytosol after their envelopes fuse with the plasma membrane at the cell surface, whereas SARS-CoV-2 take advantage of the cell’s endocytic machinery (Fig. 3) (Valle et al. 2020). The endocytosed virions are subjected to an activation step in the endosome, which is typically mediated by the acidic endosomal pH, resulting in fusion of the viral and endosomal membranes and release of the viral genome into the cytosol. After entering the cell, the viral genome is released into the cell’s cytoplasm. In the infected cell, the viral RNA is translated by the host’s replicative apparatus, which generate new viral RNA genomes. Two polyprotein precursors, pp1a and pp1ab, are translated from viral mRNA and then are cleaved by two viral proteases, chymotrypsin-like and papain-like, to generate the 16nsps (nsp1 to 16). These proteins play a role in the formation of the replication and transcription complex. The last step is the formation of new virions through the association of N- protein with the neo-synthesized viral genome. The virions are released by exocytosis (de Wit et al. 2016; Romano et al. 2020; Valle et al. 2020).
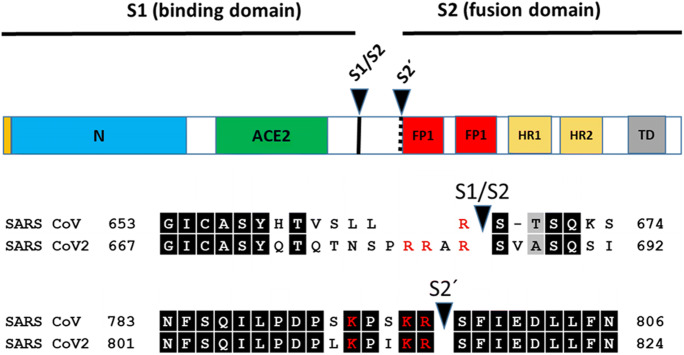
Fig. 2.
Schematic illustration of the spike protein of SARS-CoV and SARS-CoV-2 including functional domains (N-terminal region; ACE2 - receptor binding domain for angiotensin converting enzyme 2; FP1 and FP2 (two domains of fusion peptide); HR1 and HR2 (two heptad repeat regions); TD (transmembrane domain) and proteolytic cleavage sites (S1/S2, S2´). Amino acid sequences around the two protease recognition sites (red) are indicated for SARS-S and SARS-2-S (amino acid residues in black boxes indicate conserved residues; letters in red colour are positively charged amino acids in cleavage sites). Arrow heads indicate the cleavage sites
Fig. 3.
Scheme of SARS-CoV-2 penetration into recipient cells. ACE 2 is the host cell receptor responsible for mediating infection by SARS-CoV-2, the novel coronavirus responsible for coronavirus disease 2019 (COVID-19). Trimeric spike protein on the virion binds to ACE2 (angiotensin converting enzyme 2), a cell-surface protein. Furin-like serine proteases (TMPRSS2, PACE, PCSK3) are responsible for the cleavage of the S-glycoprotein in the polybasic motif and the subsequent on allowing the fusion protein (FP1 and FP2) to bind to the membrane. It helps the virion enter the cells. SARS-CoV-2 has spike protein trimers spontaneously transitioning from the closed to the open conformation
Human coronaviruses are predominantly concomitant with upper respiratory tract illnesses ranging from mild to moderate symptoms including common cold. During their lifetime, most of people may be infected with one or more of the aforementioned seven human coronaviruses (Killerby et al. 2018). If SARS-CoV-2 infection is symptomatic, symptoms range from mild to severe illness, or even to disease-related death. According to most authors, symptoms may develop in two days to two weeks following exposure to the virus. The Wuhan study found that the mean incubation period is 5.1 days with 97.5% of individuals developing symptoms within 11.5 days of infection (Lauer et al. 2020). Common symptoms of the coronavirus disease 2019 (COVID-19) include fever, muscle pain, fatigue, headache, cough or shortness of breath, loss of taste or smell, less often any gastrointestinal symptoms. Frequency of GI symptoms varied from 3.0% to 39.6%, including diarrhea: 7.5%, nausea: 4.5%, anorexia: 4.4%, vomiting: 1.3%, abdominal pain: 0.5%, belching/reflux: 0.3% (Schmulson et al. 2020) One fifth of hospitalized patients develop significant cardiovascular morbidity, characterized by troponin rise, tachyarrhythmias and thromboembolic events, which is strongly associated with mortality risk (Shi et al. 2020; Guo et al. 2020). There are also reports of acute renal and liver function impairment during SARS-CoV-2 infection. The exact mechanism is unclear, but direct viral toxicity as well as immune response consequence are possibly involved. Some of the infected individuals may have some kind of skin manifestation before the other symptoms of the viral infection begin, during the acute phase or even after the patient has already recovered. In the advanced stage of the disease, severe pneumonia and acute respiratory distress syndrome (ARDS) can develop in some patients, which is the leading cause of mortality. The elderly and obese people and those with underlying cardiovascular disease, diabetes mellitus, chronic pulmonary disorders or renal disease especially are at risk (Zhou et al. 2020). Common features of COVID-19 patients requiring hospitalization and intensive care support are severe pneumonia with hypoxic respiratory failure of subacute onset evolving into ARDS, with a clinical picture characterized by fever, lymphopenia, highly elevated C-reactive protein, proinflammatory cytokines, serum ferritin, and D-dimers. Histopathological evidence of a prominent pulmonary infiltrate dominated by monocytes and macrophages, vasculitis, and hypercoagulability is seen (Felsenstein et al. 2020). In addition, lung fibrosis can follow. Accumulating evidence suggests that a subgroup of patients with severe COVID-19 might have a hyperinflammatory syndrome characterized by a fulminant and fatal hypercytokinaemia with multiorgan failure. A cytokine profile resembling secondary haemophagocytic lymphohistiocytosis (sHLH) is associated with COVID-19 disease severity, characterized by increased interleukin (IL)-2, IL-7, granulocyte-colony stimulating factor, interferon-γ-inducible protein 10, monocyte chemoattractant protein 1, macrophage inflammatory protein 1-α, and tumour necrosis factor-α. Predictors of fatality included elevated ferritin and IL-6 suggesting that mortality might be due to virally driven hyperinflammation (Mehta et al. 2020).
