Abstract
This study aimed to identify the biological processes associated with long-term survival in high-grade serous ovarian cancer (HGSOC). HGSOC cases obtained from The Cancer Genome Atlas Ovarian Cancer (TCGA-OV) database were divided into long-term survivors (LTS) and normal-term survivors (NTS) based on survival cutoffs defined by the HGSOC cohort in the SEER database. Differentially expressed genes (DEGs) were screened using the generalized linear modeling (GLM) method. Gene Ontology (GO) functional and KEGG pathway enrichment analyses were performed using DAVID Bioinformatics Resources. DEG-related protein-protein interactions (PPI) were extracted from the STRING database and hub genes were identified using CytoHubba in the Cytoscape program. In total, 157 DEGs, including 155 upregulated and 2 downregulated genes, were identified. Upregulated genes were statistically enriched in 80 GO terms and 11 KEGG pathways related to energy and substrate metabolism, such as protein absorption, digestion, and metabolism as well as signaling pathways, including chromatin silencing, regulation of ERK1 and ERK2 cascade, and regulation of MAPKKK. ALB and POMC were the common hub genes. These findings reveal that protein anabolism is crucial to long-term survival, regulated by activation of the MAPK/ERK signaling pathway and chromatin silencing. Comprehensive understanding of the molecular mechanisms via further exploration may contribute toward an effective treatment for ovarian cancer.
Keywords: Ovarian cancer, Long-term survivor, Protein anabolism, Gene expression profiling
Highlights
-
•
Introduction of an independent grouping method based exclusively on survival time
-
•
Gene expression alternation exists in long-term survivors of ovarian cancer.
-
•
Activation of multiple nodes of protein anabolism is the key.
-
•
Activation of MAPK/ERK signal pathway and chromatin silence play a regulatory role.
-
•
Up-regulated ALB and POMC are common hub genes.
Introduction
High-grade serous ovarian cancer (HGSOC) is one of the most common types of gynecologic malignancy in the United States with 22,530 estimated new cases and 13,980 estimated deaths in 2019 [1]. Although majority of these patients are generally considered to be incurable with median survival duration of less than five years, approximately 15% of them survive for more than ten years and are seemingly cured after standard initial therapy [[2], [3], [4]]. Clinicopathologic factors associated with long-term survival in HGSOC include younger age at diagnosis, earlier stage, lower grade, absence of ascites, primary debulking surgery, normal CA-125 level prior to chemotherapy, and microscopic disease after cytoreductive surgery [2,4,5]. Since even patients with negative prognostic factors including advanced stage, platinum resistance, and recurrence can become long-term survivors (LTS), clinicopathologic criteria cannot be used on an individual basis to reliably predict who will be an LTS [1,4,6].
The advent of affordable sequencing technologies has facilitated gene expression analyses to elucidate relationships between LTS of ovarian cancer and potential causative genetic alterations. Spentzos et al. [7,8] identified two genetic signatures of ovarian cancer, one associated with prognosis and the other with chemotherapy response. Yoshihara et al. [9] reported a prognostic gene expression profile related to immune response, focusing on the antigen presentation pathway. The Cancer Genome Atlas (TCGA) uses transcriptional patterns to divide ovarian cancer into 4 subtypes that are unrelated with respect to prognosis [10]. Interestingly, there was are only a few overlapping genes among the reported signatures, suggesting that the signatures characterized distinct features of the tumors. Because of differences in research methods and clinical baselines between studies, it is difficult to obtain a consistent gene expression profile of LTS.
Accurate and consistent selection of LTS and controls is critical in identifying conserved genetic features associated with long-term survival [11]. However, limited by the small sample size and survival overlap issue, many patients probably get assigned to the wrong group based solely on survival time; therefore, any future predictions based on this model are rendered unreliable [12]. In order to address the overlap issue, a population-based approach was used to define the survival cutoffs based on the HGSOC cohort in the Surveillance, Epidemiology, and End Results (SEER) database, which is completely independent of the gene expression cohort.
In this study, we describe an in-depth analysis of transcriptomic alternations between LTS and normal-term survivors (NTS) with HGSOC to identify key genes and biological pathways. First, we divided the cases in The Cancer Genome Atlas Ovarian Cancer (TCGA-OV) hg38 dataset into the LTS and NTS groups exclusively based on overall survival time and screened the differentially expressed genes (DEGs). Next, we performed Gene Ontology (GO) functional analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis for all DEGs. Finally, we constructed a DEG-related protein-protein interaction (PPI) network and identified hub genes and functional modules. Cumulatively, these findings could help us in furthering our understanding of the molecular mechanisms related to long-term survival and ideally applying them to prolong the life of NTS.
Materials and methods
Definition of LTS and NTS
The gene expression profile matrix and clinical information of samples were downloaded from the National Cancer Institute Genomic Data Commons website (GDC, https://portal.gdc.cancer.gov/). TCGA-OV dataset provided both the ‘legacy’ GRCh37 (hg19) data and its GRCh38 (hg38) version as ‘harmonized’. The relative abundance results of these two versions were very highly concordant while restricting to a single workflow; however, the bias in absolute counts prevented direct comparison of abundance between them [13]. Therefore, only cases in the TCGA-OV hg38 dataset were included.
