Abstract
Fosfomycin and nitrofurantoin are antibiotics of choice to orally treat non-complicated urinary tract infections (UTIs) of community origin because they remain active against bacteria resistant to other antibiotics. However, epidemiologic surveillance studies have detected a reduced susceptibility to these drugs. The objective of this study was to determine possible mechanisms of resistance to these antibiotics in clinical isolates of fosfomycin- and/or nitrofurantoin-resistant UTI-producing Escherichia coli. We amplified and sequenced murA, glpT, uhpT, uhpA, ptsI, cyaA, nfsA, nfsB, and ribE genes, and screened plasmid-borne fosfomycin-resistance genes fosA3, fosA4, fosA5, fosA6, and fosC2 and nitrofurantoin-resistance genes oqxA and oqxB by polymerase chain reaction. Among 29 isolates studied, 22 were resistant to fosfomycin due to deletion of uhpT and/or uhpA genes, and 2 also possessed the fosA3 gene. Some modifications detected in sequences of NfsA (His11Tyr, Ser33Arg, Gln67Leu, Cys80Arg, Gly126Arg, Gly154Glu, Arg203Cys), NfsB (Gln44His, Phe84Ser, Arg107Cys, Gly192Ser, Arg207His), and RibE (Pro55His), and the production of truncated NfsA (Gln67 and Gln147) and NfsB (Glu54), were associated with nitrofurantoin resistance in 15/29 isolates; however, the presence of oqxAB plasmid genes was not detected in any isolate. Resistance to fosfomycin was associated with the absence of transporter UhpT expression and/or the presence of antibiotic-modifying enzymes encoded by fosA3 plasmid-mediated gene. Resistance to nitrofurantoin was associated with modifications of NfsA, NfsB, and RibE proteins. The emergence and spread of these resistance mechanisms, including transferable resistance, could compromise the future usefulness of fosfomycin and nitrofurantoin against UTIs. Furthermore, knowledge of the genetic mechanisms underlying resistance may lead to rapid DNA-based testing for resistance.
Keywords: Escherichia coli, fosfomycin, nitrofurantoin, antimicrobial resistance
1. Introduction
The high incidence of urinary tract infections (UTIs) and their usually mild character means that most patients receive empirical antibiotic treatment. However, clinicians are now faced with major challenges due to multiple factors, including population aging, the presence of allergies or adverse reactions to antibiotics, an increased number of immunodepressed patients, and, especially, high rates of multi-resistant pathogens, which can cause therapeutic failure. A good alternative option may be to return to antibiotics such as fosfomycin and nitrofurantoin [1].
The characteristics of fosfomycin and nitrofurantoin make them especially useful for UTI treatment, including their rapid oral absorption, high urine concentration, and bactericide activity against a wide range of Gram-negative and Gram-positive bacteria. Both are first-line treatments for non-complicated UTIs of community origin [2]. They have also been reported to preserve their activity against multi-resistant microorganisms, especially uropathogenic enterobacteria such as Escherichia coli and extended-spectrum beta-lactamase-producing isolates [3], although these are usually less susceptible to fosfomycin and nitrofurantoin than are non-producers [4,5].
Currently, resistance to fosfomycin or nitrofurantoin is not common in our setting, and >85% of bacteria isolated in UTIs are susceptible to these antibiotics. Nonetheless, the gradual decrease in susceptibility to these drugs may lead to their contraindication as an empirical treatment in the future [1]. Any expansion of their clinical utilization would, therefore, require the adoption of epidemiological surveillance measures to detect the possible emergence of resistance [6]. With this background, the objective of this study was to explore possible molecular mechanisms underlying the resistance of clinical isolates of UTI-producing E. coli to fosfomycin and nitrofurantoin in our setting.
2. Methods
2.1. Bacterial Isolates
The study included clinical isolates of fosfomycin- and/or nitrofurantoin-resistant E. coli with significant bacterial count selected from among urine cultures conducted for UTI analysis in the Microbiology Laboratory of Virgen de las Nieves University Hospital (Granada, Spain). They were identified by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (Bruker Daltonics, Bremen, Germany) as part of the routine microbiology laboratory workup [1]. A disk diffusion procedure (Kirby–Bauer) was also conducted on agar Mueller-Hinton plates, using McFarland 0.5 bacterial inoculum and disks with 200 μg fosfomycin supplemented with 50 μg glucose-6-phosphate (G6P) or disks with 300 μg nitrofurantoin. Each isolate was defined as "susceptible,” “intermediate,” or “resistant” according to the Clinical and Laboratory Standards Institute (CLSI) breakpoints [7]. For fosfomycin, an inhibition zone diameter ≥16 mm was considered susceptible, 13–15 mm intermediate, and ≤12 mm resistant; for nitrofurantoin, an inhibition zone diameter of ≥17 mm was considered susceptible, 15−16 mm intermediate and ≤14 mm resistant. E. coli ATCC 25922 (American Type Culture Collection, Manassas, VA, USA) was used as the control strain in the susceptibility assays.
Furthermore, in order to identify E. coli isolates producing fosfomycin resistance-mediating glutathione S-transferases, 20 μL sodium phosphonoformate (PPF) (Sigma-Aldrich, Madrid, Spain) was added at a concentration of 50 mg/mL on a second disk with 200 μg fosfomycin supplemented with 50 μg G6P, located at a distance of 30–35 mm from the first. After overnight incubation at 36 ± 1 °C, diameters of the growth inhibition zone were compared between the first disk (with PPF) and the second (without PPF). A difference of ≥5 mm between diameters was considered to confirm the phenotypic presence of the enzyme [8]. All assays were performed in duplicate.
2.2. Carbohydrate Utilization Test
All isolates were studied to determine the capacity for bacterial growth in the presence of a single source of carbon, sn-glycerol 3-phosphate (G3P), or G6P, using a previously described procedure [9]. After incubating bacteria in Mueller–Hinton broth for 24 h at 36 ± 1 °C in agitation, they were collected by centrifugation and resuspended in normal saline solution (0.9% NaCl). After five washes (to remove any remains that may act as carbon source), bacterial suspensions were then streaked onto M9 minimal medium agar supplemented with glucose (as a positive growth control), with G3P or G6P at 0.2% (w/v). Bacterial growth was determined after incubation at 36 ± 1 °C for 48 h. All assays were performed in duplicate. The absence of bacterial growth or poor growth with no colony formation in media supplemented with G3P or G6P was considered to indicate GlpT or UhpT function deficiency, respectively [9].
2.3. PCR Amplification
Polymerase chain reaction (PCR) was used to amplify murA, glpT, uhpT, uhpA, ptsI, cyaA, nfsA, nfsB, and ribE genes of E. coli, using previously reported procedures [5,9], separately amplifying two fragments (cyaA1 and cyaA2) for the cyaA gene. The primer pairs used are listed in Table 1. DNA was obtained from clinical isolates and E. coli ATCC 25922 (used as control strain) using the PureLink Microbiome DNA Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA). One microliter of the purified DNA was added to a master mix containing PCR buffer (1×), MgCl2 (2 mM), dNTPs (0.4 mM), primers (0.4 μM), and Taq polymerase (1.25 U).
Table 1.
