Abstract
The last decade has witnessed a substantial expansion in the field of microRNA (miRNA) biology, providing crucial insights into the role of miRNAs in disease pathology, predominantly in cancer progression and its metastatic spread. The discovery of tumor-suppressing miRNAs represents a potential approach for developing novel therapeutics. In this context, through miRNA microarray analysis, we examined the consequences of Prostate apoptosis response-4 (Par-4), a well-established tumor-suppressor, stimulation on expression of different miRNAs in Panc-1 cells. The results strikingly indicated elevated miR-200c levels in these cells upon Par-4 overexpression. Intriguingly, the Reverse Phase Protein Array (RPPA) analysis revealed differentially expressed proteins (DEPs), which overlap between miR200c- and Par-4-transfected cells, highlighting the cross-talks between these pathways. Notably, Phospho-p44/42 MAPK; Bim; Bcl-xL; Rb Phospho-Ser807, Ser811; Akt Phospho-Ser473; Smad1/5 Phospho-Ser463/Ser465 and Zyxin scored the most significant DEPs among the two data sets. Furthermore, the GFP-Par-4-transfected cells depicted an impeded expression of critical mesenchymal markers viz. TGF-β1, TGF-β2, ZEB-1, and Twist-1, concomitant with augmented miR-200c and E-cadherin levels. Strikingly, while Par-4 overexpression halted ZEB-1 at the transcriptional level; contrarily, silencing of endogenous Par-4 by siRNA robustly augmented the Epithelial-mesenchymal transition (EMT) markers, along with declining miR-200c levels. The pharmacological Par-4-inducer, NGD16, triggered Par-4 expression which corresponded with increased miR-200c resulting in the ZEB-1 downregulation. Noteworthily, tumor samples obtained from the syngenic mouse pancreatic cancer model revealed elevated miR-200c levels in the NGD16-treated mice that positively correlated with the Par-4 and E-cadherin levels in vivo; while a negative correlation was evident with ZEB-1 and Vimentin.
Keywords: Par-4, miR-200c, EMT, RPPA
Highlights
-
•
Prostate apoptosis response-4 (Par-4) stimulation elevates the endogenous miR-200c levels
-
•
Par-4- mediated miR-200c induction modulates the ZEB-1/miR-200c axis
-
•
Pharmacological Par-4 inducer, NGD16, boosts the miR-200c and E-cadherin levels in vivo.
-
•
Overlapping targets between miR 200c and Par-4 signaling axis highlight the cross-talks between these pathways.
Introduction
microRNAs are non-coding single-stranded RNAs that are 20–22 nucleotide long. They are predominantly synthesized as long primary transcripts by RNA polymerase II, subsequently capped and polyadenylated as pri-miRNAs. Drosha later cleaves these pri-miRNAs into pre-miRNAs that, with the help of exportin-5, are exported outside nucleus. Once in the cytoplasm, Dicer fabricates the pre-miRNAs accordingly to yield mature miRNAs. miRNAs negatively control post-transcriptional gene expression, to degrade multiple mRNAs and execute translation suppression, by base pairing with the 3′ untranslated region (3′UTR) of mRNA, [[1], [2], [3]]. miRNA dysregulation has been implicated in the etiology, pathogenesis, diagnosis and treatment of cancer [[4], [5], [6]]. Functionally, the miRNAs can be classified as oncomirs as well as tumor-suppressors depending upon their role during cancer progression [7]. A plethora of reports have demonstrated tempered levels of the miRNA-200 family members in the cells undergoing EMT [8]. One of the first miRNAs documented to have altered expression among various cancer cell lines was miR-200c [9]. Being a positive regulator of E-cadherin, miR-200c restricts the E-cadherin repressor, ZEB-1, to maintain the epithelial morphology in the cells, thus attenuating the EMT [10]. Consequently, extensive investigation has validated the role of miR-200c in cell proliferation, apoptosis, EMT, invasion, therapy-induced resistance, and metastasis in different cancer types [[11], [12], [13], [14]].
A critical event during EMT cascade is the loss of adhesion molecules that maintain the epithelial integrity of the cells, an important example being E-cadherin [15]. Several reports suggested that attenuated expression of E-cadherin directly correlates, both in vitro and in vivo, with tumor cell invasion and migration [16,17]. Various transcription factors like Snail, ZEB-1, ZEB-2, and Slug regulate E-cadherin expression and result in repressed E-cadherin protein levels specifically by binding the promoter of E-cadherin at the E-boxes domain, ultimately augmenting the EMT cascade [[18], [19], [20]]. Aberrant ZEB-1 protein expression is reported to repress E-cadherin in various cancers types, including breast, lung, prostate, colorectal, and pancreatic cancer [[21], [22], [23], [24]]. miR-200c and ZEB-1 are well-established to have a reciprocal relationship and control the EMT phenomenon in various cancers; miR-200c directly targets and impedes ZEB-1 and ZEB-2 expression [25,26]. The aberrant miR-200c loss, with a simultaneous increase in ZEB-1 expression, has been observed to promote EMT by downregulating E-cadherin in different cancers [[27], [28], [29]].
Prostate apoptosis response-4 (Par-4), well-established for its tumor-suppressing activities, is mostly inactivated or down-regulated in cancers [30,31]. In hormone-independent manner, Par-4 can augment the interaction of FAS receptor and FAS Ligand with FADD and initiate the assembly of the death-inducing signaling complex (DISC) to induce apoptosis [7]. The central core, SAC (Selective for Apoptosis of Cancer Cells) domain, of Par-4 protein is responsible for its proapoptotic function [32]. Although the SAC domain can translocate to the nucleus in both normal and cancer cells, only the excessive protein kinase A (PKA) levels found in cancer cells can activate the SAC domain [33] enabling SAC to selectively enhance apoptotic process in malignant cells. Our group previously described modulation of beta-catenin protein by Par-4 and its consequences on EMT [34]. One of our studies also implied the consistent modulation of EMT markers upon Par-4 stimulation in cancer cells by its pharmacological modulator, NGD16 [35]. Therefore, we were curious to investigate whether Par-4 upregulation could abrogate the ZEB-1-mediated EMT by enhancing the miR-200c levels in cancer cells. Notably, we studied the global proteome changes in Panc-1 cells upon ectopic stimulation with miR-200c and Par-4. We identified the common protein targets of miR-200c and Par-4 signaling that could explain how they regulate the expression of the EMT markers in a similar manner. The present work throws light upon the tumor-suppressive role of Par-4 that involves modulation of the miR-200c, in a tumor-promoting network. Current study suggests a novel role of Par-4 as a regulator of miR-200c expression.
Material and methods
Cell lines and cell culture
HCT-116, A549, and Panc-1 cell lines used in this study were purchased from the European Collection of Cell Culture (ECACC). RPMI-1640 media was used for culturing HCT-116 and A549 cells, DMEM for culturing Panc-1 cells; both RPMI-1640 and DMEM were added with streptomycin, penicillin G and 10% FBS. Sub-culturing of cells was done at about 70% confluence, and the cells were incubated in a 5% CO2 humidified incubator (Galaxy170R, New Brunswick, Germany) at 37 °C. DMSO concentration in the experiments never exceeded 0.1%. Panc-1-GR10 (Panc-1 cells resistant to 10 μM Gemcitabine) cells were generated employing constant exposure to Gemcitabine. The initial concentration of Gemcitabine used was 200 nM, which was gradually increased until we achieved the cells that procreated under continuous presence of 10 μM Gemcitabine. Dr. S. K. Batra (University of Nebraska Medical Center, USA) gifted the mouse pancreatic cancer cell line, UN-KC-6141, to Dr. S. Senapati at the [Institute of Life Sciences (ILS), Bhubaneswar, India] through a material transfer agreement. [31]. HRas stably expressing Panc-1 (HRas-Panc-1) cells were generated with mEGFP-HRas plasmid gifted by Dr. Karel Svoboda (Addgene plasmid # 18662).
