Abstract
While the number of studies related to severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) is constantly growing, it is essential to provide a framework of modeling viral infections. Therefore, this review aims to describe the background presented by earlier used models for viral studies and an approach to design an “ideal” tissue model for SARS-CoV-2 infection. Due to the previous successful achievements in antiviral research and tissue engineering, combining the emerging techniques such as bioprinting, microfluidics, and organoid formation are considered to be one of the best approaches to form in vitro tissue models. The fabrication of an integrated multi-tissue bioprinted platform tailored for SARS-CoV-2 infection can be a great breakthrough that can help defeat coronavirus disease in 2019.
Keywords: Severe acute respiratory syndrome-related coronavirus 2, Coronavirus disease 2019, Coronavirus, Bioprinting, microfluidics, Organoids, Tissue models, Viral infection, Body-on-a-chip
1 Introduction
Severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) that causes coronavirus disease 2019 (COVID-19) has become a great challenge for not only separate populations but also the whole mankind[1]. This new pathogen has almost conquered the world due to the lack of knowledge of COVID-19 pathogenesis and the absence of any vaccines or approved therapy[2].
The use of models enables the possibility to learn more about SARS-CoV-2 and infection-related conditions and to screen drugs and vaccines efficiently. Unfortunately, the number of approved models is limited, and researchers have to mainly rely on the past experience related to other viruses (SARS-CoV, Middle East respiratory syndrome coronavirus [MERS-CoV], influenza A virus [IAV], etc.) to develop new relevant models.
Models based on susceptible animals (ferrets[3,4], rhesus and cynomolgus macaques[5,6], transgenic mice[7], etc.) are highly demanded in investigations to study SARS-CoV-2 pathogenesis as well as clinical signs, and test drugs and vaccines as a part of the trials. However, high costs, virus species specificity, and ethical issues do not allow their use as a routine model. Hence, cell-based models can be a good option for screening and precise analysis of molecular pathways of COVID-19 pathogenesis. However, 2D cultures cannot provide biomimetic environment that can significantly influence virus spreading, infectivity, and drug efficiency. Therefore, 3D tissue models are of particular interest. To date, there is only one 3D model which is presented by organoids and used for studying SARS-CoV-2 infection[8]. Due to the past experience, its combination with 3D bioprinting and microfluidics, and fabrication of a multi-tissue integrated platform can help create a responsive and efficient immune-competent organism-like system tailored for SARS-CoV-2 infection (Figure 1).
Figure 1.
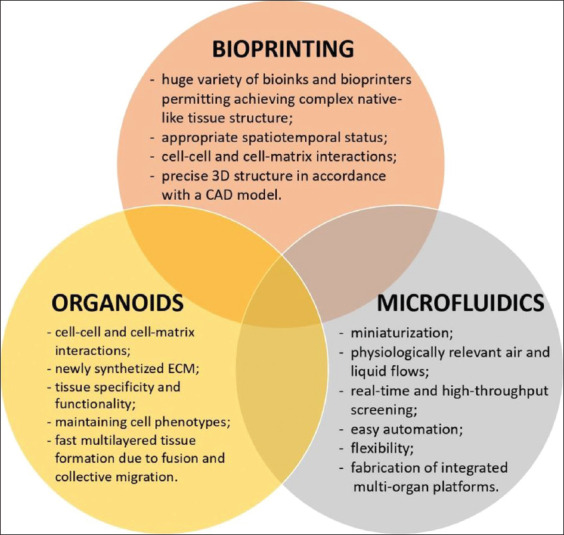
Bioprinting, microfluidics, and organoids as tools to defeat coronavirus disease 2019.
Thus, this review aims to describe the background of previously used models for viral studies and an approach to design an “ideal” tissue model to study SARS-CoV-2 infection.
The main advantages of each technique and thus, their combination can allow the fabrication of a highly responsive immune-competent organism-like platform which is tailored for SARS-CoV-2 infection and enable real-time and high-throughput screening.
2 Modeling viral infections
2.1 Humans
Nowadays, humans are not a typical object to model viral infections that is mainly caused by ethical issues. However, there is a pool of studies describing controlled human infection (CHI) trials on influenza viruses, respiratory syncytial virus (RSV), etc. For instance, the CHI model was used to assess susceptibility and resistance to Norwalk virus infection[9]. DeVincenzo et al. experimentally infected adult volunteers with wild-type RSV (it usually infects children) and showed that viral load can significantly influence the disease manifestation and its variation permit achieving the manifestation similar to natural infection[10]. Such controllable adult RSV infection model was claimed to be useful for proof-of-concept trials of antivirals candidates. Human challenge model provided valuable data on immune response to influenza infection[11-14]. Particularly, Huang et al.[14] revealed that antibody-secreting cells are virus-specific and can be the earliest marker of new influenza infection. In the case of especially dangerous infections (Ebola virus, Zika virus, yellow fever virus, etc.), CHI trials are significantly limited and almost cannot be performed. The significant efforts have been carried out to develop a dengue human infection model which has minimum harm and represents wild-type infection[15]. Hence, if such infection occurs naturally, blood, mucosa, urine, stool, tissue biopsies, etc., can serve as appropriate materials for studying lifecycle, entry, and pathogenesis of virus, and drug efficacy.
Usually, humans are involved in clinical trials of actively developed vaccines and phage-based drugs. Particularly, a novel dengue vaccine was tested using the DEN2Δ30 model[16]. Moreover, CHI models have enabled the development of the first vaccines against the influenza virus[17] and the first anti-influenza drug, amantadine[18], and other antivirals[19-23]. Phage-based drugs are particularly interesting as a therapeutic agent to treat multidrug-resistant bacteria, such as Staphylococcus aureus or Pseudomonas aeruginosa[24,25], and to modify microbiota to decrease specific microbial populations[26].
2.2 Animals
Animals are widely applied to study viral infections and antivirals. Among them, rodent models are the most common, especially used in studies to reveal features of respiratory diseases caused by IAV and RSV[27,28]. Their advantages include the possibility to use standardized animals that allow comparison and analysis of data obtained from different experiments. They are well-characterized and can be easily modified to delete particular genes or transfer them from other species that enables the extrapolation of results and detection of virus targets and pathogenesis. For instance, type-I interferon receptor-deficient mice were engineered and used to study the Zika virus entry process[29]. Moreover, handling rodents are easy and low-cost; they reproduce rapidly, have small sizes, can accustom to standard diet, and do not require much space. However, their use is limited because of species-determined differences in anatomy, pathophysiology, immune response[27], and host-determined virus infectivity[28].
Another well-established animal model is ferrets, which are susceptible to most human respiratory viruses[3,4,28,30]. Their respiratory tract is very similar to human’s, and they manifest a wide range of clinical signs[31]. Therefore, they have become a good model to test antiviral drugs. Particularly, using them, the efficacy of lopinavir-ritonavir, hydroxychloroquine sulfate, and emtricitabine-tenofovir was proven for SARS-CoV-2 treatment[3]. Nevertheless, ferret models have many limitations which include the lack of standardized strains, detailed molecular profiling, and higher handling and housing costs.
Interestingly, there is a pool of studies describing domestic animals, such as cats and dogs, as a viral model. For instance, a recent study has shown that cats are susceptible to human SARS-CoV-2[4]. Such findings provide novel insights into virus targets and its lifecycle. However, cat and dog models have similar issues as ferret ones as well as ethical concerns.
Cattle, sheep, and pigs are also used to study human viruses. Particularly, sheep and cattle are susceptible to RSV and have human-like virus spreading due to similar sizes[27]. The application of such models is limited by high handling and housing costs and biosafety considerations (e.g., pigs can be a reservoir for the reassortment and transgenic shift of influenza viruses).
The animal model closest to humans is, undoubtedly, primates that have similar genetic, anatomical, and physiological features. For instance, RSV infection of chimpanzees has all symptoms and complications (inflammation, acute respiratory distress syndrome, lung edema, etc.) that are typical in humans[32,33]. Primates are perfect candidates for preclinical studies of vaccines[27]. Despite all of the mentioned advantages, they cannot be treated as a routine viral model due to extensive economic, ethical, and logistical burdens.
However, in general, virus strains are highly selective and host-specific; therefore, only a subset of data can be extrapolated to humans. Phylogenetically close viruses often have different hosts and targets, and these viruses also cause different pathophysiologic conditions. Some drugs, whose efficacy was confirmed using animal models, failed in clinical trials[34]. One of the approaches to overcome these issues is humanization. Humanized animals (usually mice) have specific human expression profiles and are immunodeficient due to the mutation caused by severe combined immunodeficiency. They are widely used to model human immunodeficiency viruses (HIV), herpesviruses, cytomegaloviruses, dengue virus, Epstein-Barr virus, and Ebola virus infections[35,36]. However, such models require expensive specific handling and housing and are not flexible to study different viral infections.
2.3 Tissue models
Compared to the above-mentioned models, tissue models can be considered as the most flexible and ethically humane tools to study viral infections. While designing such models, a researcher can use different cell types, biomaterials, and fabrication methods (including bioprinting) that can work with a wide range of host-specific viruses. Studying viruses in vitro under controllable conditions allow better understanding of host-pathogen interactions and high-throughput screening of drug candidates.
All tissue models can be divided into three types with ten subtypes: 2D models (monolayer culture of cell lines and primary cells), 2.5D models (suspension culture using microcarriers and simple air-liquid biointerfaces), and 3D models (explants, organoids/spheroids, embedded cells, cell-seeded scaffolds, bioprinted constructs, and combined systems [organ-on-a chip]).
Monolayer (2D) cultures are the oldest and the most widely used model. Cell lines are usually applied to isolate viruses, develop new serological assays, and produce diagnostic reagents or vaccines. The current “gold-standard” cell line is Vero, an interferon-deficient aneuploid line of kidney epithelial cells[37,38]. A549 and Madin–Darby canine kidney cell lines are mostly applied for influenza viruses[39].The most common cell line for the foot-and-mouth disease virus is the mammalian baby hamster kidney cells[40] which have been used since 1964[41]. Compared to cell lines, primary cell cultures have some advantages. For instance, cells isolated from ovine pulmonary adenocarcinoma are a unique platform to reveal mechanisms of epithelial transformation in a case of the lung cancer caused by the retrovirus infection[42]. Despite of the availability of cell variety and easy handling and scaling, 2D cultures are not capable of recapitulating fully cell-cell and cell-matrix interactions.
Suspension culture is also the oldest model for viral infections and very common for the industrial production of diagnostic reagents, vaccines, etc., due to a simple scaling procedure[40] V. However, it was significantly improved by adding microcarriers – small particles of a cell adhesive substrate (e.g., Cytodex 3). Such method modification was approved for the production of a RSV vaccine[43] and research on virus-host interactions[44].