Primers used for amplification and sequencing of the Escherichia coli genes involved in fosfomycin or nitrofurantoin resistance.
| Gene | Forward Primer | Reverse Primer | Amplicon Size (bp) | Reference |
|---|---|---|---|---|
| murA | 5′-AAACAGCAGACGGTCTATGG-3′ | 5′-CCATGAGTTTATCGACAGAACG-3′ | 1542 | [9] |
| glpT | 5′-GCGAGTCGCGAGTTTTCATTG-3′ | 5′-GGCAAATATCCACTGGCACC-3′ | 1785 | |
| uhpT | 5′-TTTTTGAACGCCCAGACACC-3′ | 5′-AGTCAGGGGCTATTTGATGG-3′ | 1667 | |
| uhpA | 5′-GATCGCGGTGTTTTTTCAG-3′ | 5′-GATACTCCACAGGCAAAACC-3′ | 771 | |
| ptsI | 5′-GAAAGCGGTTGAACATCTGG-3′ | 5′-TCCTTCTTGTCGTCGGAAAC-3′ | 1908 | |
| cyaA1 | 5′-AACCAGGCGCGAAAAGTGG-3′ | 5′-TGATGGCTGATGATCGACTC-3′ | 1559 | [9] This study |
| cyaA2 | 5′-AAAGCTCAGCCGTGAACGC-3′ | 5′-ACCTTCTGGGATTTGCTGG-3′ | 1648 | |
| nfsA | 5′-ATTTTCTCGGCCAGAAGTGC-3′ | 5′-AGAATTTCAACCAGGTGACC-3′ | 1036 | [5] |
| nfsB | 5′-CTTCGCGATCTGATCAACG-3′ | 5′-CAACAGCAGCCTATGATGAC-3′ | 923 | |
| ribE | 5′-AAGGGAAGCAGCGCACGAA-3′ | 5′-GGACAACTGCCAGGAGTAGA-3′ | 634 | This study |
| fosA3 | 5′-GCGTCAAGCCTGGCATTT-3′ | 5′-GCCGTCAGGGTCGAGAAA-3′ | 282 | [10] |
| fosA4 | 5′-CTGGCGTTTTATCAGCGGTT-3′ | 5′-CTTCGCTGCGGTTGTCTTT-3′ | 230 | [11] |
| fosA5 | 5′-TATTAGCGAAGCCGATTTTGCT-3′ | 5′-CCCCTTATACGGCTGCTCG-3′ | 177 | |
| fosA6 | 5′-GCTACGGTTCAGCTTCCAGA-3′ | 5′-CGAGCGTGGCGTTTTATCAG-3′ | 242 | This study |
| fosC2 | 5′-CGTTCCGTGGAGTTCTATAC-3′ | 5′-CTTGATAGGGTTTAGACTTC-3′ | 334 | [8] |
| oqxA | 5′-GACAGCGTCGCACAGAATG-3′ | 5′-GGAGACGAGGTTGGTATGGA-3′ | 339 | [12] |
| oqxB | 5′-CGAAGAAAGACCTCCCTACCC-3′ | 5′-CGCCGCCAATGAGATACA-3′ | 240 |
PCR amplification of murA, glpT, uhpT, cyaA1, cyaA2, nfsA, nfsB, and ribE genes was performed as follows: 2 min of denaturation at 94 °C, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 2 min (1 min for nfsA, nfsB, and ribE), with a final period of extension at 72 °C for 5 min. The same conditions were used for the amplification of uhpA and ptsI genes except that the annealing temperature was 57 °C.
For the isolates in which uhpT or uhpA genes could not be detected by the aforementioned procedure, a new PCR was designed using an outer primer pair (uhpT-F2: 5′-GATGTTAATCGGTATGGCGGC-3′; uhpT-R2: 5′-CAGTCGCTGGCGGAACAAAT-3′; uhpA-F2: 5′-CGTAATTCTGGAGCTCACCG-3′; uhpA-R2: 5′-CGCCTGCGTTAGCCAGTAA-3′). Besides re-amplification with outer primers, the amplification specificity was increased by using the forward outer primer with the reverse inner primer and the forward inner primer with the reverse outer primer.
Plasmid-borne fosfomycin resistance genes fosA3, fosA4, fosA5, fosA6, and fosC2 and nitrofurantoin resistance genes oqxA and oqxB were screened by PCR amplification with the primers listed in Table 1, following previously reported procedures [8,10,11,12].
All PCR products were separated in 0.8% agarose gel and visualized under UV light after staining with ethidium bromide.
2.4. Nucleotide Sequencing
Pools of 8 and 10 amplicons were established, and each amplicon was equimolarly normalized in the pool. Each pool was tagmented (tagged and fragmented) using the Nextera XT transposome, which fragments the DNA and then tags it with adapter sequences in a single step. The tagmented DNA was amplified with 12 PCR cycles. The PCR step also adds index 1 (i7), index 2 (i5), and full adapter sequences required for cluster formation. Each DNA sample was purified using 30 µL of AMPure XP beads and was resuspended in 50 μL of water. Then, it was quantified using a Qubit® 3.0 Fluorometer (Thermo Fisher Scientific) and normalized. Pools were sequenced in a high cartridge of 300 cycles using a NextSeq platform. Data were mapped against the reference sequence of E. coli str. K-12 substr. MG1655 (NCBI Reference Sequence: NC_000913.3). A BAM file was generated, followed by a variant calling, and the most representative variants were recorded. The online Protein Variation Effect Analyzer (PROVEAN) platform (http://provean.jcvi.org/index.php) was used to predict the impact of identified amino acid substitutions on the biological function of each protein [13]. PROVEAN is able to provide predictions for any type of protein sequence variation, including single or multiple amino acid substitutions, insertions, or deletions. The platform introduces a delta alignment score based on the reference and variant versions of a protein query sequence with respect to sequence homologs collected from the NCBI protein database through BLAST. If the PROVEAN score (P-score) was equal to or below a predefined cutoff of −2.5, the protein variant was predicted to have a "deleterious" effect (potential loss of protein structure or function). If the P-score was above the threshold, the variant was predicted to have a "neutral" effect (no alteration in the structure or function of the protein).
3. Results
A total of 29 fosfomycin- and/or nitrofurantoin-resistant clinical isolates were identified: 8 were resistant to both fosfomycin and nitrofurantoin, 14 were resistant to fosfomycin and susceptible to nitrofurantoin, and 7 were susceptible to fosfomycin and resistant to nitrofurantoin. Figure 1 depicts PCR amplification of chromosomal genes murA, glpT, uhpT, uhpA, ptsI, cyaA (cyaA1 and cyaA2), nfsA, nfsB, and ribE in E. coli ATCC 25922.
Figure 1.
Electrophoresis results of polymerase chain reaction (PCR) products in Escherichia coli ATCC 25922 on 0.8% agarose gel. M: Molecular weight. Lines 1 to 10: PCR products of murA (1542 bp), glpT (1785 bp), uhpT (1667 bp), uhpA (771 bp), ptsI (1908 bp), cyaA1 (1559 bp), cyaA2 (1648 bp), nfsA (1036 bp), nfsB (923 bp), and ribE (634 bp), respectively.
3.1. Fosfomycin Resistance
Table 2 summarizes the characteristics of the 22 fosfomycin-resistant (inhibition zone diameter ≤12 mm around the disk with 200 μg fosfomycin supplemented with 50 μg glucose-6-phosphate) and 7 fosfomycin-susceptible (inhibition zone diameter ≥16 mm around the disk with 200 μg fosfomycin supplemented with 50 μg glucose-6-phosphate) clinical isolates of E. coli according to the CLSI procedure, displaying the diameter of the bacterial growth inhibition in the presence of PPF, the bacterial growth capacity in the presence of G3P or G6P as sole carbon source, and the amino acid substitutions in MurA, GlpT, UhpT, UhpA, PtsI, and CyaA proteins detected in each isolate.
Table 2.
