Abstract
Purpose:
Mutations of PIK3CA have recently been shown to play an important role in the pathogenesis and progression of breast neoplasms. The prevalence of PIK3CA in Chinese breast cancer patients may be underestimated. Therefore, we investigated the distribution of somatic PIK3CA mutation in Chinese breast cancer patients and explored their role in tumor phenotypes.
Methods:
Mutational analysis of PIK3CA was done in 113 primary breast cancers of Chinese women used Amplification refractory mutation system (ARMS). The relationship of PIK3CA mutations with several clinicopathologic characteristics was analyzed.
Results:
PIK3CA gene mutation was identified in 43(38.05%) cases and has a more significant difference between exon 9 and 20. HER2 gene amplification was 32.6% in 43 cases of PIK3CA mutation, but 37.1% in 70 cases of non-mutation (χ2 = 0.245, P > 0.05). There was no significant correlation of the age distribution, lymph node status, histological tumor grading, ER and/or PR and P53 between 2 groups (P > 0.05).
Conclusion:
A high frequency of somatic PIK3CA mutation was detected in Chinese breast cancer patients, especially in exon 20. The relationship between PIK3CA gene mutation and clinical pathological features of breast cancer needs to be further studied in a large series of patients. PIK3CA mutations seem to have the potential to be used in target treatment and as an indicator of prognosis.
Keywords: PIK3CA, ARMS, clinicopathologic characteristics, breast cancer, Chinese
Introduction
Many cell signaling pathways play a crucial role by changing a cell’s biological characteristics to cause a variety of neoplasms. The PI3K-AKT-mTOR signaling pathway is a complicated, critical intracellular pathway that plays an important role in the pathogenesis and progression of human cancers.1 AKT has been shown to be frequently activated in various types of tumors. Phosphatidylinositol 3-kinase (PI3 K) is an AKT activator, which participates in the regulation of cell growth, proliferation, survival, and motility. The PI3 K heterodimer consists of 2 subunits: regulatory subunit (P85) and catalytic subunit (p110). PIK3CA is located on chromosome 3q26.3, which encodes the p110α catalytic subunit of PI3 K. The majority of PIK3CA mutations cluster in 2 hotspot regions, including exons 9 and 20, which encode a part of the helical and kinase domains, respectively, and play an essential role in the activation of PI3 K. Therefore, PIK3CA mutations trigger the activation of oncogenes, which can act on the PI3K-AKT-mTOR signal transduction pathway, also leading to continuous AKT activation and regulating growth in several malignancies. A high frequency of PIK3CA mutations has been reported in many neoplasms, such as ovarian, colorectal, and lung cancers.2 In recent years, emerging data found that breast cancer cells have high PI3 K pathway activity, which is one of the most common mutations besides the HER2 amplification and P53 mutation.3-5 According to the Cancer Genome Atlas Network (TCGA), the percentage of PIK3CA mutations is 34%.6 Reports of the prevalence of PIK3CA mutations varied among studies from 15.6% to 28.3% in Chinese breast cancer patients.6 This mutation frequency may be underestimated. Many studies have reported on the clinicopathologic characteristics of breast cancers with HER2 amplification and P53 mutation, but thus far, only a few studies have reported on the clinicopathologic characteristics of breast cancers with PIK3CA mutation. Elucidation of the features of breast cancers with PIK3CA mutations are important for targeted treatment and potential prognoses. In this observational study, we analyzed the frequency of PIK3CA mutation in tumor tissues from 113 Chinese breast cancer patients using the Amplification refractory mutation system and explored their role in tumor phenotypes.
