Abstract
Background
Our previous study found that chicken KLF7 was an important regulator in formation of adipose tissue. In the present study, we analyzed the association for DNA methylation in chicken KLF7 with its transcripts of abdominal adipose tissue and blood metabolic indicators.
Results
The KLF7 transcripts of the adipose tissue of Chinese yellow broilers were associated with age (F = 6.67, P = 0.0035). In addition, the KLF7 transcripts were negatively correlated with blood glucose levels (r = − 0.61841, P = 0.0140). The DNA methylation levels of 26 CpG loci in the chicken KLF7 promoter and Exon 2 were studied by Sequenom MassArray. A total of 22 valid datasets were obtained. None of them was significantly different in relation to age (P > 0.05). However, the DNA methylation levels in the promoter were lower than those in Exon 2 (T = 40.74, P < 0.01). Correlation analysis showed that the DNA methylation levels of PCpG6 and E2CpG9 were significantly correlated with KLF7 transcripts and blood high-density lipoprotein levels, respectively, and many CpG loci were correlated with each other (P < 0.05). The methylation data were subjected to principal component analysis and factor analysis. The six principal components (z1–z6) were extracted and named Factors 1–6, respectively. Factor analysis showed that Factor 1 had a higher load on the loci in the promoter, and Factors 2–6 loaded highly on quite different loci in Exon 2. Correlation analysis showed that only z1 was significantly correlated to KLF7 transcripts (P < 0.05). In addition, an established regression equation between z1 and KLF7 transcripts was built, and the contribution of z1 to the variation on KLF7 transcripts was 34.29%.
Conclusions
In conclusion, the KLF7 transcripts of chicken abdominal adipose tissue might be inhibited by DNA methylation in the promoter, and it might be related to the DNA methylation level of PCpG6.
Keywords: Chicken, Adipose tissue, DNA methylation, Blood metabolic indicators, Krüppel-like factor 7
Background
Krüppel-like factor 7 (KLF7), also known as ubiquitous KLF (UKLF), is a member of the KLFs, which are characterized by three Cys2-His2 zinc fingers at the C-terminus [1]. The KLF7 gene is highly conserved among animals, especially mammalian species and birds.
Reports on mammalian specie showed that KLF7 played an important role in the differentiation of neuroectodermal and mesodermal cell lineages in vitro [2]. Gene targeting studies in mice implicated that KLF7 was involved in nervous system development [3, 4], and mice without KLF7 activity displayed hypoplastic olfactory bulbs which lack peripheral innervation [5].
KLF7 had been reported as a key regulator of human obesity [6], diabetes mellitus type 2 (T2DM) [7, 8], and blood disease [9]. In addition, KLF7 was an oncogene in several kinds of cancers, including gastric cancer [10], lung adenocarcinoma [11], glioma [12] and oral squamous cell carcinoma [13]. Additionally, Genetics analysis showed that KLF7 was associated with self-rated health [14] and neurodevelopmental disorders [15] in human. Integrative genomic studies suggested that KLF7 might be one of the core factors that regulate cardiovascular diseases [16].
Our previous study in chickens showed that the number of KLF7 transcripts of abdominal fat tissue was greater in lean broilers than in fat broilers, and KLF7 promotes the proliferation of chicken preadipocytes and inhibits their differentiation in vitro [17]. One SNP (c. A141G) in the KLF7 coding sequence might be a molecular marker for the selection of blood very-low-density lipoprotein and abdominal fat content in broilers [18]. DNA methylation is a very well-studied epigenetic phenomenon that typically correlates with gene silencing [19]. The DNA methylation in KLF7 is reported to be associated with the occurrence and development of gastric cancer in humans [20, 21]. However, the effect of KLF7 DNA methylation in the adipose tissue of birds has not been reported.
Adipose tissue is an important site for lipid storage, energy homeostasis, and insulin sensitivity in human being and animals [22]. The previous study in vitro showed that overexpression of KLF7 might induce the development of T2DM via serval ways, including suppression of insulin secretion, insulin sensitivity and adipogenesis [7, 8]. However, whether the DNA methylation of KLF7 in the adipose tissue had associations with its expression and the blood metabolic indicators, including the levels of glucose (Glu), phospholipids (PL), total cholesterol (TC), triglyceride (TG), low-density lipoprotein (LDL) and high-density lipoprotein (HDL), which were correlated with the T2DM and obesity, is still unclear. In the present study, the associations for DNA methylation in the chicken KLF7 gene with its expression and fasting blood metabolic indicators were studied. The results might improve the understanding of KLF7 expression and function in adipose tissue.
Results
KLF7 mRNA expression of chicken adipose tissues
The expression of KLF7 mRNA in the abdominal adipose tissue of Chinese fast-growing yellow broilers at the age of 2, 4, 6, and 8 weeks was studied by real-time PCR. The results showed that KLF7 mRNA was expressed in all the tissues studied. The KLF7 transcripts data conformed to a normal distribution (Shapiro–Wilk test; W = 0.957301, P = 0.4635). ANOVA analysis showed that KLF7 transcripts were significantly associated with age (F = 6.67, P = 0.0035), and at the age of 8 weeks, the level of KLF7 transcripts was significantly greater than that at the age of 2, 4, and 6 weeks (P < 0.05, Fig. 1).
Fig. 1.
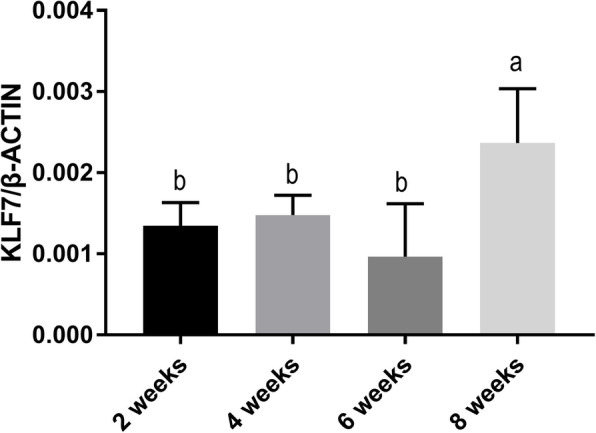
The mRNA expression pattern of KLF7 in chicken abdominal fat tissue. β-ACTIN was used as an internal control. The diagram shows the relative quantification of KLF7 expression. Error bars indicate the standard deviations from biological replicates (n ≥ 4). The different lowercase letters above columns indicate significant difference in various ages (Duncan’s multiple tests, P < 0.05)
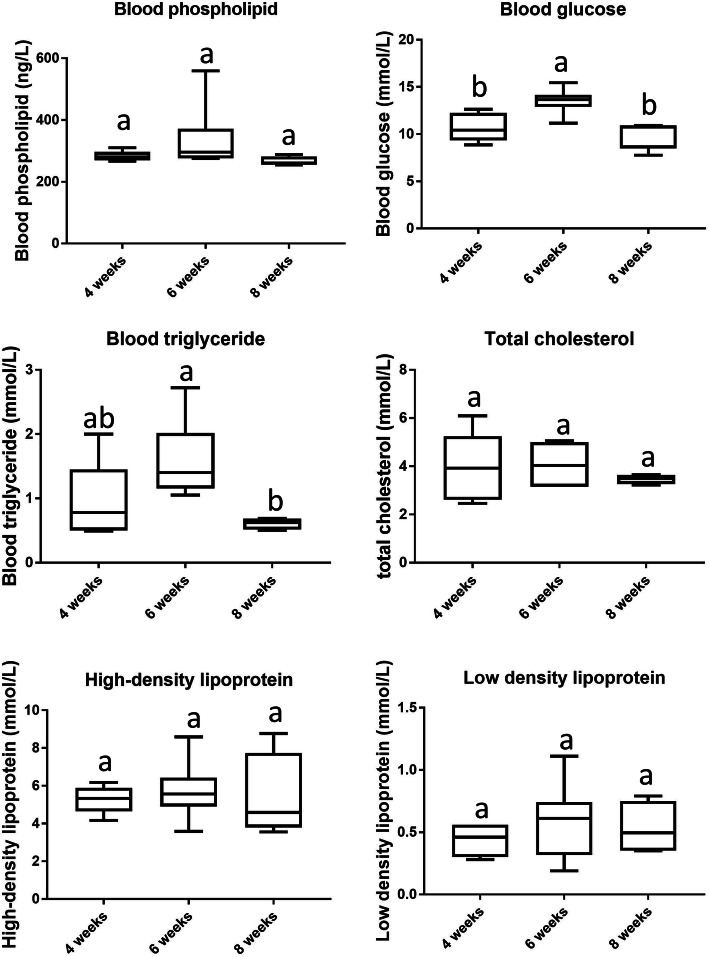
The level of blood metabolic indicators during the development and growth of broilers
The levels of Glu, PL, TC, TG, LDL and HDL in the blood of fasting Chinese fast-growing yellow broilers at the age of 4, 6, and 8 weeks were studied by spectroscopy. ANOVA or Kruskal–Wallis analysis showed there was no significant difference in the levels of PL, TC, LDL, and HDL in chicken blood among the ages of 4, 6, and 8 weeks (P > 0.05, Table 1). However, blood glucose levels in 6-week-old broilers were significantly greater than those in 4- and 8-week-old broilers (P < 0.05, Fig. 2), and the blood TG contents of 8-week-old broilers were significantly lower than those of 6-week-old broilers (P < 0.05, Fig. 2). Spearman’s correlation analysis showed that KLF7 transcripts were significantly negatively correlated with fasting blood glucose concentration (r = − 0.61841, P = 0.0140), and there was no significant association between KLF7 transcripts and other blood parameters (P > 0.05).
