Highlights
-
•
Clinical samples with ‘neat’ Ct values between 20 and 28 were all detected in pools of six.
-
•
Testing in a pool of six led to a mean drop of cycle threshold value of 2.9.
-
•
Samples with viral load around the assay's limit of detection may be missed by pooling.
-
•
All confirmed negative samples were also negative when tested in pools of 6.
Keywords: Pooling, GeneXpert, Cepheid, SARS-CoV-2
Abstract
The COVID-19 pandemic has placed unprecedented global demand on laboratory supplies required for testing. Sample pooling has been investigated by laboratories as a strategy to preserve testing capacity. We evaluate the performance of Cepheid Xpert® Xpress SARS-CoV-2 RT-PCR assay for testing samples in pools of 4 and 6. Clinical samples containing SARS-CoV-2, and confirmed negative clinical samples were used to create sample pools. Clinical samples had ‘neat’ Xpert® E gene cycle threshold values ranging between 20 and 28 and all were detected qualitatively when contained in pools of 4 or 6 samples. For these samples, pooling had a median change in cycle threshold value of 2.0 in pools of 4, and of 2.9 in pools of 6. With the use of Cepheid Xpert® Xpress SARS-CoV-2 RT-PCR assay, pooling of 4 or 6 samples may be an effective strategy to increase testing capacity.
1. Introduction
Rapid and accurate diagnostic testing for SARS-CoV-2 is central to controlling the global COVID-19 pandemic. The Cepheid Xpert® Xpress SARS-CoV-2 assay (Cepheid, Sunnyvale, CA) is a rapid, near-care, reverse-transcriptase PCR assay (RT-PCR), producing results within 45 minutes. Manufacturer instructions for use claim a limit of detection (LoD) of 250 copies/mL; however, recent work has demonstrated high analytical sensitivity of this assay, with a LoD approaching 100 viral copies/mL (Loeffelholz et al., 2020; Moran et al., 2020; Lieberman et al., 2020; Zhen et al., 2020; Wolters et al., 2020). The sheer scale of global demand for laboratory reagents, including the Xpert® SARS-CoV-2 assay, has led many laboratories to investigate alternative strategies for optimizing the use of testing supplies, including sample pooling (Torres et al., 2020; Williams et al., 2020; Wacharapluesadee et al., 2020; Lohse et al., 2020; Perchetti et al., 2020; Hogan et al., 2020). The diagnostic performance of pooling depends on several factors, including assay sensitivity, prevalence of infection in the population being tested and sample types used for pooling (Wacharapluesadee et al., 2020; Lohse et al., 2020; Perchetti et al., 2020; Hogan et al., 2020). With increased recognition that asymptomatic SARS-CoV-2 infection may contribute to transmission, the populations tested have been expanded to include asymptomatic patients: in this context when prevalence rates are <5% sample pooling can substantially increase testing capacity (Abdalhamid et al., 2020; Cherif et al., 2020; Ben-Ami et al., 2020). On July 18, 2020, the FDA issued its first Emergency Use Authorization for sample pooling in diagnostic testing for SARS-CoV-2 by RT-PCR which applies for the Quest Diagnostics test for use with pooled samples containing up to 4 individual swab specimens (Coronavirus (COVID-19) Update: FDA Issues First Emergency Authorization for Sample Pooling in Diagnostic Testing FDA, 2020). Although pooling has previously been used successfully with the Xpert® MTB/RIF assay, there are limited data on the performance of pooling using the Xpert® SARS-CoV-2 assay (Abdurrahman et al., 2015). Here, we investigated the performance of the Xpert® SARS-CoV-2 assay for detecting SARS-CoV-2 in pooled clinical samples. We chose to study pools of 4 and 6 samples based on the FDA Emergency Use Authorization authorization, available literature for other assays (Wacharapluesadee et al., 2020; Lohse et al., 2020; Perchetti et al., 2020; Hogan et al., 2020; Abdalhamid et al., 2020; Cherif et al., 2020; Ben-Ami et al., 2020) and our own experience with pooling using an in-house RT-PCR for SARS-CoV-2 (Chong et al.).