Compared to 2D ones, 3D models are highly attractive because they are more relevant to the conditions in vivo (Figure 2). Such models can be fabricated through various approaches and were approved for different viruses (Table 1). The most common technique to form 3D tissue models is cell or spheroid/organoid encapsulation (embedding).
Figure 2.
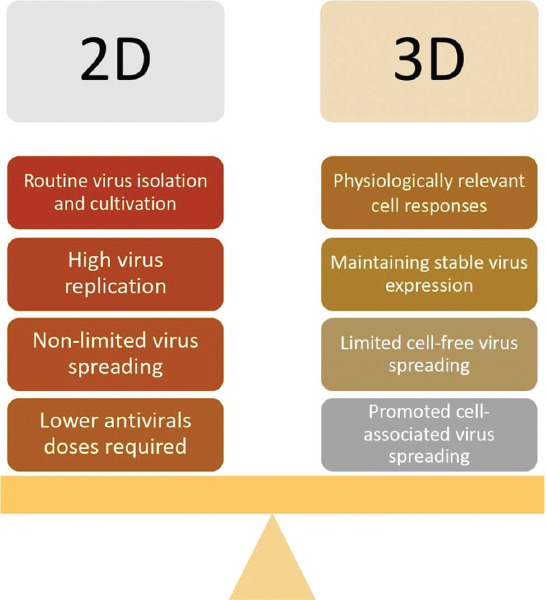
Viral infection: 2D versus 3D tissue models.
Table 1.
3D tissue models used to study various viral infections.
| Virus | Tissue model | Outcomes | Ref. | ||
|---|---|---|---|---|---|
| Cells | Biomaterial | Fabrication method | |||
| Respiratory viruses | |||||
| SARS-CoV-2 | Organoids from iPSCs and hESCs | Matrigel, collagen | Cultivation in non-adhesive well plates | • Blood vessel and human kidney organoids were susceptible to SARS-CoV-2 | [8] |
| • Viral infection can be blocked by hrsACE2 | |||||
| FCoV | BHK-21 | – | Microfluidic chip | • Virus entry caused by binding to specific receptors, protease-induced priming, and low-pH | [62] |
| • Kinetic rate parameters for the virus fusion were quantitatively measured | |||||
| MERS-CoV | hPIECs and their organoids, small intestine explants | Matrigel | Encapsulation | • Tissue models were successfully infected by the virus | [49] |
| RSV | Organoids from basal cells, multi-ciliated cells, and secretory cells | Basement membrane extract | Encapsulation | • RSV induced organoid cell motility through NS2 protein | [45] |
| RSV | Organoids from hPSCs | Matrigel | Encapsulation | • Lung bud organoids were representable and susceptible to the viral infection | [46] |
| IAV (H1N1 and H3N2) | hPSAECs | Chitosan-collagen scaffold | Air-liquid interface | • 3D model’s morphology was similar to in vivo condition | [39] |
| • 3D model had high aquaporin-5 and cytokeratin-14 expression | |||||
| • After being infected, cells had significant changes in marker protein expression and released pro-inflammatory cytokines | |||||
| IAV | A549 | Alginate, gelatin, Matrigel | Extrusion bioprinting | • The model was highly susceptible to the infection and represented the pattern similar to in vivo condition | [71] |
| • A high level of IL-29 (interferon λ1) was released | |||||
| IAV (H1N1 PR8) | A549, HeLa | – | Inkjet bioprinting | • Geometric heterogeneity influenced on cell susceptibility to the virus | [55] |
| Hepatotrophic viruses | |||||
| HCV | Huh7 | Cytodex-3 microcarrier beads | Cultivation in a rotating wall vessel bioreactor | • The phenotype of cells cultured in 3D were more hepatocyte-like | [72] |
| • 3D model is susceptible to HCV infection | |||||
| HCV | Huh7 | Matrigel | Encapsulation | • Embedded cells were polarized and formed interconnected proto-bile canaliculi structures | [73] |
| • They were easily infected by the virus and synthetized infective viral particles | |||||
| HCV | Huh7 | Mebiogel | Encapsulation | • Spheroids remained viable for 63 days and could be infected by the virus | [74] |
| Hepatotrophic viruses | |||||
| HBV | hPH | Collagen-coated polystyrene scaffolds | Microfluidic chip | • The 3D model was susceptible to infection and remained viable for up to 40 days | [75] |
| • It reproduced all steps of the viral lifecycle observed in vivo | |||||
| Herpesviruses | |||||
| HSV-1 | Human abdominal skin explant | – | Microneedle pretreatment | • The virus infection model was developed | [67] |
| • Infection-induced changes in histology and protein expression were revealed | |||||
| • Efficacy of pritelivir and acyclovir was similar | |||||
| HSV-1 | iPSC | Matrigel | Cultivation in Matrigel coated well plates | • IC50 of acyclovir in 2D cultures was lower than that in 3D cultures | [69] |
| HSV-2 | hNVECs | – | Air-liquid interface | • HNVECs and their spheres had normal morphology and expressed epithelial markers | [68] |
| • The hNVEC sphere-based model was permissive for the infection and representative for virus-induced pathological changes | |||||
| HCMV | PSCs | Basement membrane extract | Cultivation in coated well plates | • The salivary epithelial model was developed and susceptible to the virus infection | [76] |
| • The virus in lytic phase did not spread within the model and remained only in some cells. | |||||
| HCMV | Organoids from iPSCs | Matrigel | Cultivation in non-adhesive well plates; encapsulation | • Infection disrupted the activity of purinergic receptors and voltage-gated calcium channels, causing a decrease in basal calcium levels | [70] |
| • The virus disintegrated organoids, leading to insensitivity to response on stimulation | |||||
| HCMV | HFF | Collagen | Encapsulation in mini-construct chambers | • The cumulative tissue contractile force was decreased due to actin microfilament disruption | [77] |
| VZV | NHNP | Cultispher microcarrier beads | Cultivation in a rotating wall vessel bioreactor | • The model remained viable for 180 days and expressed markers of human trigeminal ganglia | [78] |
| • It was susceptible to the viral infection and could be maintained in long-term experiments | |||||
| Human immunodeficiency virus | |||||
| HIV-1 | Primary human CD4+ T-lymphocytes | Collagen | Encapsulation | • The integrated spatial model was developed | [52] |
| • Environment significantly influenced the transmission way | |||||
| • Cell motility and density determined the efficiency and way of the virus distribution | |||||
BHK-21: Baby hamster kidney-21, EBV: Epstein–Barr virus, FCoV: Feline coronavirus, HBV: Hepatitis B virus, HCMV: Human cytomegalovirus, HCV: Hepatitis C virus, hESCs: Human embryonic stem cells, HFF: Human foreskin fibroblasts, HIV-1: Human immunodeficiency virus-1, hNVECs: Human normal vaginal epithelial cells, hPH: Human primary hepatocytes, hPIECs: Human primary intestinal epithelial cells hPSAECs: Human primary small airway epithelial cells, hPSCs: Human pluripotent stem cells, hrsACE2: human recombinant soluble angiotensin converting enzyme 2, HSV-1: Herpes simplex virus-1, HSV-2: Herpes simplex virus-2, IAV: Influenza A virus, iPSCs: Induced pluripotent stem cells, MERS-CoV: Middle East respiratory syndrome-related coronavirus, NHNP: Normal human neural progenitor cells (NHNP), PSCs: Primary salivary cells, RSV: Respiratory syncytial virus, Ref: references, SARS-CoV-2: Severe acute respiratory syndrome-related coronavirus 2
Organoids and spheroids can establish cell-cell and cell-matrix interactions and are genotypically and phenotypically stable[45]. They were shown to be an efficient model to study virus infectivity and host-pathogen interaction[46-48]. For instance, using intestinal organoids, Zhou et al. confirmed that MERS-CoV might infect the gastrointestinal tract[49].
Explant cultures can also be used in studying viral infections. Their main advantage is that they are native tissues with relevant morphology. However, their application is significantly limited because of low availability and shortage of donor materials, short viability, and rapid necrosis[50].
Scaffold- and hydrogel-based models can provide a 3D microenvironment that mimics conditions in vivo for cells. Biomaterials that ensure necessary cell-matrix interactions and appropriate spatiotemporal surrounding cells are used to form a structure of such models. It was shown that they could ensure physiologically relevant cell responses to virus infection and drugs[39,51]. For instance, Bhowmick et al.[39] revealed that compared to monolayer culture, the 3D chitosan-collagen-based cell model had the native airway epithelium-like morphology and high expression and release of pro-inflammatory cytokines and chemokines after IAV infection. The virus expression in such conditions has been shown to be higher. Particularly, Archer et al.[42] found out that compared to monolayer cultures, cultures of tumor-derived alveolar type II cells on a surface coated with fibronectin and collagen type I or Matrigel exhibited efficient maintenance of reverse transcriptase activity and stable expression of Jaagsiekte sheep retrovirus. Moreover, biomaterials have been shown to significantly influence virus spreading ability and even determine its mode. For instance, Imle et al.[52] revealed that cell-laden collagen gel significantly limited the transmission of cell-free HIV and shifted it to cell-associated transmission. To fabricate complex tissue-like constructs, bioprinting is a good option[53], and bioprinted models were shown not only to be susceptible to viruses but also to recapitulate virus-associated morphological patterns similar to in vivo[54,55].
Microfluidic-based tissue models additionally allow mimicking air and fluid flows typical to in vivo conditions. Organ-on-a-chip systems consisting of various cell types, perfusion chambers, air-liquid interfaces, etc., mimic and create physiological conditions relevant to viral infection of native tissues. Microfluidic-based tissue models have many advantages. Particularly, microfluidics enables liquid handling at a microscale through a system of microchannels; therefore, the total consumption of reagents is relatively low that makes high-throughput screening easier and cheaper[56]. Such models are flexible to be automated[57,58], providing the possibility for real-time monitoring[59,60]. Moreover, they allow culturing cells in physiologically relevant dynamic conditions and controlling them[61]. Particularly, such system was tested to study the mechanism of the fusion of feline coronavirus with host cell membrane[62].
3 In vitro tissue models for modeling an infection caused by different viruses
3.1 Respiratory viruses
Tissue models that are used to study respiratory viral infections vary and include both monolayer cultures and functional airway organoids, enabling to obtain reliable data on virus infectivity, targets, and drug efficacy. Coronaviruses, a group of respiratory viruses, mostly infect epithelial cells that are used in designing relevant 3D models. The recent studies are based on organoids as a tissue model. For instance, Monteil et al.[8] revealed the efficacy of human recombinant soluble angiotensin-converting enzyme 2 against SARS-CoV-2 using infected blood vessel and kidney organoids. Moreover, intestinal organoids were used to prove that the intestine is a target organ for MERS-CoV[49]. Study of IAV using monolayer cultures fails to recapitulate the natural clustered pattern of disease transmission, but bioprinted 3D model was shown to be more relevant[54]. Using bioprinting, it was revealed that even geometrical position can significantly influence cell susceptibility to the virus[55]. Screening of drugs against RSV infection and detecting its pathogenesis was also successfully performed using airway and lung bud organoids[46,47].