Table 1.
Datasets characteristics of chicken blood metabolic indicators
| Dataset | N a | Shapiro-Wilk test | Difference test among ages | ||
|---|---|---|---|---|---|
| PL | 15 | W = 0.504656 | P < 0.0001 | H = 5.1574 | P = 0.0759 |
| GLU | 15 | W = 0.970623 | P = 0.8672 | F = 8.24 | P = 0.0056 |
| TG | 15 | W = 0.863964 | P = 0.0275 | H = 7.3490 | P = 0.0254 |
| TC | 15 | W = 0.940254 | P = 0.3856 | F = 0.43 | P = 0.6586 |
| HDL | 15 | W = 0.864245 | P = 0.0278 | H = 1.1558 | P = 0.5611 |
| LDL | 15 | W = 0.916808 | P = 0.1722 | F = 0.52 | P = 0.6077 |
Note: a represents the number of samples analysed
Fig. 2.
Levels of glucose (Glu), phospholipids (PL), total cholesterol (TC), triglyceride (TG), low-density lipoprotein (LDL), and high-density lipoprotein (HDL) in the blood of fasting Chinese fast-growing yellow broilers. The different lowercase letters above columns indicate significant difference in various ages (Duncan’s multiple tests, P < 0.05)
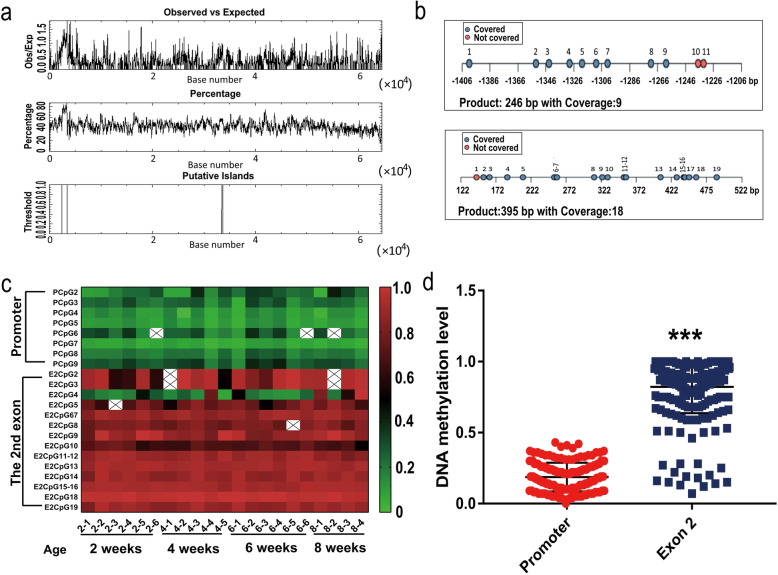
CpG density in the genomic region of chicken KLF7
The CpG density analysis showed that there were two CpG islands in the genomic region of chicken KLF7. One CpG island, with a sequence interval of − 1156 to − 164 bp upstream of the KLF7 translation initiation site, was about 993 bp in length and located in the promoter and the other was located in Exon 2. The sequence interval between 254 bp to 523 bp after the first nucleotide of Exon 2, was about 269 bp in length (Fig. 3a).
Fig. 3.
DNA methylation analysis of chicken KLF7 in adipose tissues. a. The CpG density of the genomic region of chicken KLF7 analyzed by CpGplot (Version 6.6.0). b. The CpG loci analyzed in the promoter and Exon 2. c. The levels of DNA methylation of CpG loci analyzed. d. The comparison of DNA methylation levels between the promoter and Exon 2. Asterisks indicate a significant difference between the promoter and Exon 2 (Student’s t test, ***P < 0.001)
DNA methylation of chicken KLF7 in adipose tissues
Sequenom MassArray was used to study DNA methylation in CpG-rich sequences of chicken KLF7. In the promoter region, the DNA methylation levels of nine CpG loci in the sequence interval from − 1452 bp to − 1206 bp upstream of the translation initiation site were studied (Fig. 3b), and eight valid datasets were obtained (Fig. 3c). The data were distributed normally (P > 0.05, Table 2). ANOVA analysis found no significant difference in the DNA methylation level of any of these eight loci among the ages of 2, 4, 6, and 8 weeks (P > 0.05).
Table 2.
Datasets characteristics of DNA methylation of chicken KLF7 in abdominal adipose tissues
| Dataset | N a | Shapiro-Wilk test | Difference test among ages | ||
|---|---|---|---|---|---|
| PCpG2 | 21 | W = 0.963142 | P = 0.5815 | F = 0.74 | P = 0.5405 |
| PCpG3 | 21 | W = 0.940649 | P = 0.2244 | F = 0.10 | P = 0.9566 |
| PCpG4 | 21 | W = 0.935328 | P = 0.1760 | F = 0.42 | P = 0.7418 |
| PCpG5 | 21 | W = 0.95307 | P = 0.3888 | F = 0.35 | P = 0.7874 |
| PCpG6 | 18 | W = 0.944124 | P = 0.3404 | F = 0.22 | P = 0.8776 |
| PCpG7 | 21 | W = 0.96436 | P = 0.6079 | F = 0.49 | P = 0.6906 |
| PCpG8 | 21 | W = 0.964176 | P = 0.6039 | F = 0.34 | P = 0.7965 |
| PCpG9 | 21 | W = 0.977153 | P = 0.8795 | F = 0.07 | P = 0.9753 |
| E2CpG2 | 19 | W = 0.792999 | P = 0.0009 | H = 3.4737 | P = 0.3242 |
| E2CpG3 | 19 | W = 0.792999 | P = 0.0009 | H = 3.4737 | P = 0.3242 |
| E2CpG4 | 21 | W = 0.803609 | P = 0.0007 | H = 5.5746 | P = 0.1342 |
| E2CpG5 | 20 | W = 0.937047 | P = 0.2107 | F = 0.19 | P = 0.9050 |
| E2CpG6–7 | 21 | W = 0.947677 | P = 0.3077 | F = 0.17 | P = 0.9121 |
| E2CpG8 | 20 | W = 0.943714 | P = 0.2815 | F = 0.31 | P = 0.8190 |
| E2CpG9 | 21 | W = 0.948257 | P = 0.3156 | F = 0.61 | P = 0.6192 |
| E2CpG10 | 21 | W = 0.93046 | P = 0.1406 | F = 1.38 | P = 0.2836 |
| E2CpG11–12 | 21 | W = 0.921137 | P = 0.0914 | F = 2.82 | P = 0.0703 |
| E2CpG13 | 21 | W = 0.921203 | P = 0.0917 | F = 0.42 | P = 0.7419 |
| E2CpG14 | 21 | W = 0.969337 | P = 0.7183 | F = 0.23 | P = 0.8761 |
| E2CpG15–16 | 21 | W = 0.961392 | P = 0.5446 | F = 0.98 | P = 0.4234 |
| E2CpG18 | 21 | W = 0.852523 | P = 0.0047 | H = 2.7900 | P = 0.4252 |
| E2CpG19 | 21 | W = 0.969337 | P = 0.7183 | F = 0.23 | P = 0.8761 |
Note: a represents the number of samples analysed
All the CpG loci in the CpG island of Exon 2 were studied, and highly combined CpG loci were considered as one single locus. A total of 14 valid datasets of 17 CpG loci were obtained (Fig. 3c). Not all datasets distributed normally (Table 2). ANOVA or Kruskal–Wallis analysis showed that there was no significant difference in the methylation level of loci in relation to age (P > 0.05). However, the level of DNA methylation in the promoter was significantly lower than that in Exon 2 (T = 40.74, P < 0.001, Fig. 3d).
Sequence analysis showed that, although CpG island could be both detected in the promoter of chicken and human KLF7 (supplementary Figure 1a), the DNA sequence similarity between them was low (supplementary Figure 1b). No homologous locus of CpG site was found in human KLF7 promoter. However, the DNA sequence similarity between the Exon 2 in chicken and human KLF7s was high, and the loci of E2CpG1, E2CpG4, E2CpG7, E2CpG9 and E2CpG16 were conserved between chicken and human (supplementary Figure 1c).