2. Methods
Nasopharyngeal swab samples were collected in viral transport media (Kang Jian, catalogue no KJ502-19) and initially tested for SARS-CoV-2 at the Microbiological Diagnostic Unit Public Health Laboratory, University of Melbourne using the Hologic Panther Aptima™ SARS-CoV-2 Assay. The panel consisted of 7 clinical samples containing SARS-CoV-2, and 24 confirmed negative samples used to create sample pools. Each positive sample was tested: (1) neat; (2) in a pool with 3 negative samples (pool of 4) and (3) in a pool with 5 negative samples (pool of 6). Further, the 2 samples with highest and 2 samples with lowest cycle threshold (Ct) values were tested in duplicate in the pool of 6 to study precision of the assay performance on pooled samples.
Pools were constructed by transferring 100 µL of each positive and negative sample into a sterile secondary tube and mixing this by inverting rapidly 5 times: 300 µL of this was aliquoted into the Xpert® cartridge and tested within 30 minutes of specimen addition.
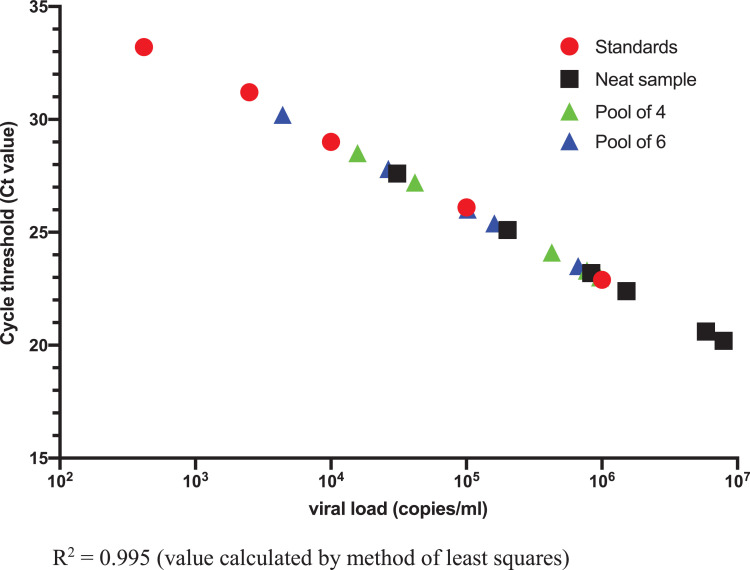
In addition, serial dilutions of gamma-irradiated SARS-CoV-2 virus were prepared in viral transport media to allow correlation of viral concentration with Xpert®Ct values (Table 1 ). These serial dilutions of inactivated virus were used to create standards for a standard curve (Fig. 1 ) from which the neat clinical sample extrapolated viral load was calculated.
Table 1.
Xpert®cycle threshold values of serial dilutions of inactivated high-titer SARS-CoV-2 virus
| Dilution | E gene | N gene | SPC |
|---|---|---|---|
| 1 × 106 copies/mL | 22.9 | 25.3 | 27.5 |
| 1 × 105 copies/mL | 26.1 | 28.4 | 27.5 |
| 1 × 104 copies/mL | 29 | 31.6 | 27.4 |
| 2.5 × 103 copies/mL | 31.2 | 34.8 | 27.7 |
| 4.17 × 102 copies/mL | 33.2 | 35.8 | 27.8 |
| 4.17 × 102 copies/mL in pool of 4 (104 copies/mL)* |
0 | 0 | 28.1 |
| 4.17 × 102 copies/mL in pool of 6 (70 copies/mL)* |
0 | 42.8 | 28 |
E = envelope; N = nucleocapsid; SPC = sample processing control.
