Abstract
Flaviviruses causes significant human disease. Recent outbreaks of the Zika virus highlight the need to develop effective therapies for this class of viruses. Previously we identified niclosamide as a broad-spectrum inhibitor for flaviviruses by targeting the interface between viral protease NS3 and its cofactor NS2B. Here, we screened a small library of niclosamide derivatives and identified a new analogue with improved pharmacokinetic properties. Compound JMX0207 showed improved efficacy in inhibition of the molecular interaction between NS3 and NS2B, better inhibition of viral protease function, and enhanced antiviral efficacy in the cell-based antiviral assay. The derivative also significantly reduced Zika virus infection on 3D mini-brain organoids derived from pluripotent neural stem cells. Intriguingly, the compound significantly reduced viremia in a Zika virus (ZIKV) animal model. In summary, a niclosamide derivative, JMX0207, was identified, which shows improved pharmacokinetics and efficacy against Zika virus both in vitro and in vivo.
Keywords: Flavivirus, Zika virus, Dengue virus, antiviral, protease inhibitor
Graphical Abstract
The genus Flavivirus is composed of more than 70 viruses. Many flaviviruses, including the four serotypes of the Dengue virus (DENV1–4), the West Nile virus, and the Zika virus (ZIKV or ZK), cause serious human diseases. Recently, significant ZIKV outbreaks have occurred worldwide.1–5 ZIKV is transmitted to humans primarily through bites from infected Aedes species mosquitoes but may also be transferred prenatally or through sexual activities or blood transfusions.6–9 ZIKV infection is associated with devastating diseases, including Guillain-Barré syndrome, a rare neurological syndrome, and congenital Zika syndrome, which is manifested by microcephaly, brain abnormalities, and other central nervous system malformations.1–5 In addition, ZIKV persists in neuronal and/or reproductive tissues of animals for up to 60 days after viral infection, leading to severe damage.10–12 Currently, there is neither a safe and effective vaccine nor a specific therapy for ZIKV. Although a DENV vaccine has been recently approved, it is only effective in children between 9 and 16 years old and poses increased risk for naive children due to antibody-dependent enhancement.13 Therefore, there is an unmet medical need to develop direct antivirals against flaviviruses.
The flavivirus genome encodes a single open reading frame, from which a polyprotein precursor is expressed.14–16 The polyprotein precursor is post-translationally processed into 10 mature proteins, including three structural and seven nonstructural proteins, by viral and host proteases in a sequential manner.14,15 The viral protease is a heterocomplex composed of viral NS3 protein and its viral cofactor NS2B.14 The viral protease has been considered a highly promising drug target.14,17–19 Various strategies have been used to develop protease inhibitors. The majority of efforts have been focused on the viral protease active site13,16–18 but with limited success in finding inhibitors with in vivo efficacy. 20 Recently, alternative strategies to target the NS2B–NS3 interaction21,22 and allosteric sites23–25 have led to the identification of potent protease inhibitors with in vivo efficacy against Zika virus.
Previously, we developed a split luciferase complementation (SLC)-based high throughput screening strategy to identify inhibitors targeting the NS2B–NS3 interface of DENV2 (DN2).21 Niclosamide was found to be a broad-spectrum inhibitor for flaviviruses by targeting the interface between viral protease NS3 and its cofactor NS2B, leading to accumulation of nonfunctional viral polyprotein precursor.21 In addition, niclosamide was found to interfere with virus entry26 and endosomal acidification.27 Moreover, niclosamide was found as a broad-spectrum antiviral against multiple viruses including SARS Cov-2, the causative agent of the current COVID-19 pandemic.28 Unfortunately, niclosamide did not show appreciable in vivo antiviral efficacy toward ZIKV (data not shown), possibly due to its poor pharmacokinetic properties26,29
In this work, we screened a small library of niclosamide derivatives. We found that one derivative, JMX0207, showed improved efficacy in the inhibition of the NS2B–NS3 interaction, better inhibition of viral protease function, and enhanced antiviral efficacy in the cell-based antiviral assay, compared to niclosamide. JMX0207 was also found to significantly reduce ZIKV infection on a 3D mini-brain organoid model derived from pluripotent neural stem cells. Intriguingly, JMX0207 significantly improved the pharmacokinetic properties and markedly reduced viremia in a ZIKV animal model.
RESULTS
Synthesis and Screening of Niclosamide Derivatives
Niclosamide was originally used as an anthelmintic drug. It was also found to have anticancer and antibacterial activities.30–34 We previously developed a small library composed of 62 niclosamide derivatives for other purposes.33–38 These niclosamide analogs have been resynthesized in-house following our published procedures for antiviral evaluation. To identify niclosamide derivatives that have equal or better antiviral efficacy and improved pharmacokinetic properties, we used our SLC-based NS2B–NS3 interaction assay to screen this library, with niclosamide as a control.
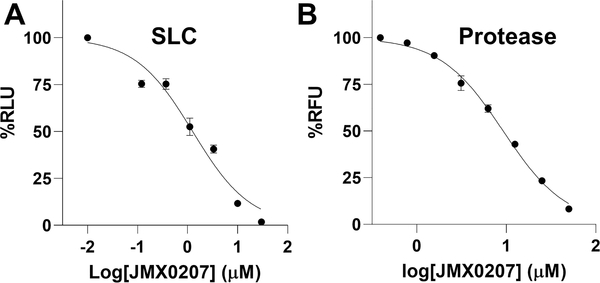
Eleven out of 62 niclosamide derivatives showed dose–response inhibition of the interaction between viral NS3 protease and its cofactor NS2B, with IC50-SLC values lower than 30 μM, the highest concentration tested (IC50-SLC/IC50-pro/EC50/CC50: compound concentrations required to reduce 50% of the split luciferase complementation signal, protease activity, virus production, and cell viability, respectively) (Table 1, Figure 1). The derivatives identified included seven compounds with O-alkylamino chains, one compound with 3-NO2 instead of 5-Cl on the salicylic ring, and three compounds with modifications on the nitro group of the aniline moiety. Of these active derivatives, compounds HJC0308 with a 3-aminopropoxy moiety, HJC0431 with a 4-aminobutoxy moiety, and JMX0207 with a 3-NO2 group showed comparable or better inhibition of the NS2B–NS3 SLC with IC50-SLC values of 4.9, 3.1, and 1.3 μM, respectively, compared to niclosamide.
Table 1.
IC50,a EC50, CC50 (all in μM), and Therapeutic Index (TI) of Niclosamide Derivatives
| Compoundb | Structure | IC50-SLC-(DN2) | IC50-pro (DN2) | EC50-DN2 (A549) | EC50-ZK (A549) | CC50 (A549) | TIc (DN2) | TI (ZK) |
|---|---|---|---|---|---|---|---|---|
| Niclosamide | 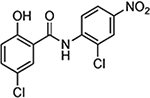 |
2.0 | 21.6 | 0.55 | 0.48 | 4.8 | 8.7 | 10.0 |
| HJC0114 | 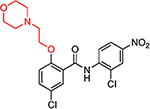 |
18.4 | 33.6 | >30 | >30 | >200 | ||
| HJC0125 | 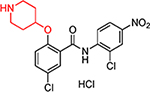 |
16.3 | 44.5 | 0.21 | 0.15 | 50.2 | 239 | 335 |
| HJC0308 | 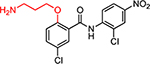 |
4.9 | 39.7 | 1.4 | 6.2 | 17.7 | 12.6 | 2.8 |
| HJC0365 | 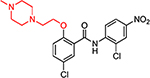 |
13.2 | >60 | 6.4 | 6.0 | 177 | 27.6 | 29.5 |
| HJC0381 | 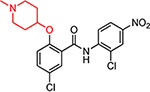 |
10.8 | 25.9 | 1.2 | 3.9 | 91.1 | 76 | 23.3 |
| HJC0390 | 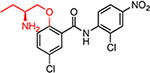 |
6.0 | 46.5 | 0.43 | <0.1 | 30.1 | 70 | >300 |
| HJC0431 | 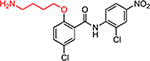 |
3.1 | 34.0 | 2.2 | 0.36 | 28.4 | 13 | 79 |
| HJC0129 | 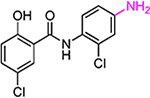 |
9.6 | 11.3 | >30 | >30 | 176 | ||
| HJC0140 | 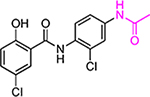 |
9.4 | 25.5 | >30 | >30 | >200 | ||
| HJC0149 | 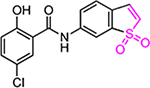 |
18.6 | 20.3 | 0.9 | 1.1 | 241 | 268 | 219 |
| JMX0207 | 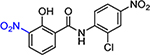 |
1.3 | 8.2 | 0.31 | 0.30 | 31.9 | 103 | 106 |
IC50-SLC/IC50-pro/EC50/CC50: Compound concentrations required to reduce 50% of the split luciferase complementation (SLC) signal, protease activity, virus production, and cell viability, respectively. All values are in micromolar (μM) except TI. DN2, Dengue virus serotype 2; ZK, Zika virus.
Original compound names from the literature and this work. According to their structures, these compounds can be classified into three groups, colored as red, magenta, and blue, respectively.
TI, therapeutic index (TI) defined as CC50/EC50.
Figure 1.
Inhibition of the NS2B–NS3 interactions and protease activity. (A) Dose-dependent inhibition of SLC upon binding of NLuc-NS2B49–66 to GST-CLuc-NS3 by JMX0207. N =3. (B) Dose–response inhibitions of the DENV2 His-NS2B/His-MBP-NS3 protease activity by JMX0207. N = 3. In both (A) and (B), the DMSO control was set as 100%. The values represent means ± standard deviation (SD) in all panels.