This study included cases diagnosed as per the International Classification of Disease for Oncology, 3rd Edition (ICD—O—3) histology code 8441 and excluded those with unknown variables in the “year of diagnosis”, “age at diagnosis”, and “vital status”, as well as gene expression information. Using these criteria, 374 cases were retrieved from the TCGA-OV hg38 dataset. These cases were diagnosed between 1992 and 2013 and grouped according to age, at five-year intervals. For deceased patients, survival in days was converted to survival in years by dividing by 365. For surviving patients, survival was defined as the time period till the last follow-up and converted to years.
In order to define reliable and accurate cutoffs for overall survival time, the present study referred to the survival landscape derived from the ovarian cancer cohort in the SEER database (www.seer.cancer.gov SEER*Stat Database: Incidence - SEER 18 Regs Research Data + Hurricane Katrina Impacted Louisiana Cases, Nov 2018 Sub 1975–2016 varying). Surveillance Research Program, National Cancer Institute SEER*Stat software (seer.cancer.gov/seerstat) version 8.3.6 was used to screen cases that met the following criteria: (1) Year of diagnosis = “1992 to 2013”; (2) Site recode ICD-O-3/WHO 2008 = “ovary”; (3) Behavior recode for analysis = “malignant”; (4) Histologic type ICD-O-3 = “8441”, and (5) Vital status recode (study cutoff used) = “dead”. After excluding 10 patients with unknown survival months, 9413 cases were included and their survival information was used to draw a survival landscape, which comprised boxplots in five-year groups.
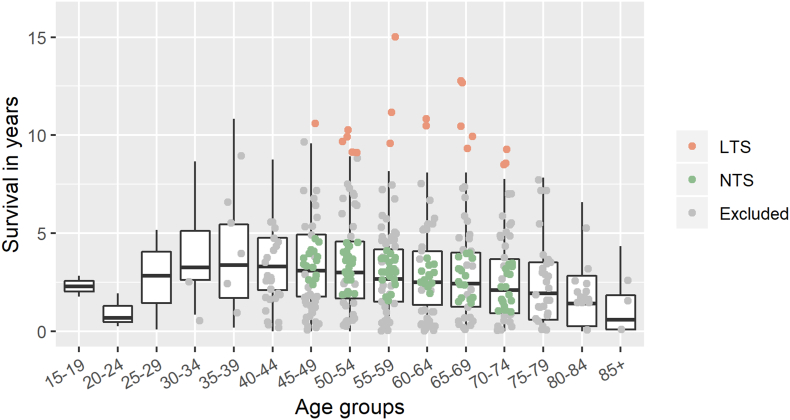
As shown in Fig. 1, the survival scatter plot of the TCGA-OV cohort was added to the survival landscape. LTS were defined as cases both alive and dead in outlier areas, while NTS were defined as only dead cases within the boxes. The cutoffs of LTS and NTS in each five-year group are shown in Table S1. Ultimately, this study included 19 LTS and 105 NTS, and their baselines were compared with no significant differences in age, histological diagnosis, FIGO stage, chemotherapy, and radiotherapy (Table 1).
Fig. 1.
Survival landscape of HGSOC and selection of long-term survivors (LTS) and normal-term survivors (NTS) from TCGA cohort. The boxplots were drawn according to the survival time of deceased HGSOC cases in SEER database. Red points represent the LTS, including both alive and deceased cases. They are distributed in the outlier region of their respective age groups. The green points represent the NTS. They are deceased cases within the box. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
clinical and demographic information of study cohort.
| Characteristics | LTS | NTS | p value |
|---|---|---|---|
| Number | 19 | 105 | |
| Age (mean ± sd), years | 60.5 ± 8.3 | 59.1 ± 8.5 | 0.480 |
| Figo stage | |||
| Stage IC | – | 1 | 0.772 |
| Stage IIB | – | 1 | |
| Stage IIC | – | 2 | |
| Stage IIIA | – | 3 | |
| Stage IIIB | – | 2 | |
| Stage IIIC | 16 | 77 | |
| Stage IV | 3 | 19 | |
| Chemotherapy | |||
| Yes | 18 | 101 | 0.571 |
| No | – | – | |
| NA | 1 | 4 | |
| Radiotherapy | |||
| Yes | 1 | 9 | 0.656 |
| No | 17 | 94 | |
| NA | 1 | 2 | |
| Vital status | |||
| Alive | 14 | – | <0.001 |
| Dead | 5 | 105 | |
| Survival (mean ± sd), years | 10.4 ± 1.6 | 2.9 ± 0.9 | <0.001 |
| Race | |||
| White | 19 | 91 | 0.174 |
| Black or african american | – | 10 | |
| Native hawaiian or other pacific islander | – | 1 | |
| Asian | – | 3 | |
| Ethnicity | |||
| Not hispanic or latino | 16 | 61 | 0.070 |
| Hispanic or latino | 0 | 1 | |
| Not reported | 3 | 43 |
Gene expression data download and processing
The harmonized gene expression profile matrix was downloaded from the National Cancer Institute Genomic Data Commons website (GDC, https://portal.gdc.cancer.gov/) via the “TCGAbiolinks” package 3.10 in R program [14]. According to the suggested workflow, an SE object included information for both genes and samples with gene expression tables of HTSeq-based counts from reads harmonized and aligned to the hg38 genome assembly. Next, an array-array intensity correlation (AAIC) was applied to pinpoint samples with low correlation (threshold of 0.6). Finally, the gene counts were normalized for GC content and quantile filtering was applied with a cutoff of 0.25.