It has been reported that the SARS-CoV and SARS-CoV-2 have similar receptors, especially the receptor-binding domain (RBD) and the receptor-binding motif (RBM) in the viral genome (Yin and Wunderink 2018; Tai et al. 2020). During SARS-CoV infection, the RBM of the S protein binds to the angiotensin-converting enzyme 2 (ACE2) receptor present in the respiratory epithelium and alveoli of the lungs, in kidneys and in the intestinal enterocytes (Phan 2020). Because of the similarity of nucleotide sequences in the RBD binding motifs of both viruses, SARS-CoV-2 may use the same mechanism as well (Lu et al. 2020). SARS-CoV-2 binds ACE2 with much higher avidity than SARS-CoV, which could be the explanation for the increased transmission rate and its capacity to affect other species easily (Vellingiri et al. 2020). Once the virus enters the cell, ACE2 gets cleaved and shed by the furin protease ADAM17 into the extra membrane space. Reduced ACE2 has been known to be concomitant with alveoli injury and increases pulmonary vascular permeability.
Concurrently, the inflammasome pathway is activated through various means by ORF3a, ORF8b, E proteins, and the NF-kB (Nuclear Factor kappa-light-chain-enhancer of activated B-cells), leading to the expression of cytokines. This results in a cytokine storm, further leading to respiratory distress (Vellingiri et al. 2020). It is not yet clear how SARS-CoV-2 affects the immune system. Most theories come from the observations of SARS-CoV pathogenesis. The CD8+ T cells account for about 80% of total inflammatory cells in the pulmonary interstitium in SARS-CoV infected patients and play a critical role in clearing coronaviruses from the infected cells and inducing immune injury (Maloir et al. 2018). The CD4 + T cells accelerate the production of viral-specific antibodies by activating T cell-dependent B cells. In addition, T helper cells produce proinflammatory cytokines via NF-kB signalling. Proinflammatory cytokines recruit monocytes and neutrophils to the infection site (Vellingiri et al. 2020). It is noteworthy that even though the intestinal epithelia produce much higher levels of ACE2 than bronchial epithelia, not all patients develop gastrointestinal symptoms and when they do, symptoms tend to be mild, and some patients remain SARS-CoV-2 positive in stool samples long after respiratory specimen became negative (Xiao et al. 2020; Felsenstein et al. 2020).
Diagnostics and therapy
The real-time reverse transcription–polymerase chain reaction (rRT-PCR) assay is recommended to be used to diagnose the virus in upper respiratory tract specimens. If tests are negative using upper respiratory tract specimens despite persistent clinical suspicion, the WHO recommends retesting using lower respiratory tract specimens, which contain higher viral loads (World Health Organization WHO 2020). Serological tests can help examine the ongoing outbreak and retrospectively assess the extent of the attack or the extent of the outbreak. There are several chest computed tomography (CT) findings in COVID-19 patients, but not specific enough to discriminate the disease from other severe pulmonary diseases. The distribution of pulmonary lesions on CT in COVID-19 pneumonia patients is mostly peripheral at all stages. With the development of the disease, the lesions gradually spread from the periphery to the centre. In most patients, the lesions are present bilaterally, in multiple lobes (Adair II and Ledermann 2020; Ding et al. 2020). The chest CT may be useful if applied in the appropriate clinical context.
Patients with mild COVID-19 without complications are treated symptomatically with antipyretics and, if needed, with supplemental oxygen. With the progression of the disease, fluid replacement therapy may be required and antimicrobial therapy of possible bacterial co-infection should be considered, while in severe hypoxemic respiratory failure cases, the advanced oxygen/ventilatory support either non-invasive or via endotracheal intubation is necessary. Further disease progression to the multiple organ failure may require other advanced organ replacement treatment approaches (World Health Organization 2020a).
At the moment, the only specific antiviral treatment has been authorised in the European Union. RNA polymerase inhibitor remdesivir has been given conditional authorisation for the treatment of COVID-19 in adults and adolescents from 12 years of age with pneumonia who require supplemental oxygen (European Medicines Agency 2020). Though remdesivir is nowadays widely used as a part of the treatment protocols, effectiveness and safety is to be assessed. There are multiple other antivirals under investigation. Because viral replication may be particularly active early in the course of the disease, antiviral therapy may have the greatest impact before the illness progresses into the hyperinflammatory state that can characterize the later stages of disease. In severe and critical COVID-19 cases, defined as oxygen saturation < 90% on room air, signs of severe respiratory distress, or meeting the criteria of ARDS, sepsis or septic shock, systemic corticosteroids are recommended for the treatment (World Health Organization 2020b). In such severe cases, the host immune response may drive the pathophysiology of disease, so dexamethasone, as a corticosteroid with strong anti-inflammatory and immunosuppressant effect, is commonly used as a part of the treatment.
Multiple agents that modulate the immune response are being under investigation in clinical trials as adjunctive treatments for the management of moderate to critical COVID-19. These agents include human blood-derived products (e.g. convalescent plasma obtained from individuals who have recovered from SARS-CoV-2 infection) and a broad spectrum of immunomodulatory therapies. Remdesivir, and dexamethasone in advanced stages of the disease, are commonly used as treatment of severe COVID-19 in the Czech Republic nowadays, together with convalescent plasma at some hospitals.
