Abstract
Rapid person-to-person transfer of viruses such as SARS-CoV-2 and their occasional mutations owing to the human activity and climate/ecological changes by the mankind led to creation of wrecking worldwide challenges. Such fast transferable pathogens requiring practical diagnostic setups to control their transfer chain and stop sever outbreaks in early stages of their appearance. Herein, we have addressed this urgent demand by designing a rapid electrochemical diagnostic kit composed of fixed/screen printed electrodes that can detect pathogenic viruses such as SARS-CoV-2 and/or animal viruses through the differentiable fingerprint of their viral glycoproteins at different voltage positions. The working electrode of developed sensor is activated upon coating a layer of coupled graphene oxide (GO) with sensitive chemical compounds along with gold nanostars (Au NS) that can detect the trace of viruses in any aquatic biological media (e.g., blood, saliva and oropharyngeal/nasopharyngeal swab) through interaction with active functional groups of their glycoproteins. The method do not require any extraction and/or biomarkers for detection of target viruses and can identify trace of different pathogenic viruses in about 1 min. The nanosensor also demonstrated superior limit of detection (LOD) and sensitivity of 1.68 × 10−22 μg mL−1 and 0.0048 μAμg.mL−1. cm−2, respectively, toward detection of SARS-CoV-2 in biological media, while blind clinical evaluations of 100 suspected samples furtherly confirmed the superior sensitivity/specificity of developed nanosystem toward rapid identification of ill people even at incubation and prodromal periods of illness.
Keywords: SARS-CoV-2, Virus, Outbreak, Electrochemical detection, Nanosensor
Graphical abstract
Highlights
A nanosensor based on graphene and gold nanostars was developed toward detection of viruses
The developed nanosensor detected diverse viruses through their viral glycoproteins
The nanosensor showed different fingerprints for each examined virus
The nanosensor detected the SARS-CoV-2 with high accuracy and low DL
1. Introduction
Viruses and their resulting infectious disease have caused severe problems for the modern mankind and animals due to their rapid distribution and mutations. Among these viruses, influenza virus, dengue virus, hepatitis virus, Japanese encephalitis virus, human immune deficiency virus (HIV) and corona viruses can be mentioned that are capable to spread very fast among living species and create worldwide scale threat for the public health. Viruses could invade the host via different mechanisms, including retaliation to the machinery and metabolism of host cells in case of their self-replication. Furthermore, human activity, artificial climate/ecological changes by the mankind, movement of pathogens between animal species to human being and demographic shifts led to creation of a wide variety of viral infectious illnesses (Fenton and Pedersen 2005). Thereby, created troubles and destructive consequences by wide speared of viral infectious diseases raised the global concern for development of state of the art diagnostic platforms capable of detecting the trace of viruses in early stages of their infection. Such approach provide the possibility to stop the main distribution chain of viruses and control them before turning into an out of control pandemic (Kaushik 2019; Mujawar et al., 2020; Spackman 2012).
For instance, novel coronavirus (SARS-CoV-2) can be mentioned as a recent viral pandemic that affected all aspects of the human life and led to a large rate of death tolls worldwide due to the lack of rapid and accurate detection platforms toward pausing its person-to-person transfer chain (Li et al., 2020; Xu et al., 2020). Similarly, the greatest problem regarding this virus is its silent stage that leads to high rate of infection worldwide which require developing a rapid/sensitive diagnostic test to detect ill people in early stages of the disease to avoid its further distribution. In Fig. S1, a view of diverse kinds of coronaviruses and their respective categories can be seen.
What is more, glycoproteins as one of the most abundant post-translational enhancement of proteins within the nature playing vital roles in various biological processes of living organisms, e.g., cell signaling, molecular recognition, immune response, cellular component and enzymatic reactions (Alley et al., 2013; Paleček et al., 2015). Glycoproteins containing glycosylated parts composed of oligosaccharide chains linked to polypeptides (Cole and Smith 1989). These useful proteins were also selected as illness biomarkers and/or therapeutic metrics for development of clinical diagnostic kits (Rudd et al., 2001). Thereby, design and fabrication of practical methods for tracing and quantifying glycoprotein-based structures in biological specimens are so important.
Quantitation/identification of glycoproteins can be used as an essential biomarker for early detection of pathologies processes, while its increasing content within biological samples can be used as a promising biological marker (Ye et al., 2014; Zhang et al., 2014). So far, various kinds of techniques have been designed/developed for identification/quantification of glycoproteins within aquatic biological matters including enzyme-linked immune-sorbent assay (ELISA), capillary electrophoresis, high-performance anion exchange chromatography and liquid chromatography (Lequin 2005; Stavenhagen et al., 2015; Wang et al., 2015; Yang et al., 2015; Zhang and Hage 2016). These techniques providing some advantages for detection of target glycoprotein-based structures, however, a majority of them suffering from expensive cost, complex specimen pretreatment, time-consuming processes, requirement for skilled personnel, poor physical or chemical stability and difficult processes for obtaining anti-bodies which restrict their applicability in time of difficulties/pandemics. Therefore, its bare bone essential to fabricate a highly sensitive/selective assay with superior robustness, ease of preparation, economic cost for consumer and long-term stability in comparison with natural biological receptors (i.e., anti-boy/anti-gen and enzyme) as a fast and reliable screening technique (Chen et al., 2016).
