Endicott et al. report that inhibition of class I PI3K activates chaperone-mediated autophagy (CMA) in cultured cells and mice. Selective inhibition of class III PI3K does not activate CMA. This work disentangles previously misunderstood roles of PI3Ks in CMA regulation.
Abstract
Chaperone-mediated autophagy (CMA) is the most selective form of lysosomal proteolysis, where individual peptides, recognized by a consensus motif, are translocated directly across the lysosomal membrane. CMA regulates the abundance of many disease-related proteins, with causative roles in neoplasia, neurodegeneration, hepatosteatosis, and other pathologies relevant to human health and aging. At the lysosomal membrane, CMA is inhibited by Akt-dependent phosphorylation of the CMA regulator GFAP. The INS-PI3K-PDPK1 pathway regulates Akt, but its role in CMA is unclear. Here, we report that inhibition of class I PI3K or PDPK1 activates CMA. In contrast, selective inhibition of class III PI3Ks does not activate CMA. Isolated liver lysosomes from mice treated with either of two orally bioavailable class I PI3K inhibitors, pictilisib or buparlisib, display elevated CMA activity, and decreased phosphorylation of lysosomal GFAP, with no change in macroautophagy. The findings of this study represent an important first step in repurposing class I PI3K inhibitors to modulate CMA in vivo.
Introduction
Chaperone-mediated autophagy (CMA) is the most selective form of lysosomal proteolysis. Individual proteins bearing a pentapeptide consensus motif resembling Lys-Phe-Glu-Arg-Gln (KFERQ) are recognized by the cytosolic chaperone HSPA8 (also called Hsc70), which, along with other chaperones, participates in the trafficking of the substrate to the lysosomal membrane (Agarraberes and Dice, 2001; Chiang et al., 1989; Dice, 1990). Cytosolic chaperones unfold the substrate and facilitate its translocation into the lysosome via the CMA translocation complex, consisting primarily of LAMP2A (encoded by splice variant A of the Lamp2 transcript; Cuervo and Dice, 1996; Kaushik and Cuervo, 2018; Agarraberes and Dice, 2001).
CMA plays a role in preventing disease pathogenesis by degrading proteins with causative roles in hepatosteatosis, neoplasia, and a number of neurodegenerative diseases (Gomes et al., 2017; Schneider et al., 2014; Kaushik and Cuervo, 2018). Notably, rats with reduced CMA in the brain quickly exhibit loss of dopaminergic neurons in the substantia nigra and display Parkinson’s-like motor symptoms (Xilouri et al., 2016). Mutated forms of α-synuclein resist degradation by CMA and bind to the LAMP2A receptor, blocking other peptides from being degraded by this pathway (Cuervo et al., 2004). CMA also regulates the abundance of Tau (MAPT), Huntingtin (HTT), amyloid precursor protein (APP), LRRK2, TDP-43 (TARDBP), and other neurodegeneration-associated peptides (Wang et al., 2009; Huang et al., 2014; Qi et al., 2012; Orenstein et al., 2013; Park et al., 2016; Kaushik and Cuervo, 2018). The clear role of CMA in controlling the abundance of neurodegeneration-related proteins suggests that pharmacological interventions for increasing CMA in vivo could have clinical use for treatment or prevention of human neurodegenerative disease.
Small molecule compounds that activate CMA in cultured cells have been identified (Finn et al., 2005; Anguiano et al., 2013). However, a lack of clinically safe and orally bioavailable compounds for activating CMA in vivo has limited the potential of using CMA for therapeutic purposes. Understanding the basic biology of CMA control is a necessary step in developing the translational potential of this important protein regulatory pathway.
CMA is regulated by Akt on the lysosomal membrane. Akt phosphorylates lysosomal glial fibrillary acidic protein (GFAP; Arias et al., 2015). In its unphosphorylated form, GFAP binds the cytosolic tail of LAMP2A and promotes the multimerization of LAMP2A into the CMA translocation complex, facilitating substrate uptake (Bandyopadhyay et al., 2010; 2008). Thus, Akt mediated GFAP phosphorylation inhibits CMA (Arias et al., 2015). Lysosomal Akt activity is regulated by the opposing action of mechanistic target of rapamycin complex 2 (mTORC2) and PHLPP1. mTORC2 phosphorylates and activates Akt. PHLPP1 dephosphorylates and deactivates Akt (Arias et al., 2015). It is unclear whether there are other lysosomal GFAP kinases (in addition to Akt), and the GFAP phosphatases relevant to CMA are unknown.
Recent characterization of mTORC2 structure and signaling activity has revealed a positive-feedback loop between Akt and mTORC2, downstream of the insulin (INS)–phosphatidylinositol 3-kinase (PI3K)–3-phosphoinositide–dependent protein kinase 1 (PDPK1) signaling axis (Yang et al., 2015). Downstream of INS signaling, class I PI3Ks phosphorylate plasma membrane PtdIns-4,5-P2 to generate PtdIns-3,4,5-P3. Through their pleckstrin homology (PH) domains, which bind to PtdIns-3,4,5-P3, PDPK1 and Akt are recruited to the plasma membrane, where their proximity allows PDPK1 to phosphorylate Akt on T308 (Fruman et al., 2017). Subsequently, Akt phosphorylates essential mTORC2 component MAPKAP1/mSIN1 on T86 (Humphrey et al., 2013; Yang et al., 2015). This increases mTORC2 activity, and mTORC2 reciprocally phosphorylates Akt on S473, leading to a full activation of Akt (Humphrey et al., 2013; Yang et al., 2015; Cameron et al., 2011). Ablation of MAPKAP1 prevents Akt phosphorylation on S473 (Frias et al., 2006). There are multiple splice variants of MAPKAP1, which have different subcellular localizations and differentially regulate mTORC2 activity (Frias et al., 2006). Some MAPKAP1 variants possess a PH domain that is important for mTORC2 membrane localization, but the role of the PH domain in the Akt feedback loop is not fully clear (Yuan et al., 2015). There is at least one MAPKAP1 splice variant that is not responsive to INS signaling, and can facilitate phosphorylation of Akt by mTORC2 even in the absence of INS (Frias et al., 2006). The activity of this variant might be regulated by other signals or by localization. While it is clear that the cross-talk between Akt and mTORC2 downstream of class I PI3Ks and PDPK1 is important for regulating Akt activity, it is not known whether phosphorylation of lysosomal GFAP, and consequently CMA activity, is regulated by INS-PI3K-PDPK1.
Previous studies have reported conflicting results about the relationship between PI3K signaling pathways and CMA. Inhibition of the class III PI3K, Vps34, perturbs vesicle trafficking and inhibits macroautophagy (Petiot et al., 2000; Zeng et al., 2006). Treatment of IMR-90 (human fetal lung) cells with three inhibitors of all classes of PI3K (wortmannin, LY294002, and 3-methyladenine) showed no effect on CMA, despite inhibition of macroautophagy (Finn et al., 2005). A later study found that 3-methyladenine activates CMA in NIH3T3 cells (Kaushik et al., 2008). The authors attributed the increase in CMA to a compensatory response to the inhibition of macroautophagy, but this study did not control for the effects of 3-methyladenine on class I PI3K signaling, i.e., Akt (Kaushik et al., 2008). These conflicting results justify a study that selectively inhibits class I or class III PI3Ks, carefully controlling for effects on Akt and macroautophagy, to disentangle the roles of these two pathways in regulating CMA.
Here, we report that selective inhibition of class I PI3Ks is sufficient to activate CMA in three cultured cell types, in the presence of serum. Inhibition of PDPK1 activates CMA, further confirming that the INS signaling cascade directly inhibits CMA. Selective inhibition of the class III PI3K, Vps34, inhibits macroautophagy, but does not block Akt activation, nor does it activate CMA. Inhibition of macroautophagy by knocking down Atg5 does activate CMA, consistent with previous reports, indicating that the mechanism of CMA activation in response to macroautophagy inhibition is more nuanced than previously understood. Finally, we treated young mice with two class I PI3K inhibitors, and found that these drugs increased CMA in liver lysosomes, accompanied by a decrease in lysosomal GFAP phosphorylation. We concluded that suppression of the class I PI3K pathway in vivo is sufficient to modulate lysosomal GFAP and promote CMA.
Results
Inhibition of class I PI3Ks activates CMA in NIH3T3 cells
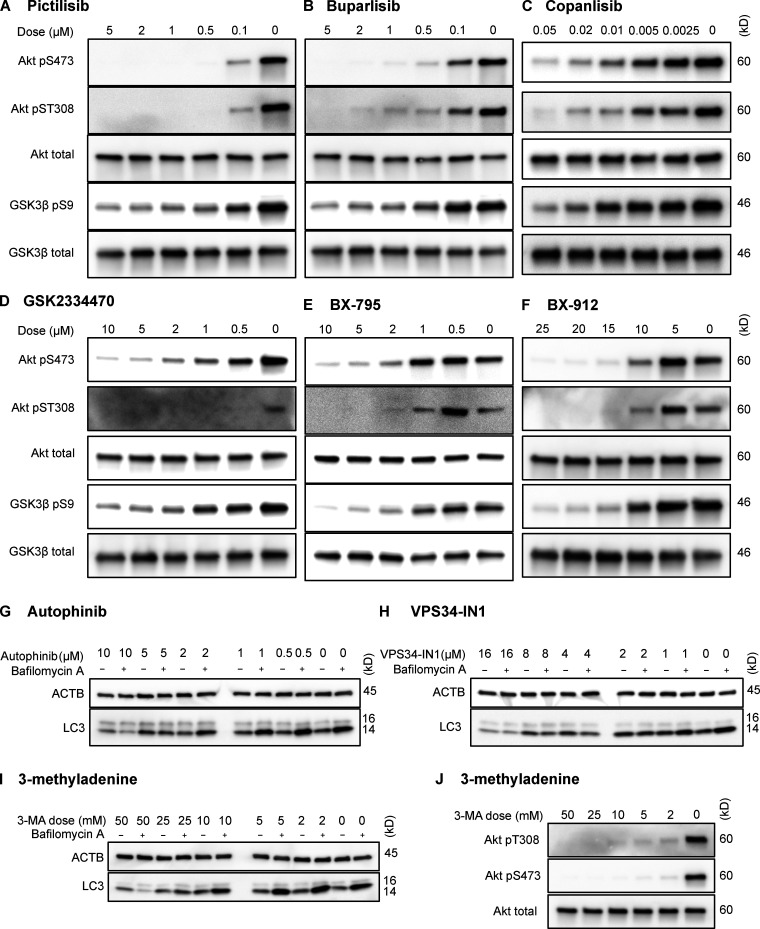
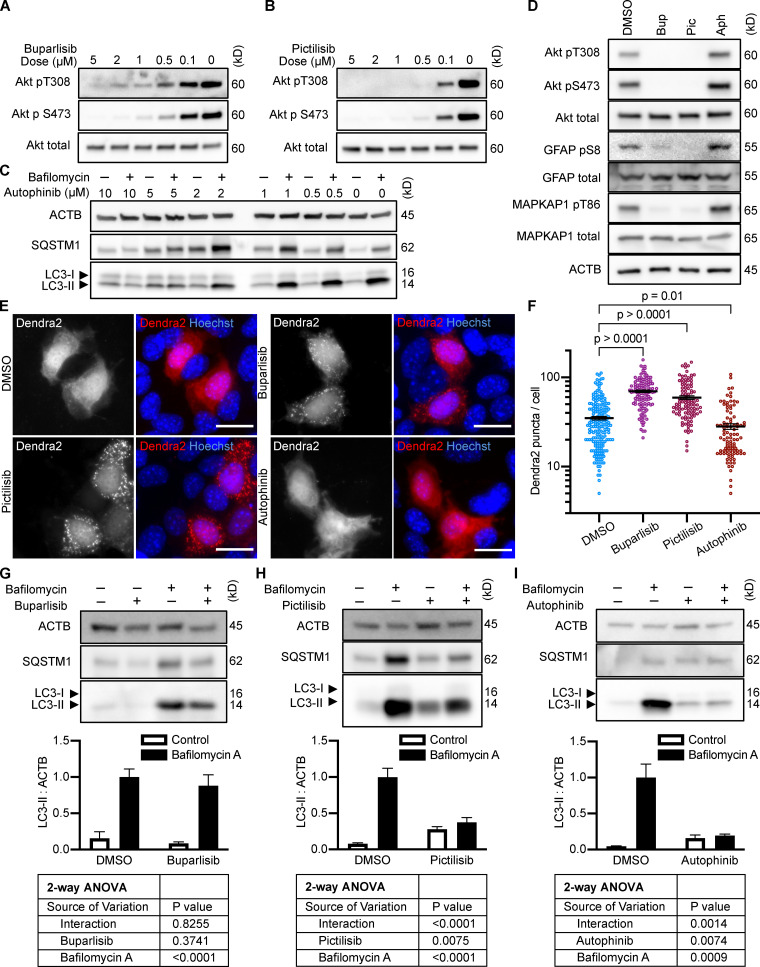
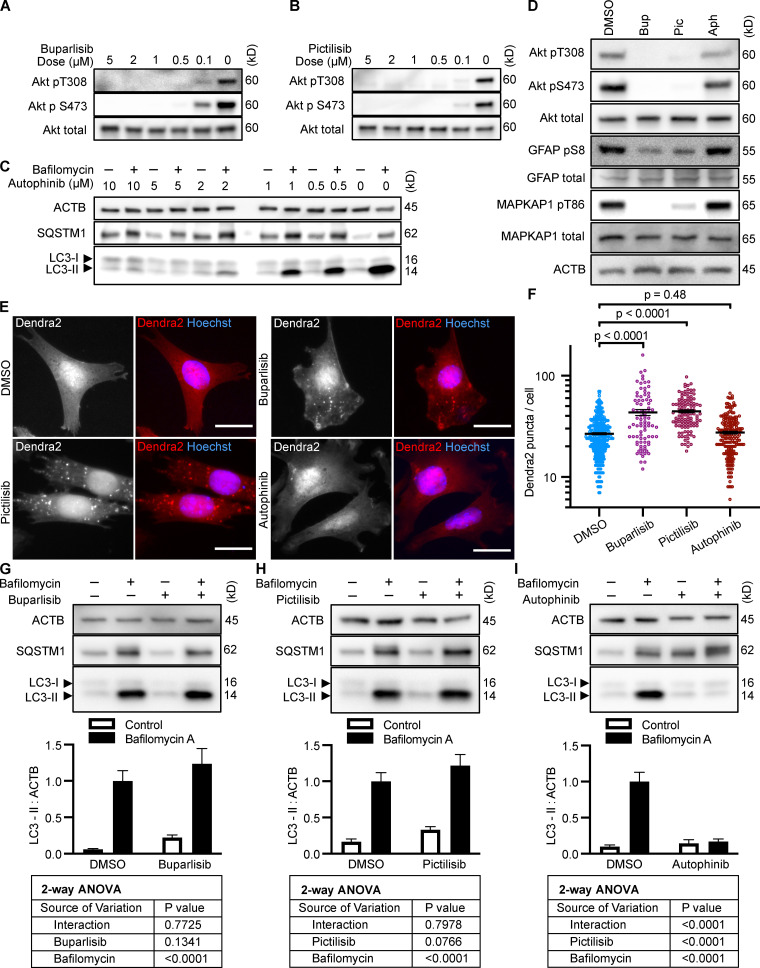
To test whether inhibition of intracellular signaling downstream of INS is sufficient to activate CMA, we selected three structurally distinct inhibitors of all four isoforms of the class I PI3K catalytic domain, p110-α, p110-β, p110-γ, and p110-δ, i.e., buparlisib, pictilisib, and copanlisib. PI3K inhibitors commonly have off-target effects, varying with the inhibitor’s structure (Crabbe et al., 2007). We reasoned that if multiple chemically distinct inhibitors consistently give the same biological effect, then it is unlikely that the measured result arises from inhibition of off-target kinases. We first performed dose curves in NIH3T3 cells to identify the lowest dose of each inhibitor that maximizes Akt inhibition (to reduce the chances of nonspecific kinase inhibition), as judged by the phosphorylation status of Akt and its own target, GSK3β (Fig. S1, A–C). Subsequent experiments in NIH3T3 cells were performed using 2 µM buparlisib, 500 nM pictilisib, and 50 nM copanlisib, unless otherwise specified.
