Dear Sir,
The first wave of COVID-19 has already infected more than 31 million patients (5 million in May 2020) in the world. In France, in May 21, 2020, 144163 confirmed cases and 28215 deaths have been recovered [1]. The Grand Est region has been one of the most affected in France (3335 deaths) with dynamics of mortality far beyond to what announced in China [1]. The outbreak has overwhelmed hospitals. A second wave has to be considered given that deconfinement may favor new cases. Screening tests on respiratory samples were one of the cornerstones for triage plans. As Kim et al., feedbacks regarding co-infection could help to propose screening strategies to face a second wave [2].
From March 1 to April 29, 2020, we screened 24257 patients for SARS-CoV-2 (RT-PCR) in two of the three Teaching Hospitals of the Grand Est region (Nancy and Reims). Of all the patients screened, 5428 (22.4 %) were positive for SARS-CoV-2. Among the hospitalized patients, we recovered available status of infection for other respiratory pathogens for 232 patients determined by multiplex PCR (Table 1 ).
Table 1.
Patient demographics according to SARS-CoV-2 status and proportions of samples positive for other respiratory pathogens in SARS-CoV-2 subsets.
| Characteristics and pathogensb | SARS-CoV-2 statusa |
|||
|---|---|---|---|---|
| Negative, n = 91 (39.2 %) |
Positive, n = 141 (60.8 %) |
|||
| Positive for other respiratory pathogenc | Negative for other respiratory pathogen | Positive for other respiratory pathogen | Negative for other respiratory pathogen | |
| No. Sample (%) | 13 (14.3) | 78 (85.7) | 7 (5) | 134 (95) |
| Age, mean (range) years | 59.1 (20–91) | 60.7 (18–101) | 66.1 (42–81) | 64.5 (29–89) |
| Male, No./total (%) | 7/13 (53.8) | 46/78 (58.9) | 6/7 (85.7) | 94/134 (70.1) |
| Site of specimen collection, No./total (%) | ||||
| ICU | 6/13 (46.2) | 50/78 (64.1) | 7/7 (100) | 122/134 (91.0) |
| Inpatient | 7/13 (53.8) | 28/78 (35.9) | 0/7 | 12/134 (9.0) |
| Influenza A | 2/91 (2.2) | 0/141 | ||
| Other Coronaviridae | 1/91 (1.1) | 0/141 | ||
| Metapneumovirus | 3/91 (3.3) | 5/141 (3.5) | ||
| Parainfluenza 1-4 | 0/91 | 1/141 (0.7) | ||
| Rhinovirus/Enterovirus | 7/91 (7.7) | 0/141 | ||
| RSV | 1/91 (1.1) | 0/141 | ||
| Mycoplasma pneumoniae | 0/91 | 1/141 (0.7) | ||
The 232 patients were tested for each respiratory pathogen. Differences in proportion were evaluated with Fisher tests (significance, P < 0.05). Analyses were performed using Prism v.7.
Influenza B, Adenovirus and C. pneumoniae were never detected.
One patient was positive for Metapneumovirus and Rhinovirus/Enterovirus.
Among the 232 patients, 141 (60.8 %) were positive for SARS-CoV-2 and 20 (8.6 %) for one or more other pathogens (Table 1). Of the 141 SARS-CoV-2-positive patients, only 7 (5.0 %) were positive for other pathogens, compared with 13 (14.3 %) of the 91 SARS-CoV-2-negative patients (difference, 9.3 % [95 % CI, - 0.3–17.5%], P < 0.05). All co-infected patients were hospitalized in intensive care unit. Metapneumovirus was the most common pathogen found in co-infected patients (3.5 %), followed by Parainfluenza (0.7 %), and M. pneumoniae (0.7 %). For SARS-CoV-2-negative-patients, different results were found, with Rhinovirus/Enterovirus (7.7 %) as the predominant pathogens, followed by other Coronaviridae (1.1 %), Influenza A virus (2.2 %), RSV (1.1 %) and Metapneumovirus (3.3 %).