Materials and Methods
Tissue Samples
A total of 113 FFPE breast specimens were included in this study. They were collected from patients aged between 30 and 88 years old (median age 49) diagnosed with invasive breast carcinoma (IBC) between 2016 and 2017. The IBC diagnosis was made by evaluating the morphology according to the WHO classification. The pathological types included non-specific invasive ductal carcinoma (110 cases), infiltrating micropapillary carcinoma (1 case), invasive lobular carcinoma (1 case), and mucinous carcinoma (1 case). There were 37 histological grade III cases in the pathological diagnosis and 51 cases of metastasis in the axillary lymph nodes. Histopathological parameters, immunophenotype results, and patient outcome data were obtained from pathology and hospital records. This study was approved by the Human Research Ethics Board of Shanxi Provincial Cancer Hospital (Approval number: 2019075). All patients provided written informed consent prior to enrollment in the study.
Amplification Refractory Mutation System
Amplification refractory mutation system (ARMS): DNA was extracted from FFPE breast tumor specimens using the QIAamp DNA FFEP Tissue Kit (Qiagen, Germany) according to the manufacturer’s instructions. ARMS analysis was performed using the LightCycler 480 Real-Time PCR system (Roche Diagnostics Ltd. Switzerland) and PIK3CA Five-Mutations Detection Kit (Amoy Diagnostics Co., Xiamen, China). The kit contains a tube of Taq DNA polymerase, a tube of positive quality control, a tube of the externally controlled reaction mixture, and 5 tubes of PIK3CA reaction mixture. The E542 K and E545 K primers were multiplexed with 0.2 µL Taq polymerase per 20 µL reaction mixture. The others were multiplexed with 0.18 µL per 20µL reaction mixture. Each reaction was performed in a final volume of 15 μL, containing 5 µL PIK3CA reaction mixture, 5µL sample (DNA concentration 2 ng/μL), and 5 µL purified water. All reaction mixture was sextuplicated on the LightCycler 480 Real-Time PCR system. The reaction mixture was incubated at 95°C for 10 min, followed by 15 cycles of 95°C for 25 s; 64°C for 20 s; and 72°C for 20 s, followed by 31 cycles of 93°C for 25 s; 60°C for 35 s; and 72°C for 20 s. The signals were collected at 60°C in the third stage.
The Ct value was obtained at the inflection point, rising from the amplification curve according to the actual situation. As the percentage of mutation in the sample varies, the Ct values are also different. At the same time, a value for the change in the threshold cycle (△Ct) = (mutation Ct − external control Ct was defined for each reaction. The cutoff △Ct value was determined to be 1 Ct below the lowest △Ct value observed in all reactions for each assay. The cutoff △Ct was defined as 11 for the H1047 R assay and 12 for the H1047 L, the E542 K, the E545 K, and the E545D assays.
Immunohistochemistry
An immunohistochemistry (IHC) assay of formalin-fixed and paraffin-embedded (FFPE) specimens was performed using Dako’s EnVision System (Glostrup, Denmark). This is a 2-step method in which the application of the primary antibody is followed by a polymeric conjugate consisting of many secondary antibodies bound directly to a dextran backbone, including ER (clone1D5, DAKO), PR (Clone pgR 1294, DAKO), P53 (Clone DO-7, DAKO) and HER2 (VENTANA anti-HER2/neu(4B5), Roche Diagnostics GmbH). Procedures were provided by the manufacturers and completed on a Roche automated IHC instrument (USA). The antibodies and IHC procedures used in this study were part of a standard IHC panel for a pathology laboratory.
Results Interpretation
HER2 testing guidelines in breast cancer, 2014 edition, China): IHC (0) were defined as no staining is observed or membrane staining that is incomplete and is faint/barely perceptible and in ≤ 10% of tumor cells; IHC 1+ were defined as incomplete membrane staining that is faint/barely perceptible and in >10% of tumor cells; IHC 2+ were defined as weak to moderate complete membrane staining observed in > 10% of tumor cells or robust and complete membrane staining observed in ≤10% of tumor cells; IHC 3+ were defined as circumferential membrane staining, that is, complete, intense, and in >10% of tumor cells.
Positive localization of ER, PR, and P53 is in the nucleus. It is the positivity of ER and/or PR (ER/PR) that more than 10% of tumor nuclei are brown or tan. The positivity of P53 was classified into wild-type and mutant-type. The wild type presented few tumor nuclei, with varying color intensity. The mutant-type includes missense and nonsense mutations. About 60% of tumor nuclei are brown with a missense mutation. A nonsense mutation means that tumor cells do not express the P53 protein.