DEG screening
We used the TCGAbiolinks package called the edgeR pipeline [15] implementing a generalized linear modeling (GLM) procedure based on the log-rank score method to screen the DEGs between LTS and NTS samples. Genes with an adjusted P value of <0.001 and |log fold change (FC)| > 1 were set as cutoff criteria for DEGs.
GO functional and KEGG pathway enrichment analyses of DEGs
The DAVID 6.8 database (https://david.ncifcrf.gov/) [16,17] is a commonly used database for gene enrichment and functional annotation analyses. This database integrates biological data and analytical tools to provide systematic and comprehensive annotation of biological functions for large-scale lists of genes or proteins with respect to three aspects: molecular function (molecular-level activities performed by gene products), cellular component (the locations relative to cellular structures in which a gene product performs a function), and biological process (the larger processes, or ‘biological programs’ accomplished by multiple molecular activities). GO annotation and KEGG pathway enrichment analyses of the identified DEGs were performed using DAVID, and the TXT files of their results were downloaded for subsequent analysis. The results were considered statistically significant if P < 0.05. A visual network analysis of the GO analysis results was performed using QuickGO, a web-based tool for GO searching (https://www.ebi.ac.uk/QuickGO/) [18].
PPI network construction and hub gene identification
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) is a search tool that can analyze the interaction relationships between proteins (https://string-db.org/) [19]. STRING was used to analyze the PPI network of DEGs to help us understand the relationships between different genes. The minimum required interaction score was set as the median confidence (0.400). The TVS file of the PPI results was downloaded for subsequent analysis.
The PPI result was imported into the Cytoscape 3.7.2 software [20]. Hub genes were screened from the PPI network using the cytoHubba plugin [21]. In this study, we explored hub genes in DEG-related PPI networks using four topological algorithms: degree, bottleneck (BN), closeness, and betweenness. The top ten ranked genes were selected as candidate hub genes in each method. Venn diagram analysis was used to identify common genes.
Authorization was not requested from a local ethics committee, as all data were available on open access from the GEO and SEER databases.
Results
Identification of DEGs
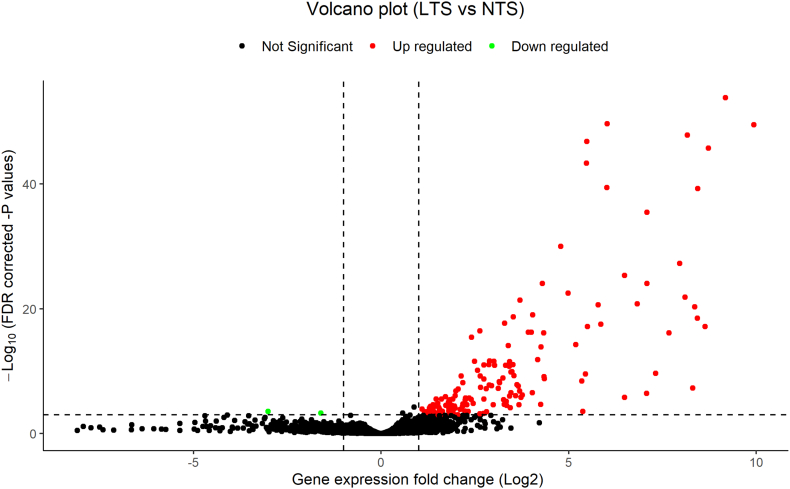
A total of 157 genes were differentially expressed with |log2FC| > 1 and FDR < 0.001, of which 155 genes were upregulated and only two were downregulated (Fig. 2). These genes identified in the tumors of LTS and NTS were significantly different (Table S2).
Fig. 2.