Reagents and constructs
DMEM, RPMI-1640, Trypsin-EDTA and Fetal bovine serum (FBS) were acquired from Invitrogen. Penicillin G, Streptomycin, Paraformaldehyde, Sodium Orthovanadate, Protease inhibitor cocktail, Phosphate buffer saline, Phenylmethylsulfonyl fluoride (PMSF), Bradford reagent, Dimethylsulphoxide (DMSO), Triton X-100; anti-Beta-actin, anti-Mouse and anti-Rabbit antibodies were from Sigma Chemicals (St. Louis, MO). Anti-E-cadherin from CST, Inc. (Danvers, MA, USA) was used in the study. Antibodies against Par-4, ZEB-1, Vimentin, and Twist-1; UltraCruz DAPI mounting medium were procured from Santa Cruz Biotechnologies (Santa Cruz, CA). Dr. Vivek Rangnekar (University of Kentucky, Lexington, KY) generously gifted the GFP and GFP-Par-4 constructs. The miR-200c construct was a kind gift from Dr. Giovanni Scambia (Catholic University of the Sacred Heart, Rome, Italy).
Cell lysate preparation and western blotting
Our previously standardized protocol for cell lysis and western blotting was followed [31]. Briefly, the cell from the respective experiments were collected and centrifuged. To the pellet, lysis buffer was added followed by centrifugation. The supernatant was collected and protein quantification was carried out. For immunoblot analysis, equivalent amount of protein was taken from each sample for SDS-PAGE. Using the enhanced chemiluminescence reagent (Millipore), the protein bands were visualized and the signals were captured onto Biomax light film (Eastman Kodak Co., Rochester, NY).
Real-time PCR
RT-PCR was performed following our previously published protocol [36]. Panc-1, A549, and HCT-116 cells were subjected to the respective treatments, following which the cell pellets were collected. Similarly, the treated/untreated tumor samples were homogenized and the suspensions were preserved. Isolation of total RNA was carried out from cell pellets as well as the tumor suspensions using TRIzol® from Invitrogen, Life Technologies, USA. RNA purity and RNA quality were determined using the NanoDrop spectrophotometer and agarose gel electrophoresis (2%), respectively. To avoid any DNA contamination, the RNA mixture was subjected to DNase treatment. miRNA qPCR assays were performed as reported earlier [37]. For qPCR of miRNA and mRNA, we employed the miScript II RT Kit (Qiagen, The Netherlands) to synthesize cDNA, using 1 μg of DNase-treated RNA. Respective primers used for miRNA and mRNA expression analysis are given in Supplementary Table 1. qPCR analysis was carried out in the LightCycler® 96 Real-Time PCR System (Hoffmann-La Roche, Switzerland). Each PCR reaction (10 μL) contained appropriately diluted cDNA, 1 μM primers and 2× LightCycler® 480 SYBR Green I Master mix (Hoffmann-La Roche, Switzerland). Thermal cycling conditions for the qPCR: preincubation at 95 °C for 10 min, followed by 45–55 cycles of 3 step amplification (95 °C for 10 s, 60 °C for 10 s and 72 °C for 10 s). Dissociation curve analysis (heating to 95 °C for10 s at normal ramping, cooling to 65 °C for 60 s at normal ramping followed by slow heating to 97 °C for 1 s at a reduced ramping rate of 0.2 °C/s) was carried out after the PCR, to test qPCR reaction specificity. All the assays were carried out in triplicates. For the calculations, the threshold cycle (Ct) of the amplification curve was applied. Ct values were normalized to the 5.8S gene and Beta-actin (BAC) for miRNA and mRNA, respectively. Using the 2−∆∆ct method, the relative expressional level was analyzed [31,36].
Reverse phase protein array
Following transient transfection with Mock/miR-200c/GFP/GFP-Par-4/GFP-Par-4/155A construct, Panc-1 cells were lysed and protein concentration was calculated. The expression of 119 proteins was then examined using the RPPA analysis as per the standard published protocol [38]. Proteins, as well as phosphoproteins, selection for analysis was done based on their involvement in crucial cellular functions, including (1) Proliferation, (2) EMT, (3) Apoptosis, and (4) Migration.
Scratch motility assay
Our previously published protocol was followed to perform the scratch motility assay [3]. Briefly, 5 × 105 Panc-1 cells/well were seeded in a six-well plate and incubated until a confluent monolayer was formed. Using a sterile pipette tip, the monolayers were scratched. Any detached or floating cells were removed by washing the wells with serum-free medium and images were captured (time = 0 h). Respective treatments were given to the cells in the presence of DMEM medium supplemented with 1% serum. After 48 h, wound closure was assessed and the images were captured using a Nikon D3100 inverted microscope camera (20× magnification).
Knockdown and overexpression studies
Small interfering RNA (siRNA) oligonucleotide duplexes against human Par-4 mRNA (siPar-4) from Sigma chemicals were used in the study. Following the manufacturer's protocol, Lipofectamine 3000 transfection kit (Invitrogen) was used for the Scramble/siPar-4, Mock/miR-200c and GFP/GFP-Par-4 transfections. Panc-1/A549 cells were seeded in an eight-well chamber slide (for immunocytochemistry) or 6-well plate (for Western blotting and RT-PCR) and transiently transfected with GFP/GFP-Par-4, Scramble/siPar-4 or Mock/miR-200c. After 48 h, the cells were respectively subjected to immunocytochemistry, western blotting or RT-PCR.
Cell scattering assay
For cell scattering assay, the previously published protocol was followed [35]. Briefly, HRas-Panc-1 cells were seeded in a six-well plate at a density of 1000 cells/well and incubated to form small, discrete colonies for 3–4 days. Respective treatments with Vehicle/NGD16 or transfections with GFP/GFP-Par-4/Mock/miR-200c were given. 48 h Post-treatment, colonies were fixed, stained and the photographs were captured using Nikon D3100 inverted microscope camera (20× magnification).
Immunocytochemistry
Previously published protocol for immunocytochemistry was carried out, with some modifications [35]. Post-treatment, the cells were incubated with anti-Cytokeratin-18, anti-FAS and anti-ZEB-1 antibodies at a dilution of 1: 100 at 4 °C overnight. After thorough washing with PBST (PBS + 0.1% Tween-20), the samples were accordingly probed with anti-rabbit Alexa Fluor-647 (anti-Cytokeratin-18) or anti-rabbit Alexa Fluor-488 (anti-FAS) or anti-mouse Alexa Fluor-488 (anti-ZEB-1) conjugated secondary antibody (1:500 dilution) for 1 h at room temperature, in the dark. After thorough washing with PBST, the cells were counterstained with DAPI containing mounting medium. Post-mounting, the slide was sealed, dried and images were captured with Floid Cell Imaging Station (Life Technologies) at 20× magnification.
Scanning electron microscopy (SEM)
The previously published procedure was followed for SEM microscopy of the GFP/GFP-Par-4-transfected Panc-1-GR10 cells [38].
Cell viability assay
The experiment was performed using the previously published protocol [3]. Panc-1-GR10 cells transfected with GFP/GFP-Par-4 were proceeded for the MTT dye uptake assay to evaluate their cell viability.
In vivo studies for miR-200c and Par-4 expression
The experimental guidelines approved by the Institutional Animal Ethics Committee were strictly adhered to for the in vivo experiments. The tumor tissue samples collected from the previous study [35], were stored at −20 °C and later processed for RT-PCR and immunoblotting.
Bioinformatics analysis
The online database String 11.0 (https://string-db.org/) was used for the analysis of the RPPA data and identifies the differentially expressed proteins and their protein network.
Statistical analysis
Data represented, were expressed as mean ± S.D. of at least three independent experiments performed and analyzed using Student's t-test and. A two-sided value of *P < 0.05 was considered significant.