Fluorescence In Situ Hybridization
Fluorescence in situ hybridization (FISH) was performed on FFPE specimens using the PathVysion HER2 DNA Probe Kit (Abbott Molecular Inc.), with 20 nuclei scored for each tissue specimen. The HER2 gene probe was a dual-color probe targeting the HER2 gene (SpectrumOrange) on chromosome 17 (SpectrumGreen). Washes were performed and slides were counterstained with 4’, 6-diamidino-2-phenylindole (phenylindole (DAPI) anti-fade solution before being analyzed under a fluorescence microscope.
Results interpretation (HER2 testing guideline in breast cancer, 2014 edition, China): Signals from 20 non-overlapping nuclei in representative fields were enumerated for the HER2 and CEP17 numbers, and the results were scored as the ratio of HER2 to CEP17 signal numbers. Positive results were defined as ratios of at least 2.0 or ratios less than 2.0, with an average HER2 copy number of 6.0 signals or more per cell. Equivocal results were considered as ratios less than 2.0, with an average HER2 copy number between 4.0 and 6.0 signals per cell. Negative results were defined as ratios less than 2.0, with an average HER2 copy number of than 4.0 signals per cell.
Statistical Analysis
All data were analyzed by SPSS 20.0 software. The statistical comparison of differences in HER2 amplification in PIK3CA mutation vs non-mutation groups is listed in Table 1; the comparison of the relationship between PIK3CA and clinic-pathological parameters is listed in Table 2 and was performed using the Pearson chi-squared test and continuity correction test. Results were considered statistically significant if the P-value was < 0.05.
Table 1.
Rationship Between PIK3CA Mutations and HER2 Amplification or p53 Mutation.
| PIK3CA mutations | χ2 | P value | ||
|---|---|---|---|---|
| Mutant | Wild | |||
| HER2 ampilification | ||||
| Positive Negative |
14 29 |
26 44 |
0.245 | 0.621 |
| P53 mutation | ||||
| Mutant-type Wild-type |
10 33 |
18 52 |
0.086 | 0.769 |
Table 2.
Relationship between PIK3CA Mutation Status and Clinical-Pathological Parameters.
| Characteristics | PIK3CA mutations | χ2 | P value | |
|---|---|---|---|---|
| Mutant | Wild | |||
| Age(years) | ||||
| ≥49 | 30 | 38 | 2.664 | 0.103 |
| <49 | 13 | 32 | ||
| Lymph node status | ||||
| Positive | 20 | 32 | 0.053 | 0.817 |
| Negative | 23 | 38 | ||
| Histological grade | ||||
| I-II | 29 | 47 | 0.001 | 0.974 |
| III | 14 | 23 | ||
| ER/PR | ||||
| Positive | 30 | 50 | 0.036 | 0.850 |
| 13 | 20 | |||
Results
Frequency and Location of PIK3CA Mutations
Mutational analysis of PIK3CA was done in 113 primary breast cancers; finally, 43 mutations were identified in total. Of these 43 mutations, 7 and 35 mutations clustered in exon 9 and 20, respectively. Furthermore, 1 out of 43 cases with aberrant PIK3CA have mutations simultaneously in exon 9 and 20 (E545K+H1047 L). The detected mutations in exon 9 are changing the codon sequences of the helical domain as follows: 4 cases with E545 K (Figure 1) and 3 cases with E542 K (Figure 2). Two mutations in exon 20 that change the codon sequences of kinase domain were also identified in the present study, including 10 cases with H1047 L (Figure 3), 24 cases with H1047 R (Figure 4), and 1 case with 1047L+1047 R. E545D from exon 9 was not detected in any tumor samples. The mutations have a more significant difference between exon 9 and 20: the mutation rate of exon 20 is much higher than exon 9. The hotspot mutation H1047 R was observed at a frequency of 21.24% (24/113), representing 68.57% (24/35) of the PIK3CA exon 20 mutations.