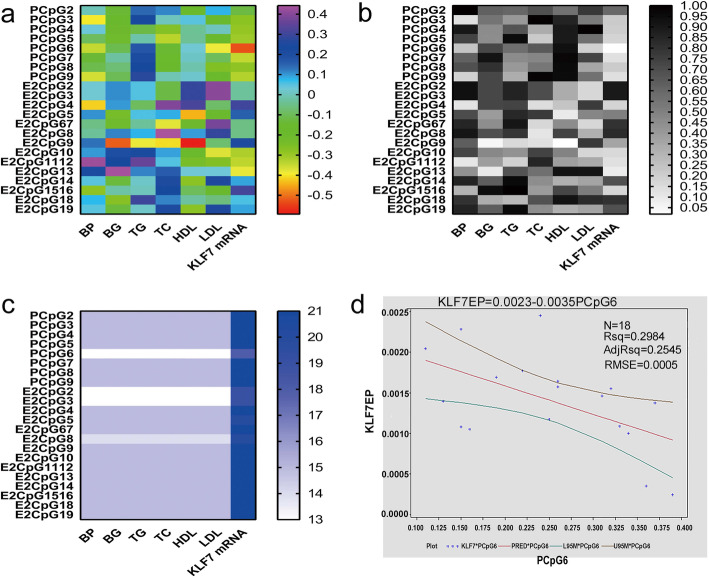
The association for DNA methylation with KLF7 transcripts and blood metabolic indicators
The association for the DNA methylation in each locus with KLF7 transcripts was studied by Spearman correlation analysis. The results showed that the DNA methylation of PCpG6 was significantly correlated to KLF7 transcripts (r = − 0.53099, P = 0.0234). However, no significant correlation between the DNA methylation of other loci and KLF7 expression was found (P > 0.05, Fig. 4).
Fig. 4.
Association analysis of DNA methylation with KLF7 transcripts and blood metabolic indicators. a. The r value of the Spearman correlation analysis. b. The P value of the Spearman correlation analysis. c. The number of samples used in the Spearman correlation analysis. d. The regression analysis between the DNA methylation of PCpG6 and KLF7 transcripts in chicken adipose tissue
In addition, the DNA methylation of E2CpG9 was significantly correlated with fasting blood glucose level (r = − 0.51706, P = 0.0484) and blood HDL (r = − 0.59373, P = 0.0196). No other significant correlation between the DNA methylation of other loci and the blood metabolic indicators measured was found (P > 0.05, Fig. 4).
Both the dataset of PCpG6 DNA methylation and KLF7 transcripts distributed normally (Shapiro–Wilk test, P > 0.05), so the regression relationship between PCpG6 DNA methylation and KLF7 transcripts was studied using a generalized linear regression model. The established regression equation, Y (KLF7 transcripts) = 0.00229–0.00351× (DNA methylation of PCpG6), was statistically significant (F = 6.80, P = 0.0190, R2 = 0.2545; Fig. 4d).
Correlation between the DNA methylation of different CpG loci
The correlations between the methylation of different CpG loci were analyzed by Spearman correlation analysis. The results showed that there were significant associations for several CpG loci (P < 0.05). The methylation levels of PCpG3-PCpG9 were positively correlated with each other (P < 0.05), and the methylation level of PCpG2 was positively correlated with that of PCpG8 (P < 0.05). In addition, the methylation levels of PCpG4, PCpG5, and PCpG8 were significantly associated with that of E2CpG4 (P < 0.05). Additionally, the methylation data onto E2CpG2 and E2CpG14 were the same to those of E2CpG3 and E2CpG19, respectively (r = 1, P < 0.0001). The methylation data onto E2CpG2, E2CpG3, and E2CpG4 was negatively correlated with that of E2CpG11–12 (P < 0.05); The methylation data onto E2CpG4 was negatively correlated with that of E2CpG18 (P < 0.05), The methylation data onto E2CpG5 was positively correlated with that of E2CpG9 (P < 0.05), and the methylation data onto E2CpG8 was positively correlated with that of E2CpG13 (P < 0.05, Table 3).
Table 3.
Correlation matrix of the CpG locus in the region of chicken KLF7
| E2CpG2 | E2CpG3 | E2CpG4 | E2CpG5 | E2CpG6–7 | E2CpG8 | E2CpG9 | E2CpG10 | E2CpG11–12 | E2CpG13 | E2CpG14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| E2CpG3 | 1.0000 a | ||||||||||
| <.0001 b | |||||||||||
| 19 c | |||||||||||
| E2CpG4 | 0.2938 | 0.2938 | |||||||||
| 0.2221 | 0.2221 | ||||||||||
| 19 | 19 | ||||||||||
| E2CpG5 | 0.0197 | 0.0197 | 0.0859 | ||||||||
| 0.9380 | 0.9380 | 0.7187 | |||||||||
| 18 | 18 | 20 | |||||||||
| E2CpG67 | 0.2824 | 0.2824 | −0.3787 | −0.0197 | |||||||
| 0.2414 | 0.2414 | 0.0905 | 0.9342 | ||||||||
| 19 | 19 | 21 | 20 | ||||||||
| E2CpG8 | 0.0366 | 0.0366 | 0.2197 | 0.2710 | 0.2730 | ||||||
| 0.8854 | 0.8854 | 0.3520 | 0.2618 | 0.2442 | |||||||
| 18 | 18 | 20 | 19 | 20 | |||||||
| E2CpG9 | −0.1636 | − 0.1636 | − 0.0599 | 0.4711 | 0.3857 | 0.1230 | |||||
| 0.5034 | 0.5034 | 0.7964 | 0.0360 | 0.0842 | 0.6054 | ||||||
| 19 | 19 | 21 | 20 | 21 | 20 | ||||||
| E2CpG10 | −0.3801 | − 0.3801 | − 0.4156 | − 0.0442 | − 0.0668 | − 0.0898 | − 0.1541 | ||||
| 0.1084 | 0.1084 | 0.0610 | 0.8532 | 0.7734 | 0.7065 | 0.5050 | |||||
| 19 | 19 | 21 | 20 | 21 | 20 | 21 | |||||
| E2CpG11–12 | −0.4571 | −0.4571 | − 0.6615 | 0.2613 | 0.0946 | 0.2131 | 0.1588 | 0.4974 | |||
| 0.0491 | 0.0491 | 0.0011 | 0.2659 | 0.6834 | 0.3671 | 0.4918 | 0.0218 | ||||
| 19 | 19 | 21 | 20 | 21 | 20 | 21 | 21 | ||||
| E2CpG13 | 0.1260 | 0.1260 | 0.1022 | 0.0637 | 0.2334 | 0.7436 | 0.2532 | −0.0261 | 0.3612 | ||
| 0.6072 | 0.6072 | 0.6595 | 0.7896 | 0.3087 | 0.0002 | 0.2680 | 0.9106 | 0.1077 | |||
| 19 | 19 | 21 | 20 | 21 | 20 | 21 | 21 | 21 | |||
| E2CpG14 | 0.3611 | 0.3611 | 0.0749 | 0.3268 | 0.3277 | 0.4237 | 0.3537 | 0.1157 | 0.1184 | 0.3634 | |
| 0.1288 | 0.1288 | 0.7469 | 0.1597 | 0.1470 | 0.0627 | 0.1158 | 0.6175 | 0.6092 | 0.1054 | ||
| 19 | 19 | 21 | 20 | 21 | 20 | 21 | 21 | 21 | 21 | ||
| E2CpG15–16 | 0.0687 | 0.0687 | 0.4151 | 0.2157 | 0.0786 | 0.3225 | 0.3674 | 0.1090 | −0.0307 | 0.3456 | 0.3779 |
| 0.7800 | 0.7800 | 0.0613 | 0.3611 | 0.7348 | 0.1655 | 0.1013 | 0.6380 | 0.8950 | 0.1249 | 0.0912 | |
| 19 | 19 | 21 | 20 | 21 | 20 | 21 | 21 | 21 | 21 | 21 | |
| E2CpG18 | −0.3398 | −0.3398 | − 0.4726 | 0.0556 | 0.2350 | −0.0527 | − 0.0214 | 0.1926 | 0.2877 | −0.3586 | − 0.1449 |
| 0.1546 | 0.1546 | 0.0305 | 0.8160 | 0.3052 | 0.8255 | 0.9265 | 0.4030 | 0.2060 | 0.1104 | 0.5310 | |
