Abstract
Protein arginine methyltransferase 5 (PRMT5) is an important type II arginine methyltransferase that can play roles in cancers in a highly tissue-specific manner, but its role in the carcinogenesis and metastasis of head and neck squamous cell carcinoma (HNSCC) remains unclear. Here, we detected PRMT5 expression in HNSCC tissues and performed series of in vivo and in vitro assays to investigate the function and mechanism of PRMT5 in HNSCC. We found that PRMT5 was overexpressed in dysplastic and cancer tissues, and associated with lymph node metastasis and worse patient survival. PRMT5 knockdown repressed the malignant phenotype of HNSCC cells in vitro and in vivo. PRMT5 specific inhibitor blocked the formation of precancerous lesion and HNSCC in 4NQO-induced tongue carcinogenesis model, prevented lymph node metastasis in tongue orthotopic xenograft model and inhibited cancer development in subcutaneous xenograft model and Patient-Derived tumor Xenograft (PDX) model. Mechanistically, PRMT5-catalyzed H3R2me2s promotes the enrichment of H3K4me3 in the Twist1 promoter region by recruiting WDR5, and subsequently activates the transcription of Twist1. The rescue experiments indicated that overexpressed Twist1 abrogated the inhibition of cell invasion induced by PRMT5 inhibitor. In summary, this study elucidates that PRMT5 inhibition could reduce H3K4me3-mediated Twist1 transcription and retard the carcinogenesis and metastasis of HNSCC.
Abbreviations: ATCC, American Type Culture Collection; Co-IP, Co-immunoprecipitation; EMT, Epithelial-mesenchymal transition; FBS, Fetal bovine serum; HNSCC, Head and neck squamous cell carcinoma; HE, Hematoxylin-eosin staining; IHC, Immunohistochemistry assay; 4NQO, 4-Nitroquinoline-1-Oxide; OSCC, Oral squamous cell carcinoma; PRMT5, Protein arginine methyltransferase 5; PCK, Pan-cytokeratin; WDR5, WD repeat containing protein 5; PDX, Patient-Derived tumor Xenograft
Keywords: Head and neck cancer, Arginine methylation, PRMT5, Carcinogenesis, H3K4me3, Twist1
Introduction
Head and neck squamous cell carcinoma (HNSCC), which includes malignant squamous lesions arising in the oral cavity, larynx and pharynx, is the sixth most common cancer in the world [1], [2]. Most HNSCC patients are diagnosed at locally advanced or metastatic stages and the 5-year survival rates is still low [3], [4]. Thus, identifying the genetic and epigenetics mechanisms driving HNSCC initiation, invasion and metastasis is a prime target for novel biomarkers and precision treatment strategies.
Protein arginine methyltransferase 5 (PRMT5), the main type II arginine methyltransferase, catalyzes most arginine symmetry dimethylation and monomethylation of histone and non-histone proteins [5], [6], [7], [8], [9], [10], [11], [12]. Aberrant expression of PRMT5 could affects cell proliferation, differentiation and movement, which leads to disease progression, especially cancer [5], [8]. Recent evidence shows that PRMT5 is frequently upregulated in hematological malignancies and some solid tumors, including squamous cell carcinoma (SCC), and highly associated with poor prognosis, but the underlying mechanisms needs further clarification [6], [13], [14], [15], [16], [17].
The association between PRMT5 and HNSCC remains limited. Although it was reported that PRMT5 was overexpressed in oral squamous cell carcinoma (OSCC) [16], its potential roles and mechanisms in the carcinogenesis of HNSCC remains unclear. On the other hand, PRMT5 becomes an attractive therapeutic target in PRMT5-dysregulated diseases. PRMT5 inhibitors display clinical potential in a variety of tumors, but data on the effects of these inhibitors on HNSCC are not available [18], [19], [20], [21]. Obviously, it is urgent to disclose the role of PRMT5 in HNSCC carcinogenesis and its potential as a therapeutic target.
In this study, we demonstrated the role of PRMT5 in the carcinogenesis and metastasis of HNSCC and assessed the therapeutic potential of a PRMT5 specific inhibitor in HNSCC using in vivo and in vitro studies. Mechanistically, we revealed that H3R2me2s, catalyzed by PRMT5, promoted the enrichment of H3K4me3 in the Twist1 promoter region by recruiting WDR5, and transcriptional activation of Twist1. Activation of this important transcription factor promoted cancer initiation and invasion. Our findings uncover a novel mechanism of PRMT5 in HNSCC and define a potential therapeutic target for HNSCC.
Methods
Patients, animals and HNSCC cell lines
All experiments involving human tissues were approved by the Ethics Committee of the Hospital of Stomatology, Sun Yat-sen University. Written informed consent was obtained in accordance with the Declaration of Helsinki. All human tissues were collected from the Department of Oral and Maxillofacial Surgery of the First Affiliated Hospital and Hospital of Stomatology, Sun Yat-sen University from 2004 to 2018.
All procedures involving BALB/c nude mice and Sprague-Dawley (SD) rats (the Animal Care Unit of Guangdong, China) were approved by Institutional Animal Care and Use Committee, Sun Yat-sen University. All animal experiments were complied with the ARRIVE guidelines and carried out in accordance with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978).
HSC6 from Professor J. Silvio Gutkind (NIH, Bethesda, MD, USA) and SCC25 from American Type Culture Collection (ATCC) (Manassas, VA, USA) were used as subjects. All cell lines were cultured as previously described [22], [23].
Animal experiments
For the subcutaneous xenograft, 6 × 107 HSC6 cells were subcutaneously inoculated into the back of 4-week-old BALB/c nude mice. For the orthotopic xenograft, 5 × 105 HSC6 cells were inoculated submucosally into the tongue of 4-week-old BALB/c nude mice as described before [24].
For 4-Nitroquinoline-1-Oxide (4NQO)-induced carcinogenetic model, 0.002% 4NQO in the drinking water was used for 12 weeks. Rats were randomly divided into 2 groups (n ≥ 8 for each group) at week 12: control and EPZ015666 (30 mg/kg). At the end of week 22, the rats were sacrificed to obtain the lesions in the tongue tissues.
For Patient-Derived tumor Xenograft (PDX) model, the primary tumor tissues from patients with tongue squamous cell carcinoma were collected, minced into 1∼2 mm3 and implanted into BALB/c nude mice (n = 4, 4 pieces/mice) and passaged on to derived as P1, P2 and subsequent passages as described before [25]. The tumor tissues of the “P2” generation were collected and equally implanted into the subcutaneous of 4-week-old BALB/c mice. When the tumors grew to 50 mm3, mice were randomly divided into two groups: Control and EPZ015666 (30 mg/kg).
Immunochemistry (IHC)
Immunochemistry was performed as previously described with the appropriate primary antibody (PRMT5 1:150, Ki67 1:400, Pan-cytokeratin 1:400, Twist1 1:250) [23]. Information of primary antibodies were listed in supplementary data Table S1. For statistical analysis, 5 fields of view were randomly selected from each sample, and the percentage of positive cells was calculated using Image J software (NIH, Bethesda, MD, USA). The proportion of positive cells was calculated as follows: 0, <25%; 1, <50%; 2, <75%; 3, ≥75%.
Lentivirus transfection and RNA interference
PRMT5-associated lentiviruses (CTRL, MYC-PRMT5, shCTRL, shPRMT5) and Twist1-associated lentiviruses (NC, Flag-Twist1) (Cyagen Biosciences, China) were transfected into HSC6 and SCC25 respectively (Target sequence of shPRMT5: GGACCTGAGAGATGATATA). After 24 hours of infection, the virus solution was removed. Then, puromycin was added to screen for stable transfected cell lines after 3 days of culture.
For siRNA transfection, 20 nM of siRNA against WDR5 (Target sequences are as follows: SiWDR5-1: GCGTGAGGATATGGGATGT; SiWDR5-2: GTCGTGATCTCAACAGCTT; SiWDR5-3: GGAAGTGCCTCAAGACTTT) or a negative control (Ribobio, China) was used. Lipofectamine RNAiMax (Invitrogen, USA) was used for the transfection according to the manufacturer’s instructions.
Proliferation and colony formation assay
Cell proliferation was examined using the Cell Counting Kit-8 (CCK-8, Sigma-Aldrich, USA). For colony formation assay, cells were digested and blown into individual cells. Each group of cells was inoculated into a 6-well (500 cells/well), gently mixed and incubated for 10–14 days. When macroscopic clones appeared, the culture was terminated. After formaldehyde fixation and crystal violet staining, the clone formation rate was calculated by directly counting the clones with naked eyes.