Figure 1.
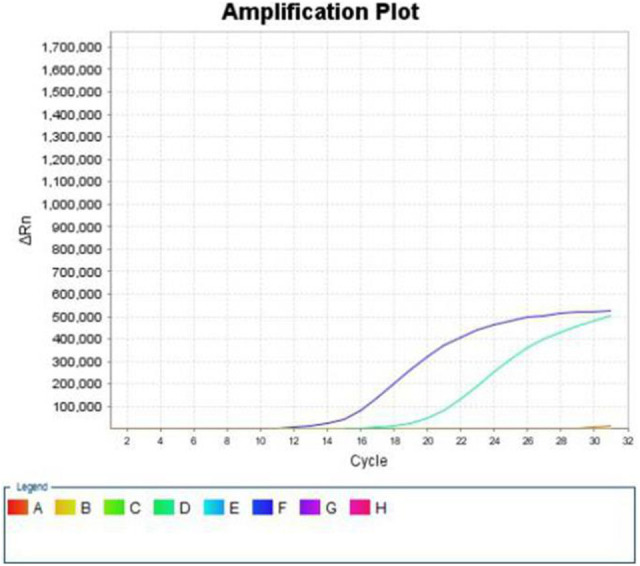
ARMS results: E545K mutation of exon 9.
Figure 2.
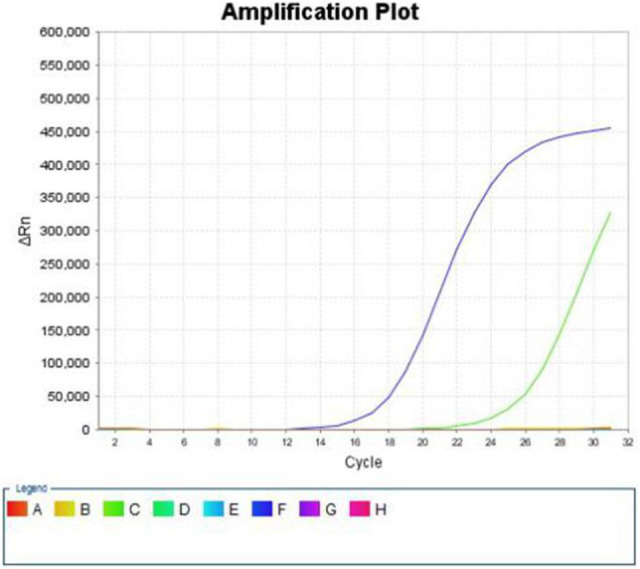
ARMS results: E542K mutation of exon 9.
Figure 3.
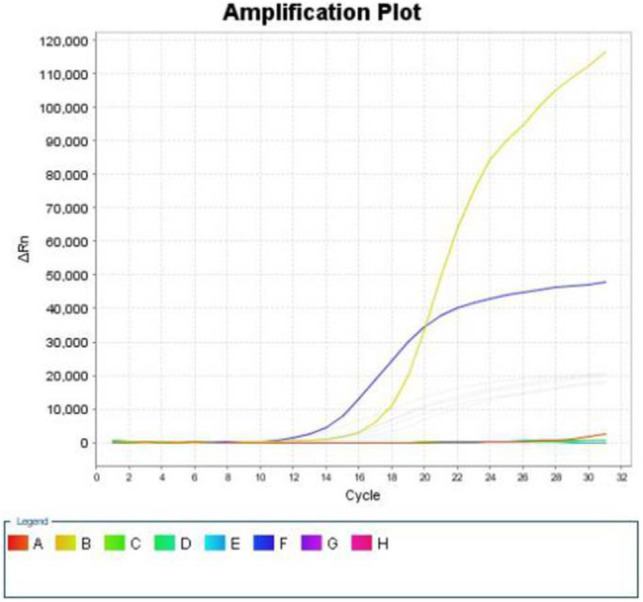
ARMS results: H1047L mutation of exon 20.
