Abstract
Objective
Individuals born with intrauterine growth retardation (IUGR) are more prone to cardio-metabolic diseases as adults, and environmental changes during the perinatal period have been identified as potentially crucial factors. We have studied in a preclinical model early-onset molecular alterations present before the development of a clinical phenotype.
Methods
We used a preclinical mouse model of induced IUGR, in which we modulated the nutrition of the pups during the suckling period, to modify their susceptibility to cardio-metabolic diseases in adulthood.
Results
Mice born with IUGR that were overfed (IUGR-O) during lactation rapidly developed obesity, hepatic steatosis and insulin resistance, by three months of age, whereas those subjected to nutrition restriction during lactation (IUGR-R) remained permanently thin and highly sensitive to insulin. Mice born with IUGR and fed normally during lactation (IUGR-N) presented an intermediate phenotype and developed insulin resistance by 12 months of age. Molecular alterations to the insulin signaling pathway with an early onset were observed in the livers of adult IUGR-N mice, nine months before the appearance of insulin resistance. The implication of epigenetic changes was revealed by ChIP sequencing, with both posttranslational H3K4me3 histone modifications and microRNAs involved.
Conclusions
These two changes lead to the coherent regulation of insulin signaling, with a decrease in Akt gene transcription associated with an increase in the translation of its inhibitor, Pten. Moreover, we found that the levels of the implicated miRNA19a-3p also decreased in the blood of young adult IUGR mice nine months before the appearance of insulin resistance, suggesting a possible role for this miRNA as an early circulating biomarker of metabolic fate of potential use for precision medicine.
Keywords: Nutrition, microRNA19a, Histone modifications, Epigenetics, Biomarker, DOHaD, IUGR
Highlights
-
•
Male mice are highly sensitive to changes in nutrition during the perinatal period.
-
•
Post-natal nutrition modulates metabolic diseases induced by IUGR in male mice.
-
•
Metabolic programming by perinatal nutrition involves epigenetic mechanisms.
-
•
Pre-symptomatic IUGR mice present molecular alterations of the insulin pathway.
-
•
Plasma miRNA19a-3p levels are associated with the development of insulin resistance.
1. Introduction
The world is currently in the grip of an epidemic of overweight and obesity, with 1.9 billion overweight adults (39% of the world's adult population) in 2016, and more than 650 million obese adults (13% of the world's adult population) [1]. Obesity and overweight are associated with numerous comorbid conditions, including cardiometabolic diseases (hypertension, diabetes, and insulin resistance) and certain types of cancers. The World Health Organization (WHO) estimates that cardiovascular diseases account for 17.7 million deaths annually and diabetes accounts for 1.6 million deaths per year [1]. The epidemic is worsening, as it is now also affecting children, with 340 million overweight or obese children and adolescents aged 5–19 years in 2016 [1]. The proportion of affected children increased from 4% in 1975 to more than 18% in 2016 [1]. Comorbidities associated with being overweight will lead to long-term health crises in the coming decades. Overweight or obese children are at a higher risk of asthma and cognitive impairment in childhood and diabetes, heart disease, some cancers, respiratory diseases, mental health, and reproductive disorders later in life [2].
Genetics and lifestyle have a major impact, but the life history of children also plays a key role in the development of cardiometabolic diseases. These chronic non-communicable diseases with long duration result from a combination of genetic, epigenetic, physiological, environmental, and behavioral factors. Environmental changes during the fetal and early post-natal periods have been identified as potentially crucial factors for programming late-onset diseases, including cardiometabolic diseases in particular. Indeed, the results of many studies on neonates born small for gestational age [3], preclinical models [4], and humans, such as a Dutch cohort of pregnant women suffering from food restrictions at the end of World War II (the Dutch Hunger cohort) [5], converge to support the concept of a developmental origin of health and adult diseases (that is, the DOHaD hypothesis). Consistent with this hypothesis, the fetal environment, and the nutritional environment in particular, have been shown to be associated with a higher susceptibility to cardiometabolic diseases during adulthood [6]. Changes in the fetal environment can lead to intrauterine growth retardation (IUGR) in the fetus, and individuals born with IUGR have a higher frequency of cardiometabolic diseases in adulthood than those born with a normal weight [5,7]. The vast majority of newborns with IUGR display catch-up growth during the first few years of life [[8], [9], [10], [11]]. However, the velocity at which this catch-up growth occurs has also been associated with the occurrence of more severe diseases and earlier time points [12]. IUGR affects 5–7% of live births in developed countries [13], increasing to up to 27% of all live births in low- and middle-income countries, with a prevalence of 10.5%–72.5% in Nepal and 12.0%–78.4% in India, depending on the reference population [[14], [15], [16], [17], [18]]. This particular population of infants born with IUGR may subsequently fuel the current worldwide epidemic of obesity and metabolic syndrome.
Most of the molecular mechanisms involved in the programming of metabolic diseases involve epigenetic modifications [4,19]. Epigenetic modifications may be associated with and modulate gene expression through the methylation of DNA at CpG sites, post-translational modifications of histone tails, and protein translation through the action of microRNAs. Histone modifications may either upregulate or downregulate transcription, whereas DNA methylation and microRNAs generally inhibit gene expression. In this study, we used a preclinical model of induced IUGR in which we modulated pup nutrition during the early post-natal period. We deciphered coherent early onset molecular alterations in the liver and blood of these mice before the development of a clinical phenotype in adulthood.
2. Materials and methods
2.1. Animal experiments
All the animal procedures were conducted in accordance with institutional directives and EU directive 2010/63/EU for animal experiments and the care of laboratory animals. All of the procedures were approved by the French National Ethics Committee 05 Charles Darwin and the French Ministry of Education and Research (project protocol agreement numbers #Ce5/2011/021, #02216.02, and #8393–2017010314035875 v3). The mice used in these studies were housed under standard SPF conditions in individual ventilated cages at 22 °C under a 12-h light/12-h dark cycle with free access to water and a standard chow diet (LASQCDiet Rod16, LASvendi, Soest, Germany) (Supplementary Table S1) unless otherwise specified.
129S2/SvPas females were mated with C57BL/6J males (Charles River Laboratories, L'Arbresle, France) to produce offspring with a stable F1 hybrid genetic background. F1 hybrids have a reproducible genetic composition and display hybrid vigor, with an absence of most of the phenotypic defects described in inbred strains. We used C57BL/6J and 129S2/SvPas mice as these are two of the most widely used strains of inbred mice worldwide. We also worked with 129S2/SvPas mice because of the reputed excellent and stable maternal behavior of this strain's females when caring for fostered pups.
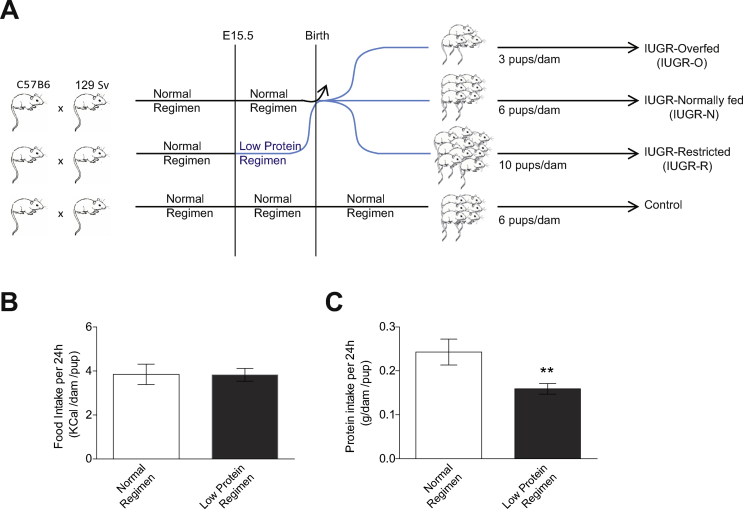
One week before mating, the mice were acclimated to control chow (A03, Safe Diets, Augy, France) (Supplementary Table S2) provided ad libitum. Pregnancy was timed from the observation of a vaginal plug. Pregnant dams were isolated and housed individually from gestational day 15. IUGR was induced by feeding the pregnant dams a low-protein/isocaloric diet (−40%) (A05, Safe Diets, Augy, France) (Supplementary Table S3) during the third week of gestation. Control litters were produced from females fed control chow throughout the entire gestation period. At birth, pups with IUGR were cross-fostered to a control female that gave birth on the same day. At our facility, 129S2/SvPas females deliver a mean of 6 ± 0.1 live newborns (n = 245 litters). IUGR litters were redistributed on day 1 such that there were six newborns per dam for normal milk feeding (IUGR-N), three per dam in the overfed group (IUGR-O), or 10 per dam in the group subjected to dietary restriction (IUGR-R) [20,21]. The mice with IUGR were compared with a control group of mice fed normally during both the fetal period and lactation. Each litter had equal numbers of male and female pups individually identified by toe clipping at five days of age. At 28 days of age, all the animals were randomly weaned, with seven animals per cage, independently of their nutritional conditions during the fetal and lactation periods. The mice were then maintained on a standard chow maintenance diet (LASQCDiet Rod16). A schematic representation of the IUGR model is provided in Supplementary Figure S1A. Five mouse cohorts were produced: one was analyzed in utero, one at 20 days of age, and one during post-natal development up to 3 months of age, and two were used for pathophysiological analyses at 3 and 12–18 months. Tissue samples were collected from 3- and 18-month-old mice under deep isoflurane anesthesia. Organs and tissues were weighed and immediately frozen on dry ice for storage at −80 °C. The pups used for all the experiments and statistical analyses originated from at least three different litters to prevent maternal bias.
2.2. Assessment of food intake and growth
Food intake by individually housed pregnant dams was measured during the third week of gestation. The mice were provided pre-weighed food ad libitum. The amount of food remaining was determined at delivery. Daily intake was calculated per animal and normalized according to the number of pups.
For body growth measurement, each pup's body weight was monitored individually between the ages of 5 and 90 days. Naso-anal length was measured at 3 months of age under isoflurane anesthesia. Body mass index (BMI) was calculated with the dedicated formula for rodents, the Lee index, as follows: [3 square root of body weight (g)/naso-anal length (cm)] x 1000.
2.3. Histological determinations
Oil Red O staining was performed on 16 μm cryostat sections of liver tissue snap frozen on dry ice. Liver sections were fixed by incubation in 4% paraformaldehyde for 20 min before staining with 0.3% Oil Red O dissolved in 60% isopropanol for 10 min. Hematoxylin counterstaining was performed before mounting. Oil Red O staining was visualized under a 20X objective with an Olympus B×612 microscope and a DP71 CCD camera (Olympus, Rungis, France). Quantification was performed by measuring the stained area with NIH ImageJ software (NIH, Bethesda, MD, USA).
Glomerulus density was determined as the number of glomeruli per mm2 in the kidneys of the adult IUGR and control mice on 7-μm-thick paraffin sections stained with hematoxylin and eosin.
2.4. Hemodynamic determination
Systolic and diastolic blood pressures were measured in 18-month-old male mice using a non-invasive tail cuff method with a Coda 6 system (Kent Scientific Corp., Torrington, CT, USA). Signals were recorded and analyzed with Kent Scientific software. We averaged 10 measurements per mouse taken after 1 week of adaptation.