Furthermore, recently developed biosensors for detection of pathogens such as viruses are mainly based on biological reagents such as antibody, DNA, antigen, cells and etc. requiring time consuming and costly processes for extraction and/or fabrication of target biological metrics/reagents (Cheng and Toh 2013). In this matter, in order to stop the transfer chain of fast transferable pathogenic viruses such as SARS-CoV-2, its vital to consider a part of viral structure that do not require extraction and can be detected via practical nanotechnological approaches.
Additionally, the nastiest thing about coronavirus is its rapid person-to-person transmission that require serious actions to prevent its further distribution among people (Chan et al., 2020). In this matter, health care authorities mostly using time consuming traditional approaches that are not match with high infection speed of SARS-CoV-2 or exhibit low sensitivity/specificity which cannot be used for practical separation of ill people from healthy ones in least time span. Herein, we have addressed this demand via developing a rapid and highly sensitive diagnostic kit that do not require any extraction or biological marker and can detect trace of different kinds of pathogenic animal/human viruses such as SARS-CoV-2, infectious bronchitis virus (IBV), avian influenza and Newcastle Disease Virus (LaSota and V4 strains) via active functional groups of their viral glycoproteins upon demonstration of differentiable fingerprints of each virus at diverse voltage positions. The active part of developed kit is consisted of decorated graphene oxide (GO) with 8-hydroxyquinoline (8H), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and N-hydroxysuccinimide (NHS) (GO-8H-EDC-NHS) coupled with gold nanostars (Au NS). Additionally, the validity of the platform was also proved via performing real blind examinations on biological samples that were examined via both fixed and/or screen printed electrodes.
2. Results and discussions
2.1. Detection of viruses within biological media
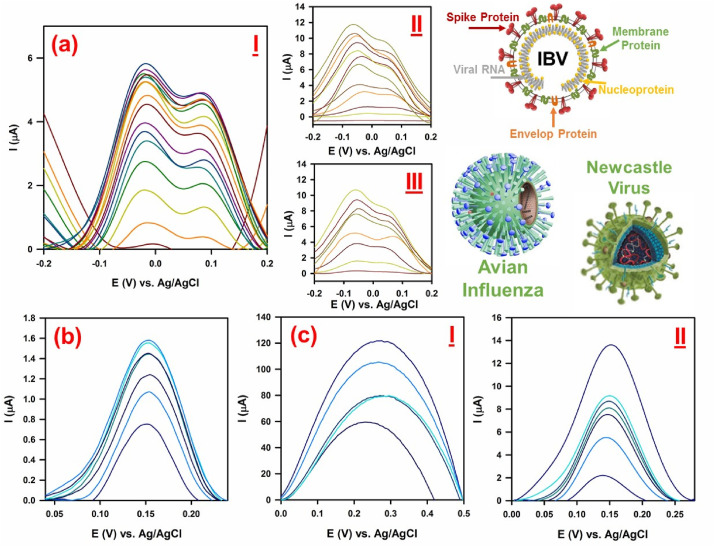
In this section, the applicability of fabricated nanosensor for rapid detection of viruses from their viral spike glycoproteins is evaluated. From Fig. 1 (a) (I) it is clearly revealed that the current signals were noticed at the surface of modified GCE by enhancing the virus concentrations from 5 × 10−16 EID50 to 5 × 10−13 EID50, which approved the performance of developed electrocatalyst for detection of viral S spike glycoproteins of gammacoronavirus (i.e., IBV) in biological samples. In this matter, twine appeared peaks at about −0.017 V and 0.086 V are related to the finger print of S spike glycoprotein of IBV.
Fig. 1.
(a) (I) DPV analysis of gammacoronavirus (i.e., Infectious bronchitis viruses (IBV)) in PBS (pH = 7.4), (II) DPV analysis of bronchitis viruses' S-spike in PBS with pH 7.4 and (III) DPV analysis of bronchitis viruses' S-spike in human plasma; (b) DPV analysis of Avian influenza in PBS with pH = 7.4; (c) Newcastle disease virus (I) LaSota strain and (II) V4 strain in PBS with pH = 7.4; DPV conditions: Pulse amplitude: 25 mV and the width of pulse: 50 ms.
For further admittance of electrode toward sensing viral S spike glycoproteins and obtaining analytical figures of merits, the calibration curve of modified electrode was obtained in PBS (pH 7.4) and human plasma sample as biological aquatic specimens. The quantitative analysis of viruses was performed by constructing differential pulse voltammograms of viral S spike glycoproteins through variating concentrations of virus in optimized condition which can be seen in Fig. 1 (a) (II) and (III); the respective calibration curves can be seen in Fig. S10 (a) and (b). As illustrated within these Figures, the currents of viral S spike glycoproteins showed a regular increase with respect to the increase in the concentration of virus, while their standard calibration curve was obtained from plotting the value of virus's concentration versus height of peak currents (Fig. S10 (a) and (b)). The calibration curves of viral S spike glycoproteins signals were obtained in the range of 1.6 × 10−16 - 2.9 × 10−14 EID50 and 4 × 10−16 - 0.8 × 10−14 EID50, in PBS and plasma samples, respectively, whereas, the corresponding DL/sensitivity was estimated to be 0.53 × 10−16 EID50/0.8 μA.EID50. cm−2 and 1.2 × 10−16 EID50/2.204 μA.EID50. cm−2 for appeared peak currents in buffer and human plasma samples, respectively. It was also found that the developed sensor based on GO-8H-EDC–NHS–Au NS exhibited a low detection limit and high sensitivity towards detection of viral S spike glycoproteins of IBV within aquatic media.
