Abstract
Our study aimed to explore the applicability of deep learning and machine learning techniques to distinguish MPE from BPE. We initially used a retrospective cohort with 726 PE patients to train and test the predictive performances of the driverless artificial intelligence (AI), and then stacked with a deep learning and five machine learning models, namely gradient boosting machine (GBM), extreme gradient boosting (XGBoost), extremely randomized trees (XRT), distributed random forest (DRF), and generalized linear models (GLM). Furthermore, a prospective cohort with 172 PE patients was applied to detect the external validity of the predictive models. The area under the curve (AUC) in the training, test and validation set were deep learning (0.995, 0.848, 0.917), GBM (0.981, 0.910, 0.951), XGBoost (0.933, 0.916, 0.935), XRT (0.927, 0.909, 0.963), DRF (0.906, 0.809, 0.969), and GLM (0.898, 0.866, 0.892), respectively. Although the Deep Learning model had the highest AUC in the training set (AUC = 0.995), GBM demonstrated stable and high predictive efficiency in three data sets. The final AI model by stacked ensemble yielded optimal diagnostic performance with AUC of 0.991, 0.912 and 0.953 in the training, test and validation sets, respectively. Using the driverless AI framework based on the routinely collected clinical data could significantly improve diagnostic performance in distinguishing MPE from BPE.
Abbreviations: MPE, malignant pleural effusion; BPE, benign pleural effusion; AI, artificial intelligence; GBM, gradient boosting machine; XGboost, extreme gradient boosting; XRT, extremely randomized trees; DFR, distributed random forest; GLM, generalized linear models; AUC, area under the curve; DL, deep learning; CEA, carcinoembryonic antigen; ADA, adenosine deaminase; LDH, lactate dehydrogenase
Keywords: Malignant pleural effusion, Differential diagnosis, Artificial intelligence, Machine learning, Deep learning
Highlights
-
•
This is the first study to apply AI to identify MPE from BPE with the largest sample size and most available variables.
-
•
Our predictive models are not only validated with a retrospective set, but also externally verified with a prospective set.
-
•
Our study provides a very effective and non-invasive diagnostic method using AI to help physicians in the management of PE.
Introduction
Pleural effusion (PE) is characterized with significant accumulations of fluid in pleural cavity, which is a common problem in clinical practice [[1], [2], [3], [4]]. There are about 1.5 million patients in the United States newly diagnosed with PE each year [5,6]. PE is related to more than 50 etiologies and could be divided into benign pleural effusion (BPE) and malignant pleural effusion (MPE) [7]. BPE is mainly caused by tuberculosis, pneumonia and chronic heart failure in China [8]. Sometimes, MPE presents as the initial or even only sign in patients with cancer, but this does not mean that MPE is a warning sign for early stage of cancer. To the contrary, MPE generally signifies an advanced stage of cancer and a worse survival [9]. Accurate identification of patients with high probability of MPE is critical to deploy optimal interventions and thus improve patients' clinical outcomes. Hence, a convenient method with a minimum invasion that can accurately identify malignancy from BPE as early as possible is highly desirable.
Several clinical markers including serum carcinoembryonic antigen (CEA), adenosine deaminase (ADA) and lactate dehydrogenase (LDH), are commonly used to differentiate MPE from BPE in a clinical setting. However, no single marker can obtain satisfactory diagnostic performance in identifying MPE from BPE. Therefore, combination of several significant variables seems to achieve better diagnostic performance than single index. A clinical study [10] from Spain revealed that combination of four serum tumor markers reached a sensitivity of 54%. Furthermore, a recent study [11] demonstrated that combination of four indexes (age, proteins, glucose, and lactic acid) selected by logistical regression to separate tuberculous pleurisy from BPE achieved 78% specificity and 93.5% sensitivity with an area under curve (AUC) of 0.915. Yang et al. [12] exploited logistic model to develop a PET-CT scoring model for the differential diagnosis of MPE and BPE, and the scoring model yielded a sensitivity of 83.3% and a specificity of 92.2% with the cut-off value of 4 points in the training group. Hence, the predictive performances of diagnostic models created by logistical regression are somewhat limited.
Recently, artificial intelligence (AI) in the medical field has become a research hotpot, and holds the promise to automatically diagnose heterogeneous diseases with high accuracy [[13], [14], [15], [16], [17], [18]]. In our previous work [19], four machine learning (ML) algorithms were successfully employed to construct and validate a quantitative histomorphometry to identify gastric cancer patients with high risk of recurrence. Chen et al. [20] created a computer-aided system of deep learning (DL) to automatically detect hyperplastic colorectal polyps with high accuracy. Kather et al. [21] successfully used DL algorithm to accurately predict microsatellite instability (MSI) from H&E histology in gastrointestinal tumors. Moreover, Yu et al. [22] demonstrated that they applied seven ML classifiers based on histopathology features to predict the survival of patients with lung cancer and the ML classifiers obtained fairly satisfactory predictive accuracy. In addition, a population-based cohort study revealed that the XGBoost model exhibited better predictability in differentiating between critically ill patients who would and would not response to fluid intake compared with a conventional logistic regression model [23]. However, no current studies have systematically assessed the predictive values of DL and ML models in the identification of MPE from BPE.