Microbiome as a barrier to virus infection
The human body has many levels of structural organization: atoms, cells, tissues, organs, and organ systems. With some exaggeration, the microbiome was deciphered as the last organ of the human body (Baquero and Nombela 2012). The human microbiome is the community of trillions of microbes – tiny fauna, fungi, viruses and other living entities – that inhabit every part of the body. For many millions of years, the community of microorganisms has evolved along with the human species. Due to this coevolution of bacteria - eukaryotic cells, genomic functional complementarity with genetic reduction could occur. Another view speaks of an association of different cells that coexist and work together to form a living consortium, the holobiont (Zilber-Rosenberg and Rosenberg 2008). In the relationships between members of the holobiont, it is necessary for the proper homeostasis of organisms to establish complex interconnections between all cells. In order for such a complex relationship to work, it is necessary to create good communication with each other. Cells and microbes interact both within and between species to survive in environments that are based on fluctuating patterns of competition and cooperation. To maintain these balances, cells transmit and receive chemical signals that modulate gene expression and cellular functions, transmit biomolecules and metabolites that improve the persistence and viability of the community, and also activate defence strategies to control species competing for the same valuable resources (Caruana and Walper 2020). Among the various types of molecules that can be used to transmit information, RNA plays a key role for biological systems, which is the link between the genome and the proteome. Eukaryotic and prokaryotic cells can transmit signals via non-coding RNAs using specific extracellular conveyors that travel to the target cell and can be converted to a regulatory response by a specialized molecular apparatus (Leitão et al. 2020). RNAs transcribed from ultraconserved region (T-UCRs) are highly evolutionarily conserved in many mammalian genomes and have been found to be important in the pathogenesis of many diseases (Wang et al. 2018). Bacterial non-coding RNAs regulate host gene expression and conversely eukaryotic miRNAs may regulate bacterial gene expression (Duval et al. 2016). It has been reported that SARS-CoV-2 replication in a human host is limited by host miRNA defence (Hosseini Rad Sm and AD 2020). Long non-coding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degradation of miRNA195 (Xiao et al. 2018) and has influence on homeostasis of the intestinal epithelial barrier (Wang et al. 2018). Epitranscriptomic modification is another level of interaction between commensal bacteria and their host. N6-methyladenosine (m6A) modification correlate with variation in the gut microbiota (Jabs et al. 2020). Communication between bacteria - eukaryotic cells is on many levels, tight and very complex. In order to completely interrupt the communication between pathogenic bacteria and the host in the pathophysiological process, it is sometimes necessary to make a drastic change. It is therefore not surprising that one of the most successful therapies that has been introduced recently is fecal transplantation (Aas et al. 2003; Polák et al. 2015). As has been shown, probiotic-based intervention can modulate the composition of the microbiome so that it can be protected from viral infection (Yitbarek et al. 2018). The microbiome, the last organ of the human body discovered, seems to be a prominent factor why the viral infection usually has little effect on its health and why sometimes life is at stake.
As noted above, approximately 81% of individuals with COVID-19 suffer from mild symptoms of the disease (Weiss and Murdoch 2020) or no symptoms at all (Azkur et al. 2020). Due to the fact that infectious dose data is missing up now, we do not have accurate information on whether a high viral load corresponds to a more severe disease (Liu et al. 2020; He et al. 2020a). A study on influenza virus showed that the higher is the dose of influenza virus given to healthy volunteers, the worse are their symptoms (Memoli et al. 2015). However, the viral load alone is not a clear predictor of disease outcome. Other studies have found that some asymptomatic patients had similar viral loads as patients with symptoms of COVID-19 (Lee et al. 2020). Whether a person develops mild symptoms or pneumonia depends not only on how much virus is in the lungs, but also on the immune response and overall health (To et al. 2020). Sekine et al. came up with evidence that T cell responses play an important role in patients with asymptomatic or very mild forms of the disease. These patients often inadvertently act as major carriers of the disease. They showed that many activated/cycling T cells in the acute phase were functionally replete and specific for SARS-CoV-2 in the absence of detectable circulating antibodies (Sekine et al. 2020). The resulting image is thus further complicated. Why, in many cases, a T cell response is sufficient to stop the infection, and sometimes a complete immune response is not enough? Leaving aside transient factors of indisposition, such as exhaustion, extreme stress or colds, we can say that good human fitness influences the immune system. An important player that significantly affects the immune system is the composition of the microbiota. The microbiota plays a fundamental role in the induction, training and function of the host immune system (Belkaid and Hand 2014). Chronic diseases that are caused by poor condition of people, such as diabetes, heart disease, stroke, respiratory diseases, but also old age or obesity, are associated with a change in the composition of microbiota (Hand et al. 2016). The causal link between the disease and the composition of the microbiota remains an issue - whether a change in the microbiota allows the disease to develop or the disease causes a change in the microbiota. Maybe the relationship is two-way. Numerous byproducts of host inflammation are growth factors for pathogenic bacteria (Huffnagle et al. 2017). For a pathogen to succeed, it must exchange its hostile environment for a favourable environment. At some point, the forced change of microbiota (viral and bacterial infections, antibiotics) will inevitably allow the pathogen to realize itself. If this logic applies, a forced positive change (dietary change, use of probiotics, faecal transplantation) can reverse this unfavourable condition.