For further evaluation of nanosensor's performance, two different viruses, i.e., avian influenza and Newcastle virus were examined. In Fig. 1 (b), DPV pattern of avian influenza can be seen; the respective calibration curve is depicts within Fig. S10 (c). As shown, avian influenza showed its fingerprint at 0.154 V with completely different pattern compared with IBV. Additionally, the nanosensor showed LOD and sensitivity of 0.342 × 10−13 EID50 and 0.0944 μA.EID50. cm−2 toward detection of avian influenza, respectively, which furtherly confirmed the superior sensitivity and specificity of developed platform toward detection of viruses. Moreover, two different strains of Newcastle virus, i.e., LaSota (Fig. 1 (c) (I)) and V4 (Fig. 1 (c) (II)) were examined. As illustrated, both strains of Newcastle virus showed their fingerprint at different positions with diverse patterns. In this regard, fingerprint of LaSota and V4 strains appeared at 0.270 and 0.149 V, respectively, with completely different patterns. As depicts in Fig. 1 (c) (I) and its respective calibration curve in Fig. S10 (d), the developed nanosensor can detect the LaSota strain with high accuracy, where the LOD and sensitivity of the nanosenor toward detection of this strain measured to be 0.014 × 10−13 EID50 and 43.12 μA.EID50. cm−2, respectively. Same thing happened for V4 strain (Fig. 1 (c) (II)), while the sensor showed much more sensitivity toward its detection with far less limit of detection. In this matter, by referring to the calibration curve of V4 strain (Fig. S10 (e)), the LOD and sensitivity of nanosensor toward this strain of Newcastle virus calculated to be 0.00956 × 10−20 EID50 and 323.868 μA.EID50. cm−2, respectively.
These outcomes showed that, the developed sensor not only can detect different types of viruses at different positions but also their diverse strains exhibiting different fingerprints. Such method can be used as a fast viral mapping platform to rapidly detect viruses based on the electrochemical pattern/fingerprint of their surface glycoproteins and/or S spike glycoproteins.
The superior catalytic performance of GCE-GO-8H-EDC–NHS–Au NS could be due to the superior adsorptive ability of hybrid Au NS and GO-8H-EDC-NHS, where the Au NS facilitate the electronic transfers and GO-8H-EDC-NHS provides higher specific surface area along with ideal sensitivity toward detection of viral spike glycoproteins owing to its introduced active functional groups upon attachment of 8H, EDC and NHS on its surface. In this regard, GO-8H-EDC–NHS–Au NS electrocatalyst can provide more adsorptive capability for viral S spike glycoproteins via various interactions such as formation of hydrogen bonds between functional groups on nano-based catalyst with functional groups of protein (amino and acidic) and probably electrostatic interactions.
For evaluating the repeatability of designed nanosensor, modified GCE with GO-8H-EDC–NHS–Au NS was applied for detection of 2.0 × 10−14 EID50 IBV through ten-fold continuous dilutions and detection in PBS with pH 7.4, where the relative standard deviation (RSD) was calculated to be 4.02%. Furthermore, two different GCE working electrodes were modified independently in identical approach and corresponding RSD of fabricated nanosensors was calculated to be 4.46%. Achieved data showed the ultra-high sensitivity of developed nanosensor that can detect gammacoronaviruses (i.e., IBV) within biological media through dividable and accurate approach based on their fingerprints.
Besides, the overall influence of typical electroactive interferences within the real biological fluid on the current responses of 2.0 × 10−14 EID50 coronavirus have been investigated via adding 0.1 mM interfering biomolecules such as ascorbic acid (AA), glucose and urea. The obtained results did not show any change in the Ip of viral S spike glycoproteins that highlighting the predominant ability of the nanosensor for precise detection of viruses upon the presence of possible interferences compounds (Fig. S11).