In our previous work [24], we succeeded to design a three dimensional scaffold microchip which could efficiently isolate individual effusion tumor cell (ETC) and ETC cluster from effusions. Then we used logistical regression analysis to create a three-marker (effusion CEA, ETC count and ETC cluster count) predictive model, and this predictive model obtained excellent diagnostic performances both in the training and validation sets. In this study, we used five ML algorithms and a DL classifier, which were automatically tuned to develop predictive models based on the most accessible clinical features and laboratory indexes to identify MPE from BPE. Next, we compared the diagnostic performances among the six computational models as well as the stacked ensemble model. Finally, we prospectively validated the seven predictive models with an independent cohort of 172 patients.
Materials and methods
Study population
An observational study of PE cases from January 2014 through April 2018 was performed in Renmin Hospital of Wuhan University (RHWU). A total of 726 patients with PE were finally included in the study and were randomly split into the training and test sets by the ratio of 8:2. To investigate the external validity, a prospective cohort containing 172 patients with PE in Wuhan Union Hospital (WUH) from August 2019 through December 2019 was used as a validation set. Both the clinical ethics committees of RHWU (No. WDRY 2019-K014) and WUH (No. 2019-S075) checked and approved the study design prior to the commencement of this clinical study. All patients were required to provide the informed consent. In addition, the prospective study conducted in WUH was also registered on the website of Clinical Trials (No. NCT03997669) prior to the initiation of this study.
The inclusion criteria:
(1) confirmed to suffer from PE by ultrasonography, chest CT or X-ray; (2) patients who underwent diagnostic thoracentesis; (3) PE patients with known etiologies after a series of examinations.
The exclusion criteria:
(1) patients without willingness to participate in this study; (2) patients younger than eighteen years old; (3) patients lack of critical clinical information; (4) PE patients with indeterminable causes.
Diagnostic criteria
A PE was determined as MPE if cancer cells were clearly found through cytological smear, cell block together with immunohistochemistry or pleural biopsy. Tuberculous pleural effusion (TPE) was diagnosed when acid-fast stain or mycobacterial culture was positive, or caseous necrosis was observed in histopathology. The diagnosis of chronic heart failure (CHF) was mainly based on medical history, a series of examinations (echocardiogram, electrocardiogram, b-type natriuretic peptide). More importantly, CHF reacts to diuretics. Additionally, parapneumonic effusion was identified when the presence of PE was associated with pneumonia, and PE was soon disappeared after antibiotic therapy. Besides, other types of BPE followed well-established diagnostic criteria.
Data collection
This study aimed to create diagnostic models based on the available electronic health record data, so the following clinical variables were collected through the electronic medical record (EMR). Data were collected at the time of admission and prior to any medical interventions. Demographic features (gender and age), objective clinical symptom (fever), radiological characteristics (volume of PE, site of PE,) blood routine [white blood cells (WBC), lymphocytes (LC), neutrophil cells (NC), red blood cell distribution width (RDW), platelets (PLT)], serum biochemical parameters [erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), serum albumin (ALB), serum LDH, serum alkaline phosphatase (ALP)], serum tumor markers [CEA, serum neuron-specific enolase (NSE), serum squamous cell carcinoma antigen (SCC)], effusion biochemical parameters [ADA, effusion ALB, effusion LDH], effusion routine [effusion WBC, percentage of lymphocytes (L%) and percentage of neutrophils (N%)] and effusion tumor marker (CEA). Furthermore, fever refers to a body temperature > 37.5 °C. The site of PE was classified into unilateral and bilateral. The volume of PE based on ultrasonography was categorized as mild PE (<500 mL), moderate PE (500–1000 mL) or severe PE (>1000 mL).
Machine learning and deep learning classifiers
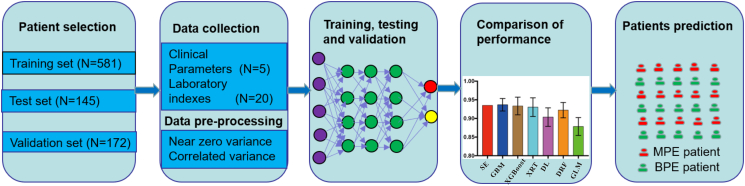
In this study, we use the driverless artificial intelligence (AI) by h2o package (version 3.28.0.4) in R, stacked with a DL and five types ML models [25], namely gradient boosting machine (GBM), extreme gradient boosting (XGBoost), extremely randomized trees (XRT), distributed random forest (DRF), and generalized linear models (GLM) to create predictive models for the differential diagnosis of MPE and BPE. The final models and the stacked ensemble model were chosen among the 100 models which were automatically trained and tuned. All available variables (N = 25) were directly taken into account as inputs to classify PE patients likely to be diagnosed with MPE. The confusion matrix contained the predicted probabilities and actual classification was applied to calculate the diagnostic performances of seven algorithmic models. 5-fold cross-validation were undertaken in patients randomly assigned to a 80% training set and a 20% test set to determine the average diagnostic performance. The workflow for establishing and validating the candidate predictive models via computational algorithms is shown in Fig. 1. Moreover, the parameters and code source of the algorithms are clearly illustrated in Table S1 and S2.
Fig. 1.
Flow chart of creating and validating predictive models using deep learning and machine learning algorithms.