Figure S1.
Dose curves for drugs used in NIH3T3 cells. Dose curves for each of the inhibitors used in this study are presented: pictilisib (A), buparlisib (B), copanlisib (C), GSK2334470 (D), BX795 (E), BX912 (F), autophinib (G), VPS34-IN1 (H), 3-methyladenine effects on macroautophagy (I), and 3-methyladenine effects on Akt (J). Drug doses are displayed above each lane in μM. ACTB, β-Actin.
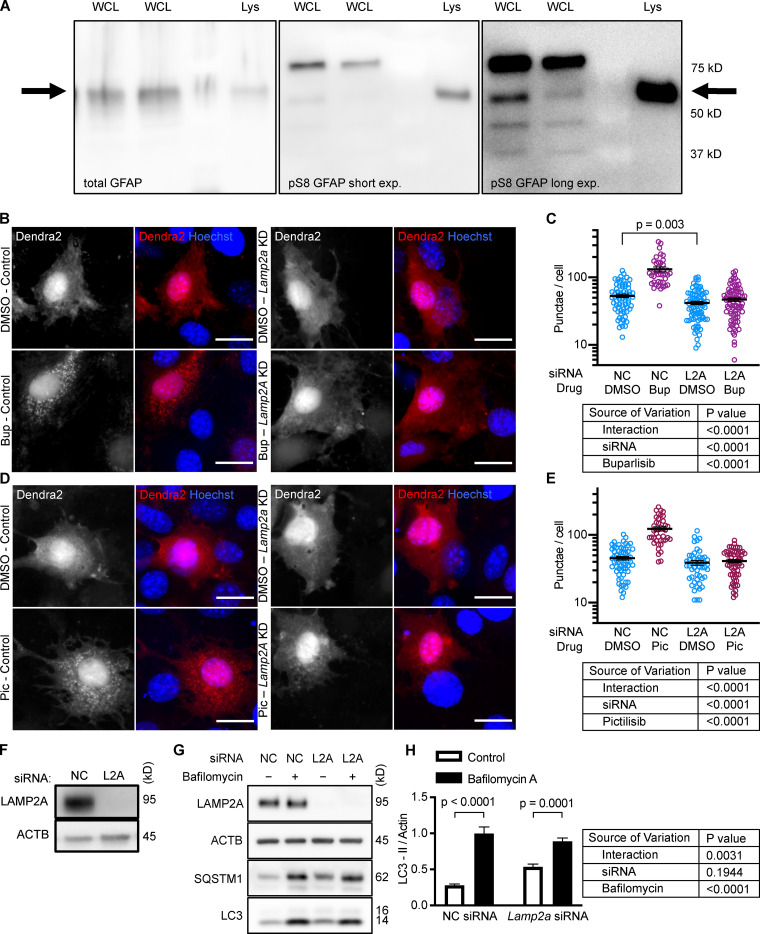
Next, we tested whether inhibiting p110 is sufficient to decrease phosphorylation of GFAP. We previously found that GFAP and GFAP phospho-S8 (pGFAP) antibodies can detect multiple bands in whole tissue lysates (Endicott et al., 2020) and whole cell lysates, but that the antibodies detect a single band on lysosomes. We found that the pGFAP band in whole cell lysate that corresponds in size to the lysosomal band correlates very tightly with lysosomal pGFAP and CMA activity (Endicott et al., 2020). We identified the lysosomal pGFAP and GFAP bands in NIH3T3 cells as the ∼55 kD bands (Fig. S2 A). For all subsequent experiments, only these bands are quantified in graphs and presented in representative images.
Figure S2.
Controls for GFAP isoforms and Dendra2 reporter. (A) Displays which band of GFAP and pGFAP from NIH3T3 whole cell lysate (WCL) is the same molecular weight as the GFAP and pGFAP from purified lysosomes (Lys). (B–E) NIH3T3 cells depleted of LAMP2A by siRNA do not form more Dendra2 puncta in response to buparlisib and pictilisib treatment. For B–E, data were pooled from at least three independent experiments. In C, NC-DMSO = 68 cells, NC-Bup = 40 cells, Lamp2a-DMSO = 80 cells, and Lamp2a-Bup = 81 cells. In E, NC-DMSO = 62 cells, NC-Pic = 41 cells, Lamp2A-DMSO = 50 cells, and Lamp2A-Pic = 64 cells. (F) Control for knockdown efficiency under conditions used in B–E. (G and H) NIH3T3 cells treated with siRNA against Lamp2a show a small but significant decrease in LC3-II flux, indicating macroautophagy impairment, n = 6. NC is negative control siRNA. P values written above brackets are derived from unpaired t tests. Two-way ANOVA tables are displayed next to graphs where appropriate. Error bars are SEM. Scale bars are 20 µm. ACTB, β-Actin; Bup, buparlisib; exp., exposure; Pic, pictilisib.
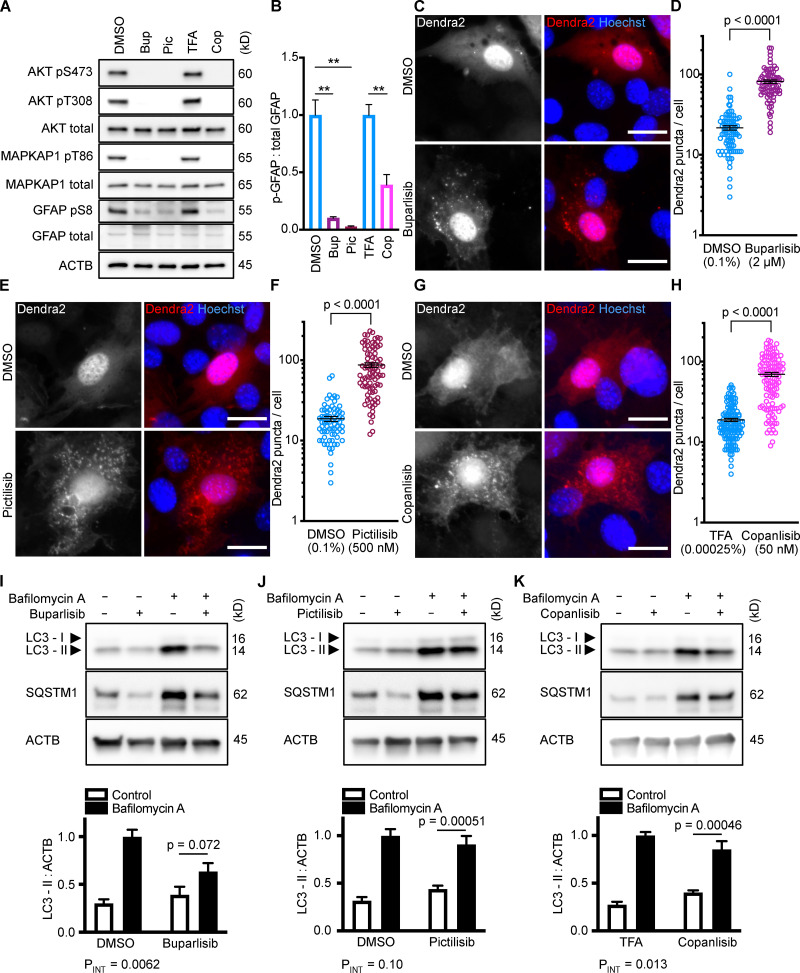
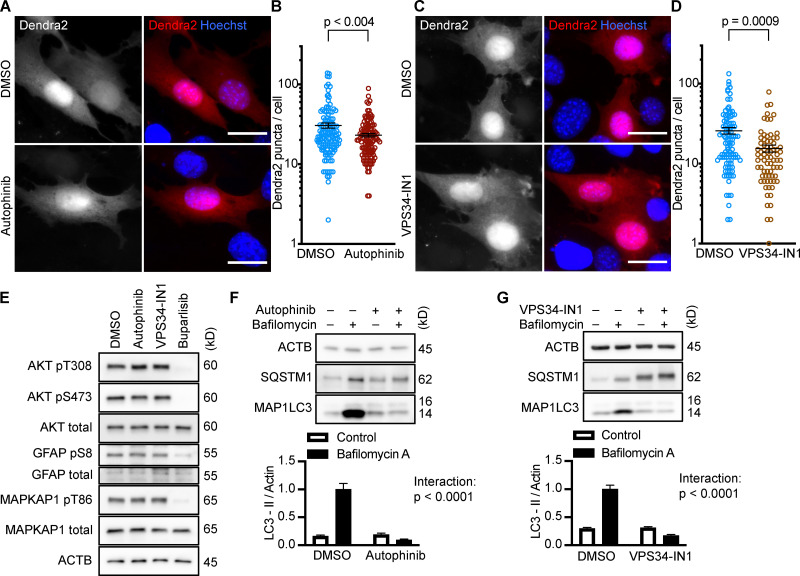
All three p110 inhibitors sharply reduced phosphorylation of Akt on T308 and S473 and MAPKAP1 on T86, as expected (Fig. 1 A). GFAP phosphorylation was also significantly reduced by all three p110 inhibitors, relevant to their solvent controls (Fig. 1, A and B). A reduction in GFAP phosphorylation suggests the hypothesis that CMA activity is increased.
Figure 1.
Inhibiting p110 activates CMA in NIH3T3 cells. (A) Western blots showing effects of selected doses of buparlisib (2 µM), pictilisib (500 nM), and copanlisib (50 nM) on phosphorylation of Akt, MAPKAP1, and GFAP. Blots are representative of six experimental replicates. DMSO is the solvent control for buparlisib and pictilisib. TFA is the solvent control for copanlisib. (B) Quantification of GFAP phosphorylation change in response to p110 inhibitors, as shown in A, n = 6. For C–H, data points are pooled from at least three independent experiments. (C and D) 10 h buparlisib treatment induces the accumulation of Dendra2 CMA reporter puncta. DMSO = 92 cells; buparlisib = 83 cells. (E and F) 10 h pictilisib treatment induces the accumulation of Dendra2 CMA reporter puncta. DMSO = 71 cells; pictilisib = 86 cells. (G and H) 10 h copanlisib treatment induces the accumulation of Dendra2 CMA reporter puncta. TFA = 132 cells; copanlisib = 116 cells. (I–K) Effects of buparlisib, pictilisib, and copanlisib on macroautophagy, as measured by LC3-II flux, n = 6. For all experiments, cells were maintained in complete growth medium with 10% serum. P values written above brackets are derived from unpaired t tests. **, P < 0.01 by unpaired t test. For charts in I–K, PINT is the interaction term P value from a two-way ANOVA. Error bars are SEM. Scale bars are 20 µm. ACTB, β-Actin; Bup, buparlisib; Cop, copanlisib; Pic, pictilisib; TFA, trifluoroacetic acid.