Results
The miR-200c expression is positively correlated with Par-4 protein levels in cancer cells
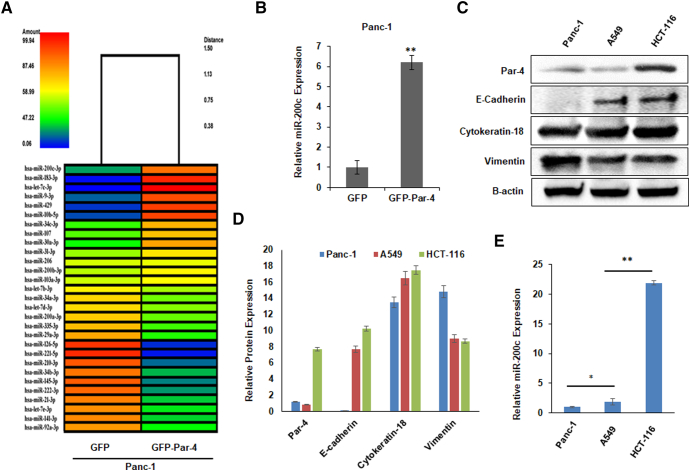
Over last few decades, various reports have highlighted the importance of miRNAs, particularly the miR-200 family members, for the abrogation of EMT [8,27,39]. Repressed expression of miR-200 family members, especially miR-200c, is a frequently noted event in various human cancers, which is well-documented to be associated with EMT [10,25]. Contrarily, Prostate apoptosis response 4 (Par-4) is a pro-apoptotic protein that possesses tumor-suppressing activities and is generally inactivated or down-regulated in cancers [31]. Restoration of Par-4 (therapy-induced) in many cancers offers superior anti-tumor effects of the test drug. Recently, the role of Par-4 in the suppression of EMT and its markers in pancreatic cancer cells has been established by our group [35]. Further, in continuation, we were curious to examine whether Par-4 restoration in pancreatic cancer cells could have any effect on the miR-200c. Heatmap of microarray analysis for a series of miRNAs was generated following transient transfection with GFP-Par-4 in Panc-1 cells. The data revealed that the miR-200c expression was significantly augmented in the GFP-Par-4-transfected cells compared to the GFP-transfected control (Fig. 1A). Fig. 1B shows the relative fold increase in the miR-200c levels upon Par-4 overexpression in Panc-1 cells. To examine any correlation between the basal Par-4 and miR-200c levels in different cancer cell lines, the cell lysates obtained from Panc-1, A549, and HCT-116 cells were analyzed by western blotting and qRT-PCR for the expression of Par-4 and miR-200c, respectively, along with other EMT markers. HCT-116 cell lysate showed the highest Par-4 basal level, followed by A549 and Panc-1 (Fig. 1C & D). The RT-PCR results were consistent with the western blotting results, wherein, HCT-116 cells with peaked Par-4 levels displayed the highest miR-200c expression followed by A549 and Panc-1 cells (Fig. 1E). Together, these results suggest a positive correlation between the Par-4 and miR-200c in the cancer cells.
Fig. 1.
Par-4-mediated induction of miR-200c expression in cancer cell lines. (A) Heatmap showing results of miRNA microarray analysis upon GFP/GFP-Par-4 transfection in Panc-1 cells. The dendrogram shows different expression levels of miRNAs among the samples. Orange indicates the highest expression of miRNA, while blue indicates the lowest expression. (B) The graph shows the fold increase in the expression level of miR-200c upon GFP/GFP-Par-4-overexpression in Panc-1 cells. (C) Western blot data of the basal protein levels of Par-4, E-cadherin, Cytokeratin-18, and Vimentin in HCT-116, Panc-1, and A549 cells. Beta-actin was used as a loading control. (D) The bar graph represents the relative protein expression levels of Par-4, E-cadherin, Cytokeratin-18, and Vimentin in HCT-116, Panc-1, and A549 cell lines. (E) The bar graph depicts the differential miR-200c expression levels in HCT-116, Panc-1, and A549, as monitored by qRT-PCR. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Par-4-induction by the pharmacological inducer, NGD16, positively regulates miR-200c levels leading to inhibition of cell motility
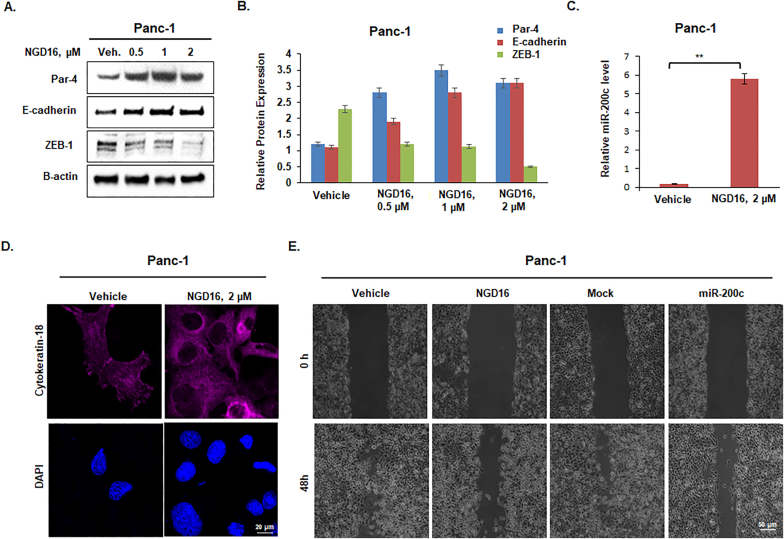
NGD16, a semi-synthetic derivative of 3,3′-Diindolylmethane (DIM), has been reported to inhibit the growth and migration of cancer cells [35]. Since we recently reported the Par-4-dependent EMT-inhibiting ability of NGD16, we were curious to investigate the outcomes of NGD16-mediated Par-4 induction on the endogenous miR-200c levels. As shown in Fig. 2A & B, Panc-1 cells treated with increasing concentrations of NGD-16 for 48 h, depicted a marked increase in the Par-4 levels concomitant with the augmented E-cadherin expression. Given that ZEB-1 is a crucial suppressor of E-cadherin and miR-200c in cancer cells, we sought whether the NGD16-mediated Par-4 induction could have any effect on the ZEB-1 level as well [[21], [22], [23], [24]]. Surprisingly, we found altered ZEB-1 level in the NGD16-treated Panc-1 cells, and the pattern was as similar as Par-4 induced lane (Fig. 2A & B). Panc-1 cells were similarly treated with Vehicle/NGD16 and employed for the qRT PCR analysis to examine miR-200c levels. As shown in Fig. 2C, there was a marked increase in the miR-200c expression of the NGD16-treated Panc-1 cells. Further, we were interested in examining the effects of NGD16 on another epithelial marker, Cytokeratin-18 [40,41]. Accordingly, Panc-1 cells were treated with Vehicle/NGD16 and subjected to immunochemistry. As depicted in Fig. 2D, the NGD16-treated Panc-1 cells displayed augmented expression of Cytokeratin-18 as compared to the respective control. Since miR-200c has been demonstrated to prevent migration of the cancer cells [42], the scratch wound healing assay was carried out to assess the effects of NGD16-mediated miR-200c upregulation on the motility of Panc-1 cells. The results unveiled a pronounced inhibition in the migration ability of NGD16-treated Panc-1 cells, similar to the miR-200c-transfected cells (Fig. 2E). The above results strongly suggest that the amplification of endogenous Par-4 levels can restore the mir-200c levels in the cancer cells, resulting in inhibition of EMT and migration.
Fig. 2.