Statistical analysis
The continuous variables between MPE and BPE groups were analyzed with either Student t-test or Mann–Whitney U test as appropriate. While, the categorical data were compared with Chi-square test or Fisher's exact test. The receiver operator characteristic (ROC) analyses were executed to evaluate the diagnostic performance of the models for predicting MPE. The area under the curve (AUC) was measured in each ROC curve and specificity together with sensitivity was also calculated to assess the diagnostic performances of the models. The above statistical analyses were implemented with R software version 3.6.1. and SPSS 20.0. Differences were regarded as statistically significant when P < 0.05 at both sides.
Results
Patients characteristics
A total of 1641 patients with PE were initially screened from both cohorts. 726 PE patients with intact clinical information from RHWU cohort and 172 cases from WUH cohort were finally enrolled in our analysis. The detailed flow chart of patient selection was clearly shown in Fig. 2. In the RHWU cohort, the differences in clinical features between MPE and BPE groups were exhibited in Table 1, and we could observe that age, gender and most laboratory indexes were statistically different between MPE and BPE groups. Moreover, the detailed disease types in RHWU and WUH cohorts were listed in Table 2. In the present study, lung cancer was the principle part of MPE both in the RHWU cohort and WUH cohort, while TPE was the main cause of BPE.
Fig. 2.
The detailed process of patient selection.
Table 1.
Clinical characteristics of the patients with PE in RHWU cohort.
| BPE (n = 391) | MPE (n = 335) | P | |
|---|---|---|---|
| Age | 57.5 ± 18.0 | 63.5 ± 13.6 | <0.001 |
| Gender, male | 270 (69.1%) | 192 (57.3%) | 0.001 |
| Male | 270(30.9%) | 192(42.7%) | |
| Female | 121 | ||
| Fever | 157 (40.2%) | 54 (16.1%) | <0.001 |
| PE volume | 0.410 | ||
| Mild | 97 (24.8%) | 92 (27.5%) | |
| Moderate | 200 (51.2%) | 168 (50.1%) | |
| Severe | 94 (24.0%) | 75 (22.4%) | |
| Primary site | 0.316 | ||
| Unilateral | 219 (56.0%) | 200 (59.7%) | |
| Bilateral | 172 (44.0%) | 135 (40.3%) | |
| WBC (109/L) | 7.5 ± 2.6 | 7.7 ± 2.3 | 0.570 |
| NC (109/L) | 5.6 ± 1.3 | 5.5 ± 1.9 | 0.788 |
| LC (109/L) | 1.2 ± 0.5 | 1.3 ± 0.7 | 0.375 |
| RDW | 45.2 ± 6.5 | 46.1 ± 7.0 | 0.069 |
| PLT (109/L) | 276.3 ± 118.2 | 263.0 ± 113.0 | 0.122 |
| CRP (mg/L) | 67.8 ± 29.4 | 35.4 ± 18.5 | <0.001 |
| ALP (U/L) | 67.1 ± 20.6 | 70.3 ± 17.6 | 0.321 |
| ALB (g/L) | 60.5 ± 17.3 | 66.7 ± 26.3 | 0.142 |
| LDH (U/L) | 280.2 ± 131.4 | 289.9 ± 147.2 | 0.571 |
| ESR (mm/h) | 65.3 ± 18.6 | 41.4 ± 12.9 | <0.001 |
| CEA (ng/mL) | 3.1 ± 1.7 | 57.3 ± 19.0 | <0.001 |
| SCC (ng/mL) | 1.3 ± 0.6 | 2.4 ± 0.7 | 0.001 |
| NSE (ng/mL) | 20.0 ± 7.7 | 27.1 ± 13.8 | <0.001 |
| Effusion WBC | 1290.0 (445.0, 2739.0) | 1158.0 (480.0, 2100.0) | 0.418 |
| Effusion N% | 20.5 ± 5.1 | 16.9 ± 8.7 | 0.029 |
| Effusion L% | 74.0 ± 26.4 | 74.4 ± 22.0 | 0.842 |
| Effusion ALB | 38.7 ± 13.7 | 40.8 ± 11.9 | 0.028 |
| Effusion ADA | 31.5 ± 11.7 | 14.3 ± 6.1 | <0.001 |
| Effusion LDH | 256.0 (133.0, 501.0) | 301.0 (184.0, 530.0) | 0.293 |
| Effusion CEA | 1.0 (0.5, 1.8) | 168.8 (6.9, 688.0) | <0.001 |
Table 2.
Origins of PE in RHWU cohort and WUH cohort.