RNA isolation and real-time quantitative PCR (RT-qPCR)
According to the manufacturer's instructions, RNA was extracted by Trizol (Invitrogen, USA) method, and then reverse transcription was performed using Takara's reverse transcription system (Takara, Shiga, Japan) to obtain cDNA. The obtained cDNA was subjected to real-time quantitative PCR by Roche's 20 μL system. The PCR primer sequences were listed in supplementary data Table S2.
Chromatin immunoprecipitation-qPCR (ChIP-qPCR)
ChIP was performed with appropriate antibody ratios (H3R2me2s 1:100; H4R3me2s 1:100; H3K4me3 1:50; WDR5 1:50; Set1 1:50) according to the instructions of a ChIP assay kit (Millipore, USA). The primer sequences were listed as follows:
Primer1: Forward TCCCCAGTTACACTTGGATGC;
Reverse GGCCAGGAACATACTGGCTT.
Primer2: Forward AGCCCTCTGAATACCAAACTGG;
Reverse GGAGGGAGGGGGCACTAATA.
Western blot and Co-immunoprecipitation (Co-IP)
Western blot analysis was performed with primary antibody concentration according to the instructions (GAPDH: 1:2000, PRMT5, Twist, E-cadherin, N-cadherin, WDR5, H3R2me2s and H3K4me3 are 1:1000) and performed as previously described. Co-immunoprecipitation was performed as previously described (H3R2me2s: 1:100; Set1:1:50; H3K4me3:1:100) [26]. Information of primary and secondary antibodies was listed in supplementary data Table S1.
Cell migration and invasion assay
Cell migration and invasion assays were performed in a 24-well plate with 8-μm pore size chamber inserts (Corning, Toledo, OH, USA) as previously described [23].
Data acquisition
Level 3 RNA-Seq data of HNSCC were downloaded from the TCGA data portal (https://portal.gdc.cancer.gov/, up to December 8, 2018). RNA-seq data of ESCC and LUSCC were downloaded from the UCSC Xena (https://xenabrowser.net/datapages/, up to September 30, 2019).
Statistical analysis
Each experiment was repeated at least three times. The experimental data obtained were presented as mean ± SD and analyzed by Graphpad Prism 5 (GraphPad Software, San Diego, CA, USA) and SPSS22.0 statistical software (SPSS, Chicago, IL, USA). The significance was analyzed using chi-square test, Student’s t test, rank sum test, Fish's exact test, one-way ANOVA and Log-rank test as described in each corresponding figure legends. Statistical significance was set at p less than 0.05.
Results
PRMT5 is overexpressed in HNSCC patients and correlates with metastasis and poor prognosis
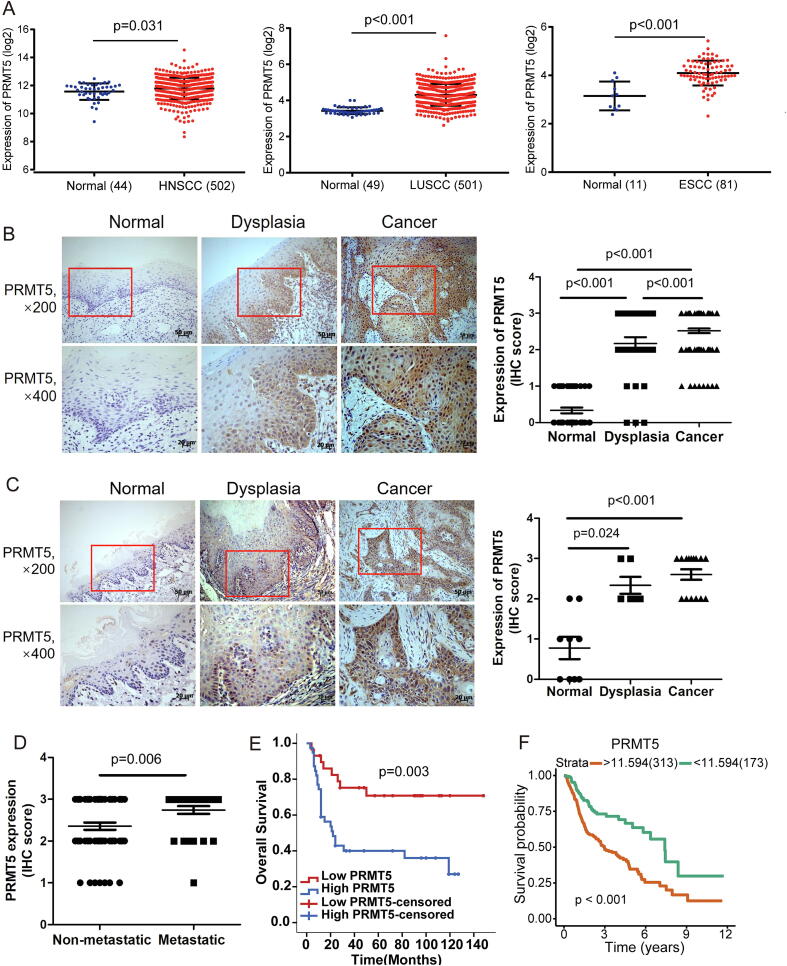
To determine the expression level of PRMT5 in squamous cell carcinoma (SCC) patients, we analyzed 502 head and neck, 501 lung and 81 esophageal SCC patients from The Cancer Genome Atlas (TCGA) database and showed that PRMT5 mRNA level was significantly upregulated in these 3 types of SCC tissues (Fig. 1A). We then examined the protein level of PRMT5 in 104 HNSCC, 30 dysplastic, and 36 normal oral mucosa tissues by immunochemistry. PRMT5 expression was low or absent in normal tissues, but upregulated in dysplastic and cancerous tissues, with positive expression in both cytoplasm and nucleus (Fig. 1B). In an experimental model of carcinogenesis, 4NQO-induced rats SCC, PRMT5 was also increased in dysplastic and cancerous tissues (Fig. 1C). These findings suggested a positive correlation between PRMT5 upregulation and HNSCC progression.
Fig. 1.
PRMT5 is upregulated in HNSCC and correlates with metastasis and patient survival. (A) Analysis of PRMT5 in HNSCC, ESCC, LUSCC and normal control in TCGA database. (Student’s t-test). (B) Representation and quantification of IHC staining of normal oral mucosa (n = 36), dysplasia tissues (n = 30) and HNSCC tissues (n = 104) using anti-PRMT5. Scale bar, 50 μm, 20 μm. (Chi-Square test). (C) Representation and quantification of IHC staining of normal oral mucosa (n = 9), dysplasia tissues (n = 6) and cancer tissues (n = 15) of rats using anti-PRMT5. Scale bar, 50 μm, 20 μm. (Chi-Square test). (D) Quantification of PRMT5 protein levels according to IHC scores in N-regional lymph nodes metastatic HNSCC tissues (n = 31) and non-metastatic HNSCC tissues (n = 62). (Student’s t test). (E) Kaplan–Meier overall survival curve of PRMT5 for HNSCC patients (n = 68) (cut-off value: score = 2). (log-rank test). (F) Kaplan–Meier overall survival curve of PRMT5 for HNSCC patients (n = 486) in TCGA database (cut-off value:11.594). (log-rank test).
Further correlation of PRMT5 expression with clinical pathological parameters in HNSCC patents indicated that high PRMT5 expression was associated with lymph node metastasis and advanced clinical stages, but not with gender, age, T stage or histological grade (Table S3, Fig. 1D). Survival analysis of this cohort showed that patients with lower expression of PRMT5 showed significantly better overall survival (OS) (Fig. 1E). Furthermore, we found a similar trend in TCGA database whereby higher PRMT5 expression indicated lower OS (Fig. 1F). These data provide evidence that PRMT5 is involved in the pathogenesis of HNSCC and could serve as a prognostic biomarker for HNSCC patients.