2.5. Evaluation of glucose homeostasis
Glucose tolerance tests (GTT) and insulin tolerance tests (ITT) were performed one week apart on the same animals after 6 h of fasting. Tail blood was collected 0, 15, 30, 60, and 120 min after the intraperitoneal (ip) injection of either 2 g of a 20% d-glucose solution (ipGTT)/kg body weight or 1 U/kg human insulin (Actrapid Penfill, Novo Nordisk, Paris, France) (ITT). The blood glucose concentration was determined with the tail vein blood and an Accu-Chek Performa glucometer (Roche, Meylan, France) with electronic probes requiring a very low sample volume (1–3 μl).
For the in vivo determination of insulin signaling pathway activation, the mice were fasted for 18 h, after which they received an ip injection of 5 IU of human insulin (Actrapid Penfill) or saline. Livers were rapidly removed, snap frozen on dry ice, and stored at −80 °C until further processing.
2.6. Western blotting
Proteins were extracted from livers homogenized in a buffer consisting of 10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, and 0.1 mM EGTA supplemented with cocktails of both protease and phosphatase inhibitors (Roche, Meylan, France) as previously described [22]. The samples were incubated for 15 min on ice. We then added 6.3 μl of 1% Nonidet P-40 per 100 μl buffer, vortexed briefly, and centrifuged the samples at 5,000×g for 1 min at 4 °C. The protein concentration of the resulting supernatant enriched in the cytosolic fraction was then determined by NanoDrop spectroscopy (Thermo Fisher Scientific, Wilmington, DE, USA). For each sample, we subjected 20 μg of cytoplasmic protein to electrophoresis in NuPAGE Bis-Tris (4–12%) gels (Life Technologies, Courtaboeuf, France) and transferred the resulting bands onto polyvinylidene difluoride (PVDF) membranes (Merck-Millipore, Saint-Quentin-en-Yvelines, France). The membranes were then incubated with primary antibodies against phospho-AKT Ser473 (p-AKT, Cell Signaling Technology Cat #4060, Danvers, MA, USA; 1:2000) and PTEN (Cell Signaling Technology Cat# 9559; 1:1000) and then an anti-rabbit horseradish peroxidase (HRP)-conjugated IgG secondary antibody (Cell Signaling Technology Cat# 7074; 1:20,000), or HRP-conjugated antibodies against AKT and β-actin (Cell Signaling Technology Cat# 8596 and Cell Signaling Technology Cat# 12620; 1:2000, respectively). Antibody binding was then detected by incubation of membrane with SuperSignal West Pico (for actin and AKT) or Femto (for p-AKT and PTEN) chemiluminescent substrates (Cat #37080 and Cat #34095, Thermo Scientific, Courtaboeuf, France), and photographed with a high-sensitivity, cooled charge-coupled device (CCD) 6 Mpx camera on a ChemiDoc Touch Imaging System (Bio-Rad, Marnes-La-Coquette, France). The relative quantification was performed by densitometric analysis with NIH ImageJ software.
2.7. Lipid determination by liquid chromatography tandem mass spectrometry (LC-MS/MS)
Homogenates were subjected to lipid extraction as described by Matyash et al. [23]. Briefly, frozen liver tissue samples were homogenized with a TissueLyser II (Qiagen) with 5 mm stainless steel beads for three 1-min cycles at 30 Hz. We added 500 μl of PBS per 5–10 mg of tissue and lipids were extracted by adding 5 ml of cold tert-butyl methyl ether/methanol (TMBE/MeOH) (10/3; v/v) solution containing butylated hydroxytoluene (BHT) (50 μg/ml) and 50 μl of internal standard solution with appropriate concentrations and incubated for 10 min before vortexing the mixture. We added an additional 2 ml of PBS and the homogenates were incubated for 15 min, with occasional mixing, followed by centrifugation and the collection of the upper organic phase. The extracted lipid phase containing TAG, DAG, and ceramides was separated by hydrophilic interaction chromatography (HILIC) on a PVA-Sil column (vinyl alcohol polymerized silica (5 μm) support; L 250 mm X ID 4 mm) (YMC, Kyoto, Japan). The PVA-Sil column was fitted to HPLC equipment (Shimadzu LXR) coupled to an electrospray ionization (ESI) source of a triple-quadrupole mass spectrometer QTRAP 6500 (AB Sciex). Ceramide species were quantified according to published procedures [24,25], and neutral lipids (DAG and TAG) were quantified by mass spectrometry as described by Leiker et al. [26].
2.8. ChIP sequencing
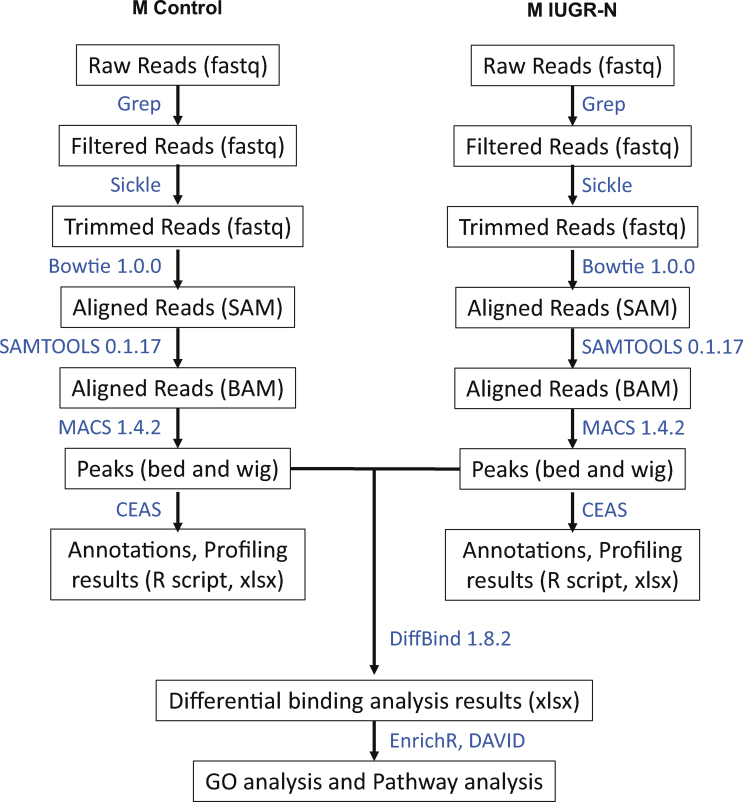
We fixed 50 mg of liver from five IUGR-N mice and six controls by incubation in 1% paraformaldehyde for 8 min, and 100 bp fragments of DNA were then generated by 25 sonication cycles of 30″ on/off at high power on a Bioruptor sonication system (Diagenode, Seraing, Belgium). Chromatin immunoprecipitation (ChIP) was performed as described by Dahl and Collas [27] against the active H3K4me3 histone mark with a specific ChIP-grade antibody (Diagenode Cat# pAb-003-050). Control immunoprecipitation was performed in parallel by conducting ChIP on an aliquot of each tissue sample to determine the background noise with normal serum-purified IgG (Abcam Cat# ab46540, Abcam, Paris, France). After DNA retrieval and purification by the phenol-chloroform method, ChIPed DNA was prepared for sequencing. Library preparation and ChIP sequencing were performed by the P3S Platform (P3S, Sorbonne Université, Paris, France). DNA ends were repaired to overhang a 3′-dA, and adapters were ligated to the end of DNA fragments including adaptor and barcode sequences for multiplexed sequencing with a NEXTflex ChIPSeq kit in accordance with manufacturer's instructions (Bio Scientific, Austin, TX, USA). DNA fragments were subjected to PCR amplification, and fragments with the correct size (100–300 bp) were sequenced. Paired-end multiplex sequencing was performed on an Illumina HiSeq 2500 platform for five IUGR and six control H3K4me3-ChIPed DNA samples and two IUGR and two control normal serum IgG ChIPed DNA samples. We obtained approximately 80 million 2 × 50 bp raw reads in FASTQ format for each ChIPSeq sample. Sequence data were filtered and trimmed to obtain good-quality reads. The preprocessed reads were then mapped to the reference genome mm9 from the University of California at Santa Cruz (UCSC, https://genome.ucsc.edu/) with Bowtie 1.0.0. The distribution of uniquely mapped reads was analyzed. The read enrichment region (peak) was scanned from the whole genome with MACS (model-based analysis of ChIPSeq). After peak scanning, the distribution of peaks was analyzed with CEAS (the cis-regulatory element annotation system), and differential binding sites between the control and IUGR groups were analyzed with the DiffBind package of R/Bioconductor. The pipeline for bioinformatics analysis is shown in Supplementary Figure S2. A differential analysis of ChIPS sequencing data was performed with edgeR, Trended_edgeR, DESeq, and DESeq2 approaches. A comparison of these four approaches revealed that outputs overlapped only partially, with significant differences detected in many peaks in the Trended_edgeR and DESeq2 analyses. Differences in histone enrichment sites were considered significant only for adjusted p values < 0.05 in both Trended_edgeR and DESeq2 analyses. Some genes were then excluded from the analysis because there were fewer than 100 reads for these genes, they were more than 4 kbp from the transcription start site (TSS, consistent with the H3K4me3 distribution profile), or the log2-fold change in enrichment was between −0.33 and + 0.33. Pathway and gene ontology (GO) function analyses were performed for the genes with differential binding sites with DAVID [28]. The STRING database (https://string-db.org) was used for protein interactions as illustrated for the subset of genes displaying > 1.5-fold differential enrichment between the IUGR-N and control groups and a high level of confidence.
2.9. Gene expression
Total RNA was isolated from liver samples by the silica membrane method (Nucleospin RNA II, Macherey–Nagel, Hoerdt, France) and quantified by NanoDrop spectroscopy. We reverse-transcribed 1 μg of RNA with random hexamers, RNAse inhibitor, and RevertAid H Minus reverse transcriptase (Fermentas, Waltham, MA, USA). Quantitative real-time PCR was performed in duplicate with SYBR Green technology on an Applied Biosystems 7300 PCR system (Applera, Courtaboeuf, France) on 10 ng of cDNA in each well. The abundance of mRNA was determined by the standard curve method and normalized against the housekeeping gene H3f3b. The sequences of the primers used are provided in Supplementary Table S4.
2.10. miRNA extraction and RT-qPCR
We isolated miRNAs from the liver and plasma samples with a Nucleospin miRNA kit (Macherey–Nagel, Hoerdt, France) in accordance with the manufacturer's instructions. We reverse-transcribed 100 ng of miRNA for the liver samples or 2 μl of miRNA extract for the plasma samples with a TaqMan MicroRNA Reverse Transcription kit (liver samples; Thermo Fisher Scientific) or a TaqMan Advanced miRNA cDNA Synthesis kit (plasma samples; Thermo Fisher Scientific). Quantitative real-time PCR assays were performed in duplicate with the TaqMan gene expression and an ABI StepOnePlus PCR system (Thermo Fisher Scientific). TaqMan miRNA assays for hsa-miR-19a-3p (#000395) and RNU6B (#001093) were used for the liver samples. TaqMan advanced miRNA assays (Thermo Fisher Scientific) for hsa-miR-19a-3p (479228_mir) and mmu-miR-195a-5p (mmu482953-mir) were used for the plasma samples. The abundance of miRNA was determined by the comparative CT method, with normalization against RNU6B and mir195a-5p abundance in the liver and plasma samples, respectively.