| Disease type | RHWU cohort (N = 726) | WUH cohort (N = 172) |
|---|---|---|
| MPE | ||
| Lung cancer | 261 (35.95%) | 51 (29.65%) |
| Breast cancer | 12 (1.65%) | 2 (1.16%) |
| Lymphoma | 18 (2.48%) | 1 (0.58%) |
| Mesothelioma | 4 (0.55%) | 1 (0.58%) |
| Ovary cancer | 6 (0.83%) | 0 (0%) |
| Other cancers | 34 (4.68%) | 4 (2.33%) |
| BPE | ||
| Tuberculous pleurisy | 186 (25.62%) | 56 (32.56%) |
| parapneumonic effusions | 151 (20.80%) | 35 (20.35%) |
| Heart failure | 30 (4.13%) | 7 (4.07%) |
| Pulmonary embolism | 1 (0.14%) | 0 (0%) |
| Empyema | 12 (1.65%) | 5 (2.91%) |
| Other benign diseases | 11 (1.52%) | 10 (5.81%) |
Model performance in the RHWU cohort
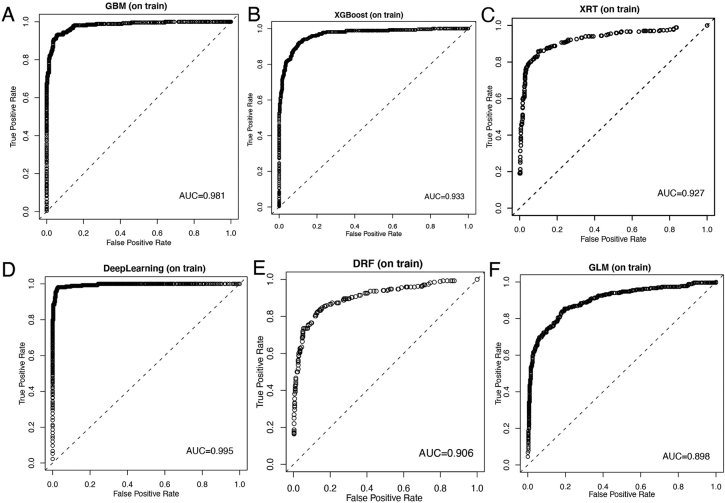
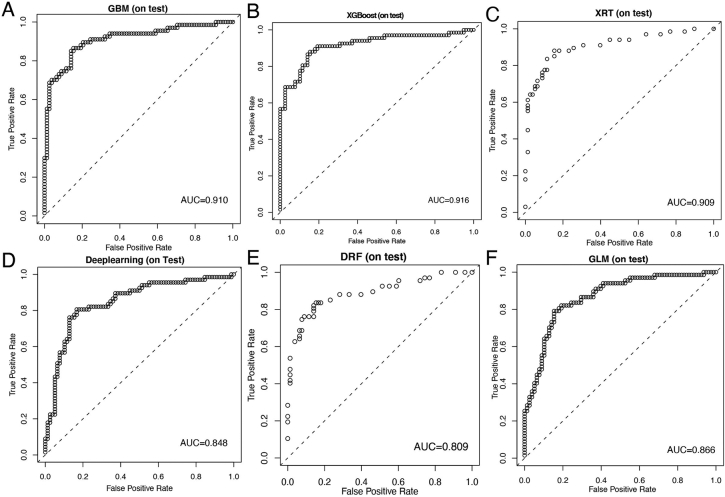
Patients in the RHWU cohort were randomly portioned to the training set (N = 581) and testing set (N = 145). Discriminative abilities of six models were evaluated using ROC analysis. As illustrated in Fig. 3, the AUCs for identification of MPE in the training set were 0.981 by the GBM, 0.933 by XGBoost, 0.927 by the XRT, 0.995 by the DL, 0.906 by the DRF and 0.898 by the GLM. Similarly, as shown in Fig. 4, the diagnostic performances in the testing set were the following: 0.910 by the GBM, 0.916 by XGBoost, 0.909 by the XRT, 0.848 by the deep learning, 0.809 by the DRF and 0.866 by the GLM. Hence, we could conclude that although the DL model had the highest AUC in the training set, GBM model achieved excellent predictive ability for the differentiation of MPE and BPE in the RHWU cohort. Furthermore, the sensitivity and specificity of each predictive models in the training, test and validation sets were listed in Table 3.
Fig. 3.
Discriminative abilities of predictive models for the identification of MPE from BPE in the training set. ROC curves of predictive model created by GBM (A); XGBoost (B); XRT (C); DL (D); DRF(E); GLM(F).
Fig. 4.
Diagnostic abilities of predictive models for the differential diagnosis of MPE and BPE in the test set. ROC curves of predictive model created by GBM (A); XGBoost (B); XRT (C); DL (D); DRF(E); GLM(F).
Table 3.
Sensitivity and specificity of seven predictive models in the training, test and validation sets.
| Algorithmic models | Training set |
Test set |
Validation set |
|||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| DL | 95.90% | 98.08% | 80.59% | 83.33% | 89.83% | 87.61% |
| GBM | 92.91% | 94.57% | 85.57% | 84.62% | 84.75% | 95.58% |
| XGBoost | 93.66% | 87.54% | 88.06% | 84.62% | 94.92% | 83.19% |
| XRT | 85.82% | 90.10% | 83.58% | 88.46% | 94.92% | 89.61% |
| DFR | 82.46% | 86.90% | 83.58% | 84.62% | 93.22% | 91.15% |
| GLM | 83.96% | 81.79% | 79.10% | 84.61% | 81.36% | 82.30% |
| Stacked Ensemble | 97.76% | 94.89% | 80.60% | 87.18% | 92.33% | 92.92% |
Validation of the models in the WUH cohort
To probe external validity of the models, we prospectively collected clinical information of 172 patients with PE from WUH cohort which was independent from the WHWU cohort. The basic features of patients from WUH cohort were listed in Table S3. In the validation set, ROC analyses revealed that the predict accuracy as measured by the AUC was 0.951 by the GBM, 0.935 by XGBoost, 0.963 by the XRT, 0.917 by the DL model, 0.969 by the DRF and 0.892by the GLM (Fig. 5). Among them, GBM model achieved the most favorable predictive performance in the WUH cohort with a sensitivity of 84.75% and a specificity of 95.58%.