Despite a low rate of SARS-CoV-2-positive patients (9.5 %), Kim et al. suggested higher rates (20.7 %) of co-infections in the USA (California) [2] compared with earlier reports from China [3]. In our hospitals, among a cohort comprising 60.8 % of SARS-CoV-2-positive-patients, co-infections with other pathogens were rare. Our results were recovered around the outbreak peak (March 23–29) [1], whereas in March 2020, the USA were still in the beginning phase of the pandemic and current RSV and non–SARS-CoV-2 Coronaviridae outbreaks were noted in California [4]. Our findings are in line with a recent study reporting 54.8 % of SARS-CoV-2-positive patients in the USA (New York City area) [5] and another study performed in Spain [6] that both described lower co-infection rates. Data regarding co-infections in France are very scarce. To our knowledge, only one report, which studied a cluster in French Alps, mentioned 1/21 case co-infected with SARS-CoV-2, Rhinovirus/Enterovirus and Influenza A virus [7]. Interestingly, it has been recently proposed that the low rate of co-infection might stem from the clearance of the SARS-CoV-2 by other respiratory viruses given its lower growth rate [8]. In our non-SARS-CoV-2 infected patients, the high rate of Rhinovirus/Enterovirus and other Coronaviridae may be due to the type I interferon-based competitive inhibition between RNA viruses [9]. A recent study has reported that Metapneumovirus accounts for 3 % of patients hospitalized for community-acquired Influenza-like illness during winter in France and affects mainly the elderly and patients with chronic conditions [10]. This could explain that we found Metapneumovirus as the most common co-infecting pathogen in SARS-CoV-2 patients since we focused on hospitalized patients.
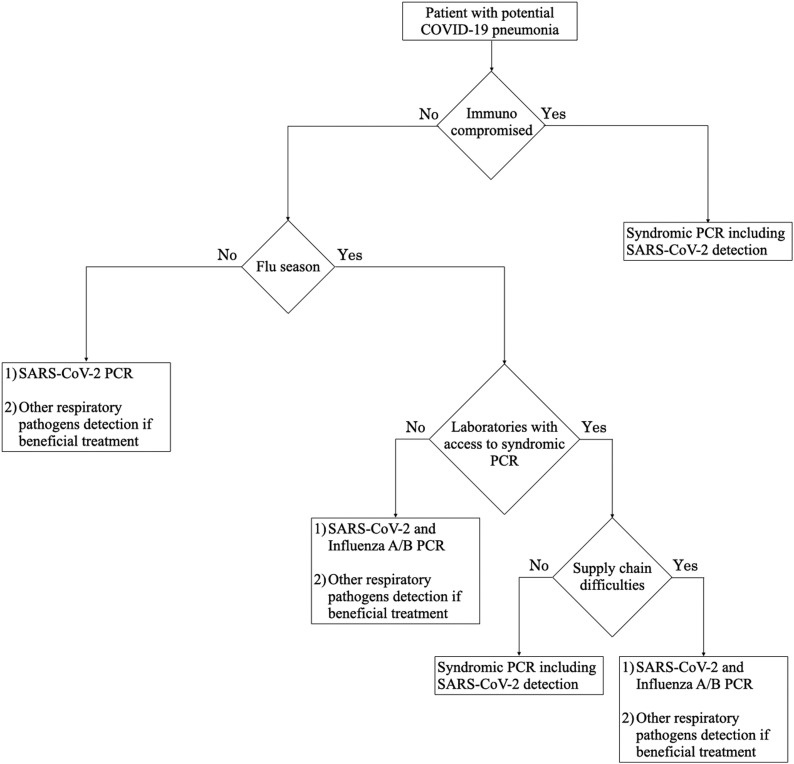
Our study has geographical (the north-east of France) and temporal (no overlap with peak flu season) limitations. However, our findings suggest that if SARS-CoV-2 resurges before the flu season, documentation of co-infections may provide limited benefit and could be proposed as a second-line screening. It is noteworthy that for immunocompromised patients, first-line testing should target several respiratory viruses given that potentially beneficial treatments exist for RSV, Influenza and Parainfluenza virus (e.g. ribavirin, intravenous immunoglobulin, or oseltamivir) [11,12].