Susceptibility to fosfomycin according to the Clinical and Laboratory Standards Institute (CLSI) procedure and supplemented with sodium phosphonoformate (PPF); bacterial growth on M9 minimal medium agar supplemented with sn-glycerol 3-phosphate (G3P) or glucose-6-phosphate (G6P); and amino acid substitutions in MurA, GlpT, UhpT, UhpA, PtsI, and CyaA proteins in 29 clinical isolates of Escherichia coli.
| Strain | Fosfomycin Disk 1 | Clinical Category 2 | Fosfomycin Disk Plus PPF 3 | G3P 4 | G6P 5 | Amino Acid Substitutions in | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MurA | GlpT | UhpT | UhpA | PtsI | CyaA | ||||||
| 11 | 6 | R | 6 | + | − | None | Leu297Phe Glu443Gln Gln444Glu Glu448Lys |
Not detected | Not detected | Arg367Lys | Asn142Ser Ala349Glu Ser352Thr Ser356Leu Gly359Glu Glu362Asp |
| 17 | 6 | R | 6 | + | − | None | Glu448Lys | Glu350Gln | Not detected | Val25Ile Arg367Lys |
Asn142Ser Asp837Glu Thr840Ala |
| 26 | 12 | R | 12 | + | − a | None | Gly84Asp Glu448Lys |
Not detected | None | Arg367Lys | Asn142Ser Ala349Glu Ser356Leu Gly359Glu Glu362Asp Ala363Ser Asp837Glu Thr840Ala |
| 66 | 6 | R | 13 | + | − | None | Glu448Lys | Glu350Gln | Not detected | Val25Ile Arg367Lys |
Asn142Ser Asp837Glu Thr840Ala |
| 302 | 29 | S | 30 | + | + | None | Glu448Lys | None | None | Arg367Lys | Asn142Ser Gly222Ser |
| 334 | 31 | S | 32 | + | + | None | Glu448Lys | None | None | Arg367Lys | Asn142Ser |
| 381 | 6 | R | 6 | + | − | None | Glu448Lys | Glu350Gln | Not detected | Val25Ile Arg367Lys |
Asn142Ser Asp837Glu Thr840Ala |
| 387 | 6 | R | 6 | + | − | None | Glu448Lys | Glu350Gln | Not detected | Val25Ile Arg367Lys |
Asn142Ser Asp837Glu Thr840Ala |
| 462 | 12 | R | 13 | + | − | None | Glu448Lys | Glu350Gln | Not detected | Val25Ile Arg367Lys |
Asn142Ser Asp837Glu Thr840Ala |
| 632 | 6 | R | 6 | + | − | None | Leu297Phe Glu443Gln Gln444Glu Glu448Lys |
Glu350Gln | Not detected | Arg367Lys | Asn142Ser Ala349Glu Ser352Thr Ser356Leu Gly359Glu Glu362Asp Ala363Gly |
| 751 | 30 | S | 32 | + | + | None | Ala16Thr Glu448Lys |
None | Arg46Cys | Ala306Thr Arg367Lys |
Asn142Ser Ala349Glu Ser356Leu Gly359Glu Glu362Asp Asp837Glu Thr840Ala |
| 752 | 11 | R | 11 | + | − | None | Pro212Leu Glu448Lys |
Glu350Gln | Not detected | Val25Ile Arg367Lys |
Asn142Ser Asp837Glu Thr840Ala |
| 757 | 6 | R | 6 | + | − a | None | Leu297Phe Glu443Gln Gln444Glu Glu448Lys |
None | Not detected | Arg367Lys | Asn142Ser Ala349Glu Ser352Thr Ser356Leu Gly359Glu Glu362Asp |
| 776 | 12 | R | 12 | + | − a | None | Glu448Lys | Glu350Gln | Not detected | Val25Ile Arg367Lys |
Asn142Ser Asp837Glu Thr840Ala |
| 789 | 6 | R | 6 | + | − | Leu370Ile | Leu297Phe Glu443Gln Gln444Glu Glu448Lys |
Glu350Gln | Not detected | Arg367Lys | Asn142Ser Ala349Glu Ser352Thr Ser356Leu Gly359Glu Glu362Asp |
| 792 | 6 | R | 6 | + | − | None | Glu448Lys | Glu350Gln | Not detected | Val25Ile Arg367Lys |
Asn142Ser Asp837Glu Thr840Ala |
| 795 | 6 | R | 6 | + | − | None | Leu297Phe Glu443Gln Gln444Glu Glu448Lys |
Glu350Gln | Not detected | Arg367Lys | Asn142Ser Ala349Glu Ser352Thr Ser356Leu Gly359Glu Glu362Asp |
| 797 | 20 | S | 21 | + | + | None | Ala16Thr Leu373Arg Glu448Lys |
None | Arg46Cys | Ala306Thr Arg367Lys |
Asn142Ser Ala349Glu Ser356Leu Gly359Glu Glu362Asp Asp837Glu Thr840Ala |
| 799 | 6 | R | 7 | + | − | None | Leu297Phe Glu443Gln Gln444Glu Glu448Lys |
None | Not detected | Arg367Lys | Asn142Ser Ala349Glu Ser352Thr Ser356Leu Gly359Glu Glu362Asp |
| 802 | 30 | S | 30 | + | + | None | Phe133Cys Gly135Trp Ala197Val Glu448Lys |
None | None | Arg367Lys | Asn142Ser |
| 809 | 6 | R | 6 | + | − | Leu370Ile | Leu297Phe Glu443Gln Gln444Glu Glu448Lys |
Glu350Gln | Not detected | Arg367Lys | Asn142Ser Ala349Glu Ser352Thr Ser356Leu Gly359Glu Glu362Asp |
| 853 | 6 | R | 8 | + | − | Leu370Ile | Leu297Phe Glu443Gln Gln444Glu Glu448Lys |
Glu350Gln | Not detected | Arg367Lys | Asn142Ser Ala349Glu Ser352Thr Ser356Leu Gly359Glu Glu362Asp |
| 854 | 6 | R | 6 | + | − | None | Glu448Lys | None | Not detected | Arg367Lys | Asn142Ser |
| 860 | 6 | R | 7 | + | − | None | Met52Leu Leu297Phe Glu443Gln Gln444Glu Glu448Lys |
None | Not detected | Arg367Lys | Asn142Ser Ala349Glu Ser352Thr Ser356Leu Gly359Glu Glu362Asp |
| 871 | 6 | R | 14 | + | − a | None | Leu297Phe Glu443Gln Gln444Glu Glu448Lys |
Glu350Gln | Not detected | Arg367Lys | Asn142Ser Ala349Glu Ser352Thr Ser356Leu Gly359Glu Glu362Asp Ala363Gly |
| 872 | 35 | S | 35 | + | + | None | Glu448Lys | None | None | Arg367Lys | Asn142Ser Asp837Glu Thr840Ala |
| 883 | 11 | R | 12 | + | − | None | Glu448Lys | Glu350Gln | Not detected | Val25Ile Arg367Lys |
Asn142Ser Asp837Glu Thr840Ala |
| 891 | 11 | R | 11 | + | − | None | Glu448Lys | Glu350Gln | Not detected | Val25Ile Arg367Lys |
Asn142Ser Asp837Glu Thr840Ala |
| 892 | 21 | S | 21 | + | + | None | Glu448Lys | None | None | Arg367Lys | Asn142Ser |
1 Diameter (in mm) of the bacterial growth inhibition halo around the disk with 200 μg fosfomycin supplemented with 50 μg glucose-6-phosphate on Mueller-Hinton agar. 2 Clinical categories of each isolate against fosfomycin according to CLSI breakpoints (S: susceptible; R: resistant). 3 Diameter (in mm) of the bacterial growth inhibition halo around the disk with 200 μg fosfomycin supplemented with 50 μg glucose-6-phosphate and 20 μL sodium phosphonoformate (PPF) in order to identify E. coli isolates producing fosfomycin resistance-mediating glutathione S-transferases (between-diameter difference of ≥5 mm considered to confirm the phenotypic presence of the enzyme). 4 Bacterial growth on M9 minimal medium agar supplemented with 0.2% sn-glycerol 3-phosphate (all isolates showed growth). 5 Bacterial growth on M9 minimal medium agar supplemented with 0.2% glucose-6-phosphate (+: bacterial growth; −: absence of bacterial growth). a Only poor growth was observed after 48 h of incubation. Not detected: gene not detected by PCR after the different combinations of two pairs of primers (loss of the entire gene). None: no amino acid substitutions found.