3.2 Hepatotropic viruses
The hepatotropic viruses include different types and hepatitis A, B, C, D, and E viruses, which are the most common causes of viral hepatitis leading to liver failure worldwide[63]. Hepatitis B virus (HBV) and hepatitis C virus (HCV) induce chronic liver inflammation that results in cirrhosis and hepatocellular carcinoma[64]. Cell polarity and micro-environmental complexity and interactions are absent in 2D culture systems. Due to the drawbacks of 2D model systems, researchers are looking for alternative 3D models. The establishment of 3D models including spheroids, organoids with multi-cellular structures, and their specific extracellular matrix (ECM), was shown to exhibit higher tissue-specific environmental complexity, more mature cells, and better physiological functionality compared to simple 2D counterparts. For instance, several studies established liver spheroid models to study the hepatotrophic virus lifecycle in liver tissue. Chong et al. generated primary human hepatocyte spheroids from uninfected liver resections and inoculated the spheroids with HCV-positive serum[65]. Data showed that spheroids have differentiated phenotype and expressed putative HCV receptors; the HCV RNA was detected in the cells as well as supernatant of culture media[65]. Moreover, Nie et al. used a coculture system of human induced pluripotent stem cell (hiPSC)-derived endoderm, human umbilical vein endothelial cells, and mesenchymal stem cells in a 3D microwells to assess the potential of liver organoids for HBV infection and virus-host interactions[66]. The cells self-organized and differentiated into the functional liver organoids. Then, organoids were infected with the HBV genome. The liver organoids exhibited more functionality and higher susceptibility to HBV infection compared to human iPSC-derived 2D hepatic-like cells. These organoids could sustain HBV propagation and produce infectious virus up to 20 days. HBV infection decreased the expression of hepatic-specific genes and increased the expression of early biomarkers for acute liver failure, alanine aminotransferase, and lactate dehydrogenase in the supernatant of infected organoids. The advantage of hiPSC-derived 3D liver organoids was that they provided HBV infection models for precision medicine[66]. In addition, other cell culture models including, specific scaffold embedded cells and single-channel microfluidic devices are promising platforms in vitro models to study hepatotropic viruses[63].
3.3 Herpesviruses
Epithelial tissue is the initial site of infection for most herpesviruses. Although they cause latent infection mainly in neurons, they are still able to infect other cells. Therefore, there are numerous tissue models available, which include various cell types to study this virus group. For example, Tajpara et al. developed a model based on the microneedle-pretreated human abdominal skin explant to test antivirals and their combinations against human simplex virus (HSV-1) infection[67]. Zhu et al. fabricated 3D air-liquid interface culture consisting of human normal vaginal epithelial cells to describe viral transmission of HSV-2 and related pathological changes[68]. To study viral effects on neural tissue and acyclovir efficacy, D’Aiuto et al. proposed a scaffold-free 3D hiPSC-based neuronal model and showed that the IC50 of acyclovir in 2D cell cultures was lower than that in 3D culture[69]. Sison et al. also used iPSCs to fabricate cortical organoids to study human cytomegalovirus infection and revealed the organoid structure disruption and alterations in specific markers expression[70].
3.4 HIV
HIV infection is one the most difficult infections to study due to high selectivity and host specificity. HIV mainly infects immune cells, and the recent study has offered a novel efficient 3D model based on CD4+ T-lymphocytes[52]. By varying density of collagen gel embedding cells, Imle et al. evaluated the virus transmission and revealed that it can be significantly influenced by 3D environment.
4 Designing an “ideal” tissue model to study SARS-CoV-2 infection
4.1 Models and their limitations
To date, there is only a limited number of models available to study the SARS-CoV-2 infection. The most susceptible animals to this coronavirus are ferrets[3,4], cats[4], and rhesus and cynomolgus macaques[5,6]. The latter is considered to be a rapidly established SARS-CoV-2 model without any additional modifications[6]. Rhesus macaques were successfully applied to test siRNA to treat and prevent SARS-CoV infection[79]. Moreover, transgenic mice expressing human ACE2 can become a great option as ACE2 is one of the main SARS-CoV-2 targets[7]. Nevertheless, almost all animal models are expensive and do not allow researchers to fully overcome species specificity for such host-specific virus as well as their use is limited because of ethical issues.
Cell-based models are a good alternative to animal models. Particularly, Vero cell line cultured in 2D conditions was used for anti-SARS-CoV-2 drug screening[80]. However, the main limitation of such cell lines is the deficiency of interferon, which is an important regulator of binding proteins involved in SARS-CoV-2 infection[81]. This requires the use of other cell types and biofabrication methods that would better recapitulate in vivo conditions, including 3D environment, flows dynamics, and immune response. Moreover, blood vessel and human kidney organoids are still the only 3D models used to study SARS-CoV-2 infection[8].
4.2 Specific targets
Despite SARS-CoV-2 is a novel virus infecting humans, understanding of its possible entry mechanisms was already pre-defined because of earlier studies on other coronaviruses, for example, SARS-CoV[82-85]. Therefore, after its appearance, most research teams have been focusing on SARS-CoV receptor ACE2 and other related enzymes. Particularly, Hoffmann et al.[86] proved that the entry of SARS-CoV-2 into a cell occurs due to binding of the viral surface spike glycoprotein (S protein) to ACE2 and its priming by the transmembrane protease serine protease 2 (TMPRSS2). Zang et al. also revealed that the transmembrane protease serine protease 4 plays a crucial role in virus entry using human small intestinal enteroids as a model[87].
Actually, the entry mechanisms are considered the main targets for designing drugs treating and preventing COVID-19. Hence, their inhibition can block the infection that was proven to be true in several studies. For instance, Monteil et al. showed that human recombinant ACE2 prevented the virus entry into cells that form blood vessel and kidney organoids[8]. Hoffmann et al. revealed that Camostat mesylate, a serine protease inhibitor, blocked SARS-CoV-2 infection in lung cell line Calu-3[86].
These target enzymes, i.e., ACE2 and TMPRSS2, are widely expressed by tissues in human organisms. ACE2 is a monocarboxypeptidase that regulates the peptide cleavage in the renin-angiotensin system, and high levels of its expression could be found in alveolar epithelial type II cells, esophagus keratinocytes, small intestine, ileum and rectum enterocytes, stomach epithelial cells, colon colonocytes, liver cholangiocyte, arterial and venous endothelial cells, arterial smooth muscle cells, myocardial cells, sustentacular cells of the olfactory epithelium, spermatogonia and Leydig and Sertoli cells, prostate epithelial cells, bladder urothelial cells, and kidney proximal tubules cells[88-94]. TMPRSS2 regulating viral uptake by S protein priming is highly expressed by sustentacular cells of the olfactory epithelium, small intestine enterocytes, bronchial transient secretory cells, prostate epithelial cells, nasal goblet and ciliated cells, etc.[87,89,95-97].
4.3 Key points for the rational design
Our lack of knowledge in the COVID-19 pathogenesis and the absence of its adequate and licensed therapy[2] have led to the need to create novel in vitro platforms that mimic in vivo conditions and are specifically tailored for the SARS-CoV-2 infection. There is no doubt that 3D tissue models are more suitable than 2D models to study any viral infections because they share the similarity to the native tissue, organ structure, and physiological functionality, and this is also applicable to COVID-19.
3D tissue-like constructs can be fabricated by a huge variety of methods that can be divided into two main groups: Scaffold-based and scaffold-free. However, scaffold-free techniques such as bioprinting and cell self-organization (spheroidogenesis and organoidogenesis) and their combinations are considered to be the most promising ones as they allow precise reproduction of tissue morphological and functional properties.
Bioprinting is a complex technique that, particularly, enables tissue fabrication using spheroids or organoids (microtissues) as building blocks. Hence, the appropriate spatiotemporal status and cell-cell and cell-matrix contacts may be achieved[98]. In bioprinting, cells distributed in a hydrogel system (“bioink”) are usually deposited by a bioprinter, which can be based on different technologies such as extrusion[99], ink jet[100], laser-induced forward transfer (LIFT)[101], stereolithography[102]. Extrusion-based bioprinting is the most widely used technique[103]; however, only LIFT bioprinter can enable precise deposition at high speed and resolution and is considered to be the best option to print minor cell populations within complex 3D tissue-like structures[101]. Regarding bioinks, the most promising cell components are spheroids or organoids establish intercellular junctions and newly synthetized ECM compared to a single cell suspension and maintain cell phenotype[22,104-106]; the biomaterial component – hydrogel system – is usually presented by natural and synthetic polymers, including their conjugates such as acellularized ECM, alginate, gelatin, fibrin, hyaluronic acid, cellulose, polyethylene glycol, and Pluronic-F127.[101,107-109].
As specific targets for SARS-CoV-2 are ACE2 and TMPRSS2, it is rational to include those tissues whose cells express these enzymes in the COVID-19 test tissue platform. Particularly, there should be 3D models of the nasal mucosa (including the olfactory neuroepithelium), lungs (particularly, the alveoli), blood vessels, heart, kidney, and intestine (Figure 3). To date, scientists have accumulated data on their fabrication through bioprinting, and this experience is shortly described further.
Figure 3.
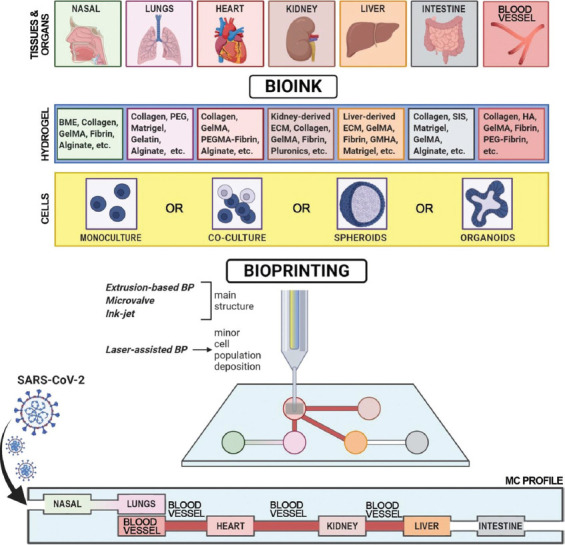
Designing an “ideal” tissue model to study severe acute respiratory syndrome-related coronavirus 2 infection. BME – basement memebrane extract; BP – bioprinting; BV – blood vessel; ECM – extracellular matrix; GelMA – gelatin methacrylated; GMHA – glycidyl methacrylated hyaluronic acid; MC – microfluidic chamber (chip); PEG – polyethylene glycol; PEGMA – polyethylene glycol monoacrylate; SARS-CoV-2 – severe acute respiratory syndrome-related coronavirus 2; SIS – small intestinal submucosa.