The human body is inhabited by a diverse microbial community, which is interconnected and forms a commensal microbiota. Interestingly, organs such as the lungs, stomach, esophagus and intestine, which are most populated by the microbiota, are embryologically derived from the endoderm (Wang et al. 2020). It is therefore not surprising that these developmentally homologous organs are connected together in the intestinal-lung axis (Fig. 4). Changes of microbiota in the gut and also in the lungs are connected to the immune response alterations and disease development (Belkaid and Hand 2014). For example, chronic obstructive pulmonary disease (COPD) typically occurs together with chronic diseases of the gastrointestinal tract. Another example is a higher risk of allergic airway disease connected to the usage of antibiotics and alteration in gut microbiota composition (Budden et al. 2017). Several studies proved the connection of orally administered bacteria and lung function. As an example, there is a correlation between the presence of probiotic bacteria Bifidobacterium longum in the gut and decreased incidence of asthma (Budden et al. 2017). Also, some environmental factors, such as dietary fibres, lead to similar changes both in the gut and in the lung microbiota. SCFAs (short-chain fatty acids) are not beneficial just for enterocytes, but they are also able to mitigate allergic respiratory diseases. The host receptor for SCFAs is FFAR2 (free fatty acids receptor 2), which is coupled with a G protein and is expressed on immune cells and connected to colitis, arthritis and asthma in the animal model. This receptor provides a connection between SCFAs and their beneficial effect on health (Budden et al. 2017).
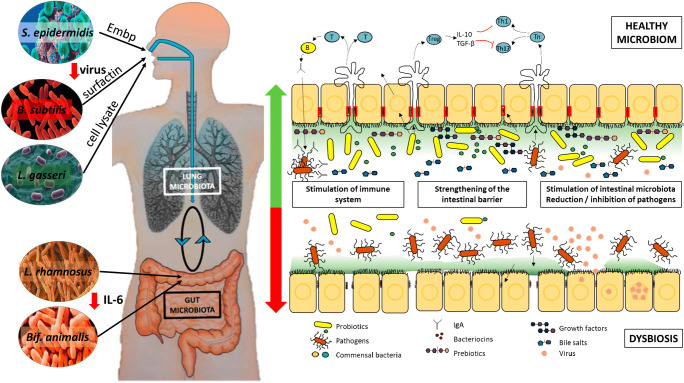
Fig. 4.
Principles of functioning of the lung - intestine axis in people with virus-infected people with a healthy microbiome and in dysbiosis. DCs promote the activation of various T cell subsets in the mesenteric lymph nodes (MLNs) and the production of various regulatory cytokines, such as IL-10, TGF-β, INFγ and IL-6. A healthy intestinal microbiota maintains homeostatic local immune responses through exposure to structural ligands, such as lipopolysaccharides (LPS) and secreted metabolites such as short-chain fatty acids - SCFA. Invasion of microorganisms and absorbed metabolites affects circulating lymphocytes and the intestinal microbiota is disrupted, leading to a change in the immune response
The upper and lower airways contain an embryologically similar surface that begins in the mouth and nose and ends in the lungs or in the case of food ends in the stomach and intestines. Therefore, it is not surprising that some skin commensal bacteria protect their host and protection begins as soon as the virus enters the nose. Supernatants of the commensal bacterium S. epidermidis significantly suppressed the infectivity of various influenza viruses. Chen et al. (2016) identified a giant extracellular matrix-binding protein (Embp) as the major component involved in the anti-influenza effect of S. epidermidis (Chen et al. 2016). One of the antimicrobial peptides, surfactin (a cyclic lipopeptide), produced by bacterium Bacillus subtilis has been found to disintegrate enveloped viruses (Fig. 4). Surfactin degrades not only the lipids of the viral envelope, but also the capsid of the virus through the formation of ion channels. Therefore, it is also effective against many other viruses, such as influenza A (strains H1N1 and H3N2), Ebola and Zika. The usage of B. subtilis peptidoglycans reduces infectivity of CoV viruses (Johnson et al. 2019). In China, Bacillus subtilis, Bacillus licheniformis and Bacillus cereus are ingested by adults, infants and newborns to relieve diarrhoea and build healthy intestinal microbiota. Bacillus strains are considered to efficiently antagonize pathogenic bacteria, while promoting the growth of Lactobacillus to balance the microbiota, and thus achieving therapeutic purposes (Yu et al. 2019). Some sceptics doubt the ability of probiotic bacteria to settle in the gut. Very often, the protective effects are provided rather by cellular components than vital bacteria or their metabolites. Probiotic bacteria Lactobacillus gasseri TMC0356 can protect mice from influenza virus infection both in the form of a living and inactivated cell (Kawase et al. 2012).
Microbiota is not only important for the reduction of pathogenic viruses or bacteria, but also has an effect on the nutrition of intestinal cells and the regulation of the immune system. One of the nutrients produced by microbiota is butyrate, which upregulates mucus production and enterocyte proliferation. Changes in microbiota composition are related to many gastrointestinal (GI) disorders such as inflammatory bowel disease (IBD), gastritis, peptic ulcer or even cancer. For example, during IBD, there is a significant decrease in Bacteroidetes and Clostridium, and an increase in the population of Enterobacterales. This leads to a reduction of growth of enterocytes because of a decrease in SCFAs (short-chain fatty acids) production and an increase in gut permeability. A mixture of some bacteria (Bacteroides thetaiotaomicron and Eubacterium rectale) induces production of mucosal glycans which are utilized by these bacteria, but not by pathogens, and this leads to growth suppression of pathogens. Probiotic bacteria can also modulate behaviour of the mucosal immune response (Galdeano and Perdigón 2006).
Besides nutrition and the ability to reduce the viability of viruses, some types of microbiota are able to significantly modulate the immune response. There is evidence of a positive effect of daily intake of probiotic strains Lactobacillus rhamnosus GG and Bifidobacterium animalis subsp. lactis on the response to Streptococcus pneumoniae and Bordetella pertussis vaccination in infants. The vaccine responses were at the same levels in both the probiotic and placebo group but the main difference was in the production of IL-6, which belongs to the group of proinflammatory cytokines, with the production significantly higher in the placebo group (Fig. 4) (Sørensen et al. 2019). In particular, interleukin IL-6 is one of the substances that are significantly elevated in patients with SARS-CoV-2 disease, especially in the non-survival group. Suppressing this pro-inflammatory interleukin could help increase patient survival (Zhou et al. 2020).