NGD16-mediated Par-4 induction resulted in elevated levels of miR-200c. (A) Western blot data of Par-4, E-cadherin, and ZEB-1 expression levels in Panc-1 cells treated with indicated concentrations of NGD16 for 48 h. Beta-actin was used as a loading control. (B) The bar graph represents the relative protein expression levels of Par-4, E-cadherin, and ZEB-1 in NGD16-treated Panc-1 cells. (C) The bar graph shows the fold increase in the expression level of miR-200c upon treatment with the indicated concentration of NGD16 for 48 h. (D) Immunocytochemistry analysis for the Cytokeratin-18 (magenta) expression in the NGD16-treated Panc-1 cells along with DAPI (blue). Photographs were taken at 60× magnification. Scale bar 20 μm. (E) The anti-migratory effect of NGD16 and miR-200c in Panc-1 cells evaluated by the wound healing assay. The images were taken at 10× magnification. Scale bar, 50 μm. For the statistical analysis, the experiments were performed at least thrice. Data were compared with untreated control. Error bars represent standard deviations (S.D.) of three independent experiments. *P < 0.05, **P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Inverse relationship of Par-4/miR-200c with ZEB-1 in pancreatic cancer cells
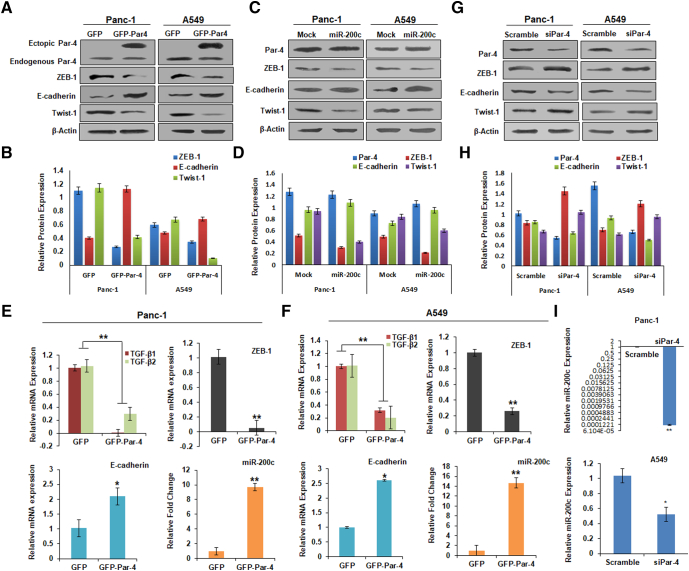
miR-200 family (miR-200s) members function as tumor-suppressors via directly targeting ZEB-1 in various cancers [25]. To study whether the Par-4-mediated miR-200c induction could modulate the ZEB-1 expression, we transfected the Panc-1 and A549 cells with GFP/GFP-Par-4/Mock/miR-200c and examined various EMT markers through immunoblotting. The results demonstrated that ectopic Par-4 attenuated the ZEB-1 protein levels along with another pro-EMT marker, Twist-1, with consistent amplification of the E-cadherin levels in both the cell lines (Fig. 3A & B). Nevertheless, miR-200c transfection had negligible effects on endogenous Par-4 levels. However, as expected, miR-200c downregulated the EMT markers and enhanced the E-cadherin protein levels in a similar way as good as Par-4 (Fig. 3C & D). TGF-beta signaling is altered in most of the cancers and aids in tumor progression. Moreover, Par-4 has been found to restore the apoptotic arm of the TGF-beta signaling pathway by rescuing Smad4 [43]. The maintenance of a stable mesenchymal phenotype requires the establishment of autocrine transforming growth factor-β (TGF-β) signaling to drive sustained ZEB-1 expression that suppresses the miR-200c expression [40]. We, therefore, investigated the effects of Par-4 overexpression on the mRNA levels of TGF- β1 and TGF- β2 in Panc-1 and A549 cells (Fig. 3E & F). Intriguingly, a clear downregulation in the mRNA levels of TGF-β1 and TGF-β2 was achieved in GFP-Par-4-transfected cells, followed by the downmodulation of ZEB-1 and robust escalation of E-cadherin mRNA levels. Since miR-200c is known to induce E-cadherin levels by inhibiting ZEB-1, we wanted to confirm whether the above E-cadherin amplification was attributed to restored miR-200c levels upon GFP-Par-4 transfection. We evaluated the miR-200c levels in a similar set of conditions wherein a drastic elevation in the miR-200c level was noted in the GFP-Par-4-transfected Panc-1/A549 cells compared to GFP-transfected cells (Fig. 3E & F). Rationally, we were interested in investigating the effects of silencing of endogenous Par-4 on ZEB-1 and miR-200c expression, along with other EMT markers. Remarkably, Par-4 silencing attributed to alter E-cadherin expression concomitant with an increase in the EMT markers- ZEB-1 and Twist-1 (Fig. 3G &H). The enhanced expression of EMT-markers upon Par-4 silencing corresponded with the impeded miR-200c levels in both Panc-1 and A549 cells, highlighting that the ZEB-1 suppression in the above conditions may be attributed to the increased miR-200c levels induced by Par-4 (Fig. 3I). The above results validate that Par-4 blocks EMT by downregulating TGF-β1, TGF-β2, and ZEB-1 and restoring the miR-200c and E-cadherin levels.
Fig. 3.
Par-4 and miR-200c show a similar effect on the EMT markers. (A) Western blot analysis GFP/GFP-Par-4-transfected Panc-1 and A549 cells for Par-4, ZEB-1, E-cadherin, and Twist-1 protein levels. Beta-actin was taken as the loading control. (B) The bar-graph represents the relative protein expression as quantified by the densiometric analysis. (C) Par-4, E-cadherin, Twist-1, Vimentin, and ZEB-1 expression levels in Mock/miR-200c-transfected Panc-1 and A549 cells as depicted by western blot, along with Beta-actin. (D) The bar-graph represents the relative protein expression as quantified by the densiometric analysis. (E) The bar graph depicts the relative mRNA expression levels of TGF-β1, TGF-β2, ZEB-1, and E-cadherin along with miR-200c levels in GFP/GFP-Par-4-transfected Panc-1 cells, as monitored by qRT-PCR analysis. (F) The bar-graph depicts the relative mRNA expression levels of TGF-β1, TGF-β2, ZEB-1, and E-cadherin along with miR-200c levels in GFP/GFP-Par-4-transfected A549 cells, as monitored by qRT-PCR analysis. (G) Western blot analysis of the whole-cell lysates of scramble (control) siRNA/siRNA Par-4 (siPar-4)-transfected Panc-1 and A549 cells to check the expression of Par-4, E-cadherin, Twist-1, Vimentin, and ZEB-1 along with Beta-actin. (H) The bar-graph represents the relative protein expression as quantified by the densiometric analysis. (I) The bar graph depicts the miR-200c levels in the Panc-1 and A549 cells transfected with scramble (control) siRNA/siRNA Par-4 (siPar-4). The data shown are representative of at least three independent experiments. Error bars represent standard deviations (S.D.) of three independent experiments. *P < 0.05, **P < 0.01.