Fig. 5.
Diagnostic performances of predictive models for the discrimination of MPE and BPE in the validation set. ROC curves of predictive model created by GBM (A); XRT (B); XGBoost (C); DL(D); DRF(E); GLM(F).
Comparison of the model performance for prediction of MPE
Stacked Ensemble is an important ML approach using multiple predictive models from different algorithms to pick out the best combination of a variety of prediction algorithms. As illustrated in Fig. 6A-C, The Stacked Ensemble model performed favorable predictive performances in the training set (AUC = 0.991), in the testing set (AUC = 0.912) and in the validation set (AUC = 0.953). Moreover, 5-fold cross validation was performed to evaluate the comprehensive predictabilities of the six models and the Stacked Ensemble model. Surprisingly, DL ranked fourth among the six models (Fig. 6D). Three machine learning models (GBM, XGBoost and XRT) achieved more favorable predictive outcomes than the DL model and only GBM model showed significantly better AUC than the DL model. Moreover, Taken together, assembling the DL and ML algorithms by Stacked Ensembles could greatly improve diagnostic significance for the differentiation of MPE and BPE.
Fig. 6.
Predictability of the model developed by Stacked Ensemble and comparison of the seven predictive models. ROC curves of the model developed by Stacked Ensemble in the training set (A); test set (B) and validation set (C). The seven algorithms were ten cross-validation to calculate the average AUC of each predictive model (D).
Important features from the models
To investigate the potential impact of each clinical feature on the discriminative abilities of the predictive models, we ranked the clinical variables from high to low in each model based on the functional contribution to the outputs. In this method [25], variables that provide important information to the trained models are ranked higher than those providing redundant information. Since the features with lower rank possessed little impacts on the predictive performance of the classification model, we only listed the first ten features. As displayed in Supplementary Fig. S1, effusion CEA ranked first in the DL and ML models, highlighting its great significance in the separation of MPE from BPE. Therefore, we exploited the ROC analyses to specifically evaluate the diagnostic accuracy of effusion CEA. In the training set, effusion CEA exhibited good diagnostic performance as measured by an AUC of 0.909 with a sensitivity of 82.09% and a specificity of 91.37% when the optimal cutoff value was set at 3.6 ng/mL (Fig. S2A). As displayed in Fig. S2B—C, effusion CEA obtained acceptable performances both in the test set (AUC = 0.883) and validation set (AUC = 0.866). With the same cutoff value, the sensitivity and specificity of effusion CEA for the differential diagnosis of MPE and BPE were 79.1% and 85.9% in the test set, and 63.41% and 89.38% in the validation set. To sum up, we concluded that the driverless AI framework, including ML and DL, offered great improvement in separating MPE from BPE over the effusion CEA.
Discussion
MPE usually represents end-stage malignancy and is closely related to poor median survival [26,27]. The high morbidity of MPE continues to rise and therefore causes a heavy health care burden [28]. An ability to distinguish between MPE and BPE with considerable accuracy is clinically significant to avoid the deferred diagnosis of MPE [[29], [30], [31], [32]]. Thus, this clinical study aiming to precisely differentiate MPE from BPE was of great public health implications. Using sophisticated DL and ML techniques, we identified some important clinical features associated with the discrimination of MPE and BPE, such as effusion CEA, serum CEA, ADA and ESR. In this study, we demonstrated that automatically tuned ML algorithms such as the GBM, XGBoost and XRT models can enrich the most informative clinical features and allow us to well construct better-performing predictive models by stacked ensemble, whose predictive accuracy was significantly superior to the DL model and effusion CEA. More importantly, a prospective cohort was applied to detect the clinical application of predictive models in different patients’ population, and the predictive models also exhibited satisfactory diagnostic accuracy. To the best of our knowledge, this is the first study to apply driverless AI to identify MPE from BPE with the largest sample size.
Patients with MPE could present similar manifestations as BPE patients, so tremendous efforts have been made to differentiate MPE from BPE in the past few decades. Rong et al. [33] detected the level of Hsp90-beta in PE and found that effusion Hsp90-beta could identify MPE with a sensitivity of 93.46% and s specificity of 79% when the optimal level of Hsp90-beta was set at 1.659 ng/mL. Jing et al. [34] demonstrated that the effusion sB7-H4 was a potentially promising biomarker in identification of MPE from BPE with a sensitivity of 81.82% and s specificity of 90.48%. Additionally, a recent meta-analysis [35] with 550 PE patients from seven case-control studies revealed that detection of serum IL-27 concentration was an accurate test for the differential diagnosis of MPE and BPE with a sensitivity of 93% and a specificity of 97%. In spite of the relatively high diagnostic accuracy, these tumor markers were not routinely detected in clinical practice.