Inhibition of the NS2B–NS3 Protease Activity
We next evaluated the inhibition efficacy of these positive derivatives against the viral NS2B–NS3 protease function as previously described.21 Previously, we generated two versions of the NS3 protease, the NS3 protease domain refolded from inclusion bodies and a soluble version of the NS3 protease domain fused to maltose-binding protein (MBP).21,22 Our data showed that the refolded and MBP-tagged NS3 proteins have indistinguishable protease activities when transactivated by cofactor NS2B. In this work, we used both versions to characterize the inhibitors and found that the IC50 values determined from both versions were similar. Therefore, only values for the refolded NS3 were shown. As shown in Table 1, most active compounds in the SLC assay except HJC0365 still showed inhibition against the viral NS2B–NS3 protease activity. However, we found that there seemed to be no strict correlation between IC50-SLC and IC50-pro values. This is not surprising because niclosamide is a known luciferase inhibitor.39 The SLC inhibition is an indirect readout of the NS2B–NS3 interactions. Direct inhibition of luciferase activity and/or direct inhibition of luciferase complementation in addition to inhibition of NS2B–NS3 interaction may account for the poor correlation between IC50-SLC and IC50-pro values. While IC50-SLC is a poor indicator of inhibition efficacy of the NS2B–NS3 interactions, it remains a valuable indicator to eliminate compounds if they failed to inhibit SLC.
Among these derivatives, compounds HJC0140 with a 4′-acetamido group (IC50-pro = 25.5 μM), HJC0149 with a benzo[b]thiophene 1,1-dioxide moiety (IC50-pro = 20.3 μM), and HJC0381 with a (1-methylpiperidin-4-yl)oxy moiety (IC50-pro = 25.9 μM) maintained the same level of potency, while compounds HJC0129 with a 4′-NH2 group (IC50-pro = 11.3 μM) and JMX0207 with a 3-NO2 group (IC50-pro = 8.2 μM) showed nearly or more than 2-fold improvement in inhibition of the viral NS2B–NS3 protease activity, compared to niclosamide (IC50-pro = 21.6 μM) (Table 1, Figure 1). Taken together, the three compounds HJC0129 (IC50-SLC = 9.6 μM and IC50-pro = 11.3 μM), HJC0140 (IC50-SLC = 9.4 μM and IC50-pro = 25.5 μM), and JMX0207 (IC50-SLC = 1.3 μM and IC50-pro = 8.2 μM) showed potent inhibitory activity against the NS2B–NS3 interactions in both NS2B–NS3 SLC and NS2B–NS3 protease assays.
Inhibition of DENV2 Viral Replication
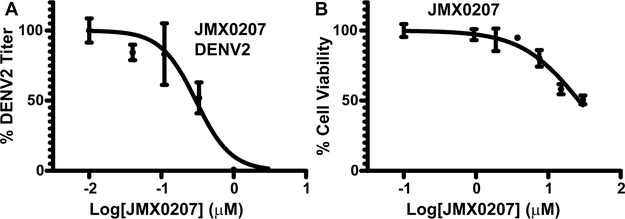
A viral plaque reduction assay was performed next to evaluate the antiviral efficacy of these derivatives. Human lung carcinoma A549 cells were infected with DENV2 in the presence of a concentration series of derivatives or a dimethyl sulfoxide (DMSO) control, and viral titers were measured at 48 h postinfection, as described previously.21,22,40 Our results indicated that 8 out 11 active derivatives showed appreciable antiviral efficacy with EC50-DN2 less than 30 μM (Table 1). The best derivative, JMX0207, showed slightly improved antiviral efficacy (EC50-DN2 of 0.31 μM), compared to niclosamide (EC50-DN2 of 0.55 μM) (Figure 2A).
Figure 2.
JMX0207 inhibits DENV. (A) Dose-dependent inhibition of DENV2 infectivity by JMX0207. N =3. (B) Cell viability assay. A549 cells were incubated with various concentrations of JMX0207 and then assayed for viability at 48 h postincubation. N =3. Error bars in both panels represent the standard deviations at each concentration.
The observed antiviral activity could result from the compound’s cytotoxicity. To address this concern, we next measured the compound’s cytotoxicity CC50 using a WST-8 cell proliferation assay, as we described previously.21 As shown in Table 1 and Figure 2B, the majority of these derivatives showed improved CC50 and therapeutic index (defined as CC50/EC50), compared to niclosamide.
Inhibition of ZIKV Replication
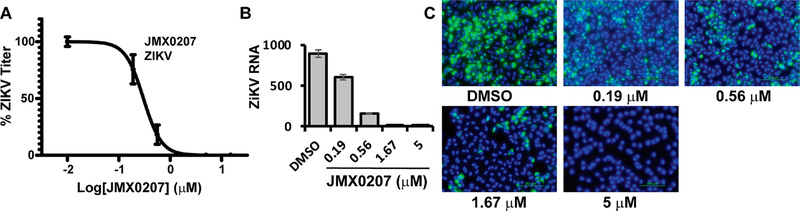
Our previous studies indicated that niclosamide is a broad-spectrum inhibitor against multiple flaviviruses.21 To determine the antiviral spectrum of the derivatives, we evaluated the antiviral potency of the derivatives against ZIKV. Our results showed that the derivatives were also potent inhibitors against ZIKV, with EC50-zk values comparable to those for DENV2 (Table 1, Figure 3A). Similarly, JMX0207 showed better inhibition efficacy toward ZIKV than niclosamide (Figure 3A).
Figure 3.
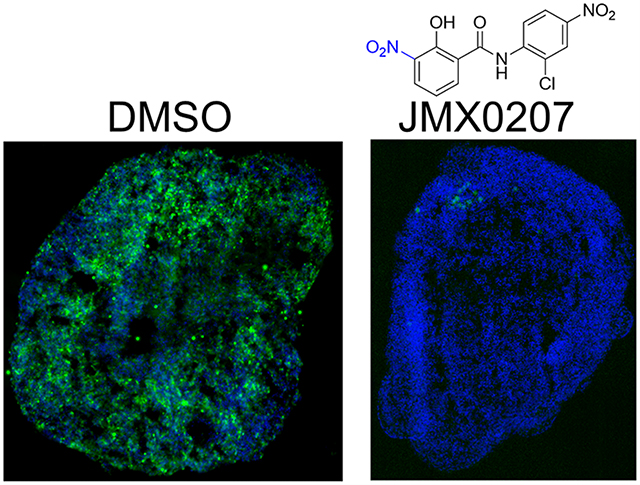
JMX0207 inhibits ZIKV. (A) Dose-dependent inhibition of ZIKV infectivity. N =3. (B) qRT-PCR analysis of the inhibition of viral RNA from ZIKV-infected A549 cells by JMX0207. N = 3. Error bars in panels (A) and (B) represent the standard deviations at each concentration. (C) Immunofluorescence assay of inhibition of viral protein production by JMX0207, using pan-flavivirus anti-E 4G2 antibody (green) (ATCC).
JMX0207 Treatment Leads to Reductions in Viral RNA Yield and Protein Production
Among the derivatives, JMX0207 showed improved efficacy in almost all categories (Table 1), compared to niclosamide. Therefore, we chose JMX0207 to further characterize whether JMX0207 treatment led to the inhibition of viral RNA synthesis and viral protein production, using qRT-PCR and immunofluorescence assay (IFA), respectively. Our results indicated that JMX0207 significantly reduced the ZIKV RNA copy number in a dose-dependent manner (Figure 3B). Using a pan flavivirus anti-E antibody 4G2, we conducted an IFA to demonstrate that JMX0207 greatly reduced ZIKV antigen production in A549 cells in a dose-dependent manner (Figure 3C), presumably because of inhibition of viral replication.
Overall, these experimental results indicated that JMX0207 showed improved antiprotease, antiviral, and cytotoxicity properties, compared to niclosamide. JMX0207 treatment results in inhibition of viral infectivity, viral RNA replication, and viral protein production and is a broad-spectrum antiviral for flaviviruses.
JMX0207 Inhibits Viral Production in Cells Relevant to ZIKV Pathogenesis
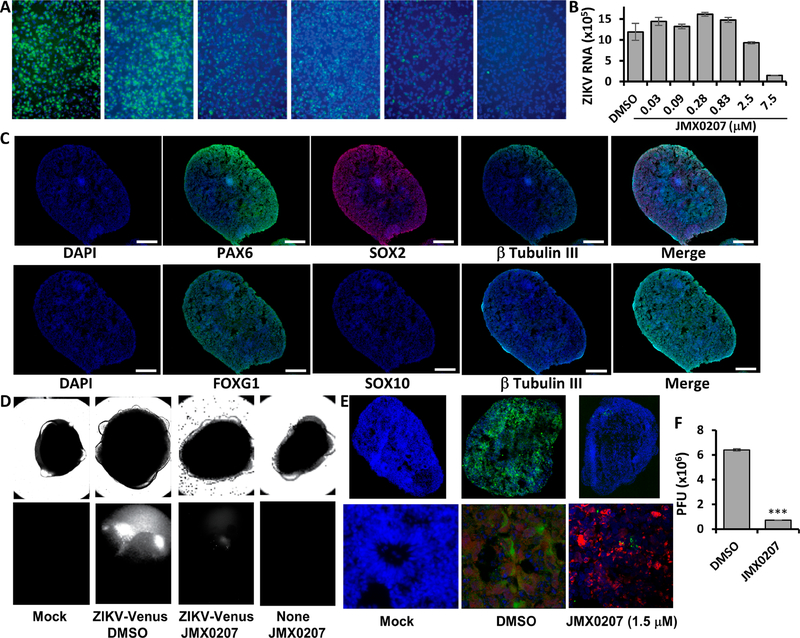
We next used human induced pluripotent stem cell (iPSC)-derived neural progenitor cells (HNPCs)21,41 to investigate whether JMX0207 is an effective inhibitor in human primary cells demonstrated to be relevant to ZIKV pathogenesis.42–46 As shown (Figure 4A,B), JMX0207 effectively inhibited ZIKV protein expression and RNA synthesis in HNPCs in dose-dependent manner. Overall, these experiments demonstrate that JMX0207 is an effective antiviral in neural progenitor cells relevant to ZIKV infection.