Figure 4.
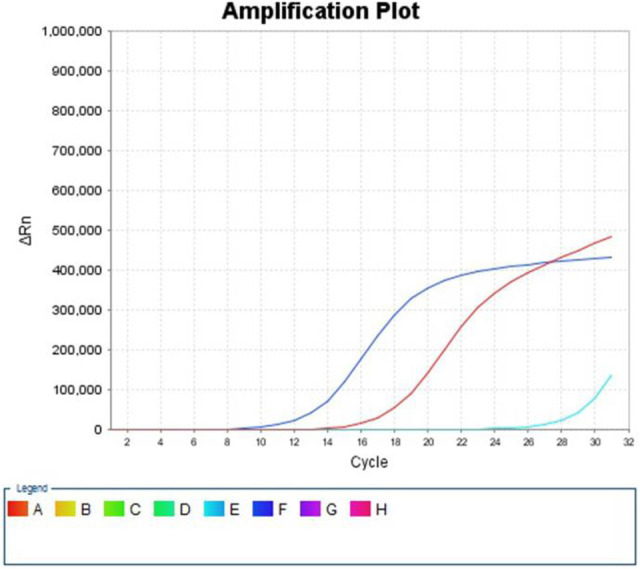
ARMS results: H1047R mutation of exon 20.
Relationship of PIK3CA Mutations With HER2 Amplification and P53 Mutation
Thirty-two out of 113 cases identified positive using the IHC for the HER2 protein. Nine in 29 cases with equivocal results. The percentage of mutant-type P53 was 24.8% identified by IHC. We studied the relationship between PIK3CA mutations and HER2 expression or P53 mutation (Table 1). HER2 expression and P53 mutation were not significantly associated with PIK3CA mutations.
PIK3CA Mutations and Clinicopathologic Characteristics of Chinese Breast Cancers
The relationship of PIK3CA mutations with clinicopathologic characteristics is shown in Table 2. There is no significant correlation between PIK3CA mutations and clinicopathologic characteristics, including age distribution, lymph node status, histological tumor grading, and ER/PR.
Discussion
PIK3CA is one of the most frequently mutated genes in breast cancer, and most somatic mutations (80–90%) are in several “hot spots”: E542 K or E545 K in exon 9 and H1047 R or H1047 L in exon 20. Samuels et al first reported PIK3CA mutation in human cancer. Since then, many studies have demonstrated the occurrence of PIK3CA gene mutations in breast cancer. These published reports vary according to the population studied and also the number of exons analyzed in the gene. The observed frequency of PIK3CA mutations is approximately 29% and 46.5% in Japan and West China Hospital (Sichuan, China), respectively.6,7 In India, the recorded frequency of somatic mutations in PIK3CA was 34%, which was similar to our results, and the same study concluded that H1047 R and H1047 L were deleterious in nature.8 In the present study, we examined 113 breast cancer samples for somatic mutations in exon 9 and exon 20 of the PIK3CA gene. Among these 113 samples, the mutation in PIK3CA was detected in 43 (38%) of the samples. The differences in the frequency of mutation across various studies may be attributed to sampling size, geographical distribution, and analysis techniques.
In several studies, a high frequency of PIK3CA mutations located in exon 9 has been found in lobular carcinoma,9 but PIK3CA mutations located in exon 20 were more common in ductal carcinoma. The hotspot in this group was H1047 R, which may be related to the high proportion of invasive ductal carcinoma. The data from Mattia Barbareschi et al’s study showed that exon 9 PIK3CA mutations are not only typical of infiltrating lobular carcinomas but also independently associated with early recurrence and death.10 It was reported simultaneously in that report that exon 20 PIK3CA mutations are associated with optimal prognosis. Later, Lai YL et al and Flavia R. Mangone FR et al reported that PIK3CA exon 20 mutations are associated with poor prognoses in breast cancer patients.11,12 Therefore, the mutation of PIK3CA in different exons has distinction and different prognostic value in various histological types. The reason for the inconsistent results may be due to differences in the specific molecular mechanisms that caused different PIK3CA mutation regions will increase the activity of PI3 K kinase, causing different biological effects. In the follow-up work this year, 4 cases were lost, and there were only 13 deaths in total, of which 9 (9/70) were in the wild type group, and 3 (3/43) were in the mutation group. Survival analysis was conducted among 109 breast cancer patients with at least 28 months follow-up. Survival was evaluated between patients with hotspot and wild type, and there was no difference in iDFS or OS. Our preliminary results need to be confirmed by a future study, including more number of patients and a longer follow-up period.