2.11. Statistical analysis
Prism 6 software (GraphPad Software, La Jolla, CA, USA) was used for all of the statistical analyses. Results are presented as means ± SEM. The duration of gestation and maternal food intake during gestation were analyzed with unpaired t-tests and Mann–Whitney U tests, respectively. Differences between diet groups were assessed by ANOVA followed by Newman–Keuls post hoc tests. Effects of diet and sex were analyzed by two-way ANOVA followed by Newman Keuls post hoc tests. In utero morphometric parameters and the post-natal growth curve were analyzed by two-way repeated measures ANOVA. A p value < 0.05 was considered significant.
3. Results and discussion
3.1. Modulation of the fetal and early post-natal diets alter growth and weight trajectories
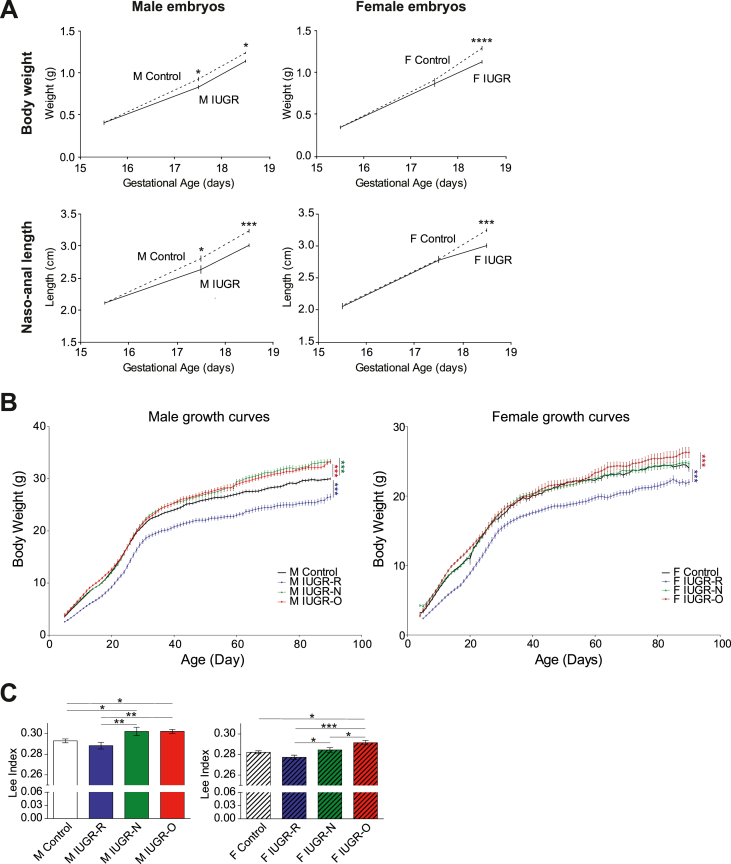
A low-protein diet during pregnancy can be used as a model for the induction of IUGR in fetuses [29,30]. We induced IUGR by decreasing the protein ratio by 40% in the diets of the pregnant mice during the third and last weeks of gestation (E15.5) (Supplementary Figure S1A). This dietary modification effectively decreased the amount of protein ingested by the pregnant dams without triggering any compensation in daily food intake (Supplementary Figure S1B and S1C) or any change in the gestation duration. Indeed, the gestation duration (19.88 ± 0.04 and 19.83 ± 0.05 days in the IUGR and control animals, respectively) remained similar between dams fed low-protein/isocaloric chow and normal chow. Interestingly, the naso-anal length and body weight measurements of the fetuses at E15.5, E17.5, and E18.5 revealed that male fetuses were more sensitive to changes in maternal nutrition than female fetuses. The male fetuses of pregnant dams fed a low-protein/isocaloric chow diet had a significantly lower naso-anal length and body weight by E17.5 (Figure 1A, left panels), whereas these effects were not visible until E18.5 in female fetuses (Figure 1A, right panels).
Figure 1.
Changes in nutrition during lactation modulate the long-term consequences of IUGR on growth in male and female mice. (A) Body weight (top panels) and naso-anal length (bottom panels) were measured at E15.5, E17.5, and E18.5 in male (left panels) and female (right panels) embryos in both the control (dashed line) (n = 8–9) and IUGR (solid line) (n = 8) groups. The impact of maternal diet on both of these parameters was more pronounced in the males, with significant differences starting at E17.5 in the males and E18.5 in the females. (B) Post-natal growth curves from days 5–90 in the male (left panel) and female (right panel) mice in the control (dark line), IUGR-R (blue line), IUGR-N (green line), and IUGR-O (red line) groups (n = 8–13 per group). The male (M) and female (F) IUGR-R mice remained lighter throughout the study's course. The IUGR-N and IUGR-O mice of both sexes displayed rapid catch-up growth. From the age of 30 days, the male IUGR-N and IUGR-O mice gained significantly more weight than the controls. The female IUGR-O mice became heavier than the controls only in adulthood. (C) BMI calculated with the Lee index for rodents in the 3-month-old control, IUGR-R, IUGR-N, and IUGR-O mice of both sexes (the males in the left panel and the females in the right panel). Note the increase in the Lee index in the male IUGR-N and IUGR-O mice relative to the control and IUGR-R mice (n = 8–14 per group). In the females, only the IUGR-O mice had a higher BMI than the control mice. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 between groups.
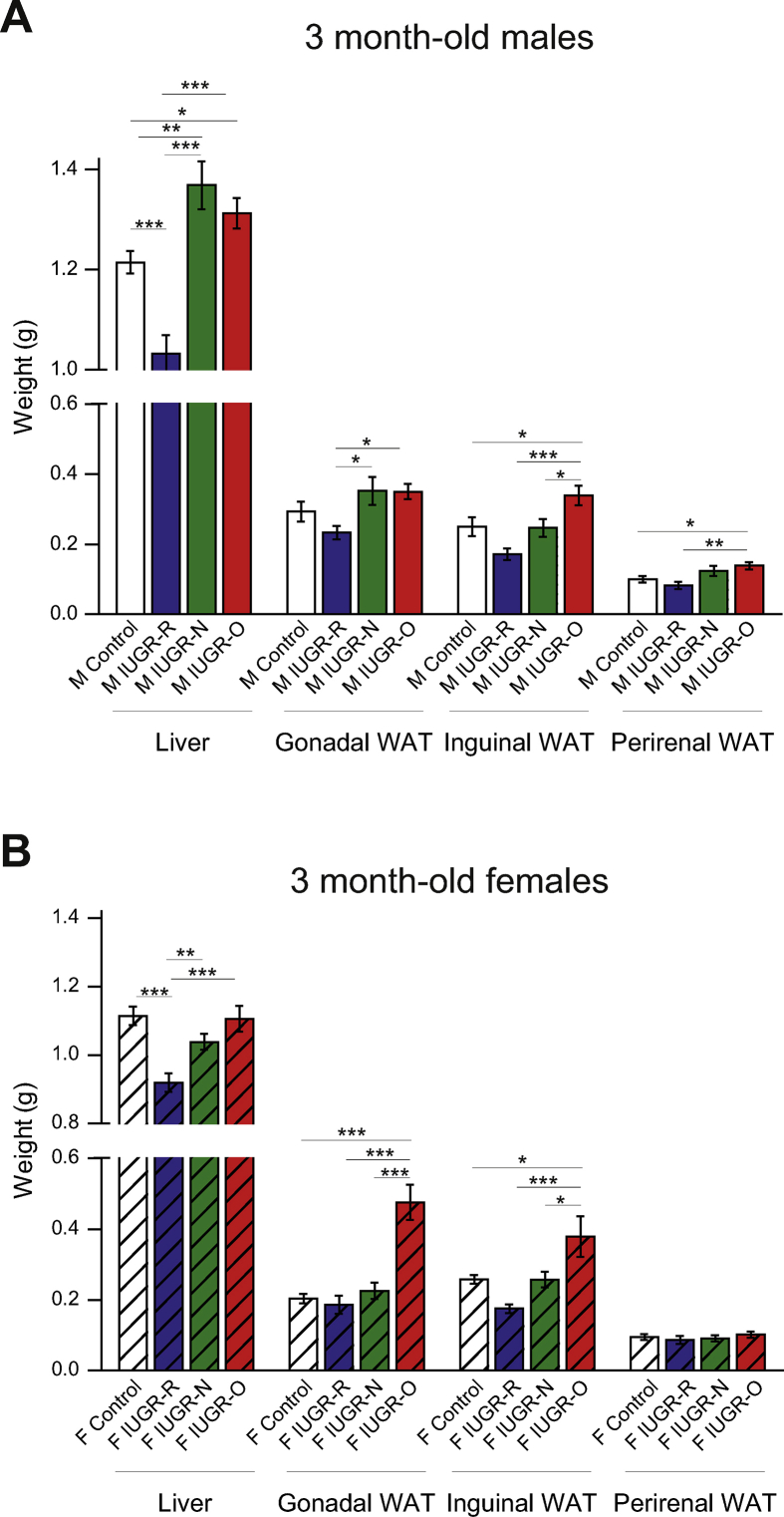
At birth, the pups with IUGR were cross-fostered to dams with a normal chow diet that had given birth on the same day to prevent any uncontrolled bias in the pups’ nutrition during lactation. Litters were normalized to 3, 6, or 10 pups per lactating dam to assess the differential effects of overfeeding (IUGR-O), normal feeding (IUGR-N), and restricted feeding (IUGR-R) of the pups, respectively, during their lactation period (Supplementary Figure S1A) [[20], [21], [22]]. The increase in pup body weight recorded between 5 and 90 days of age was compared with that of the cross-fostered pups fed normally during both gestation and lactation (Figure 1B). We began recording their weight at the age of five days to prevent any stress in the litter because pups with IUGR are very fragile. Consistent with the restriction of caloric intake during the lactation period, the IUGR-R mice of both sexes displayed no catch-up growth during lactation and remained lighter than the control mice in adulthood. In contrast, both the IUGR-N and IUGR-O mice presented rapid catch-up growth, with a normalization of body weight to the control levels before five days of age (Figure 1B) from their lower body weights observed at E17.5 and E18.5 (Figure 1A). Interestingly, by 30 days of age, the male IUGR-O and IUGR-N mice began to gain more weight than the controls (Figure 1B, left panel). This was associated with an increase in the body mass index (BMI, calculated with the dedicated Lee formula for rodents) in 3-month-old mice (Figure 1C, left panel). The effects of early post-natal diet on females seemed to be more discreet, with only the IUGR-O mice gaining weight and displaying a higher BMI in adulthood (Figure 1B,C, right panels). An analysis of body composition indicated consistent patterns of changes in organ weights, particularly in organs associated with metabolism, such as the liver and white adipose tissues (Supplementary Table S5 and Supplementary Figure S3A and S3B).
