Graphical abstract
Keywords: COVID 19 pneumonia, Artifial intelligence, Deep learning, Staging, Prognosis, Biomarker discovery, Ensemble methods
Abstract
Coronavirus disease 2019 (COVID-19) emerged in 2019 and disseminated around the world rapidly. Computed tomography (CT) imaging has been proven to be an important tool for screening, disease quantification and staging. The latter is of extreme importance for organizational anticipation (availability of intensive care unit beds, patient management planning) as well as to accelerate drug development through rapid, reproducible and quantified assessment of treatment response. Even if currently there are no specific guidelines for the staging of the patients, CT together with some clinical and biological biomarkers are used. In this study, we collected a multi-center cohort and we investigated the use of medical imaging and artificial intelligence for disease quantification, staging and outcome prediction. Our approach relies on automatic deep learning-based disease quantification using an ensemble of architectures, and a data-driven consensus for the staging and outcome prediction of the patients fusing imaging biomarkers with clinical and biological attributes. Highly promising results on multiple external/independent evaluation cohorts as well as comparisons with expert human readers demonstrate the potentials of our approach.
1. Introduction
COVID-19 emerged in December 2019 in Wuhan, China Zhu et al., 2020 caused by the SARS-CoV-2 virus, and it could lead to respiratory failure due to severe viral pneumonia Zhou et al., 2020. The disease spread worldwide leading the World Health Organization to declare it as a pandemic in March 2020. One of the important actions to handle the pandemic is the fast and robust use of imaging along with clinical and biological comorbidities for the quantification and staging of patients upon their hospital admission. Being able to identify patients that need intubation upon admission is very important and essential for the management of a hospital’s resources and the most optimal management of patients. Moreover, a robust staging of the patients could also facilitate proper selection of patients for different treatments, reducing the unnecessary use of the hospital’s intensive care units. To the best of our knowledge, currently the staging of the patients is mainly based on clinical and biological biomarkers such as age, sex and other comorbidities Guo et al., 2020, Li et al., 2020, Onder et al., 2020, Tang et al., 2020, Terpos et al., 2020, Yuan et al., 2020, Zhou et al., 2020, while the role of imaging is mainly focusing on an estimation of the disease extent from CT scans. This estimation is mainly done by medical experts and hence suffers from inter- and intra-observer variability.
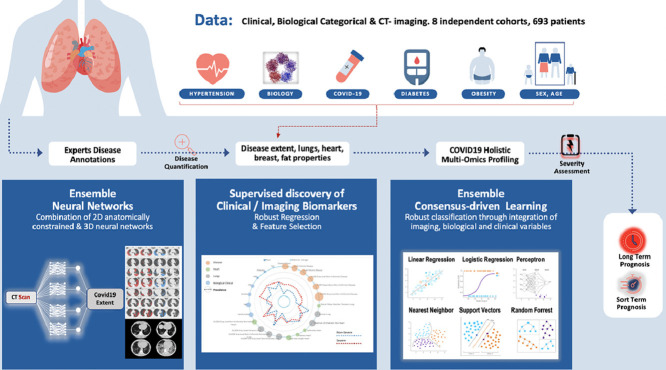
In this study, we investigated an automatic method (Fig. 1 ) for COVID-19 disease quantification and staging that extracts and selects image characteristics directly from the CTs and fuse them with known clinical and biological markers. A variety of image characteristics are proposed providing insights about their use on patient staging and better disease understanding. The contributions of this study are three-folds: (i) a tool for automatic disease quantification based on 2D & 3D deep convolutional neural networks (CNNs) is developed, facilitating severity estimation for optimal patient care, (ii) a COVID19-specific holistic, highly compact multi-omics patient signature integrating imaging, clinical, and biological data and associated comorbidities for automatic patient staging is presented, (iii) short and long-term prognosis for clinical resources optimization offering alternative/complementary means to facilitate triage are reported. To the best of our knowledge this is among a few systematic efforts to quantify disease extent, to discover low dimensional and interpretable imaging biomarkers and to integrate them to clinical variables into short and long term prognosis of COVID-19 patients.
Fig. 1.
Overview of the method for automatic quantification, staging and prognosis of COVID-19. Our study includes 8 independent cohorts, resulting in 693 COVID-19 patients in total. A variety of clinical and biological attributes were collected and combined with imaging biomarkers for short and long term prognosis of COVID-19 patients. Our study is composed by three different steps: (i) Proposing a state-of-the-art deep learning based consensus of 2D & 3D networks for automatic quantification of COVID-19 disease, reaching expert-level annotations, (ii) A radiomics study integrating interpretable features extracted from disease, lung and heart regions. A consensus-driven COVID-19 low dimensional bio(imaging)-holistic profiling and staging signature has been proposed using robust machine learning algorithms, fusing imaging, clinical and biological attributes. & (iii) An ensemble of robust linear & non-linear classification methods for the proper identification of patients that need intubation.
The paper is organized as follows: we first review related work mainly focusing on interstitial lung diseases (ILDs) diseases, which is followed by a description of all the components and implementation details of our method. We then present the acquired multi-center dataset, the evaluation setting, and the results of our experiments. Furthermore, we discuss in detail similarities and differences of our method with other recently proposed methods for quantification and staging of COVID-19. Lastly, we present possible directions for future research.
2. Related work
In this section, we provide a short review of previous studies on quantification of ILDs since COVID-19 and ILDs share a lot of similarities due to their diffuse pathological manifestations, such as ground glass opacities, band consolidations, and reticulations. Furthermore, we elaborate on studies that tackle severity or treatment response for such types of disease.
2.1. ILD quantification
There are numerous studies proposed the last years on automatic quantification of ILD diseases using CT scans. The main goal of these studies is to develop models that are able to identify one or more types of different pathological lung tissue in ILD cases (such as ground glass, consolidation, honey-combing, etc.) and successfully separate them from the healthy tissue. Initial efforts were mainly based on classification schemes. In particular, small patches including only a single tissue type were extracted and described using a number of handcrafted features focusing mainly on texture, then these features were used to train different machine learning classifiers Gangeh et al., 2010, Huber et al., 2012. Following recent advances in deep learning and especially the success of convolutional neural networks (CNNs), researchers have recently employed such tools also in thoracic imaging tasks Chassagnon et al., 2020, with ILD quantification being among them. The main advantage of CNNs is their ability to generate features automatically from the input, and create meaningful representations for the studied per time problems. In particular, a patch-based framework using a convolutional architecture is presented in Anthimopoulos et al. Anthimopoulos et al., 2016 for the automatic quantification of 5 different ILD patterns. Similarly, in Gao et al., 2018 a patch-based approach is adapted to classify them in 6 different ILD patterns. Even if the method reported higher performance than other methods based on handcrafted features, the use of patches, besides being time consuming and inefficient, does not exploit the texture of the entire lung.
Many of the already proposed CNNs have further been adapted to perform the task of semantic segmentation in an end-to-end fashion instead of only image classification. Semantic segmentation refers to the task of infering a class for each of the pixels of an image instead of a single class for the entire image. Such models can be found in literature both in 2D Badrinarayanan et al., 2017, Ronneberger et al., 2015 and 3D Çiçek et al., 2016 and have also been used for ILD quantification. The authors of Vakalopoulou et al., 2018 present the coupling of 2D fully convolutional networks with deformable registration for the automatic quantification of systemic sclerosis disease. Moreover, in Anthimopoulos et al., 2018 the authors propose the use of dilated filters for the segmentation of different ILD tissue types. Furthermore in Bermejo-Peláez et al., 2020 an ensemble of 2D, 2.5D and 3D networks is proposed for the segmentation of 8 different radiographic ILD patterns. At this point, it is important to note that since COVID-19 shares similar patterns with ILDs, these recent advances on ILD quantification are of great assistance for the development of tools for its quantification.
2.2. ILD staging
Staging of patients with ILDs is very important as it could greatly help clinicians with their daily practice, while choosing treatment options Kolb and Collard, 2014. There have been a number of studies recently that try to identify and extract biomarkers from CT scans and associate them with the severity and treatment of ILD patients. These biomarkers are usually enhanced with clinical and physiological information to provide a scoring system as survival predictor. Among the variety of biomarkers, disease extent is one of the most powerful ones providing strong associations with severity and mortality Cottin and Brown, 2019, Tomassetti et al., 2015. Visual scoring of the disease extent on CT can be time-consuming Robbie et al., 2017 highlighting the need for tools for automatic disease quantification. Moreover, except the disease extent, the location of the disease is also very important for the staging. In Depeursinge et al., 2015, Christe et al., 2019 the quantification of the disease is performed on different lung regions providing descriptive information about the severity of the ILD patients.
A variety of works report that radiomics, quantitative features extracted from the images, provide valuable information about the severity and response to treatment for different diseases including cancer Sun et al., 2018. These features could also provide very good tools for monitoring disease progression and therapeutic response Wu et al., 2019. In particular, in Bocchino et al., 2019 intensity-based characteristics such as skewness and kurtosis were used together with disease extent to distinguish between systemic sclerosis patients with and without ILD diseases. Moreover, in Lafata et al. Lafata et al., 2019 a variety of image radiomics and their relationship with the pulmonary function were investigated. Their results indicate that highthroughput radiomics data extracted from the lungs may be associated with pulmonary function as measured by common PFT metrics.
3. Methodology
In this section, we describe our AI driven scheme for the quantification of CT scans for patients suffering from COVID-19 pneumonia. Furthermore, we provide a method for the automatic selection and combination of multi-modal variables towards a holistic signature designed for the COVID-19 triage. On the basis of this interpretable, clinically relevant signature we develop advanced machine learning techniques integrating multi-modal data for severity assessment and short/long term outcome prediction. Our method endows robustness, good generalization properties, explainability and establishes causality with known clinical COVID-19 confounding factors. In the following parts of this section, we provide details for all the different components of the system.
3.1. Lung, breast and heart segmentation
Segmentation of the heart and breast were extracted by using the software ART-Plan (TheraPanacea, Paris, France). ART-Plan is a CE-marked solution for automatic annotation of organs, harnessing a combination of anatomically preserving and deep learning concepts. The segmentation of lungs was also performed using ART-Plan software, but the models used were re-trained using COVID-19 patients in order to address proper segmentation of diseased lungs. In particular, the existing lung models, providing segmentation of left and right lungs, were retrained using 50 full COVID-19 lung annotations provided by medical experts. The models were evaluated on 130 COVID-19 patients partially annotated by two different experts, reporting mean dice coefficient higher than 0.96 for both left and right lungs and mean standard deviation lower than 0.015. Moreover, the lung segmentation of the model was similar to the one provided by the medical experts with dice coefficient 0.96 versus 0.97 respectively.
3.2. Ensemble of deep architectures for disease quantification
Our proposed COVID-19 related lung damage segmentation tool was built using an ensemble method combining 2D & 3D deep learning architectures. All the COVID-19 related CT abnormalities which are similar to other ILD diseases (ground glass opacities, band consolidations, and reticulations) were segmented as a single class. The proposed method (CovidENet) borrows elements from already established fully convolutional neural network architectures Çiçek et al., 2016, Badrinarayanan et al., 2017 while it incorporates powerful design aspects such as deformable registration methods for natural data augmentation. The combination of the different CovidENet components has been performed using their scoring output (before hard decision) fusing the output of the different networks based on majority voting. This is a rather standard technique when combining prediction between multiple neural networks. Our motivation to adopt a 2D architecture was driven from the interest of exploring the spatial resolution on the axial space after mapping to a common space, while the integration of 3D networks was dictated from the interest of integrating consistency on the coronal/sagital planes.