The differentially expressed genes (DEGs) were identified between long-term survivors (LTS) and normal-term survivors (NTS) with HGSOC. The black points were not DEGs with |log2FC| ≤ 1 or FDR ≥ 0.001. The red points were up-regulated genes (log2FC > 1 and FDR < 0.001), while the green points were down-regulated genes (log2FC < −1 and FDR < 0.001). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
GO functional and KEGG pathway enrichment analyses of DEGs
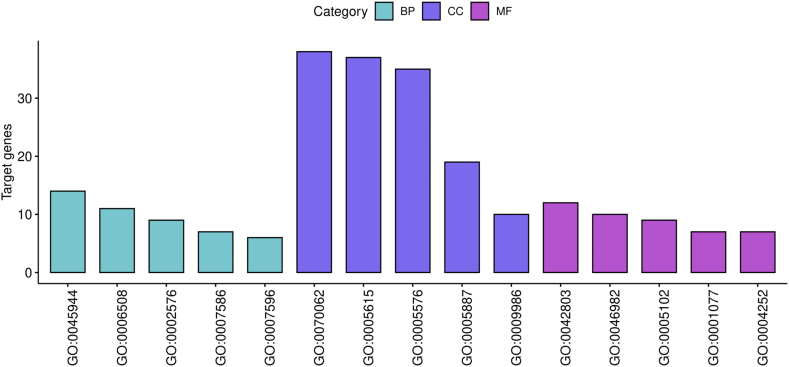
GO function annotation of the integrated DEGs was performed using the DAVID database and its online analysis tool. The GO functional analysis of the integrated differential genes was divided into the following three parts: biological process (BP), molecular function (MF), and cell component (CC). The results were considered statistically significant if P < 0.05 and yielded a total of 80 results from the GO enrichment analysis of the upregulated DEGs, as shown in Table S3. The top 15 results obtained from the GO enrichment analysis of the upregulated DEGs are shown in Fig. 3. The enrichment analysis of these genes identified the extracellular region and cell surface as the CC. Their MF comprised protein homodimerization activity, protein heterodimerization activity, receptor binding, serine-type endopeptidase activity, and transcriptional activator activity. Their BPs were positive regulation of transcription from RNA polymerase II promoter, proteolysis, platelet degranulation, digestion, and blood coagulation.
Fig. 3.
GO Biological Function Enrichment for up-regulated DEGs. Contain: Molecular Function Group (MF), Biological Process Group (BP), and Cellular Component Group (CC).
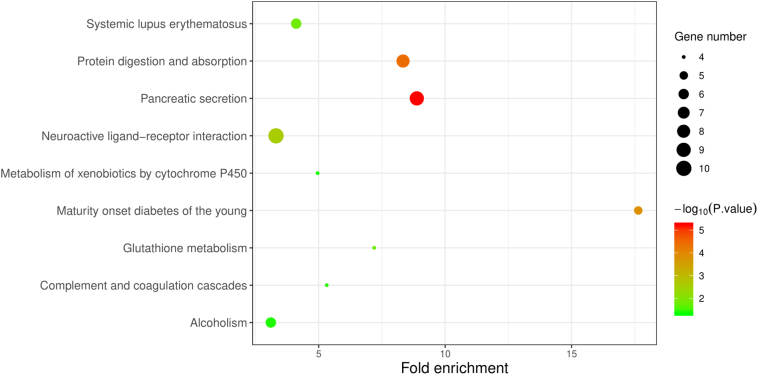
A KEGG pathway analysis of the integrated DEGs was performed using the DAVID database. The DAVID database and the results of the analysis are shown in Table S3 and Fig. 4, respectively. The upregulated DEGs were mainly enriched in two KEGG pathways, namely pancreatic secretion and protein digestion and absorption.
Fig. 4.
KEGG pathway enrichment analysis of up-regulated DEGs.
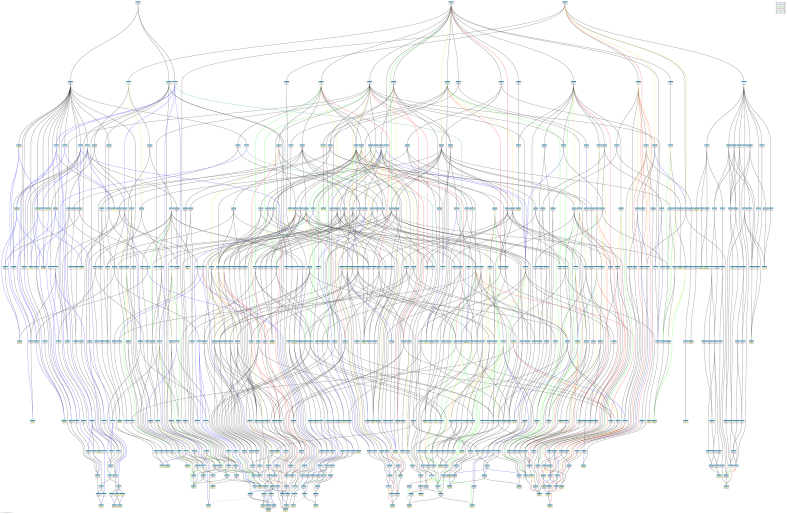
To understand the panorama of biological processes related to long-term survival, an ancestor chart for all 80 GO function terms was generated using the QuickGO online tool (Fig. S1). Furthermore, in order to make the biology clearer and more interpretable, functional annotation clustering was performed using the DAVID online tool. The classification stringency was set to ‘low’ as the default, and the result set included 10 clusters. The top 5 results obtained from the functional annotation clustering of the upregulated DEGs are shown in Table S3. The upregulated five genes positively regulate protein digestion and absorption, coagulation and fibrinolysis, carbohydrate metabolic process, regulation of endopeptidase activity, and chromatin silencing, respectively.
Fig. S1.