Most of studies focused on the role of serum CEA in the differential diagnosis of MPE and BPE, while few studies investigated the potential diagnostic significance of effusion CEA. Pan et al. [36] found that levels of effusion CEA were significantly higher in patients with MPE than that in BPE patients and thus effusion ECA was incorporated into the predictive model. Furthermore, a clinical study conducted by Zhang et al. [37] revealed that effusion CEA could be served as a promising indicator for the discrimination of MPE and with favorable diagnostic performance as reflected by an AUC of 0.924. Our study showed that effusion CEA was the most informative parameter in the deep learning and machine learning models, indicating the significant role of effusion CEA in the differential diagnosis of MPE and BPE. ROC analyses in our study also showed the diagnostic performance as reflected by AUC was 0.909 in the training set, 0.883 in the test set and 0.866 in the validation set respectively. Effusion CEA was directly secreted by exfoliative cancer cells in the process of tumor invasion and metastasis, and levels of CEA were much higher in PE than that in serum [38]. Hence, effusion CEA is a good biomarker for the identification of MPE.
Due to the limitation of algorithm, predictive model created by logistical regression analysis is unlikely to achieve adequate diagnostic accuracy compared with that by the advanced DL or ML algorithms. Compared with logistical regression, one of the advantages of DL and ML methods is handling the complex associations of a vast number of clinical variables with nonlinear interactions [39]. Currently, few clinical studies have been performed on the application of ML algorithms in the recognition of MPE from BPE. Porcel et al. [40] employed a decision tree model to selected 4 discriminant variables (age, body temperature, PE ADA and LDH) among 12 clinical features. Their predictive model obtained 98.3% specificity, 92.2% sensitivity, and AUC of the ROC curve was 0.976 for identifying TPE. Moreover, Ren et al. [41] applied 12 clinical features to design a random forest model, and this model exhibited favorable diagnostic performance for the identification of TPE with a sensitivity of 90.6% and a specificity of 92.3%. They also verified the diagnostic model in the prospective study, and the results indicated that the specificity and sensitivity were 90.0% and 100.0% respectively. Exactly speaking, no studies have specifically applied the DL algorithm to create and validate a predictive model for the differentiation between MPE and BPE. In our prognostic study containing records of 898 patients, we demonstrated that the diagnostic models constructed by ML and DL classifiers exhibited acceptable performances in the diagnosis of MPE, and GBM stands as the superior ML method.
We undertook this clinical study with the primary goal to compare five ML classifiers with DL for construction of predictive models for the identification of MPE from BPE. It is worth noting that DL demonstrated the highest AUC of 0.995 in the train set whereas the prediction efficiency was less satisfactory in the test and validation set. DL method is more sensitive to changes of sample size than ML technique [41], and DL algorithm requires sufficient samples to obtain high predictive accuracy. The sample sizes in the test and validation sets were relatively small compared with that in the training set. Therefore, ML models offered significant improvement over DL model in predicting MPE based on the most accessible clinical features in this study. The underlying reason might be due to overfitting [42] of DL in the training set in our study. Besides, the inputs of the models were relatively simple compared with large factors in dealing with complex application scenarios such as medical images, which might be more suitable for DL [43,44].
Note that a few limitations exist in this study. First, the study design in training and test sets was retrospective and thus suffered from inherent biases. Although we validated our study with a prospective cohort registered on the clinical trial website, the sample size in WUH cohort was relatively small. Second, our cohorts contained patients with PE only from one geographic region (Wuhan) of China, which might limit the generalizability of predictive models and require further validations in patients from distinct geographic regions. Finally, some other serum tumor markers, such as CA19–9, CA12–5 were not included in our study, as most of the included patients with MPE were lung cancer and few patients underwent such tests. Therefore, more prospective studies with large sample size from multiple medical centers are further warranted to verify our conclusion in the future.
In conclusion, using driverless AI to create a predictive model based on the routine clinical indexes for the identification of MPE could improve diagnostic performance. GBM is superior to DL and stacked ensemble offers the optimal combination of a collection of prediction algorithms for the discrimination of MPE and BPE, which may provide a more effective and non-invasive diagnostic method to help physicians in decision-making. Further researches are necessary to verify the feasibility and generalizability of applying the computational algorithms to accurately identify patients with MPE in clinical settings.
CRediT authorship contribution statement
Yuan Li: Conceptualization, Methodology, Software, Writing- Original draft preparation. Shan Tian: Data curation, Writing- Original draft preparation. Yajun Huang: Data collection, Investigation. Weiguo Dong: Supervision, Writing- Reviewing and Editing.
Declaration of competing interest
No potential conflicts of interest were disclosed by the authors, Yuan Li, Shan Tian, Yajun Huang, Weiguo Dong.
The funders had no influence on the study design, data collection, data analysis, data interpretation or writing of the report.
Acknowledgments
Acknowledgements
Not applicable.
Funding
This work was financially supported by the National Natural Science Fund of China (NSFC; Grant No. 81902369; 8157101984).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.100896.