3.2.1. CovidE2D component
Deep learning architectures based on 2D networks are commonly used for the segmentation of ILD diseases Anthimopoulos et al., 2018, Vakalopoulou et al., 2018 due to a lot of times limited annotated datasets that are available for the specific task and the 2D nature of the annotations. In this paper, we based the first component (CovidE2D) of our CovidENet architecture on AtlasNet 2D architecture Vakalopoulou et al., 2018. AltasNet has already been used for ILD segmentation in systemic sclerosis patients, achieving very good performance on limited annotated ILD datasets. AtlasNet couples deformable registration with deep learning performing data augmentation in a natural way while preserving the human anatomy. The main idea lies in training different deep learning classifiers (Ci) in a simplified space, after registering each sample (Si) on predefined templates/atlases (Ai). During inference (Algorithm 1 ), the final segmentation is obtained by using the inverse transformation () to back-project to the original anatomy, while a majority voting scheme is used to produce the final projection, combining the results of the different networks.
Algorithm 1.

AtlasNet inference.
For the registration of the CT scans to the templates, an elastic registration framework based on Markov Random Fields was used, providing the optimal displacements for each template Ferrante et al., 2017. In particular, the registration is performed by a non-linear transformer T, corresponding to the operator that optimizes in the continuous domain Ω the following energy,
| (1) |
where ρj corresponds to the different similarity metrics (sum of absolute difference, normalised cross correlation, etc) used to compare the source 3D volume to the target anatomy, wj are linear constraints factorizing the importance of the different metric functions and ψ( · ) is a penalty function acting on the spatial derivatives of the transformation.
Concerning the details of the architecture, in our experiments each Ci consists of a SegNet Badrinarayanan et al., 2017 based architecture. More specifically, for the CovidE2D models the CT scans were separated on the axial view. Each network included 5 convolutional blocks, each one containing two Conv-BN-ReLU layer successions. Maxpooling layers were also distributed at the end of each convolutional block for the encoding part. Upsampling operators were used on the decoding part to restore the spatial resolution of the slices together with the same successions of layers.
3.2.2. CovidE3D component
To fully exploit the 3D nature of our dataset, the second component of our proposed CovidENet is based on a 3D fully convolutional network similar to 3D-UNet Çiçek et al., 2016. In order to train this model, 3D sub-volumes of the CT scan that fully included without any downsampling either the left or right lung were extracted. The corresponding sub-volumes were also extracted from the ground truth annotation masks. To this end, we trained the model with the CT scan sub-volume as input and the annotation as target. As far as the architecture is concerned, the model consisted of five blocks with a down-sampling operation applied every two consequent Conv3D-BN-ReLU layers. Additionally, five decoding blocks were utilized for the decoding path, were at each block a transpose convolution was performed in order to up-sample the input. Skip connections were also employed between the encoding and decoding paths. The dimensions of the input that corresponded to the spatial dimensions of the CT scan and consequently the spatial dimensions of the features maps were not bound to some fixed dimension in order to feed the entire left/ right lung volumes. As such, 3D volumes of arbitrary spatial dimensions could be fed to the network and thus the batch size was fixed to 1.
3.3. Holistic multi-omics profiling & staging
In order to combine disease extent with disease characteristics and patients commodities, we investigate a variety of imaging characteristics extracted using disease, cardiac and lung segmentations. These imaging characteristics (radiomics) were then combined with meaningful clinical and biological indicators that have been reported to be associated with the prognosis of COVID-19. Patient charts were reviewed to assess short term (4 days after the chest CT) and long term prognosis (31 days after the chest CT). For the staging task, patients were divided in 2 groups: those who died, or required mechanical ventilation either at the initial or at a subsequent admission as severe cases (S), and the rest as non-severe cases (NS). For the prognosis task, three distinct subpopulations were defined: those who had a short term negative (SD = short-term deceased) outcome (deceased within 4 days after admission), those who didn’t recover (LD= long-term deceased) within 31 days after the chest CT (either died after day 4 or still intubated at day 31) and those who recovered (LR= long-term recovered). The last two groups formed the short intubated (SI) group of patients.
3.3.1. Feature extraction
Radiomics features were extracted from the CT scans using the previously described segmentations of the disease, lung and heart. As a preprocessing step, all images were resampled by cubic interpolation to obtain isometric voxels with sizes of 1 mm. Subsequently, disease, lung and heart masks were used to extract 107 radiomic features for each of them (left and right lung were considered separately both for the disease extent and entire lung). These features included first order statistics (maximum attenuation, skewness, 90th percentile etc), shape features (surface, maximum 2D diameter per slice, volume etc) and texture features (GLSZM, GLDM, GLRLM etc.). For the extraction, the open source Pyradiomics library was used Van Griethuysen et al., 2017.
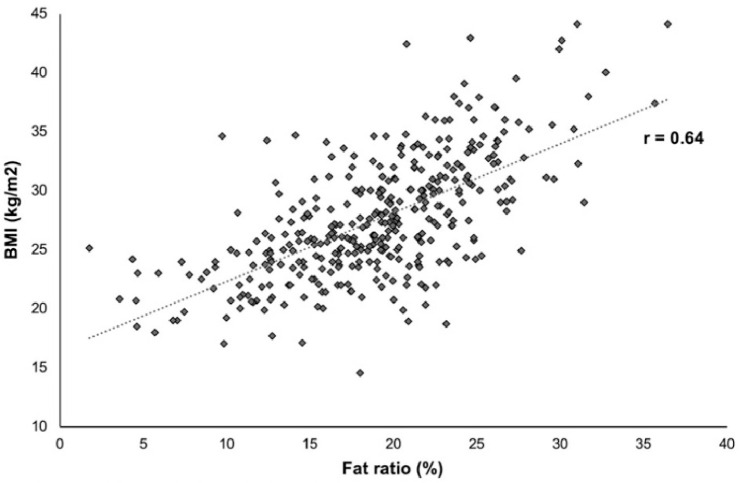
Two other image indexes were also calculated, namely disease extent and fat ratio. The disease extent was calculated as the percentage of lung affected by the disease in respect to the entire lung volume. The disease components were extracted by calculating the number of individual connected components for the entire disease regions. The fat ratio, calculated as an indicator of obesity, was used as a surrogate of the body mass index and calculated by dividing the volume of thoracic fat by the volume of the thorax. The index was defined in an unsupervised manner. To obtain fat segmentation, CT scans were smoothed using a Gaussian kernel with a standard deviation of 2. Then, a threshold of the densities in the range of was applied on the smoothed CTs to isolate the fat regions. Fat masks were calculated starting from the highest to the lowest part of the lungs. In order to avoid gender bias, we used breast segmentation to exclude breast fat. Then the volume of the fat segmentation was divided by the body volume. To validate this morphometric measurement we assessed its correlation with BMI in the 362 patients for which BMI was available and we found a strong correlation using Pearson correlation (; p < 0.001; Fig. 2 ).
Fig. 2.
Correlation between body mass index (BMI) and fat ratio.
3.3.2. Holistic biomarker selection
Using all the calculated attributes (clinical, biological, imaging) we constructed a high dimensional space of size 543, including clinical/biological variables. A min-max normalization of the attributes was performed by calculating the minimum and maximum values for the training and validation cohorts. The same values were also applied on the test set.
To prevent overfitting and discover the most informative and robust attributes for the staging and prognosis of the patients we propose a robust biomarker selection process. Feature selection is very important for classification tasks and has been used widely in literature especially for radiomics Sun et al., 2018. First, the training data set was subdivided into training and validation on the principle of 80%-20% maintaining the distribution of classes between the two subsets identical to the observed one. To perform features selection, we have created 100 subdivisions on this basis and evaluated variety of classical machine learning - using the entire feature space - classifiers such as Decision Tree Classifier, Linear Support Vector Machine, XGBoosting, AdaBoost and Lasso. These classifiers were trained and validated to distinguish between severe (S) and non severe (NS) cases. In addition to these 5 classifier-based feature selection approaches, we also considered statistics-based approaches based on Mutual Information, Chi-squared statistics and Univariate linear regression tests. Each of these methods was used to assess the importance of the features regarding outcome prediction. Features were ranked according to their prevalence, the total number of splits they were selected in, for each of the methods. Our experiments indicated that different classifiers highlight different attributes as important. In order to take advantage of the different feature selection properties, we adopted a consensus feature selection method by selecting features with the highest sum of prevalence over all methods. Besides, to maintain structural properties, we selected the features in the top 5 prevalence in each region.
3.3.3. COVID-19 multi-omics profiling signature
Using the aforementioned selection method, we have extracted 15 different radiomics features. These features belong to: features from imaging and in particular from the disease regions (5 features), lung regions (5 features) and heart features (5 features). On these radiomics features biological and clinical data were added (6 features: age, sex, high blood pressure (HBP), diabetes, lymphocyte count and CRP level) and image indexes (2 features: disease extent and fat ratio). At the end our biomarker consisted of 23 features in total.
Regarding imaging features, we identified the following features as more important for the prognosis of the COVID-19 patients. These features include both first and second order statistics together with some shape features.
-
•
Disease areas: Non- Uniformity of the Gray Level Dependence Matrix (GLDM), Dependence Non-Uniformity of the GLDM, Mesh Volume, Voxel Volume, Non-Uniformity of the Gray level Run Length Matrix (GLRLM).
-
•
Lung areas: Kurtosis, Mean Absolute Deviation, Zone Emphasis of the GLSZM, Non-Uniformity of the GLSZM, Variance of the GLSZM.
-
•
Heart areas: Maximum 2D diameter Slice, Non-Uniformity of the GLSZM, Sphericity, Flatness, Minimum Length on the Axis.
The selected disease area features capture both disease extent and disease textural heterogeneity. Disease textural heterogeneity is associated with lesions, the presence of which generates imaging pattern more complex than pure ground glass opacities usually found in mild disease. The selected lung features capture the dispersion and heterogeneity of lung densities, both of which may reflect the presence of an underlying airway disease such as emphysema but also the presence of sub-radiological disease. Lastly, the selected heart features can be seen as a surrogate for cardiomegaly and coronary calcifications.
3.3.4. Staging mechanism
The staging/prognosis component was addressed using an ensemble learning approach. Similarly to the biomarker extraction, the training data set was subdivided into training and validation sets on the principle of 80%-20%. This subdivision was performed such that the distribution of classes between the two subsets was identical to the observed one. We have used 10-fold cross validation on this basis and evaluated the average performance of the following supervised classification methods: Nearest Neighbor, {Linear, Sigmoid, Radial Basis Function (RBF), Polynomial Kernel} Support Vector Machines (SVM), Gaussian Process, Decision Trees, Random Forests, AdaBoost, XGBoosting, Gaussian Naive Bayes, Bernoulli Naive Bayes, Multi-Layer Perceptron & Quadratic Discriminant Analysis. These classifiers have been trained using the identified holistic signature. For each binary classification task a consensus model was designed selecting the top 5 classifiers with acceptable performance, > 60% in terms of balanced accuracy, as well as coherent performance between training and validation, performance decrease < 20% for the balanced accuracy. The selected models were trained and combined together through a weighted winner takes all approach to determine the optimal outcome. The weights granted to each selected classifier determined according to the rank of this classifier on validation regarding balanced accuracy weighted with a higher importance the best performing algorithms. Then, the selected classifiers were retrained using the entire training set, and their performance was reported on the external test cohort.