HER2 amplification can activate the PI3K-AKT-mTOR pathway; therefore, tumors with HER2 amplification do not require further activation of this pathway by PIK3CA mutations. Recent studies show that PIK3CA-mutated and HER2-positive breast cancer patients treated with HER2 target therapies have poor PCR compared with wild-type tumors.13-15 Using the cell models established, it showed that the co-occurrence of PIK3CA mutation and HER2 gene amplification greatly enhanced proliferation, and the treatment with 2 inhibitors is more potent in inhibiting cell proliferation than using either inhibitor alone in cells with both HER2 amplification and PIK3CA mutations.16 Moreover, Saal et al17 reported a significant positive association between PIK3CA mutations and HER2 overexpression, suggesting that more than one mechanism activating the PI3K-AKT-mTOR pathway might be necessary for carcinogenesis, progression, target treatment, and prognosis of breast cancer. In the present study, we failed to show a significant association between HER2 amplification and PIK3CA mutations. Bhuvanesh Singh et al18 and Arezoo Astanehe et al19 reported that PIK3CA mutations and P53 mutations were mutually exclusive. Inconsistent with their result, we have not found a significant association between PIK3CA mutations and P53 mutation. Our results need to be interpreted with caution and should be confirmed through further studies using a more significant number of tumors.
Now, the relationship between PIK3CA mutations and clinical-pathological features of breast cancer is still controversial. Many reports have shown that PIK3CA-mutated tumors were more frequent in hormone receptor-positive tumors.9,17,20 Mohammed A. Aleskandarany et al21 found that positive PIK3CA expression was associated with weak clinicopathological variables, including higher histopathological grade, larger tumor size, higher proliferative fraction, vascular invasion, and lymph node involvement. In this assay, no significant association was found between PIK3CA mutations and some important clinicopathologic characteristics regarding patient age distribution (≤ 49 or > 49 years), lymph node status, histological tumor grading, and ER/PR. Likewise, no significant differences were found regarding patient age, clinical stage, tumor size, lymph node metastasis, hormonal receptor status, and HER2 status by Mattia Barbareschi et al,9 and Mangone et al.12 Similarly, no significant associations were found between PIK3CA mutations and clinicopathologic characteristics in this study. The inconsistencies across various studies may be attributed to sampling selection(age, histological subtypes, molecular types, clinical stages), ethnic differences, etc. Therefore, the relationship between PIK3CA mutations and clinical-pathological characteristics of breast cancer should be confirmed in a more extensive series of patients and further studies.
The PI3K-AKT-mTOR signaling pathway–targeted therapy has become a hot topic. The resistance of endocrine therapy is a major clinical challenge. The mechanism is very complex, and one of the important mechanisms is the activation of the PI3K-AKT-mTOR signaling pathway. Everolimus, an mTOR inhibitor, has been used in clinical trials in breast cancer with hormone receptor-positive and HER2 negative, which can significantly improve progression-free survival.22 Furthermore, Chen et al demonstrated the PI3K/mTOR dual inhibitor BEZ235 and Trichostain, resulting in synergistic growth inhibition and apoptosis of tumor cell lines.23 This result highlights the unique characteristics arising from their co-occurrence, which leads us to re-think the progression mechanism and treatment of tumors. Recently, Rimawi MF et al24 showed that PI3 K pathway activation is associated with resistance to lapatinib and trastuzumab in breast cancer.