At the beginning of the pandemic, Gao and co-workers surprisingly pointed to a link between the state of the intestinal microbiome and the severity of the course of COVID-19 (Gao et al. 2020; Wong et al. 2020). A few months later, after this text has been sent to the journal Biologia, He et al. published an article directly designating the intestinal microbiota as a potential strategy to fight SARS-CoV-2 infection (He et al. 2020b). Although there is no direct clinical evidence that intestinal microbiota modulation has a therapeutic role in the treatment of COVID-19, in the following section, we suggest that targeting the intestinal microbiota may be a new therapeutic option or at least an adjuvant treatment of choice.
Opinion: A healthy microbiome could be one of the factors responsible for lower case fatality ratio on COVID-19 patients.
The coronavirus pandemic has different case fatality ratio (CFR) in different countries (which estimates this proportion of deaths among identified confirmed cases). Many hypotheses try to explain this phenomenon. Apparently, crucial are the differences in the methodology of counting disease-related deaths or the quality and number of people tested. However, CFR from COVID-19 is not a single-factor issue. The quality of health care, the age of the afflicted population and the three-generation social connection in families also play a role. One of the frequently discussed hypotheses considers the different practices in vaccination against tuberculosis (Bacillus Calmette Guérin (BCG) vaccination) as a causal link (Hegarty et al. 2020). Another theory assumes periodical contact with seasonal coronaviral diseases such as HCoV-NL63, HCoV-229E, HCoV-OC43 and HCoV-HKU1 as an important protective factor to COVID-19 (Aldridge et al. 2020; La Jolla Institute for Immunology 2020). The immune system more often exposed to HCoV could be set differently than in Mediterranean nations. The production of proteins such as interferon-inducible trans-membrane proteins IFITM3, PUSL1, TPST1 and WDR33 inhibits the invasion of viruses into cells and affects the course of the disease (Brass et al. 2009; Almén et al. 2012; Everitt et al. 2012). Schillaci proposed the idea that a linear correlation exists between the initial growth rate of COVID-19 infection and the average percentage of R1b Y-DNA haplogroup in different countries. It has been indicated that if Y-DNA haplogroup R1b does not play a directly active role, then it certainly plays a passive role in the spread of SARS-CoV-2 virus (Schillaci 2020).
In addition to these hypotheses, we suggest that the quality of the intestinal microbiota is involved in the course of the disease, because it may be more serious in people suffering from intestinal dysbiosis caused by chronic diseases, obesity or even old age. Dysbiosis can be caused by many external factors. One of the factors is the anxious effort to protect people from pathogenic microorganisms everywhere and in everything. Unnecessary overuse of antibiotics and food additives and stabilizers that keep food fresh for a long time leads to the gradual extinction of the commensal species present in the human microbiome in terms of the total number of species present. Thanks to industrial progress, we live in a hygienically perfect but almost bacterial-free environment, alone or in small families without contact with soil and livestock (Strachan 1989; Round and Mazmanian 2009; Gonzalez et al. 2011; Hunter 2012; Blum et al. 2019). There are not enough remaining bacteria to supplement the missing substances necessary for the gut - postbiotics (SCFAs, vitamins, immunomodulators, enzymes, peptides, etc.). Therefore, we cannot be surprised that the incidence of dysbiosis is increasing. In addition, microbial dysbiosis is associated with many inflammatory diseases, including the aforementioned diseases such as diabetes, heart disease, stroke, chronic obstructive pulmonary disease (COPD), asthma and cystic fibrosis (CF), but old age and obesity, are also associated with changes in the composition of the microbiota (Henao-Mejia et al. 2012; Kamada et al. 2013; Mehal 2013; Carding et al. 2015). Interestingly, obesity is one of the high-risk factors in COVID-19 disease related to its more severe course (Dietz and Santos-Burgoa 2020). In many cases, we are able to partially overcome this low diversity of symbiotic organisms by including fermented products in our diet in which essential postbiotics important for intestinal health are present. Rye-wheat sourdough bread, yoghurts and sour dairy products or sauerkraut (fermented cabbage; sauer in German means acidic, kraut - cabbage) are daily or frequently used foods in the regular diet in many countries including the Czech Republic. The traditional three-stage sourdough, typical of Central European countries (Austria, Czech Republic, Germany, Slovak Republic) is prepared before the actual production of bread from a fermentation culture, which consists of a natural mixture of yeasts (1 × 107/mL) and lactic acid bacteria (1 × 109/mL) together with rye flour and water. In addition, yoghurts and sour dairy products are becoming increasingly popular. The predominant organisms in these starter cultures are lactic acid bacteria (LAB), for example, Lactococcus lactis, Lactobacillus species, Streptococcus thermophilus, Bifidobacterium species, and Leuconostoc species (Chen et al. 2017). Sauerkraut is made by fermenting cabbage in which the bacteria produce SCFAs preserving the plant material. This food rich in vitamins and environmentally stable proteases contains bacteria such as Lactobacillus lactis and Bacillus cereus (Kotlar et al. 2012). It may seem exaggerated that fermented foods would have such an effect on a healthy microbiome. However, if we realize that the products of lactobacilli (postbiotics) are responsible for the positive effect and the recommended effective daily dose of probiotic microorganisms according to the World Gastroenterological Organization is about 20 billion bacteria (Guarner et al. 2017), then 100 g sourdough bread originally contained 100 billion bacteria in the dough. In the Czech Republic, the daily consumption of bread per capita is 100 g, yoghurt and sour milk products 40 g and sauerkraut 4 g. You could say that thanks to the bread, we have enough postbiotics to maintain the balance of the microbiome. It’s not that simple because not all people consume sourdough bread and also the popularity of the bread from frozen semi-finished products is growing, where there are more additives with antimicrobial effect than products of lactic acid bacteria. Whether there is an association between the good condition of the microbiome and the severity of COVID-19 needs to be investigated. Statistics of celiac disease, which is an autoimmune disease dependent on the condition of the intestine, confirmed that the Czech Republic had lower incidence rates (0.19%) than the world average (1–1.2%) (Hoffmanová 2019). Non conscious approach to microbiome care in the form of daily supplementation of postbiotics in the form of sourdough bread, yoghurt, sauerkraut, and other fermented foods may explain this epidemiologically significant difference during the course of the disease (across countries). This could explain why case fatality ratio from COVID-19 is relatively lower in the Czech Republic than in other European countries.