3.3.5. Prognosis mechanism
To perform the short-term deceased (SD), long-term Deceased (LD), long term recovered (LR) classification task, a SD/SI (SI: intubated at 4 days) classifier and a LD/LR classifier was applied in a hierarchical way, performing first the short-term staging and then the long-term prognosis for patients classified as in need of mechanical ventilation support. More specifically, a majority voting method was applied to classify patients into SD and SI cases. Then, another hierarchical structure was applied on the cases predicted as SI only to classify them into the ones who didn’t recover within days of mechanical ventilation (LD) and the ones who recovered with 30 days on mechanical ventilation (LR).
3.4. Implementation details
3.4.1. Deep learning segmentation
In order to train all the models, each CT scan was normalized by cropping the Hounsfield units in the range . A variety of hyperparameters including loss functions, learning rates, optimizers had been tested and in this section we report the ones with the best performance for each component. Regarding implementation details, 6 templates/ atlases (Ai) were used for the AtlasNet framework together with normalized cross correlation and mutual information as similarity metrics for the registration to each template. All 6 models of the CovidE2D networks were trained using weighted cross entropy loss. Moreover, the CovidE3D network was trained using a dice loss. CovidENet aims to fuse different training strategies (2D, 3D) as well as different loss functions to fully explore the capabilities of deep learning architectures. 2D networks have been proven to be very robust for the ILD segmentation using cross entropy as it is reported from a variety of studies Anthimopoulos et al., 2018, Vakalopoulou et al., 2018.
For the CovidE2D experiments we used classic stochastic gradient descent for the optimization with initial learning rate = 0.01, decrease of learning rate = every 10 epochs, momentum =0.9 and weight decay =. For CovidE3D experiments we used the AMSGrad and a learning rate of 0.001. TensorFlow library Abadi et al., 2016 was used for the implementation of the CovidENet components.
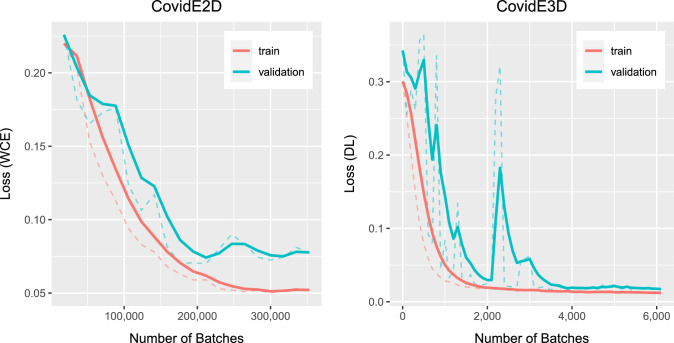
The training of a single network for both CovidE2D and CovidE3D was completed in approximately 12 hours using a GeForce GTX 1080 GPU, while the prediction for a single CT scan was done in a few seconds. Training and validation curves for one template of CovidE2D and the CovidE3D networks are shown in Fig. 3 . Early stopping has been used for ending the training process and the most appropriate model for each CovidENet component was the one that was performing the best in the validation set until this point.
Fig. 3.
Training and validation curves for one template/ atlas (Ai) of CovidE2D and the CovidE3D.
3.4.2. COVID-19 multi-omics profiling & staging
For the feature selection, features having the best combined prevalence (sum of prevalences over the 8 selection techniques) were kept. For this feature selection task, Decision Tree Classifier was taken of maximum depth 3, Linear SVM was taken with a linear kernel, a polynomial kernel function of degree 3 and a penalty parameter of 0.25, XGBoosting was used with a regression tree boosted over 30 stages, AdaBoost was used with a Decision Tree Classifier of maximum depth 2 boosted 3 times and Lasso method was used with 200 alphas along a regularization path of length 0.01 and limited to 1000 iterations.
Concerning the implementation details, to overcome the unbalanced dataset for the different classes, each class received a weight inversely proportional to its size. For the NS versus S majority voting classifier the top 5 classifiers consist of RBF SVM, Linear SVM, AdaBoost, Random forest, Decision Tree. The SVM methods were granted a polynomial kernel function of degree 3, the Linear kernel had a penalty parameter of 0.3 while the RBF SVM had a penalty parameter of 0.15. In addition, the RBF SVM was granted a kernel coefficient of 1. The Decision Tree classifier was limited to a depth of 2 to avoid overfitting. The Random Forest classifier was composed of 25 of such Decision Trees. AdaBoost classifier was based on a decision tree of maximal depth of 1 boosted 4 times. For the SI versus SD majority voting classifier the top 5 classifiers consists in polynomial SVM, Linear SVM, Decision Tree, Random Forest and AdaBoost. The Linear and Polynomial SVM were granted a polynomial kernel function of degree 2 and a penalty parameter of 0.35. The Decision Tree classifier was limited to a depth of 1 and Random Forest was composed of 50 of such trees. AdaBoost classifier was based on a decision tree of maximal depth of 1 boosted 2 times. Finally, the LR versus LD majority voting classifier was only using the 4 classifiers with balanced accuracy > 0.6 namely Linear and Sigmoid SVM, Decision Tree, and AdaBoost Classifiers. The SVM methods were defined with a kernel function of degree 3 and a penalty parameter of 1. Decision Tree was defined to a depth of 1, AdaBoost being defined with such a Decision Tree boosted 3 times. For the implementation of all the models Scikit-learn library was used Pedregosa et al., 2011.
4. Dataset
This retrospective multi-center study was approved by our Institutional Review Board (AAA-2020-08007) which waived the need for patients’ consent. Patients diagnosed with COVID-19 from March 4th to April 5th from eight large University Hospitals were eligible if they had positive reverse transcription polymerase chain reaction (PCR-RT) and signs of COVID-19 pneumonia on unenhanced chest CT. Only the CT examination that was performed at initial evaluation was included in our dataset. Exclusion criteria were (i) contrast medium injection and (ii) important motion artifacts. No patient was intubated at the time of the CT acquisition. A total of 693 patients, after all the exclusion criteria were applied, formed the full dataset (321,360 CT slices).
Chest CT exams were acquired on 4 different CT models from 3 manufacturers (Aquilion Prime from Canon Medical Systems, Otawara, Japan; Revolution HD from GE Healthcare, Milwaukee, WI; Somatom Edge and Somatom AS+ from Siemens Healthineer, Erlangen, Germany). The different acquisition and reconstruction parameters are summarized in Table 1 . CT exams were mostly acquired at 120 (; 69%) and 100 kVp (; 27%). Images were reconstructed using iterative reconstruction with a 512 × 512 matrix and a slice thickness of 0.625 or 1 mm depending on the CT equipment. Only the lung images reconstructed with high frequency kernels were used for analysis. For each CT examination, dose length product (DLP) and volume Computed Tomography Dose Index (CTDIvol) were collected.
Table 1.
Acquisition and reconstruction parameters of the dataset used in this study. Note: For quantitative variables, data are presented as mean ± standard deviation, and numbers in brackets indicate their range. CT, computed tomography; CTDIvol, volume computed tomography Dose Index; DLP, dose length product.
| Center A | Center B | Center C | Center D | Center E | Center F | Center G | Center H | |
|---|---|---|---|---|---|---|---|---|
| CT equipment | Somatom AS+ | Resolution HD | Aquilion Prime | Somatom Edge | Revolution HD | Aquilion Prime | Revolution HD | Somatom AS+ |
| Kilovoltage | 100–120 | 120 | 100–120 | 100–120 | 120–140 | 100–120 | 120 | 100–120 |
| DLP (mGy.cm) | 109 ± 42 [44–256] | 306 ± 104 [123–648] | 102 ± 30 [43–189] | 131 ± 44 [55–499] | 177 ± 48 [43–276] | 115 ± 26 [75–186] | 285 ± 108 [70–679] | 332 ± 156 [179–755] |
| CTDIvol (mGy) | 3.2 ± 1.5 [1.2–11.9] | 8.7 ± 2.8 [3.9–18.5] | 2.7 ± 0.9 [1.0–5.3] | 3.2 ± 0.9 [1.4–9.5] | 5.5 ± 1.8 [1.2–12.3] | 2.5 ± 0.6 [1.7–4.3] | 7.9 ± 2.9 [1.7–18.0] | 8.5 ± 4.0 [4.4–19.8] |
| Slice thickness | 1 mm | 0.625 mm | 1 mm | 0.625 mm | 1 mm | 1 mm | 0.625 mm | 1 mm |
| Convolution Kernel | i70 | Lung | FC51-FC52 | i50 | Lung | FC51-FC52 | Lung | i70 |
| Iterative reconstructions | SAFIRE 3 | ASIR-v 80% | IDR 3D0.67 | SAFIRE 4 | ASIR-v 60% | IDR 3D | ASIR-v 60% | SAFIRE 3 |
For the COVID-19 radiological pattern segmentation part, 50 patients from 3 centers (A: 20 patients; B: 15 patients, C: 15 patients) were included to compose a training and validation dataset, 130 patients from the remaining 3 centers (D: 50 patients; E: 50 patients, F: 30 patients) were included to compose the test dataset (Table 2 ). The patients from the training cohort were annotated slice-by-slice, while the patients from the testing cohort were partially annotated on the basis of 20 slices per exam covering in an equidistant manner the lung regions. The proportion between the CT manufacturers in the datasets was pre-determined in order to maximize the model generalizability while taking into account the data distribution.
Table 2.
Patient characteristics for the automatic quantification of COVID-19 disease. Note: For quantitative variables, data are presented as mean ± standard deviation, and numbers in brackets indicate their range. CT, computed tomography; CTDIvol, volume computed tomography dose index; DLP, dose length product.
| Training/validation dataset (Centers A+B+C; n = 50) | Test dataset (Centers D+E+F; n = 130) | p value | |
|---|---|---|---|
| Age (y) | 57 ± 17 [26–97] | 59 ± 16 [17–95] | 0.363 |
| No. of Men | 31(62) | 87(67) | 0.534 |
| Disease extent* | |||
| Manual | 18.1 ± 14.9 [0.3–68.5] | 19.5 ± 16.5 [1.1–75.7] | 0.574 |
| Automated | – | 19.9% ± 17.7 [0.5–73.2] | – |
| DLP (mGy.cm) | 180 ± 124 [43–527] | 139 ± 49.0 [43–276] | 0.026 |
| CTDIvol (mGy) | 4.9 ± 3.4 [1.0–13.0] | 4.0 ± 1.9 [1.2–12.3] | 0.064 |
For the staging (NS/S) and prognosis (short and long-term) study, 513 additional patients from centers A (121 patients), B (157 patients), D (138 patients), G (77 patients) and H (20 patients) were included. Data of 536 patients from 5 centers (A, B, C, D and H) were used for training and those of 157 patients from 3 other centers (E, F and G) composed an independent test set (Table 3 ). In addition to the CT examination - when available - patient sex, age, and body mass index (BMI), blood pressure and diabetes, lymphocyte count, CRP level and D-dimer level were also collected (Table 3).