| 19 | 19 | 21 | 20 | 21 | 20 | 21 | 21 | 21 | 21 | 21 | |
| E2CpG19 | 0.3611 | 0.3611 | 0.0749 | 0.3268 | 0.3277 | 0.4237 | 0.3537 | 0.1157 | 0.1184 | 0.3634 | 1.0000 |
| 0.1288 | 0.1288 | 0.7469 | 0.1597 | 0.1470 | 0.0627 | 0.1158 | 0.6175 | 0.6092 | 0.1054 | <.0001 | |
| 19 | 19 | 21 | 20 | 21 | 20 | 21 | 21 | 21 | 21 | 21 | |
| PCpG2 | −0.3467 | −0.3467 | −0.2836 | 0.3315 | −0.1760 | 0.0369 | 0.1558 | 0.1069 | 0.2605 | −0.0689 | −0.0848 |
| 0.1459 | 0.1459 | 0.2129 | 0.1534 | 0.4454 | 0.8774 | 0.5002 | 0.6447 | 0.2542 | 0.7667 | 0.7149 | |
| 19 | 19 | 21 | 20 | 21 | 20 | 21 | 21 | 21 | 21 | 21 | |
| PCpG3 | −0.2517 | −0.2517 | − 0.3798 | − 0.1509 | 0.2249 | − 0.0830 | − 0.1594 | 0.2155 | 0.1652 | − 0.2025 | − 0.0603 |
| 0.2987 | 0.2987 | 0.0895 | 0.5254 | 0.3271 | 0.7281 | 0.4900 | 0.3481 | 0.4742 | 0.3786 | 0.7953 | |
| 19 | 19 | 21 | 20 | 21 | 20 | 21 | 21 | 21 | 21 | 21 | |
| PCpG4 | −0.4287 | −0.4287 | −0.4807 | −0.2412 | − 0.0719 | −0.3458 | − 0.0344 | 0.4087 | 0.1869 | −0.2648 | − 0.0884 |
| 0.0670 | 0.0670 | 0.0274 | 0.3056 | 0.7566 | 0.1353 | 0.8825 | 0.0658 | 0.4172 | 0.2460 | 0.7032 | |
| 19 | 19 | 21 | 20 | 21 | 20 | 21 | 21 | 21 | 21 | 21 | |
| PCpG5 | −0.4217 | − 0.4217 | −0.6114 | − 0.2451 | 0.1056 | −0.3069 | − 0.1065 | 0.3589 | 0.2236 | −0.3098 | − 0.2323 |
| 0.0722 | 0.0722 | 0.0032 | 0.2977 | 0.6488 | 0.1882 | 0.6460 | 0.1101 | 0.3300 | 0.1717 | 0.3108 | |
| 19 | 19 | 21 | 20 | 21 | 20 | 21 | 21 | 21 | 21 | 21 | |
| PCpG6 | −0.3245 | − 0.3245 | − 0.2233 | − 0.2761 | 0.0764 | −0.1193 | − 0.2020 | 0.4222 | 0.1364 | −0.0861 | − 0.1235 |
| 0.2038 | 0.2038 | 0.3732 | 0.2833 | 0.7633 | 0.6482 | 0.4216 | 0.0809 | 0.5895 | 0.7341 | 0.6254 | |
| 17 | 17 | 18 | 17 | 18 | 17 | 18 | 18 | 18 | 18 | 18 | |
| PCpG7 | −0.3755 | −0.3755 | − 0.5032 | − 0.2610 | 0.0416 | − 0.2866 | − 0.2846 | 0.3672 | 0.1833 | − 0.3242 | − 0.4005 |
| 0.1131 | 0.1131 | 0.0201 | 0.2663 | 0.8578 | 0.2206 | 0.2111 | 0.1015 | 0.4264 | 0.1516 | 0.0720 | |
| 19 | 19 | 21 | 20 | 21 | 20 | 21 | 21 | 21 | 21 | 21 | |
| PCpG8 | −0.3497 | −0.3497 | −0.5527 | − 0.0989 | 0.1695 | −0.1919 | − 0.0984 | 0.2266 | 0.3144 | −0.2547 | − 0.2960 |
| 0.1422 | 0.1422 | 0.0094 | 0.6783 | 0.4626 | 0.4177 | 0.6714 | 0.3234 | 0.1652 | 0.2651 | 0.1926 | |
| 19 | 19 | 21 | 20 | 21 | 20 | 21 | 21 | 21 | 21 | 21 | |
| PCpG9 | −0.2245 | − 0.2245 | − 0.4057 | −0.1304 | 0.2498 | − 0.0412 | − 0.1825 | 0.2054 | 0.1496 | −0.1909 | − 0.1361 |
| 0.3555 | 0.3555 | 0.0681 | 0.5838 | 0.2749 | 0.8632 | 0.4285 | 0.3719 | 0.5175 | 0.4072 | 0.5565 | |
| 19 | 19 | 21 | 20 | 21 | 20 | 21 | 21 | 21 | 21 | 21 | |
| E2CpG14 | E2CpG15–16 | E2CpG18 | E2CpG19 | PCpG2 | PCpG3 | PCpG4 | PCpG5 | PCpG6 | PCpG7 | PCpG8 | |
| E2CpG15–16 | 0.3779 | ||||||||||
| 0.0912 | |||||||||||
| 21 | |||||||||||
| E2CpG18 | −0.1449 | −0.0417 | |||||||||
| 0.5310 | 0.8577 | ||||||||||
| 21 | 21 | ||||||||||
| E2CpG19 | 1.0000 | 0.3779 | −0.1449 | ||||||||
| <.0001 | 0.0912 | 0.5310 | |||||||||
| 21 | 21 | 21 | |||||||||
| PCpG2 | −0.0848 | −0.2857 | 0.1994 | −0.0848 | |||||||
| 0.7149 | 0.2094 | 0.3863 | 0.7149 | ||||||||
| 21 | 21 | 21 | 21 | ||||||||
| PCpG3 | −0.0603 | −0.1634 | 0.5540 | −0.0603 | 0.3050 | ||||||
| 0.7953 | 0.4792 | 0.0092 | 0.7953 | 0.1788 | |||||||
| 21 | 21 | 21 | 21 | 21 | |||||||
| PCpG4 | −0.0884 | −0.2127 | 0.2790 | −0.0884 | 0.4169 | 0.6760 | |||||
| 0.7032 | 0.3547 | 0.2207 | 0.7032 | 0.0601 | 0.0008 | ||||||
| 21 | 21 | 21 | 21 | 21 | 21 | ||||||
| PCpG5 | −0.2323 | −0.3160 | 0.4481 | −0.2323 | 0.4101 | 0.7998 | 0.8982 | ||||
| 0.3108 | 0.1628 | 0.0417 | 0.3108 | 0.0648 | <.0001 | <.0001 | |||||
| 21 | 21 | 21 | 21 | 21 | 21 | 21 | |||||
| PCpG6 | −0.1235 | −0.0142 | 0.3876 | −0.1235 | 0.1251 | 0.9250 | 0.6821 | 0.7313 | |||
| 0.6254 | 0.9555 | 0.1120 | 0.6254 | 0.6208 | <.0001 | 0.0018 | 0.0006 | ||||
| 18 | 18 | 18 | 18 | 18 | 18 | 18 | 18 | ||||
| PCpG7 | −0.4005 | −0.1929 | 0.5588 | −0.4005 | 0.3459 | 0.7831 | 0.7776 | 0.8794 | 0.8090 | ||
| 0.0720 | 0.4022 | 0.0085 | 0.0720 | 0.1245 | <.0001 | <.0001 | <.0001 | <.0001 | |||
| 21 | 21 | 21 | 21 | 21 | 21 | 21 | 21 | 18 | |||
| PCpG8 | −0.2960 | −0.3326 | 0.5817 | −0.2960 | 0.4773 | 0.8641 | 0.6400 | 0.8551 | 0.6924 | 0.7911 | |
| 0.1926 | 0.1407 | 0.0057 | 0.1926 | 0.0287 | <.0001 | 0.0018 | <.0001 | 0.0015 | <.0001 | ||
| 21 | 21 | 21 | 21 | 21 | 21 | 21 | 21 | 18 | 21 | ||
| PCpG9 | −0.1361 | −0.2181 | 0.5731 | −0.1361 | 0.3592 | 0.9684 | 0.6016 | 0.7965 | 0.8591 | 0.7927 | 0.9092 |
| 0.5565 | 0.3422 | 0.0066 | 0.5565 | 0.1098 | <.0001 | 0.0039 | <.0001 | <.0001 | <.0001 | <.0001 | |
| 21 | 21 | 21 | 21 | 21 | 21 | 21 | 21 | 18 | 21 | 21 |
Note: a, b, and c represent the r value, P value and the number of samples analysed for the Spearman correlation analysis, respectively
Principal component analysis of the methylation data
To avoid the misinterpretation of the interactions between different loci and the association of a single CpG locus with KLF7 transcripts, the methylation data onto all CpG loci were subjected to principal component analysis (PCA). Fourteen effective principal components (z1–z14) were extracted from these 22 methylation data (Table 4).
Table 4.