Further studies are necessary to investigate how to use these molecular biomarkers to identify patients who may respond to anti-HER2 treatment alone up-front, without chemotherapy. It will stimulate physicians to advise PIK3CA mutation screening in clinical practice to avoid unnecessary treatment. Furthermore, Dupont J et al’s study suggests that PIK3CA status frequently changes between primary and metastatic, and which emphasizes the necessity of assessing the PIK3CA mutations for selection of PIK3CA inhibitor therapy.25 PIK3CA mutations are essential for tumor growth, so the evaluation and search for pharmacologic inactivation of mutant forms of PIK3CA is becoming a fascinating field of research. The study results of D. Thirumal Kumar et al showed a convincing phenomenon that PIK3CA mutation occurring in the conserved region influences the stability of the protein using molecular dynamics simulation analysis, and different mutation sites were shown to have different effects on protein stability.26,27 These findings solve many mysteries associated with the development of breast cancer and continuous drug failure to treat the cause. Future studies will further enhance drug efficiency and efficacy and enhance the development of individualized medicine.
Footnotes
Authors’ Note: Jing Lian, Li-Xia Wang and Yan-Feng Xi identified case of interest and designed the study. Hui-Wen Wang and Peng Bu carried out the studies. Jing Lian and En-Wei Xu collected data and performed the statistical analysis. Jing Lian and Li-Xia Wang produced the initial draft of the manuscript. Jing Lian, Li-Xia Wang and Yan-Feng Xi contributed to revise the manuscript. All authors participated in writing, editing and reviewing the manuscript, and gave final approval for publication. Jing Lian takes responsibility for the integrity of the data and the accuracy of the data analysis.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the Institute Research Foundation of Shanxi Cancer Hospital.
ORCID iD: Jing Lian  https://orcid.org/0000-0001-9753-1980
https://orcid.org/0000-0001-9753-1980
References
- 1. Yuan TL, Cantley LC. PI3 K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27(41):5497–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jong WL, Young HS, Su YK, et al. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005;24(8):1477–1780. [DOI] [PubMed] [Google Scholar]
- 3. Hernandez-Aya LF, Gonzalez-Angulo AM. Targeting the phosphatidylinositol 3-kinase signaling pathway in breast cancer. Oncologist. 2011;16(4):404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sohei Y, Hitoshi T, Masashi T, Keichi L, Seiichi T, Osamu M. PIK3CA mutations is an early event in the development of endometriosis-associated ovarian clear cell adenocarinoma. J Pathol. 2011;225(2):189–194. [DOI] [PubMed] [Google Scholar]
- 5. Miron A, Varadi M, Carrasco D, et al. PIK3CA mutations in in situ and invasive breast carinomas. Cancer Res. 2010;70(14):5674–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Deng L, Zhu X, Sun Y, et al. Prevalence and prognosis role of PIK3CA/AKT1 mutations in Chinese breast cancer patients. Cancer Res Treat. 2019;51(1):128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maruyama N, Miyoshi Y, Taguchi T, Tamaki Y, Monden M, Noguchi S. Clinicopathologic analysis of breast cancers with PIK3CA mutations in Japanese women. Clin Cancer Biol. 2007;13(2):408–415. [DOI] [PubMed] [Google Scholar]
- 8. Sudhakar N, George PDC, Thirumal KD, Chakraborty C, Anand K, Suresh M. Deciphering the impact of somatic mutations in exon 20 and exon 9 of PIK3CA gene in breast tumors among Indian women through molecular dynamics approach. J Biomol Struct Dyn. 2015;34(1):29–41. [DOI] [PubMed] [Google Scholar]
- 9. Buttitta F, Felicioni L, Barassi F, et al. PIK3CA mutation and histological type in breast carcinoma: high frequency of mutations in lobular carcinoma. J Pathol. 2006;208(3):350–355. [DOI] [PubMed] [Google Scholar]