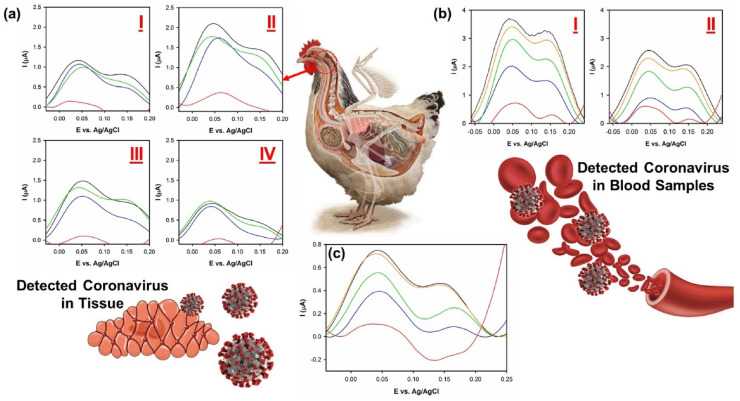
For further evaluation of developed nanosensor toward detection of gammacoronavirus (i.e., IBV) within real biological samples, seven specimens (4 oropharyngeal swabs, 2 blood samples and 1 tracheal mucosa layer) from different infected chickens with wild strain of IBV were prepared. In Fig. 2 , a view of obtained results can be seen. As shown in Fig. 2 (a) (I-IV), all of extracted oropharyngeal swabs showed fingerprint of IBV related to the pattern of S spike glycoprotein of coronavirus at around 0.04 and 0.14 V, respectively. Moreover, as can be seen in Fig. 2 (b) (I and II), both blood samples showed trace of gammacoronavirus at same peaks which are correspond to the fingerprint of virus's glycoproteins. These data showed that the developed biosensor can detect even lowest trace of coronavirus within biological specimens which is a bare bone essential feature for developing practical diagnostic setups.
Fig. 2.
Detection of wild version of IBV from (a) oropharyngeal swab, (b) blood and (c) tracheal mucosa layer of infected chickens. DPV conditions: Pulse amplitude: 25 mV and the width of pulse: 50 ms.
In addition, a sample of tracheal mucosa layer is extracted from an infected bird with wild strain of IBV. The obtained sample is placed within 1 mL PBS (pH = 7.4) and kept stationary till extraction of viruses from tissue. Thence, the sample was shook for 10 min and trace of virus was detected within the aquatic media. Surprisingly, coronavirus is successfully extracted from the tissue into the aquatic media and its fingerprint was detected via the developed nanosensor (Fig. 2 (c)); however, the concentration of coronavirus within the tissue is significantly lower than blood and swap samples. This outcome showed that the virus hide inside the tissue and duplicate itself which shows the dangerous aspect of coronaviruses. In this matter, the treated patient should be kept under watch for a specific time span even after removal of main signs of illness.
In addition, the developed nanosensor can also determine the exact population of viruses within biological samples based on EID50 unit, where the current approach determine the EID50 unit of samples much more faster than traditional approaches. In this matter, related data for examined real samples extracted from infected birds with wild version of IBV can be seen in Table S2. As shown, the lowest concentration of viruses is related to the tracheal mucosa layer, while oropharyngeal swab and blood samples showed different/higher concentrations of viruses based on the stage of illness. In this matter, by measuring the exact population of viruses within biological samples in different stages of illness, a road map for progress of disease can be drawn and the stage of illness could be exactly determined based on the extracted data from experimental evaluations via the developed nanosensor.
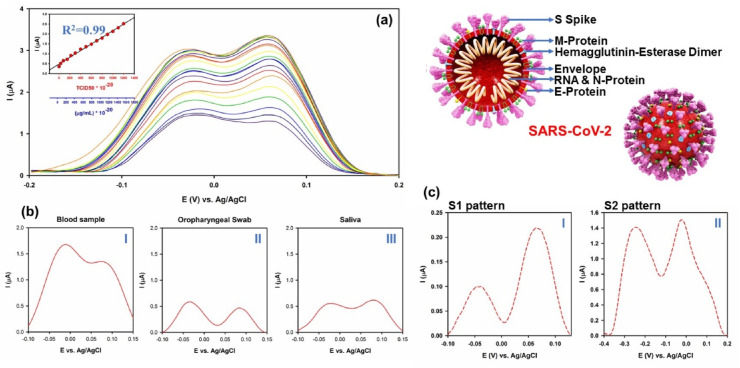
After successful detection of IBV, avian influenza and Newcastle viruses, the nanosensor is applied toward detection of SARS-CoV-2 within biological media. For this matter, antigen of the whole virus was isolated and its finger print was recorded via the electrochemical sensor. In Fig. 3 (a), the DPV pattern of SARS-CoV-2 along with its calibration curve and structure can be seen. As depicts, SARS-CoV-2 showed a twine pattern with peaks at −0.0367 and 0.0689 V which is similar to pattern of IBV but appeared at different positions. More importantly, the developed nanosensor showed fantastic LOD and sensitivity of 1.68 × 10−22 μg mL−1 and 0.0048 μAμg.mL−1. cm−2, respectively (by considering the approximate weight of each SAR-CoV-2 virus to be 85 ag). These outcomes showed the extremely high sensitivity of the nanosensor toward detection of SARS-CoV-2 than other types of viruses and their strains which can be used as an ideal and rapid platform toward detection of infected people even in the common and/or silent stages of the illness. In Fig. 3 (b) (I-III), the DPV pattern of SARS-CoV-2 in blood, swab and saliva samples of an infected person with COVID-19 can be seen. This outcome shows another great potential of the sensor that can detect trace of viruses in diverse aquatic biological media which is a vital matter for studying the behavior of virus in different stages of the disease.
Fig. 3.
(a) DPV pattern of SARS-CoV-2 virus in PBS (pH = 7.4) along with its respective calibration curve and structure of SARS-CoV-2 virus, (b) DPV pattern of SARS-CoV-2 in obtained samples from (I) blood, (II) oropharyngeal swab and (III) saliva of an infected person; and (c) obtained DPV patterns from (I) S1 and (II) S2 glycoproteins' antigen related to S spike glycoprotein of SARS-CoV-2 virus.