Par-4 and miR-200c induction reverse the effects of H-Ras-mediated oncogenic transformation
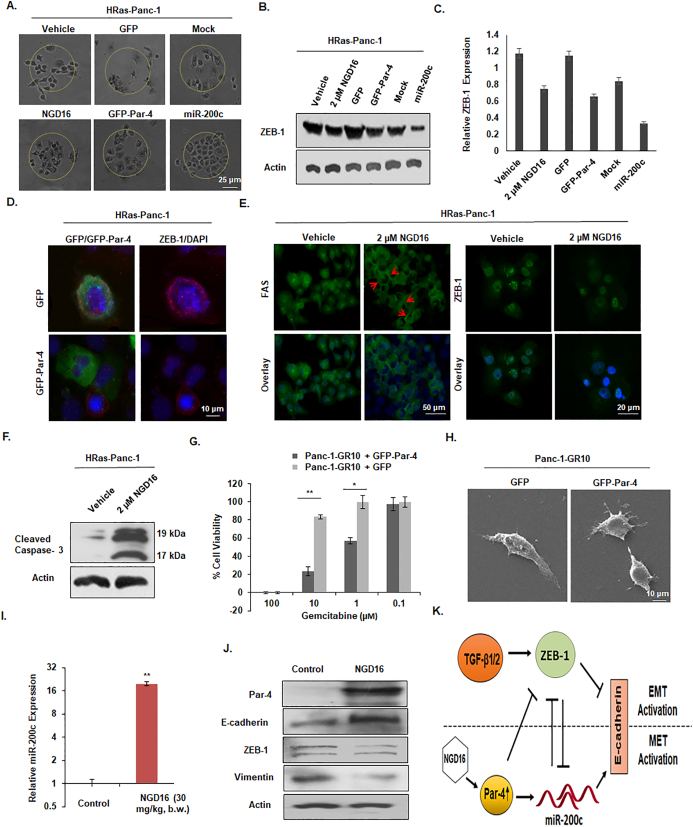
HRas is an oncogene known to be overexpressed in diverse cancers and its expression drastically promotes mesenchymal phenotype of cancer cells [44,45]. Rationally, we sought to investigate whether Par-4 stimulation, either by ectopic Par-4 or NGD-16, could abrogate the motility of HRas expressing Panc-1 cells (stable HRas-Panc-1). When these cells were subjected to Vehicle/NGD16 or transfected with GFP/GFP-Par-4/Mock/miR-200c for scattering assay, it was observed that NGD16, GFP-Par-4, and miR-200c hindered the scattering phenomena compared to the Vehicle, GFP and Mock control, respectively (Fig. 4A). Since ZEB-1 is one of the major transcription factors that drive the oncogene-mediated transformation in cancer cells, we were curious to examine whether Par-4 overexpression could alter the ZEB-1 protein levels in HRas-transformed Panc-1 cells. Indeed, the western blot data demonstrated that the HRas-Panc-1 cells subjected to NGD16/GFP-Par-4/miR-200c treatment depicted impeded ZEB-1 levels compared to their respective controls (Fig. 4B & C). Next, HRas-Panc-1 cells were transfected with GFP/GFP-Par-4 for immunocytochemistry for ZEB-1 analysis. The results showed a significant reduction in the ZEB-1 signal (red) in the GFP-Par-4-transfected cells as compared to GFP-transfected cells (Fig. 4D). Since oncogenic transformation in cancer cells makes them more resistant to apoptosis, we ought to study whether Par-4 induction in such cells could sensitize them towards apoptosis. Consequently, we treated the HRas-Panc-1cells with Vehicle/NGD16 and assessed the effect on FAS ligand trafficking. Remarkably, the data illustrated a fair increase in FAS trafficking in cells treated with NGD16 compared to the control. However, the ZEB-1 expression was appreciably downregulated in the NGD16-treated cells (Fig. 4E). The above results were also confirmed by assessing the activation of Caspase-3 in NGD16-treated HRas-Panc-1 cells. As demonstrated in Fig. 4F, the protein levels of cleaved Caspase-3 were significantly up in the NGD16-treated HRas-Panc-1 cells compared to the control, validating the activation of apoptosis cascade in these cells (Fig. 4F). Since ZEB-1 is an important EMT activator and is convincingly associated with the therapy-resistant phenotype [40], we wanted to confirm whether Par-4 restoration in the Gemcitabine resistant Panc-1-GR10 cells could augment the sensitivity of such cells to Gemcitabine. As demonstrated in Fig. 4G, the percent cell viability was decreased in the GFP-Par-4-transfected Panc-1-GR10 cells compared to the respective GFP control, indicating the re-sensitization to Gemcitabine in these cells. When the above sets of conditions were replicated for SEM analysis, we noticed the reversal of the mesenchymal phenotype in the GFP-Par-4-transfected Panc-1-GR10 cells, highlighting the role of Par-4-mediated ZEB-1 suppression in the reversal of EMT (Fig. 4H). Further, to validate our in vitro results, tumor samples collected from the control and NGD16-treated mice were analyzed for the miR-200c levels through RT-PCR. The data revealed a sharp augmentation in the miR-200c levels in the NGD-16-treated tumor sample compared to the control-treated sample (Fig. 4I). Of note, the immunoblots analysis revealed increased Par-4 expression in the tumor tissue lysates obtained from NGD16-injected mice as compared to the control. The augmented Par-4 levels were directly correlated with E-cadherin expression and inversely related to the ZEB-1 and Vimentin protein levels (Fig. 4J). The above results demonstrate that Par-4 negatively regulated the oncogene-mediated transformation, and its stimulation, either by ectopically or with its inducer (NGD16), impeded tumor progression by restoring the miR-200c levels to modulate the ZEB-1/E-cadherin axis (Fig. 4K).
Fig. 4.
Par-4 and miR-200c upregulation reverse the HRas-mediated transformation in Panc-1 cells. (A) Cell scattering ability of HRas-Panc-1 cells subjected to Vehicle/NGD16 treatment or transfected with GFP/GFP-Par-4/Mock/miR-200c is depicted. (B) Western blot analysis of HRas-Panc-1 cells, subjected to Vehicle/NGD16 treatment or transfected with GFP/GFP-Par-4/Mock/miR-200c, for the ZEB-1 protein levels. (C) The bar-graph represents the relative ZEB-1 protein expression as quantified by the densiometric analysis. (D) Immunocytochemistry analysis for the ZEB-1 (red) expression in the HRas-Panc-1 cells transiently transfected with GFP/GFP-Par-4 (green), along with DAPI (blue). Photographs were taken at 20× magnification. Scale bar 10 μm. (E) Immunocytochemistry analysis for FAS trafficking (20× magnification) and ZEB-1 (20× magnification) expression in HRas-Panc-1 cells treated with Vehicle/NGD16. Scale bar, 50 μm and 20 μm respectively. (F) Western blot analysis of HRas-Panc-1 cells, subjected to Vehicle/NGD16 treatment for the Cleaved Caspase-3 protein levels. (G) Percent Cell viability of the GFP/GFP-Par-4-transfected Panc-1-GR10 cells stimulated with different concentrations of Gemcitabine (0, 1, 10, and 100 μM). (H) SEM images of Panc-1-GR10 cells transfected with GFP/GFP-Par-4, 1000× magnification. Scale bar 10 μm. (I) The bar graph represents the relative fold increase in the miR-200c level in the NGD16-treated tumor sample compared to the control. (J) Western blotting of the tissue lysates obtained from NGD16-treated and control-treated tumor samples for Par-4, E-cadherin, ZEB-1, and Vimentin expression analysis. Beta-actin was used as a loading control. (K) The schematic diagram depicts the mechanism of miR-200c induction by Par-4 resulting in EMT suppression. Data is represented as means ± S.D. of at least three independent experiments. *P < 0.05, **P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Ectopic miR-200c and Par-4 triggers a coherent change in the expression of EMT and apoptosis-associated genes in pancreatic cancer cells
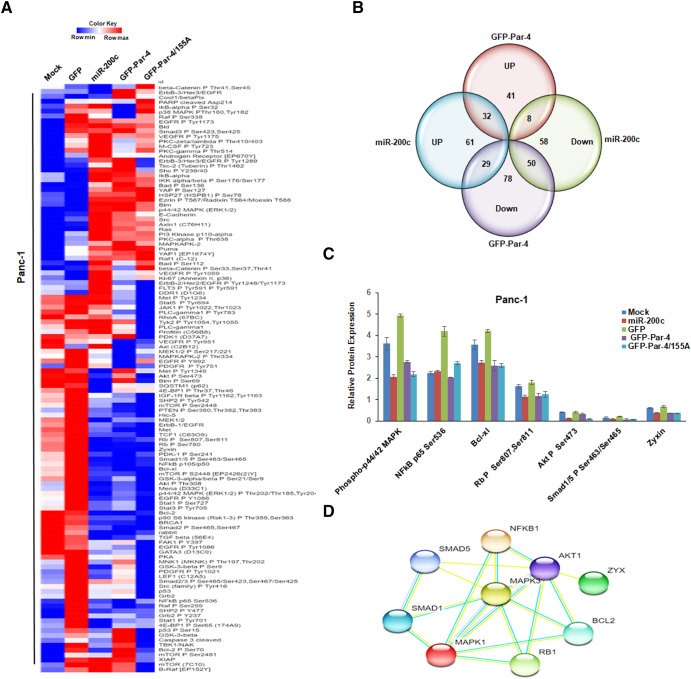
The above results validated that Par-4 restoration could amplify the expression of miR-200c in cancer cells and consequently suppress the EMT-markers like that of miR-200c. Therefore, we strongly hypothesized that Par-4 and miR-200c could indulge in adequate cross-talks that allow them to modulate the expression of various EMT markers in a similar manner. Phosphorylation of Par-4 at T155 residue is essential for nuclear translocation of Par-4 and apoptosis [33]. Moreover, nuclear translocation of Par-4 is essential for inhibition of cell survival NF-κB activity [32]. Therefore, we also employed the GFP-Par-4/155A construct, a point mutant of Par-4 made by site-directed mutagenesis to change the sequence at T155 from AGC to GCC, that unlike GFP-Par-4 could not translocate to the nucleus and inhibit the NF-κB activity. We aimed to draw a comparison between the differentially expressed proteins in the GFP-Par-4 and GFP-Par-4/155A-transfected Panc-1 cells and reveal some novel players that are crucial to inhibit NF-κB activity. To identify the proteome changes common to both Par-4, GFP-Par-4/155A and mir-200c-transfected cells, the RPPA experiment was carried out using Mock/miR-200c/GFP/GFP-Par-4/GFP-Par-4/155A-transfected Panc-1 cells. The RPPA analysis revealed many differentially expressed proteins (DEPs) between miR-200c and GFP-Par-4- transfected Panc-1 cells, as depicted by the heatmap in Fig. 5A. In total, 119 proteins were analyzed using validated antibodies. Compared to the control, Increased expression of 61 proteins was observed while 58 proteins were down-regulated in the miR-200c-transfected Panc-1 cells. In the GFP-Par-4-transfected Panc-1 cells, the expression of 41 proteins was found to be amplified while the levels of 78 proteins were revealed to have receded (Fig. 5B). As depicted by the Venn diagram analysis, 32 DEPs were commonly up-regulated while 50 DEPs were commonly down-regulated in the miR-200c and GFP-Par-4-transfected Panc-1 cells. However, 8 DEPs that were up-regulated in GFP-Par-4- transfected Panc-1 cells were down-regulated in miR-200c-transfected Panc-1 cells while 29 DEPs that were up-regulated in miR-200c-transfected Panc-1 cells were down-regulated GFP-Par-4-transfected Panc-1 cells. Next, we identified the most significant differentially expressed proteins that were common to both the miR-200c and GFP-Par-4-transfected Panc-1 cells. As shown in bar graphs (Fig. 5C), ectopic expression of both miR-200c and GFP-Par-4 led to the reduction in the levels of Phospho-p44/42 MAPK; Bcl-xl; Bim; Rb P Ser807, Ser811; Akt P Ser473; Smad1/5 P Ser463/Ser465 and Zyxin. Both GFP-Par-4 and GFP-Par-4/155A-transfected cells displayed a decrease in the NF-KB p65 Ser536 protein levels, but the effect was more significant with GFP-Par-4; However, miR-200c did not alter the NF-KB p65 Ser536 protein levels at all. Further analysis of the deduced differentially expressed proteins was performed using the String 11.0 database to identify their protein network (Fig. 5D). These results provide evidence that there could be multiple cross-talks between mir-200c and GFP-Par-4 signaling.