Three of the twenty-two fosfomycin-resistant isolates (strains 789, 809, and 853) showed a single substitution in the amino acid sequence of MurA (Leu370Ile), categorized as neutral (no alteration in structure or function of the protein) in the PROVEAN analysis (P-score: −1.995).
Twelve amino acid substitutions were detected in GlpT: Glu448Lys (P-score: 0.486, categorized as neutral), in all isolates, both resistant and susceptible; Ala16Thr (P-score: −0.713, categorized as neutral), and Phe133Cys (P-score: −5.549), Gly135Trp (P-score: −7.756), Ala197Val (P-score: −3.472), and Leu373Arg (P-score: −5.328), all categorized as deleterious (potential loss of protein structure or function), in susceptible isolates alone; and Met52Leu (P-score: −1.261), Leu297Phe (P-score: −2.375), Glu443Gln (P-score: 0.014), and Gln444Glu (P-score: −0.106), all four categorized as neutral, and Gly84Asp (P-score: −6.056) and Pro212Leu (P-score: −9.698), both categorized as deleterious, in resistant isolates alone.
No amplification product of the uhpT gene was obtained from strains 11 and 26 using the two primer pairs reported above (loss of entire gene). Amino acid substitution in UhpT (Glu350Gln), categorized as neutral (P-score: −0.016), was observed in 16 of the 22 fosfomycin-resistant isolates but in none of the susceptible isolates.
The uhpA gene was detected in all fosfomycin-susceptible isolates, and two of these showed substitution of Arg46Cys in the protein sequence, categorized as neutral (P-score: −0.268). By contrast, this gene was detected in only 1 of the 22 fosfomycin-resistant isolates: strain 26 (wild-type).
Three amino acid substitutions were detected in PtsI: Arg367Lys (P-score: 0.842), in all isolates, both resistant and susceptible; Ala306Thr (P-score: 0.030), in two susceptible isolates alone; and Val25Ile (P-score: −0.606), in 10 resistant isolates alone; and all three substitutions were categorized as neutral in the PROVEAN analysis.
Finally, 11 amino acid substitutions were detected in CyaA: Asn142Ser (P-score: 0.016, categorized as neutral), in all isolates; Gly222Ser (P-score: −3.447, categorized as deleterious), in one of the seven susceptible isolates; Ala349Glu (P-score: 2.261), Glu362Asp (P-score: −0.286), Asp837Glu (P-score: 0.123), and Thr840Ala (P-score: −0.314) all four categorized as neutral; and Ser356Leu (P-score: −2.624) and Gly359Glu (P-score: −3.077) both categorized as deleterious, in some susceptible and resistant isolates; and Ala363Ser (P-score: 0.900), Ala363Gly (P-score: −0.251), and Ser352Thr (P-score: −0.645) all three categorized as neutral, in fosfomycin-resistant isolates alone.
The effect of these amino acid substitutions on transporters GlpT and UhpT in resistant and susceptible isolates was evaluated by testing bacterial growth on M9 minimal medium agar supplemented with G3P or G6P (substrates for GlpT or UhpT, respectively). As reported in Table 2, all isolates grew on M9 medium with G3P, indicating no significant loss of GlpT function with any substitution detected in the amino acid sequence of this transporter. However, fosfomycin-resistant isolates did not grow or showed poor growth on the medium containing G6P, because of the loss of function of UhpT due to the complete deletion of uhpT (strains 11 and 26) and/or uhpA genes (strains 11, 17, 66, 381, 387, 462, 632, 752, 757, 776, 789, 792, 795, 799, 809, 853, 854, 860, 871, 883 and 891).
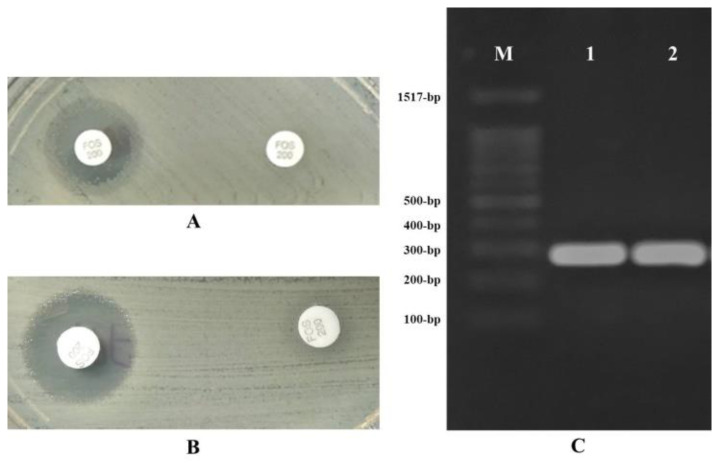
Furthermore, fosfomycin resistance-mediating glutathione S-transferase was observed in two of the fosfomycin-resistant isolates (strains 66 and 871) due to a significantly increased bacterial growth inhibition halo (≥5 mm) in the presence of PPF (Table 2). This phenotypic finding was confirmed by PCR amplification of the fosA3 gene in both isolates (Figure 2). No fosA4, fosA5, fosA6, and fosC2 plasmid genes were detected in any isolate.
Figure 2.
Detection of fosfomycin resistance-mediating glutathione S-transferase (sodium phosphonoformate test) and fosA3 gene (electrophoresis) in strains 66 and 871. (A,B) Phenotypic detection of fosfomycin resistance-mediating glutathione S-transferase in strains 66 and 871, respectively, showing an increase of ≥5 mm in growth inhibition halo around the disk of 200 μg fosfomycin supplemented with 50 μg G6P plus sodium phosphonoformate in comparison to the disk containing 200 μg fosfomycin supplemented with 50 μg G6P alone. All assays were performed in duplicate in all isolates, obtaining the same between-assay results; (C) Electrophoresis results for the PCR products of fosA3 gene (282 bp) on 0.8% agarose gel in strains 66 (line 1) and 871 (line 2). M: molecular weight.
3.2. Nitrofurantoin Resistance
Table 3 summarizes the characteristics of the 15 nitrofurantoin-resistant or intermediate isolates (inhibition zone diameter ≤14 mm or 13−15 mm, respectively, around the disk with 300 μg nitrofurantoin) and the 14 nitrofurantoin-susceptible (inhibition zone diameter ≥17 mm around the disk with 300 μg nitrofurantoin) clinical isolates of E. coli according to the CLSI procedure. Amino acid substitutions in NfsA, NfsB, or RibE proteins were detected in all isolates.
Table 3.