PRMT5 regulates proliferation and invasion of HNSCC in vitro and in vivo
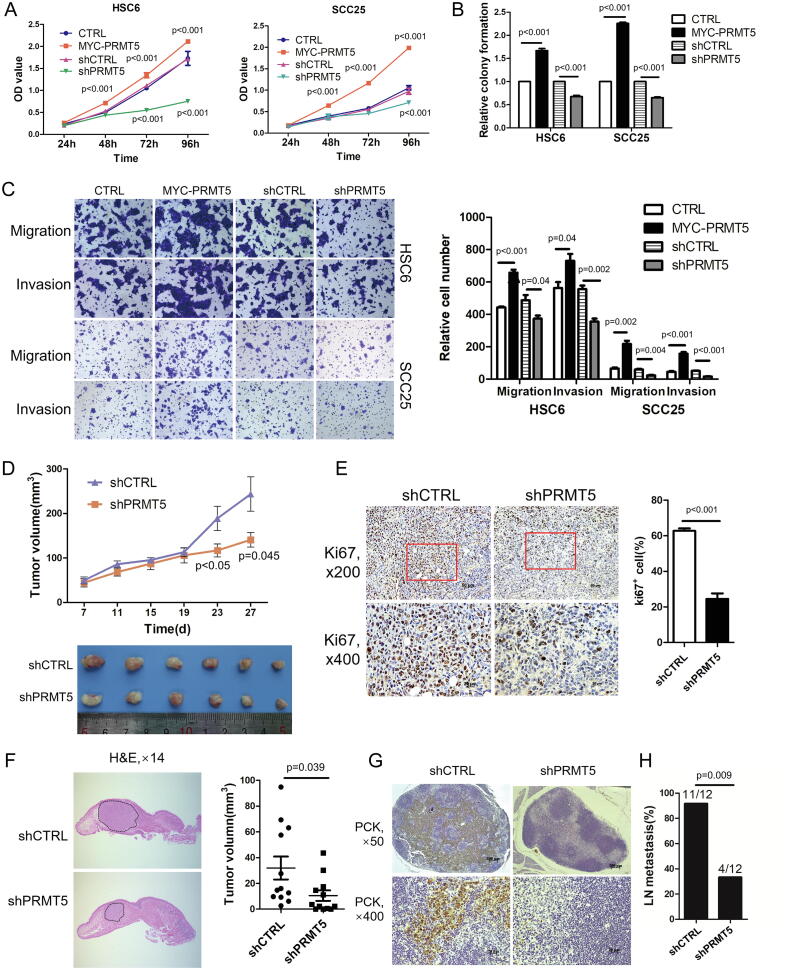
We initially determined the expression levels of PRMT5 in 7 HNSCC cell lines and Normal Oral Keratinocytes (NOK) by Western blot analysis. As showed in Fig. S1A, the expression of PRMT5 in all tested cancer cell lines was higher than that of NOK. To investigate the effects of PRMT5 overexpression on the biological behavior of HNSCC, we engineered the HNSCC cell line, SCC25, and oral SCC cell line, HSC6, to stably overexpress PRMT5 (Fig. S1B and C). In both SCCs, overexpression of PRMT5 promoted proliferation, colony formation. To validate that PRMT5 is critical for HNSCC proliferation and colony formation, we used short hairpin RNA (shRNA) to deplete PRMT5 in HSC6 and SCC25 cells. Depletion of endogenous PRMT5 significantly inhibited proliferation and colony formation in these HNSCC (Fig. 2A and B). We then investigate the role of PRMT5 in regulating HNSCC motility. Overexpression of PRMT5 promoted migration and invasion in HSC6 and SCC25 cells, and depletion of PRMT5 using shRNA inhibited migration and invasion (Fig. 2C).
Fig. 2.
PRMT5 promotes cell proliferation, migration and invasion of HNSCC in vitro and in vivo. (A) The effects of PRMT5 gain- or loss-of-function on in vitro proliferation of HNSCC cells measured by CCK8 conversion. Values are mean ± SD from three independent experiments. (Student’s t test). (B) The effects of PRMT5 gain- or loss-of-function on in vitro proliferation of HNSCC cells measured by colony formation assay. (Student’s t test). (C) Representative photos and quantification of the effects of PRMT5 gain- or loss-of-function on in vitro invasion and migration ability of HNSCC cells. (Student’s t test). (D) The appearance of HSC-6 xenografts and quantification of the volume change of HSC-6 xenografts transfected by shCTRL (n = 6) and shPRMT5 (n = 6) in BALB/c nude mice. Data represent mean ± SD. (Student’s t test). (E) Representative IHC staining of Ki-67 and quantification of Ki-67+ cells in shCTRL and shPRMT5 groups. (Student’s t test). (F) Representative HE staining of longitudinal section of tongues and quantification of the volume of orthotopic tumors in shCTRL (n = 12) and shPRMT5 groups (n = 12) in orthotopic xenografts. (Student’s t test). (G) Representative IHC staining of metastatic tumor cells in cervical lymph nodes of shCTRL and shPRMT5 groups using anti-pan-cytokeratin (PCK). Scale bar, 200 μm, 20 μm. (H) Statistical analysis of different rates in cervical lymph node metastasis between shCTRL and shPRMT5 groups. (fisher’s exact test).
To study the role of endogenous PRMT5 in cancer growth and metastasis, we used a xenograft model with subcutaneous implantation of shPRMT5-HSC6 or shCTRL-HSC6 cells on the back of BALB/c nude mice and followed up for 1–4 weeks. The shPRMT5 group developed significantly smaller tumor compared to the shCTRL group (Fig. 2D), whereas the body weight between the two groups showed no significant difference (Fig. S1D). Proliferating cells within the tumor were detected with Ki-67 immunostaining. The number of Ki-67-positive (Ki67+) cells in shPRMT5 group was significantly lower than that in the shCTRL group (Fig. 2E).
To determine effects of PRMT5 on cervical lymph node metastasis, we employed an orthotopic xenograft model implanting shPRMT5-HSC6 cells or shCTRL-HSC6 cells in the tongue of BALB/c nude mice. PRMT5 knockdown inhibited the growth of primary tumors (Fig. 2F) without significant effects on body weights (Fig. S1E), which is consistent with results from the subcutaneous xenograft model. Metastasis was detected using pan-cytokeratin (PCK) antibody staining of the cervical lymph node tissue. We showed that PRMT5 knockdown significantly reduced the rate of cervical lymph node metastasis compared with the control group (Fig. 2G and H; Fig. S1F). No distant metastasis was detected (Fig. S1G). Taken together, these findings indicate that PRMT5 promotes tumorigenesis and cervical lymph node metastasis of HNSCC.
PRMT5 inhibitor inhibits carcinogenesis, growth and metastasis of HNSCC in vivo
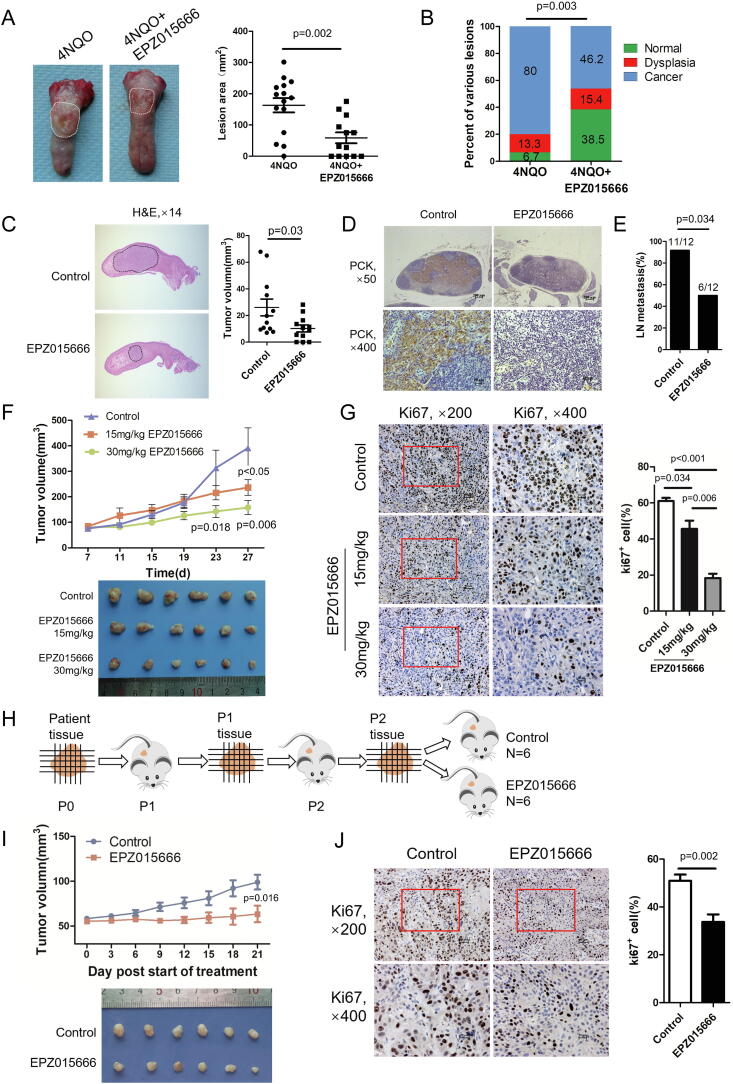
To further study the function of endogenous PRMT5 in HNSCC, we used a PRMT5 specific inhibitor that could competitively bind to the substrate of PRMT5, EPZ015666. This compound demonstrated significant anti-leukemia activity in vitro and in vivo [21]. As showed in Fig. S2A-C, EPZ015666 inhibits the proliferation, migration and invasion of HNSCC cells in vitro. To assess the effects of PRMT5 inhibition on HNSCC, we established the 4NQO-induced carcinogenesis model in Sprague Dawley (SD) rats and started administration of EPZ015666 (30 mg/kg) or vehicle control via tail vein injection at week 12 (Fig. S3A). When the experiment was terminated at week 22, we found that EPZ015666 significantly reduced the lesion area on the rat tongue surface (Fig. 3A). Each tongue was pathologically diagnosed and categorized into normal, dysplasia and cancer areas. The carcinogenesis rate of EPZ015666-treated group was significantly lower than that of control group (Fig. 3B). These data indicate that EPZ015666 inhibits carcinogenesis of HNSCC.