These serial dilutions of inactivated virus were used to create standards for standard curve in Fig. 1.
Note: Manufacturer claimed LOD for Xpert® is 250 copies/mL (lowest concentration that can be reproducibly distinguished from a negative sample ≥95% of the time with 95% confidence).
Fig. 1.
Standard curve of Xpert E gene Ct value and viral load.
All Xpert® testing was performed by 1 investigator (E.W.), blinded to previous testing results and pooling details.
3. Results
Clinical samples with ‘neat’ Xpert® E gene Ct values ranging between 20 and 28 were detected qualitatively when contained in pools of 4 or 6 samples (Table 2 ). For these samples, pooling had a median change in E gene Ct (ΔCt) value of 2.0 in pools of 4, and a ΔCt 2.9 in pools of 6. All 24 negative samples were negative for SARS-CoV-2 by Xpert® when tested in 4 pools of 6 samples to confirm a specificity of pool testing of 100%.
Table 2.
Cycle threshold (Ct) results for clinical samples with pools of 4 and 6 samples
| Neat sample extrapolatedviral load* (copies/mL) | Neat sample Ct (Xpert®) |
Four sample pool Ct (Xpert®) |
Six sample pool Ct & replicate result (Xpert®) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| E gene | N gene | SPC | E gene | N gene | SPC | E gene | N gene | SPC | |
| 7.91 × 106 | 20.2 | 23.3 | 26.9 | 23.3 | 25.9 | 27.3 | 23.3 | 25.6 | 27.3 |
| 23.1 | 25.5 | 27.6 | |||||||
| 5.86 × 106 | 20.6 | 23.2 | 27 | 23 | 25.4 | 27.1 | 23.5 | 26.3 | 27.4 |
| 23.1 | 25.8 | 27.2 | |||||||
| 1.52 × 106 | 22.4 | 24.5 | 27.1 | 24.1 | 26.2 | 27.7 | 26 | 28.3 | 27.4 |
| 8.35 × 105 | 23.2 | 25.5 | 27.4 | 25.1 | 26.9 | 27.3 | 25.4 | 28.1 | 27 |
| 4.26 × 105 | 24.1 | 26.5 | 27.2 | 26.1 | 28.8 | 27.2 | 28.3 | 29.8 | 27.8 |
| 2.01 × 105 | 25.1 | 27.7 | 27 | 27.2 | 29.7 | 27.3 | 27.8 | 30.7 | 27.4 |
| 28.1 | 31.3 | 27.3 | |||||||
| 3.09 × 104 | 27.6 | 30.1 | 27.6 | 28.5 | 31.3 | 27.4 | 30.2 | 33.9 | 27.3 |
| 30.6 | 34.5 | 27.4 | |||||||
E = envelope; N = nucleocapsid; SPC = sample processing control.
Based on Xpert® E gene Ct value: viral load for clinical samples was extrapolated from standard curve (Fig. 1).
Serial dilutions of inactivated SARS-CoV-2 virus between 4.17 × 102 copies/mL and 1 × 106 copies/mL were reliably detected by the Xpert® assay. Virus was detected when testing the lowest dilution of 4.17 × 102 copies/mL in a pool of 6 but not when tested in a pool of 4 negative samples.
4. Discussion
We have found that testing samples in pools of 4 or 6 using Xpert® assay can increase testing capacity and that virus from samples with neat Ct values of between 20 and 28 can be reliably detected during pooling. Consistent with other studies, we have found that samples with viral load around the LoD for the assay used may be missed when testing in pools (Ben-Ami et al., 2020). Since the required input into the Xpert® cartridge is fixed at 300 µL, when testing a pool size of 4, 75 µL of each original sample is tested. In comparison, for assays where RNA extraction of the pool can be performed as a separate step greater possible original sample input volumes can be accommodated to increase sensitivity. However, use of more sensitive assays such as the Xpert® to test sample pools is less likely to miss samples with low viral load when using lower sample input volumes. Furthermore, samples with low viral load, particularly in asymptomatic persons, may indicate the presence of noninfectious virus since studies have found that samples with higher Ct values are less likely to yield culturable virus (La Scola et al., 2020).