Figure 4.
Inhibition of ZIKV in cells relevant to ZIKV. (A) Immunofluorescence assay (IFA) of inhibition of viral protein expression in human neural progenitor cells by JMX0207, using pan-flavivirus anti-E 4G2 antibody (green) (ATCC). Nuclei (blue) were stained in all IFA assays by the Hoechst stain solution. (B) qRT-PCR analyses of inhibition of viral RNA from ZIKV-infected human neural progenitor cells (HNPC) by JMX0207. N =3. (C) Forebrain reginal specification of organoids. Organoids were stained positive for forebrain identity markers PAX6 (green, upper panel), FOXG1 (green, lower panel) and SOX2 (Red (Magenta after merge with DAPI (Blue)), upper panel) at 20 days. The sections were stained positive for general neuronal marker TUJI (cyan, upper and lower panels) and were negative for SOX10 (Red, lower panel). DAPI-merged data were shown. Nuclei (DAPI, Blue); scale bar 200 μm. (D) ZIKV organoid infected with ZIKV-Venus. The 3D organoids were infected with PBS (Mock), or ZIKV untreated (DMSO), or ZIKV treated with JMX0207, or Mock treated with JMX0207. Upper panel, bright field image of intact organoids. Lower panel, Venus fluorescence image (excitation 515 nm, emission 528 nm) of the intact 3D organoids. (E) Slices of organoid infected with ZIKV PRVABC59. The 3D organoids were infected with PBS (Mock), ZIKV untreated (DMSO), or ZIKV treated with JMX0207. Upper panel, IFA using anti-E 4G2 antibody (green); DAPI (blue). Lower panel, details of the signature rosette region of the 3D organoids (Mock) or infected region (DMSO/ZIKV and JMX0207/ZIKV). Red: Pax6. (F) ZIKV production from the 3D organoids at 5 dpi. Culture supernatants were collected, and virus production was quantified by the PFU assay. N = 3. Error bars in panels (B) and (F) represent the standard deviations at each concentration.
JMX0207 Protects 3D Mini-Brain Organoid from ZIKV Infection
Newborns from mothers with ZIKV infection during pregnancy have a significantly increased risk of developing microcephaly, a birth defect where a baby has smaller than normal head circumference. Babies born with microcephaly may develop many brain-related or other neurological problems. Recently, neural stem cell-derived 3D cerebral organoids were used to dissect ZIKV pathogenesis and antiviral development.45,47–49 Compared to the cultured 2D monolayer cells, the 3D brain organoids can better represent the composition, diversity, and organization of cell types found in the developing human brain.
Therefore, we used the 3D mini-brain organoid model to further investigate whether JMX0207 can protect against ZIKV-associated neurological damage. Induced pluripotent stem cells (iPSC) derived from a healthy control (Male, Caucasian) were differentiated using an established protocol to generate region-specific organoids patterned to resemble the dorsal forebrain (Figure 4C).50 Organoids were stained positive for forebrain identity markers PAX6 (dorsal forebrain progenitors; upper panel), FOXG1 (lower panel), and SOX2 (neural ectoderm marker, upper panel) at 20 days. The sections were stained positive for general neuronal marker TUJI (neuron-specific class III β-tubulin; upper and lower panels) and were negative for SOX10 (neural crest, red, lower panel).
To evaluate if JMX0207 protects organoids from ZIKV infections, we first generated a full-length infectious ZIKV clone expressing Venus fluorescent protein (ZIKV-Venus) (details to be published elsewhere). At day 20, the organoids displayed signature features of forebrain, including neural rosettes (Figure 4C–E), and were pretreated with JMX0207 (1.5 μM) or DMSO control. At day 20, the organoids were infected with either Mock or ZIKV strain PRVABC59 or ZIKV-Venus at an estimated multiplicity of infection (MOI) of 1 in the presence of 1.5 μM JMX0207 or DMSO control. ZIKV infection was quantified by a plaque forming unit (PFU) assay at 5 dpi and by fluorescence imaging at 7 dpi, respectively. Our results showed that JMX0207 at 1.5 μM concentration did not have any toxic effect on the 3D organoids, which remained intact and unchanged in morphology (Figure 4D). Compared to the DMSO control, JMX0207 treatment completely abolished ZIKV infection in the 3D organoid (Figure 4D).
The 3D mini-brain organoids were sliced for immunostaining to investigate the antiviral effects of JMX0207 (Figure 4E). As shown, untreated organoids (DMSO) were infected with ZIKV throughout all layers (Figure 4E). In contrast, JMX0207 treatment nearly completely protected the organoids from ZIKV infection (Figure 4E). Moreover, ZIKV production was also significantly inhibited by JMX0207 treatment (Figure 4F). Collectively, the results indicated that JMX0207 is an effective inhibitor to protect developing human cortical tissue from ZIKV infection.
JMX0207 Shows Improved Pharmacokinetic Properties
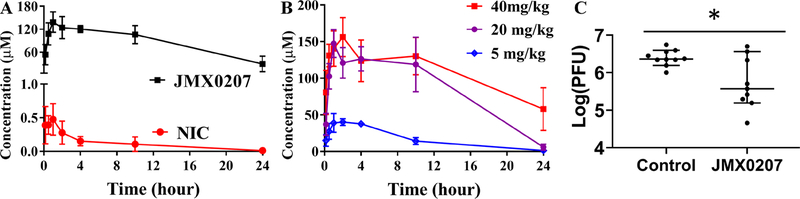
We next evaluated whether JMX0207 has favorable pharmacokinetic properties using a mouse model. As shown in Table 2 and Figure 5A, JMX0207 displayed much better pharmacokinetic properties than the lead compound niclosamide. In contrast to niclosamide, which had a short T1/2 and low Cmax (0.6 μM), when given to B6 mice orally at 40 mg/kg, JMX0207 had an excellent pharmacokinetic profile with Cmax of 145 μM (Table 2), which was much higher than the EC50 (0.3 μM) required to inhibit ZIKV. The longer T1/2 (~11 h) was also more favorable to ensure less frequent drug administration to infected animals. A further pharmacokinetic study with different doses of JMX0207 indicated that the plasma drug concentration remained at the same level after the dose was reduced to 20 mg/kg (Figure 5B). More importantly, mice with repeated oral dosing at 40 mg/kg/day for 7 days did not display any sign of toxicity (N = 6). These results indicate good bioavailability and low toxicity for the derivative JMX0207.
Table 2.
Pharmacokinetic Properties of Niclosamide and JMX0207
| Tmax ± SDa (h) | Cmax ± SD (μM) | T1/2 ± SD (h) | AUC0→∞ ± SD (μmol/L*h) | |
|---|---|---|---|---|
| niclosamide | 0.7 ± 0.4 | 0.6 ± 0.1 | 4.6 ± 1.0 | 3.5 ± 0.8 |
| JMX0207 | 1.2 ± 0.5 | 145 ± 18 | 11 ± 6.8 | 2719 ± 1018 |
SD, standard deviation.
Figure 5.
In vivo antiviral activity of JMX0207 against ZIKV. (A) Pharmacokinetic study of niclosamide and JMX0207. Adult female B6 mice were given a single oral dose of either niclosamide or JMX0207 at 40 mg/kg. Plasma was obtained at various times after dosing. Both niclosamide and JMX0207 were extracted from the plasma and then analyzed by LC-MS/MS as described in the Methods. N = 4. (B) Pharmacokinetic study of JMX0207 at different doses. N = 4–5. The values in panels (A) and (B) represent means ± SD. (C) Viremia was detected by a plaque forming unit (PFU) assay on day 3 postinfection of ZIKV with a dose of 1.7 × 105 PFU/mouse in four week-old A129 mice, which were treated with vehicle or JMX0207 though oral gavage. The difference between JMX0207 (N = 10) or vehicle (N = 10) treatment was analyzed by using the unpaired, two-tailed t test. *, P = 0.0081. Error bars represent the data range of the median with 95% confidence interval.
JMX0207 Reduces Viremia in a ZIKV Mouse Model
Finally, we evaluated the in vivo antiviral efficacy using a viremia mouse model, as we described previously.51,52 Our data showed that JMX0207 treatment at 20 mg/kg/day resulted in a significant reduction in ZIKV-induced viremia in the A129 mice inoculated with 1.7 × 105 PFU PRVABC59 ZIKV/mouse compared to the vehicle control (Figure 5C). Overall, the results indicated that JMX0207 not only inhibited viral replication in vitro but also significantly reduced viremia in an in vivo animal model.
JMX0207 Is Effective Postinfection
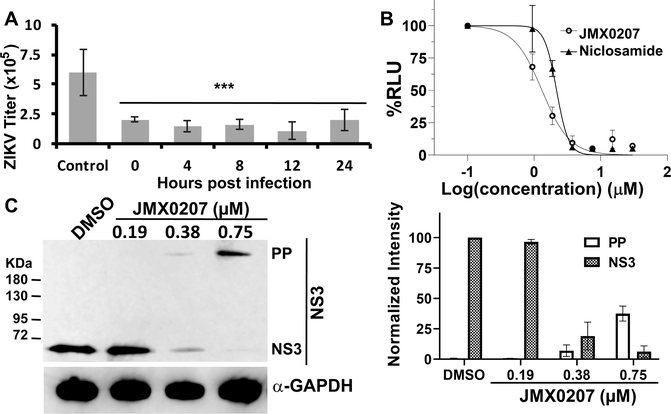
Antiviral inhibitors are generally divided into two categories, entry and postentry (replication), respectively. To determine the mode of action, we first performed a time of addition experiment by adding JMX0207 at different time points postinfection (Figure 6A). Our results showed that JMX0207 was equally effective in reducing ZIKV titer even at 24 h postinfection. The results indicated that JMX0207 inhibited viral replication instead of viral entry, which agrees with our hypothesis that JMX0207, as a protease inhibitor, attenuates viral production in the postentry replication stage.