3.2. Perinatal food modulation induces changes in energy homeostasis
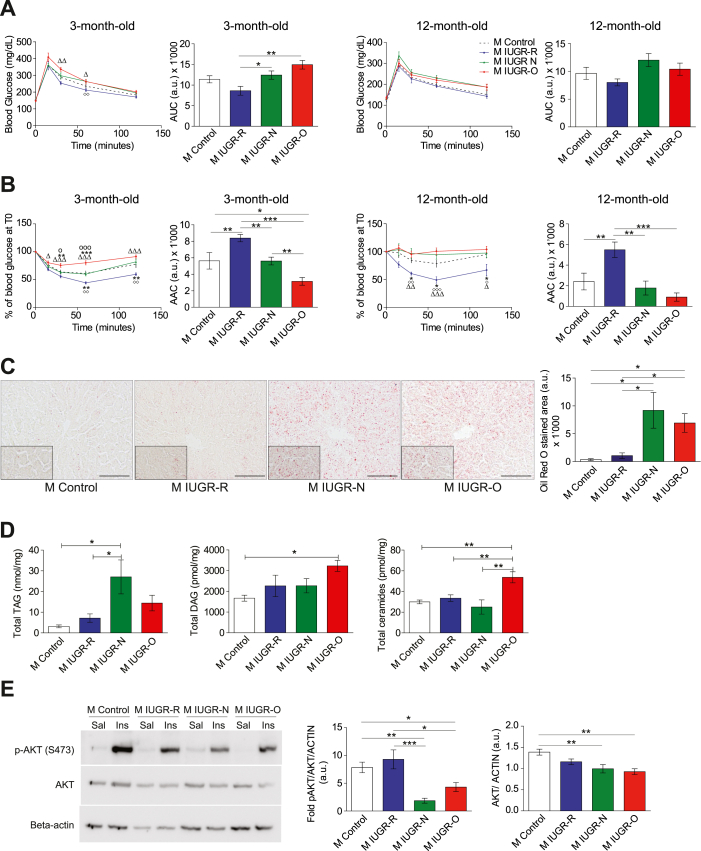
Given the greater impact on the liver and white adipose tissues of the male mice with IUGR (Supplementary Figure S3), we then checked for changes in energy homeostasis. Glucose tolerance tests indicated a non-significant trend toward a slight intolerance in the 3-month-old male IUGR-N and IUGR-O mice (Figure 2A), which was supported by plasma insulin findings and higher HOMA-IR indices (data not shown). These tendencies suggested silent alterations. Consistent with this hypothesis, insulin sensitivity tests (ITT) performed in 3- and 12-month-old mice indicated that the male IUGR-R mice were much more sensitive to insulin than the controls at both these ages (Figure 2B). In contrast, the male IUGR-O mice were already more resistant to the effects of insulin by the age of 3 months, but this difference was attenuated at 12 months, because the control male mice became less sensitive to insulin with aging (Figure 2B). Interestingly, the male IUGR-N mice presented an intermediate phenotype, with similar insulin sensitivity to the control animals at 3 months of age, but with the development of greater insulin resistance with aging, as shown by the data obtained at 12 months of age (Figure 2B). Tests on females excluded spontaneous metabolic alterations. Only the female IUGR-O mice displayed slightly higher glucose intolerance and insulin resistance at 12 months of age (Supplementary Figure S4).
Figure 2.
Energy homeostasis is affected by changes in nutrition during lactation in male mice with IUGR. (A) Blood glucose concentration during an intraperitoneal glucose tolerance test (ipGTT) in the male (M) young adult (3-month-old mice, left panels, n = 7–12 per group) or old (12-month-old mice, right panels, n = 5–9 per group) control (dashed line), IUGR-R (blue line), IUGR-N (green line), and IUGR-O (red line) mice. The calculation of the area under the curve (AUC) for the young males showed differences between the IUGR-R and IUGR-N/IUGR-O mice. No significant differences in AUC values were observed in the older males. (B) Percent decrease in the glucose levels during an insulin tolerance test (ITT) in the male young adult (3 months old, left panels, n = 9–12 per group) or old (12 months old, right panels, n = 6–10 per group) control, IUGR-R, IUGR-N, and IUGR-O mice. The area above the curve (AAC) indicates better insulin sensitivity in the 3-month-old male IUGR-R mice, an effect that persisted at 12 months of age. Conversely, lower insulin sensitivity was observed in the IUGR-O mice at 3 months, which persisted at 12 months of age. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 for the control vs IUGR, with statistical significance reported with the following comparison symbols Δ: IUGR-O vs IUGR-R, o: IUGR-O vs IUGR-N, and ◊: IUGR-N vs IUGR-R. The corresponding results for the females are shown in Supplementary Figure S4. (C) Representative photomicrographs of Oil Red O-stained sections of livers from the male 3-month-old control, IUGR-R, IUGR-N, and IUGR-O mice (scale bar, 200 μm), with a higher magnification shown in the inset. Quantifications (right panel) indicated a stronger accumulation of lipids in the IUGR-N and IUGR-O mice than in the control and IUGR-R mice (n = 4 per group). (D) Hepatic triacylglycerol (TAG) (left panel), diacylglycerol (DAG) (middle panel), and total ceramide (right panel) contents were determined in the 3-month-old male mice by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The TAG levels were higher in the IUGR-N and IUGR-O mice than in the control and IUGR-R mice, whereas DAG and total ceramide contents were higher in the male IUGR-O mice than in the male IUGR-N and control mice. (E) Representative immunoblots of Ser473-phosphorylated AKT (p-AKT), total AKT, and β-actin in the livers of the 3-month-old male mice after saline (Sal) or insulin (Ins) injections (left panel). The increase in the p-AKT/AKT ratio (normalized against β-actin) in the insulin-stimulated mice compared to the saline-injected mice was lower in the IUGR-N and IUGR-O mice than in the control and IUGR-R mice (n = 6–7 blots per group) (middle panel). The total AKT protein content of the liver was lower in the 3-month-old IUGR-N and IUGR-O mice after saline injections than in the control mice (n = 5–8 blots per group) (right panel). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 between groups.
3.3. Perinatal food modulation is associated with arterial hypertension
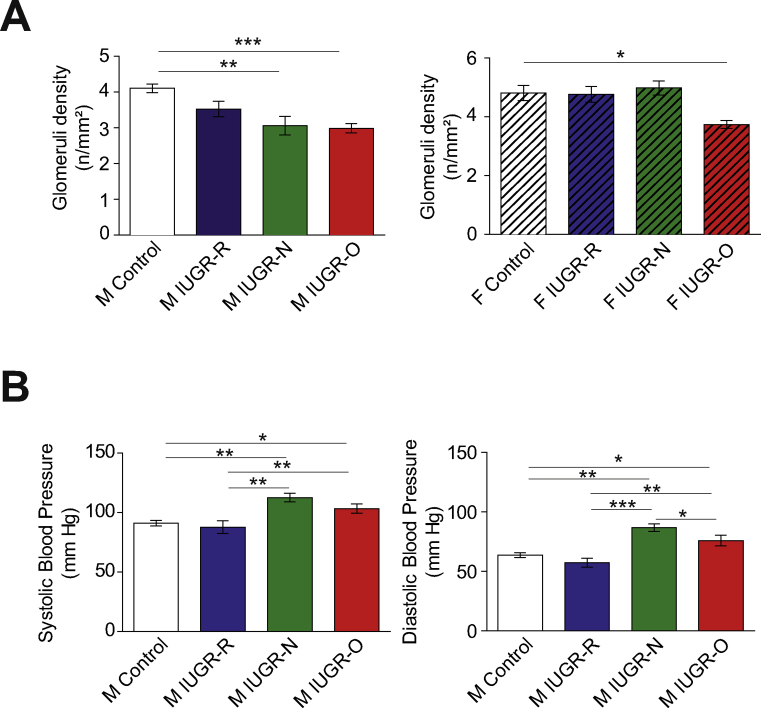
Regarding the other organs affected (Supplementary Table S5) and highly associated with IUGR [31], we found that a decrease in kidney weight was associated with a lower glomerulus density in the male IUGR-N and IUGR-O mice, with a smaller effect in the female mice with IUGR (Supplementary Figure S5A). The male mice, thus, had a stronger phenotype in response to IUGR induced by maternal protein restriction. We therefore kept these mice until the age of 18 months for the measurement of arterial blood pressure. Consistent with the predictive adaptive response [4,32,33], both the IUGR-N and IUGR-O mice, which were subjected to a richer environment during the early post-natal period than anticipated during the fetal period, spontaneously developed systolic and diastolic arterial hypertension with age, whereas the IUGR-R mice seemed to be protected against hypertension (Supplementary Figure S5B).
3.4. Perinatal food modulation is associated with changes in hepatic metabolism
These data confirm the crucial role of nutrition throughout the perinatal period in the growth of mice and clearly indicate that males are much more affected by the nutritional environment than females, from the fetal period onward [[34], [35], [36], [37], [38]]. We therefore investigated the mechanisms associated with the programming of metabolic alterations in adulthood solely in males.
The male mice with IUGR displayed major differences from the controls in terms of both liver weight and insulin sensitivity (Figure 2B and Supplementary Figure S3). We therefore focused on this key metabolic organ. Liver steatosis was observed by the age of 3 months in the IUGR-O and IUGR-N mice as demonstrated by the quantification of Oil Red O staining (Figure 2C). Liver steatosis was confirmed by determining the hepatic triacylglycerol (TAG) content, which displayed a similar profile (Figure 2D, left panel). Moreover, the 3-month-old male IUGR-O mice also had higher liver diacylglycerol (DAG) (Figure 2D, middle panel) and total ceramide (Figure 2D, right panel) content. Increases in these two parameters have been associated with insulin resistance [39,40], consistent with the ITT profile observed in the IUGR-O mice at the same age (Figure 2B, left panel).
We thus focused on the insulin signaling pathway and assessed its activation in the male mice with IUGR following an in vivo pulse of insulin. As expected, the level of AKT phosphorylation (the phospho-AKT/total-AKT ratio) induced after an in vivo pulse of insulin was much lower in the 3-month-old IUGR-O mice (Figure 2E, left and middle panels). The induction of metabolic diseases after IUGR is consistent with the findings of many studies and the DOHaD hypothesis, particularly in instances of overfeeding during lactation, as in the IUGR-O mice [12]. Interestingly, AKT-phosphorylation capacity was also much lower in the 3-month-old IUGR-N mice (Figure 2E, left and middle panels). By modulating nutrition levels during the lactation period, we were therefore able to change the age at which metabolic diseases appeared, thus highlighting, in the IUGR-N mice, molecular alterations occurring at the very start of the pathological process. These data suggest that the programming of metabolic diseases such as insulin resistance and diabetes may be associated with very early molecular alterations potentially detectable in organisms with no overt clinical phenotype. Moreover, the alterations highlighted in the young IUGR-N mice preceded the well-known increase in hepatic DAG and ceramide levels, suggesting that it may be possible to detect insulin resistance earlier, and that mechanisms other than those involving ceramides may also be involved.