The nose is one of the main ports of SARS-CoV-2 infection, and the nasal mucosa is the first barrier tissue for the virus[81]. Among the existing models mimicking the nasal mucosa, none of them was fabricated through bioprinting. They are mainly presented by monolayers, air-fluid single or multilayered biointerfaces, and scaffold-based and explant-based cultures[110]. As the olfactory neuroepithelium located in the nasal mucosa is involved in virus entry and smell dysfunction[89,111], the rational model should contain sustentacular cells expressing high levels of ACE2 and TMPRSS2 and olfactory receptor neurons expressing these enzymes at lower levels. There are numerous efficient protocols to form olfactory neurospheres and to differentiate olfactory neuroepithelial cells[112-115]; therefore, these cells and their self-aggregates can be a perfect cell component for a bioink to print a “smell-sensitive” nasal mucosa construct.
The lungs, particularly the alveoli, are the main target for the SARS-CoV-2 infection and remain technically challenging. Only a limited number of studies achieved success in the 3D reconstruction of alveolar epithelial-endothelial barrier, and most scientists attempt to model only the air-cell and fluid-cell biointerfaces. For instance, Horvath et al.[116] bioprinted the epithelial/endothelial cell barrier system on a porous membrane and showed that it is possible to create reproducible thin homogenous cell layers. To date, the most complex lung-like structure was fabricated by Grigoryan et al. using a stereolithographic bioprinter[117]. To reproduce the alveoli scale and morphology, particularly their epithelial side, Lewis et al. created hollow epithelial cysts using the microsphere-based approach[118]. Such cysts as a cell component of a bioink can be easily hierarchically structured through bioprinting to achieve lung-like constructs.
Blood vessels containing ACE-expressing endothelial cells are a common object for bioprinting because they ensure the proper survivability and engraftment of tissue-engineered constructs. Different approaches varying in fabrication method and bioink blends were offered and can be classified as follows: Sacrificial and core/shell techniques. The choice depends on the required shape and sizes; vessels with bigger diameter can be fabricated using an extrusion-based bioprinter, vessels with smaller diameter using a laser-assisted bioprinter, and multibranched vessels using stereolithographic bioprinter[101,117,119,120].
Another target organ for COVID-19 that should be included in an integrated platform is the heart. Engineering cardiac tissues requires the restoration of their functionality. To date, there is a number of successful studies that can be used as a base. Particularly, Maiullari et al. bioprinted a cardiac vascularized construct from hiPSC-derived cardiomyocytes and umbilical vein endothelial cells. This construct had native tissue-like morphology and successfully grafted with host tissues and vasculature[121]. Zhang et al. showed a complex approach to fabricate a vascularized-myocardium-on-a-chip, which was able to contract, using the combination of both bioprinting and microfluidics[122].
The kidneys play a crucial role in COVID-19[123,124], but its biofabrication remains an appealing goal. For sure, scientists have achieved particular success which is mostly related to engineering miniaturized kidney models[125-129]. Particularly, Homan et al. bioprinted renal proximal tubules placed into a microfluidic chip and showed that such model had the typical epithelial morphology and was sensitive to cyclosporine A[127]. King et al. fabricated an in vitr o multicellular model consisting of renal fibroblasts, endothelial cells, and proximal tubule epithelial cells and revealed its susceptibility to cisplatin in a dose-dependent manner and response to TGFβ[130].
The liver is also challenging for bioprinting mainly because of its complex structure that includes microvasculature and innervation[131]. However, there is a number of studies achieved good results in the restoration of the liver morphology and functionality. For instance, Yanagi et al. developed an approach to fabricate liver-like tissues based on the fusion of the bioprinted spheroids[132]. In addition, Bhise et al. designed a bioprinted liver-on-a-chip and showed its full functionality for 30 days and sensitivity to acetaminophen-induced toxicity[133].
The intestine is highly susceptible to the SARS-CoV-2 infection because its epithelial cells express ACE2[90] involved in amino acid homeostasis[134]. Therefore, it is essential to include it as a target organ in the designed tissue model platform. There are numerous approaches for fabrication of intestinal models, bioprinting is considered to be a promising approach[135]. Particularly, Madden et al. showed that it is possible to fabricate a two-layered construct consisting of epithelial cells and myofibroblasts through bioprinting[136]. Such construct had clear morphology, and cells expressed villin, E-cadherin, ZO-1, and enzymes and proteins participating in xenobiotics metabolism (cytochrome P450 2C9, multidrug resistance protein 1, breast cancer resistance protein, etc.)
To mimic air and liquid flow for recapitulating the in vivo conditions, microfluidics can be used as a tool. Such systems can be fabricated using bioprinting[137-139] and have been already approved as both single organ (organ-on-a-chip)[131,140-143] and integrated (body-on-a-chip) platforms[144-146]. Multi-organ model systems are more physiologically relevant and permits better detection of complex virus-host effects than the first ones. Particularly, Maschmeyer et al. fabricated a four-organ-chip representing the intestine, the liver, the skin, and the kidney[144]. Later, Vernetti et al. offered a more complex system reproducing the microphysiology of coupled intestine, liver, kidney proximal tubule, blood–brain barrier, and skeletal muscle models[145]. However, there are only several platforms which were fabricated using bioprinting because the combination of these emerging techniques is a relatively new approach. For instance, Skardal et al. developed a three-tissue system consisting of functional lung, cardiac, and liver modules and proved its applicability for drug testing[146]. Compared to such 2D models, bioprinted models ensure complex cell-cell and cell-matrix interactions that are crucial in studying COVID-19 pathogenesis.
The most challenging aspect of designing COVID-19 tissue platforms is modeling immune response relevant for this disease[2]. Hence, the “ideal” system should represent effects of dendritic cells and macrophages that secret inflammatory cytokines and chemokines (Interleukin [IL]-6, IL-8, IL-12, tumor necrosis factor-α, monocyte chemoattractant protein-1, Granulocyte-macrophage colony-stimulating factor, etc.) and cytotoxic T cells (CD4+ and CD8+ T cells). Hence, two approaches might be applied. The first one is designing immune-competent models[147,148] or integrated platforms by including lymph node models[149,150]. The second approach is based on the perfusion of immune cells suspension through a chip.
The integrated platform (Figure 3) includes six target tissues/organs (nasal tissue, lungs, heart, kidney, liver, and intestine), including blood vessels. They connect to each other using microfluidic channels ensuring virus transmission, cell supply with nutrients and oxygen, cell migration, etc. Each particular model is bioprinted to achieve native-like morphology and functionality. Bioink consists of cell (yellow) and hydrogel (blue) components and is tissue-specific. The best option for a bioink cell component is organoids/spheroids as they can perform cell-cell and cell-matrix interactions. To model immune response, two approaches might be applied: Designing immune-competent models/including lymph node models into such integrated platform and perfusing immune cells suspension through it.
5 Conclusion
To defeat COVID-19, the mankind should create new tools combining the emerging techniques such as bioprinting, microfluidics, and organoid formation. To date, our understanding of the fabrication of tissue models for different viruses and tissue engineering is growing, and they can be applied in designing an integrated multi-tissue bioprinted platform tailored for SARS-CoV-2 infection. Despite that none has yet tested such complex systems to study this virus and perform drug screening, this multidisciplinary approach can be a new chapter in antiviral research in view of the outstanding achievements described in this review.
Acknowledgments
This work was supported by the Russian Science Foundation (18-15-00407, Introduction and Section 4.1), Russian Foundation for Basic Research (18-29-06059, Section 2; 20-04-60063, Section 4.2 and 4.3), and Russian academic excellence project 5–100 (Section 3 and Conclusions).
Conflict of interest
The authors declare that they have no conflict of interest.
Author contributions
A.Sh., E.G., V.M., and P.S. outlined the manuscript. P.B. and M.P. contributed to Section 2.3 and 3 (except 3.2), D.B. – Section 2.1, N.K. – Section 2.2, M.V., E.Z., and N.K.R. – Section 3.2, and A.S. – Section 4 and 5, A.Sv. – Section 1 and 2.3. I.Z. drafted and prepared Figure 3, including its description; D.B., E.Z., and N.K.R. – Figures 1 and 2. A.S. drafted the manuscript with primary editing and revision support from A.Sv., P.S., and V.M. A.Sh., A.Sv., D.B., E.Z., E.G., and N.K.R. prepared the paper’s revision to address reviewers’ comments. A.Sv., P.S., and E.G. coordinated the manuscript preparation. All authors read and approved the final manuscript.