Strategies to optimize the performance of pooling include limiting its use to low prevalence situations such as testing of asymptomatic populations. By implementing pooling in low prevalence settings, the work of ‘de-coupling’ of pools for individual testing in the event of a positive pool is minimized. For a test with assumed sensitivity and specificity of 99% and a SARS-CoV-2 prevalence of under 5%, the expected number of PCR reactions required for testing of 1000 samples in pools of 4 is under 500, inclusive of initial pool testing and deconstruction of positive pools (Chong et al.). The Xpert® assay is authorized to be used in patient care settings outside of the clinical laboratory environment and it has been widely used in low-resource settings particularly for the diagnosis of tuberculosis (Abdurrahman et al., 2015). Its ease of use, including for pool testing, makes it an ideal assay for use in settings where expertise for SARS-CoV-2 testing may not be readily available. However, in view of high global demand for test kits the supply of cartridges does not always meet demand in some settings, which further supports the use of pooling with this assay.
During our study, we developed laboratory strategies to mitigate against errors during the pooling process. These included: (1) oversight of the pool assembly process by a second staff member; (2) checking of manual transcriptions by a second staff member; (3) holding back reporting of negative pooled samples until individual testing of positive pools is complete; (4) use of standardized worksheets for recording 2 identifiers (specimen number and patient name) of each specimen in the pool, and (5) requirement for operator signatures at each step of the testing process.
Here, we demonstrate that with the use of a sensitive and specific molecular assay that is easy to use, pooling of 4 or 6 samples is an effective strategy to increase testing capacity. With limited testing resources, sample pooling may preserve testing capacity; findings of our study may be particularly valuable for low-resource settings.
Authors' contribution
Maryza Graham: Data curation; Formal analysis; Investigation; Methodology; Supervision; Validation; Visualization; Roles/Writing - original draft; Writing - review & editing.
Eloise Williams: Data curation; Formal analysis; Investigation; Writing - review & editing.
Nicole Isles: Data curation; Formal analysis; Investigation; Methodology; Validation; Writing - review & editing.
Eka Buadromo: Conceptualization; Methodology; Resources; Writing - review & editing.
Tebuka Toatu: Conceptualization; Methodology; Resources; Writing - review & editing.
Julian Druce: Methodology; Supervision; Validation; Visualization; Writing - review & editing.
Mike Catton: Formal analysis; Methodology; Supervision; Validation; Writing - review & editing.
Chantel Lin: Funding acquisition; Project administration; Resources; Writing - review & editing.
Benjamin P. Howden: Conceptualization; Funding acquisition; Supervision; Writing - review & editing.
Deborah A. Williamson: Conceptualization; Formal analysis; Funding acquisition; Methodology; Project administration; Resources; Supervision; Validation; Visualization; Writing - review & editing.
Funding
This project was supported by the Australian Government Department of Foreign Affairs and Trade Indo-Pacific Centre for Health Security.
Declarations of interest
None.
Acknowledgments
We would like to thank the Australian Government Department of Foreign Affairs and Trade Indo-Pacific Centre for Health Security for developing the project concept, providing advice and guidance for the evaluation and providing support for resources to undertake this project.
We thank Professor Patrick Reading (The Peter Doherty Institute) for discussions during the planning stages and assistance with proofreading of the draft manuscript.of the draft manuscript.