- 10. BarBareschi M, Buttitta F, Felicioni L, et al. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res. 2007;13(20):6064–6069. [DOI] [PubMed] [Google Scholar]
- 11. Lai YL, Mau BL, Cheng WH, Chen HM, Chiu HH, Tzen CY. PIK3CA exon 20 mutation is independently associated with a poor prognosis in breast cancer patients. Ann Surg Oncol. 2008;15(4):1064–1069. [DOI] [PubMed] [Google Scholar]
- 12. Mangone FR, Bobrovnitchaia IG, Salaorni S, Manuli E, Nagai MA. PIK3CA exon 20 mutations are associated with poor prognosis in breast cancer patients. Clinics. 2012;67(11):1285–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loibl S, Majewski I, Guarner V, et al. PIK3CA mutations are associated with reduced pathological complete response rates in primary HER2-positive breast cancer-pooled analysis of 967 patients from five prospective trials investigating lapatinib and trastuzumab. Ann Oncol. 2016;27(8):1519–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loibl S, Von MG, Schneeweiss A, et al. PIK3CA mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2(HER2) therapy in primary HER2-overexpressing breast cancer. J Clin Oncol. 2014;32(29):3212–3220. [DOI] [PubMed] [Google Scholar]
- 15. Majewski IJ, Nuciforo P, Mittempergher L, et al. PIK3CA mutations are associated with decreased benefit to neoadjuvant human epidermal growth factor receptor 2-targeted therapies in breast cancer. J Clin Oncol. 2015;33(12):1334–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dong L, Meng F, WU L, et al. Cooperative oncogenic effect and cell signaling crosstalk of co-occurring HER2 and mutant PIK3CA in mammary epithelial cells. Int J Oncol. 2017;51(4):1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saal LH, Holm K, Maurer M, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65(7):2554–2559. [DOI] [PubMed] [Google Scholar]
- 18. Singh B, Reddy PG, Goberdhan A, et al. p53 regulates cell survival by inhibiting PIK3CA in squamous cell carcinomas. Genes Dev. 2002;16(8):984–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Astanehe A, Arenillas D, Wasserman WW, et al. Mechanisms underlying p53 regulation of PIK3CA transcription in ovarian surface epithelium and in ovarian cancer. J Cell Sci. 2008;121(pt 5):664–674. [DOI] [PubMed] [Google Scholar]
- 20. Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68(15):6084–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aleskandarany MA, Rakha EA, Ahmed MA, et al. PIK3CA expression in invasive breast cancer: a biomarker of poor prognosis. Breast Cancer Res Treat. 2010;122(1):45–53. [DOI] [PubMed] [Google Scholar]
- 22. Rugo HS, Keck S. Reversing hormone resistance: have we found the golden key? J Clin Oncol. 2012;30(22):2707–2709. [DOI] [PubMed] [Google Scholar]
- 23. Chen L, Jin T, Zhu K, et al. PI3K/mTOR dual inhibitor BEZ235 and histone deacetylase inhibitor trichostain a synergistically exert anti-tumor activity in breast cancer. Oncotarget. 2017;8(7):11937–11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rimawi MF, De Angelis C, Contreras A, et al. Low PTEN levels and PIK3CA mutations predict resistance to neoadjuvant lapatinib and trastuzumab without chemotherapy in patients with HER2 over-expressing breast cancer. Breast Cancer Res Treat. 2018;167(3):731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dupont JJ, Laenkholm AV, Knoop A, et al. PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin Cancer Res. 2011; 17(4): 667–677. [DOI] [PubMed] [Google Scholar]
- 26. Thirumal KD, George PDC. Investigating the inhibitory effect of wortmannin in the hotspot mutation at codon 1047 of PIK3CA kinase domain: a molecular docking and molecular dynamics approach. Adv Protein Chem Struct Biol. 2016;102:267–297. [DOI] [PubMed] [Google Scholar]
- 27. Thirumal KD, George PDC. Role of E542 and E545 missense mutations of PIK3CA in breast cancer: a comparative computational approach. J Biomol Struct Dyn. 2017;35(12):2745–2757. [DOI] [PubMed] [Google Scholar]