References
- 1.Choong YYC, Tan HW, Patel DC, et al. The Global Rise of 3D Printing During the COVID-19 Pandemic. Nat Rev Mater. 2020:1–3. doi: 10.1038/s41578-020-00234-3. DOI: 10.1038/s41578-020-00234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hossein-Khannazer N, Shokoohian B, Shpichka A, et al. Novel Therapeutic Approaches for Treatment of COVID-19. J Mol Med. 2020;3:1–15. doi: 10.1007/s00109-020-01927-6. DOI:10.1007/s00109-020-01927-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park SJ, Yu KM, Kim YI, et al. Antiviral Efficacies of FDA-Approved Drugs against SARS-COV-2 Infection in Ferrets. MBio. 2020;11(3):ne01114–20. doi: 10.1128/mBio.01114-20. DOI:10.1128/mbio.01114-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi J, Wen Z, Zhong G, et al. Susceptibility of Ferrets, Cats, Dogs, and other Domesticated Animals to SARS-coronavirus 2. Science. 2020;368(6494):1016–20. doi: 10.1126/science.abb7015. DOI:10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Bras A. SARS-CoV-2 Causes COVID-19-Like Disease in Cynomolgus Macaques. Lab Anim. 2020;49(6):174. doi: 10.1038/s41684-020-0571-8. DOI:10.1038/s41684-020-0571-8. [DOI] [PubMed] [Google Scholar]
- 6.Rockx B, Kuiken T, Herfst S, et al. Comparative Pathogenesis of COVID-19, MERS, and SARS in a Nonhuman Primate Model. Science. 2020;368(6494):1012–5. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lutz C, Maher L, Lee C, et al. COVID-19 Preclinical Models:Human Angiotensin-Converting Enzyme 2 Transgenic Mice. Hum Genomics. 2020;14:20. doi: 10.1186/s40246-020-00272-6. DOI:10.1186/s40246-020-00272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monteil V, Kwon H, Prado P, et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell. 2020;181(4):905–13. doi: 10.1016/j.cell.2020.04.004. DOI:10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindesmith L, Moe C, Marionneau S, et al. Human Susceptibility and Resistance to Norwalk Virus Infection. Nat Med. 2003;9(5):548–53. doi: 10.1038/nm860. DOI:10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- 10.DeVincenzo JP, Wilkinson T, Vaishnaw A, et al. Viral Load Drives Disease in Humans Experimentally Infected with Respiratory Syncytial Virus. Am J Respir Crit Care Med. 2010;182(10):1305–14. doi: 10.1164/rccm.201002-0221OC. DOI:10.1164/rccm.201002-0221oc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Memoli MJ, Shaw PA, Han A, et al. Evaluation of Antihemagglutinin and Antineuraminidase Antibodies as Correlates of Protection in an Influenza A/H1N1 Virus Healthy Human Challenge Model. MBio. 2016;7(2):1–12. doi: 10.1128/mBio.00417-16. DOI:10.1128/mbio.00417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkinson TM, Li CK, Chui CS, et al. Preexisting Influenza-Specific CD4 +T Cells Correlate with Disease Protection against Influenza Challenge in Humans. Nat Med. 2012;18(2):274–80. doi: 10.1038/nm.2612. DOI:10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 13.He XS, Holmes TH, Zhang C, et al. Cellular Immune Responses in Children and Adults Receiving Inactivated or Live Attenuated Influenza Vaccines. J Virol. 2006;80(23):11756–66. doi: 10.1128/JVI.01460-06. DOI:10.1128/jvi.01460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang KY, Li CK, Clutterbuck E, et al. Virus-Specific Antibody Secreting Cell, Memory B-Cell, and Sero-antibody Responses in the Human Influenza Challenge Model. J Infect Dis. 2014;209(9):1354–61. doi: 10.1093/infdis/jit650. DOI:10.1093/infdis/jit650. [DOI] [PubMed] [Google Scholar]
- 15.Larsen CP, Whitehead SS, Durbin AP. Dengue Human Infection Models to Advance Dengue Vaccine Development. Vaccine. 2015;33(50):7075–82. doi: 10.1016/j.vaccine.2015.09.052. DOI:10.1016/j.vaccine.2015.09.052. [DOI] [PubMed] [Google Scholar]
- 16.Kirkpatrick BD, Whitehead SS, Pierce KK, et al. The Live Attenuated Dengue Vaccine TV003 Elicits Complete Protection Against Dengue in a Human Challenge Model. Sci Transl Med. 2016;8(330):330ra36. doi: 10.1126/scitranslmed.aaf1517. DOI:10.1126/scitranslmed.aaf1517. [DOI] [PubMed] [Google Scholar]
- 17.Henle W, Henle G, Stokes J., Jr Demonstration of the Efficacy of Vaccination Against Influenza Type A by Experimental Infection of Human Beings. J Immunol. 1943;46(3):163–75. [Google Scholar]
- 18.Jackson GG, Muldon RL, Akers LW. Serological Evidence for Prevention of Influenzal Infection in Volunteers by an Anti-influenzal Drug Adamantanamine Hydrochloride. Antimicrob Agents Chemother. 1963;161:703–7. [PubMed] [Google Scholar]
- 19.Calfee DP, Peng AW, Cass LM, et al. Safety and Efficacy of Intravenous Zanamivir in Preventing Experimental Human Influenza A Virus Infection. Antimicrob Agents Chemother. 1999;43(7):1616–20. doi: 10.1128/aac.43.7.1616. DOI:10.1128/aac.43.7.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayden FG, Zylidnikov DM, Iljenko VI, et al. Comparative Therapeutic Effect of Aerosolized and Oral Rimantadine HC1 in Experimental Human Influenza A Virus Infection. Antiviral Res. 1982;2(3):147–53. doi: 10.1016/0166-3542(82)90016-x. DOI:10.1016/0166-3542(82)90016-x. [DOI] [PubMed] [Google Scholar]
- 21.Hayden FG, Treanor JJ, Fritz RS, et al. Use of the Oral Neuraminidase Inhibitor Oseltamivir in Experimental Human Influenza Randomized Controlled Trials for Prevention and Treatment. JAMA. 1999;282(13):1240-6. doi: 10.1001/jama.282.13.1240. DOI:10.1001/jama.282.13.1240. [DOI] [PubMed] [Google Scholar]
- 22.Mironov V, Visconti R, Kasyanov V, et al. Organ Printing:Tissue Spheroids as Building Blocks. Biomaterials. 2009;30(12):2164–74. doi: 10.1016/j.biomaterials.2008.12.084. DOI:10.1016/j.biomaterials.2008.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gubareva LV, Kaiser L, Matrosovich MN, et al. Selection of Influenza Virus Mutants in Experimentally Infected Volunteers Treated with Oseltamivir. J Infect Dis. 2001;183(4):523–31. doi: 10.1086/318537. DOI:10.1086/31⅙. [DOI] [PubMed] [Google Scholar]
- 24.McCallin S, Sarker SA, Sultana S, et al. Metagenome Analysis of Russian and Georgian Pyophage Cocktails and a Placebo-Controlled Safety Trial of Single Phage Versus Phage Cocktail in Healthy Staphylococcus aureus Carriers. Environ Microbiol. 2018;20(9):3278–93. doi: 10.1111/1462-2920.14310. DOI:10.1111/1462-2920.14310. [DOI] [PubMed] [Google Scholar]
- 25.Jault P, Leclerc T, Jennes S, et al. Efficacy and Tolerability of a Cocktail of Bacteriophages to Treat Burn Wounds Infected by Pseudomonas aeruginosa (PhagoBurn):A Randomised, Controlled, Double-Blind Phase 1/2 Trial. Lancet Infect Dis. 2019;19(1):35–45. doi: 10.1016/S1473-3099(18)30482-1. DOI:10.1016/s1473-3099(18)30482-1. [DOI] [PubMed] [Google Scholar]
- 26.Febvre HP, Rao S, Gindin M, et al. PHAGE Study:Effects of Supplemental Bacteriophage Intake on Inflammation and Gut Microbiota in Healthy Adults. Nutrients. 2019;11(3):666. doi: 10.3390/nu11030666. DOI:10.3390/nu11030666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bem RA, Domachowske JB, Rosenberg HF. Animal Models of Human Respiratory Syncytial Virus Disease. Am J Physiol Lung Cell Mol Physiol. 2011;301:L148-56. doi: 10.1152/ajplung.00065.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radigan KA, Misharin AV, Chi M, et al. Modeling Human Influenza Infection in the Laboratory. Infect Drug Resist. 2015;8:311–20. doi: 10.2147/IDR.S58551. DOI:10.2147/idr.s58551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dowall SD, Graham VA, Rayner E, et al. A Susceptible Mouse Model for Zika Virus Infection. PLoS Negl Trop Dis. 2016;10(5):ne0004658. doi: 10.1371/journal.pntd.0004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ter Meulen J, Bakker AB, Van Den Brink EN, et al. Human Monoclonal Antibody as Prophylaxis for SARS Coronavirus Infection in Ferrets. Lancet. 2004;363(9427):2139–41. doi: 10.1016/S0140-6736(04)16506-9. DOI:10.1016/s0140-6736(04)16506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng PS, Böhm R, Hartley-Tassell LE, et al. Ferrets Exclusively Synthesize Neu5Ac and Express Naturally Humanized Influenza A Virus Receptors. Nat Commun. 2014;5(1):1–9. doi: 10.1038/ncomms6750. DOI:10.1038/ncomms6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hancock GE, Smith JD, Heers KM. Serum Neutralizing Antibody Titers of Seropositive Chimpanzees Immunized with Vaccines Coformulated with Natural Fusion and Attachment Proteins of Respiratory Syncytial Virus. J Infect Dis. 2000;181(5):1768–71. doi: 10.1086/315475. DOI:10.1086/315475. [DOI] [PubMed] [Google Scholar]
- 33.Clarke CJ, Watt NJ, Meredith A, et al. Respiratory Syncytial Virus-associated Bronchopneumonia in a Young Chimpanzee. J Comp Pathol. 1994;110(2):207–12. doi: 10.1016/s0021-9975(08)80191-0. DOI:10.1016/s0021-9975(08)80191-0. [DOI] [PubMed] [Google Scholar]
- 34.Perrin S. Make Mouse Studies Work. Nature. 2014;517:423–5. doi: 10.1038/507423a. [DOI] [PubMed] [Google Scholar]
- 35.Ito R, Takahashi T, Ito M. Humanized Mouse Models:Application to Human Diseases. J Cell Physiol. 2018;233:3723–8. doi: 10.1002/jcp.26045. DOI:10.1002/jcp.26045. [DOI] [PubMed] [Google Scholar]
- 36.Walsh NC, Kenney LL, Jangalwe S, et al. Humanized Mouse Models of Clinical Disease. Annu Rev Pathol Mech Dis. 2017;12(1):187–215. doi: 10.1146/annurev-pathol-052016-100332. DOI:10.1146/annurev-pathol-052016-100332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kistner O, Barrett PN, Mundt W, et al. Development of a Mammalian Cell (Vero) Derived Candidate Influenza Virus Vaccine. Vaccine. 1998;16(9-10):960–8. doi: 10.1016/s0264-410x(97)00301-0. DOI:10.1016/s0264-410x(97)00301-0. [DOI] [PubMed] [Google Scholar]