3.5. IUGR is associated with epigenetic modifications in the liver of young adult mice
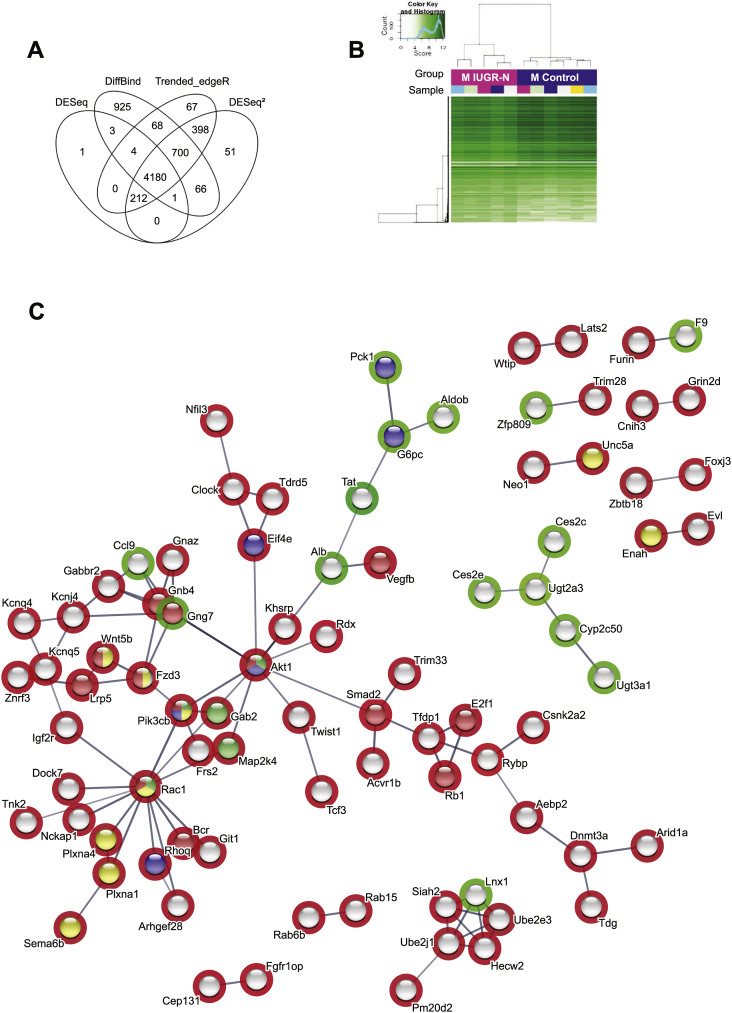
Fetal programming has been strongly associated with late-onset metabolic diseases and at least partly involves epigenetic mechanisms [4,6,19]. We evaluated epigenetic alterations by performing ChIP sequencing experiments on the livers of the pre-symptomatic young adult (3 months old) IUGR-N and control animals. We used the H3K4me3 (trimethylated histone H3 on lysine 4) histone mark, which is known to display a well-defined distribution around the proximal promoter of genes and is strongly associated with gene transcription [[41], [42], [43], [44]]. In accordance with the bioinformatics pipeline (Supplementary Figure S2), paired-end sequencing reads were trimmed on the basis of quality to 92–93% in the control (n = 6) and 92–94% in the IUGR-N (n = 5) mice; the percent alignment to the reference genome mm9 (UCSC) was approximately 75–81% and 76–79%, respectively. The global immunoprecipitation rate was approximately 68% (vs. the background signal), and the global signal obtained in ChIP sequencing experiments made it possible to achieve a 2.61% enrichment of the genome. Four different statistical methods (DESq, DiffBind, Trended_EdgeR, and DESq2) were used to identify differentially expressed histones. Trended_EdgeR and DESq2 provided the best ratios of significant sites and the lowest rates of false positives (Figure 3A). We therefore focused in downstream analyses on the 5490 histone peaks identified as significant (adjusted p value < 0.05) in both the Trended_EdgeR and DESq2 analyses. The resulting heatmap indicated that the differential binding sites between the control and IUGR-N mice clustered separately (Figure 3B).
Figure 3.
Epigenetic modifications in the livers of male young adult IUGR-N mice. (A) The four statistical analysis approaches indicated overlapping significant differential H3K4me3 peaks (adjusted p value < 0.05) between the 3-month-old male IUGR-N and control mice (n = 5–6 per group) as revealed by ChIP sequencing. Statistically significant differential peaks common to Trended_edgeR and DESeq2 (5490 peaks) were conserved for further analysis. (B) The heatmap of differential H3K4me3 peaks showed that the male controls (blue group) and IUGR-N (pink group) mice were segregated. (C) Protein interactions with high confidence illustrated with the STRING database for the subset of genes displaying differential enrichment (>1.5-fold) between the IUGR-N and control groups. Each node corresponds to one protein, and relevant connections are illustrated with connecting lines. The red core indicates proteins related to pathways in cancer, the blue core denotes insulin signaling pathway proteins, the yellow core represents axon guidance proteins, and the green core demonstrates Fc epsilon RI-mediated signaling pathway proteins according to the KEGG pathway analysis with the STRING database. The green circles indicate greater enrichment in the H3K4me3 mark in the IUGR-N group and the red circles indicate higher enrichment in the control group.
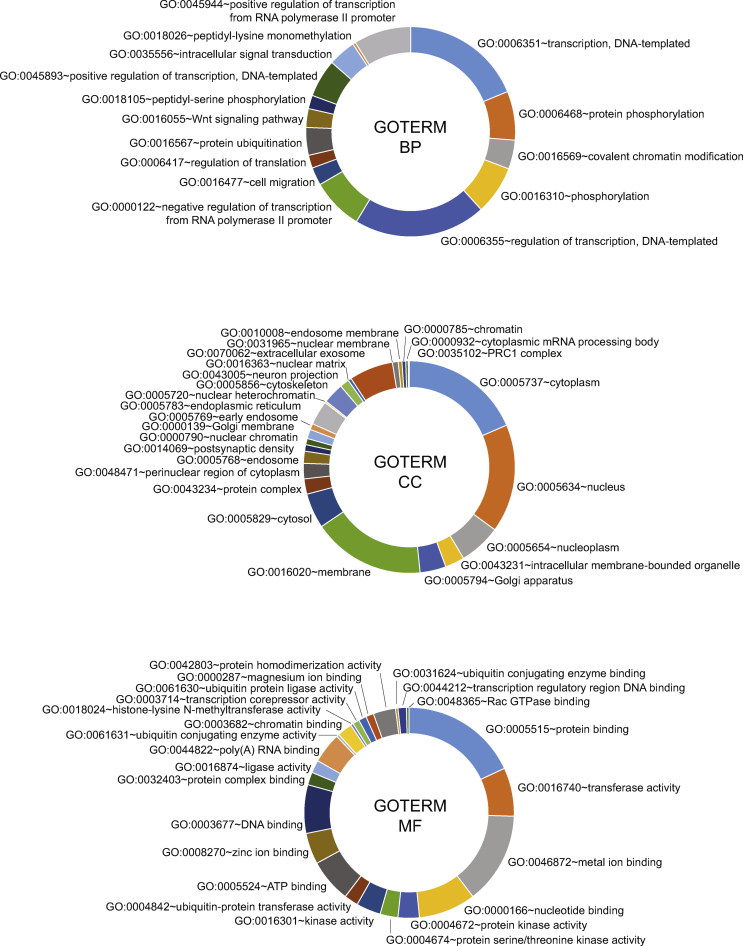
After the additional sorting of sites displaying significant differences in binding on the basis of fold changes in enrichment, read numbers, and site localization (see the methods section for details), we retained 262 genes for further analysis. We analyzed pathway enrichment with the Database for Annotation, Visualization, and Integrated Discovery (DAVID v6.8) [28], which identified genes involved in transcriptional regulation, particularly in relation to polymerase II promoters, chromatin modifications, and protein phosphorylation (Supplementary Figure S6, top panel). These gene products were evenly distributed between the three cellular compartments (cytoplasm, nucleus, and membrane) (Supplementary Figure S6, middle panel). A particular enrichment in genes encoding proteins with transferase activity and nucleotide (DNA/RNA) binding capacity was observed. For these proteins, we observed an enrichment in several gene ontology (GO) terms relating to epigenetics (nucleotide binding, poly(A) RNA binding, DNA binding, chromatin binding, and histone-lysine N-methyltransferase activity) and signaling pathways (protein kinase activity, kinase activity, and protein serine/threonine kinase activity) for biological processes (Supplementary Figure S6, bottom panel), confirming the involvement of epigenetic modifications in the programming of adult metabolism by the perinatal nutritional environment.
Promoters were particularly enriched in the H3K4me3 histone mark targeted by ChIPSeq. We therefore checked for any potential overrepresentation of transcription factor binding sites. A particular enrichment in the EGR1 (24 times, p < 10−4), EGR1-pwm (25 times, p < 0.05), and Zic3 (15 times, p < 0.01) sites was observed in the total datasets of the IUGR-N and control groups. Zic 3 is best known for its role in the initial establishment of left-right symmetry during early development, and we thus did not further pursue studies of its role in metabolic alterations during adulthood associated with IUGR. EGR1, also known as NGF-1a, is a transcription factor that is strongly expressed in the liver. It is expressed with circadian rhythms and plays a crucial role in regulating liver metabolism. It has also been implicated in cell cycle regulation and fibrogenesis via the TNF-α and IL6 pathways [45,46]. However, a large proportion of the known EGR1 target genes were not found in our ChIPSeq dataset and did not display differential enrichment between the IUGR-N and control groups at the thresholds applied. Moreover, Sirius red staining was not consistent with higher levels of liver fibrosis in the 12-month-old IUGR mice than in the age-matched control mice (data not shown). Taken together, these data suggest that, despite its crucial role in the liver, EGR1 does not play a key role in metabolic dysregulation associated with IUGR.
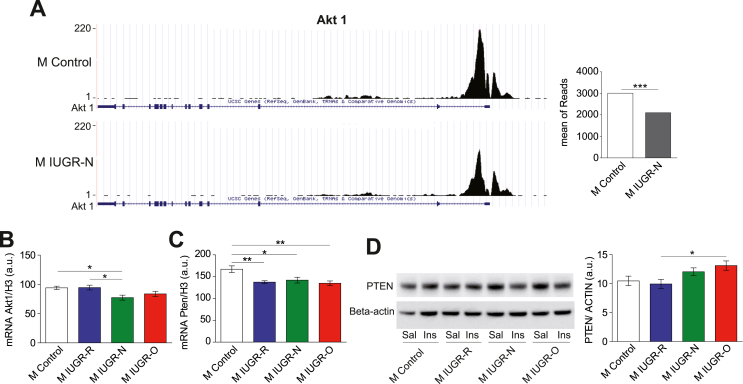
We then used the STRING database to identify potential interactions (https://string-db.org). Interactions for genes with significantly differentially enriched H3K4me3 histone marks, with a >1.5-fold difference between the IUGR-N and control groups and filtered with a high confidence setting, are illustrated. There were significantly more interactions than expected (enrichment in protein–protein interactions: p = 3.31 × 10−6) involving many genes associated with cancer biology, axon guidance, and metabolism according to STRING's Reactome and KEGG pathways (Figure 3C). Interestingly, the Akt1 gene was identified as the main network's principal node. Among GO terms related to signaling pathways for biological processes (Supplementary Figure S6), the Akt1 gene was associated with eight different GO terms for molecular functions and the detection of 12 different Mapk genes. The implication of the PI3K-AKT and MAPK signaling pathways was confirmed by the KEGG pathway analysis (Benjamini–Hochberg test, p = 0.003). Given the role of AKT in the development of insulin resistance and the alterations detected in both the IUGR-O and IUGR-N mice, we further explored the molecular alterations of this gene. The H3K4me3 profile around the Akt1 promoter was significantly weaker in the young adult (3 months old) IUGR-N mice than in the age-matched control animals (Figure 4A). The lower levels of Akt1 mRNA in the liver were confirmed in the IUGR-N mice (Figure 4B). A similar tendency was observed in the IUGR-O mice, whereas the IUGR-R mice presented no difference relative to the controls. The significantly lower (p < 0.01) AKT levels in the livers of the 3-month-old IUGR-N and IUGR-O mice was then confirmed by protein analysis (Figure 2E, right panel), which suggested that changes in the total AKT levels in the young IUGR-N mice without visible diseases (with a normal ITT response; see Figure 2B, left panel) might facilitate the subsequent development of insulin resistance before any change in ceramide levels, as in the IUGR-O mice. Moreover, this change in AKT synthesis might further decrease the total amount of phosphorylated AKT produced in response to insulin, thereby aggravating the decrease in activation capacity observed in the insulin signaling pathway (see Figure 2E, middle panel), leading to an increase in the susceptibility of the IUGR-N and IUGR-O mice to the development of insulin resistance with age. Nevertheless, the changes in AKT phosphorylation in the IUGR-N and IUGR-O mice may not have been due to changes in the total AKT levels alone. Indeed, such changes were also observed when the pAKT levels were normalized against the total AKT levels (the pAKT/AKT ratio). Thus, other mechanisms directly affecting AKT phosphorylation may also have been involved.