References
- Loeffelholz M.J., Alland D., Butler-Wu S.M., Pandey U., Perno C.F., Nava A. Multicenter evaluation of the Cepheid Xpert Xpress SARS-CoV-2 test. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00926-20. e00926-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran A., Beavis K.G., Matushek S.M., Ciaglia C., Francois N., Tesic V. Detection of SARS-CoV-2 by use of the Cepheid Xpert Xpress SARS-CoV-2 and Roche Cobas SARS-CoV-2 assays. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00772-20. e00772-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman J.A., Pepper G., Naccache S.N., Huang M.-.L., Jerome K.R., Greninger A.L. Comparison of commercially available and laboratory-developed assays for in vitro detection of SARS-CoV-2 in clinical laboratories. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00821-20. e00821-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen W., Smith E., Manji R., Schron D., Berry G.J. Clinical evaluation of three sample-to-answer platforms for detection of SARS-CoV-2. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00783-20. e00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters F., van de Bovenkamp J., van den Bosch B., van den Brink S., Broeders M., Chung N.H. Multi-center evaluation of Cepheid Xpert® Xpress SARS-CoV-2 point-of-care test during the SARS-CoV-2 pandemic. J Clin Virol. 2020;128 doi: 10.1016/j.jcv.2020.104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres I., Albert E., Navarro D. Pooling of nasopharyngeal swab specimens for SARS‐CoV‐2 detection by RT‐PCR. J Med Virol. 2020 doi: 10.1002/jmv.25971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020;58(8) doi: 10.1128/JCM.00776-20. e00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S., Kaewpom T., Ampoot W., Ghai S., Khamhang W., Worachotsueptrakun K. Evaluating the efficiency of specimen pooling for PCR-based detection of COVID-19. J Med Virol. 2020 doi: 10.1002/jmv.26005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse S., Pfuhl T., Berkó-Göttel B., Rissland J., Geiβler T., Gärtner B. Pooling of samples for testing for SARS-CoV-2 in asymptomatic people. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchetti G.A., Sullivan K.W., Pepper G., Huang M.L., Breit N., Mathias P. Pooling of SARS-CoV-2 samples to increase molecular testing throughput. J Clin Virol. 2020;131 doi: 10.1016/j.jcv.2020.104570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C.A., Sahoo M.K., Pinsky B.A. Sample pooling as a strategy to detect community transmission of SARS-CoV-2. JAMA. 2020;323:1967. doi: 10.1001/jama.2020.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdalhamid B., Bilder C.R., McCutchen E.L., Hinrichs S.H., Koepsell S.A., Iwen P.C. Assessment of specimen pooling to conserve SARS CoV-2 testing resources. Am J Clin Pathol. 2020;153:715–718. doi: 10.1093/ajcp/aqaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherif A., Grobe N., Wang X., Kotanko P. Simulation of pool testing to identify patients with coronavirus disease 2019 under conditions of limited test availability. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ami R., Klochendler A., Seidel M., Sido T., Gurel-Gurevich O., Yassour M. Large-scale implementation of pooled RNA extraction and RT-PCR for SARS-CoV-2 detection. Clin Microbiol Infect. 2020;26(9):1248–1253. doi: 10.1016/j.cmi.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus (COVID-19) Update: FDA Issues First Emergency Authorization for Sample Pooling in Diagnostic Testing FDA (2020) https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-first-emergency-authorization-sample-pooling-diagnostic
- Abdurrahman S.T., Mbanaso O., Lawson L., Oladimeji O., Blakiston M., Obasanya J. Testing pooled sputum with Xpert MTB/RIF for diagnosis of pulmonary tuberculosis to increase affordability in low-income countries. J Clin Microbiol. 2015;53:2502–2508. doi: 10.1128/JCM.00864-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B., Le Bideau M., Andreani J., Hoang V.T., Grimaldier C., Colson P. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong B.S.W., Tran T., Druce J., Ballard SA, Simpson JA, Catton M, et al. Sample pooling is a viable strategy for SARS-CoV-2 detection in low-prevalence settings. medrxiv.org/content/10.1101/2020.08.26.20181719v1 [DOI] [PMC free article] [PubMed]