- 38.de Lang A, Osterhaus AD, Haagmans BL. Interferon-? and Interleukin-4 Downregulate Expression of the SARS Coronavirus Receptor ACE2 in Vero E6 Cells. Virology. 2006;353(2):474–81. doi: 10.1016/j.virol.2006.06.011. DOI:10.1016/j.virol.2006.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhowmick R, Derakhshan T, Liang Y, et al. A Three-Dimensional Human Tissue-Engineered Lung Model to Study Influenza A Infection. Tissue Eng Part A. 2018;24(19-20):1468–80. doi: 10.1089/ten.tea.2017.0449. DOI:10.1089/ten.tea.2017.0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dill V, Hoffmann B, Zimmer A, et al. Influence of Cell Type and Cell Culture Media on the Propagation of Foot-and-Mouth Disease Virus with Regard to Vaccine Quality. Virol J. 2018;15(1):1–11. doi: 10.1186/s12985-018-0956-0. DOI:10.1186/s12985-018-0956-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radlett PJ, Pay TW, Garland AJ, et al. The Use of BHK Suspension Cells for the Commercial Production of Foot and Mouth Disease Vaccines Over a Twenty Year Period. Dev Biol Stand. 1985;60:163–70. [PubMed] [Google Scholar]
- 42.Archer F, Jacquier E, Lyon M, et al. Alveolar Type II Cells Isolated from Pulmonary Adenocarcinoma:A Model for JSRV Expression In Vitro. Am J Respir Cell Mol Biol. 2007;36(5):534–40. doi: 10.1165/rcmb.2006-0285OC. DOI:10.1165/rcmb.2006-0285oc. [DOI] [PubMed] [Google Scholar]
- 43.Hayle AJ. Culture of Respiratory Syncytial Virus Infected Diploid Bovine Nasal Mucosa Cells on Cytodex 3 Microcarriers. Arch Virol. 1986;89(1–4):81–8. doi: 10.1007/BF01309881. DOI:10.1007/bf01309881. [DOI] [PubMed] [Google Scholar]
- 44.Gardner JK, Herbst-Kralovetz MM. Three-Dimensional Rotating Wall Vessel-Derived Cell Culture Models for Studying Virus-Host Interactions. Viruses. 2016;8:304. doi: 10.3390/v8110304. DOI:10.3390/v8110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sachs N, Papaspyropoulos A, Zomer-van Ommen DD, et al. Long-term Expanding Human Airway Organoids for Disease Modeling. EMBO J. 2019;38(4):ne100300. doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen YW, Huang SX, De Carvalho AL, et al. A Three-dimensional Model of Human Lung Development and Disease from Pluripotent Stem Cells. Nat Cell Biol. 2017;19(5):542–9. doi: 10.1038/ncb3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J, Li C, Sachs N, et al. Differentiated Human Airway Organoids to Assess Infectivity of Emerging Influenza Virus. Proc Natl Acad Sci USA. 2018;115(26):6822–7. doi: 10.1073/pnas.1806308115. DOI:10.1073/pnas.1806308115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ettayebi K, Crawford SE, Murakami K, et al. Replication of Human Noroviruses in Stem Cell-derived Human Enteroids. Science. 2016;353(6306):1387–93. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou J, Li C, Zhao G, et al. Human Intestinal Tract Serves as an Alternative Infection Route for Middle East Respiratory Syndrome Coronavirus. Sci Adv. 2017;3(11):eaao4966. doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grivel JC, Margolis L. Use of Human Tissue Explants to Study Human Infectious Agents. Nat Protoc. 2009;4(2):256–69. doi: 10.1038/nprot.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koban R, Neumann M, Daugs A, et al. A Novel Three-dimensional Cell Culture Method Enhances Antiviral Drug Screening in Primary Human Cells. Antiviral Res. 2018;150:20–9. doi: 10.1016/j.antiviral.2017.12.005. DOI:10.1016/j.antiviral.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 52.Imle A, Kumberger P, Schnellbächer ND, et al. Experimental and Computational Analyses Reveal that Environmental Restrictions Shape HIV-1 Spread in 3D Cultures. Nat Commun. 2019;10(1):2144. doi: 10.1038/s41467-019-09879-3. DOI:10.1038/s41467-019-09879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shokoohian B. Bio-printing Damaged Tissues:A Novel Approach in Regenerative Medicine. Mod Med Lab J. 2018;2(1):1–5. DOI:10.30699/mmlj17.2.1.1. [Google Scholar]
- 54.Berg J, Hiller T, Kissner MS, et al. Optimization of Cell-laden Bioinks for 3D Bioprinting and Efficient Infection with Influenza A Virus. Sci Rep. 8(1):1–13. doi: 10.1038/s41598-018-31880-x. DOI:10.1038/s41598-018-31880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park JA, Yoon S, Kwon J, et al. Freeform Micropatterning of Living Cells into Cell Culture Medium Using Direct Inkjet Printing. Sci Rep. 2017;7(1):14610. doi: 10.1038/s41598-017-14726-w. DOI:10.1038/s41598-017-14726-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du G, Fang Q, den Toonder JM. Microfluidics for Cell-Based High Throughput Screening Platforms a Review. Anal Chim Acta. 2016;903:36–50. doi: 10.1016/j.aca.2015.11.023. DOI:10.1016/j.aca.2015.11.023. [DOI] [PubMed] [Google Scholar]
- 57.Montanez-Sauri SI, Sung KE, Puccinelli JP, et al. Automation of Three-Dimensional Cell Culture in Arrayed Microfluidic Devices. J Lab Autom. 2011;16(3):171–85. doi: 10.1016/j.jala.2011.02.003. DOI:10.1016/j.jala.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kane KI, Moreno EL, Hachi S, et al. Automated Microfluidic Cell Culture of Stem Cell Derived Dopaminergic Neurons. Sci Rep. 2019;9(1):1–12. doi: 10.1038/s41598-018-34828-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Super A, Jaccard N, Marques MP, et al. Real-Time Monitoring of Specific Oxygen Uptake Rates of Embryonic Stem Cells in a Microfluidic Cell Culture Device. Biotechnol J. 2016;11(9):1179–89. doi: 10.1002/biot.201500479. DOI:10.1002/biot.201500479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vergani M, Carminati M, Ferrari G, et al. Multichannel Bipotentiostat Integrated with a Microfluidic Platform for Electrochemical Real-time Monitoring Of Cell Cultures. IEEE Trans Biomed Circuits Syst. 2012;6(5):498–507. doi: 10.1109/TBCAS.2012.2187783. DOI:10.1109/tbcas.2012.21≊3. [DOI] [PubMed] [Google Scholar]
- 61.Yum K, Hong SG, Healy KE, et al. Physiologically Relevant Organs on Chips. Biotechnol J. 2013;9(1):16–27. doi: 10.1002/biot.201300187. DOI:10.1002/biot.201300187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costello DA, Millet JK, Hsia CY, et al. Single Particle Assay of Coronavirus Membrane Fusion with Proteinaceous Receptor-embedded Supported Bilayers. Biomaterials. 2013;34(32):7895–904. doi: 10.1016/j.biomaterials.2013.06.034. DOI:10.1016/j.biomaterials.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poortahmasebi V, Baghi HB. Living in the Shadows of Hepatitis. Lancet Infect Dis. 2019;19(11):1171–2. doi: 10.1016/S1473-3099(19)30534-1. DOI:10.1016/s1473-3099(19)30534-1. [DOI] [PubMed] [Google Scholar]
- 64.Ringehan M, McKeating JA, Protzer U. Viral Hepatitis and Liver Cancer. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160274. doi: 10.1098/rstb.2016.0274. DOI:10.1098/rstb.2016.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chong TW, Smith RL, Hughes MG, et al. Primary Human Hepatocytes in Spheroid Formation to Study Hepatitis C Infection. J Surg Res. 2006;130(1):52–7. doi: 10.1016/j.jss.2005.04.043. [DOI] [PubMed] [Google Scholar]
- 66.Nie YZ, Zheng YW, Miyakawa K, et al. Recapitulation of Hepatitis B Virus Host Interactions in Liver Organoids from Human Induced Pluripotent Stem Cells. EBioMedicine. 2008;35:114–23. doi: 10.1016/j.ebiom.2018.08.014. DOI:10.1016/j.ebiom.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tajpara P, Mildner M, Schmidt R, et al. A Preclinical Model for Studying Herpes Simplex Virus Infection. J Invest Dermatol. 2019;139:673–82. doi: 10.1016/j.jid.2018.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu Y, Yang Y, Guo J, et al. Ex vivo2D and 3D HSV-2 Infection Model Using Human Normal Vaginal Epithelial Cells. Oncotarget. 2017;8(9):15267–82. doi: 10.18632/oncotarget.14840. DOI:10.18632/oncotarget.14840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.D'Aiuto L, Naciri J, Radio N, et al. Generation of Three-dimensional Human Neuronal Cultures:Application to Modeling CNS Viral Infections. Stem Cell Res Ther. 2018;9(1):134. doi: 10.1186/s13287-018-0881-6. DOI:10.1186/s13287-018-0881-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sison SL, O'Brien BS, Johnson AJ, et al. Human Cytomegalovirus Disruption of Calcium Signaling in Neural Progenitor Cells and Organoids. J Virol. 2019;93(17):954. doi: 10.1128/JVI.00954-19. DOI:10.1128/jvi.00954-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berg J, Hiller T, Kissner MS, et al. Optimization of Cell-laden Bioinks for 3D Bioprinting and Efficient Infection with Influenza A Virus. Sci Rep. 2018;8(1):13877. doi: 10.1038/s41598-018-31880-x. DOI:10.1038/s41598-018-31880-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sainz B, Tencate V, Uprichard SL. Three-Dimensional Huh7 Cell Culture System for the Study of Hepatitis C Virus Infection. Virol J. 2009;6:103. doi: 10.1186/1743-422X-6-103. DOI:10.1186/1743-422x-6-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Molina-Jimenez F, Benedicto I, Thi VL, et al. Matrigel-Embedded 3D Culture of Huh-7 Cells as a Hepatocyte-Like Polarized System to Study Hepatitis C Virus Cycle. Virology. 2012;425(1):31–9. doi: 10.1016/j.virol.2011.12.021. DOI:10.1016/j.virol.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 74.Rajalakshmy AR, Malathi J, Madhavan HN, et al. Mebiolgel, a Thermoreversible Polymer as a Scaffold for Three Dimensional Culture of Huh7 Cell Line with Improved Hepatocyte Differentiation Marker Expression and HCV Replication. Indian J Med Microbiol. 2015;33(4):554–9. doi: 10.4103/0255-0857.167330. DOI:10.4103/0255-0857.167330. [DOI] [PubMed] [Google Scholar]