An ancestor chart for all 80 GO function terms
PPI network and hub gene identification
The STRING online database was used to analyze all 157 DEGs and construct a PPI network. The PPI network included 143 nodes and 262 edges with an average node degree of 3.66. The PPI enrichment p-value was less than 1.0 × 10−16. The results were downloaded and analyzed via the cytoHubba plug in Cytoscape 3.7.2 software. Based on their scores calculated using four topological algorithms, the genes were ranked and the top 10 genes were filtered as hub genes (Table 2). A Venn diagram was constructed to identify common hub genes. ALB and POMC were common according to four algorithms, and SERPINF2, AFP, SOX2, and HNF4A were common according to three algorithms.
Table 2.
Hub genes for DEGs, highly expressed genes ranked in cytoHubba.
| Catalog | Rank methods in cytoHubba |
|||
|---|---|---|---|---|
| Degree | BottleNeck | Closeness | Betweenness | |
| Gene symbol top 10 | ALB | ALB | ALB | ALB |
| FGA | SOX2 | AFP | SOX2 | |
| CPA1 | SERPINF2 | FGA | GAD1 | |
| AFP | AFP | SERPINF2 | POMC | |
| FGG | GAD1 | POMC | PRSS1 | |
| POMC | PLA2G1B | FGG | REN | |
| SERPINF2 | HIST2H2AA | HNF4A | HIST2H2AA | |
| APOH | POMC | SOX2 | HIST2H2AA3 | |
| ITIH2 | HNF4A | PRSS1 | HNF4A | |
| AMBP | PRSS3P2 | AMBP | ONECUT1 | |
Discussion
In this study, we developed a population-based approach to divide HGSOC cases in the TCGA-OV dataset into LTS and NTS groups based exclusively on overall survival time. The gene expression profile between the two groups should be almost identical because of the exact same histopathology and clinical baseline, except a small difference attributable to the difference in survival times. In the present study, we identified 157 differentially expressed genes between LTS and NTS, consisting of 155 upregulated genes and 2 downregulated genes. These upregulated genes are comprehensively enriched in multiple metabolic processes and homeostasis as well as regulatory pathways, including chromosome silencing, MAPK/ERK signaling pathway, fibrinolytic system, and regulation of endopeptidase activity. Additionally, two common hub genes ALB and POMC were identified. Interpretation of the GO annotation panorama shows that protein anabolism is the key to long-term survival of patients with HGSOC, against cancer-related malnutrition and cachexia.
Serum albumin has been shown to be an accurate predictor of malnutrition and subsequent survival in ovarian cancer [22,23]. Serum albumin is exclusively synthesized in the liver and its synthesis is modulated by dietary factors such as amino acid and protein intake [24]. One study discovered a direct correlation between serum albumin levels and survival, wherein people with lower levels showed poor survival. Low preoperative serum albumin is also associated with poor survival in patients undergoing optimal debulking [25] and cytoreductive surgery [26]. Although tumor-expressed albumin and normal serum albumin have different functions, it has been proven that tumor cells can produce albumin. In the present study, ALB, a common hub gene, was upregulated in LTS. Given that most ovarian cancer cases in this study were FIGO stage 3 and 4 with relatively high tumor burden, it is reasonable to expect that the upregulated ALB expression in tumors has a positive effect on serum albumin levels. Further studies are needed to reveal the correlation between tumor ALB expression and serum albumin levels in ovarian cancer [27].
The present study found that activation of the MAPK/ERK signaling pathway in LTS functions as a tumor suppressor by promoting protein anabolism. Recent evidence indicates that the MAPK/ERK signaling node demonstrates both oncogenic and tumor suppressor effects depending on the tissue-specific tumor microenvironment [28]. In a gynecologic oncology group study, a phase II trial of the MEK inhibitor selumetinib did not support the preponderant role of the MAPK/ERK pathway as a targeting oncogenic driver in low-grade serous ovarian tumors [29]. Another study regarding sorafenib did not demonstrate the biochemical activity of reduced ERK activation in high serous histology recurrent ovarian cancer patients, indicating the lack of genomic events in the MAPK/ERK pathway [30]. These results suggest that ovarian cancer patients cannot benefit from MAPK/ERK inhibition. On the contrary, the results of this study demonstrate that activation of the MAPK/ERK pathway is associated with long overall survival. The MAPK/ERK pathway, a convergent signaling node receiving inputs from numerous stimuli, promotes proliferation and invasion in cancer [31]. During the growth stage of cell proliferation, cells synthesize new DNA and proteins required for cell division. In the present study, the activation of anabolism, which promotes protein synthesis, is associated with long-term survival, but new DNA synthesis and invasion are antagonized by other biological processes.
The present study found that activation of new DNA synthesis in the MAPK/ERK pathway is antagonized by chromatin silencing. Chromatin silencing is the repression of transcription by altering the structure of chromatin to an inaccessible state [32]. In the present study, upregulation of HIST1H2AB, HIST1H2AH, HIST2H2AA3, and HIST2H2AA4 is associated with long-term survival. Histone 2A monoubiquitination is more often associated with gene silencing [33]. Some other mechanisms may also be involved in the regulation of chromosome silencing, but remain unclear.