Figure 6.
Mechanism of action studies. (A) Time of addition. JMX0207 (0.75 μM) was added at indicated time points postinfection. Viral titers were quantified using the plaque forming assay 48 h postinfection. DMSO was added as the control. N =3. ***, P < 0.001. (B) Dose-dependent inhibition of DENV2 replicon by niclosamide and JMX0207. N = 3. (C) WB analysis of dose response inhibition of ZIKV NS3 production by JMX0207 treatment (left). (Right) Band intensities of PP and NS3 normalized to the GAPDH control (N = 3). Preseeded A549 cells in 6-well plates were treated with JMX0207 or DMSO control and infected with ZIKV with a MOI of 0.1, as described previously.21 At 48 h postinfection, cells were washed, harvested, and protected by protease inhibitor cocktail prior to lysis by SDS-PAGE loading buffer. Upon incubation at 95 °C for 10 min, the sample was subjected to Western blot analysis with anti-ZIKV NS3 (GTX133309, GeneTex, Inc.) and anti-GAPDH (CB1001, EMD Millipore) as primary antibodies. Error bars in all panels represent the standard deviations at each condition.
JMX0207 Inhibits Viral Replication Using a DENV2 Replicon Cell Line
To further investigate the mechanism of action, we next used a DENV2 replicon cell line53 to investigate if JMX0207 inhibited flavivirus replication without an entry step. Our results indicated that both niclosamide and JMX0207 showed dose–response inhibition of DENV2 replication using the DENV2 replicon cell line (Figure 6B). JMX0207 has slightly improved antiviral efficacy (EC50-DN2 of 1.1 μM) compared to niclosamide (EC50-DN2 of 2.0 μM) (Figure 6B). Overall, the results indicate that JMX0207 is an effective antiviral to inhibit viral replication instead of entry.
JMX0207 Treatment Inhibits Viral Polyprotein Precursor (PP) Processing
We carried out Western blot (WB) analysis using an antibody recognizing ZIKV NS3 protein (Figure 6C). Our results showed that JMX0207 treatment led to dose-dependent reduction of viral NS3 protein production, presumably due to inhibition of viral production by JMX0207. In addition to reduced NS3 protein production, we observed a dose-dependent increase of a protein with high molecular weight (MW) (Figure 6C). It was known from our previous studies that the high MW protein is the unprocessed viral polyprotein precursor (PP), which can be recognized by the anti-NS3 antibody.21,23 The results are consistent with the mechanism by which JMX0207 inhibits viral protease function, leading to inhibition of viral PP processing, accumulation of nonfunctional viral PP, and finally virus reduction.
JMX0207 Directly Binds to the NS2B–NS3 Interface
As a niclosamide derivative, JMX0207 is expected to directly bind to the viral NS3 protease, as did niclosamide.21 To demonstrate binding, we first carried out a protein thermal shift assay (PTSA). Our data indicated that the binding of JMX0207 stabilized the viral NS3 protease, leading to a 0.75 °C increase in Tm of the viral protein. The data suggests that JMX0207 directly binds the viral NS3 protease.
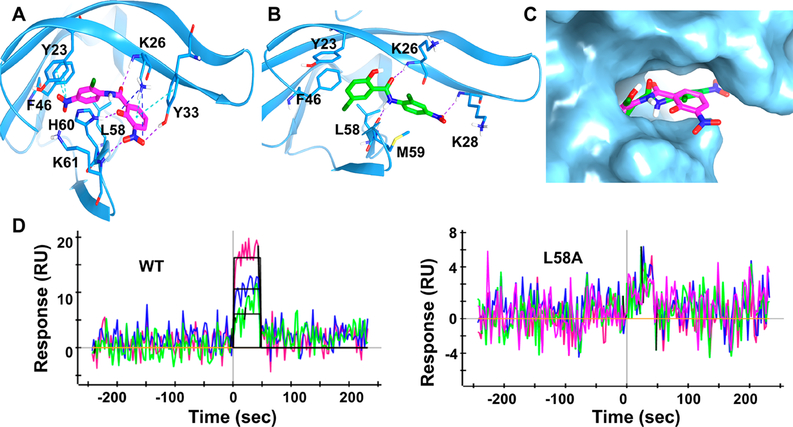
To investigate the molecular interaction between JMX0207 and viral NS3 protease, we docked JMX0207 to the NS3 protease structure of DENV2 (PDB ID: 2FOM) using the induced fit docking (IFD) protocol, as we described previously.21,22 The docked pose (Figure 7A) depicting JMX0207 can be well docked into the same 2B53 pocket identified from the previous study.21 One NO2 group of JMX0207 interacts with Y23 and F46 through π cations, while another NO2 group forms H bonds with Y33 and K61 and a salt bridge with K26. The phenyl ring on one side of JMX0207 interacts with L58 through a hydrophobic interaction, and the other phenyl ring engages with Y33 with a π–π interaction. The phenol forms an H bond with H60. The O atom on the carbonyl group of JMX02027 forms an additional H bond with K26 to further stabilize the binding. We superimposed the IFD docking pose of JMX0207 and the IFD docking pose of niclosamide. The overlay analysis demonstrates that JMX0207 binds at the 2B53 pockets on NS3 protease of ZIKV in a similar manner to that of niclosamide (Figure 7B,C), while an additional nitro group of JMX0207 also forms critical binding interactions with the protease.
Figure 7.
Docking and mutagenesis. (A) Predicted docking pose of JMX0207 (magenta) docking into NS3pro of DENV2 (PDB code: 2FOM). NS3pro is in a blue ribbon representation, and binding site key interaction residues are highlighted in stick presentation. π cation and π stacking are shown as cyan dotted lines; H bonds are in purple and salt bridges, in blue. (B) Predicted binding pose of niclosamide (green) docked into NS3pro of DENV2. H bonds are shown as purple dotted lines. (C) JMX0207 and niclosamide superimposed at the predicted binding site in surface representation. JMX0207 is shown as magenta sticks and niclosamide, in green. (D) Representative SPR analysis of JMX0207 binding to the MBP-NS3 protease wild-type and mutant L58A.
To further investigate JMX0207 binding to viral NS3 protein, we used surface plasmon resonance (SPR) to measure the binding affinity of JMX0207 to the viral NS3 protease. Because we previously generated several MBP-NS3 mutants,22 we used the MBP-NS3 fusion protein in the SPR analyses for better comparison. As shown in Figure 7D, JMX0207 bound the viral NS3 protease with a binding affinity of 1.1 μM. Collectively, our data demonstrated that JMX0207 directly binds to the viral NS3 protease with an affinity better than that for niclosamide (6.4 μM).21
In a previous study, we generated five NS3 mutants, some of which had a significant impact on the binding of small molecule inhibitors to the NS2B–NS3 interface and on viral protease activity.22 We measured the binding affinity of JMX0207 to these mutants as well. Our data showed that L58A abolished the binding of JMX0207 (Table 3, Figure 7D). Two other mutations, I25A and H60A, reduced the binding affinity 2- to 3-fold, whereas mutations Y23A and F46A do not have any effect.
Table 3.
Binding Affinity of JMX0207 to MBP-NS3 Wild-Type (WT) or Mutant
| WT | Y23A | I25A | F46A | L58A | H60A | |
|---|---|---|---|---|---|---|
| KD (μM) | 1.1 | 1.2 | 2.3 | 1.2 | NDa | 2.9 |
ND, no detectable binding.
DISCUSSION
Dengue and Zika viruses cause significant human disease, for which there are no specific vaccines or therapies. Therefore, development of effective antivirals is urgent. Previously, we identified an existing FDA-approved drug, niclosamide, as a potent antiviral against DENV and ZIKV. However, the poor pharmacokinetic properties of niclosamide prevent its clinical use to treat DENV and ZIKV infections.
In this study, we screened a small in-house compound library of niclosamide derivatives and identified JMX0207 as a candidate inhibitor with improved properties. JMX0207 inhibited the viral NS2B–NS3 protease activity of DENV2 with better efficacy in vitro than niclosamide. In cell culture, JMX0207 significantly inhibited the growth of representative flaviviruses, including DENV2 and ZIKV, with EC50 in the nanomolar range. By using IFA, WB, and qRT-PCR analyses, we showed that JMX0207 also significantly inhibited viral RNA synthesis and protein production. More importantly, compared to niclosamide, JMX0207 has a reduced cytotoxicity profile toward human cells, indicating a larger therapeutic window toward these flaviviruses. In addition, we showed that JMX0207 could rescue ZIKV-relevant neural progenitor cells from viral infection in a dose-dependent manner. Moreover, JMX0207 could almost completely protect 3D cortical organoids originating from neural stem cells from ZIKV infection. The findings establish the efficiency of JMX0207 to eliminate infection of ZIKV from human neural progenitor cells and provide a path forward to minimize the risk of fetal acquired microcephaly resulting from ZIKV infection of pregnant women.54 Furthermore, we showed that JMX0207 displayed significantly improved pharmacokinetic properties, compared to the parent compound niclosamide. The improved pharmacokinetics allowed us to carry out in vivo antiviral efficacy studies using an animal model for ZIKV infection. Our data showed that JMX0207 treatment markedly reduced viral viremia in the A129 ZIKV mouse model infected with clinical strain ZIKV PRVABC59. Mechanistically, we showed that JMX0207 directly binds to the viral NS3 protease with improved binding affinity, compared to its lead drug niclosamide. We also showed that JMX0207 treatment led to dose-dependent inhibition of viral polyprotein precursor processing, a direct consequence of inhibition of viral protease activity.