Despite that flu vaccination has to be promoted in the coming winter, a second wave has to be considered during the flu season. Thereby, it would impose co-screening of Influenza A/B. Although extended multiplex panels that include SARS-CoV-2 are now available (i.e. Biofire respiratory panel RP2.1), endorsing such a strategy may avoid clinical laboratories to be strained either because having no access to expensive tests or facing supply chain difficulties (Fig. 1 ).
Fig. 1.
Proposed SARS-CoV-2 screening strategy algorithm for co-infection with other respiratory pathogens.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
We thank Prof Christophe de Champs (MD, PhD), Dr Véronique Vernet-Garnier (PharmD, PhD), Dr Véronique Brodard (PharmD), Dr Clément Lier (PharmD) and Prof Bruno Mourvillier (MD, PhD) affiliated to the Reims Teaching Hospital and Prof Alain Lozniewski (MD, PhD), Prof Bruno Levy (MD, PhD) and Prof Gérard Audibert (MD, PhD) affiliated to the Nancy Teaching Hospital, who are all either laboratories or ICU staffs involved in this study.
References
- 1.2020. Gouvernement Français. COVID-19.https://www.gouvernement.fr/info-coronavirus/carte-et-donnees (Accessed May 29, 2020) [Google Scholar]
- 2.Kim D., Quinn J., Pinsky B., Shah N.H., Brown I. Rates of co-infection between SARS-CoV-2 and other respiratory pathogens. JAMA. 2020 doi: 10.1001/jama.2020.6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Diseases Control and Prevention . 2020. COVID View Weekly Summary.https://www.cdc.gov/coronavirus/2019-ncov/index.html (Accessed 5 May 2020) [Google Scholar]
- 5.Nowak M.D., Sordillo E.M., Gitman M.R., Paniz Mondolfi A.E. Co-infection in SARS-CoV-2 infected patients: where are influenza virus and rhinovirus/enterovirus? J. Med. Virol. 2020 doi: 10.1002/jmv.25953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasco M.L., Buesa J., Colomina J., Forner M.J., Galindo M.J., Navarro J. Co-detection of respiratory pathogens in patients hospitalized with Coronavirus viral disease-2019 pneumonia. J. Med. Virol. 2020 doi: 10.1002/jmv.25922. [DOI] [PubMed] [Google Scholar]
- 7.Danis K., Epaulard O., Bénet T., Gaymard A., Campoy S., Botelho-Nevers E. Cluster of coronavirus disease 2019 (COVID-19) in the French Alps, February 2020. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinky L., Dobrovolny H.M. SARS‐CoV‐2 coinfections: could influenza and the common cold be beneficial? J. Med. Virol. 2020 doi: 10.1002/jmv.26098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lancaster K.Z., Pfeiffer J.K. Mechanisms controlling virulence thresholds of mixed viral populations. J. Virol. 2011 doi: 10.1128/JVI.00355-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loubet P., Mathieu P., Lenzi N., Galtier F., Lainé F., Lesieur Z. Characteristics of human metapneumovirus infection in adults hospitalized for community-acquired influenza-like illness in France, 2012-2018: a retrospective observational study. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch H.H., Martino R., Ward K.N., Boeckh M., Einsele H., Ljungman P. Fourth European Conference on Infections in Leukaemia (ECIL-4): guidelines for diagnosis and treatment of human respiratory syncytial virus, parainfluenza virus, metapneumovirus, rhinovirus, and coronavirus. Clin. Infect. Dis. 2013 doi: 10.1093/cid/cis844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanson K.E., Azar M.M., Banerjee R., Chou A., Colgrove R.C., Ginocchio C.C. Molecular testing for acute respiratory tract infections: clinical and diagnostic recommendations from the IDSA’s Diagnostics Committee. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa508. [DOI] [PMC free article] [PubMed] [Google Scholar]