In conclusion, the new coronavirus SARS-CoV-2 causes primarily a respiratory infection, while tropism also affects the intestine. As a result, intestinal microbiome dysbiosis, which often occurs in the elderly, obese people and those with underlying chronic disease may be responsible for higher case fatality ratio (CFR). Microbiome, the last discovered organ of the human body seems to be a significant factor that plays a major role in the COVID-19 epidemic. An unconscious approach to microbiome care in the form of daily supplementation of postbiotics in the form of fermented foods may explain this epidemiologically significant difference in the course of the disease.
Acknowledgments
The work was supported by the Ministry of Agriculture of the Czech Republic, project RO0518.
Abbreviations
- ARDS
Acute respiratory distress syndrome
- COPD
Chronic obstructive pulmonary disease
- COVID-19
The coronavirus disease 2019
- CFR
Case fatality ratio
- LAB
Lactic acid bacteria
- LPS
Lipopolysaccharides
- SARS-CoV-2
Severe acute respiratory syndrome Coronavirus 2
- SCFAs
Short-chain fatty acids
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aas J, Gessert CE, Bakken JS. Recurrent Clostridium difficile colitis: case series involving 18 patients treated with donor stool administered via a nasogastric tube. Clin Infect Dis. 2003;36:580–585. doi: 10.1086/367657. [DOI] [PubMed] [Google Scholar]
- Adair LB, II, Ledermann EJ. Chest CT findings of early and progressive phase COVID-19 infection from a US patient. Radiol Case Rep. 2020;15:819–824. doi: 10.1016/j.radcr.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge R, Lewer D, Beale S, Johnson A, Hayward A, Fragaszy E. Dataset: Seasonality and immunity to laboratory-confirmed seasonal coronaviruses (HCoV-NL63, HCoV-OC43, and HCoV-229E): results from the Flu Watch cohort study. Wellcome Opn Res. 2020;5:52. doi: 10.14324/000.ds.10093909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almén MS, Bringeland N, Fredriksson R, Schiöth BH. The Dispanins: a novel gene family of ancient origin that contains 14 human members. PLoS One. 2012;7(2):e31961. doi: 10.1371/journal.pone.0031961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azkur AK, Akdis M, Azkur D, Sokolowska M, van de Veen W, Brüggen MC, O'Mahony L, Gao Y, Nadeau K, Akdis CA. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy. 2020;75(7):1564–1581. doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquero F, Nombela C. The microbiome as a human organ. Clin Microbiol Infect. 2012;18(Suppl 4):2–4. doi: 10.1111/j.1469-0691.2012.03916.x. [DOI] [PubMed] [Google Scholar]
- Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum WEH, Zechmeister-Boltenstern S, Keiblinger KM. Does soil contribute to the human gut microbiome? Microorganisms. 2019;7(9):287. doi: 10.3390/microorganisms7090287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Huang IC, Benita Y, et al. The IFITM proteins mediate cellular resistance to influenza a H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139(7):1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun E, Sauter D. Furin-mediated protein processing in infectious diseases and cancer. Clin Transl Immunology. 2019;8(8):e1073. doi: 10.1002/cti2.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, Hansbro PM. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. 2017;15(1):55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. 2015;26:26191. doi: 10.3402/mehd.v26.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruana JC, Walper SA. Bacterial membrane vesicles as mediators of microbe - microbe and microbe - host community interactions. Front Microbiol. 2020;11:432. doi: 10.3389/fmicb.2020.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HW, Liu PF, Liu YT, Kuo S, Zhang XQ, Schooley RT, Rohde H, Gallo RL, Huang CM. Nasal commensal Staphylococcus epidermidis counteracts influenza virus. Sci Rep. 2016;6:27870. doi: 10.1038/srep27870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Zhao S, Hao G, Yu H, Tian H, Zhao G. Role of lactic acid bacteria on the yogurt flavour. Int J Food Prop. 2017;20(sup1):S316–S330. doi: 10.1080/10942912.2017.1295988. [DOI] [Google Scholar]
- Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz W, Santos-Burgoa C. Obesity and its implications for COVID-19 mortality. Obesity (Silver Spring) 2020;28(6):1005. doi: 10.1002/oby.22818. [DOI] [PubMed] [Google Scholar]
- Ding X, Xu J, Zhou J, Long Q. Chest CT findings of COVID-19 pneumonia by duration of symptoms. Eur J Radiol. 2020;127:109009. doi: 10.1016/j.ejrad.2020.109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M, Cossart P, Lebreton A. Mammalian microRNAs and long noncoding RNAs in the host-bacterial pathogen crosstalk. Semin Cell Dev Biol. 2016;65:11–19. doi: 10.1016/j.semcdb.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Medicines Agency (2020) Treatments and vaccines for COVID-19 [online]. European Medicines Agency. https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/treatments-vaccines-covid-19. Accessed Juny 14, 2020