- 75.Ortega-Prieto AM, Skelton JK, Wai SN, et al. 3D Microfluidic Liver Cultures as a Physiological Preclinical Tool for Hepatitis B Virus Infection. Nat Commun. 2018;9(1):682. doi: 10.1038/s41467-018-02969-8. DOI:10.1038/s41467-018-02969-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morrison KM, Beucler MJ, Campbell EO, et al. Development of a Primary Human Cell Model for the Study of Human Cytomegalovirus Replication and Spread within Salivary Epithelium. J Virol. 2018;93(3):ne01608–18. doi: 10.1128/JVI.01608-18. DOI:10.1128/jvi.01608-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lam V, Bigley T, Terhune SS, et al. A Method for Quantifying Mechanical Properties of Tissue Following Viral Infection. PLoS One. 2012;7(8):ne42197. doi: 10.1371/journal.pone.0042197. DOI:10.1371/journal.pone.0042197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goodwin TJ, McCarthy M, Osterrieder N, et al. Three-Dimensional Normal Human Neural Progenitor Tissue-Like Assemblies:A Model of Persistent Varicella-Zoster Virus Infection. PLoS Pathog. 2013;9(8):ne1003512. doi: 10.1371/journal.ppat.1003512. DOI:10.1371/journal.ppat.1003512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li BJ, Tang Q, Cheng D, et al. Using siRNA in Prophylactic and Therapeutic Regimens against SARS Coronavirus in Rhesus Macaque. Nat Med. 2005;11(9):944–51. doi: 10.1038/nm1280. DOI:10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jeon S, Ko M, Lee J, et al. Identification of Antiviral Drug Candidates against SARS-CoV-2 from FDA-approved Drugs. Antimicrob Agents Chemother. 2020;64(7):ne00819–20. doi: 10.1128/AAC.00819-20. DOI:10.1128/aac.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ziegler CG, Allon SJ, Nyquist SK, et al. SARS-CoV-2 Receptor ACE2 is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181(5):1016–35. doi: 10.1016/j.cell.2020.04.035. DOI:10.3410/f.737831436.793574366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li W, Moore MJ, Vasilieva N, et al. Angiotensin-Converting Enzyme 2 is a Functional Receptor for the SARS Coronavirus. Nature. 2003;426(6965):450–4. doi: 10.1038/nature02145. DOI:10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Glowacka I, Bertram S, Muller MA, et al. Evidence that TMPRSS2 Activates the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Membrane Fusion and Reduces Viral Control by the Humoral Immune Response. J Virol. 2011;85(9):4122–34. doi: 10.1128/JVI.02232-10. DOI:10.1128/jvi.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li F, Li W, Farzan M, et al. Structural Biology:Structure of SARS Coronavirus Spike Receptor-binding Domain Complexed with Receptor. Science. 2005;309(5742):1864–8. doi: 10.1126/science.1116480. DOI:10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 85.Shirato K, Kawase M, Matsuyama S. Middle East Respiratory Syndrome Coronavirus Infection Mediated by the Transmembrane Serine Protease TMPRSS2. J Virol. 2013;87(23):12552–61. doi: 10.1128/JVI.01890-13. DOI:10.1128/jvi.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. DOI:10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zang R, Castro MF, McCune BT, et al. TMPRSS2 and TMPRSS4 Promote SARS-CoV-2 Infection of Human Small Intestinal Enterocytes. Sci Immunol. 2020;5(47):1–15. doi: 10.1126/sciimmunol.abc3582. DOI:10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hamming I, Timens W, Bulthuis ML, et al. Tissue Distribution of ACE2 Protein, the Functional Receptor for SARS Coronavirus. A First Step in Understanding SARS Pathogenesis. J Pathol. 2004;203(2):631–7. doi: 10.1002/path.1570. DOI:10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bilinska K, Jakubowska P, Von Bartheld CS, et al. Expression of the SARS-CoV-2 Entry Proteins, ACE2 and TMPRSS2, in Cells of the Olfactory Epithelium:Identification of Cell Types and Trends with Age. ACS Chem Neurosci. 2020;11(11):1555–62. doi: 10.1021/acschemneuro.0c00210. DOI:10.1021/acschemneuro.0c00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harmer D, Gilbert M, Borman R, et al. Quantitative mRNA Expression Profiling of ACE 2, a Novel Homologue of Angiotensin Converting Enzyme. FEBS Lett. 2002;532(1–2):107–10. doi: 10.1016/s0014-5793(02)03640-2. DOI:10.1016/s0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- 91.Wang Z, Xu X. scRNA-seq Profiling of Human Testes Reveals the Presence of the ACE2 Receptor, A Target for SARS-CoV-2 Infection in Spermatogonia, Leydig and Sertoli Cells. Cells. 2020;9:920. doi: 10.3390/cells9040920. DOI:10.3390/cells9040920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Song H, Seddighzadeh B, Cooperberg MR, et al. Expression of ACE2, the SARS-CoV-2 Receptor, and TMPRSS2 in Prostate Epithelial Cells. Eur Urol. 2020;78(2):296–8. doi: 10.1016/j.eururo.2020.04.065. DOI:10.1101/2020.04.24.056259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qi F, Qian S, Zhang S, et al. Single Cell RNA Sequencing of 13 Human Tissues Identify Cell Types and Receptors of Human Coronaviruses. Biochem Biophys Res Commun. 2020;526(1):135–40. doi: 10.1016/j.bbrc.2020.03.044. DOI:10.1101/2020.02.16.951913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zou X, Chen K, Zou J, et al. Single-Cell RNA-seq Data Analysis on the Receptor ACE2 Expression Reveals the Potential Risk of Different Human Organs Vulnerable to 2019-nCoV Infection. Front Med. 2020;14(2):185–92. doi: 10.1007/s11684-020-0754-0. DOI:10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lukassen S, Chua RL, Trefzer T, et al. SARS -CoV-2 Receptor ACE 2 and TMPRSS 2 are Primarily Expressed in Bronchial Transient Secretory Cells. EMBO J. 2020;39(10):1–15. doi: 10.15252/embj.20105114. DOI:10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen YW, Lee MS, Lucht A, et al. TMPRSS2, a Serine Protease Expressed in the Prostate on the Apical Surface of Luminal Epithelial Cells and Released into Semen in Prostasomes, is Misregulated in Prostate Cancer Cells. Am J Pathol. 2010;176(6):2986–96. doi: 10.2353/ajpath.2010.090665. DOI:10.2353/ajpath.2010.090665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sungnak W, Huang N, Bécavin C, et al. SARS-CoV-2 Entry Factors are Highly Expressed in Nasal Epithelial Cells Together with Innate Immune Genes. Nat Med. 2020;26(5):681–7. doi: 10.1038/s41591-020-0868-6. DOI:10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ng WL, Chua CK, Shen YF. Print Me An Organ!Why We Are Not There Yet. Prog Polym Sci. 2019;97:101145. DOI:10.1016/j.progpolymsci.2019.101145. [Google Scholar]
- 99.Ozbolat IT, Hospodiuk M. Current Advances and Future Perspectives in Extrusion-Based Bioprinting. Biomaterials. 2016;76:321–43. doi: 10.1016/j.biomaterials.2015.10.076. DOI:10.1016/j.biomaterials.2015.10.076. [DOI] [PubMed] [Google Scholar]
- 100.Saunders RE, Derby B. Inkjet Printing Biomaterials for Tissue Engineering:Bioprinting. Int Mater Rev. 2014;59(8):430–48. DOI:10.1179/1743280414y.0000000040. [Google Scholar]
- 101.Antoshin AA, Churbanov SN, Minaev NV, et al. LIFT-Bioprinting, is it Worth it? Bioprinting. (e00052) 2019;15 DOI:10.1016/j.bprint.2019e00052. [Google Scholar]
- 102.Ng WL, Lee JM, Zhou M, et al. Vat Polymerization-Based Bioprinting Process, Materials, Applications and Regulatory Challenges. Biofabrication. 2020;12(2):22001. doi: 10.1088/1758-5090/ab6034. DOI:10.1088/1758-5090/ab6034. [DOI] [PubMed] [Google Scholar]
- 103.Matai I, Kaur G, Seyedsalehi A, et al. Progress in 3D Bioprinting Technology for Tissue/Organ Regenerative Engineering. Biomaterials. 2020;226:119536. doi: 10.1016/j.biomaterials.2019.119536. DOI:10.1016/j.biomaterials.2019.119536. [DOI] [PubMed] [Google Scholar]
- 104.Zurina I, Shpichka A, Saburina I, et al. 2D/3D Buccal Epithelial Cell Self-Assembling as a Tool for Cell Phenotype Maintenance and Fabrication of Multilayered Epithelial Linings In Vitro. Biomed Mater. 2018;13(5):054104. doi: 10.1088/1748-605X/aace1c. DOI:10.1088/1748-605x/aace1c. [DOI] [PubMed] [Google Scholar]
- 105.Moldovan NI, Hibino N, Nakayama K. Principles of the Kenzan Method for Robotic Cell Spheroid-Based Three-Dimensional Bioprinting. Tissue Eng Part B Rev. 2017;23(3):237–44. doi: 10.1089/ten.TEB.2016.0322. DOI:10.1089/ten.teb.2016.0322. [DOI] [PubMed] [Google Scholar]
- 106.Gorkun AA, Shpichka AI, Zurina IM, et al. Angiogenic Potential of Spheroids from Umbilical Cord and Adipose-Derived Multipotent Mesenchymal Stromal Cells within Fibrin Gel. Biomed Mater. 2018;13(4):44108. doi: 10.1088/1748-605X/aac22d. DOI:10.1088/1748-605x/aac22d. [DOI] [PubMed] [Google Scholar]
- 107.Shpichka A, Osipova D, Efremov Y, et al. Fibrin-based Bioinks:New Tricks from an Old Dog. Int J Bioprinting. 2020;6(3):1–14. doi: 10.18063/ijb.v6i3.269. DOI:10.18063/ijb.v6i3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kornev VA, Grebenik EA, Solovieva AB, et al. Hydrogel-assisted Neuroregeneration Approaches Towards Brain Injury Therapy:A State-of-the-Art Review. Comput Struct Biotechnol J. 2018;16:488–502. doi: 10.1016/j.csbj.2018.10.011. DOI:10.1016/j.csbj.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shpichka AI, Konarev PV, Efremov YM, et al. Digging Deeper:Structural Background of PEGylated fi Brin Gels in Cell Migration and lumenogenesis. RSC Adv. 2020;10:4190–200. doi: 10.1039/c9ra08169k. DOI:10.1039/c9ra08169k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.De Rudder C, Arroyo MC, Lebeer S, et al. Modelling Upper Respiratory Tract Diseases:Getting Grips on Host-microbe Interactions in Chronic Rhinosinusitis Using In Vitro Technologies. Microbiome. 2018;6(1):75. doi: 10.1186/s40168-018-0462-z. DOI:10.1186/s40168-018-0462-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang L, Shen Y, Li M, et al. Clinical Manifestations and Evidence of Neurological Involvement in 2019 Novel Coronavirus SARS-CoV-2:A Systematic Review and Meta-analysis. J Neurol 2020. 2020:1–13. doi: 10.1007/s00415-020-09974-2. DOI:10.1007/s00415-020-09974-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li ST, Young TH, Huang TW. Poly (ethylene-co-vinyl alcohol) is a Suitable Substrate for Human Olfactory Neuroepithelial Cell Differentiation In Vitro through a Defined Regulatory Pathway. Acta Biomater. 2018;68:204–13. doi: 10.1016/j.actbio.2017.12.029. DOI:10.1016/j.actbio.2017.12.029. [DOI] [PubMed] [Google Scholar]