CONCLUSION
We conclude that the niclosamide derivative JMX0207 is a valuable candidate for further study, due to its demonstrated ability to inhibit several activities required for ZIKV propagation. When paired with its reduced cytotoxicity profile and superior pharmacokinetic properties, these positive attributes indicate that JMX0207 might effectively be used to combat viral progression following initial infection with ZIKV. Successful development of this compound would be a valuable tool in the prevention of negative ZIKV-related outcomes, including Guillain-Barré syndrome, neuropathy, and myelitis, as well as pregnancy-related outcomes including miscarriage, preterm birth, and congenital Zika syndrome.
METHODS
Synthesis of Niclosamide Derivatives
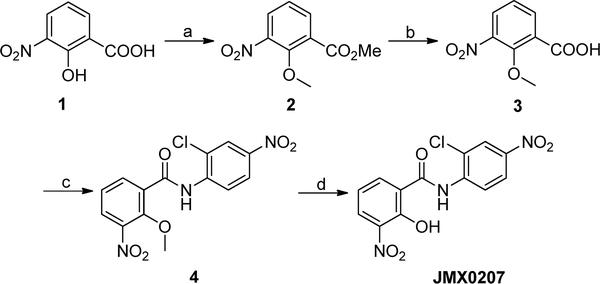
The synthetic route of compound JMX0207 was described as below (Scheme 1), and the synthesis of other niclosamide derivatives was reported in our previous publications.34–36,38 The structures and purity of all synthesized compounds were confirmed by 1H and 13C NMR, HRMS, and HPLC analysis, and all biologically evaluated compounds are >95% pure.
Scheme 1. JMX0207 Synthesis Routea.
aReagents and conditions: (a) CH3I, K2CO3, DMF, 50 °C, 24 h, 95%; (b) NaOH, H2O/MeOH, r.t., 1 h, 93%; (c) 2-chloro-4-nitroaniline, POCl3, pyridine, DCM, 0 °C to r.t., 8 h, 67%; (d) BBr3, DCM, 0 °C to r.t., 12 h, 96%.
JMX0207 Synthesis. Methyl 2-Methoxy-3-nitrobenzoate (2)
CH3I (3.9 g, 27.3 mmol) was added to a solution of 3-nitrosalicylic acid (1.0 g, 5.5 mmol) and K2CO3 (2.3 g, 16.4 mmol) in 20 mL of DMF. The mixture was stirred at 50 °C for 24 h and then diluted with 300 mL of AcOEt. The resulting mixture was washed with water (3 × 70 mL) and brine (50 mL), dried over Na2SO4, and concentrated in vacuo. The residue was purified by a silica gel column (Hexane/EtOAc = 2/1) to afford compound 2 (1.1 g, 95%) as a yellow oil. 1H NMR (300 MHz, CDCl3) δ 8.01–7.89 (m, 1H), 7.85–7.78 (m, 1H), 7.23–7.09 (m, 1H), 3.91 (s, 3H), 3.87 (s, 3H). 13C NMR (75 MHz, CDCl3) δ 164.6, 153.1, 145.4, 135.5, 128.3, 127.2, 123.7, 64.1, 52.6.
2-Methoxy-3-nitrobenzoic acid (3)
NaOH (1.1 g, 27.3 mmol, in 6 mL of H2O) was added to a solution of compound 2 (1.1 g, 5.5 mmol) in 10 mL of MeOH. The mixture was stirred at r.t. for 1 h, and then, the pH value was adjusted to 5–6 with 1 M HCl (aq.). The mixture was extracted with EtOAc (2 × 120 mL) and washed with water (80 mL) and brine (60 mL), dried over Na2SO4, and concentrated to give acid 3 (1.0 g, 93%) as a light yellow solid for direct use in the next step.
N-(2-Chloro-4-nitrophenyl)-2-methoxy-3-nitrobenzamide (4)
POCl3 (3.9 g, 25.4 mmol) was added slowly at 0 °C to a solution of compound 3 (1.0 g, 5.1 mmol), 2-chloro-4-nitroaniline (876 mg, 5.1 mmol), and pyridine (8.0 g, 101.4 mmol) in 50 mL of DCM. After addition, the mixture was stirred at r.t. for 8 h and then poured into 200 mL of ice-water. Compound 4 (1.2 g, 67%) was isolated by filtration as a yellow solid. 1H NMR (300 MHz, CDCl3) δ 10.58 (s, 1H), 8.93 (d, J = 9.3 Hz, 1H), 8.46 (dd, J = 7.8, 1.8 Hz, 1h), 8.37 (d, J = 2.7 Hz, 1H), 8.24 (dd, J = 9.3, 2.4 Hz, 1H), 8.06 (dd, J = 8.1, 1.8 Hz, 1H), 7.46 (t, J = 8.1 Hz, 1H), 4.12 (s, 3H).13C NMR (75 MHz, CDCl3) δ 161.9, 151.7, 144.4, 143.6, 140.7, 137.0, 129.9, 128.2, 125.3, 125.1, 123.8, 123.1, 121.1, 64.9.
N-(2-Chloro-4-nitrophenyl)-2-hydroxy-3-nitrobenzamide (JMX0207)
BBr3 (7.4 mL, 7.40 mmol, 1 M in DCM) was added dropwise at 0 °C to a solution of compound 4 (1.3 g, 3.70 mmol) in 250 mL of DCM. The mixture was stirred at r.t. for 2 h. Then, the mixture was poured into 200 mL of ice water. The yellow precipitate JMX0207 was isolated by filtration. The organic layer was separated and concentrated. Then, 50 mL of MeOH was added, and the mixture was stirred at r.t. for 20 min. The yellow solid was isolated by filtration. The two parts of yellow solids were combined to afford 1.2 g (96%) of compound JMX0207 in total. HPLC purity 99.9% (tR = 18.75 min). 1H NMR (300 MHz, DMSO-d6) δ 12.10 (s, 1H), 8.65 (d, J = 9.3 Hz, 1H), 8.42 (d, J = 2.4 Hz, 1H), 8.34–8.24 (m, 2H), 8.13 (d, J = 8.1 Hz, 1H), 7.03 (t, J = 7.8 Hz, 1H). 13C NMR (75 MHz, DMSO-d6) δ 164.2, 154.1, 143.1, 141.3, 139.1, 136.3, 129.8, 124.8, 124.1, 123.6, 122.5, 122.4, 117.3, HRMS (ESI) calcd for C13H9ClN3O6 338.0180 (M + H)+, found 338.0172.
Cloning, Expression, and Purification
All clones and proteins were generated as previously described.21,22
Split Luciferase Complementation (SLC) Assay
The SLC assay was performed as previously described.21,22,55
Protease Inhibition Assay
The protease inhibition assay was performed using the refolded DENV2 NS3 fusion protein (50 nM) and a fluorescence resonance energy transfer (FRET) peptide substrate (100 μM) (Abz-RRRRSAG-nTyr (Neo-Biolab)), as described previously.21–23,55 Substrate cleavage was monitored at excitation/emission wavelengths of 360 nm/420 nm (Abz substrate) by a BioTek Flx800. The rate of increase in relative fluorescence unit (RFU) over time was calculated in the linear range and normalized as a percent of the DMSO control. IC50/CC50/EC50 was determined by fitting the dose–response curve with a nonlinear regression function using GraphPad Prism 8 (San Diego, CA). All experiments were performed in triplicates.
Cytotoxicity Assay
Cytotoxicity was measured by a WST-8 cell proliferation assay kit (Dojindo Molecular Technologies, Inc.) as previously described.21 All experiments were performed in triplicates.
Viral Titer Reduction Assay
A viral titer reduction assay was used to determine the compounds’ effect on DENV2 and ZIKV strains, as described previously.21,22 Human A549 lung carcinoma cells and human primary neural progenitor cells were used, as described previously.21,22 All experiments were performed in triplicates.
Immunofluorescence Assay
The immunofluorescence assay was performed using ZIKV-infected cells treated with DMSO or JMX0207, as described previously.21,22 A mouse monoclonal pan anti-E antibody 4G2 (ATCC) and a DyLight 488 goat antimouse IgG (ImmunoReagents, Inc.) were used to monitor viral E protein expression, as described previously.21,22
Quantitative qRT-PCR
Quantitative qRT-PCR was performed as described previously.21,22 ZIKV primers CCGCTGCCCAACACAAG and CCACTAAYGTTCTTTTGCAGACAT with ZIKV probe Cy5-AGCCTACCT/TAO/TGACAAGCAGTCAGACACTCAA-IAbRQSp were used. The 2–ΔΔCT (“delta–delta Ct”) method was used to quantify samples. All experiments were performed in triplicates.
Western Blot
Western blot was performed using anti-ZIKV NS3 (GTX133309, GeneTex, Inc.) and anti-GAPDH (CB1001, EMD Millipore) antibodies, as described previously.21,23 All experiments were performed in triplicates.
DENV2 Replicon Assay
BHK-21 cells stably expressing DENV2 replicon with a Renilla luciferase (Rluc) reporter gene53 were seeded into white 96-well plates at a density of 2 × 105 cells per well. The cells were incubated at 37 °C in a humidified incubator with 5% CO2 for 48 hours in a 100 μL medium containing Minimal Essential Medium (MEM) supplemented with 10% fetal bovine serum (FBS), 100 IU/mL penicillin, 100 μg/mL streptomycin, and 300 μg/mL G418. Upon 24 h of incubation, the culture medium was discarded. Fresh culture medium (100 μL) with a concentration series of compounds or DMSO control was added to the cells in triplicates. The culture was further incubated for 48 h. The cells were then washed twice with 100 μL of phosphate-buffered saline (PBS), followed by the addition of 25 μL of a lysis buffer containing 1× PBS and 1% Triton X-100. The mixture was incubated at room temperature with gentle shaking (50 rpm) for 30 min. Assay buffer (125 μL) containing 1× PBS, 0.05% (3-((3-cholamidopropyl)dimethylammonio)-1-propanesulfonate) (CHAPS), and 0.1% BSA was then added to each well. In the meantime, a fresh 4× working substrate coelenterazine was prepared by diluting 1 μL of stock coelenterazine (2.4 mM) dissolved in ethanol into 10 mL of assay buffer (10 000-fold dilution). Finally, 50 μL of 4× substrate coelenterazine was added to each well with a final substrate concentration of 0.06 μM, using a Veritas luminometer. The luminescence was recorded immediately using the Veritas luminometer. The luminescence data was normalized to DMSO control. The EC50 was determined by nonlinear regression fitting of normalized experimental data in GraphPad Prism 8.0 (San Diego, CA). All experiments were performed in triplicates.