The present study also found that activation of cell invasion, associated with tumor metastasis, was inhibited by functions of the only two downregulated genes, HMCN2 and ARHGAP6. HMCN2 encodes an evolutionarily conserved protein that belongs to the fibulin family of extracellular matrix proteins, which regulate tissue adhesion and cell migration [34]. A human cell model for epithelial breast cell invasion revealed HMCN2 as one of the most strongly upregulated genes in invasive cells [35]. A study on ovarian cancer demonstrated that ARHGAP10, a member of the RhoGAP family of proteins, was downregulated in ovarian cancer and suppressed tumorigenicity of ovarian cancer cells by inhibiting cell adhesion, migration, and invasion [36]. ARHGAP6, another member of the RhoGAP family of proteins, has a function similar to that of ARHGAP10 as a GTPase activator for the Rho-type GTPases by converting them to an inactive GDP-bound state. However, it demonstrated the opposite effect in the present study. This result is caused by the other function of ARHGAP6 as a cytoskeletal protein that promotes actin remodeling [37]. Therefore, it is possible that downregulation of ARHGAP6 can reduce cell invasion more than its upregulation in ovarian cancer.
The present study found that upregulated genes SERPINF2, APOH, FGG, and FGA were enriched in the fibrinolytic system, namely fibrinolysis, and negatively regulated fibrinolysis. Fibrinogen and fibrin are essential for hemostasis and are major factors in thrombosis, wound healing, and several other biological functions and pathological conditions. Upon cleavage of fibrinopeptides by thrombin, fibrinogen is converted to fibrin monomers [38]. Fibrin promotes cell migration by providing a matrix for tumor cell migration and interacting with adhesive molecules and integrins [39]. Plasminogen activator inhibitor 1 (PAI-1), a component of the fibrinolytic system, inhibits apoptosis and increases tumor cell survival, representing a strong biomarker for tumor aggressiveness and poor prognosis. A mouse model of cerebral infarction demonstrated that aspirin was beneficial in thrombolysis by decreasing PAI-1 expression [40]. Therefore, use of aspirin may improve ovarian cancer prognosis by downregulating PAI-1 expression.
The present study found that upregulated POMC, encoding proopiomelanocortin peptide, is one of the common hub genes associated with long-term survival. POMC, a precursor of ACTH and β-LPH, is responsible for central melanocortin signaling in the control of food intake and energy homeostasis [41,42]. Excess cortisol secretion, caused by pituitary tumors that predominantly secrete ACTH precursors or by nonpituitary or ectopic tumors, is defined as Cushing's syndrome [41]. However, the expression of POMC is relatively low in peripheral tissues including normal and ovarian tumor tissues [43,44], and it cannot produce active POMC-derived peptides. Therefore, upregulated POMC expression in LTS with HGSOC may not contribute to appetite regulation. In vivo investigations in breast cancer demonstrated that POMC interference regulated tumor proliferation via modulating protein phosphorylation mediated by G-protein-coupled estrogen receptors [45]. Accordingly, POMC-expressing HGSOCs may enhance the biological processes related to proliferation.
Conclusion
LTS have distinct biology and molecular features and differ from NTS despite having the same histopathology type (HGSOC) and similar clinical baselines. The present study used bioinformatics methods and revealed that protein anabolism is the core biological process associated with long-term overall survival in HGSOC, that is comprehensively regulated by the MAPK/ERK pathway, chromatin silencing, and the fibrinolytic system. These results suggest that the long-term overall survival of patients with HGSOC might be related to these hub genes and biological pathways, and a more comprehensive interpretation of the molecular mechanisms by further exploration might contribute to the development of an effective treatment for ovarian cancer.
The following are the supplementary data related to this article.
Cut-offs of long-term survivors and normal term survivors defined by SEER cohort.
One hundred and fifty-seven genes composing the long-term survival related profile.
Enriched GO and KEGG categories among the integrated differentially expressed genes.
CRediT authorship contribution statement
Lingxiang Wang &Tao Sun: Conceptualization, Methodology, Software, Formal analysis, Write original draft. Shumei Li: Data curation, and Resources. Zhengmao Zhang: Validation, Investigation, and Visualization. Jingde Jia: Validation, Formal analysis, and Visualization. Baoen Shan: Conceptualization, Supervision, Project administration, and Writing - Review & Editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin L.A., Huang B., Miller R.W., Tucker T., Goodrich S.T., Podzielinski I. Ten-year relative survival for epithelial ovarian cancer. Obstet. Gynecol. 2012;120:612–618. doi: 10.1097/AOG.0b013e318264f794. [DOI] [PubMed] [Google Scholar]