To test for CMA activation in response to buparlisib, pictilisib, and copanlisib, we used a genetically encoded CMA reporter, consisting of the N terminus of RNase A fused to the photo-switchable fluorescent protein Dendra2, similar to what has been reported by other studies (Koga et al., 2011; Arias et al., 2015; Anguiano et al., 2013). The N terminus of RNase A contains a CMA targeting motif, and when CMA is active, the reporter accumulates on lysosomal membranes (Cuervo et al., 1994; Koga et al., 2011). This can be visualized as an accumulation of fluorescent puncta in the cytoplasm. We found that all three p110 inhibitors caused a very clear increase in the number of cytosolic Dendra2 puncta, suggesting an increase in CMA (Fig. 1, C–H).
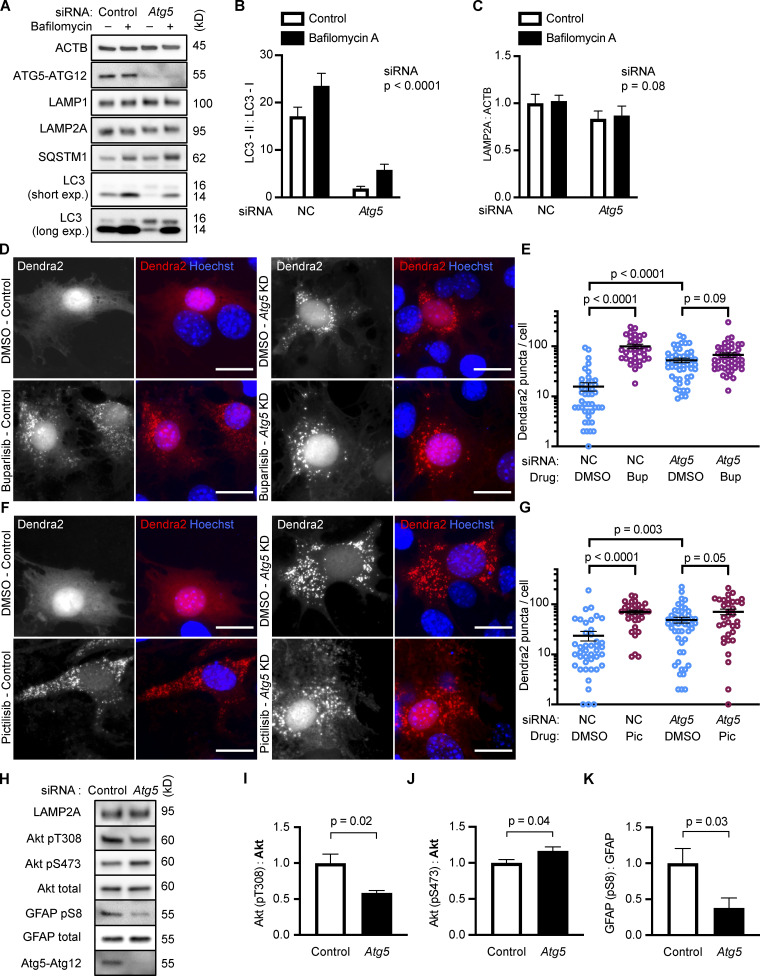
To verify that the increase in puncta is not attributable to another translocation event, we repeated the experiment in cells treated with siRNA targeting exon 8A of the Lamp2 transcript (exon 8A is unique to the Lamp2a transcript, and not found in Lamp2b or Lamp2c) or a nontargeting siRNA control. We tested buparlisib and pictilisib in the siRNA-treated cells, finding that the increase in the number of Dendra2 puncta in response to both drugs was completely blocked in cells with Lamp2a knocked down (Fig. S2, B–E). A representative Western blot controlling for successful LAMP2A protein depletion is shown in (Fig. S2 F). We note that knocking down Lamp2a does not completely abolish the presence of Dendra2 puncta, even in the DMSO-treated cells, potentially due to a trace amount of residual LAMP2A protein. But because knocking down Lamp2a completely blocks the response to the drugs, we conclude that Dendra2 puncta formation in response to the drugs is LAMP2A-dependent, suggesting that this well-established assay (Koga et al., 2011; Arias et al., 2015) is valid under the experimental conditions used here. We note that under our conditions, Lamp2a knockdown caused a small but statistically significant decrease in LC3-II flux, suggesting a small decrease in macroautophagy (Fig. S2, G and H).
Because CMA is reported to increase when macroautophagy is inhibited, we measured LC3-II flux (an established method for estimating macroautophagic flux) in NIH3T3 cells in parallel to the CMA experiments, using the same drug doses and treatment time. Cells were treated with a p110 inhibitor or its appropriate solvent control, either with or without bafilomycin A, to inhibit macroautophagy. Here, LC3-II flux is defined as the difference in the amount of LC3-II between the control and bafilomycin A–treated groups. To determine if the drugs had an effect on LC3-II flux, we used a two-way ANOVA to determine if the effects of the drugs interacted with the effects of bafilomycin A on LC3-II levels. We found that buparlisib caused a significant reduction in LC3-II flux, as indicated by a two-way ANOVA interaction term p value (PINT) = 0.0062 (Fig. 1 I). Pictilisib did not cause a significant effect on LC3-II flux (Fig. 1 J). Copanlisib caused a small but significant decrease in LC3-II flux (two-way ANOVA PINT = 0.013), although flux was clearly still intact (Fig. 1 K). Together, these data suggest that inhibiting p110 causes an increase in CMA, whether or not macroautophagy is also inhibited.
Inhibition of PDPK1 activates CMA in NIH3T3 cells
Translocation of PDPK1 to the plasma membrane in response to the generation of PtdIns-3,4,5-P3 by p110 is an essential step in the Akt activation cascade. To evaluate whether the effects of inhibiting p110 on CMA are mediated by PDPK1, as opposed to another PtdIns-3,4,5-P3–related signaling event, we next inhibited PDPK1 with three selective inhibitors, BX795, BX912, and GSK2334470. As before, we performed dose curves to identify the lowest dose of each drug that maximized the inhibition of Akt under our culture conditions (Fig. S1, D–F). For all subsequent experiments in NIH3T3 cells, we used doses of 5 µM BX795, 15 µM BX912, and 5 µM GSK2334470.
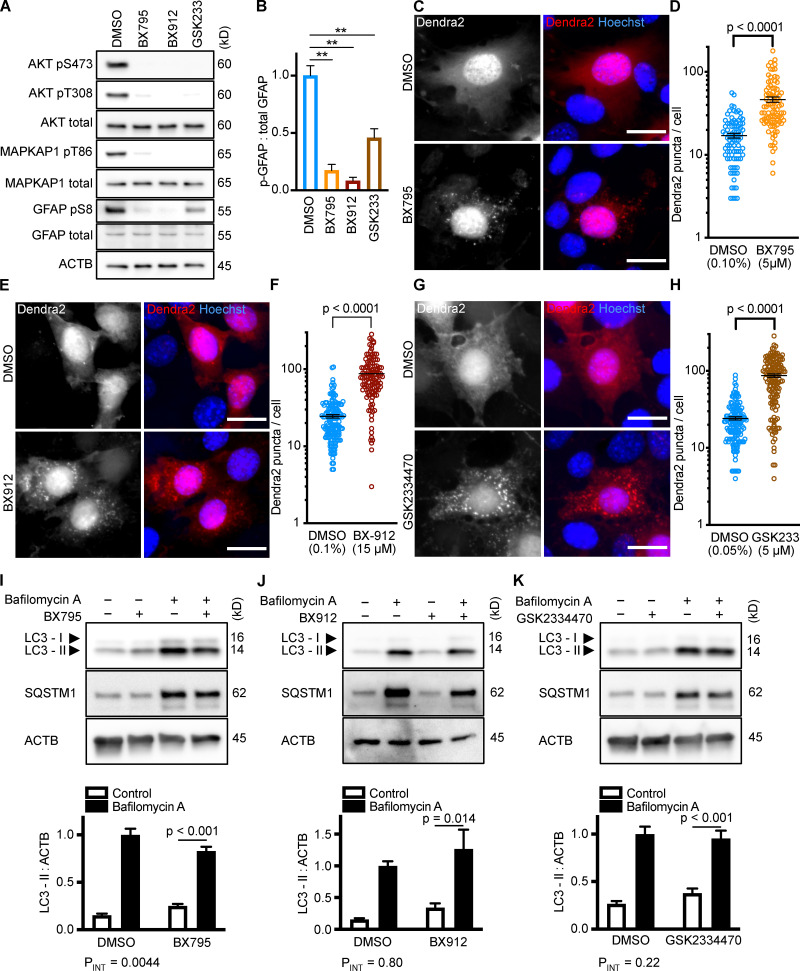
As expected, each PDPK1 inhibitor sharply reduced the phosphorylation of Akt on S473 and T308 and of MAPKAP1 on T86 (Fig. 2 A). Furthermore, each drug significantly reduced GFAP phosphorylation (Fig. 2, A and B). Each drug significantly increased the accumulation of Dendra2 puncta, suggesting that all three PDPK1 inhibitors activate CMA in NIH3T3 cells (Fig. 2, C–H). As before, we conducted LC3-II flux experiments in parallel to the Dendra2 assay, using the same 10-h time point and the same drug concentrations. BX795 caused a small but significant reduction in LC3-II flux (two-way ANOVA PINT = 0.0044), indicating a slight decrease in macroautophagy (Fig. 2 I). BX912 and GSK2334470 had no significant effect on LC3-II flux (Fig. 2, J and K). This suggests that inhibiting PDPK1 activates CMA, regardless of the state of macroautophagy.
Figure 2.
Inhibiting PDPK1 activates CMA in NIH3T3 cells. (A) Western blots showing effects of selected doses of BX795 (5 µM), BX912 (15 µM), and GSK2334470 (5 µM) on phosphorylation of Akt, MAPKAP1, and GFAP. Blots are representative of six experimental replicates. DMSO is the solvent control for all of the drugs. (B) Quantification of GFAP phosphorylation change in response to PDPK1 inhibitors, as shown in A, n = 6. In A and B, GSK2334470 is abbreviated for space, as GSK233. For C–H, data points are pooled from at least three independent experiments. (C and D) 10 h BX795 treatment induces the accumulation of Dendra2 CMA reporter puncta. DMSO = 88 cells; BX795 = 90 cells. (E and F) 10 h BX912 treatment induces the accumulation of Dendra2 CMA reporter puncta. DMSO = 155 cells; BX912 = 120 cells. (G and H) 10 h GSK2334470 treatment induces the accumulation of Dendra2 CMA reporter puncta. DMSO = 145 cells; GSK2334470 = 144 cells. (I–K) Effects of BX795, BX912, and GSK233470 on macroautophagy, as measured by LC3-II flux, n = 6. For all experiments, cells were maintained in complete growth medium with 10% serum. P values written above brackets are derived from unpaired t tests. **, P < 0.01 by unpaired t test. For charts in I–K, PINT is the interaction term P value from a two-way ANOVA. Error bars are SEM. Scale bars are 20 µm. ACTB, β-Actin.
Selective inhibition of the class III PI3K, Vps34, does not activate CMA in NIH3T3 cells
After showing that inhibition of the PI3K-PDPK1 signaling cascade activates CMA, we wanted to test whether inhibiting Vps34 activates CMA. All previous studies testing the effects of PI3K inhibition on CMA used nonselective inhibitors that are known to block both INS signaling and macroautophagy (Finn et al., 2005; Kaushik et al., 2008), but it is unclear if the effects of these drugs on CMA result from inhibition of class I, class III, or a combination of these varieties of PI3Ks. We performed dose curves in NIH3T3 cells with two selective Vps34 inhibitors, autophinib and VPS34-IN1, as well as 3-methyladenine, one of the nonselective inhibitors used in previous studies (Fig. S1, G–I). To find an effective dose of the Vps34 inhibitors, we used LC3-II flux as a readout. For all subsequent experiments, we used the lowest dose for autophinib (10 µM) and VPS34-IN1 (4 µM) that completely ablated LC3-II flux. At the doses selected, autophinib and VPS34-IN1 had no effect on Akt or GFAP phosphorylation (Fig. 3 E). However, 3-methyladenine dramatically reduced Akt phosphorylation, even at doses that were insufficient to block LC3-II flux (Fig. S1, I and J). Thus, we concluded that 3-methyladenine is not a suitable drug to understand the effects of inhibiting macroautophagy on CMA.
Figure 3.
Inhibiting Vps34 does not activate CMA in NIH3T3 cells. (A and B) 10 h treatment with autophinib (10 µM) does not induce the formation of Dendra2 CMA reporter puncta. DMSO = 131 cells; autophinib = 126 cells. (C and D) 10 h treatment with VPS34-IN1 (4 µM) does not induce the formation of Dendra2 CMA reporter puncta. DMSO = 102 cells; VPS34-IN1 = 75 cells. For A–D, data were pooled from at least three independent experiments. (E) Autophinib and VPS34-IN1 do not inhibit Akt, MAPKPA1, or GFAP phosphorylation. DMSO is the solvent control, and buparlisib (2 µM) is the positive control for Akt inhibition. Blots are representative of six experimental replicates. (F and G) Autophinib and VPS34-IN1 completely block LC3-II flux, n = 6. For all experiments, cells were maintained in complete growth medium with 10% serum. P values written above brackets are derived from unpaired t tests. For F and G, the interaction P value to the right of each graph is derived by two-way ANOVA. Error bars are SEM. Scale bars are 20 µm. ACTB, β-Actin.