- Everitt AR, Clare S, Pertel T, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484(7395):519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein S, Herbert JA, McNamara PS, Hedrich CM. COVID-19: immunology and treatment options. Clin Immunol. 2020;215:108448. doi: 10.1016/j.clim.2020.108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdeano CM, Perdigón G. The probiotic bacterium Lactobacillus casei induces activation of the gut mucosal immune system through innate immunity. Clin Vaccine Immunol. 2006;13(2):219–226. doi: 10.1128/CVI.13.2.219-226.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao QY, Chen YX, Fang JY. 2019 novel coronavirus infection and gastrointestinal tract. J Dig Dis. 2020;21(3):125–126. doi: 10.1111/1751-2980.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Clemente JC, Shade A, et al. Our microbial selves: what ecology can teach us. EMBO Rep. 2011;12(8):775784. doi: 10.1038/embor.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner F, Sanders ME, Eliakim R, Fedorak R, Gangl A, Garisch J, Kaufmann P, Karakan T, Khan AG, Kim N, De Paula JA, Ramakrishna B, Shanahan F, Szajewska H, Thomson A, Mair AL, Merenstein D, Salminen S (2017) WGO Practice Guideline - Probiotics and Prebiotics. https://www.worldgastroenterology.org/UserFiles/file/guidelines/probiotics-and-prebiotics-english-2017.pdf
- Guo T, Fan Y, Chen M, Wu X, Zhang L, He T, Wang H, Wan J, Wang X, Lu Z. (2020) Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19). JAMA Cardiol e201017. 10.1001/jamacardio.2020.1017 [DOI] [PMC free article] [PubMed]
- Hand TW, Vujkovic-Cvijin I, Ridaura VK, Belkaid Y. Linking the microbiota, chronic disease, and the immune system. Trends Endocrinol Metab. 2016;27(12):831–843. doi: 10.1016/j.tem.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26(5):672–675. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- He LH, Ren LF, Li JF, et al. Intestinal Flora as a potential strategy to fight SARS-CoV-2 infection. Front Microbiol. 2020;11:1388. doi: 10.3389/fmicb.2020.01388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty PK, Kamat A, Zafirakis H, Dinardo A (2020) BCG vaccination may be protective against Covid-19. 10.13140/RG.2.2.35948.10880
- Henao-Mejia J, Elinav E, Jin C, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482(7384):179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmanová I. Celiakie. Praha: Mladá fronta; 2019. [Google Scholar]
- Hosseini Rad Sm A, AD ML. Implications of SARS-CoV-2 Mutations for Genomic RNA Structure and Host microRNA Targeting. Int J Mol Sci. 2020;7;21(13):4807. doi: 10.3390/ijms21134807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle GB, Dickson RP, Lukacs NW. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. 2017;10(2):299–306. doi: 10.1038/mi.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit RJ, de Haan CA, Bosch BJ. Coronavirus spike protein and tropism changes. Adv Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter P. The changing hypothesis of the gut. The intestinal microbiome is increasingly seen as vital to human health. EMBO Rep. 2012;13(6):498–500. doi: 10.1038/embor.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabs S, Biton A, Bécavin C, Nahori MA, Ghozlane A, Pagliuso A, Spanò G, Guérineau V, Touboul D, Giai Gianetto Q, Chaze T, Matondo M, Dillies MA, Cossart P. Impact of the gut microbiota on the m6A epitranscriptome of mouse cecum and liver. Nat Commun. 2020;12;11(1):1344. doi: 10.1038/s41467-020-15126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Hage A, Kalveram B, Mears M, Plante JA, Rodriguez SE, Ding Z, LuoX BD, Bradrick SS, Freiberg AN, Popov V, Rajsbaum R, Rossi S, Russell WK, MenacheryVD Peptidoglycan-associated cycliclipopeptide disrupts viral infectivity. J Virol. 2019;93(22):e01282–e01219. doi: 10.1128/JVI.01282-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Seo SU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- Kawase M, He F, Kubota A, Yoda K, Miyazawa K, Hiramatsu M. Heat-killed Lactobacillus gasseri TMC0356 protects mice against influenza virus infection by stimulating gut and respiratory immune responses. FEMS Immunol Med Microbiol. 2012;64(2):280–288. doi: 10.1111/j.1574-695X.2011.00903.x. [DOI] [PubMed] [Google Scholar]
- Killerby ME, Biggs HM, Haynes A, Dahl RM, Mustaquim D, Gerber SI, Watson JT. Human coronavirus circulation in the United States 2014–2017. J Clin Virol. 2018;101:52–56. doi: 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotlar CA, Agüero MV, Ponce AG, Borla OP, Roura SI (2012) Fresh and fermented vegetables as a source of Proteolytic Bacteria. Handbook of Plant-Based Fermented Food and Beverage Technology. Second Edition. Edited by Hui. ISBN 9781439849040
- La Jolla Institute for Immunology (2020) Exposure to common cold coronaviruses can teach the immune system to recognize SARS-CoV-2: researchers caution: it is too soon to say whether pre-existing immune cell memory affects COVID-19 clinical outcomes. Sci Dly Sci Dily, 4 August 2020. www.sciencedaily.com/releases/2020/08/200804100219.htm
- Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, Azman AS, Reich NG, Lessler J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172(9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kim T, Lee E, et al (2020) Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS-CoV-2 infection in a community treatment Center in the Republic of Korea. JAMA intern med. Published online august 06, 10.1001/jamainternmed.2020.3862 [DOI] [PMC free article] [PubMed]
- Leitão AL, Costa MC, Gabriel AF, Enguita FJ. Interspecies communication in Holobionts by non-coding RNA exchange. Int J Mol Sci. 2020;27;21(7):2333. doi: 10.3390/ijms21072333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MY, Li L, Zhang Y, Wang XS. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty. 2020;9(1):45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xiao X, Wei X, Li J, Yang J, Tan H, Zhu J, Zhang Q, Wu J, Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J Med Virol. 2020;92(6):595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, Niu P, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloir Q, Ghysen K, von Frenckell C, Louis R, Guiot J. Détresse respiratoire aiguë révélatrice d’un syndrome des antisynthétases [Acute respiratory distress revealing antisynthetase syndrome] Rev Med Liege. 2018;73(7–8):370–375. [PubMed] [Google Scholar]