Principal Component Analysis of methylation data
| Eigenvalue | Difference | Proportion | Cumulative | |
|---|---|---|---|---|
| 1 | 8.58397411 | 4.89652080 | 0.3902 | 0.3902 |
| 2 | 3.68745330 | 1.31306607 | 0.1676 | 0.5578 |
| 3 | 2.37438723 | 0.54826523 | 0.1079 | 0.6657 |
| 4 | 1.82612200 | 0.30800856 | 0.0830 | 0.7487 |
| 5 | 1.51811344 | 0.34269849 | 0.0690 | 0.8177 |
| 6 | 1.17541496 | 0.21863636 | 0.0534 | 0.8712 |
| 7 | 0.95677860 | 0.22254876 | 0.0435 | 0.9146 |
| 8 | 0.73422984 | 0.26289337 | 0.0334 | 0.9480 |
| 9 | 0.47133647 | 0.24226708 | 0.0214 | 0.9694 |
| 10 | 0.22906939 | 0.03795582 | 0.0104 | 0.9799 |
| 11 | 0.19111357 | 0.03848379 | 0.0087 | 0.9885 |
| 12 | 0.15262978 | 0.07853660 | 0.0069 | 0.9955 |
| 13 | 0.07409317 | 0.04880904 | 0.0034 | 0.9989 |
| 14 | 0.02528414 | 0.02528414 | 0.0011 | 1.0000 |
Factor analysis of new variables z1–z6
Six principal components (z1–z6) were extracted from 14 effective principal components by factor extraction (Eigenvalue > 1). The total variance contribution rate of the six new variables (z1–z6) was more than 85% of the total data. The new variables (z1–z6) obtained by PCA were subjected to factor analysis and named Factors 1–6. The results indicate that the loads of Factors 1–6 (z1–6) on each CpG locus were quite different. Factor 1 had positive loads on most loci in the promoter and negative loads on most loci in Exon 2 and had a higher load on the loci of PCpG3–PCpG9 and E2CpG4. Factor 2 had a high load on several loci in Exon 2, including E2CpG6–7, E2CpG8, E2CpG9, E2CpG13, E2CpG14, E2CpG15–16, and E2CpG19. Factor 3 had a high load on E2CpG2, E2CpG3, and E2CpG11–12. Factor 4 had a high load on E2CpG5. Factor 5 had a high load on PCpG2 (Table 5). Factor 6 had no obvious high load on any locus.
Table 5.
Factor analysis of methylation data (Factor be retained by the MINEIGEN criterion)
| Factor1 | Factor2 | Factor3 | Factor4 | Factor5 | Factor6 | |
|---|---|---|---|---|---|---|
| PCpG5 | 0.92834 | 0.11879 | 0.17878 | 0.10131 | 0.22061 | 0.03886 |
| PCpG7 | 0.91141 | 0.03225 | 0.32208 | − 0.00031 | − 0.06598 | − 0.02675 |
| PCpG8 | 0.89855 | 0.10258 | 0.13508 | −0.08460 | 0.17272 | 0.23556 |
| PCpG9 | 0.85571 | 0.22213 | 0.33021 | − 0.12622 | 0.12066 | 0.20036 |
| PCpG3 | 0.84517 | 0.23869 | 0.31027 | −0.08282 | 0.12230 | 0.11663 |
| PCpG4 | 0.75354 | 0.08572 | 0.05237 | 0.29781 | 0.31393 | −0.32154 |
| PCpG6 | 0.73199 | 0.26981 | 0.49523 | −0.02023 | −0.07731 | − 0.09970 |
| E2CpG10 | 0.59539 | 0.24021 | 0.03243 | 0.30738 | −0.48003 | −0.34490 |
| E2CpG15–16 | −0.57427 | 0.52467 | 0.34678 | 0.08436 | −0.28139 | − 0.29456 |
| E2CpG4 | −0.84494 | − 0.17986 | 0.16464 | −0.09869 | 0.26624 | −0.11090 |
| E2CpG14 | −0.34301 | 0.80962 | 0.00112 | 0.42052 | 0.04567 | −0.13228 |
| E2CpG19 | −0.34301 | 0.80962 | 0.00112 | 0.42052 | 0.04567 | −0.13228 |
| E2CpG13 | −0.29532 | 0.69629 | −0.05477 | −0.48668 | 0.35375 | −0.13615 |
| E2CpG6–7 | −0.02327 | 0.65169 | 0.04899 | −0.31275 | − 0.19246 | 0.49352 |
| E2CpG9 | −0.24675 | 0.63233 | −0.42568 | 0.25199 | 0.12345 | 0.40308 |
| E2CpG8 | −0.31118 | 0.58151 | −0.04129 | − 0.54707 | 0.14842 | − 0.16275 |
| E2CpG2 | −0.59702 | 0.02401 | 0.71111 | 0.06984 | −0.00995 | 0.14808 |
| E2CpG3 | −0.59702 | 0.02401 | 0.71111 | 0.06984 | −0.00995 | 0.14808 |
| E2CpG11–12 | 0.52934 | 0.34446 | −0.53169 | −0.32012 | − 0.10485 | −0.14235 |
| E2CpG5 | −0.27250 | 0.03287 | −0.16176 | 0.57029 | 0.08492 | 0.39362 |
| PCpG2 | 0.50184 | −0.02296 | −0.21338 | 0.31024 | 0.57542 | −0.06843 |
| E2CpG18 | 0.57945 | 0.17351 | −0.25425 | 0.02533 | −0.60630 | 0.15100 |
Factors were rotated by the quartimax method, and the results showed that Factor 1 mainly loads highly on the loci in the promoter, and Factors 2–6 load highly on the loci in Exon 2 (Table 6).
Table 6.
Factor analysis of methylation data (Rotated Factor Pattern by Quartimax)
| Factor1 | Factor2 | Factor3 | Factor4 | Factor5 | Factor6 | |
|---|---|---|---|---|---|---|
| PCpG9 | 0.96036 | − 0.02374 | − 0.08436 | 0.07716 | 0.11869 | − 0.10607 |
| PCpG5 | 0.94787 | −0.16833 | − 0.03008 | − 0.08018 | −0.16020 | − 0.09249 |
| PCpG3 | 0.94185 | −0.04449 | −0.04795 | 0.08003 | 0.05205 | −0.05010 |
| PCpG7 | 0.92477 | −0.08229 | − 0.20002 | − 0.11142 | − 0.00909 | 0.16138 |
| PCpG8 | 0.90745 | −0.19428 | − 0.12816 | − 0.02476 | 0.05945 | − 0.21645 |
| PCpG6 | 0.87870 | 0.11854 | −0.02292 | 0.07802 | 0.02889 | 0.27673 |
| PCpG4 | 0.72331 | −0.22681 | 0.08899 | −0.08498 | − 0.52419 | 0.06339 |
| E2CpG4 | −0.71389 | 0.49487 | −0.04194 | 0.22560 | −0.20288 | −0.13401 |
| E2CpG2 | −0.24561 | 0.89519 | 0.09107 | 0.03695 | 0.12860 | 0.04127 |
| E2CpG3 | −0.24561 | 0.89519 | 0.09107 | 0.03695 | 0.12860 | 0.04127 |
| E2CpG18 | 0.40771 | −0.51341 | 0.03221 | −0.25990 | 0.47540 | 0.31167 |
| E2CpG11–12 | 0.32049 | −0.76497 | 0.02842 | 0.31551 | 0.13195 | 0.09815 |
| E2CpG14 | −0.10283 | 0.12752 | 0.92871 | 0.21458 | −0.03918 | 0.18065 |
| E2CpG19 | −0.10283 | 0.12752 | 0.92871 | 0.21458 | −0.03918 | 0.18065 |
| E2CpG9 | −0.16828 | −0.22207 | 0.77846 | 0.07591 | 0.26525 | −0.35127 |
| E2CpG5 | −0.23208 | 0.08102 | 0.46715 | −0.45566 | 0.02841 | −0.31904 |
| E2CpG13 | −0.10304 | 0.00649 | 0.33738 | 0.90219 | 0.04859 | −0.12158 |
| E2CpG8 | −0.16643 | −0.00372 | 0.19605 | 0.83363 | 0.15129 | 0.02258 |
| E2CpG6–7 | 0.16568 | 0.02996 | 0.34104 | 0.36178 | 0.71717 | −0.12719 |
| PCpG2 | 0.42708 | −0.29521 | 0.13624 | −0.12019 | −0.54346 | − 0.36336 |
| E2CpG10 | 0.52357 | −0.28669 | 0.17966 | −0.23173 | −0.00468 | 0.64404 |
| E2CpG15–16 | −0.30850 | 0.44691 | 0.47334 | 0.30862 | 0.11909 | 0.51961 |
Correlation and regression analysis of z1–z6 with KLF7 expression and blood metabolic indexes
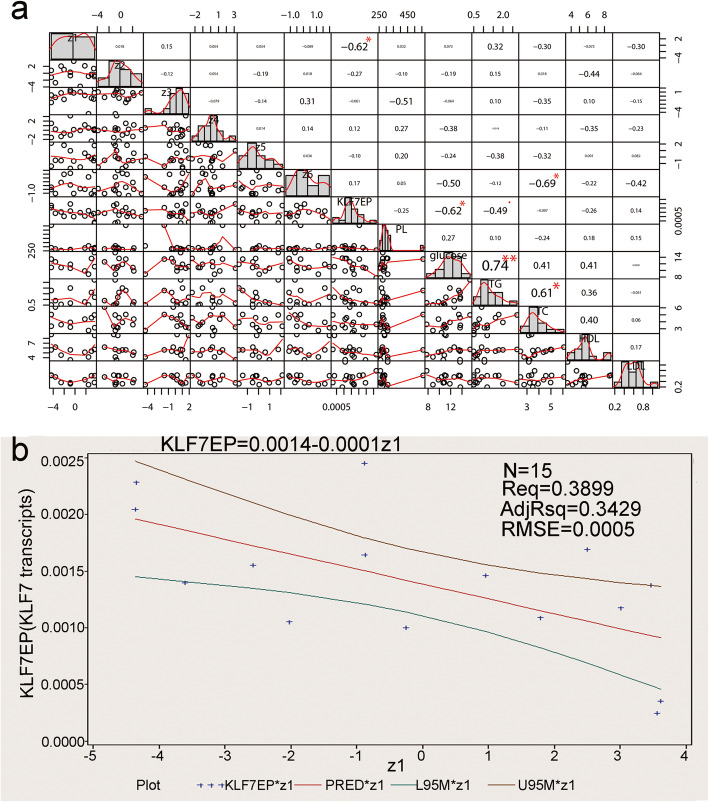
Spearman correlation analysis showed that the new variable z1 was negatively correlated with KLF7 transcripts (r = − 0.62439, P = 0.0128), while the new variables z2–z6 had no significant correlation with KLF7 transcripts (P > 0.05). There was no significant correlation between z1–z6 and blood metabolic indexes but z6 was significantly correlated with TC level in the blood (Fig. 5a).