Docking
The crystal structure of NS3pro of DENV2 (PDB code: 2FOM) was downloaded from RCSB PDB bank. After excluding the cofactor NS2B peptide, the structure was preprocessed and optimized with Schrödinger Protein Preparation Wizard using default settings. The 3D structures of JMX0207 and niclosamide were created using Schrödinger Maestro and prepared with LigPrep to generate a low energy conformation suitable for docking. The Induced Fit Docking (IFD) protocol of Schrödinger Small-Molecule Drug Discovery Suite was employed in this docking study. The grid box for docking was centered on the 2B53 binding site. The box size was set to 20 Å on each side. Selected side chains of R24, K26, and M59 were temporarily trimmed (the equivalent of being mutated to alanine) during the initial IFD process and were restored later. The receptor–ligand complex structures generated from IFD docking were imported into Schrödinger Maestro for visualization and analysis of binding site interactions.
Surface Plasmon Resonance
Surface plasmon resonance (SPR) was used to determine the affinity and kinetic analyses of the interactions between JMX0207 and the MBP-NS3 proteins at 25 °C using a ProteOn XPR36 SPR instrument (Bio-Rad). The MBP-NS3 wild-type or mutant proteins were immobilized onto a ProteOn GLH sensor chip (~9000 RU) (Bio-Rad). A 2-fold dilution series of compounds were injected as the analytes. A blank surface blocked by ethanolamine was used as the control surface. The experiment was carried out at a flow rate of 100 μL/min using a PBSTD buffer containing 1× PBS, 0.005% surfactant P20, and 5% DMSO. Association (ka) and dissociation (kd) rates, as well as the dissociation constant (KD), were obtained by global fitting of the SPR data from multiple concentrations to a simple 1:1 Langmuir binding model, using the ProteOn Manager software suite (Bio-Rad).
Culturing of Human iPSCs
Human iPSC F11350.1 culture was maintained in feed-free conditions using mTeSR1 medium (StemCell Technologies #85851) with 5× supplement (StemCell Technologies #85852). Cells were maintained on 6-well plates coated with human embryonic stem cell-qualified Matrigel at a 1:60 dilution (Corning Catalog #354277) and passaged with ReLeSR (StemCell Technologies #05872). Cortical 3D organoids were generated on the basis of the protocol described by Yoon et al.50 Briefly, iPSCs were dissociated with accutase (ThermoFisher #A1110501) and then plated at a density of 3 × 106 cells per well in an AggreWell 800 24-well plate (StemCell Technologies #34811) in Essential 8 medium (ThermoFisher #A1517001) supplemented with 10 μM rock inhibitor (Tocris #Y27632). The plate was centrifuged at 100g for 3 min and incubated at 37 °C overnight. The next day (culture day 0), the organoids were transferred to a low attachment 10 cm plate (Corning #3262) and maintained in 10 mL of neural induction medium (Essential 6 medium, Life Technologies, #A1516401) supplemented with two SMAD pathway inhibitors, 10 μM SB431541 (R&D #1614/50) and 2.5 μM dorsomorphin (Tocris #3093), and a Wnt pathway inhibitor 2.5 μM XAV-939 (Tocris #XAV-939). Media were exchanged daily. On day 6, the medium was changed to neural expansion medium (Neuralbasal A medium, ThermoFisher #10888) supplemented with 2% B27 (without Vit A; ThermoFisher #12587001), 1% antibiotic–antimycotic (ThermoFisher #15240062), 1% glutaMAX (ThermoFisher # 35050061), 20 ng/mL FGF2 (R&D #233-FB-500), and 20 ng/mL EGF2 (PeproTech #AF-100–15). Media were exchanged daily.
Cryopreservation
Organoids were fixed in 4% paraformaldehyde (Santa Cruz) overnight at 4 °C. They were then washed three times in PBS and transferred to 30% sucrose solution for 72 h. Organoids were then transferred into a plastic cryomold (10 × 10 × 5 mm; Tissue Tek cat. no. 4565) with embedding medium (Tissue-Tek OCT compound, Sakura Finetek 4583) and stored at −80 °C. For immunohistochemistry, 20 μm thick sections were cut with a Leica cryostat (model CM3050S).
Immunohistochemistry
Cryosections were blocked in 3% bovine serum albumin (BSA), 10% normal goat serum (NGS), and 0.3% Triton X–100 diluted in PBS for 1 h at room temperature. The sections were incubated overnight at 4 °C with primary antibodies diluted in blocking solution. Sections were washed three times with PBS and then incubated with appropriate secondary antibodies diluted in blocking solution for 1 h. The following primary antibodies were used for immunohistochemistry: anti-Pax-6 (1:200; Biolegend #901301), anti-Sox2 (1:100; Santa Cruz #sc-365823), anti-FoxG1 (1:500; Takara #M227), Sox10 (1:100; Santa Cruz # sc369692), and antitubulin III (1:1000; Sigma #T8660).
Pharmacokinetics
Two to three month-old female B6 mice were used for the study. Four to five mice per group were given niclosamide (40 mg/kg in 1% DMSO and 0.5% carboxymethylcellulose) or JMX0207 in the same vehicle at the indicated dose by oral gavage. Sample preparation and liquid chromatography with tandem mass spectrometry (LC-MS/MS) detection of niclosamide and JMX0207 were essentially the same as described in a previous study and were performed using a Sciex 4000 Q-Trap mass spectrometer (AB SCIEX, Framingham, MA) with the Agilent 1200 high-performance liquid chromatography system (Agilent Technologies, Santa Clara, CA).29 JMX0207 was monitored at m/z 336/171. Declustering potential, entrance potential, collision energy, and collision cell exit potential were optimized for detection and quantification of JMX0207 at −45, −10, −35, and −14 V, respectively. Data from 4 or 5 mice at each time point were averaged and used to calculate pharmacokinetic parameters, using a pharmacokinetic solver (Microsoft, Redmond, WA) by assuming a noncompartmental model. Statistical significance of various data comparisons was determined with the use of GraphPad Prism (GraphPad Software, La Jolla, CA). The Student’s t test was used. P values < 0.05 were considered statistically significant.
In Vivo Protection Efficacy
All animal studies involving infectious ZIKV were conducted at an Animal Biosafety Level 2 (ABSL-2) facility at the Wadsworth Center with Institutional Biosafety and Animal Welfare Committee approval. The in vivo antiviral activity of JMX0207 was evaluated in a viremia animal model.
A group of four week-old A129 mice were infected by subcutaneous injection with 1.7 × 105 PFU of the PRVABC59 strain. Then, the infected mice were administered JMX0207 at 20 mg/kg of body weight (N = 10) or a vehicle control (N = 10) every day for 3 consecutive days postinfection (dpi). Mice were observed daily for signs of illness and mortality. Viremia on day 3 postinfection (pi) was determined by the plaque forming assay, and statistical analysis was performed using the unpaired, two-tailed t test (Prism).
ACKNOWLEDGMENTS
We thank Dr. Padmanabhan at Georgetown University for the gift of the DENV2 replicon BHK-21 cell line. We thank the Wadsworth Center Tissue Culture Core facility for help with cell cultures and media preparation, the Biochemistry Core for SPR assistance, the animal facility for animal care, the Advanced Light Microscopy Core for immunofluorescence/confocal imaging assistance, and Dr. McClive-Reed of Heath Research, Inc. for helpful suggestions. J.Z. is also supported by the John D. Stobo, M.D. Distinguished Chair Endowment Fund. This work was partially supported by NIH grants AI131669, AI133219, AI140491, and AI134568.
ABBREVIATIONS
- ABSL-2
Animal Biosafety Level 2
- BSA
bovine serum albumin
- CHAPS
((3-((3-cholamidopropyl)-dimethylammonio)-1-propanesulfonate))
- DENV
dengue virus
- DMSO
dimethyl sulfoxide
- DN2
dengue virus serotype 2
- dpi
days postinfection
- FBS
fetal bovine serum
- FRET
fluorescence resonance energy transfer
- HNPC
human neural progenitor cells
- IFA
immunofluorescence assay
- iPCS
induced pluripotent stem cell
- LC-MS/MS
liquid chromatography with tandem mass spectrometry
- MEM
Minimum Essential Medium
- MOI
multiplicity of infection
- NGS
normal goat serum
- PBS
phosphate-buffered saline
- PFU
plaque forming unit
- PTSA
protein thermal shift assay
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- RFU
relative fluorescence unit
- RLU
relative luminescence unit
- SLC
split luciferase complementation
- SPR
surface plasmon resonance
- WB
Western blot
- ZIKV/ZK
Zika virus
Footnotes
The authors declare no competing financial interest.
Contributor Information
Zhong Li, Wadsworth Center, New York State Department of Health, Albany, New York 12208, United States.
Jimin Xu, Chemical Biology Program, Department of Pharmacology and Toxicology, University of Texas Medical Branch, Galveston, Texas 77555, United States.
Yuekun Lang, Wadsworth Center, New York State Department of Health, Albany, New York 12208, United States.
Xiaoyu Fan, Department of Pharmacology and Toxicology, College of Pharmacy, University of Arizona, Tucson, Arizona 85721, United States.