Furthermore, DPV pattern of isolated S1 and S2 parts of SARS-CoV-2 S spike glycoproteins were also evaluated which can be seen in Fig. 3 (c) (I) and (II), respectively. As illustrated, the position and pattern of SARS-CoV-2 (Fig. 3 (a)) is more likely similar to the pattern of S1 which shows that the virus is attach to the sensor via the S1 part of its S spike glycoprotein.
2.2. Detection mechanism of viral glycoproteins
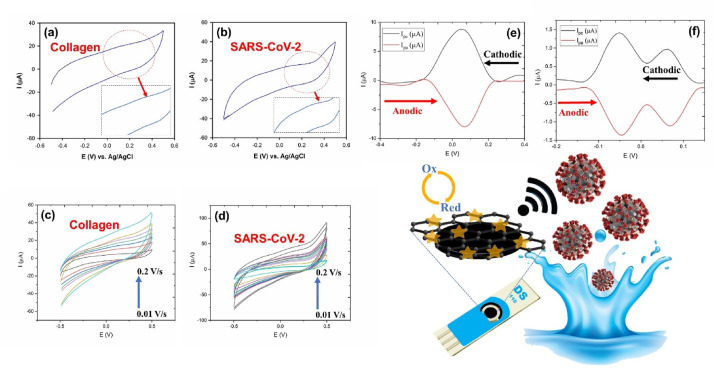
The possible mechanism for adsorption of collagen and/or SARS-CoV-2 on the modified electrode was investigated by CV measurements. The CV procedure is one of the most common diagnostic approaches in characterization of target chemical compounds and evaluation of electrochemical mechanism of chemical reactions. To investigate the catalytic mechanism of the proposed method, the impact of potential scan rate on the electrocatalytic processes of type I collagen of human as well as viral structure of SARS-CoV-2 was first surveyed using CV technique. As illustrated in Fig. 4 (a) and (b), with enhancing the scan rate from 0.01 to 0.2 V s−1, very weak current signal (zoomed insert of parts (a) and (b)) are hardly observed. Variations of the anodic currents vs. scan rate showed that the reaction is governed via confined-surface reactions and adsorption electron transfer process on the surface of electrode.
Fig. 4.
CV analysis of (a) human type I collagen and (b) SARS-CoV-2; CV curves based on different scan rates ranging from 0.01 V s−1 to 0.2 V s−1 related to (c) type I collagen of human and (d) SARS-CoV-2; comparative DPV of cathodic and anodic scans of (e) type I collagen of human and (f) SARS-CoV-2.
Additionally, Fig. 4 (c) and (d) represent CV plots of GO-8H-EDC–NHS–Au NS modified electrode loaded with collagen type I of human (4 μM) and SARS-CoV-2, respectively, recorded at PBS 0.1 M with different scan rates ranging from 10 to 200 mV s−1; respective calibration curves of these Figures can be seen in Fig. S12. As can be understood from these Figures and their calibration curves, the anodic peak currents varied linearly by increasing the scan rate. In this regard, the process was followed by surface-confined mechanism and the reaction is controlled through adsorption process, while it exhibited the direct electron transfer process.
Since the CV method has several limitations due to its inherent low resolution and low sensitivity, it is favored to use DPV as a more sensitive voltammetric method along with a higher resolution for this aim. Thereby, better diagnostic criteria was obtained by DPV analysis in order to distinguish different types of electrode mechanisms (Chen et al., 2020; Kim et al., 2003). In this case, to probe the redox mechanism of functional groups present on viral glycoproteins, the anodic and cathodic peak currents related to the DPV pattern of SARS-CoV-2 are carefully analyzed.
Fig. 4 (e) shows redox processes obtained in the cathodic and anodic scans in PBS solution (pH 7.4) related to collagen type I of human. The labeled redox peak observed at about 0.05 V is correspond to the electrochemical behavior on the surface of electrode. The DPV voltammograms exhibited a relatively low peak separation of about ~26 mV between the anodic and cathodic peaks along with ipa/ipc~1, indicating reversible behavior and attributed to the adsorption and interaction of amine/hydroxyl functional groups of glycoproteins with the more available NHS activated hydroxyl/hydrogen-based/hydrogen-linkable groups on the surface of electrode. These observations are in well agreement with electrochemical (E) mechanism (Kim et al., 2003). Likewise, as illustrated within Fig. 4 (f), SARS-CoV-2 is also exhibited same mechanism compared with collagen with ipa/ipc~1. The adsorption of glycoproteins on the surface of electrode is carried out via activation of hydrogen binding at physiological pH value using NHS, increase in the number of active OH− sites upon addition of 8H and electrostatic interactions between the hydroxyl and amine functional groups of glycoproteins and the hydroxyl and amine groups of deposited sorbents on the surface of electrode. Besides, the facilitated redox reaction by Au NS and interaction of its amine functional groups with target pathogens improved the capability of the nanosensor toward accurate/sensitive detection of viruses in biological/non-biological media through their viral glycoproteins.