Fig. 5.
Association analysis of z1-z6 with KLF7 transcripts and blood metabolic indicators. a. The Spearman correlation analysis of the z1–z6 with KLF7 transcripts and blood metabolic indicators. b. The regression analysis between z1 and KLF7 transcripts
The data onto z1 and KLF7 transcripts distributed normally (Shapiro–Wilk test, P > 0.05), and a regression relationship between z1 and KLF7 transcripts was studied using generalized linear regression model. The results showed that the established regression equation was statistically significant (F = 8.31, P = 0.0128, R2 = 0.3429), and the regression equation was: Y (KLF7 transcripts) = 0.00139–0.00013144 × z1 (Fig. 5b).
Discussion
KLF7 is a highly conserved gene in humans and animals [23, 24]. Previous reports on mammalian species showed that KLF7 regulated neuroectodermal and mesodermal development [2] and played a role in obesity [6], T2DM [7, 8], and blood disease [9]. Our previous study showed that KLF7 was an important regulator in chicken adipose tissue development [17, 18]. Currently, the results showed that KLF7 transcripts of the adipose tissue of Chinese fast-growing yellow broilers were associated with age, in line with our previous report in white broilers [17]. Although there was no biologically significant difference in KLF7 transcripts of the weeks of 2, 4, and 6, there was a downward trend at the week of 6. The decline in KLF7 expression during the weeks of 2, 4, and 6 might be due to the suppression function of KLF7 in the formation of adipose tissue at the early stage [17]. In addition, the increase in KLF7 transcripts at the age of 8 weeks suggested that chicken KLF7 might have a function in mature adipose tissue, similar to the report on its orthology in human in vitro [7, 8].
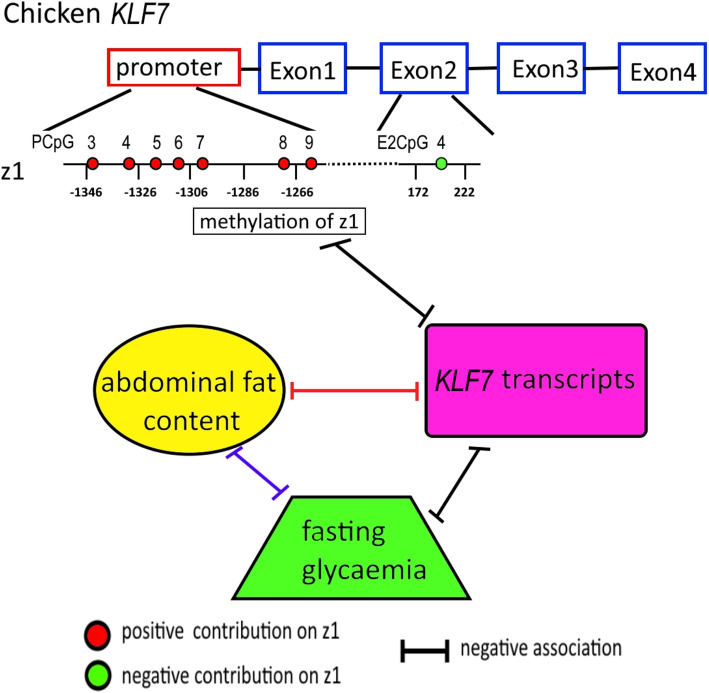
The level of blood glucose was changeable and associated with insulinaemia in chicken [25]. Here, the results showed that chicken KLF7 transcripts were correlated with fasting blood glucose level, in line with our previous reports in white broilers that the chicken KLF7 was involved in the regulation of adipogenesis and blood metabolic indicators [17, 18], and provided additional evidence for the role of KLF7 in metabolic syndrome from the perspective of non-rodent model animals. In addition, the previous study showed that chickens selected for low fasting glycaemia (LG) were fattier than their counterparts selected for high fasting glycaemia (HG) [25]. The negative correlation of KLF7 expression to the fasting glycaemia in chicken and abdominal fat content [17, 18], suggested that a feedback regulation among KLF7 expression, glycaemia, and obesity might exist in chicken (Fig. 6).
Fig. 6.
Schematic representation of the relationship of DNA methylation, KLF7 transcripts, glycaemia and abdominal fat content in chicken. The negative associations marked red and blue were confirmed by the reference [17, 25], respectively
DNA methylation is important to the regulation of gene expression and function in animals [19]. The previous studies in humans showed that DNA methylation of KLF7 was associated with the occurrence and development of gastric cancer [20, 21], however, there is no report on the DNA methylation of KLF7 in adipose tissue and birds. Sequence analysis showed that there was a similar distribution of CpG-rich sequence in chicken KLF7 as that of human KLF7, suggested that the chicken KLF7 might be also regulated by DNA methylation.
Sequenom MassArray were used to study the DNA methylation in the promoter and Exon 2 of chicken KLF7. A total of 22 valid datasets were obtained, and the level of DNA methylation in the promoter was lower than those in Exon 2. This was probably because it is a consecutively expressed gene during adipogenesis [17]; therefore, the promoter of chicken KLF7 could not be strongly silenced by a long-term mechanism like DNA methylation in adipose tissue.
In addition, none of the loci detected were significantly different among the ages of 2, 4, 6, and 8 weeks, indicated that the DNA methylation might not be a main regulation method of the KLF7 expression in adipose tissue during development.
The association analysis showed that only the methylation of PCpG6 was significantly associated with KLF7 transcripts in chicken abdominal adipose tissue, and the contribution of PCpG6 to the variation on KLF7 transcripts was 0.2545. Sequence analysis showed that there were several binding sites of transcriptional factors at the locus of PCpG6, including TFAP2C and TFAP2A (supplementary Table 1), The negative correlation between DNA methylation of PCpG6 with KLF7 transcripts might be mediated by transcriptional factor. However, further investigation is needed to verify this hypothesis.
Our previous study showed that one SNP (c. A141G) in the KLF7 coding sequence was associated with blood very-low-density lipoprotein and abdominal fat content in broilers [18] and chicken KLF7 regulated the promoter activity of lipoprotein lipase (LPL) [17]. In the current study, the result showed that the methylation of E2CpG9 was significantly associated with blood HDL level, further suggested that KLF7 might play a role in the fat transport in chicken. In addition, the E2CpG9 was conserved between chicken and human KLF7s (supplementary Figure 1C), this result might provide a clue to the function of KLF7 in human.
To avoid the misinterpretation of the interaction effect of different loci and to discover the relationship between general DNA methylation and KLF7 transcripts, the methylation data were subjected to PCA. Fourteen effective principal components (z1–z14) were extracted from these 22 methylation data. Six principal components (z1–z6) were extracted from 14 principal components by factor extraction, and named Factors 1–6, respectively.
Factor analysis showed that Factor 1 had a higher load on the loci of PCpG3–PCpG9 and E2CpG4, indicating that Factor 1 (z1) mainly represents the effect of DNA methylation in the promoter. Factors 2–6 loaded highly with quite different loci in Exon 2 and PCpG2. Therefore, Factors 2–6 might represent the effect of DNA methylation in Exon 2, and there was a large difference in them.