Lili Kuo, Wadsworth Center, New York State Department of Health, Albany, New York 12208, United States.
Lianna D’Brant, The Neural Stem Cell Institute, Rensselaer, New York 12144, United States.
Saiyang Hu, Wadsworth Center, New York State Department of Health, Albany, New York 12208, United States.
Subodh Kumar Samrat, Wadsworth Center, New York State Department of Health, Albany, New York 12208, United States.
Nicole Trudeau, The Neural Stem Cell Institute, Rensselaer, New York 12144, United States.
Anil M. Tharappel, Wadsworth Center, New York State Department of Health, Albany, New York 12208, United States
Natasha Rugenstein, The Neural Stem Cell Institute, Rensselaer, New York 12144, United States.
Cheri A. Koetzner, Wadsworth Center, New York State Department of Health, Albany, New York 12208, United States
Jing Zhang, Wadsworth Center, New York State Department of Health, Albany, New York 12208, United States.
Haiying Chen, Chemical Biology Program, Department of Pharmacology and Toxicology, University of Texas Medical Branch, Galveston, Texas 77555, United States.
Laura D. Kramer, Wadsworth Center, New York State Department of Health, Albany, New York 12208, United States; Department of Biomedical Sciences, School of Public Health, University at Albany, Albany, New York 12201, United States
David Butler, The Neural Stem Cell Institute, Rensselaer, New York 12144, United States.
Qing-Yu Zhang, Department of Pharmacology and Toxicology, College of Pharmacy, University of Arizona, Tucson, Arizona 85721, United States.
Jia Zhou, Chemical Biology Program, Department of Pharmacology and Toxicology, University of Texas Medical Branch, Galveston, Texas 77555, United States.
Hongmin Li, Wadsworth Center, New York State Department of Health, Albany, New York 12208, United States; Department of Biomedical Sciences, School of Public Health, University at Albany, Albany, New York 12201, United States.
REFERENCES
- (1).Calvet G, Aguiar RS.,Melo AS, Sampaio SA, de Filippis I, Fabri A, Araujo ES, de Sequeira PC, de Mendonca MC, de Oliveira L, Tschoeke DA, Schrago CG, Thompson FL, Brasil P, Dos Santos FB, Nogueira RM, Tanuri A, and de Filippis AM (2016) Detection and sequencing of Zika virus from amniotic fluid of fetuses with microcephaly in Brazil: a case study. Lancet Infect. Dis. 16, 653–660. [DOI] [PubMed] [Google Scholar]
- (2).Thomas DL, Sharp TM, Torres J, Armstrong PA, Munoz-Jordan J, Ryff KR, Martinez-Quinones A, Arias-Berrios J, Mayshack M, Garayalde GJ, Saavedra S, Luciano CA, Valencia-Prado M, Waterman S, and Rivera-Garcia B (2016) Local Transmission of Zika Virus - Puerto Rico, November 23, 2015-January 28, 2016. MMWR Morb Mortal Wkly Rep 65 (6), 154–8. [DOI] [PubMed] [Google Scholar]
- (3).Martines RB, Bhatnagar J, Keating MK, Silva-Flannery L, Muehlenbachs A, Gary J, Goldsmith C, Hale G, Ritter J, Rollin D, Shieh WJ, Luz KG, Ramos AM, Davi HP, Kleber de Oliveria W, Lanciotti R, Lambert A, and Zaki S (2016) Notes from the Field: Evidence of Zika Virus Infection in Brain and Placental Tissues from Two Congenitally Infected Newborns and Two Fetal Losses - Brazil, 2015. MMWR Morb Mortal Wkly Rep 65 (6), 159–60. [DOI] [PubMed] [Google Scholar]
- (4).Rodriguez-Morales AJ (2016) Zika and microcephaly in Latin America: An emerging threat for pregnant travelers? Travel Med. Infect Dis 14, 5–6. [DOI] [PubMed] [Google Scholar]
- (5).Li C, Xu D, Ye Q, Hong S, Jiang Y, Liu X, Zhang N, Shi L, Qin CF, and Xu Z (2016) Zika Virus Disrupts Neural Progenitor Development and Leads to Microcephaly in Mice. Cell Stem Cell 19, 672. [DOI] [PubMed] [Google Scholar]
- (6).Patino-Barbosa AM, Medina I, Gil-Restrepo AF, and Rodriguez-Morales AJ (2015) Zika: another sexually transmitted infection? Sex Transm Infect 91 (5), 359. [DOI] [PubMed] [Google Scholar]
- (7).Musso D, Roche C, Robin E, Nhan T, Teissier A, and Cao-Lormeau VM (2015) Potential sexual transmission of Zika virus. Emerging Infect. Dis. 21 (2), 359–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Musso D, Nhan T, Robin E, Roche C, Bierlaire D, Zisou K, Shan Yan A, Cao-Lormeau VM, and Broult J (2014) Potential for Zika virus transmission through blood transfusion demonstrated during an outbreak in French Polynesia, November 2013 to February 2014. Euro Surveill 19 (14), 20761. [DOI] [PubMed] [Google Scholar]
- (9).Foy BD, Kobylinski KC, Chilson Foy JL, Blitvich BJ, Travassos da Rosa A, Haddow AD, Lanciotti RS, and Tesh RB (2011) Probable non-vector-borne transmission of Zika virus, Colorado, USA . Emerging Infect. Dis. 17 (5), 880–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Duggal NK, Ritter JM, Pestorius SE, Zaki SR, Davis BS, Chang GJ, Bowen RA, and Brault AC (2017) Frequent Zika Virus Sexual Transmission and Prolonged Viral RNA Shedding in an Immunodeficient Mouse Model. Cell Rep 18 (7), 1751–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Hirsch AJ, Smith JL, Haese NN, Broeckel RM, Parkins CJ, Kreklywich C, DeFilippis VR, Denton M, Smith PP, Messer WB, Colgin LM, Ducore RM, Grigsby PL, Hennebold JD, Swanson T, Legasse AW, Axthelm MK, MacAllister R, Wiley CA, Nelson JA, and Streblow DN (2017) Zika Virus infection of rhesus macaques leads to viral persistence in multiple tissues. PLoS Pathog. 13 (3), e1006219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Govero J, Esakky P, Scheaffer SM, Fernandez E, Drury A, Platt DJ, Gorman MJ, Richner JM, Caine EA, Salazar V, Moley KH, and Diamond MS (2016) Zika virus infection damages the testes in mice. Nature 540 (7633), 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Aguiar M, Stollenwerk N, and Halstead SB (2016) The Impact of the Newly Licensed Dengue Vaccine in Endemic Countries . PLoS Neglected Trop. Dis. 10 (12), e0005179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Brecher M, Zhang J, and Li H (2013) The flavivirus protease as a target for drug discovery. Virol. Sin. 28 (6), 326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Lindenbach B, and Rice CM (2001) Flaviviridae: The virus and Their Replication, Fourth ed., Lippincott William & Wilkins. [Google Scholar]
- (16).Liu L, Dong H, Chen H, Zhang J, Ling H, Li Z, Shi PY, and Li H (2010) Flavivirus RNA cap methyltransferase: structure, function, and inhibition. Front. Biol 5 (4), 286–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kang C, Keller TH, and Luo D (2017) Zika Virus Protease: An Antiviral Drug Target. Trends Microbiol. 25 (10), 797–808. [DOI] [PubMed] [Google Scholar]
- (18).Nitsche C (2019) Proteases from dengue, West Nile and Zika viruses as drug targets. Biophys. Rev. 11 (2), 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Noble CG, Chen YL, Dong H, Gu F, Lim SP, Schul W, Wang QY, and Shi PY (2010) Strategies for development of Dengue virus inhibitors. Antiviral Res. 85 (3), 450–62. [DOI] [PubMed] [Google Scholar]
- (20).Yuan S, Chan JF, den-Haan H, Chik KK, Zhang AJ, Chan CC, Poon VK, Yip CC, Mak WW, Zhu Z, Zou Z, Tee KM, Cai JP, Chan KH, de la Pena J, Perez-Sanchez H, Ceron-Carrasco JP, and Yuen KY (2017) Structure-based discovery of clinically approved drugs as Zika virus NS2B-NS3 protease inhibitors that potently inhibit Zika virus infection in vitro and in vivo. Antiviral Res. 145, 33–43. [DOI] [PubMed] [Google Scholar]
- (21).Li Z, Brecher M, Zhang J, Sakamuru S, Liu B, Huang R, Koetzner CA, Allen CA, Jones SA, Deng YQ, Chen H, Gao F, Qin CF, Lin Q, Banavali N, Zhou J, Boles N, Xia MK, Kramer LD, and Li HM (2017) Existing drugs as broad-spectrum and potent inhibitors for Zika virus by targeting NS2B-NS3 interaction. Cell Res. 27, 1046–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Li Z, Sakamuru S, Huang R, Brecher M, Koetzner CA, Zhang J, Chen H, Qin CF, Zhang QY, Zhou J, Kramer LD, Xia M, and Li H (2018) Erythrosin B is a potent and broad-spectrum orthosteric inhibitor of the flavivirus NS2B-NS3 protease. Antiviral Res. 150, 217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Brecher M, Li Z, Liu B, Zhang J, Koetzner CA, Alifarag A, Jones SA, Lin Q, Kramer LD, and Li H (2017) A conformational switch high-throughput screening assay and allosteric inhibition of the flavivirus NS2B-NS3 protease. PLoS Pathog. 