Fig. 5.
Whole proteome changes upon transfection with Par-4 and miR-200c. (A) Heatmap of the differentially expressed proteins (DEPs) in Mock, miR-200c, GFP, GFP-Par-4 and GFP-Par-4/155A-transfected Panc-1 cells, as identified by RPPA analysis. (B) Venn diagram of DEPs in miR-200c and GFP-Par-4-transfected Panc-1 cells. (C) The bar graph depicts the protein targets that were significantly altered in the miR-200c, GFP-Par-4, and GFP-Par-4/155A-transfected Panc-1 cells. (D) Protein interaction network of the significant DEPs common to miR-200c, GFP-Par-4, and GFP-Par-4/155A-overexpressed samples. The network was built using a String program (http://www.string-db.org/), with confidence greater than 0.5. Every node represents a protein, and the line color indicates the type of interaction evidence. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion
miR-200c belongs to the miR-200 family which is closely linked with metastasis of diverse tumors, particularly by abrogating EMT cascades through negative correlation with ZEB-1 protein [10,27]. In breast cancer, melanoma, and pancreatic cancer, miR-200c has been found to stall tumor promotion/invasion [29,46,47], but there is no report about any functional consequences between a vital tumor suppressor Par-4 and miRNA 200c. From that aspect, this study probably, for the first time, unearthed a novel link of Par-4 in the modulation of evolutionarily significant ZEB-1/E-cadherin axis via miR-200c.
Decelerated Par-4 levels have been reported in several types of cancers; various strategies have been suggested and tested to restore its apoptosis-inducing levels in cancer cells. In in vivo settings, Par-4 levels have been augmented using the adenoviral Par-4 constructs as well as by nano-liposomal delivery of Par-4 constructs [48]. Diverse natural/semi-synthetic pharmacophores have been explored for their Par-4-inducing properties. Our group recently unveiled two such natural product derivatives, viz. NGD16 and 3-AWA, which augmented endogenous Par-4 in multiple cancer cells [34,35]. In this study, the results clearly showed a direct correlation between NGD16-mediated Par-4 upregulation that corresponded to a miR-200c elevation in vitro and in vivo. Nevertheless, by transient knockdown of endogenous Par-4 with the siPar-4, we not only noticed a marked decrease in the epithelial marker, E-cadherin, with a concomitant upregulation of the mesenchymal markers, viz. Twist-1, Vimentin, and ZEB-1, but also a sharp drop in the miR-200c levels. According to previous research, increased expression of miR-200c confers negative regulation on its gene target ZEB-1, consequently upregulating E-cadherin expression to impede EMT [42]. Recent findings also implied that ZEB-1 could control the microRNA expression by interfering with the microRNA promoter activity and facilitating a reciprocal feedback loop in managing EMT [49]. It was found that ZEB-1 expression was inversely correlated with miR-200c/GFP-Par-4 expression in pancreatic/lung cancer cells. The above results underscore that Par-4 might be modulating the ZEB-1/miR-200c stoichiometric balance, by augmenting the miR-200c expression and abrogating the ZEB-1 protein, simultaneously at mRNA as well as protein levels, as hypothesized in Fig. 4K. RPPA analysis for the whole proteome of miR-200c and GFP-Par-4-transfected Panc-1 cells revealed proteins that were overlapping in both the sample sets. 7 of these proteins, Phospho-p44/42 MAPK; Bcl-xl; Bim; Rb P Ser807, Ser811; Akt P Ser473; Smad1/5 P Ser463/Ser465 and Zyxin were most significant DEPs in the two data sets. The expressional changes in these distinct proteins might be exerted independently by different arms of the miR-200c and Par-4 signaling pathways. Aberrant levels of Phospho-Akt (Ser473) are reported in many cancer types and are known to aid tumor growth and progression. Phosphorylated AKT has been reported to upregulate the anti-apoptotic protein, Bcl-xL, while downregulating the pro-apoptotic protein, Bim [33,36,43,50]. Our RPPA results show that both miR-200c and Par-4 decrease Phospho-Akt (Ser473), which could explain the upregulated Bim and downregulated Bcl-xL levels, activating the apoptotic signaling in these cells. ERK1/2 is a major component of MAPK pathway that controls the proliferative signaling (TGF, EGF) in the cancer cells [51,52]. The Downregulation of Phospho-p44/42 MAPK (ERK1/2) Thr202/Thr185, Tyr204/Tyr187 levels could be responsible for the anti-proliferative effects of both Par-4 and mir-200c in Panc-1 cells. RB1 is a tumor-suppressor that controls the G1/S phase progression by binding to E2F family of transcription factors. However, phosphorylation of RB1, catalyzed by cyclin-dependent protein kinases, inhibits its protein-binding abilities [53]. Moreover, RB phosphorylation on serine-807 is reported to be essential for its association with the anti-apoptotic protein, Bax [54]. Our results demonstrated a decline in the phosphorylated RB levels upon miR-200c and Par-4 overexpression, which could adequately contribute to their pro-apoptotic effects. Zyxin is a crucial focal adhesion protein, and its involvement is reported in many cancers [55]. Recently, it has been demonstrated to promote cell migration and invasion [38,56]. From our RPPA results, it is evident that the anti-migratory and anti-invasive effects of miR-200c and Par-4 could be considered for the reduced Zyxin level. Phosphorylated Smad1/5 (Ser463/Ser465), a downstream of TGF-Beta signaling, was also significantly downregulated upon miR-200c and Par-4 induction. Of note, the protein network analysis using the String 11.0 database displayed an intense cross-talk between all the significant DEPs (Fig. 5D).Since these proteins are involved majorly in cell proliferation, migration, and apoptosis, their downregulation by Par-4 unveils novel mechanisms exerted by Par-4 to accomplish its anti-proliferative as well as anti-invasive functions. Although Par-4 is universally accepted for its apoptosis-inducing properties, an EMT inhibitory perspective of this tumor-suppressor is emerging. We have demonstrated that extracellular Par-4, secreted by a classical BFA-sensitive pathway, inhibits tumor growth, and metastasis by modulating MMP-2 [57]. A study in a tail vein metastatic model revealed that Recombinant Par-4 (TRX-Par-4) and SAC (TRX-SAC) proteins are efficient in inhibit the formation of lung nodules [58]. Amin et al. showed EMT abrogation and invasion by Par-4 through modulation of beta-catenin. All these reports unveil the anti-metastatic role of Par-4, and similarly, the current finding elicits a novel link of Par-4 activation leading to augmentation of miR-200c levels, which might have immense therapeutic potential.