Correlation analysis showed that the new variable z1 was negatively correlated with KLF7 transcripts, whereas none of these z2–z6 were significant correlated with KLF7 transcripts. In addition, the regression relationship between z1 and KLF7 transcripts was studied, and the contribution of z1 to the variation on KLF7 transcripts was 0.3429, which was greater than the contribution of the single locus PCpG6. Additionally, the ratio of the slope to truncation was about 9.5%, indicating the greatest effect that Factor 1 (z1) had on the KLF7 transcripts was about 9.5%. This was reasonable for effect of DNA methylation on gene expression, indicated that the KLF7 transcripts of chicken abdominal adipose tissue might be inhibited by DNA methylation in promoter. Sequence analysis showed that there were many binding sites of transcription factors at the loci of PCpG3–PCpG9 in chicken KLF7 promoter, respectively (supplementary Table 1), The inhibitory effect of DNA methylation on KLF7 expression might be achieved in part by affecting the binding of transcription factors to the KLF7 promoter, like the report on chicken ApoA-I [26].
There was no significant correlation between z1 and blood metabolic indexes. This might be because DNA methylation in chicken KLF7 does not directly take part in the regulation of blood metabolic indexes. However, further investigation is needed into whether an indirect association exists on DNA methylation in chicken KLF7 and blood metabolic indexes.
Conclusions
In conclusion, the KLF7 transcripts of chicken abdominal adipose tissue might be inhibited by DNA methylation in the promoter, and it might be related to the DNA methylation level of PCpG6 (Fig. 6).
Methods
Experimental birds and management
Animal work was conducted according to the guidelines on the care and use of experimental animals established by the Ministry of Science and Technology of the People’s Republic of China (approval number 2006–398) and was approved by the Animal Experimental Ethical Committee of the First Affiliated Hospital, Shihezi University School of Medicine. A total of 21 one-day-old male Chinese fast-growing yellow broilers were used in the current study, they were obtained from the Guangdong Wiz Agricultural Science & Technology Co. Ltd. (Guangzhou, China). All birds were kept in similar environmental conditions and had free access to feed and water until sacrificed.
Tissues
Several male birds (n = 4–6) were sacrificed 6 h after a meal at 2, 4, 6, and 8 weeks of age by cutting the carotid artery after ether anaesthesia, and the abdominal fat tissue was collected immediately. The collected tissues were flash-frozen and stored in liquid nitrogen until the extraction of RNA. At the age of 4, 6, and 8 weeks, the peripheral blood was also collected from wing vein to measure blood metabolic indicators.
RNA isolation
Total RNA of abdominal fat tissue (each 100 mg) was extracted using Trizol (Invitrogen) following the manufacturer’s protocol.
RT-qPCR
The RNA quality was assessed by denaturing formaldehyde agarose gel electrophoresis. Reverse transcription was performed using 1 μg of total RNA, an oligo (dT) anchor primer, and ImProm-II reverse transcriptase (Promega, Madison, USA). Reverse transcription conditions for each cDNA amplification were 25 °C for 5 min, 42 °C for 60 min, and 70 °C for 15 min. Real-time quantitative PCR (qPCR) was used to detect KLF7 and β-ACTIN using SYBR Premix Ex Taq (Takara) and a 7500 Real-Time PCR System (Applied Biosystems, Foster City, USA). Chicken β-ACTIN was used as an internal reference. The primers used are presented in Table 7. Samples (1 μL) from each reverse transcription reaction product were amplified in a 20-μl. PCR reaction. Reaction mixtures were incubated in an ABI Prism 7500 sequence detection system (Applied Biosystems) programmed for 1 cycle at 95 °C for 30 s and 40 cycles at 95 °C for 5 s, and at 60 °C for 34 s. Dissociation curves were analyzed using Dissociation Curve 1.0 software (Applied Biosystems) for each PCR to detect and eliminate possible primer–dimer artifacts. All reactions were performed in triplicate. The relative amounts of the KLF7 transcripts were calculated by the comparative cycle-time method.
Table 7.
Oligonucleotides used in the current study
| Gene | Application | Primer (5′-3′) |
|---|---|---|
| β-ACTIN | RT-qPCR | F:CCTGGCACCTAGCACAATGA |
| R:CCTGCTTGCTGATCCACATC | ||
| KLF7 | RT-qPCR | F:TGCCATCCTTGGAGGAGAAC |
| R:AGGCATGAAGGAAGCAGTCC | ||
| KLF7 | methylation analysis in promoter | F: aggaagagagTTAGTTTGTTTTTGTTGTTGTGGAG |
| R:cagtaatacgactcactatagggagaaggctAAATTACCTTTTCCAAAACTAAATCC | ||
| KLF7 | methylation analysis in Exon 2 | F:aggaagagagTGGATTTTATTTTTTTGTTAGTGGAG |
| R:cagtaatacgactcactatagggagaaggctATTTTCTAAAAATACTTCCAATCCC |
Sequenom MassARRAY methylation analysis
Sequenom MassARRAY methylation analysis was carried out by Beijing Compass Biotechnology Co., Ltd. (Beijing, China). Briefly, DNA from adipose tissues was isolated using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instruction. Genomic DNA quantification was detected by NanoDrop spectrophotometer (GE Healthcare Life Science, Uppsala, Sweden), then bisulfite conversation of the DNA was performed using EpiTect Bisulfite Kit (Qiagen) according to the manufacturer’s protocol. Two sets of primers were designed by EpiDesigner software (http://epidesigner.com; Table 7). The methylation level of individual units was measured by Quantitative Methylation Analysis (MassARRAY EpiTYOER, Sequenom, San Diego, CA). The data of MassARRAY methylation can be achieved in Supplementary Table 2.
Bioinformatics analysis
The CpG density of the genomic region of chicken KLF7 and human KLF7 analyzed by CpGplot (Version 6.6.0). The genomic sequence of chicken KLF7 was obtained from the NCBI and UCSC databases referenced to the promoter sequence (GenBank accession number: JX290203) and the full-length coding sequence of chicken KLF7 (GenBank accession number: JQ736790). The sequence analyzed was 64,584 bp in length and included the 3000 bp sequence upstream of the predicted transcription start site and the 100 bp sequence downstream of the predicted 3′UTR of chicken KLF7. The pattern of transcriptional factor binding sites in DNA sequence was analyzed by JASPAR2020.
Statistical analysis
All statistical analyses were performed using the SAS software system (version 9.2; SAS Inst. Inc., Cary, NC, USA). The Shapiro–Wilk test was used to test the normality of data. The difference in groups was analyzed by ANOVA or Kruskal–Wallis tests. The comparison of data was performed by PROC GLM procedure followed by Duncan’s multiple tests. With the following models:
where Y is the dependent variable, μ is the population mean, F is the fixed effect of age and e is the random error.
The data that conformed to the normal distribution was directly applied to the models, and the data that did not conform to the normal distribution were sorted rank firstly, and the ranks were then applied to the models. The correlation was analyzed by Spearman’s correlation test. The unpaired student t test was used to analyze the difference between two groups. The PCA and factor analysis was conducted by the PROC PRINCOMP and FACTOR procedure, respectively. Linear regression is performed with the PROG REG command of SAS. The P-value < 0.05 was considered statistically significant.
Supplementary information
Additional file 1: Supplementary Figure 1. The sequence analysis of chicken and human KLF7s. a. The CpG density in the genomic region of human KLF7 analyzed by CpGplot (Version 6.6.0). b. The dot plot analysis between the sequence studied for DNA methylation in chicken KLF7 promoter and human KLF7 promoter (3000 bp upstream of NM_003709.4). c. The alignment of the sequences of Exon 2 between chicken and human KLF7s.
Additional file 2: Supplementary Table 1. The prediction of transcription factor binding sites at the CpG loci in chicken KLF7 promoter (analyzed using JASPAR2020, Search profile = ChIP-seq, Relative profile score threshold = 0.8). Supplementary Table 2. The DNA methylation data of CpG loci in the chicken KLF7 gene detected by Sequenom MassArray. Supplementary Table 3. The mRNA expression data of KLF7 in adipose tissue and phenotype data.
Acknowledgements
Not applicable.
Abbreviations
- ANOVA
Analysis of variance
- GLM
A general linear model
- PCA
Principal component analysis
- qRT-PCR
Quantitative real-time PCR
- SNP
Single nucleotide polymorphism
- Glu
Glucose
- PL
Phospholipids
- TC
Total cholesterol
- TG
Triglyceride
- LDL
Low-density lipoprotein
- HDL
High-density lipoprotein
- KLF7
Krüppel-like factor 7
- UTR
Untranslated Region
Authors’ contributions
ZZ designed this study, performed the data analysis and prepared manuscript. CN performed blood metabolic indexes analysis. YC performed the molecule biology experiments. YD and TL collected the animal samples. All authors read and approved the final manuscript.
Funding
National Natural Science Foundation of China (No. 31501947) and Youth Innovative Talents Project of Shihezi University (No. CXBJ201905) funded all the process of this study, including the experiment, sampling, data analysis and writing of paper.
Availability of data and materials
The datasets analysed during the current study are available in the supplementary Table 2 and 3.
Ethics approval and consent to participate
All animal experiments were conducted according to the guidelines on the care and use of experimental animals established by the Ministry of Science and Technology of the People’s Republic of China (Approval number: 2006–398) and was approved by the Animal Experimental Ethical Committee of the First Affiliated Hospital, Shihezi University School of Medicine.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12863-020-00923-6.