Table 3.
Patient characteristics for the automatic staging and prognosis tools. Note: For quantitative variables, data are presented as mean ± standard deviation, and numbers in brackets indicate their range. For qualitative variables, data are numbers of patients, and numbers in parentheses are percentages. CT, computed tomography; CTDIvol, volume computed tomography dose index; DLP, dose length product. *Available clinical data: for diabetes and high blood pressure(leading to 0.19% of missing data on the training set), for lymphocyte count (leading to 2.05% and 5.10% of missing data on the training and test sets respectively), for CRP (leading to 4.66% and 8.92% of missing data on the training and test sets respectively), for Body Mass Index, and for D-dimers. **Percentage of lung volume on the whole CT. ***Data available for 688 patients.
| Training/validation dataset (Centers A + B + C+ D + H; ) | Test Dataset (Centers E + F + G; ) | p value | |
|---|---|---|---|
| Age (y) | 63 ± 16 [22–98] | 62 ± 17 [17–98] | 0.495 |
| No. of Men | 374(70) | 103(78) | 0.321 |
| High blood pression* | 235 (44) | 71 (45) | 0.773 |
| Diabetes* | 97 (18) | 37 (24) | 0.888 |
| Body mass index (kg/m2)* | 27.7 ± 5.1 [17.0–44.1] | 27.1 ± 5.1 [14.5–42.7] | 0.390 |
| Lymphocyte count ( × 109/L)* | 1.3 ± 2.7 [0.1–48.5] | 1.3 ± 3.3 [0.23–41.0] | 0.915 |
| CRP (mg/L)* | 104.3 ± 82.9 [1.0–430.7] | 94.2 ± 74.8 [2.0–342] | 0.166 |
| D-dimers (microg/L)* | 2458 ± 6533 [181–86248] | 815 ± 924 [168–6138] | < 0.001 |
| Disease extent** | 22.2 ± 18.4 [0.0–89.8] | 24.0 ± 18.7 [1.1–89.8] | |
| Fat ratio on CT | 18.6 ± 5.9 [1.7–42.3] | 18.3 ± 5.5 [2.7–30.6] | 0.589 |
| Short-term outcome | 0.994 | ||
| Deceased | 28(5) | 8(5) | |
| Intubated | 80(15) | 23(15) | |
| Alive and Not Intubated | 428(80) | 126(80) | |
| Follow-up duration | |||
| Worsening during follow-up*** | 0.554 | ||
| Deceased | 69(13) | 17(11) | |
| Intubated | 68(13) | 22(14) | |
| DLP (mGy.cm) | 181 ± 115 [43–755] | 218 ± 106 [ 43–679 ] | < 0.001 |
| CTDIvol (mGy) | 4.9 ± 3.2 [1.0–19.8] | 6.1 ± 3.0 [1.2–18.0] | < 0.001 |
For short-term outcome assessment, patients were divided into 2 groups: those who died or were intubated in the 4 days following the CT scan composed the severe short-term outcome subgroup, while the others composed the non-severe short-term outcome subgroup. For long-term outcome, medical records were reviewed from May 7th to May 10th, 2020 to determine if patients died or had been intubated during the period of at least one month following the CT examination. The data associated with each patient (holistic profiling including radiomics, biological and clinical attributes), as well as the corresponding outcomes both in terms of severity assessment as well as in terms of final outcome have been made publicly available (https://github.com/ebattistella/Covid-MedIA).
Fifteen radiologists (GC, TNHT, SD, EG, NH, SEH, FB, SN, CH, IS, HK, SB, AC, GF and MB) with 1 to 7 years of experience in chest imaging participated in the data annotation which was conducted over a 2-week period. For the training and validation set for the COVID-19 radiological pattern segmentation, the whole CT examinations were manually annotated slice by slice. On each of the 50 cases (23,423 axial slices) composing this dataset, all the COVID-19 related CT abnormalities (ground glass opacities, band consolidations, and reticulations) were segmented as a single class. Additionally, the whole lung was segmented to create another class (lung). To facilitate the collection of the ground truth for the lung anatomy, a preliminary lung segmentation was performed with Myrian XP-Lung software (version 1.19.1, Intrasense, Montpellier, France) and then manually corrected. For the test cohort, 20 CT slices equally spaced from the superior border of aortic arch to the lowest diaphragmatic dome were selected in a total of 130 patients composing a 2600 images dataset. Each of these images were systematically annotated by 2 out of the 15 participating radiologists who independently performed the annotation. Annotation consisted of manual delineation of the disease and manual segmentation of the lung without using any preliminary lung segmentation.
Furthermore, 3 radiologists, an internationally recognized expert with years of experience in thoracic imaging (ReaderA), a thoracic radiologist with years of experience (ReaderB) and a resident with 6-month experience in thoracic imaging (ReaderC) were asked to perform a triage (severe versus non-severe cases) and for the severe cases (short-term deceased versus short-term intubated) prognosis process to predict the short-term outcome.
5. Experimental results
5.1. Statistical analysis
The dice similarity score (DSC) was calculated to assess the similarity between the 2 manual segmentations of each CT exam of the test dataset and between manual and automated segmentations. The Hausdorff distance (HD) was also calculated to evaluate the quality of the automated segmentations in a similar manner. Disease extent was calculated by dividing the volume of diseased lung by the lung volume and expressed in percentage of the total lung volume. Disease extent measurement between automated and manual segmentations were compared using paired Student’s t-tests. Similarly, correlation between disease extent measurents from Covid2D, Covid3D, CovidENet and manual segmentations were compared using Spearman correlation coefficient.
For the stratification of the dataset into the different categories, classic machine learning metrics, namely balanced accuracy, weighted precision, and weighted specificity and sensitivity were utilized.
5.2. Disease quantification
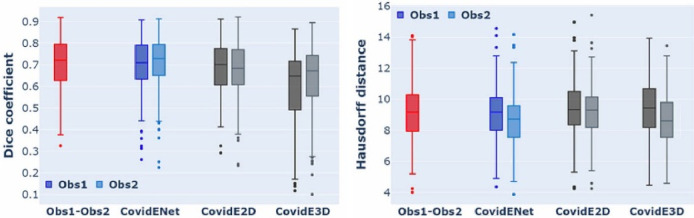
The evaluation of CovidENet together with its components and the comparison with the 2 independent experts is summarised in Table 4 . CovidE2D component performed better than the CovidE3D for the segmentation of COVID-19 disease. This is indicated by the higher DSC and HD values achieved by the CovidE2D component (Fig. 4 ). However, their fusion led to a significant improvement, comparable to human readers. Moreover, CovidENet performed equally well compared to trained radiologists in terms of DSC and better in terms of HD (Fig. 4, Fig. 6 and Table 4). The mean/median DSCs between the two expert annotations on the test dataset were 0.70/0.72 for disease segmentation while DSCs between CovidENet and the manual segmentations were 0.69/0.71 and 0.70/0.73. In terms of HDs, the average expert distance was 9.16 mm while it was 8.96 mm between CovidENet and the experts.
Table 4.
Quantitative evaluation of the CovidENet and its components CovidE2D & CovidE3D architectures in terms of Dice Coefficient and Hausdorff Distance. In particular, the mean, median and standard deviation for each of the developed tools are presented together with comparison with the 2 independent experts. With bold we indicate the highest values per metric.
| Methods |
Dice |
Hausdorff distance |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean |
Median |
STD |
Mean |
Median |
STD |
|||||||
| Obs1 | Obs2 | Obs1 | Obs2 | Obs1 | Obs2 | Obs1 | Obs2 | Obs1 | Obs2 | Obs1 | Obs2 | |
| CovidE2D | 0.69 | 0.67 | 0.70 | 0.68 | ± 0.13 | ± 0.13 | 9.40 | 9.23 | 9.33 | 9.30 | ± 1.83 | ± 1.80 |
| CovidE3D | 0.62 | 0.65 | 0.67 | 0.70 | ± 0.17 | ± 0.16 | 9.43 | 8.70 | 9.43 | 8.60 | ± 1.87 | ± 1.81 |
| CovidENet | 0.69 | 0.70 | 0.71 | 0.73 | ± 0.13 | ± 0.13 | 9.18 | 8.75 | 9.16 | 8.72 | ± 1.86 | ± 1.78 |
| Obs1-Obs2 | 0.70 | 0.72 | ± 0.12 | 9.16 | 9.16 | ± 1.83 | ||||||
| CovidENet | 0.70 | 0.72 | ± 0.12 | 8.96 | 8.94 | ± 1.82 | ||||||
Fig. 4.
Box-Plot in terms of DSC and HD between CovidENet and its individual components, Obs1 & Obs2. One can observe that CovidENet (blue) performs better and closer to Obs1-Obs2 (red) DSC and HD metrics than its individual components CovidE2D & CovidE3D. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
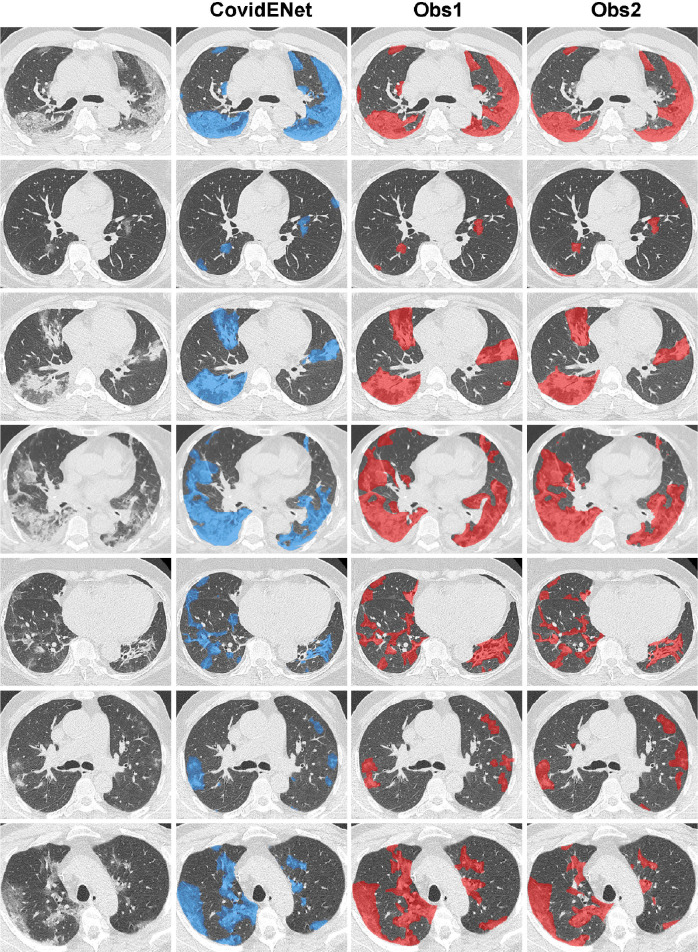
Fig. 6.