Achieved advantages could be due to the superior conductivity and also electrocatalytic effects of Au NS's hybridization with GO-8H-EDC-NHS which not only increases the electron transfer rate and effective surface area of the nanosensor, but also catalyzed the redox reaction of active substances on GO-8H-EDC–NHS–Au NS. Similarly, increase in the active surface are of the nanosensor provide higher amount of reactive hydroxyl functional groups for possible binding with glycoprotein-based structures. Accordingly, glycoproteins can be traced through oxidation signals of Au NS, which was deposited on the surface of modified electrode. The electrocatalytic behavior of Au NS created weak current peaks appeared at about 0.05 V, which could be due to the absorption of probe groups on either collagen or SARS-CoV-2 that was observed in DPV curves.
2.3. Real samples evaluation
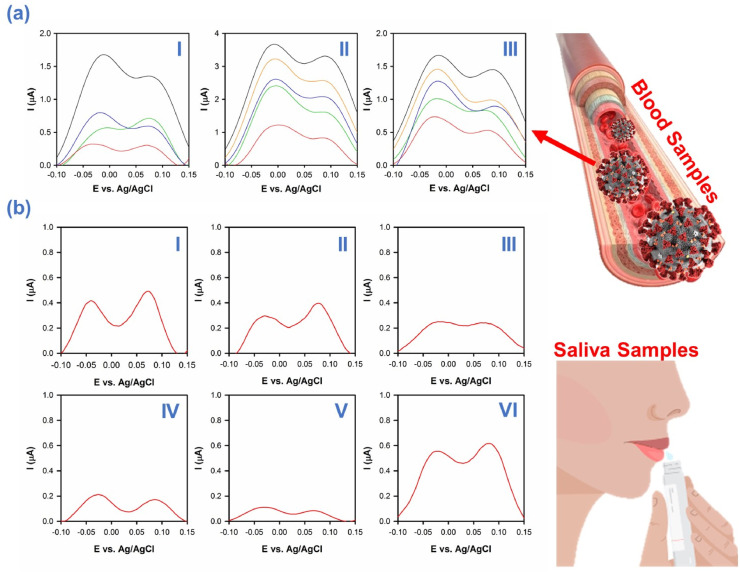
After examination of developed biosensor and confirmation of its successful performance for detection of divers kinds of viruses within biological/non-biological aquatic media, the fabricated biosensor is applied toward detection of SARS-CoV-2 in real human samples extracted from blood, saliva and nasopharyngeal swab using fixed triple electrodes. In this matter, blood and saliva samples were extracted from COVI-19's confirmed people using RT-PCR evaluation along with consideration of clinical signs.
A view of these results can be seen in Fig. 5 (a) (I-III). As depicts within these images, all three samples showed the finger print of SARS-CoV-2 in its exact position using fixed triple electrode system which attributed to the attachment of virus to the sensor via the S1 part of S spike glycoprotein of SARS-CoV-2. These peaks can be considered as the fingerprint of SARS-CoV-2 in blood and be used as a metric for its fast detection within diverse biological samples using fixed triple electrode system. Likewise, the developed platform also showed remarkable performance in detection of SARS-CoV-2 in saliva samples of infected people with COVID-19. As depicts within Fig. 5 (b) (I-VI), saliva samples of all of infected people exhibited the fingerprint of SARS-CoV-2 at its exact location with diver intensities which correspond to the viral load of each sample, while differences in the intensity of peaks could be due to the stage of illness. With such monitoring approach, presence of SARS-CoV-2 can be detected even at earliest stages of the disease that could help doctors to decline the overall rate of death tolls of this viral illness. Furthermore, the fastest way to take sample from infected people is to take saliva from them. Monitoring the level of SARS-CoV-2 in saliva samples can help health care authorities to isolate infected people from healthy ones more rapidly. Achieved evidence showed the ideal capability of developed nanosensor for detection of coronavirus within both blood and saliva samples which can be used for rapid detection of suspected people and monitoring the behavior of viral infections.
Fig. 5.
(a) (I-III) DPV patterns of SARS-CoV-2 detected in blood samples of infected people with COVID-19 and (b) (I-VI) traced SARS-CoV-2 in saliva samples of infected people with COVID-19.
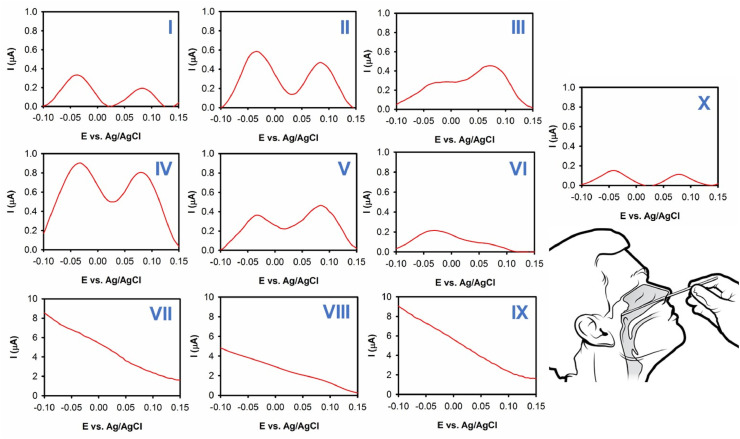
For more validation of the proposed method, 10 suspected patients were selected and their nasopharyngeal swab samples were evaluated via the developed nanosensor (Fig. 6 (I-X)). In this matter, in 7 of samples the trace of SARS-CoV-2 was successfully detected, while 3 of samples did not show any trace of coronavirus. These data was confirmed with their clinical signs and showed the superior performance of developed biosensor toward rapid detection of SARS-CoV-2 within aquatic biological specimens. Besides, the nanosensor is also capable to detect people in their common stages in which traditional approaches are incapable to detect viruses in biological samples. For instance, samples I, VI and X showed the trace of SARS-CoV-2 with intensity less than 0.2 μA and patients did not disclose any clinical signs which reveal that they are in the common stage of illness. Such approach can be used for fast screening in crowded places such as airports to stop the transfer chain of this viral infection.