Abbreviations
- Par-4
Prostate apoptosis response-4
- miR-200c
micro RNA-200c
- DEPs
Differentially Expressed Proteins
- RPPA
Reverse Phase Protein Array
- EMT
Epithelial-mesenchymal transition
- qRT PCR
Quantitative Real-time Polymerase chain reaction
- SEM
Scanning Electron Microscopy
The following is the supplementary data related to this article.
Primer sequences for qPCR performed in the study.
CRediT authorship contribution statement
Archana Katoch: Conceptualization, Methodology, experimentation and Writing - Original draft preparation. Vijay Lakshmi Jamwal: Methodology, experimentation and Writing - Original draft preparation. Mir Mohd Faheem: Software and Experimentation. Sriram Kumar: Software, Validation. Shantibhusan Senapati: in vivo experimentation. Govind Yadav: in vivo experiment design and consultation. Sumit G Gandhi: Methodology, validation and Writing - Original draft. Anindya Goswami: Conceptualization, Data curation, Writing - Reviewing and Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
Our thanks to the Director, Dr. R.A. Vishwakarma, for encouraging us to accomplish this work.
Funding
The work was supported by Department of Biotechnology (DBT), Government of India, Grant No. BT/PR6768/MED/30/893/2012 to AG. The authors are also grateful to the Department of Biotechnology, Govt. of India, and the Council of Scientific and Industrial Research (CSIR) for providing fellowship to the research scholars.
Availability of data and materials
The datasets analyzed and/or used in the current study remain available, on request, from the corresponding author.
Contributor Information
Sumit G. Gandhi, Email: sumit@iiim.ac.in, sumit@iiim.res.in.
Anindya Goswami, Email: agoswami@iiim.ac.in, agoswami@iiim.res.in.
References
- 1.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 2.Sood P., Krek A., Zavolan M., Macino G., Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li C., Feng Y., Coukos G., Zhang L. Therapeutic microRNA strategies in human cancer. AAPS J. 2009;11:747. doi: 10.1208/s12248-009-9145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 7.Zhang B., Pan X., Cobb G.P., Anderson T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 8.Park S.-M., Gaur A.B., Lengyel E., Peter M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peter M.E. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–852. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., Goodall G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 11.Cittelly D.M., Dimitrova I., Howe E.N., Cochrane D.R., Jean A., Spoelstra N.S., Post M.D., Lu X., Broaddus R.R., Spillman M.A. Restoration of miR-200c to ovarian cancer reduces tumor burden and increases sensitivity to paclitaxel. Mol. Cancer Ther. 2012;11(12):2556–2565. doi: 10.1158/1535-7163.MCT-12-0463. (0463.2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cochrane D.R., Spoelstra N.S., Howe E.N., Nordeen S.K., Richer J.K. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol. Cancer Ther. 2009 doi: 10.1158/1535-7163.MCT-08-1046. (1535-7163. MCT-08-1046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S., Tetzlaff M.T., Cui R., Xu X. miR-200c inhibits melanoma progression and drug resistance through down-regulation of BMI-1. Am. J. Pathol. 2012;181(5):1823–1835. doi: 10.1016/j.ajpath.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mutlu M., Raza U., Saatci Ö., Eyüpoğlu E., Yurdusev E., Şahin Ö. miR-200c: a versatile watchdog in cancer progression, EMT, and drug resistance. J. Mol. Med. 2016;94:629–644. doi: 10.1007/s00109-016-1420-5. [DOI] [PubMed] [Google Scholar]
- 15.Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- 16.Comijn J., Berx G., Vermassen P., Verschueren K., van Grunsven L., Bruyneel E., Mareel M., Huylebroeck D., Van Roy F. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell. 2001;7:1267–1278. doi: 10.1016/s1097-2765(01)00260-x. [DOI] [PubMed] [Google Scholar]
- 17.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cano A., Pérez-Moreno M.A., Rodrigo I., Locascio A., Blanco M.J., del Barrio M.G., Portillo F., Nieto M.A. The transcription factor snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000;2:76. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 19.Giroldi L.A., Bringuier P.-P., de Weijert M., Jansen C., van Bokhoven A., Schalken J.A. Role of E boxes in the repression of E-cadherin expression. Biochem. Biophys. Res. Commun. 1997;241:453–458. doi: 10.1006/bbrc.1997.7831. [DOI] [PubMed] [Google Scholar]
- 20.Hajra K.M., Chen D.Y., Fearon E.R. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- 21.Hashiguchi M., Ueno S., Sakoda M., Iino S., Hiwatashi K., Minami K., Ando K., Mataki Y., Maemura K., Shinchi H. Clinical implication of ZEB-1 and E-cadherin expression in hepatocellular carcinoma (HCC) BMC Cancer. 2013;13:572. doi: 10.1186/1471-2407-13-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang M.-T., Wei P.-L., Liu J.-J., Liu D.-Z., Huey-Chun H., An J., Wu C.-C., Wu C.-H., Ho Y.-S., Yang Y.-Y. Knockdown of thrombomodulin enhances HCC cell migration through increase of ZEB1 and decrease of E-cadherin gene expression. Ann. Surg. Oncol. 2010;17:3379–3385. doi: 10.1245/s10434-010-1163-4. [DOI] [PubMed] [Google Scholar]
- 23.Kurahara H., Takao S., Maemura K., Mataki Y., Kuwahata T., Maeda K., Ding Q., Sakoda M., Iino S., Ishigami S. Epithelial–mesenchymal transition and mesenchymal–epithelial transition via regulation of ZEB-1 and ZEB-2 expression in pancreatic cancer. J. Surg. Oncol. 2012;105:655–661. doi: 10.1002/jso.23020. [DOI] [PubMed] [Google Scholar]
- 24.Bae G.-Y., Choi S.-J., Lee J.-S., Jo J., Lee J., Kim J., Cha H.-J. Loss of E-cadherin activates EGFR-MEK/ERK signaling, which promotes invasion via the ZEB1/MMP2 axis in non-small cell lung cancer. Oncotarget. 2013;4:2512. doi: 10.18632/oncotarget.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pieraccioli M., Imbastari F., Antonov A., Melino G., Raschellà G. Activation of miR200 by c-Myb depends on ZEB1 expression and miR200 promoter methylation. Cell Cycle. 2013;12:2309–2320. doi: 10.4161/cc.25405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korpal M., Lee E.S., Hu G., Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 2008;283(22):14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphries B., Yang C. The microRNA-200 family: small molecules with novel roles in cancer development, progression and therapy. Oncotarget. 2015;6:6472. doi: 10.18632/oncotarget.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurteau G.J., Spivack S.D., Brock G.J. Potential mRNA degradation targets of hsa-miR-200c. Cell Cycle. 2006;5:1951–1956. doi: 10.4161/cc.5.17.3133. [DOI] [PubMed] [Google Scholar]
- 30.Goswami A., Burikhanov R., de Thonel A., Fujita N., Goswami M., Zhao Y., Eriksson J.E., Tsuruo T., Rangnekar V.M. Binding and phosphorylation of par-4 by akt is essential for cancer cell survival. Mol. Cell. 2005;20:33–44. doi: 10.1016/j.molcel.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 31.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.El-Guendy N., Rangnekar V.M. Apoptosis by Par-4 in cancer and neurodegenerative diseases. Exp. Cell Res. 2003;283:51–66. doi: 10.1016/s0014-4827(02)00016-2. [DOI] [PubMed] [Google Scholar]