Appendix A. Supplementary data
Supplementary material
References
- 1.McGrath E.E., Anderson P.B. Diagnosis of pleural effusion: a systematic approach. American journal of critical care : an official publication. American Association of Critical-Care Nurses. 2011;20:119–128. doi: 10.4037/ajcc2011685. [DOI] [PubMed] [Google Scholar]
- 2.Light R.W. Clinical practice. Pleural effusion. N. Engl. J. Med. 2002;346:1971–1977. doi: 10.1056/NEJMcp010731. [DOI] [PubMed] [Google Scholar]
- 3.Porcel J.M., Azzopardi M., Koegelenberg C.F., Maldonado F., Rahman N.M., Lee Y.C.G. The diagnosis of pleural effusions. Expert Rev Resp Med. 2015;9:801–815. doi: 10.1586/17476348.2015.1098535. [DOI] [PubMed] [Google Scholar]
- 4.Bhatnagar R., Maskell N. The modern diagnosis and management of pleural effusions. BMJ (Clinical Research Ed.) 2015;351:h4520. doi: 10.1136/bmj.h4520. [DOI] [PubMed] [Google Scholar]
- 5.Ferreiro L., Toubes M.E., San José M.E., Suárez-Antelo J., Golpe A., Valdés L. Advances in pleural effusion diagnostics. Expert Rev Resp Med. 2020;14:51–66. doi: 10.1080/17476348.2020.1684266. [DOI] [PubMed] [Google Scholar]
- 6.Feller-Kopman D., Light R. Pleural disease. N. Engl. J. Med. 2018;378:740–751. doi: 10.1056/NEJMra1403503. [DOI] [PubMed] [Google Scholar]
- 7.Porcel J.M., Esquerda A., Vives M., Bielsa S. Etiology of pleural effusions: analysis of more than 3,000 consecutive thoracenteses. Arch. Bronconeumol. 2014;50:161–165. doi: 10.1016/j.arbres.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Walker S., Maskell N. Identification and management of pleural effusions of multiple aetiologies. Curr. Opin. Pulm. Med. 2017;23:339–345. doi: 10.1097/MCP.0000000000000388. [DOI] [PubMed] [Google Scholar]
- 9.Zamboni M.M., Da Silva C.T.J., Baretta R., Cunha E.T., Cardoso G.P. Important prognostic factors for survival in patients with malignant pleural effusion. Bmc Pulm Med. 2015;15:29. doi: 10.1186/s12890-015-0025-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porcel J.M., Vives M., Esquerda A., Salud A., Pérez B., Rodríguez-Panadero F. Use of a panel of tumor markers (carcinoembryonic antigen, cancer antigen 125, carbohydrate antigen 15-3, and cytokeratin 19 fragments) in pleural fluid for the differential diagnosis of benign and malignant effusions. Chest. 2004;126:1757–1763. doi: 10.1378/chest.126.6.1757. [DOI] [PubMed] [Google Scholar]
- 11.González A., Fielli M., Ceccato A., Luna C. Score for differentiating pleural tuberculosis from malignant effusion. Medical Sciences (Basel, Switzerland) 2019;7:36. doi: 10.3390/medsci7030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang M., Tong Z., Wang Z., Zhang Y., Xu L., Wang X. Development and validation of the PET-CT score for diagnosis of malignant pleural effusion. Eur J Nucl Med Mol I. 2019;46:1457–1467. doi: 10.1007/s00259-019-04287-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J., An J.Y., Choi M.G., Park S.H., Kim S.T., Lee J.H. Deep learning-based survival analysis identified associations between molecular subtype and optimal adjuvant treatment of patients with gastric cancer. JCO Clinical Cancer Informatics. 2018;2:1–14. doi: 10.1200/CCI.17.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coudray N., Ocampo P.S., Sakellaropoulos T., Narula N., Snuderl M., Fenyö D. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 2018;24:1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saltz J., Gupta R., Hou L., Kurc T., Singh P., Nguyen V. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181–193.e7. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elfiky A.A., Pany M.J., Parikh R.B., Obermeyer Z. Development and application of a machine learning approach to assess short-term mortality risk among patients with cancer starting chemotherapy. JAMA Netw. Open. 2018;1:e180926. doi: 10.1001/jamanetworkopen.2018.0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajkomar A., Dean J., Kohane I. Machine learning in medicine. N. Engl. J. Med. 2019;380:1347–1358. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 18.Goldenberg S.L., Nir G., Salcudean S.E. A new era: artificial intelligence and machine learning in prostate cancer. Nature Reviews. Urology. 2019;16:391–403. doi: 10.1038/s41585-019-0193-3. [DOI] [PubMed] [Google Scholar]
- 19.Ji M., Yuan L., Jiang X., Zeng Z., Zhan N., Huang P. Nuclear shape, architecture and orientation features from H&E images are able to predict recurrence in node-negative gastric adenocarcinoma. J. Transl. Med. 2019;17:92. doi: 10.1186/s12967-019-1839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen P., Lin M., Lai M., Lin J., Lu H.H., Tseng V.S. Accurate classification of diminutive colorectal polyps using computer-aided analysis. Gastroenterology. 2018;154:568–575. doi: 10.1053/j.gastro.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Kather J.N., Pearson A.T., Halama N., Jäger D., Krause J., Loosen S.H. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019;25:1054–1056. doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu K., Zhang C., Berry G.J., Altman R.B., Ré C., Rubin D.L. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 2016;7:12474. doi: 10.1038/ncomms12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z., Ho K.M., Hong Y. Machine learning for the prediction of volume responsiveness in patients with oliguric acute kidney injury in critical care. Critical Care (London, England) 2019;23:112. doi: 10.1186/s13054-019-2411-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian S., Cheng S., Guo Y., Xie M., Zhan N., Zeng Z. High efficient isolation of tumor cells by a three dimensional scaffold chip for diagnosis of malignant effusions. ACS Applied Bio Materials. 2020;3:2177–2184. doi: 10.1021/acsabm.0c00031. [DOI] [PubMed] [Google Scholar]