We tested the ability of autophinib and VPS34-IN1 to induce CMA with the Dendra2 reporter assay. We found that the number of puncta was reduced in cells treated with autophinib and VPS34-IN1 relative to the DMSO-treated control cells (Fig. 3, A–D). These data suggest that inhibiting Vps34 reduces CMA. LC3-II flux experiments performed in parallel to the Dendra2 assay are shown in Fig. 3, F and G. Vps34 plays an essential role in the nucleation step of macroautophagy, but it also serves important roles in the regulation of protein sorting, Golgi membrane trafficking, and the biology of endosomes/multivesicular bodies (Schu et al., 1993; Hall et al., 2006; Burda et al., 2002; Futter et al., 2001). Thus, the role of Vps34 in macroautophagy may or may not be related to the role of Vps34 in CMA.
NIH3T3 cells with reduced Atg5 have increased CMA
The result that inhibiting Vps34 also inhibits CMA is a surprising contradiction to previous work asserting that inhibition of macroautophagy activates CMA (Kaushik et al., 2008). To test whether inhibition of macroautophagy independent of Vps34 activates CMA, we used siRNA to knock down Atg5 in NIH3T3 cells, which significantly reduced LC3-I to LC3-II conversion, as expected (Fig. 4, A and B). atg5−/− mouse embryonic fibroblasts (MEFs) have enhanced CMA (Kaushik et al., 2008). We found an increase in Dendra2 CMA reporter puncta in NIH3T3 cells depleted of Atg5, consistent with previous results using atg5−/− MEFs (Fig. 4, D–G). We treated cells depleted of Atg5 with either buparlisib or pictilisib, which produced a small increase in the number of reporter puncta per cell, beyond what was induced by knocking down Atg5 alone; however, this did not reach statistical significance for either drug (Fig. 4, E and G). Taken together, these data suggest that reducing macroautophagy does indeed enhance CMA, if Vps34 function is left intact. The increase in Dendra2 puncta in Atg5 knockdown cells was not attributable to an increase in LAMP2A (Fig. 4 C), suggesting that the increase in CMA is not resulting from a compensatory increase in LAMP2A expression.
Figure 4.
NIH3T3 cells with reduced Atg5 have increased CMA. Knocking down Atg5 significantly reduced LC3-I to LC3-II conversion (A and B), but did not significantly change LAMP2A levels (A and C), n = 6. NIH3T3 cells were treated with a negative control (NC) or Atg5 siRNA before being treated with DMSO, buparlisib (D and E), or pictilisib (F and G). For D–G, data were pooled from at least three independent experiments. Knocking down Atg5 was sufficient to strongly induce the formation of Dendra2 CMA reporter puncta, and this did not have a significant additive effect with buparlisib treatment (D and E) or with pictilisib treatment (F and G). In E, NC-DMSO = 45 cells, NC-Bup = 34 cells, Atg5-DMSO = 50 cells, and Atg5-Bup = 49 cells. In G, NC-DMSO = 43 cells, NC-Pic = 35 cells, Atg5-DMSO = 50 cells, and Atg5-Pic = 34 cells. Knocking down Atg5 caused changes to Akt phosphorylation (H–J) and a significant reduction in GFAP phosphorylation (H and K). For all experiments, cells were maintained in complete growth medium with 10% serum. P values above brackets are derived from unpaired t tests. For F and G, the siRNA P value to the right of each graph is derived by two-way ANOVA. Error bars are SEM. Scale bars are 20 µm. ACTB, β-Actin; Bup, buparlisib; exp., exposure; Pic, pictilisib.
If inhibition of class I PI3Ks does not significantly activate CMA beyond what is found with Atg5 knockdown alone, then it is possible that Atg5 knockdown is acting on a different step in the same pathway as the class I PI3K inhibitors. We measured Akt and GFAP phosphorylation in cells treated with siRNA for Atg5, finding that Atg5 knockdown alone was sufficient to reduce GFAP phosphorylation (Fig. 4, H and K). Interestingly, Atg5 knockdown had complex effects on Akt phosphorylation. Akt phosphorylation on T308 was significantly reduced, while there was a small, but significant, increase in Akt S473 phosphorylation (Fig. 4, H–J). These data suggest that both mechanisms of enhancing CMA, Atg5 knockdown and class I PI3K inhibition, converge on GFAP phosphorylation, although further work will be necessary to elucidate the details as to how reducing Atg5 affects GFAP.
Selective inhibition of class I PI3Ks, but not Vps34, activates CMA in multiple cell types
We considered the possibility that the results we obtained with the small molecule inhibitors used here might be specific to cell type or culture conditions. We therefore tested two p110 inhibitors and one Vps34 inhibitor for their effects on CMA and macroautophagy in two additional cultured cell types.
We performed dose curves for buparlisib, pictilisib, and autophinib in AML12 mouse hepatocytes to identify the minimum dose that showed the maximum effect of the drugs (Fig. S3, A–C). For subsequent experiments in AML12 cells, we used doses of 5 µM buparlisib, 2 µM pictilisib, and 5 µM autophinib. At these doses, buparlisib and pictilisib sharply lowered phosphorylation of Akt on S473 and T308, MAPKAP1 on T86, and GFAP on S8 (Fig. S3 D). Autophinib treatment did not decrease the phosphorylation of Akt, MAPKAP1, or GFAP, even when macroautophagy was completely inhibited (Fig. S3, D and I). Using the Dendra2 assay, we observed that inhibiting p110 with buparlisib or pictilisib significantly increased the formation of Dendra2 puncta (Fig. S3, E and F). Inhibiting Vps34 with autophinib did not induce Dendra2 puncta formation, but rather reduced them compared with baseline DMSO-treated cells (Fig. S3, E and F). These data are consistent with what we observed in NIH3T3 cells, suggesting that inhibiting the INS-PI3K-PDPK1 signaling axis is sufficient to activate CMA, even when cells are grown in the presence of serum. These data also indicate that inhibition of Vps34 does not activate CMA, and may in fact be inhibitory. We measured the effects of buparlisib and pictilisib on LC3-II flux in parallel to the Dendra2 assay, which revealed that in AML12 cells, buparlisib has no significant effect on LC3-II flux, but pictilisib does significantly lower LC3-II flux (Fig. S3, G and H). Taken together, these data indicate that CMA is enhanced when PI3K-PDPK1 is inhibited, independent of macroautophagy.
Figure S3.
Effects of PI3K inhibitors on CMA and macroautophagy in AML12 cells. (A–C) Dose curves in AML12 cells for buparlisib, pictilisib, and autophinib, respectively. (D) Western blots demonstrating the effects of buparlisib (5 µM), pictilisib (2 µM), and autophinib (5 µM) on phosphorylation of Akt, MAPKAP1, and GFAP. Blots are representative of six experimental replicates. DMSO is the solvent control for all drugs. (E and F) 10 h treatment with buparlisib or pictilisib induces the accumulation of Dendra2 CMA reporter puncta, but treatment with autophinib does not. Data were pooled from at least three independent experiments. DMSO = 202 cells, buparlisib = 122 cells, pictilisib = 104 cells, and autophinib = 95 cells. (G–I) Effects of buparlisib, pictilisib, and autophinib on macroautophagy, as measured by LC3-II flux, n = 6. For all experiments, cells were maintained in complete growth medium (see Materials and methods). P values written above brackets are derived from unpaired t tests. Two-way ANOVA tables are displayed below graphs where appropriate. Error bars are SEM. Scale bars are 20 µm. ACTB, β-Actin; Aph, autophinib; Bup, buparlisib; Pic, pictilisib.
The effects of buparlisib, picitilisib, and autophinib on CMA and macroautophagy were also tested in mouse kidney inner medullary collecting duct cells (mIMCD3). Dose curves showed that the minimum doses that maximized the effects of the drugs were 1 µM buparlisib, 1 µM pictilisib, and 5 µM autophinib (Fig. S4, A–C). Buparlisib and pictilisib treatment blocked phosphorylation of Akt on S473 and T308, MAPKAP1 on T86, and GFAP on S8 (Fig. S4 D). Autophinib treatment did not decrease the phosphorylation of Akt, MAPKAP1, or GFAP, even when macroautophagy was completely inhibited (Fig. S4, D and I). In mIMCD3 cells, buparlisib and pictilisib clearly stimulated the formation of Dendra2 puncta, suggesting an increase in CMA, while they had no effect on LC3-II flux (Fig. S4, E–H). Autophinib completely blocked LC3-II flux but had no effect on Dendra2 puncta formation (Fig. S3, E, F, and I).
Figure S4.
Effects of PI3K inhibitors on CMA and macroautophagy in mIMCD3 cells. (A–C) Dose curves in mIMCD3 cells for buparlisib, pictilisib, and autophinib, respectively. (D) Western blots demonstrating the effects of buparlisib (1 µM), pictilisib (1 µM), and autophinib (5 µM) on phosphorylation of Akt, MAPKAP1, and GFAP. Blots are representative of six experimental replicates. DMSO is the solvent control for all drugs. (E and F) 10 h treatment with buparlisib or pictilisib induces the accumulation of Dendra2 CMA reporter puncta, but treatment with autophinib does not. Data were pooled from at least three independent experiments. DMSO = 373 cells, buparlisib = 85 cells, pictilisib = 112 cells, and autophinib = 204 cells. (G–I) Effects of buparlisib, pictilisib, and autophinib on macroautophagy, as measured by LC3-II flux, n = 6. For all experiments, cells were maintained in complete growth medium (see Materials and methods). P values written above brackets are derived from unpaired t tests. Two-way ANOVA tables are displayed below graphs where appropriate. Error bars are SEM. Scale bars are 20 µm. ACTB, β-Actin; Aph, autophinib; Bup, buparlisib; Pic, pictilisib.
While there are clear differences among cell types concerning the effects of buparlisib and pictilisib on macroautophagy, it is clear that inhibiting p110 consistently activates CMA. Inhibiting Vps34 consistently inhibits macroautophagy without activating CMA.
Inhibiting class I PI3Ks causes a decrease in the abundance of CMA target proteins
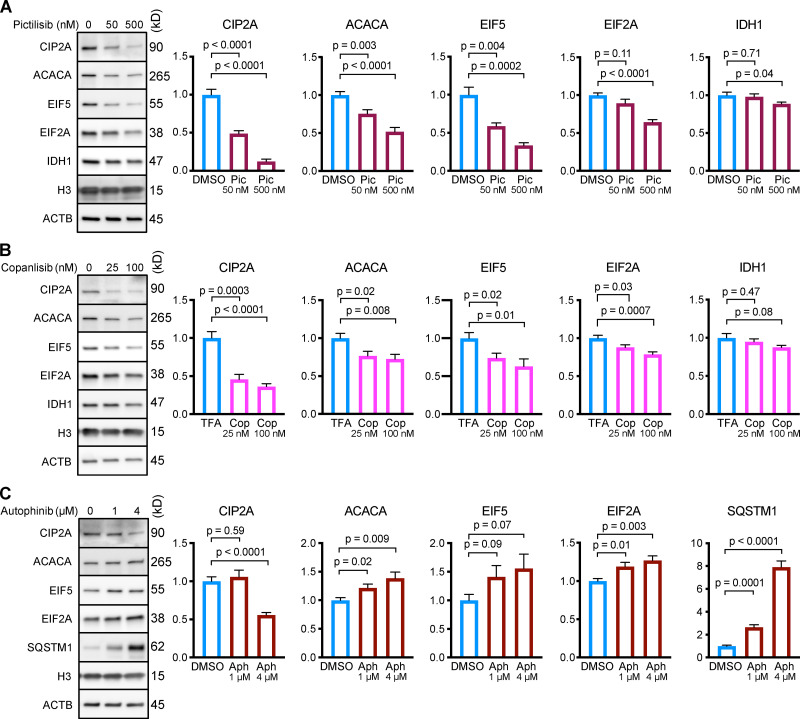
After showing that p110 inhibitors, but not Vps34 inhibitors, consistently activate CMA, we tested whether treating NIH3T3 cells with p110 inhibitors could cause a reduction in the abundance of CMA target proteins. While up to 75% of the proteome contains at least one KFERQ motif (Kirchner et al., 2019), not all proteins bearing a KFERQ domain are CMA-sensitive, and some KFERQ domains are only activated by posttranslational modification (Hao et al., 2019; Kirchner et al., 2019). Moreover, CMA target proteins show a range of sensitivities to CMA activity. Based on previous studies, we selected five CMA targets that we expected to represent a range of sensitivities to CMA: CIP2A, acetyl-coA carboxylase alpha (ACACA), EIF2A, EIF5, and IDH1 (Kirchner et al., 2019; Hao et al., 2019; Gomes et al., 2017; Schneider et al., 2014). NIH3T3 cells were treated with two doses of either pictilisib or copanlisib for 24 h, and then harvested for analysis by Western blot. Pictilisib treatment caused a significant reduction in all five CMA substrates (Fig. 5 A), while copanlisib treatment caused a significant reduction in four out of five CMA substrates (Fig. 5 B).