- Matsuyama S, Nao N, Shirato K, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A. 2020;117(13):7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehal WZ. The Gordian knot of dysbiosis, obesity and NAFLD. Nat Rev Gastroenterol Hepatol. 2013;10(11):637–644. doi: 10.1038/nrgastro.2013.146. [DOI] [PubMed] [Google Scholar]
- Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memoli MJ, Czajkowski L, Reed S, et al. Validation of the wild-type influenza a human challenge model H1N1pdMIST: an a(H1N1)pdm09 dose-finding investigational new drug study. Clin Infect Dis. 2015;60(5):693–702. doi: 10.1093/cid/ciu924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Novel coronavirus: From discovery to clinical diagnostics. Infect Genet Evol. 2020;79:104211. doi: 10.1016/j.meegid.2020.104211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polák P, Freibergerová M, Husa P, et al. Fekální bakterioterapie v léčbě rekurentní kolitidy způsobené Clostridium difficile na klinice infekčních chorob fakultní nemocnice Brno v letech 2010–2014 – prospektivní studie. Epidemiol Mikrobiol Imunol. 2015;64(4):232–235. [PubMed] [Google Scholar]
- Romano M, Ruggiero A, Squeglia F, Maga G, Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;20;9(5):1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillaci S (2020) Possible correlation between COVID-19 contagion and Y-DNA Haplogroup R1b. 10.31219/osf.io/yv8kc
- Schmulson M, Dávalos MF, Berumen J. Beware: gastrointestinal symptoms can be a manifestation of COVID-19. Rev Gastroenterol Mex. 2020;85(3):282–287. doi: 10.1016/j.rgmxen.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine T, Perez-Potti A, Rivera-Ballesteros et al (2020) Karolinska COVID-19 study group, robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell 183:158–168. 10.1016/j.cell.2020.08.017 [DOI] [PMC free article] [PubMed]
- Shi S, Qin M, Shen B, Cai Y, Liu T, Yang F, Gong W, Liu X, Liang J, Zhao Q, Huang H, Yang B, Huang C. Association of Cardiac Injury with Mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol. 2020;5(7):802–810. doi: 10.1001/jamacardio.2020.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen CA, Fuglsang E, Jørgensen CS, et al. Probiotics and the immunological response to infant vaccinations; a double-blind randomized controlled trial. Clin Microbiol infect. 2019;25(4):511.e1–511.e7. doi: 10.1016/j.cmi.2018.07.031. [DOI] [PubMed] [Google Scholar]
- Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, Zhou Y, Du L (2020) Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol 1–8. 10.1038/s41423-020-0400-4 [DOI] [PMC free article] [PubMed]
- To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle C, Martin B, Touret F, et al (2020) Drugs against SARS-CoV-2: what do we know about their mode of action? Rev med Virol. e2143. 10.1002/rmv.2143 [DOI] [PMC free article] [PubMed]
- Vellingiri B, Jayaramayya K, Iyer M, et al. COVID-19: a promising cure for the global panic. Sci Total Environ. 2020;725:138277. doi: 10.1016/j.scitotenv.2020.138277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Cui YH, Xiao L. Regulation of intestinal epithelial barrier function by Long noncoding RNA uc.173 through interaction with MicroRNA 29b. Mol Cell Biol. 2018;38(13):e00010–e00018. doi: 10.1128/MCB.00010-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhou D, Feng Y, Chen G, Li N. T-UCRs with digestive and respiratory diseases. Bioorg med Chem Lett. 2020;15;30(16):127306. doi: 10.1016/j.bmcl.2020.127306. [DOI] [PubMed] [Google Scholar]
- Weiss P, Murdoch DR. Clinical course and mortality risk of severe COVID-19. Lancet. 2020;395(10229):1014–1015. doi: 10.1016/S0140-6736(20)30633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong SH, Lui RN, Sung JJ. Covid-19 and the digestive system. J Gastroenterol Hepatol. 2020;35(5):744–748. doi: 10.1111/jgh.15047. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2020a) Clinical management of COVID-19 [online]. 27 May 2020. https://apps.who.int/iris/rest/bitstreams/1278777/retrieve. Accessed Juny 14, 2020
- World Health Organization (2020b) Corticosteroids for COVID-19 [online]. https://wwwwhoint/publications/i/item/WHO-2019-nCoV-Corticosteroids-202012. Accessed September 2020
- World Health Organization WHO (2020) Laboratory testing for coronavirus disease (COVID-19) in suspected human cases: interim guidance. https://www.who.int/publications/i/item/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117. Accessed 19 March 2020
- Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterolo. 2020;158(6):1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L, Wu J, Wang JY, Chung HK, Kalakonda S, Rao JN, Gorospe M, Wang JY (2018) Long noncoding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degradation of MicroRNA 195. Gastroenterology 154(3):599–611. 10.1053/j.gastro.2017.10.009 [DOI] [PMC free article] [PubMed]
- Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23(2):130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yitbarek A, Taha-Abdelaziz K, Hodgins DC, et al. Gut microbiota-mediated protection against influenza virus subtype H9N2 in chickens is associated with modulation of the innate responses. Sci Rep. 2018;8(1):13189. doi: 10.1038/s41598-018-31613-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Kong J, Zhang L, Gu X, Wang M, Guo T. New crosstalk between probiotics Lactobacillus plantarum and Bacillus subtilis. Sci Rep. 2019;9(1):13151. doi: 10.1038/s41598-019-49688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilber-Rosenberg I, Rosenberg E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution, FEMS microbiology reviews. Volume. 2008;32(5):723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]