Susceptibility to nitrofurantoin according to the Clinical and Laboratory Standards Institute (CLSI) procedure and amino acid substitutions in NfsA, NfsB, or RibE proteins of 29 clinical isolates of Escherichia coli.
| Strain | Nitrofurantoin Disk 1 | Clinical Category 2 | Amino Acid Substitutions in | ||
|---|---|---|---|---|---|
| NfsA | NfsB | RibE | |||
| 11 | 11 | R | Ile117Thr Gly126Arg Lys141Glu Gln147Arg Gly187Asp |
Truncated at Glu54 | None |
| 17 | 27 | S | Ile117Thr Lys141Glu Gly187Asp |
Gly66Asp Val93Ala Ala174Glu |
None |
| 26 | 24 | S | Ile117Thr Lys141Glu Gly187Asp |
Gly66Asp Val93Ala Ala174Glu |
None |
| 66 | 14 | R | Ile117Thr Lys141Glu Gly187Asp |
Gly66Asp Val93Ala Ala174Glu Arg207His |
None |
| 302 | 14 | R | None | Gly66Asp Met75Ile Val93Ala Ala174Glu Arg207His |
None |
| 334 | 15 | I | Gln67Leu | Gly66Asp Met75Ile Val93Ala Arg107Cys |
Pro55His |
| 381 | 25 | S | Ile117Thr Lys141Glu Gly187Asp |
Gly66Asp Val93Ala Ala174Glu |
None |
| 387 | 21 | S | Ile117Thr Lys141Glu Gly187Asp |
Gly66Asp Val93Ala Ala174Glu |
None |
| 462 | 14 | R | Cys80Arg Ile117Thr Lys141Glu Gly187Asp |
Gly66Asp Val93Ala Ala174Glu Gly192Ser |
None |
| 632 | 20 | S | Glu58Asp Ile117Thr Lys141Glu Gln147Arg Gly187Asp |
Val93Ala | None |
| 751 | 14 | R | Ile117Thr Lys141Glu Gly187Asp Arg203Cys |
Gly66Asp Val93Ala Ala174Glu |
Val51Ile |
| 752 | 22 | S | Ile117Thr Lys141Glu Gly187Asp |
Gly66Asp Val93Ala Ala174Glu |
None |
| 757 | 16 | I | Glu58Asp Truncated at Gln67 |
Val93Ala Lys122Arg |
None |
| 776 | 19 | S | Ile117Thr Lys141Glu Gly187Asp |
Gly66Asp Val93Ala Ala174Glu |
None |
| 789 | 24 | S | Glu58Asp Ile117Thr Lys141Glu Gln147Arg Gly187Asp |
Val93Ala | None |
| 792 | 25 | S | Ile117Thr Lys141Glu Gly187Asp |
Gly66Asp Val93Ala Ala174Glu |
None |
| 795 | 16 | I | His11Tyr Glu58Asp Ile117Thr Lys141Glu Gln147Arg Gly187Asp |
Val93Ala | None |
| 797 | 12 | R | Ile117Thr Lys141Glu Gly154Glu Gly187Asp |
Leu22Ile Gly66Asp Val93Ala Ala174Glu |
Val51Ile |
| 799 | 13 | R | Ile117Thr Lys141Glu Gln147Arg Gly187Asp |
Val93Ala | None |
| 802 | 12 | R | Truncated at Gln147 | Met75Ile Val93Ala |
None |
| 809 | 28 | S | Glu58Asp Ile117Thr Lys141Glu Gln147Arg Gly187Asp |
Val93Ala | None |
| 853 | 20 | S | Glu58Asp Ile117Thr Lys141Glu Gln147Arg Gly187Asp |
Val93Ala | None |
| 854 | 16 | I | None | Gly66Asp Met75Ile Val93Ala |
None |
| 860 | 20 | S | Ile117Thr Lys141Glu Gln147Arg Gly187Asp |
Val93Ala | None |
| 871 | 24 | S | Glu58Asp Ile117Thr Lys141Glu Gln147Arg Gly187Asp |
Val93Ala | None |
| 872 | 15 | I | Asp19Asn Ser33Arg Ile117Thr Lys141Glu Gly187Asp |
Gln44His Gly66Asp Val93Ala Ala174Glu |
None |
| 883 | 13 | R | Ile117Thr Lys141Glu Gly187Asp |
Gly66Asp Phe84Ser Val93Ala Ala174Glu |
None |
| 891 | 23 | S | Ile117Thr Lys141Glu Gly187Asp |
Gly66Asp Val93Ala Ala174Glu |
None |
| 892 | 10 | R | None | Gly66Asp Met75Ile Val93Ala |
None |
1 Diameter (in mm) of bacterial growth inhibition halo around the disk with 300 μg nitrofurantoin. 2 Clinical categories of each isolate against nitrofurantoin according to CLSI breakpoints (S: susceptible; I: intermediate; R: resistant). None: no amino acid substitutions were found.
Among the 15 nitrofurantoin-resistant or nitrofurantoin-intermediate and 14 nitrofurantoin-susceptible isolates, 14 amino acid substitutions were detected in the NfsA protein: Glu58Asp (P-score: −1.866), Ile117Thr (P-score: −0.634), Lys141Glu (P-score: 1.207), Gln147Arg (P-score: −1.170), and Gly187Asp (P-score: 1.554) all of these categorized as neutral in the PROVEAN analysis (no alteration in structure or function of the protein), in susceptible isolates; Asp19Asn (P-score: −2.091) and Ser180Asn (P-score: 0.071) both categorized as neutral, and His11Tyr (P-score: −5.746), Ser33Arg (P-score: −2.526), Gln67Leu (P-score: −5.860), Cys80Arg (P-score: −11.148), Gly126Arg (P-score: −7.544), Gly154Glu (P-score: −7.608), and Arg203Cys (P-score: −7.090) all of these categorized as deleterious (potential loss of protein structure or function), only in isolates with some level of resistance (resistant or intermediate). In addition, a single nucleotide mutation in the nfsA gene was detected in strains 757 and 802, leading to truncation of the NfsA sequence in Gln67 (Gln67stop; CAA to TAA) and Gln147 (Gln147stop; CAG to TAG), respectively. These mutations produced 66 and 146 amino acid long proteins, respectively, instead of a wild-type protein with 240 amino acids.
Eleven amino acid substitutions were detected in NfsB: Gly66Asp (P-score: −1.775), Val93Ala (P-score: 2.155), and Ala174Glu (P-score: 1.621) all of these categorized as neutral, in susceptible isolates; and Leu22Ile (P-score: 0.334), Met75Ile (P-score: 2.094), and Lys122Arg (P-score: −0.179) all of these categorized as neutral, and Gln44His (P-score: −4.800), Phe84Ser (P-score: −5.862), Arg107Cys (P-score: −7.863), Gly192Ser (P-score: −5.961), and Arg207His (P-score: −4.966) all of these categorized as deleterious, in isolates with some level of resistance (resistant or intermediate). In addition, a single nucleotide mutation in the nfsB gene (GAA to TAA) was detected in strain 11, leading to a truncation of the NfsB sequence in Glu54.
Two amino acid substitutions in RibE were detected in three isolates with some level of resistance: Val51Ile (P-score: −0.363, categorized as neutral) and Pro55His (P-score: −8.840, categorized as deleterious). Finally, no oqxAB plasmid gene was detected in any isolate.
4. Discussion
4.1. Mechanisms of Resistance to Fosfomycin in E. coli
Mechanisms of resistance to fosfomycin described in various bacteria include the modification or overexpression of target molecule MurA, a reduced permeability, and irreversible antibiotic modification. The first two mechanisms are chromosomal, whereas the third can be chromosomal or encoded in transferable multi-resistance plasmids [14].