Figure 5.
Inhibiting p110, but not Vps34, causes a reduction in the protein abundance of CMA substrates in NIH3T3 cells. NIH3T3 cells were treated for 24 h with the indicated drug doses or solvent controls. Representative Western blots with quantifications are shown for pictilisib (A), copanlisib (B), or autophinib (C). In C, SQSTM1 is shown as a positive control. Histone H3 and ACTB were used as loading controls for all experiments. Protein abundances were normalized to the geometric mean of H3 and ACTB, and then adjusted for plotting with the mean of the solvent control set to 1. n = 8 for all experiments and treatment groups. P values above brackets are derived from unpaired t tests with Welch’s correction for unequal SD. Error bars are SEM. ACTB, β-Actin; Aph, autophinib; Cop, copanlisib; H3, histone 3; Pic, pictilisib; TFA, trifluoroacetic acid.
To further evaluate the role of Vps34 in regulating CMA activity, we measured the abundance of CMA target proteins in NIH3T3 cells treated for 24 h with 1 µM or 4 µM of autophinib. Inhibiting Vps34 significantly increased the abundance of ACACA and EIF2A, and led to a nonsignificant increase in EIF5 (Fig. 5 C). The levels of CIP2A did not change at the low dose, but significantly decreased at the high dose of autophinib (Fig. 5 C). While these results are complex, they strongly support the conclusion that inhibiting Vps34 does not activate CMA.
Mice treated with two class I PI3K inhibitors have enhanced hepatic CMA
The most commonly used method for estimating CMA in rodent tissues is to isolate lysosomes and perform a substrate binding and uptake assay (Aniento et al., 1993; Kaushik and Cuervo, 2018; Xilouri et al., 2013). To test if p110 inhibitors could activate CMA in vivo, we treated normal 3-mo-old male and female mice with buparlisib (50 mg/kg), pictilisib (100 mg/kg), or solvent control by oral gavage, once per day for 7 d, using doses that have been previously reported to inhibit Akt without causing observable toxicity (Burger et al., 2011; Raynaud et al., 2009; Nanni et al., 2012; Ortega-Molina et al., 2015). 2 h after the final dose, mice were humanely euthanized and rapidly dissected, and lysosomes were isolated from the liver by density centrifugation, as we have reported previously (Endicott et al., 2020).
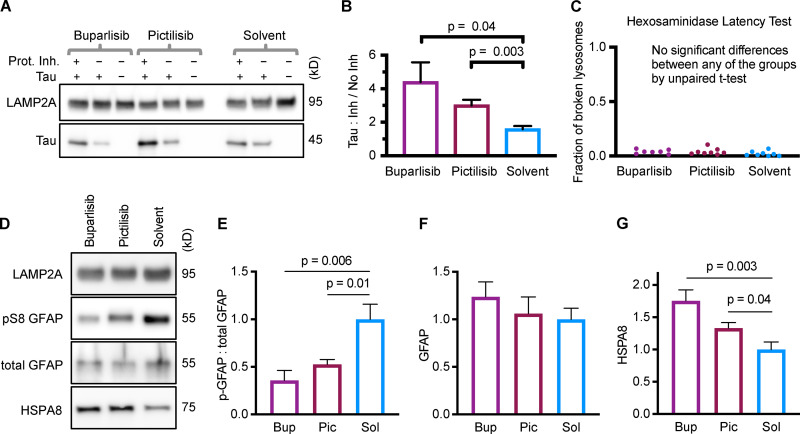
Purified lysosomes were incubated with CMA substrate Tau (MAPT), with or without protease inhibitors. The ratio of Tau accumulation in protease inhibitor–treated lysosomes to accumulation in untreated lysosomes represents the amount of Tau “substrate uptake.” Liver lysosomes from mice treated with buparlisib or pictilisib had a much higher substrate uptake ratio than lysosomes from the solvent-treated control mice (Fig. 6, A and B). We also performed a hexosaminidase latency test, a standard method for estimating the percentage of broken lysosomes in the samples. We found no difference in the fraction of broken lysosomes between the treatment groups (Fig. 6 C).
Figure 6.
Liver lysosomes from buparlisib- or pictilisib-treated mice have increased CMA substrate uptake and decreased GFAP phosphorylation. (A and B) Lysosomes from buparlisib- and pictilisib-treated mice showed enhanced uptake of CMA substrate Tau, compared with lysosomes from solvent-treated control mice. (C) There was no difference in the fraction of broken lysosomes between the treatment groups, used for uptake assays shown in A and B. (D–F) GFAP phosphorylation was reduced on lysosomes from buparlisib- and pictilisib-treated mice, but total GFAP levels were not significantly changed. (D and G) Lysosomal HSPA8 levels were increased by both buparlisib and pictilisib treatment. P values written above brackets are derived from unpaired t tests. For each experiment, the buparlisib treatment group has lysosomes from three female and four male mice. The pictilisib and solvent control treatment groups have lysosomes from four female and four male mice each. Data from males and females were pooled, after showing no sex difference in the measured endpoints. Error bars are SEM. Bup, buparlisib; Pic, pictilisib; Prot. Inh., protease inhibitors; Sol, solvent.
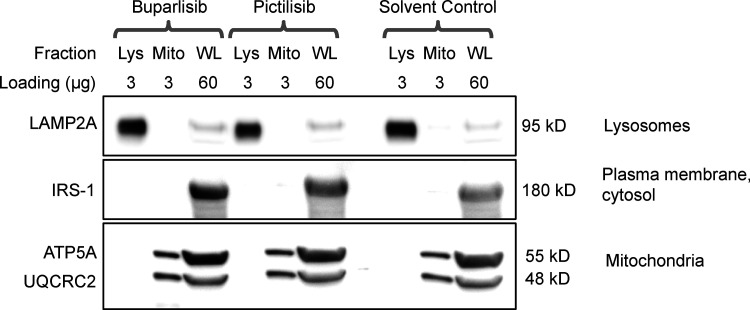
We measured GFAP, pGFAP, and HSPA8 on liver lysosomes isolated from buparlisib- and pictilisib-treated mice. We found that buparlisib and pictilisib both caused a significant reduction in lysosomal GFAP phosphorylation, without affecting the amount of total lysosomal GFAP (Fig. 6, D–F). Levels of lysosomal HSPA8 were increased by buparlisib and pictilisib treatment, consistent with an increase in CMA (Fig. 6, D and G; Cuervo et al., 1995). Our lysosomal fraction had no detectable contamination with IRS-1 (Fig. S5), a component of the INS signaling cascade at the plasma membrane. This suggests that inhibiting the INS-PI3K-PDPK1 axis at the plasma membrane is sufficient to communicate a signal that affects lysosomal GFAP phosphorylation.
Figure S5.
Lysosomal purification quality control. There was no detectable difference in lysosomal enrichment or purity between any of the treatment groups. The subcellular localization of each protein blotted for is written to the right of the representative image. The amount of protein loaded in each well, in micrograms, is written above each lane. Lys, lysosomal fraction; Mito, mitochondrial fraction; WL, whole liver lysate.
We and others have previously performed in vivo substrate uptake assays to assess lysosomal uptake of endogenous proteins (Schneider et al., 2014; Endicott et al., 2020). Here, we used this assay to determine if treatment with buparlisib and pictilisib caused an increase in the uptake of endogenous CMA substrates into mouse liver lysosomes. 30 min after mice were treated with buparlisib, pictilisib, or the solvent control by oral gavage, the mice were injected with leupeptin to block lysosomal proteolysis. 2 h after the leupeptin administration, the mice were rapidly dissected, and lysosomes were isolated from liver tissue by density centrifugation.
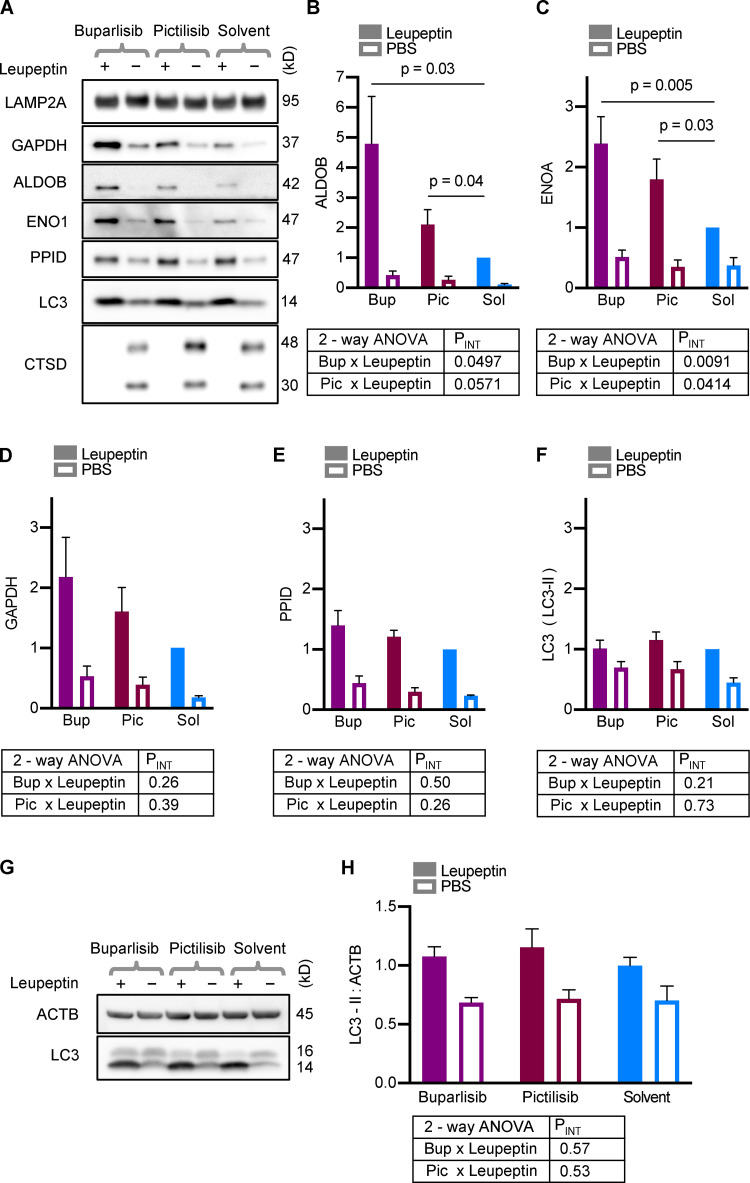
Cathepsin D (CTSD) decreases in lysosomes in response to leupeptin treatment, through an unknown mechanism (Schneider et al., 2014; Endicott et al., 2020). Thus, we used CTSD depletion from lysosomes as a positive control for successful leupeptin administration, and all samples included in the analysis showed the expected CTSD abundance corresponding to leupeptin treatment (Fig. 7 A). We found that buparlisib and pictilisib treatment caused a significant increase in the uptake of endogenous aldolase B (ALDOB) and enolase A (ENOA) into mouse liver lysosomes (Fig. 7, A–C). There was also a slight increase in the uptake of GAPDH in response to these drugs, but it did not reach statistical significance (Fig. 7, A and D). Peptidylprolyl isomerase D (PPID), a substrate of endosomal microautophagy, showed similar uptake in lysosomes from buparlisib- and pictilisib-treated mice as the solvent-treated controls (Fig. 7, A and E). Uptake of LC3, a marker of macroautophagy, was not affected by buparlisib or pictilisib treatment (Fig. 7, A and F). These data suggest that buparlisib and pictilisib treatment stimulate the uptake of CMA substrates, but not microautophagy or macroautophagy substrates, by liver lysosomes.
Figure 7.
Buparlisib or pictilisib treatment increases lysosomal uptake of endogenous CMA substrates. (A) Representative Western blots of liver lysosomes from mice that were treated with buparlisib, pictilisib, or solvent control, and subsequently injected with leupeptin or sterile PBS. CTSD is a control to show successful leupeptin administration. LAMP2A is a loading control. Mice treated with buparlisib or pictilisib showed more uptake of the endogenous CMA substrates ALDOB (B) and ENOA (C). There was a nonsignificant trend toward more uptake of GAPDH by lysosomes from buparlisib- (P = 0.09) and pictilisib- (P = 0.15) treated mice (D). Buparlisib and picitilisib treatment did not cause a significant change in the uptake of endosomal microautophagy substrate PPID (E), or macroautophagy substrate LC3 (F). Whole liver LC3-II flux was also unchanged by buparlisib or pictilisib treatment (G and H). For every experimental group in A–F, there are lysosomes from four female and four male mice. For G and H, n = 8 (four females and four males). Data from males and females were pooled, after showing no sex difference in the measured endpoints. P values written above brackets are derived from unpaired t tests. PINT is the interaction term P value derived from two-way ANOVA. Error bars are SEM. ACTB, β-Actin; Bup, buparlisib; Pic, pictilisib; Sol, solvent.
To further confirm that macroautophagy is unaffected by buparlisib and pictilisib treatment in vivo, we examined whole-liver LC3 flux, using liver lysates from mice treated with buparlisib or pictilisib, and then subjected to the leupeptin assay. Just as with the lysosomes, LC3-II flux was unchanged by buparlisib or pictilisib treatment (Fig. 7, G and H). Together, these data suggest that p110 inhibitors reduce lysosomal GFAP phosphorylation and increase CMA, without affecting macroautophagy.