- 25.Alakwaa F.M., Chaudhary K., Garmire L.X. Deep learning accurately predicts estrogen receptor status in breast cancer metabolomics data. J. Proteome Res. 2018;17:337–347. doi: 10.1021/acs.jproteome.7b00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas J.M., Musani A.I. Malignant Pleural Effusions A Review. 2013;34:459–471. doi: 10.1016/j.ccm.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Asciak R., Rahman N.M. Malignant pleural effusion: from diagnostics to therapeutics. Clin. Chest Med. 2018;39:181–193. doi: 10.1016/j.ccm.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 28.McCracken D.J., Porcel J.M., Rahman N.M. Malignant pleural effusions: management options. Semin Resp Crit Care. 2018;39:704–712. doi: 10.1055/s-0038-1676572. [DOI] [PubMed] [Google Scholar]
- 29.Psallidas I., Kalomenidis I., Porcel J.M., Robinson B.W., Stathopoulos G.T. Malignant pleural effusion: from bench to bedside. European Respiratory Review : An Official Journal of the European Respiratory Society. 2016;25:189–198. doi: 10.1183/16000617.0019-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grosu H.B., Kazzaz F., Vakil E., Molina S., Ost D. Sensitivity of initial thoracentesis for malignant pleural effusion stratified by tumor type in patients with strong evidence of metastatic disease. Respiration; International Review of Thoracic Diseases. 2018;96:363–369. doi: 10.1159/000490732. [DOI] [PubMed] [Google Scholar]
- 31.Porcel J.M., Pardina M., Bielsa S., González A., Light R.W. Derivation and validation of a CT scan scoring system for discriminating malignant from benign pleural effusions. Chest. 2015;147:513–519. doi: 10.1378/chest.14-0013. [DOI] [PubMed] [Google Scholar]
- 32.Wahla A.S., Uzbeck M., El Sameed Y.A., Zoumot Z. Managing malignant pleural effusion. Clev Clin J Med. 2019;86:95–99. doi: 10.3949/ccjm.86a.17095. [DOI] [PubMed] [Google Scholar]
- 33.Biaoxue R., Min L., Tian F., Wenlong G., Hua L. Elevated Hsp90-beta contributes to differential diagnosis of pleural effusion caused by lung cancer and correlates with malignant biological behavior of lung cancer. Bmc Pulm Med. 2018;18:188. doi: 10.1186/s12890-018-0752-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jing X., Wei F., Li J., Dai L., Wang X., Jia L. Diagnostic value of soluble B7-H4 and carcinoembryonic antigen in distinguishing malignant from benign pleural effusion. Clin. Respir. J. 2018;12:986–990. doi: 10.1111/crj.12615. [DOI] [PubMed] [Google Scholar]
- 35.Liu Q., Yu Y., Wang X., Wang Z., Wang Z. Diagnostic accuracy of interleukin-27 between tuberculous pleural effusion and malignant pleural effusion: a meta-analysis. Respiration; International Review of Thoracic Disseases. 2018;95:469–477. doi: 10.1159/000486963. [DOI] [PubMed] [Google Scholar]
- 36.Pan Y., Bai W., Chen J., Mao Y., Qian X., Xu K. Diagnosing malignant pleural effusion using clinical and analytical parameters. J. Clin. Lab. Anal. 2019;33:e22689. doi: 10.1002/jcla.22689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang F., Hu L., Wang J., Chen J., Chen J., Wang Y. Clinical value of jointly detection serum lactate dehydrogenase/pleural fluid adenosine deaminase and pleural fluid carcinoembryonic antigen in the identification of malignant pleural effusion. J. Clin. Lab. Anal. 2017;31 doi: 10.1002/jcla.22106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu Y., Zhai K., Shi H. Clinical value of tumor markers for determining cause of pleural effusion. Chinese Med J-Peking. 2016;129:253–258. doi: 10.4103/0366-6999.174501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamain-de Ruiter M., Kwee A., Naaktgeboren C.A., de Groot I., Evers I.M., Groenendaal F. External validation of prognostic models to predict risk of gestational diabetes mellitus in one Dutch cohort: prospective multicentre cohort study. BMJ. 2016;354:i4338. doi: 10.1136/bmj.i4338. [DOI] [PubMed] [Google Scholar]
- 40.Porcel J.M., Alemán C., Bielsa S., Sarrapio J., Fernández De Sevilla T., Esquerda A. A decision tree for differentiating tuberculous from malignant pleural effusions. Resp Med. 2008;102:1159–1164. doi: 10.1016/j.rmed.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 41.Ren Z., Hu Y., Xu L. Identifying tuberculous pleural effusion using artificial intelligence machine learning algorithms. Resp Res. 2019;20:220. doi: 10.1186/s12931-019-1197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mummadi S.R., Al-Zubaidi A., Hahn P.Y. Overfitting and use of mismatched cohorts in deep learning models: preventable design limitations. Am J Resp Crit Care. 2018;198:544–545. doi: 10.1164/rccm.201802-0350LE. [DOI] [PubMed] [Google Scholar]
- 43.Carin L., Pencina M.J. On deep learning for medical image analysis. JAMA. 2018;320:1192–1193. doi: 10.1001/jama.2018.13316. [DOI] [PubMed] [Google Scholar]
- 44.Kermany D.S., Goldbaum M., Cai W., Valentim C.C.S., Liang H., Baxter S.L. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122–1131.e9. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material