Qualitative analysis for the comparison between manual and the proposed CovidENet disease quantification. Delineation of the diseased areas on chest CT in different slices of COVID-19 patients. From left to right: Input, CovidENet-segmentation, Obs1-segmentation, Obs2-segmentation.
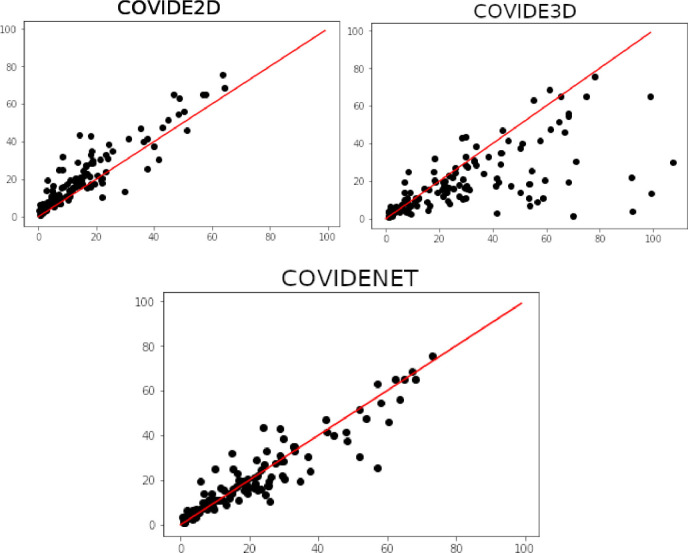
Furthermore, the superiority of CovidENet is indicated by the disease extent evaluated on the test dataset. In particular, no significant difference was observed between disease extent evaluated by the CovidENet and the manual segmentations’ average ( vs. ; ). As shown in Fig. 5 correlation to disease extent from manual segmentations was better when using CovidENet ( p < 0.001) compared to Covid3D ( p < 0.001) or Covid2D ( p < 0.001) which oversegmented the disease.
Fig. 5.
Plots indicating the correlation between the average disease extent measured from CovidE2D, CovidE3D and CovidENet respectively and the manual segmentation. Disease extent is expressed as the percentage of lung affected by the disease. The red line shows a perfect correlation (Spearman ). Spearman correlation coefficients are displayed for each comparison. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Examples of disease segmentations are presented in Fig. 6 . One can observe that the segmentations provided by CovidENet are very close to the ones generated by the experts. In particular, the algorithm detects the diseased regions even in the case that they are relatively small capturing all the different opacities of COVID-19 such as ground glass and consolidation.
5.3. COVID-19 holistic multi-omics profiling & staging
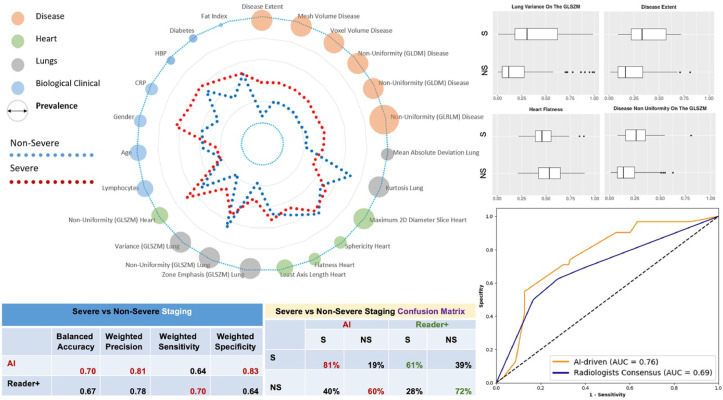
The holistic COVID-19 pneumonia signature is presented in (Table 5 ) along with the correlations with outcome. The average signature for the severe and non-severe case in the test set are presented in Fig. 7 . Consensus ensemble learning through majority voting was used to determine the subset of AI methods that have robust, reproducible performance with good generalization properties. Human “Reader+++” was used as a reference through consensus among three chest radiologists (resident, years of experience, years of experience in thoracic imaging). Our method aiming to separate patients with S/NS outcomes had a balanced accuracy of 70% (vs. 67% for human readers consensus), a weighted precision of 81% (vs. 78%), a weighted sensitivity of 64% (vs. 70%) and specificity of 77% (vs. 64%) and outperformed the consensus of human readers (Fig. 7, Table 6 ). Our method successfully predicted 81% of the severe/critical cases opposed to only 61% for the consensus reader. The superiorty of our approach is also indicated by the higher AUC reported (0.76), in comparison with the one achieved by the different readers (0.69). Severe cases as depicted in Fig. 7 referred to diabetic men, with higher level of volume/heterogeneity of disease and C-reactive protein levels. Moreover, as indicated in Fig. 7 the non-uniformity on GLRLM for both lung and disease together with the disease extent seems to contribute considerable to the classification of the patients to NS versus S cases.
Table 5.
Correlation between outcome and the 23 features of the holistic COVID19 signature. Note: GLSZM, gray level size zone matrix; GLRLM, gray level run length matrix; GLDM, gray level dependence matrix; LD, long-term-deceased; LR, long-term deceased; NS, non severe; S, severe; SI, short-term intubation; SD, short-term deceased.
| Features | Correlation |
||||||
|---|---|---|---|---|---|---|---|
| S/NS | SI/SD | LR/LD | |||||
| Age | 0.067 | 0.674 | 0.334 | ||||
| Sex | 0.132 | −0.049 | −0.059 | ||||
| CRP | 0.002 | 0.015 | 0.018 | ||||
| HBP | 0.033 | 0.293 | 0.332 | ||||
| Diabetes | 0.065 | −0.130 | −0.061 | ||||
| Lymphocytes | 0.033 | 0.020 | 0.012 | ||||
| Fat Index | 0.055 | −0.192 | 0.122 | ||||
| Disease Extent | 0.328 | −0.069 | 0.214 | ||||
| Heart | Non-uniformity on the GLSZM | 0.067 | −0.137 | −0.112 | |||
| Sphericity | −0.161 | −0.246 | −0.101 | ||||
| Flatness | −0.126 | −0.039 | −0.110 | ||||
| Minimum Length on the Axis | 0.044 | 0.067 | −0.083 | ||||
| Left | Right | Left | Right | Left | Right | ||
| Lung | Kurtosis | −0.284 | −0.289 | 0.077 | 0.009 | 0.005 | 0.006 |
| Mean Absolute Deviation | 0.305 | 0.322 | −0.003 | −0.001 | 0.017 | −0.026 | |
| Zone Emphasis on the GLSZM | 0.299 | 0.318 | −0.023 | 0.045 | 0.213 | 0.199 | |
| Non-Uniformity on the GLSZM | −0.305 | −0.305 | −0.018 | −0.031 | −0.174 | −0.138 | |
| Variance on the GLSZM | 0.305 | 0.348 | 0.018 | 0.031 | 0.174 | 0.138 | |
| Disease | Mesh Volume | 0.297 | 0.363 | −0.087 | 0.024 | 0.209 | 0.125 |
| Volume Volume | 0.297 | 0.363 | −0.087 | 0.024 | 0.209 | 0.125 | |
| Dependence Non-Uniformity on the GLDM | 0.266 | 0.338 | −0.067 | 0.202 | 0.168 | ||
| Non-Uniformity on the GLDM | 0.287 | 0.363 | −0.079 | 0.017 | 0.203 | 0.142 | |
| Non-uniformity on the GLRLM | 0.284 | 0.340 | −0.076 | 0.037 | 0.194 | 0.123 | |
Fig. 7.
COVID-19 Holistic Multi-Omics Signature & Staging: Spider chart representing average profiles (average values of the variables after normalization between 0 and 1) with respect to severe versus non-severe separation are shown along with prevalence of biomarkers (diameter of the circle). The prevalence of the biomarker corresponds here to the number of selections of the biomarker during the feature selection process. Classification performance, confusion matrices and area under the curve with respect to the proposed method and the consensus of expert readers (reader+) are reported on the right side. Selective associations of features with outcome (NS/S) are shown at the top right of the figure (box plots).
Table 6.
Prognosis of medical experts and their consensus for the Non Severe (NS) versus Severe (S), Intubated (SI) versus Deceased (SD) and NS/SI/SD patients Note: Classification Performance ReaderA (Senior), ReaderB (Established), ReaderC (Resident), Reader (Consensus among Human Readers), Reader (Average performance of Human Readers).
| Balanced accuracy | Weighted precision | Weighted sensitivity | Weighted specificity | |
|---|---|---|---|---|
| NS/SI/SD | ||||
| ReaderA | 0.62 | 0.77 | 0.68 | 0.69 |
| ReaderB | 0.59 | 0.75 | 0.67 | 0.65 |
| ReaderC | 0.61 | 0.76 | 0.68 | 0.62 |
| Reader | 0.63 | 0.77 | 0.70 | 0.67 |
| Reader | 0.61 ± 0.01 | 0.76 ± 0.01 | 0.68 ± 0.01 | 0.66 ± 0.03 |
| Proposed | 0.67 | 0.81 | 0.63 | 0.80 |
| NS/S | ||||
| ReaderA | 0.69 | 0.79 | 0.70 | 0.67 |
| ReaderB | 0.66 | 0.77 | 0.70 | 0.62 |
| ReaderC | 0.65 | 0.76 | 0.70 | 0.60 |
| Reader | 0.67 | 0.78 | 0.70 | 0.64 |
| Reader | 0.67 ± 0.01 | 0.77 ± 0.01 | 0.70 ± 0.01 | 0.63 ± 0.03 |
| Proposed | 0.70 | 0.81 | 0.64 | 0.77 |
| SI/SD | ||||
| ReaderA | 0.81 | 0.87 | 0.88 | 0.75 |
| ReaderB | 0.79 | 0.84 | 0.84 | 0.74 |
| ReaderC | 0.81 | 0.87 | 0.88 | 0.75 |
| Reader | 0.81 | 0.87 | 0.88 | 0.75 |
| Reader | 0.81 ± 0.01 | 0.87 ± 0.01 | 0.87 ± 0.01 | 0.75 ± 0.03 |
| Proposed | 0.88 | 0.94 | 0.94 | 0.81 |
5.4. Prognosis & staging
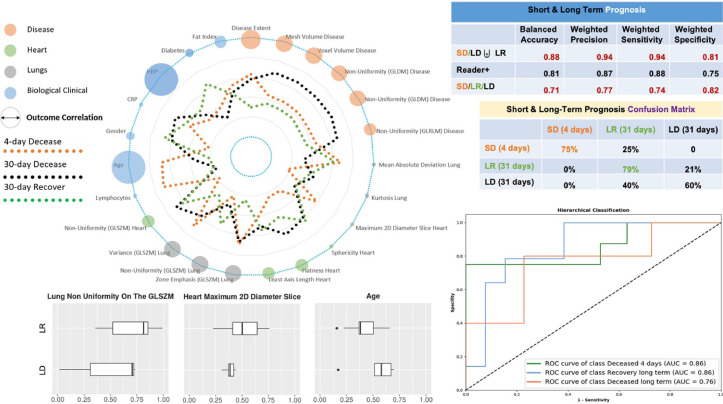
The COVID-19 pneumonia pandemic spiked hospitalizations, while exerting extreme pressure on intensive care units. In the absence of a cure, staging and prognosis is crucial for clinical decision-making for resource management and experimental outcome assessment, in a pandemic context. Our objective was to predict patient outcomes prior to mechanical ventilation support. The proposed ensemble classifier aiming to predict the SD/(LD or LR) had a balanced accuracy of 88% (vs. 81% for human readers consensus), a weighted precision of 94% (vs. 87%), a weighted sensitivity of 94% (vs. 88%) and specificity of 81% (vs. 75%) and outperformed consensus of human readers (Table 6). Our method for prognosis of SD/ LD/ LR had a balanced accuracy of 71%, a weighted precision of 77%, a weighted sensitivity of 74% and specificity of 82% to provide full prognosis (Fig. 8 ). Concerning the performance of our method for the classification of LD and LR patients (Table 7 ), our ensemble classifier reports a balance accuracy of 69%, a weighted precision of 76% a weighted sensitivity of 74% and a weighted specificity of 65%. As indicated also in Fig. 8 the performance of our method reach an AUC of 0.86 for the SD, a 0.86 for the LR and 0.76 for the LD classes. Moreover, the age, HBP and lung non uniformity on the GLSZM seems to associate better for this task.