Fig. 3.
PRMT5 inhibition represses carcinogenesis and metastasis of HNSCC in vivo. (A) Representative appearance and quantification of lesion area of rat tongue tissues in 4NQO group (n = 15) and 4NQO + EPZ015666 group (30 mg/kg, n = 13). (Student’s t test). (B) Quantification of various lesions between 4NQO group (n = 15) and 4NQO + EPZ015666 group (30 mg/kg, n = 13) of rat. (Rank sum test). (C) Representative HE staining of longitudinal section of tongues and quantification of orthotopic tumor in control group (n = 12) and EPZ015666 group (30 mg/kg, n = 12) in orthotopic xenografts of BALB/c nude mice. (Student’s t test). (D) Representative IHC staining of metastatic tumor cells in cervical lymph nodes of BALB/c mice using anti-pan-cytokeratin (PCK). Scale bar, 200 μm, 20 μm. (E) Statistical analysis of differences in cervical lymph node metastasis rates between Control group (n = 12) and EPZ015666 group (30 mg/kg, n = 12) in orthotopic xenografts. (fisher’s exact test). (F) The appearance of HSC-6 xenografts and quantification of the volume change of HSC-6 xenografts in BALB/c nude mice affected by different concentration of EPZ015666 (15 mg/kg, 30 mg/kg). Data represent mean ± SD. (one-way ANOVA). (G) Representative IHC staining of Ki-67 and quantification of Ki-67+ cells in control and different concentration of EPZ015666 (15 mg/kg, 30 mg/kg). (Student’s t test). (H) Schematic diagram of PDX. Briefly, the primary tumor tissues were collected, minced and implanted into BALB/c nude mice and passaged on to derived as P1 and P2. The “P2” tissues were collected and equally implanted into 4-week-old BALB/c mice which were randomly divided into two groups: Control and EPZ015666 (30 mg/kg). (I) The appearance of PDX xenografts and quantification of the volume change of PDX xenografts in BALB/c nude mice affected by 30 mg/kg EPZ015666. Data represent mean ± SD. (Student’s t test). (J) Representative immunostaining of Ki-67 and quantification of Ki-67+ cells in control (n = 6) and EPZ015666 groups (30 mg/kg, n = 6) in PDX model. (Student’s t test).
To determine if EPZ015666 exerts an effect on cervical lymph node metastasis, we employed the orthotopic xenograft model using HSC6 implantation in the tongue of BALB/c nude mice. EPZ015666 inhibited the growth of primary tumors (Fig. 3C). Similarly, EPZ015666 significantly reduced the rate of cervical lymph node metastasis when compared with the control group (Fig. S3D; Fig. 3D and E). Moreover, EPZ015666 had no significant effect on the body weights of nude mice and showed no obvious toxicity or distant metastasis (Fig. S3B–E). We also tested the effects of EPZ015666 in subcutaneous xenograft models using HSC6 in BALB/c nude mice. EPZ015666 significantly decreased tumor volume compared to the control group and a more dramatic inhibitory effect was observed when a higher dose of EPZ015666 was used (Fig. 3F). Additionally, EPZ015666 showed no significant effect on the body weight of nude mice and no toxic effects on major organ (Fig. S4A–C). EPZ015666 inhibited HNSCC cell proliferation, as the number of Ki67+ cells in the EPZ015666 group was significantly lower than that of the control group (Fig. 3G). These results demonstrate that EPZ015666 inhibit tumorigenesis and cervical lymph node metastasis of HNSCC.
To determine if EPZ015666 is effective for HNSCC patients, we established a Patient-Derived tumor Xenograft (PDX) model using patient tumor tissue expanded in BALB/c nude mice (Fig. 3H; Fig. S4D). EPZ015666 significantly inhibited the growth of PDX tumors and showed no significant effect on body weight or liver/kidney toxicity (Fig. 3I; Fig. S4E–G). In addition, EPZ015666 significantly reduced the number of Ki67+ cells in PDX tumors (Fig. 3J). These data present EPZ015666 as a promising therapeutic reagent for HNSCC patients.
PRMT5 inhibition downregulates Twist1 expression
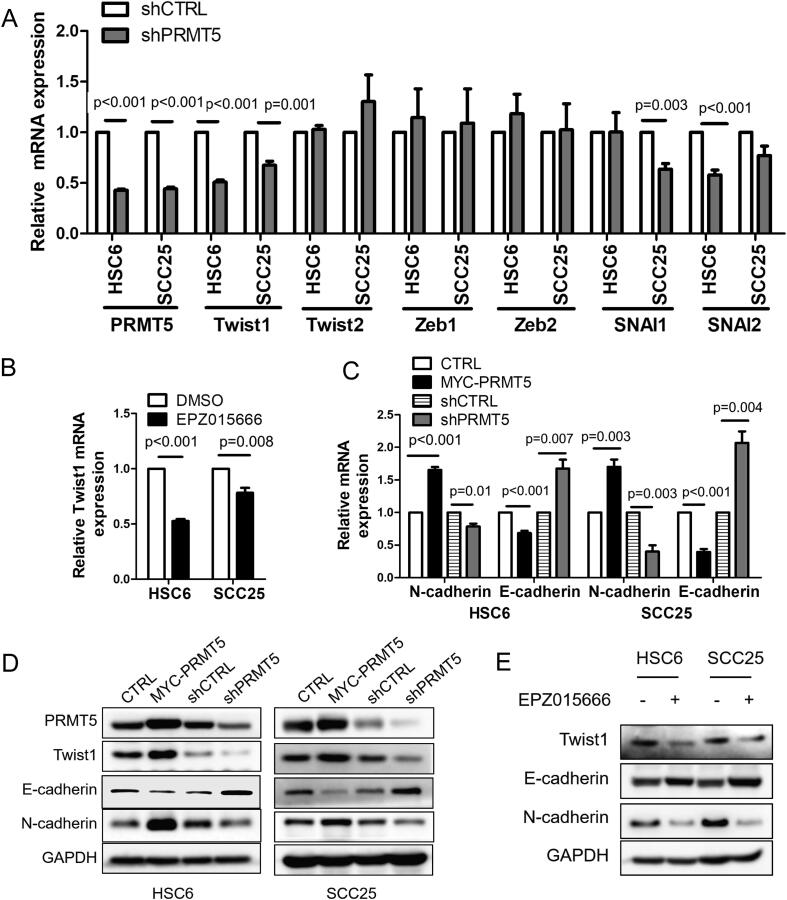
SCC initiation and metastasis are associated with a cellular process called epithelial-mesenchymal transition (EMT), in which epithelial SCCs gradually lose their epithelial characteristics while acquiring mesenchymal features. Cancer cells can exist as a transitional or hybrid EMT state during cancer progression [27], [28]. We examined EMT-related transcription factors in response to PRMT5 knockdown and identified Twist1 as a downstream effector for PRMT5 in both HSC6 and SCC25 cell, while Zeb1/2, SNAI1/2 and Twist2 did not change significantly or consistently in two cell lines (Fig. 4A). Twist1 transcription is also repressed by PRMT5 inhibitor EPZ015666 (Fig. 4B). We further examined Twist1 expression at the protein level and demonstrated that overexpressing PRMT5 enhanced Twist1 level, while depleting PRMT5 reduced Twist1 level (Fig. 4D). Consistently, overexpressing PRMT5 promoted EMT, as indicated by the decrease of epithelial marker E-cadherin and increase of mesenchymal marker N-cadherin. On the contrary, depleting PRMT5 increased epithelial marker E-cadherin expression and decreased mesenchymal marker N-cadherin expression (Fig. 4C and D). PRMT5 inhibitor EPZ015666 exhibit similar effects as PRMT5 depletion (Fig. 4E). These findings demonstrate that PRMT5, endogenous or overexpressed, drives HNSCC towards the mesenchymal state along the EMT process.
Fig. 4.