Discussion
At least three signaling pathways have been shown to modulate CMA: the T cell receptor pathway, the retinoic acid receptor pathway, and the Akt pathway. Activation of the T cell antigen receptor on T cells promotes CMA by increasing the transcription of Lamp2a and by increasing total lysosomal biogenesis, dependent on transcription factor EB (TFEB; Valdor et al., 2014). Mouse fibroblasts treated with siRNA targeting retinoic acid receptor α (RARα) had enhanced CMA, and small molecule inhibitors of RARα increase Lamp2a mRNA expression and promote CMA (Anguiano et al., 2013). Neither the T cell antigen receptor pathway nor the RARα pathway appears to directly regulate lysosomal biology, but instead they indirectly affect lysosomes through transcriptional responses. Treating purified lysosomes with inhibitors of the mechanistic target of rapamycin (mTOR) or Akt is sufficient to increase the uptake of CMA substrates, as evaluated by in vitro uptake assays, indicating a direct role for these kinases in controlling CMA in the lysosomal compartment (Arias et al., 2015), independently of transcriptional effects. Treating cells with small molecule inhibitors of Akt or mTOR also increased CMA reporter puncta (Arias et al., 2015), but the role of upstream PI3K-PDPK1 signaling was not examined in this earlier study.
Fms-like tyrosine kinase 3 (FLT3) is a receptor tyrosine kinase that is commonly duplicated in acute myeloid leukemia, and is an appealing target for cancer therapy (Kayser and Levis, 2014). FLT3 activation promotes the activity of multiple pro-growth pathways, including PI3K/Akt, MAPK/extracellular signal–related kinase, and STAT5 (Kayser and Levis, 2014). Inhibition of FLT3 with quizartinib (AC220) in Sum159 breast cancer cells or ES-2 ovarian cancer cells is sufficient to activate CMA (Xia et al., 2015). FLT3 inhibition is accompanied by a decrease in Akt phosphorylation on S473 (Xia et al., 2015). However, it is not known whether inhibiting FLT3 activates CMA by affecting GFAP activity, activating transcription of lysosomal genes, or a combination of both. The result that inhibiting FLT3 activates CMA, when taken together with the data presented by this study, suggests the hypothesis that inhibiting FLT3 activates CMA by decreasing Akt activity through INS-independent signaling. In cancerous cell types where inhibiting FLT3 is sufficient to lower Akt activity, CMA is likely to be activated as a result of decreased GFAP phosphorylation, but this needs to be experimentally verified. Future work that selectively inhibits other pathways modified by FLT3 signaling will also be necessary to completely understand how FLT3 activity regulates CMA.
Most studies of CMA regulation have been conducted in cultured cells and isolated lysosomes (Bandyopadhyay et al., 2008; 2010; Arias et al., 2015; Valdor et al., 2014; Xia et al., 2015). For this reason, little is known about the endocrine control of CMA. We recently reported that CMA is constitutively elevated in the liver of mutant mice with lifelong reductions in growth hormone signaling, i.e., Snell dwarf (pit1/pou1f1 loss of function point mutation) and ghr knockout (GHRKO). However, this is not directly attributable to growth hormone effects on liver cells, because mice with a liver-specific ghr knockout (Li-GHRKO) do not show increased hepatic CMA (Endicott et al., 2020). This suggests that a signaling event resulting from a whole-body reduction in growth hormone signaling is responsible for the up-regulation of CMA in the liver of Snell and GHRKO mice. Both Snell and GHRKO mice have reduced circulating INS, compared with controls, in fasted and fed states (Brooks et al., 2007; Al-Regaiey et al., 2005). Li-GHRKO mice have increased circulating INS when fasting, and elevated glucose levels in both fasting and fed states (List et al., 2014), suggesting poor INS sensitivity. Because circulating INS is decreased in Snell and GHRKO (which have enhanced CMA), but INS is not decreased in Li-GHRKO (which has some endocrine changes that overlap with Snell and GHRKO, but does not have enhanced CMA), it is reasonable to hypothesize that INS is a critical regulator of CMA in endocrine mutant mice.
Rats and mice fasted for prolonged periods of time (16 h or more) have elevated CMA (Cuervo et al., 1995; Kaushik and Cuervo, 2018), but it has been unclear if the increase in CMA in response to nutrient deprivation is a result of reduced circulating INS or another of the many other physiological changes induced by fasting. Based on the correlation between reduced INS signaling and increased CMA in rodent liver, we hypothesized that inhibiting intracellular signaling events downstream of INS signaling would activate CMA. Here, using assays to measure CMA activation in both mice and cultured cells, we have found that selective small molecule inhibitors of the INS-PI3K-PDPK1 pathway robustly activate CMA, independently of the activation of macroautophagy. Future work will be needed to disentangle how the INS-PI3K-PDPK1 and FLT3 pathways affect CMA in different conditions, and to determine if suppression of Akt activity is necessary or only sufficient to enhance CMA.
There is currently great scientific interest in modulating CMA for medical purposes. CMA decreases with age in rodents (Cuervo and Dice, 2000; Zhang and Cuervo, 2008), and declines in CMA have been associated with neurodegeneration, metabolic dysfunction, and neoplasia (Schneider et al., 2014; Gomes et al., 2017; Xia et al., 2015; Xilouri et al., 2016). Here, we have presented the first evidence of effective use of p110 inhibitors to activate CMA in mice. Small molecule inhibitors of p110 are clinically safe, and many of them are orally bioavailable (Crabbe et al., 2007). Not all p110 inhibitors penetrate the blood-brain barrier, but some of them do, including buparlisib (de Gooijer et al., 2018), suggesting that these drugs hold therapeutic promise for stimulating CMA in the brain, where enhancing CMA might play an important anti-neurodegenerative role (Wang et al., 2009; Huang et al., 2014; Qi et al., 2012; Orenstein et al., 2013; Park et al., 2016; Kaushik and Cuervo, 2018). In support of the idea that enhancing CMA can delay the pathogenesis of neurodegenerative disease, rats with transgenic overexpression of LAMP2A in the brain are resistant to α-synuclein–mediated Parkinson’s-like disease (Xilouri et al., 2013). Future work is necessary to repurpose p110 inhibitors to treat mouse models of neurodegenerative disease, to determine whether or not these drugs hold promise for human therapy.
In addition to its role in disease pathogenesis, CMA has been speculated to play a role in healthy aging. The age-related decline in CMA in rodents is largely attributed to a decrease in the stability of LAMP2A at the lysosomal membrane, as lysosomal membrane dynamics change with age (Kaushik et al., 2006; Kiffin et al., 2007). A transgenic mouse overexpressing LAMP2A in the liver is resistant to the age-related decline in hepatic CMA and shows improved liver function with age, with reduced accumulation of oxidized proteins and protein aggregates (Zhang and Cuervo, 2008). While lifespan studies of mice with CMA enhanced (or at least restored to youthful levels in aged mice) have not yet been published, it has been speculated that these animals might show improved health in aging (Kaushik and Cuervo, 2018).
Snell and GHRKO mice, which have enhanced CMA, live ∼40% longer than their wild-type siblings (Flurkey et al., 2001; Coschigano et al., 2003). We previously reported that two proteins whose abundances are sensitive to CMA activity (CIP2A, a direct CMA target, and MYC transcription factor, indirectly affected by CMA) are decreased in multiple tissues of these mice (Endicott et al., 2020). These data suggest the provocative hypothesis that CMA might be playing an important role in shaping the proteomes of long-lived mice with reduced INS signaling early in life in a way that has pro-longevity effects, but further work is needed to fully test this possibility.
In conclusion, the finding that CMA is directly regulated by the INS-PI3K-PDPK1 pathway is an important step in understanding how this pathway is regulated independently of macroautophagy. The disease implications of CMA justify future work studying how pharmacological manipulation of CMA affects tissue abundance of disease-associated peptides. Because the INS-PI3K-PDPK1 pathway is so important for cancer research, new tools are regularly developed for manipulating this pathway, with continuous improvements to target specificity, potentially reducing side effects. Thus, we believe that boosting CMA through the INS-PI3K-PDPK1 pathway holds great promise for treating CMA-related human pathology.
Materials and methods
Antibodies
Antibodies were acquired as follows: ACACA/ACC1 (Cell Signaling Technology; 4190S; rabbit host), ACTB/β-Actin (Cell Signaling Technology; 8457L; rabbit host), ALDOB (AbCam; 137628; rabbit host), AKT (Cell Signaling Technology; 4691S; rabbit host), pT308 AKT (Cell Signaling Technology; 13038S; rabbit host), pS473 AKT (Cell Signaling Technology; 4060S; rabbit host), ATG5 (Cell Signaling Technology; 12994S; rabbit host), CIP2A (Santa Cruz; sc-80659; mouse host), CTSD/cathepsin D (AbCam; 75852; rabbit host), EIF2A (Cell Signaling Technology; 9722S; rabbit host), EIF5 (Cell Signaling Technology; 13894S; rabbit host), ENO1 (Cell Signaling Technology; 3810S; rabbit host), GAPDH (Cell Signaling Technology; 2118S; rabbit host), GSK3β (Cell Signaling Technology; 12456S; rabbit host), pS9 GSK3β (Cell Signaling Technology; 9323S; rabbit host), GFAP (AbCam; 7260; rabbit host), pS8 GFAP (Thermo Fisher Scientific; PA5-12991; rabbit host), HSPA8/Hsc70 (AbCam; 154415; rabbit host), IDH1 (AbCam; 172964; rabbit host), IRS1 (Cell Signaling Technology; 2382S; rabbit host) LAMP1 (AbCam; 24170; rabbit host), LAMP2A (AbCam; 125068; rabbit host), MAP1LC3B/LC3 (Cell Signaling Technology; 2775S; rabbit host), MAPKAP1/mSIN1 (AbCam; 64188; rabbit host), pT86 MAPKAP1/mSIN1 (Cell Signaling Technology; 14716S; rabbit host), MAPT/Tau (Cell Signaling Technology; 46687S; rabbit host), OXPHOS (oxidative phosphorylation complex) cocktail (AbCam; 110413; mouse host), PPID/cyclophilin 40 (AbCam; 181983; rabbit host), and SQSTM1/p62 (Cell Signaling Technology; 5114S; rabbit host).
For Western blots, most antibodies detected a single band at the expected molecular weight. For GFAP antibodies, multiple bands were detected in lysates from cultured cells. Only the band corresponding to the molecular weight of the lysosomal GFAP band is shown in the figures (as explained by Fig. S2). For MAPKAP1, multiple bands were detected, as expected, due to the presence of many splice variants. The band corresponding in molecular weight to the ∼65 kD pT86 MAPKAP1 was invariant in expression across treatment groups, and regarded as the loading control for the pT86 65 kD band. ATG5 is predominantly present as a covalently joined heterodimer with ATG12 (55 kD band). The ATG5-ATG12 heterodimer band is shown in Western images, as indicated on the figures.
Cell culture
For all cell culture experiments, no cell line was passaged more than 20 times, after thawing the original stock from the American Type Culture Collection (ATCC). Cells were checked for mycoplasma contamination upon thaw, returning a negative result each time. All cells were maintained in a humidified 37°C incubator with 10% CO2 and ambient O2. Recipes for cell culture growth media were obtained from the ATCC website.
NIH3T3 cells were obtained from ATCC (CRL-1658) and maintained in DMEM (Gibco; 11965092) supplemented with 10% FBS (Corning; 35–016-CV) and penicillin-streptomycin (Gibco; 15070063).
AML12 cells were obtained from ATCC (CRL-2254) and maintained in DMEM:F12 (Gibco; 11320033), supplemented with INS-selenium-transferrin (Gibco; 51500–056), 10% FBS (Corning; 35–016-CV), 40 ng/ml dexamethasone (Sigma-Aldrich; D4902), and penicillin-streptomycin (Gibco; 15070063).
mIMCD3 cells were obtained from ATCC (CRL-2123) and maintained in DMEM:F12 (Gibco; 11320033), supplemented with 10% FBS (Corning; 35–016-CV) and penicillin-streptomycin (Gibco; 15070063).
Drugs for cell culture experiments
Drugs for cell culture experiments were obtained as follows: buparlisib (SelleckChem; S2247), pictilisib (SelleckChem; S1065), copanlisib (SelleckChem; S2802), BX795 (SelleckChem; S1274), BX912 (SelleckChem; S1275), GSK2334470 (Tocris; 4143), bafilomycin A (Sigma-Aldrich; SML1661), 3-methyladenine (SelleckChem; S2767), autophinib (SelleckChem; S8596), and VPS34-IN1 (SelleckChem; 7980).
Bafilomycin A was used at a concentration of 160 nM for all experiments. All other drug doses and incubation times are indicated in the figure legends or directly on the figures. Drugs were all dissolved in DMSO (Sigma-Aldrich; D2650), except for copanlisib and 3-methyladenine. Copanlisib, which is not DMSO-soluble, was initially solubilized in a 10% trifluoroacetic acid (Sigma-Aldrich; T6508)/90% water solution to a concentration of 2 mM and then diluted to appropriate working stock concentrations. 3-Methyladenine solutions were prepared fresh each time. 3-Methyladenine powder was dissolved directly into cell culture medium to produce a 50 mM stock. 3-Methyladenine solutions were heated to 55°C for 5 min, vortexed until clear, diluted to the appropriate concentration, and used immediately.