Fig. 6.
(I–X) Outcome of nasopharyngeal swab samples' evaluation related to 10 different suspected people.
For further validation of nanosensor's performance, carbon-based screen printed electrodes (Metrohm DropSens DRP C110 L) were used to evaluate 100 blind nasopharyngeal swab samples taken from suspected people. The samples were also examined via RT-PCR essay as gold standard and their responses compared with each other. In this matter, screen printed electrodes were first sterilized and their working electrode is decorated with 8 μL of GO-8H-EDC–NHS–Au NS suspension (5 mg mL−1) followed by drying electrodes in a clean BSL 2 laminar hood. The prepared electrodes were then attached to the PGSTAT 302 N device and used to examine swab samples. In this matter, 100 μL of each swab sample is mixed with 100 μL of PBS (pH 7.3, 0.1 M KCl) and poured on the active area of the nanosensor. The sample was thence evaluated via DPV technique and trace of coronavirus was detected in swab sample of infected people, while negative samples did not show any trace. In Table S3, outcome of evaluations by the nanosensor and RT-PCR test can be seen in detail along with the fingerprint pattern of SARS-CoV-2 related to swab samples of infected people. The obtained data compared with RT-PCR as reference can be seen in Table S4. As shown, the nanosensor showed great performance compared with RT-PCR as gold standard. In this regard, the nanosensor assembled on screen printed electrode showed superior sensitivity of about 95% along with very low miss rate of 5% (by considering results of RT-PCR as gold standard) which happens due to the failure of screen printed electrodes and/or detachment of nanosensor from the working electrode; such failure can be minimized to zero upon stablishing an accurate electrode inspection strategy. In fact, the nanosensor only showed 3 false negative outcomes out of 40 negative samples which is a remarkable achievement compared with traditional methods.
Besides, the specificity of the nanosensor compared with RT-PCR measured to be 60% which is due to higher amount of positive outcomes by the nanosensor compared with RT-PCR. In fact, the miss rate of RT-PCR lies between range of 30–40% at optimum conditions and even in some cases it becomes more than that which is mainly depend on the quality of technique/samples and extraction methods (Wikramaratna et al., 2020). In this matter, the current nanosensor can cover the hidden majority of infected people owing to its superiorly high sensitivity and low detection limit that cannot be detected via traditional approaches such as RT-PCR. In this case, the device showed 40% more positive outcomes compared with RT-PCR which is correspond to the infected population that was missed with RT-PCR. Last but not least, usage of such fast, accurate, sensitive and novel techniques can help healthcare authorities to face viral pandemics, control their transfer chain, detect people in the common stages of illness and stop their rapid person-to-person transfer chain via practical isolation strategies.
3. Conclusions
In current study, a novel electrochemical diagnostic setup based on GO-8H-EDC–NHS–Au NS was developed that can detect different kinds of pathogenic viruses in any aquatic biological media (i.e., saliva, blood and swab) through interaction with active functional groups of viral glycoprotein and present a differentiable DPV pattern as a fingerprint for each virus at different voltage positions. The nanosystem showed superior detection limit and sensitivity toward detection/quantification of glycoprotein-based structures and found to be an ideal platform for rapid detection of viruses. More importantly, the setup do not require any extraction prior to analysis and can detect trace of viruses in about 1 min without using any biological marker. The electrochemical detection mechanism is governed with surface-confined mechanism through adsorption process which leads to differentiable detection of pathogens within selected aquatic biological media. Clinical evaluation of blind SARS-CoV-2 samples revealed the ideal performance of this nanosystem upon exhibiting sensitivity and miss rate of 95 and 5% compared with RT-PCR as gold standard. Such approach can be used as a reliable and fast diagnostic platform for detection of viral disease even in their silent stages and check the progress of illnesses via monitoring the concentration of viruses within biological fluids. Moreover, the nanosensor can cover the missed infected population by RT-PCR which is a great advantage for controlling the pandemic and returning the society into normal conditions.