PRMT5 inhibition induces downregulation of Twist1. (A) Quantitative RT-PCR analysis of EMT transcriptional factor (Twist1, Twist2, Zeb1, Zeb2, SNAI1, SNAI2) mRNA levels in HNSCC cells when PRMT5 knockdown in vitro. Values are mean ± SD from three independent experiments. (Student’s t test). (B) Quantitative RT-PCR analysis of Twist1 mRNA levels in HNSCC when treated with 10 μM EPZ015666 for 4 days in vitro. Values are mean ± SD from three independent experiments. (Student’s t test). (C) Quantitative RT-PCR analysis of E-cadherin and N-cadherin mRNA levels in HNSCC after PRMT5 gain- or loss-of-function in vitro. Values are mean ± SD from three independent experiments. (Student’s t test). (D) Immunoblot of E-cadherin, N-cadherin and Twist1 when PRMT5 gain- or loss-of-function in vitro. (E) Immunoblot of E-cadherin, N-cadherin and Twist1 after treated with 10 μM EPZ015666 for 4 days in vitro.
PRMT5 promotes H3K4me3 by recruiting WDR5 through H3R2me2s to activate Twist1
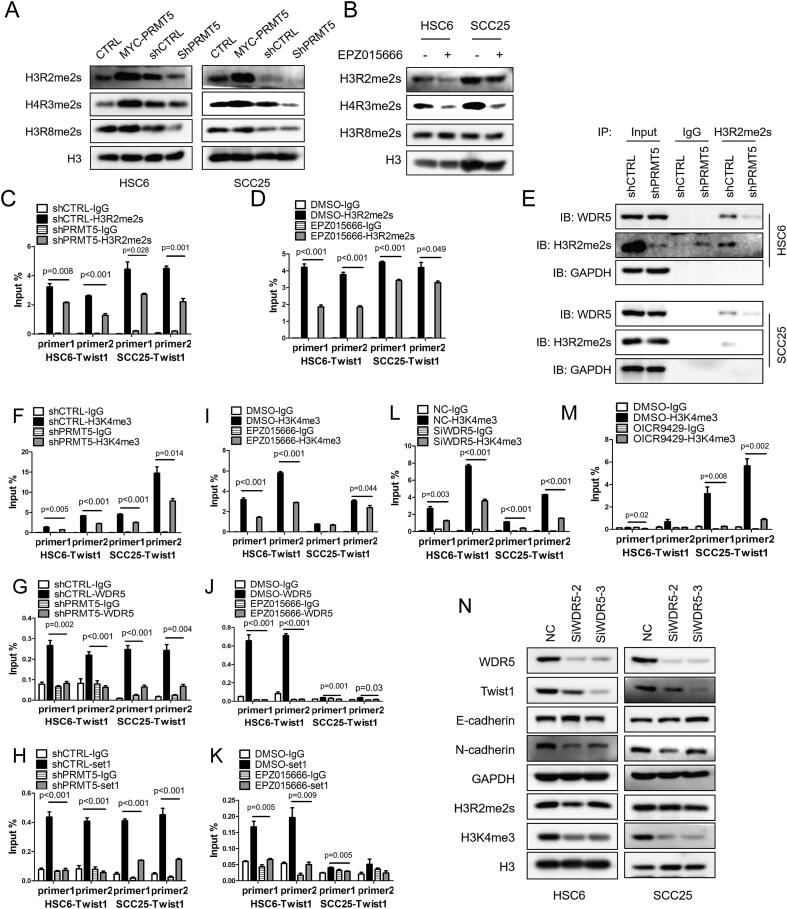
PRMT5 is a major type II arginine methyltransferase, which is responsible for symmetric methylation of arginine residues in histone tails [5]. PRMT5 methylates H3R2, H4R3 and H3R8, through which controls transcription activation or repression [5], [6], [7], [9], [13]. We surveyed the symmetric methylation level of H3R2, H4R3 and H3R8. H3R2me2s, H4R3me2s and H3R8me2s were all significantly decreased by depletion or inhibition of PRMT5 (Fig. 5A and B). H3R2me2s and H4R3me2s was significantly increased by overexpression of PRMT5, but H3R8me2s displayed no obvious change (Fig. 5A and B). We further showed that PRMT5 depletion or inhibition resulted in a significant decrease of H3R2me2s enrichment in Twist1 promoter region (Fig. 5C and D) without a significant effect on H4R3me2s enrichment in the same region (Fig. S5A).
Fig. 5.
PRMT5 promotes Twist1 through H3R2me2s-WDR5-H3K4me3 axis. (A) Immunoblot of histone H3R2me2s, H4R3me2s and H3R8me2s after PRMT5 gain- or loss-of-function in vitro. (B) Immunoblot of histone H3R2me2s, H4R3me2s and H3R8me2s after treated with 10 μM EPZ015666 for 4 days in vitro. (C, D) Quantification of the binding levels of H3R2me2s in Twist1 promoter region after PRMT5 knockdown (C) or treated with 10 μM EPZ015666 for 4 days in vitro (D) in HNSCC cells. Values are mean ± SD from three independent experiments. (Student’s t test). (E) Representative co-IP of WDR5 and H3R2me2s when knockdown PRMT5 in HNSCC cells. (F–K) Quantification of the binding levels of H3K4me3, WDR5 and Set1 in Twist1 promoter region after PRMT5 knockdown (F–H) or treated with 10 μM EPZ015666 for 4 days in vitro (I–K) in HNSCC cells. Values are mean ± SD from three independent experiments. (Student’s t test). (L and M) Quantification of the binding levels of H3K4me3 in Twist1 promoter region after WDR5 knockdown (L) or using 20 μM OICR-9429 (M) in HNSCC cells. Values are mean ± SD from three independent experiments. (Student’s t test). (N) Immunoblot of EMT-related proteins and histone H3K4me3 after WDR5 knockdown in vitro.
H3R2me2s enhances the recruitment of WDR5, a core subunit of the human MLL/Set1 complex (COMPASS) [29], [30]. To examine whether PRMT5-regulated H3R2me2s controls WDR5 recruitment in HNSCC, we performed Co-immunoprecipitation (Co-IP) assay and showed that PRMT5 knockdown reduced H3R2me2s and its associated WDR5 without changing WDR5 expression (Fig. 5E). The WDR5-associated MLL/Set1 complex “write” the H3K4me3 mark at promoter regions, which enables chromatin remodeling and transcription activation [30], [31]. We found that overexpression of PRMT5 increased trimethylation of H3K4, H3K4me3, while knockdown of PRMT5 decreased the level of H3K4me3 in HNSCC cells (Fig. S5B). Focusing on the Twist1 promoter region, we demonstrated that PRMT5 knockdown or inhibition reduced the enrichment of H3K4me3, as well as the occupancy of WDR5 and Set1 at Twist1 promoter region (Fig. 5F–K). The MLL complex not only “write” the H3K4me3 mark, but also “read” this mark, and its binding to H3K4me3 is required for MLL-activated transcription [32]. We showed that WDR5-associated MLL/Set1 complex also bound to H3K4me3 in HNSCC cells (Fig. S5C).
To determine the requirement of WDR5 in PRMT5-elicited deposition of H3K4me3, we depleted WDR5 using small interfering RNA (SiRNA) and showed that WDR5 knockdown significantly reduced the enrichment of H3K4me3 at Twist1 promoter (Fig. 5L). We also used a pharmacological inhibitor of WDR5, OICR9429, which blocks WDR5 interaction with MLL and histone 3 [33]. OICR9429 also reduced the enrichment of H3K4me3 at Twist1 promoter (Fig. 5M). As expected from the above findings, WDR5 knockdown reduced Twist transcription and protein expression (Fig. 5N, S5E). Furthermore, WDR5 knockdown or using OICR-9429 in HSC6 and SCC25 cells decreased the total level of H3K4me3 and expression of mesenchymal marker N-cadherin while increasing the expression of E-cadherin (Figs. 5N, S5F).
Taken together, these data demonstrate that PRMT5-deposited H3R2me2s on Twist1 promoter recruits the MLL/Set1/WDR5 complex via WDR5. Subsequently, the MLL/Set1/WDR5 complex catalyzes H3K4me3 to activate Twist1 transcription and promote EMT in HNSCCs.