Fig. 8.
Short & Long Term Prognosis. Spider chart representing average profiles (average values of the variables after normalization between 0 and 1) with respect to the short deceased (SD), long deceased (LD) and long recovered (LR) classes are shown along with their correlations with the outcome (diameter of the circle). The presented correlation corresponds to Pearson Correlation for LR/LD outcome (Table 5). Classification performance, confusion matrices and area under the curve with respect to the proposed method and - when feasible - the consensus of expert readers (reader+) are reported on the right side. ROC curves correspond to one-vs-all classification of the SD/LR/LD patients. Selective associations of features with final outcome (LD/LR) are shown at the bottom of the figure (box plots).
Table 7.
Performance for the Deceased (LD) and Recovered (LR) in the long-term outcome for each of the selected classifiers and their ensemble. Note: P-SVM, support vector machine with a polynomial kernel; S-SVM, support vector machine with a sigmoid kernel.
| Classifier | Balanced accuracy |
Weighted pecision |
Weighted sensitivity |
Weighted specificity |
||||
|---|---|---|---|---|---|---|---|---|
| Train | Test | Train | Test | Train | Test | Train | Test | |
| L-SVM | 0.77 | 0.62 | 0.81 | 0.7 | 0.74 | 0.63 | 0.81 | 0.61 |
| S-SVM | 0.63 | 0.69 | 0.71 | 0.76 | 0.56 | 0.63 | 0.7 | 0.74 |
| AdaBoost | 0.82 | 0.69 | 0.84 | 0.76 | 0.8 | 0.74 | 0.83 | 0.65 |
| Decision Tree | 0.7 | 0.72 | 0.8 | 0.78 | 0.6 | 0.68 | 0.81 | 0.76 |
| Ensemble Classifier | 0.82 | 0.69 | 0.84 | 0.76 | 0.8 | 0.74 | 0.83 | 0.65 |
Moreover, in order to assess the impact of each feature category on the implemented models we performed an ablation study by successively removing one category of features from the 6 categories defined for each classification task. Results are presented in Table 8 . The feature categories were identified as follows: a) D0: disease extent, b) D1: disease variables that are shape/geometry related, c) D2: disease variables that are tissue/texture, d) O1: heart/lungs variables that are shape/geometry related, e) O2: heart/lungs variables that are tissue/texture, f) B1: age, gender, biological/obesity/diabetes/fat/high blood pressure. One can observe that the Clinical Only category contributes a lot to the separation of SD/LD/LR while for the NS/S cases their contribution is marginal, in contrary to the other imaging characteristics.
Table 8.
An ablation study of the different selected features. A leave-one-out method has been applied by removing one feature sequentially to test the features importance and the performance robustness. Note: a) D0: disease extent, b) D1: disease variables that are shape/geometry related, c) D2: disease variables that are tissue/texture, d) O1: heart/lungs variables that are shape/geometry related, e) O2: heart/lungs variables that are tissue/texture, f) B1: age, gender, biological/obesity/diabetes/fat/high blood pressure. LD, long-term-deceased; LR, long-term deceased; NS, non severe; S, severe; SI, short-term intubation; SD, short-term deceased.
| Study case | Task | Balanced accuracy |
Weighted precision |
Weighted sensitivity |
Weighted specificity |
||||
|---|---|---|---|---|---|---|---|---|---|
| Training | Test | Training | Test | Training | Test | Training | Test | ||
| All Features | NS/S | 0.73 | 0.70 | 0.82 | 0.81 | 0.67 | 0.64 | 0.80 | 0.77 |
| SI/SD | 0.90 | 0.88 | 0.92 | 0.94 | 0.92 | 0.94 | 0.88 | 0.81 | |
| LD/LR | 0.82 | 0.69 | 0.84 | 0.76 | 0.8 | 0.74 | 0.83 | 0.65 | |
| SD/LD/LR | 0.77 | 0.71 | 0.8 | 0.77 | 0.78 | 0.74 | 0.9 | 0.82 | |
| Without D0 | NS/S | 0.73 | 0.7 | 0.82 | 0.8 | 0.68 | 0.65 | 0.79 | 0.74 |
| SI/SD | 0.89 | 0.88 | 0.92 | 0.94 | 0.92 | 0.94 | 0.88 | 0.81 | |
| LD/LR | 0.56 | 0.5 | 0.74 | 0.54 | 0.74 | 0.74 | 0.39 | 0.26 | |
| SD/LD/LR | 0.65 | 0.58 | 0.73 | 0.64 | 0.76 | 0.74 | 0.79 | 0.72 | |
| Without D1 | NS/S | 0.74 | 0.69 | 0.82 | 0.8 | 0.67 | 0.64 | 0.8 | 0.74 |
| SI/SD | 0.89 | 0.88 | 0.91 | 0.93 | 0.91 | 0.93 | 0.88 | 0.81 | |
| LD/LR | 0.56 | 0.5 | 0.74 | 0.54 | 0.74 | 0.74 | 0.39 | 0.26 | |
| SD/LD/LR | 0.65 | 0.58 | 0.73 | 0.64 | 0.76 | 0.74 | 0.79 | 0.72 | |
| Without D2 | NS/S | 0.73 | 0.69 | 0.82 | 0.8 | 0.67 | 0.64 | 0.8 | 0.74 |
| SI/SD | 0.89 | 0.88 | 0.91 | 0.93 | 0.91 | 0.93 | 0.88 | 0.81 | |
| LD/LR | 0.58 | 0.5 | 0.74 | 0.54 | 0.76 | 0.74 | 0.48 | 0.26 | |
| SD/LD/LR | 0.67 | 0.58 | 0.73 | 0.64 | 0.76 | 0.74 | 0.82 | 0.72 | |
| Without O1 | NS/S | 0.73 | 0.7 | 0.82 | 0.79 | 0.72 | 0.73 | 0.75 | 0.67 |
| SI/SD | 0.89 | 0.88 | 0.91 | 0.93 | 0.91 | 0.93 | 0.88 | 0.81 | |
| LD/LR | 0.58 | 0.5 | 0.73 | 0.54 | 0.74 | 0.74 | 0.42 | 0.26 | |
| SD/LD/LR | 0.66 | 0.58 | 0.72 | 0.64 | 0.76 | 0.74 | 0.81 | 0.72 | |
| Without O2 | NS/S | 0.75 | 0.69 | 0.83 | 0.8 | 0.67 | 0.62 | 0.82 | 0.76 |
| SI/SD | 0.89 | 0.88 | 0.91 | 0.93 | 0.91 | 0.93 | 0.88 | 0.81 | |
| LD/LR | 0.78 | 0.59 | 0.82 | 0.68 | 0.83 | 0.68 | 0.72 | 0.5 | |
| SD/LD/LR | 0.74 | 0.65 | 0.78 | 0.73 | 0.79 | 0.7 | 0.87 | 0.78 | |
| Without B1 | NS/S | 0.73 | 0.71 | 0.82 | 0.81 | 0.67 | 0.66 | 0.79 | 0.77 |
| SI/SD | 0.67 | 0.58 | 0.74 | 0.65 | 0.74 | 0.67 | 0.6 | 0.48 | |
| LD/LR | 0.74 | 0.53 | 0.79 | 0.64 | 0.79 | 0.68 | 0.7 | 0.37 | |
| SD/LD/LR | 0.58 | 0.41 | 0.59 | 0.48 | 0.59 | 0.48 | 0.73 | 0.66 | |
| Clinical Only | NS/S | 0.71 | 0.58 | 0.8 | 0.73 | 0.68 | 0.58 | 0.73 | 0.58 |
| SI/SD | 0.89 | 0.88 | 0.91 | 0.93 | 0.91 | 0.93 | 0.88 | 0.81 | |
| LD/LR | 0.73 | 0.53 | 0.79 | 0.64 | 0.8 | 0.68 | 0.65 | 0.37 | |
| SD/LD/LR | 0.72 | 0.6 | 0.77 | 0.7 | 0.78 | 0.7 | 0.85 | 0.74 | |
6. Discussion
AI-enhanced imaging, clinical and biological information proved capable to identify patients with severe short/long-term outcomes, bolstering healthcare resources under the extreme pressure of the current COVID-19 pandemic. The information obtained from our AI staging and prognosis could be used as an additional element at admission to assist decision making.
Variety of studies have reported the use of deep learning for the diagnosis and quantification of COVID-19 with CT scans. In particular, studies have already reported on deep learning diagnosing COVID-19 pneumonia on chest CTs. In Li et al., 2020 the authors proposed the use of a deep learning architecture based on ResNet50 for the diagnosis of COVID-19 reporting very high performances, while they investigated the attention maps produced from their network. A very similar method is presented in Mei et al. Mei et al., 2020 reporting the use of deep learning on COVID-19 diagnosis. Moreover, in Huang et al., 2020 the authors propose the use of a UNet architecture for the quantification of the disease using 14,482 slices for training and 5,303 slices for test, reporting a median DSC of 0.8481. Since their dataset is not publicly available, it is not possible to perform a direct comparison. A 3D deep learning architecture (DenseUNet) is proposed in Christe et al., 2020 for the quantification of COVID-19 disease. The segmentation is then used to regress a number of scores proposed in that study such as lung high opacity, lung severity, percentage of high opacity and percentage of opacity. Again, a direct comparison could not be reported, as the evaluation of the method was not performed using DSC or HD, since the performance was measured on the ability to regress the proposed scores. Finally, recently Tilborghs et al., 2020 presents a comparable study of deep learning based methods for the automatic quantification of COVID-19.