4.1.1. Modification or Overexpression of the Target (MurA)
The main action mechanism of fosfomycin is inhibition of the first step of peptidoglycan synthesis. Its chemical structure is analogous to that of phosphoenolpyruvate (PEP), therefore blocking the active center of enzyme UDP-N-acetylglucosamine enolpyruvyl transferase (MurA), covalently binding to the residue of cysteine Cys115 and preventing the binding of the substrate with the enzyme. In E. coli, amino acid substitutions in the active center of MurA, specifically Cys115Asp, are related to fosfomycin resistance [15] but are not common in clinical isolates of this species due to a drastic reduction in bacterial cell viability [16]. Only a few reports have associated amino acid substitutions in the MurA sequence of E. coli with resistance, especially Asp369Asn and Leu370Ile [9]. The latter was detected in 3 of the 22 fosfomycin-resistant isolates in the present study (strains 789, 809, and 853) but the protein variant was predicted to have a neutral effect in the PROVEAN analysis, with no alteration in the structure or function of the protein. Although previous crystallization studies found that leucine in position 370 of MurA does not interfere with its binding to fosfomycin, the fact that it is a highly preserved residue suggests an important role in the binding of PEP and therefore fosfomycin to the active site of the enzyme [9].
4.1.2. Permeability Reduction
Fosfomycin can use two transport systems to access the bacterial cytoplasm: glycerol-3-phosphate transporter (GlpT) and hexose phosphate transporter (UhpT). They are induced by the presence of their substrates (G3P and G6P, respectively) and require high levels of cyclic AMP (cAMP), whose synthesis depends on the enzyme adenylate cyclase (CyaA) and is regulated by the phosphoenolpyruvate-protein phosphotransferase (PtsI) system. The expression of GlpT is determined by a repressor gene, glpR, given that the interaction of GlpR with G3P increases transcription of the glpT gene. The expression of UhpT is in turn controlled by various regulating genes (uhpA, uhpB, and uhpC) [14]. This mechanism of action is unique; it does not confer cross-resistance to other antibiotics and it favors additive action with beta-lactams, aminoglycosides, glycopeptides, and fluoroquinolones, among others [17].
GlpT and UhpT are transporters with an extensive amino acid sequence homology that appear in several bacterial species with a high degree of conservation [14]. Various studies of E. coli have identified modifications of these proteins and/or proteins that regulate their expression (UhpA, PtsI, and CyaA) due to gene mutations or complete loss [9,18,19,20,21]. However, although the most important fosfomycin-resistance mechanism in this bacterium, modifications in chromosomal genes uhpT, glpT, uhpA, ptsI, or cyaA are reported to carry a high fitness cost, and clinical isolates with this resistance are known to be outcompeted by isolates susceptible to fosfomycin [22].
In the present study, all clinical isolates of E. coli presented substitutions in the amino acid sequence of GlpT. Some of them were detected in fosfomycin-susceptible isolates (Ala16Thr, Phe133Cys, Gly135Trp, Ala197Val, Leu373Arg, and Glu448Lys). Hence, these substitutions do not appear to be related per se to an alteration in GlpT function or resistance to the antibiotic. Other substitutions were solely detected in resistant isolates (Met52Leu, Leu297Phe, Glu443Gln, and Gln444Glu) but were classified as neutral in the PROVEAN analysis and would have no impact on the biological function of this protein. According to the PROVEAN analysis, only Gly84Asp and Pro212Leu substitutions could be significantly related to an alteration of GlpT functionality; however, the two isolates with this substitution (strains 26 and 752) proved able to grow in the presence of G3P. Hence, all isolates grew on M9 medium with G3P, indicating no significant loss of GlpT function with any substitution detected in the amino acid sequence of this transporter.
Among the 22 fosfomycin-resistant E. coli isolates, 4 showed no amino acid substitution in UhpT, 16 showed one substitution (Glu350Gln) and 2 were defective in UhpT due to gene loss (strains 11 and 26). We highlight that the uhpA gene was detected in strain 26 alone and that none of the 22 fosfomycin-resistant isolates were able to grow in the presence of G6P. In the UhpA sequence, the only substitution was Arg46Cys, which was only detected in two fosfomycin-susceptible isolates; therefore, it does not appear to be related per se to an alteration in the function of these proteins or to antibiotic resistance. According to our findings, all of the resistant isolates analyzed were defective in the UhpT transport system due to uhpT and/or uhpA deletion and showed no growth or only poor growth in a medium containing G6P as sole carbon source. Therefore, this finding supports the hypothesis that fosfomycin resistance in E. coli is most frequently attributable to blockage of the entry pathway of the antibiotic into the bacteria, mainly due to modifications in the UhpT transporter or its regulating proteins [18,19].
All of the E. coli clinical isolates in the present study showed substitutions in PtsI and CyaA. Given that some of these were detected in fosfomycin susceptible isolates (Ala306Thr and Arg367Lys in PtsI; Asn142Ser, Gly222Ser, Ala349Glu, Ser356Leu, Gly359Glu, Glu362Asp, Asp837Glu, and Thr840Ala in CyaA), they do not appear to be related per se to an alteration in the function of these proteins or to antibiotic resistance. Some other substitutions in these proteins were only detected in resistant isolates (Val25Ile in PtsI; Ser352Thr, Ala363Ser, and Ala363Gly in CyaA), as in previous studies [9]; nevertheless, their contribution to antibiotic resistance in these isolates cannot be affirmed, given that they were categorized as neutral in the PROVEAN analysis and there was no alteration in the function of GlpT, which was permeable to G3P. Therefore, it cannot be affirmed that amino acid substitutions in PtsI and CyaA contributed to resistance to fosfomycin in the clinical isolates of E. coli in the present study.
4.1.3. Enzymatic Modification of Fosfomycin
Two mechanisms may underlie fosfomycin resistance due to the action of modifying enzymes: epoxide ring opening, catalyzed by FosA enzymes (glutathione S-transferase), FosB (L-cysteine thiol transferase), or FosX (hydrolase epoxide); or antibiotic phosphorylation by FomA, FomB, or FosC enzymes [23]. Among these enzymes, FosA3 is the most widely described in E. coli plasmids, largely in Eastern Asia countries, although its detection is infrequent in Europe [11]. To our knowledge, this is the first time that the fosA3 gene has been detected in clinical isolates of E. coli in Spain (strains 66 and 871). Both isolates were also defective in the UhpT transport system due to uhpA deletion; however, the importance of this finding is that this plasmid-mediated gene may accelerate the dissemination of fosfomycin resistance in the near future.
4.2. Mechanisms of Resistance to Nitrofurantoin in E. coli
Nitrofurantoin is a prodrug of the nitrofuran family and exerts its antibiotic activity via multiple mechanisms of action, although none have been fully elucidated. It is known to inhibit: (i) protein synthesis, (ii) aerobic metabolism, (iii) nucleic acid synthesis, and (iv) cell wall synthesis. Its active form is generated within the bacterium by the action of nitroreductase enzymes, which reduce the nitro group coupled to the furan heterocyclic ring, giving rise to active intermediate metabolites that inhibit the synthesis of proteins involved in DNA, RNA, and carbohydrate metabolism.
Various studies have attributed resistance to nitrofurantoin in E. coli to the loss of intracellular nitroreductase activity via sequential mutations in nfsA and nfsB genes, which encode oxygen-insensitive nitroreductases, as well as to deletions affecting the active center of ribE, although the latter have not yet been reported in clinical isolates. Mutations in genes encoding oxygen-sensitive nitroreductases have not yet been described [5,24]. However, as in the case of fosfomycin, this nitrofurantoin resistance is reported to confer a high biological cost, and clinical isolates with this resistance are known to be outcompeted by susceptible isolates, reducing the likelihood of its detection in clinical isolates [5].