Overexpression of Twist1 abrogates the effects of EPZ015666 on HNSCC cells
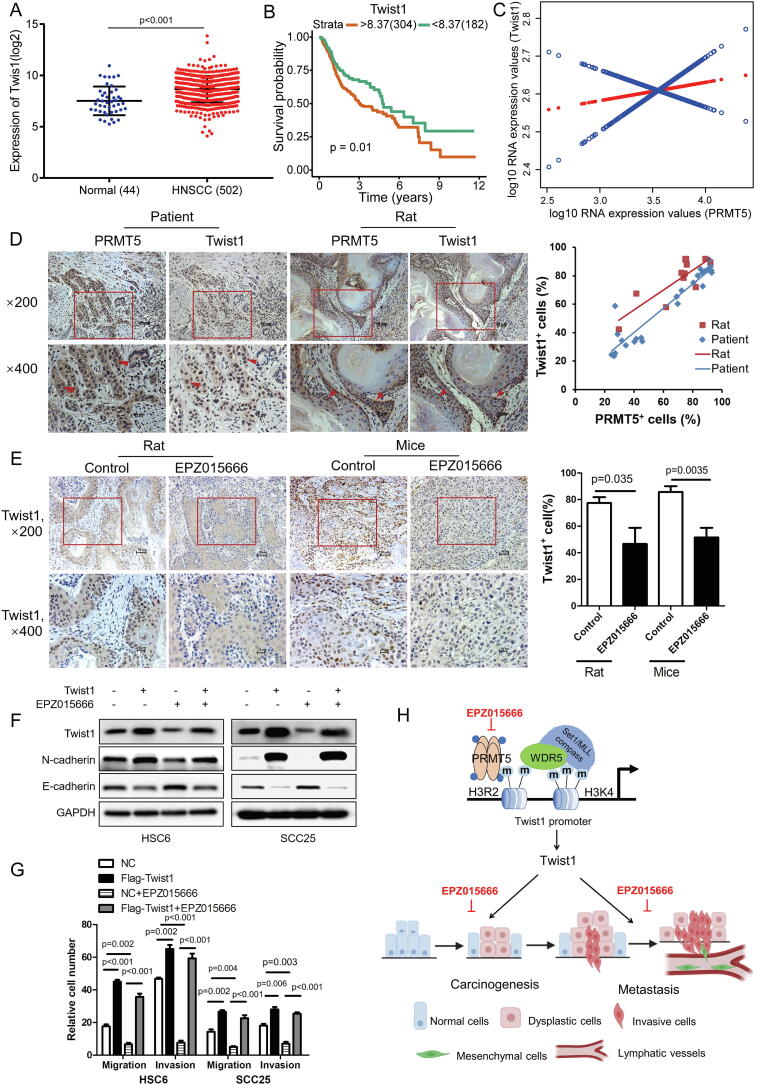
Twist1 was significantly upregulated in HNSCC patient tissues as demonstrated by analysis of the TCGA database (Fig. 6A). Higher expression of Twist1 correlated with significantly lower probability of survival (Fig. 6B). There was also a positive correlation between PRMT5 and Twist1 at the transcriptional level from TCGA database analysis (Fig. 6C). To validate this positive correlation, immunochemistry of PRMT5 and Twist1was conducted in serial sections of 25 human HNSCC tissues and 11 rat tongue cancer tissues. The number of PRMT5-positive cells positively correlated with Twist1-positive cells with a co-efficiency of 0.9463 in patient tissues and 0.8477 in rat tissues (Fig. 6D). In addition, EPZ015666 reduced the number of Twist1-positive cells in tumors developed from subcutaneously-implanted HSC6 cells and 4NQO-induced cancerous tissues (Fig. 6E).
Fig. 6.
Overexpression of Twist1 restores effects of EPZ015666 on HNSCC cells. (A) Analysis of Twist1 in HNSCC and Normal control in TCGA database. (Student’s t test). (B) Kaplan–Meier overall survival curve of Twist1 for HNSCC patients (n = 486) in TCGA database. (log-rank test). (C) A smooth curve fitting for the relationship between PRMT5 and Twist1. (D) Representative IHC staining and correlation analysis of PRMT5 and Twist1 in the serial section of HNSCC tissues (n = 25, blue line, r = 0.9463, p < 0.001, Pearson’s correlation) and tongue cancer tissues from rats induced by 4NQO (n = 11, red line, r = 0.8477, p < 0.001, Pearson’s correlation). (E) Representative IHC staining and quantitative analysis of Twist1 in Control and EPZ015666 group of Rats (n = 11/6) and Mice (n = 6/6) Data represent mean ± SD. (Student’s t test). (F) Immunoblot of Twist1, N-cadherin and E-cadherin in Twist1 overexpression cells with or without treated with 10 μM EPZ015666 for 4 days in vitro. (G) Quantitative analysis of migration and invasion in Twist1 overexpression cells with or without treated with 10 μM EPZ015666 for 4 days in vitro. Values are mean ± SD from three independent experiments. (one-way ANOVA). (H) The schematic diagram of targeting PRMT5 represses carcinogenesis and metastasis of HNSCC.
HSC6 and SCC25 cell lines with stable overexpression of a flag-tagged Twist1 (Flag-Twist1) were generated to perform rescue experiments. As expected, overexpression of Twist1 promoted EMT in HSC6 and SCC25 cells, as shown by the repression of epithelial E-cadherin and promotion of mesenchymal N-cadherin (Fig. 6F). These HNSCCs with Twist1 overexpression were resistant to EPZ015666′s inhibitory effects on EMT (Fig. 6F). Similarly, Twist1-overexpressing HNSCCs were resistant to EPZ015666′s inhibitory effects on migration and invasion (Fig. 6G).
Discussion
PRMT5 was overexpressed in a variety of tumors and is closely related to poor prognosis, but the role of PRMT5 in cancer initiation is not fully understood [13], [14], [19], [21]. Amino et al reported that there was no statistical difference between the expression of PRMT5 in epithelial dysplastic and OSCC tissues although PRMT5 expression in these two tissues is significantly higher than in normal tissues [16]. Our data showed that PRMT5 expression was significantly increased in dysplastic and cancerous tissues compared with normal tissues when its expression in cancer was significantly higher than dysplasia. Meanwhile, higher levels of PRMT5 expression in HNSCC were associated with more advanced clinical stages, higher incidence of lymph node metastasis, and poor OS. Our findings indicated that PRMT5 played important roles in the initiation of HNSCC and closely related to the malignant phenotype.
Metastasis remains the primary challenge in cancer treatment. The most common site of metastasis in HNSCC is cervical lymph node, because drainage channel of the neck is rich in lymph nodes with more than 400 in number [34]. More than 30% of patients with HNSCC have cervical lymph node metastasis at the time of diagnosis, and cervical lymph node metastasis is heavily associated poor prognosis [35]. Our results suggested that patients with a high level of PRMT5 expression are more prone to cervical lymph node metastasis. Experiments using rodent tumor models also confirmed that PRMT5 loss-of-function inhibited cervical lymph node metastasis. Our findings suggested high PRMT5 expression as a potential marker to predict whether a patient needs cervical lymph node dissection.
As the latest potent and selective inhibitor for PRMT5, EPZ015666 offers highly anticipated therapeutic strategies in mantle cell lymphoma, multiple myeloma, glioblastoma and triple-negative breast cancer [18], [19], [20], [21]. However, till now, the role of EPZ015666 in HNSCC is rarely studied. In the present study, we provide the most comprehensive study on the inhibitory effects of EPZ015666 in HNSCC. EPZ015666 can not only inhibit the proliferation of HNSCC in vivo, but also inhibit the regional cervical lymph node metastasis in vivo. What’s more important, EPZ015666 could inhibit the 4NQO-induced carcinogenesis of rats and the patient-derived tumors of HNSCC. These suggest that targeted intervention of PRMT5 can benefit patients with HNSCC during the tumor initiation, development and metastasis stages. EPZ015666 is expected to become a promising antitumor drug in HNSCC.
Activation of the epithelial-mesenchymal transition (EMT) program is considered a key mechanism by which cancer epithelial cells acquire a malignant phenotype [36], [37]. PRMT5 promotes EMT probably via EGFR/AKT/β-catenin pathway in pancreatic cancer [38]. PRMT5/MEP50 are necessary to maintain EMT markers and enhance epigenetic mechanisms of TGF-β response [7]. In our study, we found that overexpressed PRMT5 could repress expression of epithelial gene E-cadherin, and induce expression of mesenchymal gene N-cadherin. Twist1, a basic helix-loop-helix (bHLH) transcription factor, was responsible for the initiation, EMT and metastasis of HNSCC in our study. Twist1 is originally thought to play a key regulatory role in embryogenesis, but silenced in most adult tissues [39]. Recent studies identified that Twist1 is an oncogene that is overexpressed in tumor tissues and plays multiple roles in tumor invasion and metastasis [40], [41]. In the present study, Twist1 overexpression was positively correlated with PRMT5 overexpression, and correlated with poor prognosis of HNSCC patients.