For all drug treatments, the cells were maintained in complete growth medium, with serum, according to the recipes specified above.
Drugs for mouse experiments
Drugs used in mice were acquired as follows: buparlisib (MedChem Express; HY-70063), pictilisib (MedChem Express; HY-50094), and leupeptin (Sigma-Aldrich; 108976).
All drug solutions were prepared in a biosafety cabinet, using a sterile technique. Doses are reported in milligrams per kilograms of body weight.
Buparlisib and pictilisib were administered at doses of 50 mg/kg and 100 mg/kg, respectively. Buparlisib and pictilisib were dissolved in 10% N-methylpyrrolidone and 90% polyethylene glycol 300 to concentrations of 15 mg/ml and 30 mg/ml, respectively, so that the final volume delivered to each mouse was ∼100 µl, similar to previous studies (Nanni et al., 2012; Ortega-Molina et al., 2015). To dissolve the drugs, solutions were passed through a 23-gauge needle repeatedly, until solutions were clear. Drugs were then aliquoted and stored at −80°C, then thawed immediately before use. Drugs were administered by oral gavage at approximately the same time every day for 1 wk.
Leupeptin was dissolved in sterile PBS to a concentration of 25 mg/ml and administered by intraperitoneal injection at a dose of 100 mg/kg, in a final volume of ∼100 µl, as previously described (Endicott et al., 2020).
Fluorescent CMA reporter
The KFERQ-Dendra2 plasmid was a gift of A.M. Cuervo. The reporter consists of the N terminus of RNase A fused to the photoswitchable fluorescent protein Dendra2, in a third-generation lentiviral backbone, pCCL, similar to what has been reported in other studies (Koga et al., 2011; Arias et al., 2015; Anguiano et al., 2013). Lentivirus for transgene delivery was created by the University of Michigan Biomedical Research Core Facilities Vector Core. Cells were transduced with virus, using standard techniques to generate stable lines. Counting fluorescent puncta in individual cells is not feasible when 100% of cells express the reporter, due to overlap between the cells, so we intentionally titrated down the dose of virus used in the transfection to generate a more sparse labeling amenable to accurate quantitation.
Cells carrying the reporter gene were seeded in 24-well plates with a cover glass (Thermo Fisher Scientific; 1254580) dropped into each well. For photoconversion, plates were exposed to violet light (400–415 nm) with an irradiance of ∼100 mW/cm2 for 5 min, using a Nightsea DFP-VR lamp equipped with 440–460 nm and 400–415 nm filters.
Cells were treated with drugs for 10 h, washed with PBS, and fixed in a solution of 1% paraformaldehyde and 1% sucrose, in PBS, pH 7.2, for 7 min at room temperature. Cells were rinsed twice with PBS, stained with Hoechst (Sigma-Aldrich; 94403), and mounted onto microscope slides (Thermo Fisher Scientific; 12–550-343), using fluorescent protein mounting medium (GBI Labs; E18-18). Slides dried overnight and were imaged the next day. At every step after the photoconversion, the cells were protected from light.
Image analysis
Dendra2 puncta were counted blind, with images analyzed in random order. First, images were subjected to the ImageJ Max Entropy thresholding algorithm to identify cells with sufficient fluorescent protein expression to be counted. Then the number of puncta per cell was manually counted in unadjusted images and recorded (because of uneven background, automated counting is reliably less accurate than manual counting). Cells were excluded from analysis if they were on the edge of the image (such that a portion of the cytosol was outside of the image, preventing an accurate count), or if their cytoplasm overlapped substantially with another cell, preventing accurate quantification. Final results were unblinded and tabulated by a different person than the one performing the analysis. For each experiment, data were pooled from at least three independent replicates.
Lysosome isolation and CMA evaluation
To control for known effects of circadian rhythms on autophagic processes, mice were dissected at approximately the same time for each experiment (between 9 and 10 a.m., with the dark period ending at 6 a.m.). CMA activity is sensitive to feeding state. Upon dissection, mice were qualitatively assessed for the presence of food in the stomach (all mice used in the study had food in the stomach).
Freshly dissected liver tissue was immediately washed in ice-cold PBS until no visible blood remained. The tissue was diced with scissors and gently Dounce-homogenized in commercially available fractionation buffers (Thermo Fisher Scientific; 89839). All subsequent steps were performed at 4°C unless otherwise stated. Nuclei, extracellular matrix, and unbroken cells were removed by 10-min centrifugation at 500 g. The postnuclear sample was mixed with iodixanol/OptiPrep to a concentration of 15% OptiPrep and loaded onto a discontinuous 17–30% OptiPrep (Sigma-Aldrich; D1556) density gradient in ultracentrifuge tubes (Beckman Coulter; 344057) for a 55Ti-SW rotor (Beckman Coulter; 342194). A 10% Optiprep layer was laid over the sample, and the samples were subjected to ultracentrifugation at 145,000 g for 2 h in a Beckman Coulter L-70 ultracentrifuge. During ultracentrifugation, the sample resolved into visible bands on the density gradient. The visible band at the 10–15% gradient interface is highly enriched for lysosomal markers and was used for all experiments. After the lysosome-containing band was removed from the density gradient, the sample was diluted in PBS to decrease the OptiPrep density, allowing the lysosomes to be pelleted, washed, and resuspended. Protein concentrations were measured by bicinchoninic acid (BCA) assay (Thermo Fisher Scientific; 23225), and all samples were adjusted to the same protein concentration. Lysosomes generated by this method have been rigorously tested by our group for enrichment of lysosomal markers and purity from contamination with other subcellular compartments (Endicott et al., 2020).
For CMA substrate uptake assays, isolated lysosomes were incubated in uptake assay buffer: 300 mM sucrose, 10 mM MOPS, pH 7.2, 10 mM ATP (Sigma-Aldrich; A26209), and 10 mg/ml recombinant HSPA8 (AbCam; ab78431). Recombinant Tau (Sino Biological; 10058-H07E) and/or protease inhibitors (Sigma-Aldrich; 11836153001) were added as indicated. Lysosomes were incubated at 37°C for 20 min, pelleted, and then washed with 300 mM sucrose, 10 mM MOPS, pH 7.2. They were then prepared for analysis by Western blotting.
To assess lysosomal breakage, lysosomes were diluted in uptake assay buffer: 300 mM sucrose, 10 mM MOPS, pH 7.2, and 10 mM ATP, and treated to the same conditions as the lysosomes used in the substrate uptake assay, in parallel to that experiment. At the end, the lysosomes were pelleted (no washes), and the supernatant was collected. The pellets and supernatants were assessed for hexosaminidase activity using a 4-nitrophenyl-N-acetyl-b-D-glucosaminide colorimetric assay, as described by Wendeler and Sandhoff (2009).
For analysis of lysosomal uptake of endogenous substrates, mice were injected with 100 mg/kg body weight of leupeptin, 30 min after gavage with the appropriate drug or control treatment. Lysosomes were isolated by the same methods described above, and then analyzed by Western blotting.
Microscope image acquisition
Microscopy was performed in the University of Michigan Biomedical Research Core Facilities Microscopy Core. Images were acquired with a Zeiss Axioplan2 microscope equipped with Zeiss ApoTome for optical sectioning. Image capture was performed with a Zeiss AxioCam MRm camera. The objective lens used was a Zeiss Plan-NEOFLUAR 40×, with a numerical aperture of 0.75 (this lens requires no immersion medium). The microscope was operated with Zeiss Axiovision software. All microscopy was performed on fixed samples at room temperature. Postacquisition image adjustment was performed with ImageJ software. Images presented in figures are z-projections of the depth of the entire cell, using the maximum intensity algorithm for both channels, generated in ImageJ from image stacks acquired with a 0.5 µm z-plane slice-distance.
Mouse stocks and husbandry
All mouse experiments were performed in 3-mo-old mice of the genetically heterogeneous UM-HET3 mouse stock, as previously described (Miller et al., 1999, 2007). The UM-HET3 stock is produced by crossing female CByB6F1/J (JAX stock no. 100009) to male C3D2F1/J (JAX stock no. 100004). Each of the parents are F1 crosses. The CByB6F1/J mothers are generated by crossing BALB/cByJ females to C57BL/6J males. The C3D2F1/J fathers are generated by crossing C3H/HeJ females to DBA/2J males. All test animals are the grandchildren of the same four inbred isogenic mouse lines. All animal experiments were approved by the University of Michigan Institutional Animal Care and Use Committee. Mice were housed in specific pathogen-free facilities, with sentinel animals checked quarterly for infection (all tests were negative). Mice had free access to food and water during the course of all experiments.
RNA interference
siRNAs were acquired as follows: Medium GC content negative control (Invitrogen; 465372), Atg5 (Invitrogen; Atg5MSS247019, 44009549), and Lamp2A (cocktail of two custom-designed Stealth siRNA duplexes from Invitrogen; sequences: 5′-CAGCUCUGGGAGGAGUACUUAUUCU-3′ and 5′-CAAGCGCCAUCAUACUGGAUAUGAG-3′, complexed to the reverse complement antisense sequences). Transfections were performed using Lipofectamine RNAi MAX (Invitrogen; 56532), with transfections prepared in OptiMEM (Gibco; 31985–062), according to the manufacturer’s instructions.
Statistical analysis
All statistical analysis and graph generation were performed with GraphPad Prism 8. Results of t tests and two-way ANOVAs are reported directly on the figures or in the figure legends. Unless otherwise stated, all t tests are unpaired. For data from mice, endpoints measured from male and female mice were compared by two-way ANOVA to check for a sex effect. When sex effects were not identified, the data were pooled.
Online supplemental material
Fig. S1 shows dose curves for drugs used in NIH3T3 cells. Fig. S2 shows controls for GFAP isoforms and Dendra2 reporter function. Fig. S3 shows the effects of PI3K inhibitors on CMA and macroautophagy in AML12 hepatocytes. Fig. S4 shows the effects of PI3K inhibitors on CMA and macroautophagy in mIMCD3 epithelial cells. Fig. S5 shows lysosomal purification quality control.
Acknowledgments
Mouse husbandry was conducted by Roxann Alonzo, Asuman “Ilkim” Erturk, Natalie Perry, Lori Roberts, and Jacob Sheets. The KFERQ-Dendra2 plasmid was a gift of Ana Maria Cuervo (Albert Einstein College of Medicine, New York, NY). Thomas Lanigan (University of Michigan) provided assistance with designing experiments using viral vectors. Sasha Meshinchi (University of Michigan) provided assistance with microscopy training and experimental design. Kristen Verhey (University of Michigan) shared an ultracentrifuge rotor for critical experiments.
This work was supported by National Institutes of Health grant AG022303, National Institutes of Health training grant 5T32AG000114-34, and the Glenn Foundation for Medical Research.
The authors declare no competing financial interests.
Author contributions: S.J. Endicott and R.A. Miller conceived the project and designed the experiments. S.J. Endicott supervised the experiments. S.J. Endicott, Z.J. Ziemba, L.J. Beckmann, and D.N. Boynton performed the experiments and data analysis. S.J. Endicott made the figures. S.J. Endicott and R.A. Miller wrote the paper. R.A. Miller is the laboratory director.