Assessing the severity of COVID-19 patients is also a very quickly evolving topic in the medical community with some methods being currently under review. Extracting valuable information from the imaging using recent advances is very important and could potentially facilitate the clinical practice. Starting with, disease extent is known to be associated with severity Li et al., 2020, Yuan et al., 2020 as well as that the disease textural heterogeneity reflects more the presence of heterogeneous lesions than pure ground glass opacities observable in mild cases. In Li et al., 2020c, the authors proposed the use of Siamese networks for the severity assessment of COVID-19 directly from CT scans. In Bai et al., 2020, the authors proposed a deep learning pipeline based on LSTMs using 2D CT slices and a fusion of imaging and clinical information to assess the severity and progression of COVID-19 patients. The proposed method reports an accuracy of 89.1% on a test cohort of 80 patients, outperforming classical machine learning techniques. Besides having a smaller test cohort, our method explores features that are interpretable helping better understanding of the disease and providing additional information for the staging of the patients. Moreover, recently Lassau et al., 2020 proposed the assessment of severity using a deep learning tool. Again, even if we can not perform a direct comparison our method reports similar performance in a completely independent cohort, while it is based on interpretable features extracted from different regions. Finally, in He et al., 2020 a 2D deep learning based approach using a multi-task learning is presented in order to separate COVID-19 patients to severe and non severe cases.
6.1. Clinical impact
To the best of our knowledge this study is the first to have developed a robust, holistic COVID-19 multi-omics signature for disease staging and prognosis demonstrating an equivalent/superior-to-human-reader performance on a multi-centric data set. Our approach complied appropriate data collection and methodological testing requirements beyond the existing literature Mei et al., 2020. The proposed holistic signature harnessed imaging descriptors of disease, underlying lung, heart and fat as well as biological and clinical data. Among them, disease extent is known to be associated with severity Li et al., 2020, Yuan et al., 2020, disease textural heterogeneity reflects more the presence of heterogenous lesions than pure ground glass opacities observable in mild cases. Heart features encode cardiomegaly and cardiac calcifications. Lung features show patients with severe disease having greater dispersion and heterogeneity of lung densities, reflecting the presence of an underlying airway disease such as emphysema and the presence of sub-radiological disease. Among clinical variables, a higher CRP level, lymphopenia and a higher prevalence of hypertension and diabetes were associated with a poorer outcome, consistent with previous reports Guo et al., 2020, Terpos et al., 2020, Zhou et al., 2020. Interestingly, age was less predictive of disease severity than of poor outcome in severe patients. This is linked to the fewer therapeutic possibilities for these generally more fragile patients. Lastly, the average body mass index (BMI) in both non-severe and severe groups corresponded to overweight. Despite being correlated with BMI, the fat ratio measured on the CT scanner was only weakly associated with outcome. Several studies have reported obesity to be associated with severe outcomes Huang et al., 2020, Christe et al., 2020 and an editorial described the measurement of anthropometric characteristics as crucial to better estimate the risk of complications Stefan et al., 2020. However a meta-analysis showed that whereas being associated with an increased risk of COVID-19 pneumonia, obesity was paradoxically associated with reduced pneumonia mortality Wynants et al., 2020. Overall, the combination of clinical, biological and imaging features demonstrates their complementary value for staging and prognosis.
6.2. Future work
In conclusion, we show that the combination of chest CT and artificial intelligence can provide tools for fast, accurate and precise disease extent quantification as well as the identification of patients with severe short-term outcomes. This could be of great help in the current context of the pandemic with healthcare resources under extreme pressure. Beyond the diagnostic value of CT for COVID-19, our study suggests that AI should be part of the triage process. Our methodology designed a deep learning-based pipeline that provides disease quantification comparable to the human experts, while it explores interpretable image characteristics, fusing them with clinical and biological data in order to perform staging of the patients to non severe, needed intubation and deceased. Our prognosis and staging method achieved state of the art results through the deployment of a highly robust ensemble classification strategy with the use of image characteristics and patients’ characteristics available within the image’ metadata. In terms of future work, we are planning to investigate and generate tools for the multiclass disease segmentation and investigate in depth the characteristics of each class and their association with severity. Our findings could have a strong impact in terms of (i) patient stratification with respect to the different therapeutic strategies, (ii) accelerated drug development through rapid, reproducible and quantified assessment of treatment response through the different mid/end-points of the trial, and (iii) continuous monitoring of patient’s response to treatment.
The use of deep features towards unsupervised discovery is also an interesting direction. Despite the absence of reported results in the paper, it should be noted that advanced deep learning techniques were considered both for classification/severity assessment (deep neural networks with attention, deep features from mid-level lung/disease 3D disease quantification networks) as well as for outcome prediction with explicit integration of clinical/biological variables. The interest of these methods was tested for biomarker discovery - subsequently fed to the ensemble learning method presented in the paper - and in an end-to-end setting towards automatic quantification, staging and outcome prediction. Despite interesting performance on training, both approaches failed to produce similar equivalent performance on the test cohorts, while their results were clearly inferior in terms of overall performance, explicability, robustness and generalizability with respect to the reported solution. This could be explained from the relatively low number of samples in the training which is a known bottleneck for deep representations. Access to a significantly larger cohort with at least one order of magnitude higher order number of samples is under examination within the Assistance Publique - Hopitaux de Paris hospitals network. The use of such a cohort could be of great interest for confirming the outcomes of the presented study as well as for reviving the interest of deep features and holistic end-to-end integration of deep features with biological/clinical and imaging data for staging and short/long term outcome prediction.
CRediT authorship contribution statement
Guillaume Chassagnon: Conceptualization, Methodology, Data curation, Formal analysis, Investigation, Writing - original draft. Maria Vakalopoulou: Conceptualization, Methodology, Software, Formal analysis, Investigation, Writing - original draft. Enzo Battistella: Methodology, Software, Formal analysis, Investigation, Writing - original draft. Stergios Christodoulidis: Methodology, Software, Formal analysis, Investigation, Writing - original draft. Trieu-Nghi Hoang-Thi: Data curation, Writing - review & editing. Severine Dangeard: Data curation, Writing - review & editing. Eric Deutsch: Validation, Writing - review & editing. Fabrice Andre: Validation, Writing - review & editing. Enora Guillo: Data curation, Writing - review & editing. Nara Halm: Data curation, Writing - review & editing. Stefany El Hajj: Data curation, Writing - review & editing. Florian Bompard: Data curation, Writing - review & editing. Sophie Neveu: Data curation, Writing - review & editing. Chahinez Hani: Data curation, Writing - review & editing. Ines Saab: Data curation, Writing - review & editing. Aliénor Campredon: Data curation, Writing - review & editing. Hasmik Koulakian: Data curation, Writing - review & editing. Souhail Bennani: Data curation, Writing - review & editing. Gael Freche: Data curation, Writing - review & editing. Maxime Barat: Data curation, Writing - review & editing. Aurelien Lombard: Software, Writing - review & editing. Laure Fournier: Data curation, Writing - review & editing. Hippolyte Monnier: Data curation, Writing - review & editing. Téodor Grand: Data curation, Writing - review & editing. Jules Gregory: Data curation, Writing - review & editing. Yann Nguyen: Data curation, Writing - review & editing. Antoine Khalil: Data curation, Writing - review & editing. Elyas Mahdjoub: Data curation, Writing - review & editing. Pierre-Yves Brillet: Data curation, Writing - review & editing. Stéphane Tran Ba: Data curation, Writing - review & editing. Valérie Bousson: Data curation, Writing - review & editing. Ahmed Mekki: Data curation, Writing - review & editing. Robert-Yves Carlier: Data curation, Writing - review & editing. Marie-Pierre Revel: Conceptualization, Methodology, Supervision, Data curation, Validation, Investigation, Writing - original draft. Nikos Paragios: Conceptualization, Methodology, Supervision, Validation, Investigation, Formal analysis, Writing - original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Mihir Sahasrabudhe and Norbert Bus for their valuable comments. This project was partially supported from the European Union’s Horizon 2020 research and innovation programme under grant agreement no.880314, the Fondation pour la Recherche M\'edicale (FRM; no. DIC20161236437), the Swiss National Science Foundation Grant no.188153 and benefited from methodological developments done in the context of Dr. Guillaume Chassagnon thesis (2016–2019) supported from GE Healthcare.
Appendix A. Tables for the prognosis and staging
Tables A.9 and A.10 summarise the performance of our top-5 classifiers and their ensemble for the staging of the N/NS and SI/SD patients.
Table A.9.
Performance for the Severe (S) and Non-Severe (NS) short-term outcome for each of the top-5 selected classifiers and their ensemble presented in Section 5. Note: L-SVM, support vector machine with a linear kernel; RBF-SVM, support vector machine with a radial basis function kernel.
| Classifier | Balanced accuracy |
Weighted pecision |
Weighted sensitivity |
Weighted specificity |
||||
|---|---|---|---|---|---|---|---|---|
| Train | Test | Train | Test | Train | Test | Train | Test | |
| L-SVM | 0.7 | 0.67 | 0.79 | 0.78 | 0.71 | 0.71 | 0.69 | 0.64 |
| RBF-SVM | 0.75 | 0.68 | 0.82 | 0.79 | 0.7 | 0.67 | 0.79 | 0.7 |
| Decision Tree | 0.71 | 0.67 | 0.82 | 0.82 | 0.61 | 0.53 | 0.81 | 0.81 |
| Random Forest | 0.72 | 0.68 | 0.81 | 0.79 | 0.69 | 0.69 | 0.75 | 0.68 |
| AdaBoost | 0.72 | 0.67 | 0.83 | 0.82 | 0.63 | 0.54 | 0.82 | 0.81 |
| Ensemble Classifier | 0.73 | 0.7 | 0.82 | 0.81 | 0.67 | 0.64 | 0.8 | 0.77 |
Table A.10.
Performance for the Intubated (SI) and Deceased (SD) patients in the short-term outcome outcome for each of the top-5 selected classifiers and their ensemble presented in Section 5. Note: P-SVM, support vector machine with a polynomial kernel.