PRMT5 is a major type II PRMTs that symmetrically di-methylate histone tails at H4R3, H3R8 and H3R2. H4R3me2s and H3R8me2s are associated with transcriptional repression, while H3R2me2s is associated with transcriptional activation [6], [13], [29]. Actually, methylation of histones by PRMT5 does not always occur simultaneously. In epidermal squamous cell carcinoma, H4R3me2s and H3R8me2s formation depended upon environment [42]. In lung carcinoma, knockdown PRMT5 could not resulted in downregulation of H3R8me2s but H4R3me2s and H3R2me2s [7]. Our study demonstrated that PRMT5 symmetrically di-methylated H4R3 and H3R2, but not H3R8, in HNSCC. Further results showed that PRMT5 loss-of-function caused a significant decrease of H3R2me2s enrichment in Twist1 promoter region, suggesting that PRMT5 promotes transcriptional activation of Twist1 through deposition of H3R2me2s. H3R2me2s is a novel histone marker associated with gene transcriptional activation and that WDR5 can be recruited by it [29]. WDR5, a core subunit of the human MLL and Set1 complex (COMPASS), strongly stimulates the catalytic activity of MLL1 for the deposition of H4K4me3 [43], [44], [45]. Chen et al declared that H3R2me1 but not H3R2me2s could recruit WDR5 and activate snail in A549 cell lines [7]. In our study, we confirmed that H3R2me2s enhances binding of WDR5 to the Twist1 promoter. This suggests that the role of PRMT5 in regulating histones and downstream mechanisms may be different in different cell lines.
It has been reported that WDR5 was related to EMT, and was confirmed in colorectal cancer and breast cancer [46], [47], [48]. WDR5 could form a complex with Twist1 and lncRNA HOTTIP in the promoter region of HOXA9 to promote the metastasis of prostate cancer [49]. In our results, WDR5 loss-of-function leads to the inhibition of EMT of HNSCC cells. What’s more, WDR5, Set1, H3K4me3 all enriched in Twist1 promoter to trans-activate Twist1. H3K4me3 is a nearly universal chromatin modification of the transcription initiation site of active genes in eukaryotes, and its level is highly correlated with the amount of transcription [50]. Therefore, we concluded that the PRMT5-catalyzed H3R2me2s induced transcriptional activation of Twist1 by recruiting the WDR5, MLL, Set1 complex and subsequent H3K4 trimethylation at Twist1 promoter.
Conclusions
In summary, our studies uncovered that PRMT5 could promote H3K4me3-mediated Twist1 transcription by which PRMT5 induced carcinogenesis and metastasis in HNSCC. Our study also presented PRMT5 as a potential biomarker that predicts patient survival and an attractive therapeutic target in HNSCC.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81671000, 81870769) and Guangdong Financial fund for High-Caliber Hospital Construction (174-2018-XMZC-0001-03-0125/D-05).
CRediT authorship contribution statement
Zhaona Fan: Conceptualization, Methodology, Formal analysis, Investigation, Writing - original draft. Lihong He: Methodology, Formal analysis, Investigation, Writing - original draft. Mianxiang Li: Methodology, Validation, Investigation, Resources. Ruoyan Cao: Investigation, Resources. Miao Deng: Investigation. Fan Ping: Resources. Xueyi Liang: Validation. Yuan He: Validation. Tong Wu: Formal analysis, Writing - review & editing. Xiaoan Tao: Formal analysis, Writing - review & editing. Jian Xu: Writing - review & editing. Bin Cheng: Conceptualization, Writing - review & editing, Supervision. Juan Xia: Conceptualization, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Professor Xiaohua Chen (Hospital of Stomatology, Sun Yat-sen University) for histopathological diagnosis and Meng Zhao (Zhongshan School of Medicine, Sun Yat-sen University) for valuable comments on the article modification.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2020.09.004.
Contributor Information
Bin Cheng, Email: chengbin@mail.sysu.edu.cn.
Juan Xia, Email: xiajuan@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chi A.C., Day T.A., Neville B.W. Oral cavity and oropharyngeal squamous cell carcinoma–an update. CA Cancer J Clin. 2015;65(5):401–421. doi: 10.3322/caac.21293. [DOI] [PubMed] [Google Scholar]
- 3.Hedberg M.L., Goh G., Chiosea S.I., Bauman J.E., Freilino M.L., Zeng Y., Wang L., Diergaarde B.B., Gooding W.E., Lui V.W. Genetic landscape of metastatic and recurrent head and neck squamous cell carcinoma. J Clin Invest. 2016;126(1):169–180. doi: 10.1172/JCI82066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marur S., Forastiere A.A. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2016;91(3):386–396. doi: 10.1016/j.mayocp.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 5.Stopa N., Krebs J.E., Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci. 2015;72(11):2041–2059. doi: 10.1007/s00018-015-1847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang K., Zielinska A.E., Shaaban A.M., Sanchez-Bailon M.P., Jarrold J., Clarke T.L., Zhang J., Francis A., Jones L.J., Smith S. PRMT5 is a critical regulator of breast cancer stem cell function via histone methylation and FOXP1 expression. Cell Rep. 2017;21(12):3498–3513. doi: 10.1016/j.celrep.2017.11.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H., Lorton B., Gupta V., Shechter D. A TGFβ-PRMT5-MEP50 axis regulates cancer cell invasion through histone H3 and H4 arginine methylation coupled transcriptional activation and repression. Oncogene. 2017;36(3):373–386. doi: 10.1038/onc.2016.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y., Bedford M.T. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13(1):37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- 9.Pal S., Vishwanath S.N., Erdjument-Bromage H., Tempst P., Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24(21):9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bandyopadhyay S., Harris D.P., Adams G.N., Lause G.E., McHugh A., Tillmaand E.G., Money A., Willard B., Fox P.L., Dicorleto P.E. HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Mol Cell Biol. 2012;32(7):1202–1213. doi: 10.1128/MCB.05977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu J.M., Chen C.T., Chou C.K., Kuo H.P., Li L.Y., Lin C.Y., Lee H.J., Wang Y.N., Liu M., Liao H.W. Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat Cell Biol. 2011;13(2):174–181. doi: 10.1038/ncb2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger S.L. Out of the jaws of death: PRMT5 steers p53. Nat Cell Biol. 2008;10(12):1389–1390. doi: 10.1038/ncb1208-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan F., Alinari L., Lustberg M.E., Martin L.K., Cordero-Nieves H.M., Banasavadi-Siddegowda Y., Virk S., Barnholtz-Sloan J., Bell E.H., Wojton J. Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res. 2014;74(6):1752–1765. doi: 10.1158/0008-5472.CAN-13-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibrahim R., Matsubara D., Osman W., Morikawa T., Goto A., Morita S., Ishikawa S., Aburatani H., Takai D., Nakajima J. Expression of PRMT5 in lung adenocarcinoma and its significance in epithelial-mesenchymal transition. Hum Pathol. 2014;45(7):1397–1405. doi: 10.1016/j.humpath.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Jin Y., Zhou J., Xu F., Jin B., Cui L., Wang Y., Du X., Li J., Li P., Ren R. Targeting methyltransferase PRMT5 eliminates leukemia stem cells in chronic myelogenous leukemia. J Clin Invest. 2016;126(10):3961–3980. doi: 10.1172/JCI85239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amano Y., Matsubara D., Yoshimoto T., Tamura T., Nishino H., Mori Y., Niki T. Expression of protein arginine methyltransferase-5 in oral squamous cell carcinoma and its significance in epithelial-to-mesenchymal transition. Pathol Int. 2018;68(6):359–366. doi: 10.1111/pin.12666. [DOI] [PubMed] [Google Scholar]