All E. coli clinical isolates in the present study showed substitutions in the amino acid sequence of NfsA and/or NfsB. As reported above, some were detected in nitrofurantoin-susceptible isolates (Glu58Asp, Ile117Thr, Lys141Glu, Gln147Arg, and Gly187Asp in NfsA; and Gly66Asp, Val93Ala, and Ala74Glu in NfsB). Although some of these (positions Ile117 and Lys141 in NfsA; Gly66 and Val93 in NfsB) have been associated with resistance in other studies [5,24,25], they were all classified as neutral in the PROVEAN analysis. Hence, none of these substitutions appear to be related per se to an alteration in the function of these proteins or to resistance to the antibiotic.
Other substitutions were detected in resistant isolates alone (His11Tyr, Asp19Asn, Ser33Arg, Gln67Leu, Cys80Arg, Gly126Arg, Gly154Glu, Ser180Asn, Arg203Cys, and truncation at Gln67 and Gln147 in NfsA; Leu22Ile, Gln44His, Met75Ile, Phe84Ser, Arg107Cys, Lys122Arg, Gly192Ser, Arg207His, and truncation at Glu54 in NfsB; Val51Ile and Pro55His in RibE). Some of these (His11, Ser33, Gln67, Gln147, and Arg203 in NfsA; Gln44, Met75, Arg107, Lys122, Gly192, and Arg207 in NfsB) have been associated with nitrofurantoin resistance in other studies [5,24,25,26]. According to the PROVEAN analysis, His11Tyr, Ser33Arg, Gln67Leu, Cys80Arg, Gly126Arg, Gly154Glu, and Arg203Cys in NfsA; Gln44His, Phe84Ser, Arg107Cys, Gly192Ser, and Arg207His in NfsB; and Pro55His in RibE were predicted to have a deleterious impact on the protein structure. Production of truncated NfsA (Gln67 and Gln147) or NfsB (Glu54) may have resulted in the inability or reduced ability of nitrofurantoin-resistant isolates to reduce the nitrofurantoin and produce active intermediates from the compound. Hence, these amino acid substitutions and/or truncated proteins would be related to nitrofurantoin resistance.
According to various studies, NfsA inactivation followed by NfsB inactivation is the main mechanism for high-level nitrofurantoin resistance in E. coli [5,26]. However, several of our nitrofurantoin-resistant clinical isolates did not show any modification in the NfsA sequence compatible with resistance (strains 66, 302, 799, 854, 883, and 892). Among these six isolates, we only detected substitutions in the NfsB sequence compatible with resistance (Arg207His) in the first two. However, we cannot affirm its association with resistance in the other four, although they presented various amino acid substitutions. Therefore, the mechanism that produces nitrofurantoin resistance in these four isolates is yet to be elucidated. Some authors have affirmed that NfsB inactivation in the presence of a wild-type nfsA gene cannot be associated with resistance [26]; in contrast, according to our findings, certain NfsB modifications requiring no previous NfsA alterations may be responsible for the functional alteration of bacterial nitroreductases, as also previously reported [24].
More recently, it has been reported that the presence of OqxAB (a plasmid-encoded multidrug efflux pump that confers reduced susceptibility to quinolones, tigecycline, chloramphenicol, trimethoprim, and disinfectants such as quaternary ammonium compounds) would also enhance nitrofurantoin resistance via an active antibiotic expulsion mechanism in E. coli isolates with previous nitroreductase modifications, because it has not been possible to relate the presence of OqxAB per se in the bacterium to antibiotic resistance levels [27]. This plasmid has been widely detected in E. coli and other enterobacteria, both in human and animal isolates, mainly in China [25,27]; although its presence has also been reported in Europe [28,29], including Spain [30]. However, this plasmid was not detected in any of our series of isolates, indicating that nitrofurantoin resistance must involve mechanisms other than antibiotic extrusion.
Finally, as in the present study, there have been reports of nitrofurantoin-resistant E. coli isolates with no amino acid substitutions in NfsA, NfsB, and RibE, or presence of the oqxAB plasmid, indicating the need to identify new mechanisms that explain nitrofurantoin resistance in this bacterium [27].
5. Conclusions
These results suggest that the emergence of fosfomycin resistance in clinical isolates of E. coli in our setting is largely attributable to the absence of expression of transporter UhpT due to complete deletion of the uhpT and/or uhpA regulating genes, reducing the permeability of the bacterium to the antibiotic. To our knowledge, we report for the first time the presence in Spain of the plasmid gene fosA3, responsible for the enzyme glutathione S-transferase, which inactivates the antibiotic. We consider this finding to be of major epidemiological importance, given its potential dissemination not only in E. coli but also other bacteria. Nitrofurantoin resistance can be explained, at least in part, by the presence of specific modifications in NfsA, NfsB, or RibE proteins. The presence of oqxAB plasmid genes does not appear to represent an important resistance mechanism among E. coli clinical isolates in our setting at the present time. The emergence and spread of these resistance mechanisms, including transferable resistance, could compromise the future usefulness of fosfomycin and nitrofurantoin against UTIs.
Acknowledgments
The authors are grateful to Concepción Gimeno Cardona, Head of the Microbiology Laboratory of the University General Hospital of Valencia, Associate Professor of Microbiology at Valencia University and Coordinator of the External Quality Control Program of the Spanish Society of Infectious Diseases and Clinical Microbiology (Spanish abbreviation: SEIMC), for critically reviewing the article and providing input into the final paper.