- 113.Li ST, Young TH, Wang CT, et al. Chitosan Films Promote Formation of Olfactory Neurospheres and Differentiation of Olfactory Receptor Neurons. Rhinology. 2018;56(4):336–42. doi: 10.4193/Rhin17.155. DOI:10.4193/rhin17.155. [DOI] [PubMed] [Google Scholar]
- 114.Du L, Zou L, Wang Q, et al. A Novel Biomimetic Olfactory Cell-based Biosensor with DNA-Directed Site-Specific Immobilization of Cells on a Microelectrode Array. Sens Actuators B Chem. 2015;217:186–92. DOI:10.1016/j.snb.2014.08.054. [Google Scholar]
- 115.Skaat H, Ziv-Polat O, Shahar A, et al. Enhancement of Migration, Growth and Differnatiation of Nasal Olfactory Mucosa Cells by Growth Factor-conjugated Fluorescent-maghemite Nanoparticles. Bioconjugate Chem. 2011;22(12):2600–10. doi: 10.1021/bc200454k. DOI:10.1021/bc200454k. [DOI] [PubMed] [Google Scholar]
- 116.Horvath L, Umehara Y, Jud C, et al. Engineering an In Vitro Air-blood Barrier by 3D Bioprinting. Sci Rep. 2015;5:7974. doi: 10.1038/srep07974. DOI:10.1038/srep07974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Grigoryan B, Paulsen SJ, Corbett DC, et al. Multivascular Networks and Functional Intravascular Topologies within Biocompatible Hydrogels. Science. 2019;364(6439):458–64. doi: 10.1126/science.aav9750. DOI:10.1126/science.aav9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lewis KJ, Tibbitt MW, Zhao Y, et al. In Vitro Model Alveoli from Photodegradable Microsphere Templates. Biomater Sci. 2015;3(6):821–32. doi: 10.1039/c5bm00034c. DOI:10.1039/c5bm00034c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ozbolat IT, Moncal KK, Gudapati H. Evaluation of Bioprinter Technologies. Addit Manuf. 2017;13:179–200. DOI:10.1016/j.addma.2016.10.003. [Google Scholar]
- 120.Miri AK, Nieto D, Iglesias L, et al. Microfluidics-Enabled Multimaterial Maskless Stereolithographic Bioprinting. Adv Mater. 2018;30(27):ne1800242. doi: 10.1002/adma.201800242. DOI:10.1002/adma.201⇾01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Maiullari F, Costantini M, Milan M, et al. A Multi-cellular 3D Bioprinting Approach for Vascularized Heart Tissue Engineering Based on HUVECs and iPSC-Derived Cardiomyocytes. Sci Rep. 2018;8(1):1–15. doi: 10.1038/s41598-018-31848-x. DOI:10.1038/s41598-018-31848-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang YS, Arneri A, Bersini S, et al. Bioprinting 3D Microfibrous Scaffolds for Engineering Endothelialized Myocardium and Heart-on-a-chip. Biomaterials. 2016;110:45–59. doi: 10.1016/j.biomaterials.2016.09.003. DOI:10.1016/j.biomaterials.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pan XW, Xu D, Zhang H, et al. Identification of a Potential Mechanism of Acute Kidney Injury During the COVID-19 Outbreak:A Study Based on Single-Cell Transcriptome Analysis. Intensive Care Med. 2020;46:1114-1116. doi: 10.1007/s00134-020-06026-1. DOI:10.1007/s00134-020-06026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ronco C, Reis T. Kidney Involvement in COVID-19 and Rationale for Extracorporeal Therapies. Nat Rev Nephrol. 2020;16(6):308–10. doi: 10.1038/s41581-020-0284-7. DOI:10.1038/s41581-020-0284-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Subramanian B, Rudym D, Cannizzaro C, et al. Tissue-Engineered Three-Dimensional In Vitro Models for Normal and Diseased Kidney. Tissue Eng Part A. 2010;16(9):2821–31. doi: 10.1089/ten.tea.2009.0595. DOI:10.1089/ten.tea.2009.0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sochol RD, Gupta NR, Bonventre JV. A Role for 3D Printing in Kidney-on-a-Chip Platforms. Curr Transplant Rep. 2016;3(1):82–92. doi: 10.1007/s40472-016-0085-x. DOI:10.1007/s40472-016-0085-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Homan KA, Kolesky DB, Skylar-Scott MA, et al. Bioprinting of 3D Convoluted Renal Proximal Tubules on Perfusable Chips. Sci Rep. 2016;6:34845. doi: 10.1038/srep34845. DOI:10.1038/srep34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ali M, Kumar A, Yoo JJ, Zahran F, et al. A Photo-Crosslinkable Kidney ECM-Derived Bioink Accelerates Renal Tissue Formation. Adv Healthc Mater. 2019;8(7):1800992. doi: 10.1002/adhm.201800992. DOI:10.1002/adhm.201800992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chuah JKC, Zink D. Stem Cell-Derived Kidney Cells and Organoids:Recent Breakthroughs and Emerging Applications. Biotechnol Adv. 2017;35(2):150–67. doi: 10.1016/j.biotechadv.2016.12.001. DOI:10.1016/j.biotechadv.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 130.King SM, Higgins JW, Nino CR, et al. 3D Proximal Tubule Tissues Recapitulate Key Aspects of Renal Physiology to Enable Nephrotoxicity Testing. Front Physiol. 2017;8(1):1–18. doi: 10.3389/fphys.2017.00123. DOI:10.3389/fphys.2017.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Heydari Z, Najimi M, Mirzaei H, et al. Tissue Engineering in Liver Regenerative Medicine:Insights into Novel Translational Technologies. Cells. 2020;9(2):304. doi: 10.3390/cells9020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yanagi Y, Nakayama K, Taguchi T, et al. In Vivo and Ex Vivo Methods of Growing a Liver Bud Through Tissue Connection. Sci Rep. 2017;7(1):1–15. doi: 10.1038/s41598-017-14542-2. DOI:10.1038/s41598-017-14542-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bhise NS, Manoharan V, Massa S, et al. A Liver-on-a-Chip Platform with Bioprinted Hepatic Spheroids. Biofabrication. 2016;8(1):14101. doi: 10.1088/1758-5090/8/1/014101. [DOI] [PubMed] [Google Scholar]
- 134.Hashimoto T, Perlot T, Rehman A, et al. ACE2 Links Amino Acid Malnutrition to Microbial Ecology and Intestinal Inflammation. Nature. 2012;487(7408):477–81. doi: 10.1038/nature11228. DOI:10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Williams CF, Walton GE, Jiang L, et al. Comparative Analysis of Intestinal Tract Models. Annu Rev Food Sci Technol. 2015;6(1):329–50. doi: 10.1146/annurev-food-022814-015429. DOI:10.1146/annurev-food-022814-015429. [DOI] [PubMed] [Google Scholar]
- 136.Madden LR, Nguyen TV, Garcia-Mojica S, et al. Bioprinted 3D Primary Human Intestinal Tissues Model Aspects of Native Physiology and ADME/Tox Functions. iScience. 2018;2:156–67. doi: 10.1016/j.isci.2018.03.015. DOI:10.1016/j.isci.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ma J, Wang Y, Liu J. Bioprinting of 3D Tissues/Organs Combined with Microfluidics. RSC Adv. 2018;8(39):21712–27. doi: 10.1039/c8ra03022g. DOI:10.1039/c8ra03022g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yu F, Choudhury D. Microfluidic Bioprinting for Organ-on-a-Chip Models. Drug Discov Today. 2019;24(6):1248–57. doi: 10.1016/j.drudis.2019.03.025. DOI:10.1016/j.drudis.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 139.Miri AK, Mostafavi E, Khorsandi D, et al. Bioprinters for Organs-on-Chips. Biofabrication. 2019;11(4):42002. doi: 10.1088/1758-5090/ab2798. DOI:10.1088/1758-5090/ab2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Koroleva A, Deiwick A, Nguyen A, et al. Hydrogel-Based Microfluidics for Vascular Tissue Engineering. BioNanoMaterials. 2016;17(1–2):19–32. DOI:10.1515/bnm-2015-0026. [Google Scholar]
- 141.Ashammakhi N, Wesseling-Perry K, Hasan A, et al. Kidney-on-a-Chip:Untapped Opportunities. Kidney Int. 2018;94(6):1073–86. doi: 10.1016/j.kint.2018.06.034. DOI:10.1016/j.kint.2018.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ribas J, Sadeghi H, Manbachi A, et al. Cardiovascular Organ-on-a-Chip Platforms for Drug Discovery and Development. Appl Vitr Toxicol. 2016;2(2):82–96. doi: 10.1089/aivt.2016.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jahromi MA, Abdoli A, Rahmanian M, et al. Microfluidic Brain-on-a-Chip:Perspectives for Mimicking Neural System Disorders. Mol Neurobiol. 2019;56(12):8489–512. doi: 10.1007/s12035-019-01653-2. DOI:10.1007/s12035-019-01653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Maschmeyer I, Lorenz AK, Schimek K, et al. A Four-Organ-Chip for Interconnected Long-Term Co-culture of Human Intestine, Liver, Skin and Kidney Equivalents. Lab Chip. 2015;15(12):2688–99. doi: 10.1039/c5lc00392j. DOI:10.1039/c5lc00392j. [DOI] [PubMed] [Google Scholar]
- 145.Vernetti L, Gough A, Baetz N, et al. Functional Coupling of Human Microphysiology Systems:Intestine, Liver, Kidney Proximal Tubule, Blood-Brain Barrier and Skeletal Muscle. Sci Rep. 2017;7(1):1–14. doi: 10.1038/srep42296. DOI:10.1038/srep44517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Skardal A, Murphy SV, Devarasetty M, et al. Multi-Tissue Interactions in an Integrated Three-Tissue Organ-on-a-Chip Platform. Sci Rep. 2017;7(1):1–16. doi: 10.1038/s41598-017-08879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ramadan Q, Ting FC. In Vitro Micro-Physiological Immune-Competent Model of the Human Skin. Lab Chip. 2016;16(10):1899–908. doi: 10.1039/c6lc00229c. DOI:10.1039/c6lc00229c. [DOI] [PubMed] [Google Scholar]
- 148.Harrington H, Cato P, Salazar F, et al. Immunocompetent 3D Model of Human Upper Airway for Disease Modeling and In Vitro Drug Evaluation. Mol Pharm. 2014;11(7):2082–91. doi: 10.1021/mp5000295. DOI:10.1021/mp5000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gopalakrishnan N, Hannam R, Casoni GP, et al. Infection and Immunity on a Chip:A Compartmentalised Microfluidic Platform to Monitor Immune Cell Behaviour in Real Time. Lab Chip. 2015;15(6):1481–7. doi: 10.1039/c4lc01438c. DOI:10.1039/c4lc01438c. [DOI] [PubMed] [Google Scholar]
- 150.Rosa PM, Gopalakrishnan N, Ibrahim H, et al. The Intercell Dynamics of T Cells and Dendritic Cells in a Lymph Node-on-a-Chip Flow Device. Lab Chip. 2016;16(19):3728–40. doi: 10.1039/c6lc00702c. DOI: 10.1039/c6lc00702c. [DOI] [PubMed] [Google Scholar]