References
- 1.Matsumoto N, Laub F, Aldabe R, Zhang W, Ramirez F, Yoshida T, Terada M. Cloning the cDNA for a new human zinc finger protein defines a group of closely related Kruppel-like transcription factors. J Biol Chem. 1998;273(43):28229–28237. doi: 10.1074/jbc.273.43.28229. [DOI] [PubMed] [Google Scholar]
- 2.Caiazzo M, Colucci-D'Amato L, Esposito MT, Parisi S, Stifani S, Ramirez F, di Porzio U. Transcription factor KLF7 regulates differentiation of neuroectodermal and mesodermal cell lineages. Exp Cell Res. 2010;316(14):2365–2376. doi: 10.1016/j.yexcr.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 3.Lei L, Ma L, Nef S, Thai T, Parada LF. mKlf7, a potential transcriptional regulator of TrkA nerve growth factor receptor expression in sensory and sympathetic neurons. Development. 2001;128(7):1147–1158. doi: 10.1242/dev.128.7.1147. [DOI] [PubMed] [Google Scholar]
- 4.Laub F, Aldabe R, Friedrich VJ, Ohnishi S, Yoshida T, Ramirez F. Developmental expression of mouse Kruppel-like transcription factor KLF7 suggests a potential role in neurogenesis. Dev Biol. 2001;233(2):305–318. doi: 10.1006/dbio.2001.0243. [DOI] [PubMed] [Google Scholar]
- 5.Laub F, Dragomir C, Ramirez F. Mice without transcription factor KLF7 provide new insight into olfactory bulb development. Brain Res. 2006;1103(1):108–113. doi: 10.1016/j.brainres.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 6.Zobel DP, Andreasen CH, Burgdorf KS, Andersson EA, Sandbaek A, Lauritzen T, Borch-Johnsen K, Jorgensen T, Maeda S, Nakamura Y, et al. Variation in the gene encoding Kruppel-like factor 7 influences body fat: studies of 14 818 Danes. Eur J Endocrinol. 2009;160(4):603–609. doi: 10.1530/EJE-08-0688. [DOI] [PubMed] [Google Scholar]
- 7.Kawamura Y, Tanaka Y, Kawamori R, Maeda S. Overexpression of Kruppel-like factor 7 regulates adipocytokine gene expressions in human adipocytes and inhibits glucose-induced insulin secretion in pancreatic beta-cell line. Mol Endocrinol. 2006;20(4):844–856. doi: 10.1210/me.2005-0138. [DOI] [PubMed] [Google Scholar]
- 8.Kanazawa A, Kawamura Y, Sekine A, Iida A, Tsunoda T, Kashiwagi A, Tanaka Y, Babazono T, Matsuda M, Kawai K, et al. Single nucleotide polymorphisms in the gene encoding Kruppel-like factor 7 are associated with type 2 diabetes. Diabetologia. 2005;48(7):1315–1322. doi: 10.1007/s00125-005-1797-0. [DOI] [PubMed] [Google Scholar]
- 9.Schuettpelz LG, Gopalan PK, Giuste FO, Romine MP, van Os R, Link DC. Kruppel-like factor 7 overexpression suppresses hematopoietic stem and progenitor cell function. Blood. 2012;120(15):2981–2989. doi: 10.1182/blood-2012-02-409839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao J, Zhang H, Liu C, Chen S, Qian R, Zhao K. miR-450b-3p inhibited the proliferation of gastric cancer via regulating KLF7. Cancer Cell Int. 2020;20(47). 10.1186/s12935-020-1133-2. [DOI] [PMC free article] [PubMed]
- 11.Niu R, Tang Y, Xi Y, Jiang D. High expression of Krüppel-like factor 7 indicates unfavorable clinical outcomes in patients with lung adenocarcinoma. J Surg Res. 2020;250:216–223. doi: 10.1016/j.jss.2019.12.053. [DOI] [PubMed] [Google Scholar]
- 12.Guan F, Kang Z, Zhang J, Xue N, Yin H, Wang L, Mao B, Peng W, Zhang B, Liang X, et al. KLF7 promotes polyamine biosynthesis and glioma development through transcriptionally activating ASL. Biochem Biophys Res Commun. 2019;514(1):51–57. doi: 10.1016/j.bbrc.2019.04.120. [DOI] [PubMed] [Google Scholar]
- 13.Ding X, Wang X, Gong Y, Ruan H, Sun Y, Yu Y. KLF7 overexpression in human oral squamous cell carcinoma promotes migration and epithelial-mesenchymal transition. Oncol Lett. 2017;13(4):2281–2289. doi: 10.3892/ol.2017.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarah EH, Saskia PH, Gail ED, David HW, David CL, Stuart JR, Riccardo EM, METASTROKE consortium ICFB, CHARGE consortium aging and longevity group CCCG. Cathie LS, et al. Molecular genetic contributions to self-rated health. Int J Epidemiol. 2017;46(3):994–1009. doi: 10.1093/ije/dyw219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powis Z, Petrik I, Cohen J, Escolar D, Burton J, van Ravenswaaij-Arts C, Sival D, Stegmann A, Kleefstra T, Pfundt R, et al. De novo variants in KLF7 are a potential novel cause of developmental delay/intellectual disability, neuromuscular and psychiatric symptoms. Clin Genet. 2018;93(5):1030–1038. doi: 10.1111/cge.13198. [DOI] [PubMed] [Google Scholar]
- 16.Makeeva OA, Sleptsov AA, Kulish EV, Barbarash OL, Mazur AM, Prokhorchuk EB, Chekanov NN, Stepanov VA, Puzyrev VP. Genomic study of cardiovascular continuum comorbidity. Acta Nat. 2015;7(3):89–99. doi: 10.32607/20758251-2015-7-3-89-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Wang H, Sun Y, Li H, Wang N. Klf7 modulates the differentiation and proliferation of chicken preadipocyte. Acta Biochim Biophys Sin Shanghai. 2013;45(4):280–288. doi: 10.1093/abbs/gmt010. [DOI] [PubMed] [Google Scholar]
- 18.Zhang ZW, Wang ZP, Zhang K, Wang N, Li H. Cloning, tissue expression and polymorphisms of chicken Kruppel-like factor 7 gene. Anim Sci J. 2013;84(7):535–542. doi: 10.1111/asj.12043. [DOI] [PubMed] [Google Scholar]
- 19.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacol. 2013;38(1):23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang M, Kim HS, Cho MY. Different methylation profiles between intestinal and diffuse sporadic gastric carcinogenesis. Clin Res Hepatol Gastroenterol. 2014;38(5):613–620. doi: 10.1016/j.clinre.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 21.Choi J, Cho M, Jung S, Jan KM, Kim HS. CpG Island methylation according to the histologic patterns of early gastric adenocarcinoma. J Pathol Transl Med. 2011;45(5):469–476. [Google Scholar]
- 22.Sarjeant K, Stephens JM. Adipogenesis. Cold Spring Harb Perspect Biol. 2012;4(9):a8417. doi: 10.1101/cshperspect.a008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Wei H, Zhang Z. Research progress of Krüppel-like factor 7. Acta Physiologica Sinica. 2016;6(68):809–815. [PubMed] [Google Scholar]
- 24.Sun Y, Wang W, Zhang Z, Shao S, Li B, Wang N. Advance in biological function of Kriippel-like factor 7(KLF7) Prog Biochem Biophys. 2017;44(11):972–980. [Google Scholar]
- 25.Simon J, Guillaumin S, Chevalier B, Derouet M, Leclercq B. Plasma glucose-insulin relationship in chicken lines selected for high or low fasting glycaemia. Br Poult Sci. 2000;41(4):424–429. doi: 10.1080/713654969. [DOI] [PubMed] [Google Scholar]
- 26.Wu C, Wang Y, Gong P, Wang L, Liu C, Chen C, Jiang X, Dong X, Cheng B, Li H. Promoter methylation regulates ApoA-I gene transcription in chicken abdominal adipose tissue. J Agr Food Chem. 2019;67:4535–4544. doi: 10.1021/acs.jafc.9b00007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figure 1. The sequence analysis of chicken and human KLF7s. a. The CpG density in the genomic region of human KLF7 analyzed by CpGplot (Version 6.6.0). b. The dot plot analysis between the sequence studied for DNA methylation in chicken KLF7 promoter and human KLF7 promoter (3000 bp upstream of NM_003709.4). c. The alignment of the sequences of Exon 2 between chicken and human KLF7s.
Additional file 2: Supplementary Table 1. The prediction of transcription factor binding sites at the CpG loci in chicken KLF7 promoter (analyzed using JASPAR2020, Search profile = ChIP-seq, Relative profile score threshold = 0.8). Supplementary Table 2. The DNA methylation data of CpG loci in the chicken KLF7 gene detected by Sequenom MassArray. Supplementary Table 3. The mRNA expression data of KLF7 in adipose tissue and phenotype data.
Data Availability Statement
The datasets analysed during the current study are available in the supplementary Table 2 and 3.