Figure 4.
Altered insulin signaling pathway in the livers of male young adult IUGR mice. (A) Representative ChIPSeq data for the H3K4me3 peak at the Akt1 gene in the control and IUGR-N mice. The mean number of reads was lower in the male IUGR-N mice than in the controls (n = 5–6 per group) and (B) this result was confirmed by the RNA analysis (n = 6–8 per group). (C) The levels of Pten mRNA were lower in all of the IUGR groups irrespective of the nutritional conditions during the lactation period (n = 5–8 per group). (D) Representative immunoblots for PTEN and β-actin in the livers of the mice after the injection of saline (Sal) or insulin (Ins) (left panel). The hepatic PTEN content was higher in the IUGR-O mice than in the IUGR-R mice (n = 7–8 per group) after saline injection. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 between groups.
3.6. Changes in AKT activation are associated with changes in PTEN levels through miRNA regulation
We then further investigated the molecular mechanisms underlying early changes in AKT phosphorylation and studied changes in the levels of PTEN, one of its principal inhibitors. Interestingly, the Pten mRNA levels were slightly lower in the three groups of adult IUGR mice than in the controls, regardless of their post-natal diets (Figure 4C). However, the PTEN protein levels were higher in the IUGR-O mice (Figure 4D). Given that PTEN is a well-known inhibitor of AKT phosphorylation, the higher levels of this protein may partly explain the early onset of molecular disturbances of AKT phosphorylation. The apparent discrepancy between the PTEN mRNA and protein levels suggested post-transcriptional regulatory mechanisms and, potentially, the involvement of microRNAs (miRNAs).
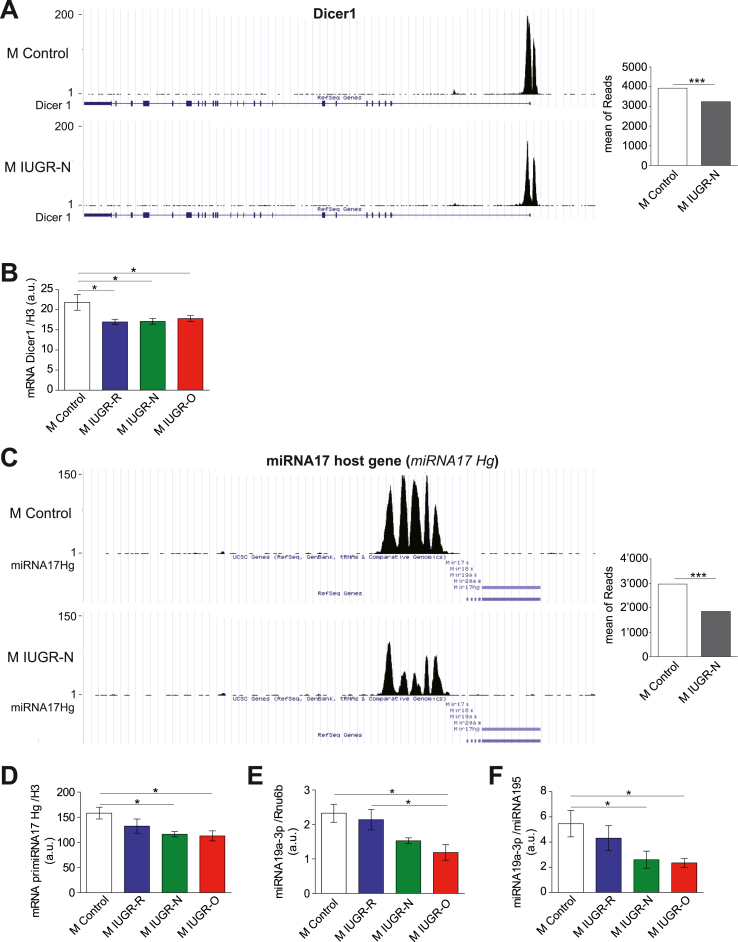
miRNAs are involved in multiple developmental processes and various diseases. They are small (20–25 bp) non-coding RNAs that modulate the expression of various proteins via post-transcriptional inhibition following association with the Ago2 protein. They are synthesized as a pri-miRNA from long non-coding RNAs and processed into mature miRNAs by Drosha, DGCR8, and Dicer [47,48]. Consistent with a possible role of miRNAs in PTEN deregulation in IUGR mice, the DAVID analysis of ChIPSeq experiments indicated that Dicer was highlighted in five different GO molecular function terms, together with Dgcr8 (twice) and Ago2 (three times). Accordingly, enrichment in the positive histone mark H3K4me3 was significantly weaker in the proximal promoter of Dicer in the 3-month-old IUGR-N mice (Figure 5A). An analysis of the liver Dicer1 gene expression in the young adult IUGR mice indicated that the mRNA levels of this gene were lower in all three IUGR groups than in the controls, regardless of diet during lactation (Figure 5B).
Figure 5.
Plasma levels and liver contents of miRNA19a-3p are lower in male young adult IUGR mice. (A) Representative ChIPSeq data for the H3K4me3 peak at the Dicer1 gene in the control and IUGR-N mice. The mean number of reads was lower in the male IUGR-N mice than in the control mice (n = 5–6 per group). (B) The Dicer1 mRNA levels were lower in all of the 3-month-old male mice with IUGR irrespective of their nutritional conditions during the lactation period than in the control animals (n = 7 per group). (C) Representative ChIPSeq data for the H3K4me3 peak in the miRNA 17 host gene (miRNA17Hg) in the control and IUGR-N mice. The mean number of reads was lower in the male IUGR-N mice than in the control mice (n = 5–6 per group). (D) The levels of primiRNA17Hg non-coding RNA (also called primiRNA17-92) were significantly lower in the livers of the young adult (3 months old) IUGR-N and IUGR-O mice than in the control mice (n = 5–8 per group). (E) The levels of mature miRNA19a-3p were lower in the livers of the male IUGR-O mice than in the control and IUGR-R mice (n = 5–8 per group). (F) The circulating miRNA19a-3p levels were lower in the plasma of the 3-month-old male IUGR-N and IUGR-O mice than in the control animals (n = 7–8 per group). ∗p < 0.05 and ∗∗p < 0.001 between groups.
Several miRNAs could potentially regulate Pten expression, including miRNA19a-3p. This miRNA species is produced, along with four others, by processing the same pri-miRNA from the miRNA17 host gene (miRNA17hg and ENSMUSG00000089726, also called primiRNA17-92). This pri-miRNA belongs to the intergenic lincRNA class and is transcribed and regulated similar to classic protein-coding genes [49,50]. Interestingly, the ChIPSeq analysis highlighted miRNA17hg as a gene displaying significantly (adjusted p = 1.5 × 10−7) lower enrichment for the histone mark H3K4me3 in the IUGR-N mice than in the controls (Figure 5C). The lower levels of pri-miRNA suggested by the ChIPSeq analysis were confirmed by mRNA quantification in the livers of the 3-month-old IUGR-N and IUGR-O mice by RT-qPCR relative to the controls (Figure 5D). Similar results were obtained for mature miRNA19a-3p levels in the liver, which tended to be lower in the IUGR-N group and were significantly lower in the IUGR-O mice (Figure 5E). Consistent with our data, a potential role for miRNAs in liver diseases has also been suggested by studies using the Dicer knockout mouse model [51]. Specifically, miRNA19a-3p was first associated with the regulation of PTEN in stem cell reprogramming and was reported to inhibit PTEN expression in bladder cancer [52,53]. Furthermore, the role of miRNA19a-3p in PTEN regulation was recently confirmed in hepatocytes, particularly during the regulation of gluconeogenesis and glycogen synthesis [[54], [55], [56]].
As miRNAs have recently been shown to be circulating molecules [57,58], we quantified the plasma miRNA19a-3p levels in the 3-month-old mice to determine whether they correlated with early onset molecular alterations. The 3-month-old IUGR-O male mice had circulating levels of miRNA19a-3p 2.5-fold lower than those in the control animals (Figure 5F), demonstrating the feasibility of determining the levels of this circulating miRNA in the blood and suggesting that it might be associated with ongoing disease. The 3-month-old male IUGR-N mice also had significantly lower circulating levels of miRNA19a-3p despite an absence of any clear glucose intolerance or insulin resistance (Figure 2A,B) and an absence of changes in the DAG and ceramide levels (Figure 2D). These data suggest that miRNA19a-3p is a potential early biomarker of the deregulation of glucose homeostasis.
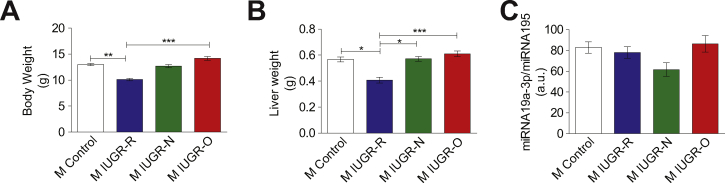
We investigated this possible role further by determining the circulating miRNA19a-3p levels in the prepubertal 20-day-old male mice, a few days after the end of the lactation period. At this age, the male IUGR-N and IUGR-O mice had a weight similar to that of the male control mice (Supplementary Figure S7A) and a similar liver weight (Supplementary Figure S7B). The changes in circulating miRNA19a-3p levels in the IUGR-N and IUGR-O mice did not differ significantly from those in the control mice at this age (Supplementary Figure S7C). Thus, changes in circulating miRNA19a-3p levels may be useful early markers that precede disturbances in glucose homeostasis, but they may not be of predictive value for liver disease development.
Globally, in our mouse model based on the modulation of nutrition during the early post-natal period, we were able to modify the susceptibility of the mice born with intrauterine growth retardation to cardiometabolic diseases later in life. This susceptibility displayed strong sexual dimorphism. Such dimorphism has been previously reported [59,60], and our data suggest that it may originate from the fetal period. Susceptibility to the development of cardiometabolic diseases in adulthood has been shown to be closely related to the developmental period (conception, first, second, and third trimester), the severity of the insult, and how it is sensed by the fetus [4]. Moreover, the higher susceptibility of the adult males in our mouse model is consistent with the observed higher susceptibility of male fetuses than female fetuses to growth retardation in response to a maternal low-protein diet (Figure 1A). The male and female fetuses studied herein were inbred and shared the same uterine horns and maternal environment, which suggests that placental function may play a crucial role, consistent with numerous studies highlighting the sexual dimorphism of placental function, including its capacity to adapt to nutritional changes [59,61]. High-throughput approaches have revealed epigenetic deregulations involving changes in DNA methylation and the levels of miRNAs associated with gene expression modifications in mouse fetuses displaying IUGR [62]. This previous study, reporting a number of differentially methylated regions similar to that described herein, suggested that miRNA plays a crucial role in placental development and function, influencing fetal growth.