- 17.Kumar B., Yadav A., Brown N.V., Zhao S., Cipolla M.J., Wakely P.E., Schmitt A.C., Baiocchi R.A., Teknos T.N., Old M. Nuclear PRMT5, cyclin D1 and IL-6 are associated with poor outcome in oropharyngeal squamous cell carcinoma patients and is inversely associated with p16-status. Oncotarget. 2017;8(9):14847–14859. doi: 10.18632/oncotarget.14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gulla A., Hideshima T., Bianchi G., Fulciniti M., Kemal Samur M., Qi J., Tai Y.T., Harada T., Morelli E., Amodio N. Protein arginine methyltransferase 5 has prognostic relevance and is a druggable target in multiple myeloma. Leukemia. 2018;32(4):996–1002. doi: 10.1038/leu.2017.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vinet M., Suresh S., Maire V., Monchecourt C., Nemati F., Lesage L., Pierre F., Ye M., Lescure A., Brisson A. Protein arginine methyltransferase 5: a novel therapeutic target for triple-negative breast cancers. Cancer Med. 2019;8(5):2414–2428. doi: 10.1002/cam4.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes B., Benavides-Serrato A., Saunders J.T., Landon K.A., Schreck A.J., Nishimura R.N., Gera J. The protein arginine methyltransferase PRMT5 confers therapeutic resistance to mTOR inhibition in glioblastoma. J Neurooncol. 2019;145(1):11–22. doi: 10.1007/s11060-019-03274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan-Penebre E., Kuplast K.G., Majer C.R., Boriack-Sjodin P.A., Wigle T.J., Johnston L.D., Rioux N., Munchhof M.J., Jin L., Jacques S.L. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11(6):432–437. doi: 10.1038/nchembio.1810. [DOI] [PubMed] [Google Scholar]
- 22.Tsompana M., Gluck C., Sethi I., Joshi I., Bard J., Nowak N.J., Sinha S., Buck M.J. Reactivation of super-enhancers by KLF4 in human head and neck squamous cell carcinoma. Oncogene. 2020;39(2):262–277. doi: 10.1038/s41388-019-0990-4. [DOI] [PubMed] [Google Scholar]
- 23.Wang H., Liang X., Li M., Tao X., Tai S., Fan Z., Wang Z., Cheng B., Xia J. Chemokine (CC motif) ligand 18 upregulates Slug expression to promote stem-cell like features by activating the mammalian target of rapamycin pathway in oral squamous cell carcinoma. Cancer Sci. 2017;108(8):1584–1593. doi: 10.1111/cas.13289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen D., Wu M., Li Y., Chang I., Yuan Q., Ekimyan-Salvo M., Deng P., Yu B., Yu Y., Dong J. Targeting BMI1+ cancer stem cells overcomes chemoresistance and inhibits metastases in squamous cell carcinoma. Cell Stem Cell. 2017;20(5):621–634 e626. doi: 10.1016/j.stem.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seshadri M., Merzianu M., Tang H., Rigual N.R., Sullivan M., Loree T.R., Popat S.R., Repasky E.A., Hylander B.L. Establishment and characterization of patient tumor-derived head and neck squamous cell carcinoma xenografts. Cancer Biol Ther. 2009;8(23):2275–2283. doi: 10.4161/cbt.8.23.10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Zhang X., Wang Z., Hu Q., Wu J., Li Y., Ren X., Wu T., Tao X., Chen X. LncRNA-p23154 promotes the invasion-metastasis potential of oral squamous cell carcinoma by regulating Glut1-mediated glycolysis. Cancer Lett. 2018;434:172–183. doi: 10.1016/j.canlet.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 27.Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., Van Keymeulen A., Brown D., Moers V., Lemaire S. Identification of the tumour transition states occurring during EMT. Nature. 2018;556(7702):463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 28.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Migliori V., Muller J., Phalke S., Low D., Bezzi M., Mok W.C., Sahu S.K., Gunaratne J., Capasso P., Bassi C. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat Struct Mol Biol. 2012;19(2):136–144. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- 30.Song J.J., Kingston R.E. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J Biol Chem. 2008;283(50):35258–35264. doi: 10.1074/jbc.M806900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dharmarajan V., Lee J.H., Patel A., Skalnik D.G., Cosgrove M.S. Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases. J Biol Chem. 2012;287(33):27275–27289. doi: 10.1074/jbc.M112.364125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang P.Y., Hom R.A., Musselman C.A., Zhu L., Kuo A., Gozani O., Kutateladze T.G., Cleary M.L. Binding of the MLL PHD3 finger to histone H3K4me3 is required for MLL-dependent gene transcription. J Mol Biol. 2010;400(2):137–144. doi: 10.1016/j.jmb.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grebien F., Vedadi M., Getlik M., Giambruno R., Grover A., Avellino R., Skucha A., Vittori S., Kuznetsova E., Smil D. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPalpha N-terminal leukemia. Nat Chem Biol. 2015;11(8):571–578. doi: 10.1038/nchembio.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dedhia A., Gosavi S., Sharma B., Pagey R. Low lymphatic vessel density correlates with lymph node metastasis in oral squamous cell carcinoma. J Dent. 2018;19(1):6–14. [PMC free article] [PubMed] [Google Scholar]
- 35.Sano D., Myers J.N. Metastasis of squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev. 2007;26(3–4):645–662. doi: 10.1007/s10555-007-9082-y. [DOI] [PubMed] [Google Scholar]
- 36.Chaffer C.L., San Juan B.P., Lim E., Weinberg R.A. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35(4):645–654. doi: 10.1007/s10555-016-9648-7. [DOI] [PubMed] [Google Scholar]
- 37.Nieto M.A., Huang R.Y., Jackson R.A., Thiery J.P. Emt: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 38.Ge L., Wang H., Xu X., Zhou Z., He J., Peng W., Du F., Zhang Y., Gong A., Xu M. PRMT5 promotes epithelial-mesenchymal transition via EGFR-beta-catenin axis in pancreatic cancer cells. J Cell Mol Med. 2020;24(2):1969–1979. doi: 10.1111/jcmm.14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sozen B., Pehlivanoglu S., Demir N. Differential expression pattern of Twist1 in mouse preimplantation embryos suggests its multiple roles during early development. J Assist Reprod Genet. 2016;33(11):1533–1540. doi: 10.1007/s10815-016-0794-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y., Lee D.K., Feng Z., Xu Y., Bu W., Li Y., Liao L., Xu J. Breast tumor cell-specific knockout of Twist1 inhibits cancer cell plasticity, dissemination, and lung metastasis in mice. Proc Natl Acad Sci U S A. 2017;114(43):11494–11499. doi: 10.1073/pnas.1618091114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beck B., Lapouge G., Rorive S., Drogat B., Desaedelaere K., Delafaille S., Dubois C., Salmon I., Willekens K., Marine J.C. Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. Cell Stem Cell. 2015;16(1):67–79. doi: 10.1016/j.stem.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Saha K., Fisher M.L., Adhikary G., Grun D., Eckert R.L. Sulforaphane suppresses PRMT5/MEP50 function in epidermal squamous cell carcinoma leading to reduced tumor formation. Carcinogenesis. 2017;38(8):827–836. doi: 10.1093/carcin/bgx044. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 43.Dou Y., Milne T.A., Ruthenburg A.J., Lee S., Lee J.W., Verdine G.L., Allis C.D., Roeder R.G. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13(8):713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 44.Li Y., Han J., Zhang Y., Cao F., Liu Z., Li S., Wu J., Hu C., Wang Y., Shuai J. Structural basis for activity regulation of MLL family methyltransferases. Nature. 2016;530(7591):447–452. doi: 10.1038/nature16952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rojas A., Sepulveda H., Henriquez B., Aguilar R., Opazo T., Nardocci G., Bustos F., Lian J.B., Stein J.L., Stein G.S. Mll-COMPASS complexes mediate H3K4me3 enrichment and transcription of the osteoblast master gene Runx2/p57 in osteoblasts. J Cell Physiol. 2019;234(5):6244–6253. doi: 10.1002/jcp.27355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Punzi S., Balestrieri C., D'Alesio C., Bossi D., Dellino G.I., Gatti E., Pruneri G., Criscitiello C., Lovati G., Meliksetyan M. WDR5 inhibition halts metastasis dissemination by repressing the mesenchymal phenotype of breast cancer cells. Breast Cancer Res. 2019;21(1):123. doi: 10.1186/s13058-019-1216-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan X., Chen S., Wu J., Lin J., Pan C., Ying X., Pan Z., Qiu L., Liu R., Geng R. PI3K/AKT-mediated upregulation of WDR5 promotes colorectal cancer metastasis by directly targeting ZNF407. Cell Death Dis. 2017;8(3) doi: 10.1038/cddis.2017.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu M.Z., Tsai Y.P., Yang M.H., Huang C.H., Chang S.Y., Chang C.C., Teng S.C., Wu K.J. Interplay between HDAC3 and WDR5 is essential for hypoxia-induced epithelial-mesenchymal transition. Mol Cell. 2011;43(5):811–822. doi: 10.1016/j.molcel.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 49.Malek R., Gajula R.P., Williams R.D., Nghiem B., Simons B.W., Nugent K., Wang H., Taparra K., Lemtiri-Chlieh G., Yoon A.R. TWIST1-WDR5-Hottip regulates Hoxa9 chromatin to facilitate prostate cancer metastasis. Cancer Res. 2017;77(12):3181–3193. doi: 10.1158/0008-5472.CAN-16-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Howe F.S., Fischl H., Murray S.C., Mellor J. Is H3K4me3 instructive for transcription activation? Bioessays. 2017;39(1):1–12. doi: 10.1002/bies.201600095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.