- 33.Gurumurthy S., Goswami A., Vasudevan K.M., Rangnekar V.M. Phosphorylation of Par-4 by protein kinase A is critical for apoptosis. Mol. Cell. Biol. 2005;25:1146–1161. doi: 10.1128/MCB.25.3.1146-1161.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amin H., Nayak D., ur Rasool R., Chakraborty S., Kumar A., Yousuf K., Sharma P.R., Ahmed Z., Sharma N., Magotra A. Par-4 dependent modulation of cellular β-catenin by medicinal plant natural product derivative 3-azido Withaferin A. Mol. Carcinog. 2016;55:864–881. doi: 10.1002/mc.22328. [DOI] [PubMed] [Google Scholar]
- 35.Katoch A., Suklabaidya S., Chakraborty S., Nayak D., Rasool R.U., Sharma D., Mukherjee D., Faheem M.M., Kumar A., Sharma P.R. Dual role of Par-4 in abrogation of EMT and switching on Mesenchymal to Epithelial Transition (MET) in metastatic pancreatic cancer cells. Mol. Carcinog. 2018;57(9):1102–1115. doi: 10.1002/mc.22828. [DOI] [PubMed] [Google Scholar]
- 36.Awasthi P., Mahajan V., Jamwal V.L., Kapoor N., Rasool S., Bedi Y.S., Gandhi S.G. Cloning and expression analysis of chalcone synthase gene from Coleus forskohlii. J. Genet. 2016;95:647–657. doi: 10.1007/s12041-016-0680-8. [DOI] [PubMed] [Google Scholar]
- 37.Seah K.S., Loh J.Y., Nguyen T.T.T., Tan H.L., Hutchinson P.E., Lim K.K., Dymock B.W., Long Y.C., Lee E.J.D., Shen H.-M. SAHA and cisplatin sensitize gastric cancer cells to doxorubicin by induction of DNA damage, apoptosis and perturbation of AMPK-mTOR signalling. Exp. Cell Res. 2018;370:283–291. doi: 10.1016/j.yexcr.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 38.Sperry R.B., Bishop N.H., Bramwell J.J., Brodeur M.N., Carter M.J., Fowler B.T., Lewis Z.B., Maxfield S.D., Staley D.M., Vellinga R.M. Zyxin controls migration in epithelial–mesenchymal transition by mediating actin-membrane linkages at cell–cell junctions. J. Cell. Physiol. 2010;222:612–624. doi: 10.1002/jcp.21977. [DOI] [PubMed] [Google Scholar]
- 39.Shimono Y., Zabala M., Cho R.W., Lobo N., Dalerba P., Qian D., Diehn M., Liu H., Panula S.P., Chiao E. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bitler B.G., Fink L.S., Wei Z., Peterson J.R., Zhang R. A high-content screening assay for small-molecule modulators of oncogene-induced senescence. J. Biomol. Screen. 2013;18:1054–1061. doi: 10.1177/1087057113491827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L., de Oliveira R.L., Wang C., Neto J.M.F., Mainardi S., Evers B., Lieftink C., Morris B., Jochems F., Willemsen L. High-throughput functional genetic and compound screens identify targets for senescence induction in cancer. Cell Rep. 2017;21:773–783. doi: 10.1016/j.celrep.2017.09.085. [DOI] [PubMed] [Google Scholar]
- 42.Wang X., Chen X., Wang R., Xiao P., Xu Z., Chen L., Hang W., Ruan A., Yang H., Zhang X. microRNA-200c modulates the epithelial-to-mesenchymal transition in human renal cell carcinoma metastasis. Oncol. Rep. 2013;30:643–650. doi: 10.3892/or.2013.2530. [DOI] [PubMed] [Google Scholar]
- 43.MohdFaheem M., Rasool R., Ahmad S.M., Jamwal V.L., Chakraborty S., Katoch A., Gandhi S.G., Bhagat M., Goswami A. Par-4 mediated Smad4 induction in PDAC cells restores canonical TGF-β/Smad4 axis driving the cells towards lethal EMT. Eur. J. Cell Biol. 2020;99(4):151076. doi: 10.1016/j.ejcb.2020.151076. [DOI] [PubMed] [Google Scholar]
- 44.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 45.Vuoriluoto K., Haugen H., Kiviluoto S., Mpindi J., Nevo J., Gjerdrum C., Tiron C., Lorens J., Ivaska J. Vimentin regulates EMT induction by slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene. 2011;30:1436. doi: 10.1038/onc.2010.509. [DOI] [PubMed] [Google Scholar]
- 46.Ahmad A., Aboukameel A., Kong D., Wang Z., Sethi S., Chen W., Sarkar F.H., Raz A. Phosphoglucose isomerase/autocrine motility factor mediates epithelial-mesenchymal transition regulated by miR-200 in breast cancer cells. Cancer Res. 2011;71(9):3400–3409. doi: 10.1158/0008-5472.CAN-10-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu J., Ohuchida K., Mizumoto K., Sato N., Kayashima T., Fujita H., Nakata K., Tanaka M. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Mol. Cancer. 2010;9:169. doi: 10.1186/1476-4598-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qin T.-J., Ma W., Liu S.-X., Yang G.-X., Wang Q.-Y., Zhao X.-H. Secretory expression of Par-4 SAC-HA2TAT following adeno-associated virus-mediated gene transfer induces apoptosis in HepG2 cells. Mol. Med. Rep. 2010;3:749–757. doi: 10.3892/mmr.2010.343. [DOI] [PubMed] [Google Scholar]
- 49.Wong T.-S., Gao W., Chan J.Y.-W. Transcription regulation of E-cadherin by zinc finger E-box binding homeobox proteins in solid tumors. Biomed. Res. Int. 2014;2014 doi: 10.1155/2014/921564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kreisberg J.I., Malik S.N., Prihoda T.J., Bedolla R.G., Troyer D.A., Kreisberg S., Ghosh P.M. Phosphorylation of Akt (Ser473) is an excellent predictor of poor clinical outcome in prostate cancer. Cancer Res. 2004;64:5232–5236. doi: 10.1158/0008-5472.CAN-04-0272. [DOI] [PubMed] [Google Scholar]
- 51.Ma J., Zeng S., Zhang Y., Deng G., Qu Y., Guo C., Yin L., Han Y., Cai C., Li Y. BMP4 promotes oxaliplatin resistance by an induction of epithelial-mesenchymal transition via MEK1/ERK/ELK1 signaling in hepatocellular carcinoma. Cancer Lett. 2017;411:117–129. doi: 10.1016/j.canlet.2017.09.041. [DOI] [PubMed] [Google Scholar]
- 52.Mann K.M., Ying H., Juan J., Jenkins N.A., Copeland N.G. KRAS-related proteins in pancreatic cancer. Pharmacol. Ther. 2016;168:29–42. doi: 10.1016/j.pharmthera.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 53.Knudsen E.S., Wang J. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol. Cell. Biol. 1997;17:5771–5783. doi: 10.1128/mcb.17.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Antonucci L.A., Egger J.V., Krucher N.A. Phosphorylation of the retinoblastoma protein (Rb) on serine-807 is required for association with Bax. Cell Cycle. 2014;13:3611–3617. doi: 10.4161/15384101.2014.964093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kotb A., Hyndman M.E., Patel T.R. The role of zyxin in regulation of malignancies. Heliyon. 2018;4 doi: 10.1016/j.heliyon.2018.e00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamamura M., Noguchi K., Nakano Y., Segawa E., Zushi Y., Takaoka K., Kishimoto H., Hashimoto-Tamaoki T., Urade M. Functional analysis of zyxin in cell migration and invasive potential of oral squamous cell carcinoma cells corrigendum in/10.3892/ijo. 2016.3702. Int. J. Oncol. 2013;42:873–880. doi: 10.3892/ijo.2013.1761. [DOI] [PubMed] [Google Scholar]
- 57.Rah B., Amin H., Yousuf K., Khan S., Jamwal G., Mukherjee D., Goswami A. A novel MMP-2 inhibitor 3-azidowithaferin A (3-azidoWA) abrogates cancer cell invasion and angiogenesis by modulating extracellular Par-4. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao Y., Burikhanov R., Qiu S., Lele S.M., Jennings C.D., Bondada S., Spear B., Rangnekar V.M. Cancer resistance in transgenic mice expressing the SAC module of Par-4. Cancer Res. 2007;67:9276–9285. doi: 10.1158/0008-5472.CAN-07-2124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences for qPCR performed in the study.
Data Availability Statement
The datasets analyzed and/or used in the current study remain available, on request, from the corresponding author.