The placenta expresses a large number of miRNAs, for many of which alterations have been reported in cases of fetal growth restriction, suggesting a potential role in this context. Consistent with the potential excretion/secretion of miRNA into the bloodstream, numerous placenta-specific miRNAs have been observed in the blood of pregnant mothers [63]. However, most of these studies found no significant differences in placenta-specific miRNA levels between the blood of pregnant women carrying fetuses displaying in utero growth restriction and blood from pregnant women without complications [64]. Increasing evidence suggests that epigenetic regulation is important in the placenta, but the nature of this regulation, which is affected by all environmental factors (for example, maternal nutrition, toxins, endocrine disruptors, drugs, parental age, and health), may account for conflicting results between studies. Further studies in large cohorts are required to identify the important miRNAs. The detection, by miRSeq, of changes in miRNA levels in the bloodstream during childhood may provide the best opportunity to identify predictive biomarkers, because the nutritional environment during the early post-natal period (for example, the suckling period) also appears to be crucial.
The correction of nutrition to “normal” levels during lactation (6 pups/dam) in the pups born with IUGR was associated with the same cardiometabolic diseases as overfeeding. This suggests that any increase in the food supply during lactation, after a restriction, may be sensed as overfeeding, thereby increasing susceptibility to cardiometabolic diseases, but the absolute amount of food may also be important, as these effects occurred later in the IUGR-N mice than in the IUGR-O mice. This delay in the IUGR-N mice enabled to detect molecular modifications before the appearance of obvious physiological alterations, concerning glucose homeostasis in particular. These modifications, which appeared before changes to the DAG and ceramide levels, may be associated with epigenetic changes in the liver, highlighting the crucial role of this particular mechanism. The ChIPSeq dataset highlighted a few hundred genes displaying differential enrichment in the H3K4me3 histone mark between the young adult control and male IUGR-N mice due to the stringency of the post-bioinformatic analysis. This analysis revealed alterations in the expression of genes involved in various pathways in cancer, coagulation, axon guidance, metabolism, and the insulin signaling pathway as suggested by the STRING database. The potential influence of perinatal nutrition in cancers was illustrated, in particular, by the Reactome pathway, suggesting that challenges in mice born with IUGR could lead to differences in sensitivity to liver cancer development.
We focused on modifications associated with metabolic alterations. The STRING network centered on the Akt gene suggested an important role for this gene, and, more globally, for early changes to the insulin signaling pathway in the pre-symptomatic 3-month-old IUGR-N mice. Consistent with this finding, we observed highly coherent modifications affecting various levels of this pathway (Figure 6). The STRING network also suggested a potential involvement of the Clock genes system, notably Clock and Rev1, which may be regulated by EGR1 [46]. In addition to indications of the early development of insulin resistance, the livers of the IUGR-N mice also presented signs of early onset steatosis. However, our dataset did not indicate any significant differential enrichment in the H3K4me3 histone mark on the promoter of peroxisome proliferator-activated receptor (PPAR) genes in the young adult IUGR-N mice relative to the controls. The alpha, beta, delta, and gamma 1,3-PPAR genes are expressed in the liver and encode proteins acting as lipid sensors in metabolism [65,66]. PPAR gamma has also been implicated in the M1 to M2 transition of macrophages in response to fatty acids [67], thereby affecting hepatocyte metabolism by modulating local inflammatory processes. Moreover, the potential regulation of PPAR gamma expression at the DNA levels (through methylation) and the direct role of this regulation in the placenta in fetal growth restriction both suggest a crucial role in the regulation of metabolism after IUGR [68,69]. However, although both the Ppara and Ppargc1b genes were identified in the raw dataset, both these genes were filtered out as they did not satisfy the applied threshold. Thus, despite the essential role of PPARs in liver metabolism, we were unable to associate changes in their expression with IUGR in our mouse model. Multiple high-throughput approaches combining RNASeq, MeDIPSeq, and various ChIPSeqs and MIRSeq techniques during fetal and post-natal development would be required to reveal the precise molecular mechanisms underlying these first alterations inducing the development of metabolic diseases during adulthood following IUGR. We performed a ChIPSeq analysis of the H3K4me3 histone mark, the deposition of which in chromatin is a result of transcription activity. Unlike other technical approaches, including ATACSeq, the method used herein did not allow an analysis of genome-wide chromatin accessibility or the mapping transcription factor binding sites. Further studies of this regulatory aspect are required to determine the underlying regulatory mechanisms and understand the pathophysiology of early onset programmed metabolic diseases.
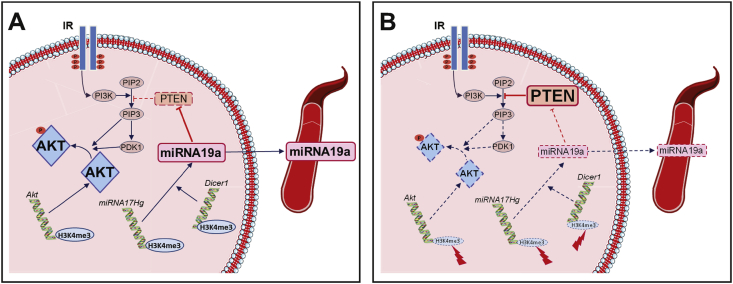
Figure 6.
Graphical summary. Relative to the control conditions (A), perinatal changes in nutrition in mice induce multiple and coherent epigenetic alterations in hepatocytes (B), notably affecting key genes involved in insulin signaling: lower amounts of AKT are synthesized, potentially leading to a predisposition to developing insulin resistance with age. This effect is aggravated by complementary epigenetic alterations to the primiRNA17Hg and Dicer1 genes, leading to a decrease in miRNA19a-3p levels. The decrease in miRNA19a-3p leads to an increase in the translation of PTEN, a well-known inhibitor of AKT activation, and may also contribute to a predisposition to developing insulin resistance with age. The change in miRNA19a-3p levels in the asymptomatic adult mice with IUGR was also detectable in the blood, making it possible to envisage using this miRNA as an early biomarker of a higher risk of developing insulin resistance.
The data reported herein indicate the coherent additional implication of miRNAs in metabolic programming, particularly for miRNA19a-3p (see graphical abstract in Figure 6). Interestingly, in addition to its role in hepatocytes, miRNA19a-3p displays changes in circulating levels in the bloodstream, although the tissue origin is unclear. It is now widely accepted that miRNAs can be found in all biological fluids, including the blood, despite uncertainties about their role in these compartments [70]. miRNAs are good candidates for use as biomarkers because they are highly resistant to degradation and their levels are stable and reproducible in serum and consistent between individuals of the same species [57,58]. They were first used as potential biomarkers for patient stratification regarding the potential benefit of various adjuvant therapies for certain cancers [71,72]. Consistent with such use, kits for their determination are now commercially available. Circulating miRNAs are beginning to be used as biomarkers for metabolic alterations [73]. Interestingly, precocious miRNA biomarkers of non-alcoholic fatty liver disease (NAFLD) development have recently been proposed as a means of enhancing the predictability of non-alcoholic steatohepatitis in combination with clinical variables [74]. In line with the DOHaD hypothesis, the fetal programming of cardiometabolic diseases may provide opportunities to assess circulating precocious biomarkers for the risk of developing particular diseases in later life, as shown herein for miRNA19a-3p and insulin resistance. Consistent with this, variations in the amounts of miRNA19a-3p circulating in the bloodstream were shown herein to precede the increase in hepatic DAG and ceramide contents known to lead to insulin resistance. Many additional studies are required to confirm the utility of this miRNA as a tool and its advantages for use in humans in both prospective and adult patient cohorts. In prospective cohorts, MIRSeq determinations in childhood are of particular importance, as this period has been shown to be crucial in strategies designed to promote a healthy life expectancy. Life-course studies suggest that interventions in early life, when biological systems are more adaptive, are likely to have sustained effects on health, particularly because they can influence responses to lifestyle factors in later life [2].
Data availability
The datasets generated during and/or analyzed in this study are available from the figshare website repository (https://figshare.com) (DOI under embargo: 10.6084/m9.figshare.8015906; private link for reviewers: https://doi.org/10.6084/m9.figshare.8015906). The ChIPSeq data supporting this study's findings were deposited in the NCBI's Sequence Read Archive with the BioProject ID: PRJNA533940.
Authors’ contributions
S.S, R.C., L.D., C.G., M.C., P.L., M.M., Y.L.B., and L.K. designed and conducted the physiological, molecular, histological, and biochemical experiments. R.C., M.L.E., C.P., B.M.O., D.T., and L.K. designed, performed, and analyzed the ChIPSeq experiments. L.M. and M.L. conducted and analyzed the histological kidney studies. S.S., L.D., C.G., L.D., and L.K. designed and produced the experimental cohorts. M.L., D.T., M.M., B.F., Y.L.B., and L.K. designed the study. S.S., C.G., M.M., B.F., Y.L.B., and L.K. wrote the manuscript. All of the authors revised, commented on, and approved the manuscript.
Financial disclosure
This study was supported by grants from Inserm (L.K. and Y.L.B.), Sorbonne Université (L.K. and Y.L.B.), IHU ICAN ANR-10-IAHU-05 (L.K.), the Premup Foundation (L.K., L.M., M.L, and Y.L.B.), the Fondation pour la Recherche Médicale FRM (Y.L.B.), and an SFD-MSD research grant (L.K.). S.S. holds fellowships from Premup/Sorbonne Université and SFEDP-Sandoz.
Acknowledgments
The authors thank T. Ledent and the staff at the SPF experimental animal housing platform (PHEA) at the Saint Antoine Research Center for animal care, S. Placier and C. Chatziantoniou from the hemodynamic platform at Tenon Hospital for help during blood pressure measurements, the P3S platform staff at the UMS 29 OMICS for ChIP sequencing, and Alex Edelman and Associates for English corrections.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101083.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
figs6.
figs7.
References
- 1.WHO Obesity and overweight. 2018. http://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight 16 February 2018.