References
- Agarraberes, F.A., and Dice J.F.. 2001. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J. Cell Sci. 114:2491–2499. [DOI] [PubMed] [Google Scholar]
- Al-Regaiey, K.A., Masternak M.M., Bonkowski M., Sun L., and Bartke A.. 2005. Long-lived growth hormone receptor knockout mice: interaction of reduced insulin-like growth factor i/insulin signaling and caloric restriction. Endocrinology. 146:851–860. 10.1210/en.2004-1120 [DOI] [PubMed] [Google Scholar]
- Anguiano, J., Garner T.P., Mahalingam M., Das B.C., Gavathiotis E., and Cuervo A.M.. 2013. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat. Chem. Biol. 9:374–382. 10.1038/nchembio.1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aniento, F., Roche E., Cuervo A.M., and Knecht E.. 1993. Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J. Biol. Chem. 268:10463–10470. [PubMed] [Google Scholar]
- Arias, E., Koga H., Diaz A., Mocholi E., Patel B., and Cuervo A.M.. 2015. Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy. Mol. Cell. 59:270–284. 10.1016/j.molcel.2015.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay, U., Kaushik S., Varticovski L., and Cuervo A.M.. 2008. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol. Cell. Biol. 28:5747–5763. 10.1128/MCB.02070-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay, U., Sridhar S., Kaushik S., Kiffin R., and Cuervo A.M.. 2010. Identification of regulators of chaperone-mediated autophagy. Mol. Cell. 39:535–547. 10.1016/j.molcel.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, N.L., Trent C.M., Raetzsch C.F., Flurkey K., Boysen G., Perfetti M.T., Jeong Y.-C., Klebanov S., Patel K.B., Khodush V.R., et al. 2007. Low utilization of circulating glucose after food withdrawal in Snell dwarf mice. J. Biol. Chem. 282:35069–35077. 10.1074/jbc.M700484200 [DOI] [PubMed] [Google Scholar]
- Burda, P., Padilla S.M., Sarkar S., and Emr S.D.. 2002. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J. Cell Sci. 115:3889–3900. 10.1242/jcs.00090 [DOI] [PubMed] [Google Scholar]
- Burger, M.T., Pecchi S., Wagman A., Ni Z.-J., Knapp M., Hendrickson T., Atallah G., Pfister K., Zhang Y., Bartulis S., et al. 2011. Identification of NVP-BKM120 as a Potent, Selective, Orally Bioavailable Class I PI3 Kinase Inhibitor for Treating Cancer. ACS Med. Chem. Lett. 2:774–779. 10.1021/ml200156t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, A.J.M., Linch M.D., Saurin A.T., Escribano C., and Parker P.J.. 2011. mTORC2 targets AGC kinases through Sin1-dependent recruitment. Biochem. J. 439:287–297. 10.1042/BJ20110678 [DOI] [PubMed] [Google Scholar]
- Chiang, H.-L., Terlecky S.R., Plant C.P., and Dice J.F.. 1989. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 246:382–385. 10.1126/science.2799391 [DOI] [PubMed] [Google Scholar]
- Coschigano, K.T., Holland A.N., Riders M.E., List E.O., Flyvbjerg A., and Kopchick J.J.. 2003. Deletion, but not antagonism, of the mouse growth hormone receptor results in severely decreased body weights, insulin, and insulin-like growth factor I levels and increased life span. Endocrinology. 144:3799–3810. 10.1210/en.2003-0374 [DOI] [PubMed] [Google Scholar]
- Crabbe, T., Welham M.J., and Ward S.G.. 2007. The PI3K inhibitor arsenal: choose your weapon! Trends Biochem. Sci. 32:450–456. 10.1016/j.tibs.2007.09.001 [DOI] [PubMed] [Google Scholar]
- Cuervo, A.M., and Dice J.F.. 1996. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 273:501–503. [DOI] [PubMed] [Google Scholar]
- Cuervo, A.M., and Dice J.F.. 2000. Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 275:31505–31513. 10.1074/jbc.M002102200 [DOI] [PubMed] [Google Scholar]
- Cuervo, A.M., Terlecky S.R., Dice J.F., and Knecht E.. 1994. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by isolated rat liver lysosomes. J. Biol. Chem. 269:26374–26380. [PubMed] [Google Scholar]
- Cuervo, A.M., Knecht E., Terlecky S.R., and Dice J.F.. 1995. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am. J. Physiol. 269:C1200–C1208. [DOI] [PubMed] [Google Scholar]
- Cuervo, A.M., Stefanis L., Fredenburg R., Lansbury P.T., and Sulzer D.. 2004. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 305:1292–1295. 10.1126/science.1101738 [DOI] [PubMed] [Google Scholar]
- de Gooijer, M.C., Zhang P., Buil L.C.M., Çitirikkaya C.H., Thota N., J.H. Beijnen, and van Tellingen O.. 2018. Buparlisib is a brain penetrable pan-PI3K inhibitor. Sci. Rep . 8:10784. doi:. 10.1038/s41598-018-29062-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice, J.F. 1990. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem. Sci. 15:305–309. [DOI] [PubMed] [Google Scholar]
- Endicott, S.J., Boynton D.N. Jr., Beckmann L.J., and Miller R.A.. 2020. Long-lived mice with reduced growth hormone signaling have a constitutive upregulation of hepatic chaperone-mediated autophagy. Autophagy. 00:1–14. 10.1080/15548627.2020.1725378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, P.F., Mesires N.T., Vine M., and Dice J.F.. 2005. Effects of small molecules on chaperone-mediated autophagy. Autophagy. 1:141–145. 10.4161/auto.1.3.2000 [DOI] [PubMed] [Google Scholar]
- Flurkey, K., Papaconstantinou J., Miller R.A., and Harrison D.E.. 2001. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc. Natl. Acad. Sci. USA. 98:6736–6741. 10.1073/pnas.111158898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias, M.A., Thoreen C.C., Jaffe J.D., Schroder W., Sculley T., Carr S.A., and Sabatini D.M.. 2006. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 16:1865–1870. 10.1016/j.cub.2006.08.001 [DOI] [PubMed] [Google Scholar]
- Fruman, D.A., Chiu H., Hopkins B.D., Bagrodia S., Cantley L.C., and Abraham R.T.. 2017. The PI3K Pathway in Human Disease. Cell. 170:605–635. 10.1016/j.cell.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futter, C.E., Collinson L.M., Backer J.M., and Hopkins C.R.. 2001. Human VPS34 is required for internal vesicle formation within multivesicular endosomes. J. Cell Biol. 155:1251–1264. 10.1016/S0898-6568(98)00011-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, L.R., Menck C.F.M., and Cuervo A.M.. 2017. Chaperone-mediated autophagy prevents cellular transformation by regulating MYC proteasomal degradation. Autophagy. 13:928–940. 10.1080/15548627.2017.1293767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, B.S., Gabernet-Castello C., Voak A., Goulding D., Natesan S.K., and Field M.C.. 2006. TbVps34, the trypanosome orthologue of Vps34, is required for Golgi complex segregation. J. Biol. Chem. 281:27600–27612. [DOI] [PubMed] [Google Scholar]
- Hao, Y., Kacal M., Ouchida A.T., Zhang B., Norberg E., and Vakifahmetoglu-Norberg H.. 2019. Targetome analysis of chaperone-mediated autophagy in cancer cells. Autophagy. 15:1558–1571. 10.1080/15548627.2019.1586255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.C., Bose J.K., Majumder P., Lee K.H., Huang J.T.J., Huang J.K., and Shen C.K.J.. 2014. Metabolism and mis-metabolism of the neuropathological signature protein TDP-43. J. Cell Sci. 127:3024–3038. 10.1242/jcs.136150 [DOI] [PubMed] [Google Scholar]
- Humphrey, S.J., Yang G., Yang P., Fazakerley D.J., Stöckli J., Yang J.Y., and James D.E.. 2013. Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17:1009–1020. 10.1016/j.cmet.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik, S., and Cuervo A.M.. 2018. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19:365–381. 10.1038/s41580-018-0001-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik, S., Massey A.C., and Cuervo A.M.. 2006. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 25:3921–3933. 10.1038/sj.emboj.7601283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik, S., Massey A.C., Mizushima N., and Cuervo A.M.. 2008. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol. Biol. Cell. 19:2179–2192. 10.1091/mbc.e07-11-1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser, S., and Levis M.J.. 2014. FLT3 tyrosine kinase inhibitors in acute myeloid leukemia: clinical implications and limitations. Leuk. Lymphoma. 55:243–255. 10.1200/JCO.2005.02.2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiffin, R., Kaushik S., Zeng M., Bandyopadhyay U., Zhang C., Massey A.C., Martinez-Vicente M., and Cuervo A.M.. 2007. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J. Cell Sci. 120:782–791. 10.1242/jcs.001073 [DOI] [PubMed] [Google Scholar]
- Kirchner, P., Bourdenx M., Madrigal-Matute J., Tiano S., Diaz A., Bartholdy B.A., Will B., and Cuervo A.M.. 2019. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. PLoS Biol. 17:e3000301. 10.1371/journal.pbio.3000301.s019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, H., Martinez-Vicente M., Macian F., Verkhusha V.V., and Cuervo A.M.. 2011. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat. Commun. 2:386. 10.1038/ncomms1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- List, E.O., Berryman D.E., Funk K., Jara A., Kelder B., Wang F., Stout M.B., Zhi X., Sun L., White T.A., et al. 2014. Liver-specific GH receptor gene-disrupted (LiGHRKO) mice have decreased endocrine IGF-I, increased local IGF-I, and altered body size, body composition, and adipokine profiles. Endocrinology. 155:1793–1805. 10.1210/en.2013-2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R.A., Burke D., and Nadon N.. 1999. Announcement: four-way cross mouse stocks: a new, genetically heterogeneous resource for aging research. J. Gerontol. A Biol. Sci. Med. Sci. 54:B358–B360. [DOI] [PubMed] [Google Scholar]
- Miller, R.A., Harrison D.E., Astle C.M., Floyd R.A., Flurkey K., Hensley K.L., Javors M.A., Leeuwenburgh C., Nelson J.F., Ongini E., et al. 2007. An Aging Interventions Testing Program: study design and interim report. Aging Cell. 6:565–575. 10.1111/j.1748-1716.1994.tb09816.x [DOI] [PubMed] [Google Scholar]
- Nanni, P., Nicoletti G., Palladini A., Croci S., Murgo A., Ianzano M.L., Grosso V., Stivani V., Antognoli A., Lamolinara A., et al. 2012. Multiorgan metastasis of human HER-2+ breast cancer in Rag2−/−;Il2rg−/− mice and treatment with PI3K inhibitor. PLoS One. 7:e39626. 10.1371/journal.pone.0039626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein, S.J., Kuo S.-H., Tasset I., Arias E., Koga H., Fernandez-Carasa I., Cortes E., Honig L.S., Dauer W., Consiglio A., et al. 2013. Interplay of LRRK2 with chaperone-mediated autophagy. Nat. Neurosci. 16:394–406. 10.1038/nn.3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Molina, A., Lopez-Guadamillas E., Mattison J.A., Mitchell S.J., Muñoz-Martin M., Iglesias G., Gutierrez V.M., Vaughan K.L., Szarowicz M.D., González-García I., et al. 2015. Pharmacological inhibition of PI3K reduces adiposity and metabolic syndrome in obese mice and rhesus monkeys. Cell Metab. 21:558–570. 10.1016/j.cmet.2015.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.-S., Kim D.-H., and Yoon S.-Y.. 2016. Regulation of amyloid precursor protein processing by its KFERQ motif. BMB Rep. 49:337–342. 10.5483/BMBRep.2016.49.6.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petiot, A., Ogier-Denis E., Blommaart E.F.C., Meijer A.J., and Codogno P.. 2000. Distinct classes of phosphatidylinositol 3¢-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275:992–998. 10.1074/jbc.275.2.992 [DOI] [PubMed] [Google Scholar]
- Qi, L., Zhang X.-D., Wu J.-C., Lin F., Wang J., DiFiglia M., and Qin Z.-H.. 2012. The role of chaperone-mediated autophagy in huntingtin degradation. PLoS One. 7:e46834. 10.1371/journal.pone.0046834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud, F.I., Eccles S.A., Patel S., Alix S., Box G., Chuckowree I., Folkes A., Gowan S., De Haven Brandon A., Di Stefano F., et al. 2009. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol. Cancer Ther. 8:1725–1738. 10.1158/1535-7163.MCT-08-1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, J.L., Suh Y., and Cuervo A.M.. 2014. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 20:417–432. 10.1016/j.cmet.2014.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu, P.V., Takegawa K., Fry M.J., Stack J.H., Waterfield M.D., and Emr S.D.. 1993. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 260:88–91. 10.1126/science.8385367 [DOI] [PubMed] [Google Scholar]
- Valdor, R., Mocholi E., Botbol Y., Guerrero-Ros I., Chandra D., Koga H., Gravekamp C., Cuervo A.M., and Macian F.. 2014. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat. Immunol. 15:1046–1054. 10.1038/ni.3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Martinez-Vicente M., Krüger U., Kaushik S., Wong E., Mandelkow E.-M., Cuervo A.M., and Mandelkow E.. 2009. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum. Mol. Genet. 18:4153–4170. 10.1093/hmg/ddp367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendeler, M., and Sandhoff K.. 2009. Hexosaminidase assays. Glycoconj. J. 26:945–952. 10.1007/s10719-008-9137-5 [DOI] [PubMed] [Google Scholar]
- Xia, H.-G., Najafov A., Geng J., Galan-Acosta L., Han X., Guo Y., Shan B., Zhang Y., Norberg E., Zhang T., et al. 2015. Degradation of HK2 by chaperone-mediated autophagy promotes metabolic catastrophe and cell death. J. Cell Biol. 210:705–716. 10.1083/jcb.201503044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xilouri, M., Brekk O.R., Landeck N., Pitychoutis P.M., Papasilekas T., Papadopoulou-Daifoti Z., Kirik D., and Stefanis L.. 2013. Boosting chaperone-mediated autophagy in vivo mitigates α-synuclein-induced neurodegeneration. Brain. 136:2130–2146. 10.1093/brain/awt131 [DOI] [PubMed] [Google Scholar]
- Xilouri, M., Brekk O.R., Polissidis A., Chrysanthou-Piterou M., Kloukina I., and Stefanis L.. 2016. Impairment of chaperone-mediated autophagy induces dopaminergic neurodegeneration in rats. Autophagy. 12:2230–2247. 10.1080/15548627.2016.1214777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, G., Murashige D.S., Humphrey S.J., and James D.E.. 2015. A Positive Feedback Loop between Akt and mTORC2 via SIN1 Phosphorylation. Cell Rep. 12:937–943. 10.1016/j.celrep.2015.07.016 [DOI] [PubMed] [Google Scholar]
- Yuan, Y., Pan B., Sun H., Chen G., Su B., and Huang Y.. 2015. Characterization of Sin1 Isoforms Reveals an mTOR-Dependent and Independent Function of Sin1γ. PLoS One. 10:e0135017. 10.1371/journal.pone.0135017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, X., Overmeyer J.H., and Maltese W.A.. 2006. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J. Cell Sci. 119:259–270. 10.1242/jcs.02735 [DOI] [PubMed] [Google Scholar]
- Zhang, C., and Cuervo A.M.. 2008. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat. Med. 14:959–965. 10.1038/nm.1851 [DOI] [PMC free article] [PubMed] [Google Scholar]