Author Contributions
Conceptualization: A.S.-P. and J.G.-F.; Methodology: A.S.-P., I.L.-M., M.A.-C., L.J.M.-G. and J.G.-F.; Formal Analysis: A.S.-P. and I.L.-M.; Investigation: A.S.-P., I.L.-M. and M.A.-C.; Validation: A.S.-P. and J.G.-F.; Writing—Original Draft Preparation: A.S.-P.; Writing—Review & Editing: A.S.-P., I.L.-M. and J.G.-F. All authors have read and agreed to the published version of the manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Sorlozano A., Jimenez-Pacheco A., de Dios Luna Del Castillo J., Sampedro A., Martinez-Brocal A., Miranda-Casas C., Navarro-Marí J.M., Gutiérrez-Fernández J. Evolution of the resistance to antibiotics of bacteria involved in urinary tract infections: A 7-year surveillance study. Am. J. Infect. Control. 2014;42:1033–1038. doi: 10.1016/j.ajic.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Vardakas K.Z., Legakis N.J., Triarides N., Falagas M.E. Susceptibility of contemporary isolates to fosfomycin: A systematic review of the literature. Int. J. Antimicrob. Agents. 2016;47:269–285. doi: 10.1016/j.ijantimicag.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Falagas M.E., Kastoris A.C., Kapaskelis A.M., Karageorgopoulos D.E. Fosfomycin for the treatment of multidrug-resistant, including extended-spectrum beta-lactamase producing, Enterobacteriaceae infections: A systematic review. Lancet Infect. Dis. 2010;10:43–50. doi: 10.1016/S1473-3099(09)70325-1. [DOI] [PubMed] [Google Scholar]
- 4.Sánchez-García J.M., Sorlózano-Puerto A., Navarro-Marí J.M., Gutiérrez Fernández J. Evolution of the antibiotic-resistance of microorganisms causing urinary tract infections: A 4-year epidemiological surveillance study in a hospital population. Rev. Clin. Esp. 2019;219:116–123. doi: 10.1016/j.rce.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Sandegren L., Lindqvist A., Kahlmeter G., Andersson D.I. Nitrofurantoin resistance mechanism and fitness cost in Escherichia coli. J. Antimicrob. Chemother. 2008;62:495–503. doi: 10.1093/jac/dkn222. [DOI] [PubMed] [Google Scholar]
- 6.Giske C.G. Contemporary resistance trends and mechanisms for the old antibiotics colistin, temocillin, fosfomycin, mecillinam and nitrofurantoin. Clin. Microbiol. Infect. 2015;21:899–905. doi: 10.1016/j.cmi.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 7.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2017. CLSI supplement M100. [Google Scholar]
- 8.Nakamura G., Wachino J., Sato N., Kimura K., Yamada K., Jin W., Shibayama K., Yagi T., Kawamura K., Arakawa Y. Practical agar-based disk potentiation test for detection of fosfomycin-nonsusceptible Escherichia coli clinical isolates producing glutathione S-transferases. J. Clin. Microbiol. 2014;52:3175–3179. doi: 10.1128/JCM.01094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahata S., Ida T., Hiraishi T., Sakakibara S., Maebashi K., Terada S., Muratani T., Matsumoto T., Nakahama C., Tomono K. Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int. J. Antimicrob. Agents. 2010;35:333–337. doi: 10.1016/j.ijantimicag.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 10.Hou J., Huang X., Deng Y., He L., Yang T., Zeng Z., Chen Z., Liu J.H. Dissemination of the fosfomycin resistance gene fosA3 with CTX-M β-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob. Agents Chemother. 2012;56:2135–2138. doi: 10.1128/AAC.05104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benzerara Y., Gallah S., Hommeril B., Genel N., Decré D., Rottman M., Arlet G. Emergence of plasmid-mediated fosfomycin-resistance genes among Escherichia coli isolates, France. Emerg. Infect. Dis. 2017;23:1564–1567. doi: 10.3201/eid2309.170560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X., Zhang W., Pan W., Yin J., Pan Z., Gao S., Jiao X. Prevalence of qnr, aac(6=)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob. Agents Chemother. 2012;56:3423–3427. doi: 10.1128/AAC.06191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi Y., Chan A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castañeda-García A., Blázquez J., Rodríguez-Rojas A. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics. 2013;2:217–236. doi: 10.3390/antibiotics2020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D.H., Lees W.J., Kempsell K.E., Lane W.S., Duncan K., Walsh C.T. Characterization of a Cys115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry. 1996;35:4923–4928. doi: 10.1021/bi952937w. [DOI] [PubMed] [Google Scholar]
- 16.Herring C.D., Blattner F.R. Conditional lethal amber mutations in essential Escherichia coli genes. J. Bacteriol. 2004;186:2673–2681. doi: 10.1128/JB.186.9.2673-2681.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falagas M.E., Vouloumanou E.K., Samonis G., Vardakas K.Z. Fosfomycin. Clin. Microbiol. Rev. 2016;29:321–347. doi: 10.1128/CMR.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucas A.E., Ito R., Mustapha M.M., McElheny C.L., Mettus R.T., Bowler S.L., Kantz S.F., Pacey M.P., Pasculle A.W., Cooper V.S., et al. Frequency and mechanisms of spontaneous fosfomycin nonsusceptibility observed upon disk diffusion testing of Escherichia coli. J. Clin. Microbiol. 2017;56:e01368-17. doi: 10.1128/JCM.01368-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tseng S.P., Wang S.F., Kuo C.Y., Huang J.W., Hung W.C., Ke G.M., Lu P.L. Characterization of fosfomycin resistant extended-spectrum β-lactamase-producing Escherichia coli isolates from human and pig in Taiwan. PLoS ONE. 2015;10:e0135864. doi: 10.1371/journal.pone.0135864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wachino J., Yamane K., Suzuki S., Kimura K., Arakawa Y. Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob. Agents Chemother. 2010;54:3061–3064. doi: 10.1128/AAC.01834-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y., Zheng B., Li Y., Zhu S., Xue F., Liu J. Antimicrobial susceptibility and molecular mechanisms of fosfomycin resistance in clinical Escherichia coli isolates in Mainland China. PLoS ONE. 2015;10:e0135269. doi: 10.1371/journal.pone.0135269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nilsson A.I., Berg O.G., Aspevall O., Kahlmeter G., Andersson D.I. Biological costs and mechanisms of fosfomycin resistance in Escherichia coli. Antimicrob. Agents Chemother. 2003;47:2850–2858. doi: 10.1128/AAC.47.9.2850-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karageorgopoulos D.E., Wang R., Yu X.H., Falagas M.E. Fosfomycin: Evaluation of the published evidence on the emergence of antimicrobial resistance in Gram-negative pathogens. J. Antimicrob. Chemother. 2012;67:255–268. doi: 10.1093/jac/dkr466. [DOI] [PubMed] [Google Scholar]
- 24.Vervoort J., Xavier B.B., Stewardson A., Coenen S., Godycki-Cwirko M., Adriaenssens N., Kowalczyk A., Lammens C., Harbarth S., Goossens H., et al. An in vitro deletion in ribE encoding lumazine synthase contributes to nitrofurantoin resistance in Escherichia coli. Antimicrob. Agents Chemother. 2014;58:7225–7233. doi: 10.1128/AAC.03952-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X., Zhang Y., Wang F., Wang C., Chen L., Liu H., Lu H., Wen H., Zhou T. Unravelling mechanisms of nitrofurantoin resistance and epidemiological characteristics among Escherichia coli clinical isolates. Int. J. Antimicrob. Agents. 2018;52:226–232. doi: 10.1016/j.ijantimicag.2018.04.021. [DOI] [PubMed] [Google Scholar]
- 26.Whiteway J., Koziarz P., Veall J., Sandhu N., Kumar P., Hoecher B., Lambert I.B. Oxygen-insensitive nitroreductases: Analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J. Bacteriol. 1998;180:5529–5539. doi: 10.1128/JB.180.21.5529-5539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho P.L., Ng K.Y., Lo W.U., Law P.Y., Lai E.L., Wang Y., Chow K.H. Plasmid-mediated oqxAB is an important mechanism for nitrofurantoin resistance in Escherichia coli. Antimicrob. Agents Chemother. 2015;60:537–543. doi: 10.1128/AAC.02156-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campos J., Mourão J., Marçal S., Machado J., Novais C., Peixe L., Antunes P. Clinical Salmonella Typhimurium ST34 with metal tolerance genes and an IncHI2 plasmid carrying oqxAB-aac(6′)-Ib-cr from Europe. J. Antimicrob. Chemother. 2016;71:843–845. doi: 10.1093/jac/dkv409. [DOI] [PubMed] [Google Scholar]
- 29.Dotto G., Giacomelli M., Grilli G., Ferrazzi V., Carattoli A., Fortini D., Piccirillo A. High prevalence of oqxAB in Escherichia coli isolates from domestic and wild lagomorphs in Italy. Microb. Drug Resist. 2014;20:118–123. doi: 10.1089/mdr.2013.0141. [DOI] [PubMed] [Google Scholar]
- 30.Rodríguez-Martínez J.M., Díaz de Alba P., Briales A., Machuca J., Lossa M., Fernández-Cuenca F., Rodríguez Baño J., Martínez-Martínez L., Pascual Á. Contribution of OqxAB efflux pumps to quinolone resistance in extended-spectrum-β-lactamase-producing Klebsiella pneumoniae. J. Antimicrob. Chemother. 2013;68:68–73. doi: 10.1093/jac/dks377. [DOI] [PubMed] [Google Scholar]