13 (5), e1006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yao Y, Huo T, Lin YL, Nie S, Wu F, Hua Y, Wu J, Kneubehl AR, Vogt MB, Rico-Hesse R, and Song Y (2019) Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease. J. Am. Chem. Soc. 141 (17), 6832–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Shiryaev SA, Farhy C, Pinto A, Huang CT, Simonetti N, Elong Ngono A, Dewing A, Shresta S, Pinkerton AB, Cieplak P, Strongin AY, and Terskikh AV (2017) Characterization of the Zika virus two-component NS2B-NS3 protease and structure-assisted identification of allosteric small-molecule antagonists. Antiviral Res. 143, 218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Mazzon M, Ortega-Prieto AM, Imrie D, Luft C, Hess L, Czieso S, Grove J, Skelton JK, Farleigh L, Bugert JJ, Wright E, Temperton N, Angell R, Oxenford S, Jacobs M, Ketteler R, Dorner M, and Marsh M (2019) Identification of Broad-Spectrum Antiviral Compounds by Targeting Viral Entry. Viruses 11 (2), 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kao JC, HuangFu WC, Tsai TT, Ho MR, Jhan MK, Shen TJ, Tseng PC, Wang YT, and Lin CF (2018) The antiparasitic drug niclosamide inhibits dengue virus infection by interfering with endosomal acidification independent of mTOR. PLoS Neglected Trop. Dis. 12 (8), e0006715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Xu J, Shi PY, Li H, and Zhou J (2020) Broad Spectrum Antiviral Agent Niclosamide and Its Therapeutic Potential . ACS Infect. Dis. 6, 909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Fan X, Li H, Ding X, and Zhang QY (2019) Contributions of Hepatic and Intestinal Metabolism to the Disposition of Niclosamide, a Repurposed Drug with Poor Bioavailability. Drug Metab. Dispos. 47 (7), 756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Burock S, Daum S, Keilholz U, Neumann K, Walther W, and Stein U (2018) Phase II trial to investigate the safety and efficacy of orally applied niclosamide in patients with metachronous or sychronous metastases of a colorectal cancer progressing after therapy: the NIKOLO trial. BMC Cancer 18 (1), 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Barbosa EJ, Lobenberg R, de Araujo GLB, and Bou-Chacra NA (2019) Niclosamide repositioning for treating cancer: Challenges and nano-based drug delivery opportunities. Eur. J. Pharm. Biopharm. 141, 58–69. [DOI] [PubMed] [Google Scholar]
- (32).Chen W, Mook RA Jr., Premont RT, and Wang J (2018) Niclosamide: Beyond an antihelminthic drug. Cell. Signalling 41, 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Fan X, Xu J, Files M, Cirillo JD, Endsley JJ, Zhou J, and Endsley MA (2019) Dual activity of niclosamide to suppress replication of integrated HIV-1 and Mycobacterium tuberculosis (Beijing). Tuberculosis (Oxford, U. K.) 116S, S28–S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Xu J, Pachon-Ibanez ME, Cebrero-Cangueiro T, Chen H, Sanchez-Cespedes J, and Zhou J (2019) Discovery of niclosamide and its O-alkylamino-tethered derivatives as potent antibacterial agents against carbapenemase-producing and/or colistin resistant Enterobacteriaceae isolates. Bioorg. Med. Chem. Lett. 29 (11), 1399–1402. [DOI] [PubMed] [Google Scholar]
- (35).Chen H, Yang Z, Ding C, Xiong A, Wild C, Wang L, Ye N, Cai G, Flores RM, Ding Y, Shen Q, and Zhou J (2014) Discovery of potent anticancer agent HJC0416, an orally bioavailable small molecule inhibitor of signal transducer and activator of transcription 3 (STAT3). Eur. J. Med. Chem. 82, 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Chen H, Yang Z, Ding C, Chu L, Zhang Y, Terry K, Liu H, Shen Q, and Zhou J (2013) Discovery of O-Alkylamino Tethered Niclosamide Derivatives as Potent and Orally Bioavailable Anticancer Agents. ACS Med. Chem. Lett. 4 (2), 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wang Y, Wang S, Wu Y, Ren Y, Li Z, Yao X, Zhang C, Ye N, Jing C, Dong J, Zhang K, Sun S, Zhao M, Guo W, Qu X, Qiao Y, Chen H, Kong L, Jin R, Wang X, Zhang L, Zhou J, Shen Q, and Zhou X (2017) Suppression of the Growth and Invasion of Human Head and Neck Squamous Cell Carcinomas via Regulating STAT3 Signaling and the miR-21/beta-catenin Axis with HJC0152. Mol. Cancer Ther. 16 (4), 578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Chen H, Yang Z, Ding C, Chu L, Zhang Y, Terry K, Liu H, Shen Q, and Zhou J (2013) Fragment-based drug design and identification of HJC0123, a novel orally bioavailable STAT3 inhibitor for cancer therapy. Eur. J. Med. Chem. 62, 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Thorne N, Shen M, Lea WA, Simeonov A, Lovell S, Auld DS, and Inglese J (2012) Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter. Chem. Biol. 19 (8), 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Vernekar SK, Qiu L, Zhang J, Kankanala J, Li H, Geraghty RJ, and Wang Z (2015) 5′-Silylated 3′−1,2,3-triazolyl Thymidine Analogues as Inhibitors of West Nile Virus and Dengue Virus. J. Med. Chem. 58 (9), 4016–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, and Studer L (2009) Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol 27 (3), 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Tabata T, Petitt M, Puerta-Guardo H, Michlmayr D, Wang C, Fang-Hoover J, Harris E, and Pereira L (2016) Zika Virus Targets Different Primary Human Placental Cells, Suggesting Two Routes for Vertical Transmission. Cell Host Microbe 20 (2), 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, Christian KM, Didier RA, Jin P, Song H, and Ming GL (2016) Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell 18 (5), 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zhang F, Hammack C, Ogden SC, Cheng Y, Lee EM, Wen Z, Qian X, Nguyen HN, Li Y, Yao B, Xu M, Xu T, Chen L, Wang Z, Feng H, Huang WK, Yoon KJ, Shan C, Huang L, Qin Z, Christian KM, Shi PY, Xia M, Zheng W, Wu H, Song H, Tang H, Ming GL, and Jin P (2016) Molecular signatures associated with ZIKV exposure in human cortical neural progenitors. Nucleic Acids Res. 44, 8610–8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon KJ, Jeang W, Lin L, Li Y, Thakor J, Berg DA, Zhang C, Kang E, Chickering M, Nauen D, Ho CY, Wen Z, Christian KM, Shi PY, Maher BJ, Wu H, Jin P, Tang H, Song H, and Ming GL (2016) Brain-Region-Specific Organoids Using Minibioreactors for Modeling ZIKV Exposure. Cell 165 (5), 1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, and Rehen SK (2016) Zika virus impairs growth in human neurospheres and brain organoids. Science 352 (6287), 816–818. [DOI] [PubMed] [Google Scholar]
- (47).Qian X, Nguyen HN, Jacob F, Song H, and Ming GL (2017) Using brain organoids to understand Zika virus-induced microcephaly. Development 144 (6), 952–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Xu YP, Qiu Y, Zhang B, Chen G, Chen Q, Wang M, Mo F, Xu J, Wu J, Zhang RR, Cheng ML, Zhang NN, Lyu B, Zhu WL, Wu MH, Ye Q, Zhang D, Man JH, Li XF, Cui J, Xu Z, Hu B, Zhou X, and Qin CF (2019) Zika virus infection induces RNAi-mediated antiviral immunity in human neural progenitors and brain organoids. Cell Res. 29 (4), 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Li C, Deng YQ, Wang S, Ma F, Aliyari R, Huang XY, Zhang NN, Watanabe M, Dong HL, Liu P, Li XF, Ye Q, Tian M, Hong S, Fan J, Zhao H, Li L, Vishlaghi N, Buth JE, Au C, Liu Y, Lu N, Du P, Qin FX, Zhang B, Gong D, Dai X, Sun R, Novitch BG, Xu Z, Qin CF, and Cheng G (2017) 25-Hydroxycholesterol Protects Host against Zika Virus Infection and Its Associated Microcephaly in a Mouse Model. Immunity 46 (3), 446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Yoon S-J, Elahi LS, Pasca AM, Marton RM, Gordon A, Revah O, Miura Y, Walczak EM, Holdgate GM, Fan HC, Huguenard JR, Geschwind DH, and Pasca SP (2019) Reliability of human cortical organoid generation. Nat. Methods 16 (1), 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Zhang NN, Tian M, Deng YQ, Hao JN, Wang HJ, Huang XY, Li XF, Wang YG, Zhao LZ, Zhang FC, and Qin CF (2016) Characterization of the contemporary Zika virus in immunocompetent mice. Hum. Vaccines Immunother. 12 (12), 3107–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Larocca RA, Abbink P, Peron JP, Zanotto PM, Iampietro MJ, Badamchi-Zadeh A, Boyd M, Ng’ang’a D, Kirilova M, Nityanandam R, Mercado NB, Li Z, Moseley ET, Bricault CA, Borducchi EN, Giglio PB, Jetton D, Neubauer G, Nkolola JP, Maxfield LF, De La Barrera RA, Jarman RG, Eckels KH, Michael NL, Thomas SJ, and Barouch DH (2016) Vaccine protection against Zika virus from Brazil. Nature 536 (7617), 474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Boonyasuppayakorn S, Reichert ED, Manzano M, Nagarajan K, and Padmanabhan R (2014) Amodiaquine, an antimalarial drug, inhibits dengue virus type 2 replication and infectivity. Antiviral Res. 106, 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Lessler J, Chaisson LH, Kucirka LM, Bi Q, Grantz K, Salje H, Carcelen AC, Ott CT, Sheffield JS, Ferguson NM, Cummings DA, Metcalf CJ, and Rodriguez-Barraquer I (2016) Assessing the global threat from Zika virus. Science 353 (6300), aaf8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Lang Y, Li Z, and Li H (2019) Analysis of Protein-Protein Interactions by Split Luciferase Complementation Assay. Curr. Protoc Toxicol 82 (1), e90. [DOI] [PMC free article] [PubMed] [Google Scholar]