- 3.Gockley A., Melamed A., Bregar A.J., Clemmer J.T., Birrer M., Schorge J.O. Outcomes of women with high-grade and low-grade advanced-stage serous epithelial ovarian cancer. Obstet. Gynecol. 2017;129:439–447. doi: 10.1097/AOG.0000000000001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cress R.D., Chen Y.S., Morris C.R., Petersen M., Leiserowitz G.S. Characteristics of long-term survivors of epithelial ovarian cancer. Obstet. Gynecol. 2015;126:491–497. doi: 10.1097/AOG.0000000000000981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaern J., Aghmesheh M., Nesland J.M., Danielsen H.E., Sandstad B., Friedlander M. Prognostic factors in ovarian carcinoma stage III patients. Can biomarkers improve the prediction of short- and long-term survivors? Int. J. Gynecol. Cancer. 2005;15:1014–1022. doi: 10.1111/j.1525-1438.2005.00185.x. [DOI] [PubMed] [Google Scholar]
- 6.Petrillo M., Fagotti A., Ferrandina G., Fanfani F., Costantini B., Vizzielli G. Ovarian cancer patients with localized relapse: clinical outcome and prognostic factors. Gynecol. Oncol. 2013;131:36–41. doi: 10.1016/j.ygyno.2013.06.020. [DOI] [PubMed] [Google Scholar]
- 7.Spentzos D., Levine D.A., Ramoni M.F., Joseph M., Gu X., Boyd J. Gene expression signature with independent prognostic significance in epithelial ovarian cancer. J. Clin. Oncol. 2004;22:4700–4710. doi: 10.1200/JCO.2004.04.070. [DOI] [PubMed] [Google Scholar]
- 8.Spentzos D., Levine D.A., Kolia S., Otu H., Boyd J., Libermann T.A. Unique gene expression profile based on pathologic response in epithelial ovarian cancer. J. Clin. Oncol. 2005;23:7911–7918. doi: 10.1200/JCO.2005.02.9363. [DOI] [PubMed] [Google Scholar]
- 9.Yoshihara K., Tsunoda T., Shigemizu D., Fujiwara H., Hatae M., Fujiwara H. High-risk ovarian cancer based on 126-gene expression signature is uniquely characterized by downregulation of antigen presentation pathway. Clin. Cancer Res. 2012;18:1374–1385. doi: 10.1158/1078-0432.CCR-11-2725. [DOI] [PubMed] [Google Scholar]
- 10.N. Cancer Genome Atlas Research, Integrated genomic analyses of ovarian carcinoma, Nature, 474 (2011) 609–615. doi: 10.1038/nature10166 [DOI] [PMC free article] [PubMed]
- 11.Hoppenot C., Eckert M.A., Tienda S.M., Lengyel E. Who are the long-term survivors of high grade serous ovarian cancer? Gynecol. Oncol. 2018;148:204–212. doi: 10.1016/j.ygyno.2017.10.032. [DOI] [PubMed] [Google Scholar]
- 12.Bair E., Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao G.F., Parker J.S., Reynolds S.M., Silva T.C., Wang L.B., Zhou W. Before and after: comparison of legacy and harmonized TCGA genomic data commons' data. Cell Syst. 2019;9:24–34. doi: 10.1016/j.cels.2019.06.006. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D. TCGAbiolinks: An R/bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.W. da Huang, B.T. Sherman, R.A. Lempicki, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists, Nucleic Acids Res, 37 (2009) 1–13. doi: 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed]
- 17.W. da Huang, B.T. Sherman, R.A. Lempicki, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nat Protoc, 4 (2009) 44–57. doi: 10.1038/nprot.2008.211 [DOI] [PubMed]
- 18.Binns D., Dimmer E., Huntley R., Barrell D., O'Donovan C., Apweiler R. QuickGO: a web-based tool for gene ontology searching. Bioinformatics. 2009;25:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.C.H. Chin, S.H. Chen, H.H. Wu, C.W. Ho, M.T. Ko, C.Y. Lin, cytoHubba: identifying hub objects and sub-networks from complex interactome, BMC Syst Biol, 8 Suppl 4 (2014) S11. doi: 10.1186/1752-0509-8-S4-S11 [DOI] [PMC free article] [PubMed]
- 22.Asher V., Lee J., Bali A. Preoperative serum albumin is an independent prognostic predictor of survival in ovarian cancer. Med. Oncol. 2012;29:2005–2009. doi: 10.1007/s12032-011-0019-5. [DOI] [PubMed] [Google Scholar]
- 23.Schwartz G.G., Tretli S., Vos L., Robsahm T.E. Prediagnostic serum calcium and albumin and ovarian cancer: A nested case-control study in the Norwegian Janus Serum Bank Cohort. Cancer Epidemiol. 2017;49:225–230. doi: 10.1016/j.canep.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Wada Y., Takeda Y., Kuwahata M. Potential role of amino acid/protein nutrition and exercise in serum albumin redox state. Nutrients. 2017;10:17. doi: 10.3390/nu10010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayhan A., Gunakan E., Alyazici I., Haberal N., Altundag O., Dursun P. The preoperative albumin level is an independent prognostic factor for optimally debulked epithelial ovarian cancer. Arch. Gynecol. Obstet. 2017;296:989–995. doi: 10.1007/s00404-017-4511-9. [DOI] [PubMed] [Google Scholar]