| Classifier | Balanced accuracy |
Weighted pecision |
Weighted sensitivity |
Weighted specificity |
||||
|---|---|---|---|---|---|---|---|---|
| Train | Test | Train | Test | Train | Test | Train | Test | |
| P-SVM | 0.88 | 0.7 | 0.89 | 0.76 | 0.84 | 0.74 | 0.92 | 0.67 |
| Decision Tree | 0.9 | 0.88 | 0.92 | 0.94 | 0.92 | 0.94 | 0.88 | 0.81 |
| Random Forest | 0.9 | 0.81 | 0.92 | 0.91 | 0.92 | 0.9 | 0.88 | 0.81 |
| AdaBoost | 0.9 | 0.88 | 0.92 | 0.94 | 0.92 | 0.94 | 0.88 | 0.81 |
| Gaussian Process | 0.95 | 0.77 | 0.96 | 0.83 | 0.96 | 0.84 | 0.94 | 0.7 |
| Ensemble Classifier | 0.9 | 0.88 | 0.92 | 0.94 | 0.92 | 0.94 | 0.88 | 0.81 |
References
- Abadi, M., Agarwal, A., Barham, P., Brevdo, E., Chen, Z., Citro, C., Corrado, G. S., Davis, A., Dean, J., Devin, M., Ghemawat, S., Goodfellow, I. J., Harp, A., Irving, G., Isard, M., Jia, Y., Józefowicz, R., Kaiser, L., Kudlur, M., Levenberg, J., Mané, D., Monga, R., Moore, S., Murray, D. G., Olah, C., Schuster, M., Shlens, J., Steiner, B., Sutskever, I., Talwar, K., Tucker, P. A., Vanhoucke, V., Vasudevan, V., Viégas, F. B., Vinyals, O., Warden, P., Wattenberg, M., Wicke, M., Yu, Y., Zheng, X., 2016. Tensorflow: large-scale machine learning on heterogeneous distributed systems. CoRR abs/1603.04467
- Anthimopoulos M., Christodoulidis S., Ebner L., Christe A., Mougiakakou S. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans. Med. Imaging. 2016;35(5):1207–1216. doi: 10.1109/TMI.2016.2535865. [DOI] [PubMed] [Google Scholar]
- Anthimopoulos M., Christodoulidis S., Ebner L., Geiser T., Christe A., Mougiakakou S. Semantic segmentation of pathological lung tissue with dilated fully convolutional networks. IEEE J. Biomed. Health Inform. 2018;23(2):714–722. doi: 10.1109/JBHI.2018.2818620. [DOI] [PubMed] [Google Scholar]
- Badrinarayanan V., Kendall A., Cipolla R. Segnet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017;39(12):2481–2495. doi: 10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
- Bai, X., Fang, C., Zhou, Y., Bai, S., Liu, Z., Xia, L., Chen, Q., Xu, Y., Xia, T., Gong, S., et al., 2020. Predicting COVID-19 malignant progression with ai techniques. [DOI] [PMC free article] [PubMed]
- Bermejo-Peláez D., Ash S.Y., Washko G.R., Estépar R.S.J., Ledesma-Carbayo M.J. Classification of interstitial lung abnormality patterns with an ensemble of deep convolutional neural networks. Sci. Rep. 2020;10(1):1–15. doi: 10.1038/s41598-019-56989-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocchino M., Bruzzese D., D’Alto M., Argiento P., Borgia A., Capaccio A., Romeo E., Russo B., Sanduzzi A., Valente T., et al. Performance of a new quantitative computed tomography index for interstitial lung disease assessment in systemic sclerosis. Sci. Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-45990-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti, S., Balachandran, A., Chabin, G., Cohen, S., Flohr, T., Georgescu, B., Grenier, P., Grbic, S., Liu, S., Mellot, F., et al., 2020. Quantification of tomographic patterns associated with COVID-19 from chest CT. arXiv preprint arXiv:2004.01279 [DOI] [PMC free article] [PubMed]
- Chassagnon G., Vakalopolou M., Paragios N., Revel M.-P. Deep learning: definition and perspectives for thoracic imaging. Eur. Radiol. 2020;30:2021–2030. doi: 10.1007/s00330-019-06564-3. [DOI] [PubMed] [Google Scholar]
- Christe A., Peters A.A., Drakopoulos D., Heverhagen J.T., Geiser T., Stathopoulou T., Christodoulidis S., Anthimopoulos M., Mougiakakou S.G., Ebner L. Computer-aided diagnosis of pulmonary fibrosis using deep learning and CT images. Investig. Radiol. 2019;54(10):627. doi: 10.1097/RLI.0000000000000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çiçek Ö., Abdulkadir A., Lienkamp S.S., Brox T., Ronneberger O. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2016. 3D U-Net: learning dense volumetric segmentation from sparse annotation; pp. 424–432. [Google Scholar]
- Cottin V., Brown K.K. Interstitial lung disease associated with systemic sclerosis (SSC-ILD) Respir. Res. 2019;20(1):13. doi: 10.1186/s12931-019-0980-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depeursinge A., Chin A.S., Leung A.N., Terrone D., Bristow M., Rosen G., Rubin D.L. Automated classification of usual interstitial pneumonia using regional volumetric texture analysis in high-resolution CT. Investig. Radiol. 2015;50(4):261. doi: 10.1097/RLI.0000000000000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante E., Dokania P.K., Marini R., Paragios N. International Workshop on Machine Learning in Medical Imaging. Springer; 2017. Deformable registration through learning of context-specific metric aggregation; pp. 256–265. [Google Scholar]
- Gangeh M.J., Sørensen L., Shaker S.B., Kamel M.S., de Bruijne M., Loog M. In: Medical Image Computing and Computer-Assisted Intervention – MICCAI 2010. Jiang T., Navab N., Pluim J.P.W., Viergever M.A., editors. Springer Berlin Heidelberg; Berlin, Heidelberg: 2010. A texton-based approach for the classification of lung parenchyma in CT images; pp. 595–602. [DOI] [PubMed] [Google Scholar]
- Gao M., Bagci U., Lu L., Wu A., Buty M., Shin H.-C., Roth H., Papadakis G.Z., Depeursinge A., Summers R.M., et al. Holistic classification of CT attenuation patterns for interstitial lung diseases via deep convolutional neural networks. Comput. Methods Biomech. Biomed. Eng. 2018;6(1):1–6. doi: 10.1080/21681163.2015.1124249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, K., Zhao, W., Xie, X., Ji, W., Liu, M., Tang, Z., Shi, F., Gao, Y., Liu, J., Zhang, J., et al., 2020. Synergistic learning of lung lobe segmentation and hierarchical multi-instance classification for automated severity assessment of COVID-19 in CT images. arXiv preprint arXiv:2005.03832 [DOI] [PMC free article] [PubMed]
- Guo W., Li M., Dong Y., Zhou H., Zhang Z., Tian C., Qin R., Wang H., Shen Y., Du K., et al. Diabetes is a risk factor for the progression and prognosis of COVID-19. Diabetes/Metab. Res. Rev. 2020;36(7):e3319. doi: 10.1002/dmrr.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Han R., Ai T., Yu P., Kang H., Tao Q., Xia L. Serial quantitative chest CT assessment of COVID-19: deep-learning approach. Radiology. 2020;2(2):e200075. doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M.B., Bunte K., Nagarajan M.B., Biehl M., Ray L.A., Wismüller A. Texture feature ranking with relevance learning to classify interstitial lung disease patterns. Artif. Intell. Med. 2012;56(2):91–97. doi: 10.1016/j.artmed.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb M., Collard H.R. Staging of idiopathic pulmonary fibrosis: past, present and future. Eur. Respir. Rev. 2014;23(132):220–224. doi: 10.1183/09059180.00002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafata K.J., Zhou Z., Liu J.-G., Hong J., Kelsey C.R., Yin F.-F. An exploratory radiomics approach to quantifying pulmonary function in CT images. Sci. Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-48023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassau, N., Ammari, S., Chouzenoux, E., Gortais, H., Herent, P., Devilder, M., Soliman, S., Meyrignac, O., Talabard, M.-P., Lamarque, J.-P., et al., 2020. Ai-based multi-modal integration of clinical characteristics, lab tests and chest CTs improves COVID-19 outcome prediction of hospitalized patients. medRxiv.
- Li K., Fang Y., Li W., et al. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19) Eur. Radiol. 2020;30:4407–4416. doi: 10.1007/s00330-020-06817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. D., Arun, N. T., Gidwani, M., Chang, K., Deng, F., Little, B. P., Mendoza, D. P., Lang, M., Lee, S. I., O’Shea, A., et al., 2020c. Automated assessment of COVID-19 pulmonary disease severity on chest radiographs using convolutional siamese neural networks. medRxiv. [DOI] [PMC free article] [PubMed]
- Li L., Qin L., Xu Z., et al. Using Artificial Intelligence to Detect COVID-19 and Community-acquired Pneumonia Based on Pulmonary CT. Radiology. 2020;296(2):E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei X., Lee H.-C., Diao K.-y., Huang M., Lin B., Liu C., Xie Z., Ma Y., Robson P.M., Chung M., et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020;26:1–5. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020;323(18):1775–1776. doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 2011;12(Oct):2825–2830. [Google Scholar]
- Robbie H., Daccord C., Chua F., Devaraj A. Evaluating disease severity in idiopathic pulmonary fibrosis. Eur. Respir. Rev. 2017;26(145):170051. doi: 10.1183/16000617.0051-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger O., Fischer P., Brox T. International Conference on Medical image computing and computer-assisted intervention. Springer; 2015. U-Net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- Stefan N., Birkenfeld A.L., Schulze M.B., et al. Obesity and impaired metabolic health in patients with COVID-19. Nat. Rev. Endocrinol. 2020;16:341–342. doi: 10.1038/s41574-020-0364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R., Limkin E.J., Vakalopoulou M., Dercle L., Champiat S., Han S.R., Verlingue L., Brandao D., Lancia A., Ammari S., et al. A radiomics approach to assess tumour-infiltrating cd8 cells and response to anti-pd-1 or anti-pd-l1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19(9):1180–1191. doi: 10.1016/S1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
- Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020;18(4):844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilborghs, S., Dirks, I., Fidon, L., Willems, S., Eelbode, T., Bertels, J., Ilsen, B., Brys, A., Dubbeldam, A., Buls, N., et al., 2020. Comparative study of deep learning methods for the automatic segmentation of lung, lesion and lesion type in CT scans of COVID-19 patients. arXiv preprint arXiv:2007.15546
- Terpos E., Ntanasis-Stathopoulos I., Elalamy I., Kastritis E., Sergentanis T.N., Politou M., Psaltopoulou T., Gerotziafas G., Dimopoulos M.A. Hematological findings and complications of COVID-19. Am. J. Hematol. 2020;95(7):834–847. doi: 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomassetti S., Ryu J.H., Poletti V. Staging systems and disease severity assessment in interstitial lung diseases. Curr. Opin. Pulm. Med. 2015;21(5):463–469. doi: 10.1097/MCP.0000000000000198. [DOI] [PubMed] [Google Scholar]
- Vakalopoulou M., Chassagnon G., Bus N., Marini R., Zacharaki E.I., Revel M.-P., Paragios N. In: Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Frangi A.F., Schnabel J.A., Davatzikos C., Alberola-López C., Fichtinger G., editors. Springer International Publishing; Cham: 2018. Atlasnet: multi-atlas non-linear deep networks for medical image segmentation; pp. 658–666. [Google Scholar]
- Van Griethuysen J.J., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., Beets-Tan R.G., Fillion-Robin J.-C., Pieper S., Aerts H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Kim G.H., Salisbury M.L., Barber D., Bartholmai B.J., Brown K.K., Conoscenti C.S., De Backer J., Flaherty K.R., Gruden J.F., et al. Computed tomographic biomarkers in idiopathic pulmonary fibrosis. the future of quantitative analysis. Am. J. Respir. Critical Care Med. 2019;199(1):12–21. doi: 10.1164/rccm.201803-0444PP. [DOI] [PubMed] [Google Scholar]
- Wynants L., Van Calster B., Bonten M.M., Collins G.S., Debray T.P., De Vos M., Haller M.C., Heinze G., Moons K.G., Riley R.D., et al. Prediction models for diagnosis and prognosis of COVID-19 infection: systematic review and critical appraisal. BMJ. 2020;369 doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Yin W., Tao Z., Tan W., Hu Y. Association of radiologic findings with mortality of patients infected with 2019 novel coronavirus in Wuhan, China. PLoS One. 2020;15(3):e0230548. doi: 10.1371/journal.pone.0230548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;385(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in china, 2019. New Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]