CRediT authorship contribution statement
Seyyed Alireza Hashemi: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization, Supervision, Project administration, Funding acquisition. Nader Ghaleh Golab Behbahan: Methodology, Validation, Formal analysis, Resources. Sonia Bahrani: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Writing - original draft, Visualization. Seyyed Mojtaba Mousavi: Conceptualization, Validation, Formal analysis, Investigation, Resources, Visualization, Funding acquisition. Ahmad Gholami: Formal analysis, Resources. Seeram Ramakrishna: Formal analysis, Writing - review & editing, Supervision, Project administration. Mohammad Firoozsani: Formal analysis, Resources, Funding acquisition. Mohsen Moghadami: Methodology, Validation, Formal analysis, Resources. Kamran Bagheri Lankarani: Validation, Formal analysis. Navid Omidifar: Formal analysis.
Declaration of competing interest
Authors of this essay declaring that we do not have any conflict of interest regarding publication of this paper. As corresponding author, I declare and confirm this statement on behalf of my teammates.
Acknowledgement
Details of materials and methods along with characterization outcomes of developed nanomaterials can be seen within the supporting information. Besides, details regarding detection of type I collagen of human and pig along with CV and EIS evaluations of developed nanomaterials can be seen in supporting information. This research is conducted with full logistic and financial support of MEbtekar Nano Sanat Kimia company (located in Shiraz Science and Technology Park, Shiraz, Iran) under supervision of Mr. Yousef Amrollahi, Mr. Seyyed Hadi Ahmadi and Mr. Mohammad Mehdi Screwchi as heads of company and commercial matters, respectively. Clinical part of this essay was conducted via considering all of ethical standards and registered as a part of project with ethic code of IR. SUMS.REC.1399.314 in the Shiraz University of Medical Sciences. Results of 100 blind swab samples along with their RT-PCR outcomes and respective DPV patterns can be seen in the supporting information (Table(s) S3 and S4).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112731.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Alley W.R., Jr., Mann B.F., Novotny M.V. Chem. Rev. 2013;113(4):2668–2732. doi: 10.1021/cr3003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Yuan S., Kok K.H., To K.K.W., Chu H., Yang J., Xing F., Liu J., Yip C.C.Y., Poon R.W.S. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Wang X., Lu W., Wu X., Li J. Chem. Soc. Rev. 2016;45(8):2137–2211. doi: 10.1039/c6cs00061d. [DOI] [PubMed] [Google Scholar]
- Chen P., De Meulenaere E., Deheyn D.D., Bandaru P.R. Sci. Rep. 2020;10(1):4033. doi: 10.1038/s41598-020-60640-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M.S., Toh C.S. Analyst. 2013;138(21):6219–6229. doi: 10.1039/c3an01394d. [DOI] [PubMed] [Google Scholar]
- Cole C.R., Smith C.A. Biochem. Educ. 1989;17(4):179–189. [Google Scholar]
- Fenton A., Pedersen A.B. Emerg. Infect. Dis. 2005;11(12):1815. doi: 10.3201/eid1112.050306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik A. Frontiers in Nanotechnology. 2019;1:1. [Google Scholar]
- Kim M.H., Yan L., Birke R.L., Czae M.Z. Electroanalysis. 2003;15(19):1541–1553. [Google Scholar]
- Lequin R.M. Clin. Chem. 2005;51(12):2415–2418. doi: 10.1373/clinchem.2005.051532. [DOI] [PubMed] [Google Scholar]
- Li Q.H., Ma Y.H., Wang N., Hu Y., Liu Z.Z. Asian Toxicology Research. 2020;2(1):1–7. [Google Scholar]
- Mujawar M.A., Gohel H., Bhardwaj S.K., Srinivasan S., Hickman N., Kaushik A. Materials Today Chemistry. 2020:100306. doi: 10.1016/j.mtchem.2020.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek E., Tkac J., Bartosik M., Bertok T.S., Ostatna V., Palecek J. Chem. Rev. 2015;115(5):2045–2108. doi: 10.1021/cr500279h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd P.M., Elliott T., Cresswell P., Wilson I.A., Dwek R.A. Science. 2001;291(5512):2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- Spackman E. Avian Pathol. 2012;41(3):251–258. doi: 10.1080/03079457.2012.675051. [DOI] [PubMed] [Google Scholar]
- Stavenhagen K., Plomp R., Wuhrer M. Anal. Chem. 2015;87(23):11691–11699. doi: 10.1021/acs.analchem.5b02366. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang Y., Gao M., Zhang X., Yang P. ACS Appl. Mater. Interfaces. 2015;7(29):16011–16017. doi: 10.1021/acsami.5b04295. [DOI] [PubMed] [Google Scholar]
- Wikramaratna P., Paton R.S., Ghafari M., Lourenco J. 2020. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Sci. China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Boysen R.I., Chowdhury J., Alam A., Hearn M.T. Anal. Chim. Acta. 2015;872:84–94. doi: 10.1016/j.aca.2015.02.055. [DOI] [PubMed] [Google Scholar]
- Ye J., Chen Y., Liu Z. Angew. Chem. Int. Ed. 2014;53(39):10386–10389. doi: 10.1002/anie.201405525. [DOI] [PubMed] [Google Scholar]
- Zhang C., Hage D.S. J. Chromatogr. A. 2016;1475:102–109. doi: 10.1016/j.chroma.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Liu W., Li P., Xiao H., Wang H., Tang B. Angew. Chem. Int. Ed. 2014;53(46):12489–12493. doi: 10.1002/anie.201405634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.