- 26.Ataseven B., du Bois A., Reinthaller A., Traut A., Heitz F., Aust S. Pre-operative serum albumin is associated with post-operative complication rate and overall survival in patients with epithelial ovarian cancer undergoing cytoreductive surgery. Gynecol. Oncol. 2015;138:560–565. doi: 10.1016/j.ygyno.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Graner M.W., Likhacheva A., Davis J., Raymond A., Brandenberger J., Romanoski A. Cargo from tumor-expressed albumin inhibits T-cell activation and responses. Cancer Res. 2004;64:8085–8092. doi: 10.1158/0008-5472.CAN-04-1871. [DOI] [PubMed] [Google Scholar]
- 28.Burotto M., Chiou V.L., Lee J.M., Kohn E.C. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120:3446–3456. doi: 10.1002/cncr.28864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farley J., Brady W.E., Vathipadiekal V., Lankes H.A., Coleman R., Morgan M.A. Selumetinib in women with recurrent low-grade serous carcinoma of the ovary or peritoneum: an open-label, single-arm, phase 2 study. Lancet Oncol. 2013;14:134–140. doi: 10.1016/S1470-2045(12)70572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matei D., Sill M.W., Lankes H.A., DeGeest K., Bristow R.E., Mutch D. Activity of sorafenib in recurrent ovarian cancer and primary peritoneal carcinomatosis: a gynecologic oncology group trial. J. Clin. Oncol. 2011;29:69–75. doi: 10.1200/JCO.2009.26.7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Luca A., Maiello M.R., D'Alessio A., Pergameno M., Normanno N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin. Ther. Targets. 2012;16(Suppl. 2):S17–S27. doi: 10.1517/14728222.2011.639361. [DOI] [PubMed] [Google Scholar]
- 32.Straight A.F., Shou W., Dowd G.J., Turck C.W., Deshaies R.J., Johnson A.D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 33.Schwertman P., Bekker-Jensen S., Mailand N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 2016;17:379–394. doi: 10.1038/nrm.2016.58. [DOI] [PubMed] [Google Scholar]
- 34.Feitosa N.M., Zhang J., Carney T.J., Metzger M., Korzh V., Bloch W. Hemicentin 2 and Fibulin 1 are required for epidermal-dermal junction formation and fin mesenchymal cell migration during zebrafish development. Dev. Biol. 2012;369:235–248. doi: 10.1016/j.ydbio.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moon A., Yong H.Y., Song J.I., Cukovic D., Salagrama S., Kaplan D. Global gene expression profiling unveils S100A8/A9 as candidate markers in H-ras-mediated human breast epithelial cell invasion. Mol. Cancer Res. 2008;6:1544–1553. doi: 10.1158/1541-7786.MCR-08-0189. [DOI] [PubMed] [Google Scholar]
- 36.Luo N., Guo J., Chen L., Yang W., Qu X., Cheng Z. ARHGAP10, downregulated in ovarian cancer, suppresses tumorigenicity of ovarian cancer cells. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2015.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prakash S.K., Paylor R., Jenna S., Lamarche-Vane N., Armstrong D.L., Xu B. Functional analysis of ARHGAP6, a novel GTPase-activating protein for RhoA. Hum. Mol. Genet. 2000;9:477–488. doi: 10.1093/hmg/9.4.477. [DOI] [PubMed] [Google Scholar]
- 38.Weisel J.W., Litvinov R.I. Fibrin formation, structure and properties. Subcell. Biochem. 2017;82:405–456. doi: 10.1007/978-3-319-49674-0_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwaan H.C., Lindholm P.F. Fibrin and fibrinolysis in cancer. Semin. Thromb. Hemost. 2019;45:413–422. doi: 10.1055/s-0039-1688495. [DOI] [PubMed] [Google Scholar]
- 40.Wang X., Shen B., Sun D., Cui X. Aspirin ameliorates cerebral infarction through regulation of TLR4/NFkappaBmediated endoplasmic reticulum stress in mouse model. Mol. Med. Rep. 2018;17:479–487. doi: 10.3892/mmr.2017.7879. [DOI] [PubMed] [Google Scholar]
- 41.Harno E., Gali Ramamoorthy T., Coll A.P., White A. POMC: the physiological power of hormone processing. Physiol. Rev. 2018;98:2381–2430. doi: 10.1152/physrev.00024.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chretien M., Mbikay M. 60 YEARS OF POMC: From the prohormone theory to pro-opiomelanocortin and to proprotein convertases (PCSK1 to PCSK9) J. Mol. Endocrinol. 2016;56:T49–T62. doi: 10.1530/JME-15-0261. [DOI] [PubMed] [Google Scholar]
- 43.Gallinelli A., Garuti G., Matteo M.L., Genazzani A.R., Facchinetti F. Expression of pro-opiomelanocortin gene in human ovarian tissue. Hum. Reprod. 1995;10:1085–1089. doi: 10.1093/oxfordjournals.humrep.a136099. [DOI] [PubMed] [Google Scholar]
- 44.Katchman B.A., Chowell D., Wallstrom G., Vitonis A.F., LaBaer J., Cramer D.W. Autoantibody biomarkers for the detection of serous ovarian cancer. Gynecol. Oncol. 2017;146:129–136. doi: 10.1016/j.ygyno.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin X., Chen W., Wei F., Zhou B.P., Hung M.C., Xie X. POMC maintains tumor-initiating properties of tumor tissue-derived long-term-cultured breast cancer stem cells. Int. J. Cancer. 2017;140:2517–2525. doi: 10.1002/ijc.30658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cut-offs of long-term survivors and normal term survivors defined by SEER cohort.
One hundred and fifty-seven genes composing the long-term survival related profile.
Enriched GO and KEGG categories among the integrated differentially expressed genes.