- 2.WHO . 2016. Consideration of the evidence on childhood obesity for the Commission on Ending Childhood Obesity: report of the ad hoc working group on science and evidence for ending childhood obesity. [Google Scholar]
- 3.Hales C.N., Barker D.J. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia. 1992;35(7):595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- 4.McMillen I.C., Robinson J.S. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiological Reviews. 2005;85(2):571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- 5.Roseboom T., de Rooij S., Painter R. The Dutch famine and its long-term consequences for adult health. Early Human Development. 2006;82(8):485–491. doi: 10.1016/j.earlhumdev.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Hanson M.A., Gluckman P.D. Early developmental conditioning of later health and disease: physiology or pathophysiology? Physiological Reviews. 2014;94(4):1027–1076. doi: 10.1152/physrev.00029.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaquet D., Gaboriau A., Czernichow P., Levy-Marchal C. Insulin resistance early in adulthood in subjects born with intrauterine growth retardation. Journal of Clinical Endocrinology & Metabolism. 2000;85(4):1401–1406. doi: 10.1210/jcem.85.4.6544. [DOI] [PubMed] [Google Scholar]
- 8.Karlberg J., Albertsson-Wikland K. Growth in full-term small-for-gestational-age infants: from birth to final height. Pediatric Research. 1995;38(5):733–739. doi: 10.1203/00006450-199511000-00017. [DOI] [PubMed] [Google Scholar]
- 9.Karlberg J., Lawrence C., Albertsson-Wikland K. Prediction of final height in short, normal and tall children. Acta Paediatrica - Supplement. 1994;406:3–9. doi: 10.1111/j.1651-2227.1994.tb13411.x. discussion 10. [DOI] [PubMed] [Google Scholar]
- 10.Albertsson-Wikland K., Karlberg J. Postnatal growth of children born small for gestational age. Acta Paediatrica - Supplement. 1997;423:193–195. doi: 10.1111/j.1651-2227.1997.tb18413.x. [DOI] [PubMed] [Google Scholar]
- 11.Hochberg Z., Feil R., Constancia M., Fraga M., Junien C., Carel J.C. Child health, developmental plasticity, and epigenetic programming. Endocrine Reviews. 2011;32(2):159–224. doi: 10.1210/er.2009-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ong K.K., Ahmed M.L., Emmett P.M., Preece M.A., Dunger D.B. Association between postnatal catch-up growth and obesity in childhood: prospective cohort study. BMJ. 2000;320(7240):967–971. doi: 10.1136/bmj.320.7240.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romo A., Carceller R., Tobajas J. Intrauterine growth retardation (IUGR): epidemiology and etiology. Pediatric Endocrinology Reviews. 2009;6(Suppl 3):332–336. [PubMed] [Google Scholar]
- 14.Ananth C.V., Balasubramanian B., Demissie K., Kinzler W.L. Small-for-gestational-age births in the United States: an age-period-cohort analysis. Epidemiology. 2004;15(1):28–35. doi: 10.1097/01.ede.0000100288.37475.19. [DOI] [PubMed] [Google Scholar]
- 15.Black R.E. Global prevalence of small for gestational age births. Nestle Nutrition Institute of Workshop Series. 2015;81:1–7. doi: 10.1159/000365790. [DOI] [PubMed] [Google Scholar]
- 16.Katz J., Wu L.A., Mullany L.C., Coles C.L., Lee A.C., Kozuki N. Prevalence of small-for-gestational-age and its mortality risk varies by choice of birth-weight-for-gestation reference population. PloS One. 2014;9(3) doi: 10.1371/journal.pone.0092074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawn J.E., Blencowe H., Oza S., You D., Lee A.C., Waiswa P. Every Newborn: progress, priorities, and potential beyond survival. Lancet. 2014;384(9938):189–205. doi: 10.1016/S0140-6736(14)60496-7. [DOI] [PubMed] [Google Scholar]
- 18.Liu L., Johnson H.L., Cousens S., Perin J., Scott S., Lawn J.E. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet. 2012;379(9832):2151–2161. doi: 10.1016/S0140-6736(12)60560-1. [DOI] [PubMed] [Google Scholar]
- 19.Gatford K.L., Simmons R.A., De Blasio M.J., Robinson J.S., Owens J.A. Review: placental programming of postnatal diabetes and impaired insulin action after IUGR. Placenta. 2010;31(Suppl):S60–S65. doi: 10.1016/j.placenta.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiorotto M.L., Burrin D.G., Perez M., Reeds P.J. Intake and use of milk nutrients by rat pups suckled in small, medium, or large litters. American Journal of Physiology. 1991;260(6 Pt 2):R1104–R1113. doi: 10.1152/ajpregu.1991.260.6.R1104. [DOI] [PubMed] [Google Scholar]
- 21.Kappeler L., De Magalhaes Filho C., Leneuve P., Xu J., Brunel N., Chatziantoniou C. Early postnatal nutrition determines somatotropic function in mice. Endocrinology. 2009;150(1):314–323. doi: 10.1210/en.2008-0981. [DOI] [PubMed] [Google Scholar]
- 22.Decourtye L., Mire E., Clemessy M., Heurtier V., Ledent T., Robinson I.C. IGF-1 induces GHRH neuronal axon elongation during early postnatal life in mice. PloS One. 2017;12(1) doi: 10.1371/journal.pone.0170083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matyash V., Liebisch G., Kurzchalia T.V., Shevchenko A., Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. The Journal of Lipid Research. 2008;49(5):1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaner R.L., Allegood J.C., Park H., Wang E., Kelly S., Haynes C.A. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. The Journal of Lipid Research. 2009;50(8):1692–1707. doi: 10.1194/jlr.D800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sullards M.C., Liu Y., Chen Y., Merrill A.H., Jr. Analysis of mammalian sphingolipids by liquid chromatography tandem mass spectrometry (LC-MS/MS) and tissue imaging mass spectrometry (TIMS) Biochimica et Biophysica Acta. 2011;1811(11):838–853. doi: 10.1016/j.bbalip.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leiker T.J., Barkley R.M., Murphy R.C. Analysis of diacylglycerol molecular species in cellular lipid extracts by normal-phase LC-electrospray mass spectrometry. International Journal of Mass Spectrometry. 2011;305(2–3):103–109. doi: 10.1016/j.ijms.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dahl J.A., Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nature Protocols. 2008;3(6):1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- 28.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Dahri S., Snoeck A., Reusens-Billen B., Remacle C., Hoet J.J. Islet function in offspring of mothers on low-protein diet during gestation. Diabetes. 1991;40(Suppl 2):115–120. doi: 10.2337/diab.40.2.s115. [DOI] [PubMed] [Google Scholar]
- 30.Langley-Evans S.C. Critical differences between two low protein diet protocols in the programming of hypertension in the rat. International Journal of Food Sciences & Nutrition. 2000;51(1):11–17. doi: 10.1080/096374800100859. [DOI] [PubMed] [Google Scholar]
- 31.Vehaskari V.M., Aviles D.H., Manning J. Prenatal programming of adult hypertension in the rat. Kidney International. 2001;59(1):238–245. doi: 10.1046/j.1523-1755.2001.00484.x. [DOI] [PubMed] [Google Scholar]
- 32.Bateson P., Barker D., Clutton-Brock T., Deb D., D'Udine B., Foley R.A. Developmental plasticity and human health. Nature. 2004;430(6998):419–421. doi: 10.1038/nature02725. [DOI] [PubMed] [Google Scholar]
- 33.Gluckman P.D., Hanson M.A. The developmental origins of the metabolic syndrome. Trends in Endocrinology and Metabolism. 2004;15(4):183–187. doi: 10.1016/j.tem.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Nugent B.M., O'Donnell C.M., Epperson C.N., Bale T.L. Placental H3K27me3 establishes female resilience to prenatal insults. Nature Communications. 2018;9(1):2555. doi: 10.1038/s41467-018-04992-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugden M.C., Holness M.J. Gender-specific programming of insulin secretion and action. Journal of Endocrinology. 2002;175(3):757–767. doi: 10.1677/joe.0.1750757. [DOI] [PubMed] [Google Scholar]
- 36.Zambrano E., Bautista C.J., Deas M., Martinez-Samayoa P.M., Gonzalez-Zamorano M., Ledesma H. A low maternal protein diet during pregnancy and lactation has sex- and window of exposure-specific effects on offspring growth and food intake, glucose metabolism and serum leptin in the rat. Journal of Physiology. 2006;571(Pt 1):221–230. doi: 10.1113/jphysiol.2005.100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lucas A., Baker B.A., Desai M., Hales C.N. Nutrition in pregnant or lactating rats programs lipid metabolism in the offspring. British Journal of Nutrition. 1996;76(4):605–612. doi: 10.1079/bjn19960066. [DOI] [PubMed] [Google Scholar]
- 38.Manning J., Vehaskari V.M. Low birth weight-associated adult hypertension in the rat. Pediatric Nephrology. 2001;16(5):417–422. doi: 10.1007/s004670000560. [DOI] [PubMed] [Google Scholar]
- 39.Piccolis M., Bond L.M., Kampmann M., Pulimeno P., Chitraju C., Jayson C.B.K. Probing the global cellular responses to lipotoxicity caused by saturated fatty acids. Molecular Cell. 2019;74(1):32–44. doi: 10.1016/j.molcel.2019.01.036. e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roden M. Does endurance training protect from lipotoxicity? Diabetes. 2012;61(10):2397–2399. doi: 10.2337/db12-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Rivera C.M., Ren B. Mapping human epigenomes. Cell. 2013;155(1):39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang X., Gao X., Li W., Jiang S., Li R., Hong H. Stable H3K4me3 is associated with transcription initiation during early embryo development. Bioinformatics. 2019;35(20):3931–3936. doi: 10.1093/bioinformatics/btz173. [DOI] [PubMed] [Google Scholar]
- 44.Howe F.S., Fischl H., Murray S.C., Mellor J. Is H3K4me3 instructive for transcription activation? BioEssays. 2017;39(1):1–12. doi: 10.1002/bies.201600095. [DOI] [PubMed] [Google Scholar]
- 45.Magee N., Zhang Y. Role of early growth response 1 in liver metabolism and liver cancer. Hepatoma Research. 2017;3:268–277. doi: 10.20517/2394-5079.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tao W., Wu J., Zhang Q., Lai S.S., Jiang S., Jiang C. EGR1 regulates hepatic clock gene amplitude by activating Per1 transcription. Scientific Reports. 2015;5:15212. doi: 10.1038/srep15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hutvagner G., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 48.Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 49.Saini H.K., Enright A.J., Griffiths-Jones S. Annotation of mammalian primary microRNAs. BMC Genomics. 2008;9:564. doi: 10.1186/1471-2164-9-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozsolak F., Poling L.L., Wang Z., Liu H., Liu X.S., Roeder R.G. Chromatin structure analyses identify miRNA promoters. Genes & Development. 2008;22(22):3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mori M.A., Ludwig R.G., Garcia-Martin R., Brandao B.B., Kahn C.R. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metabolism. 2019;30(4):656–673. doi: 10.1016/j.cmet.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He X., Cao Y., Wang L., Han Y., Zhong X., Zhou G. Human fibroblast reprogramming to pluripotent stem cells regulated by the miR19a/b-PTEN axis. PloS One. 2014;9(4) doi: 10.1371/journal.pone.0095213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng Y., Liu J., Kang Y., He Y., Liang B., Yang P. miR-19a acts as an oncogenic microRNA and is up-regulated in bladder cancer. Journal of Experimental & Clinical Cancer Research. 2014;33:67. doi: 10.1186/s13046-014-0067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qian Y.Y., Liu Z.S., Zhang Z., Levenson A.S., Li K. Pterostilbene increases PTEN expression through the targeted downregulation of microRNA-19a in hepatocellular carcinoma. Molecular Medicine Reports. 2018;17(4):5193–5201. doi: 10.3892/mmr.2018.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dou L., Meng X., Sui X., Wang S., Shen T., Huang X. MiR-19a regulates PTEN expression to mediate glycogen synthesis in hepatocytes. Scientific Reports. 2015;5:11602. doi: 10.1038/srep11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dou L., Wang S., Huang X., Sun X., Zhang Y., Shen T. MiR-19a mediates gluconeogenesis by targeting PTEN in hepatocytes. Molecular Medicine Reports. 2018;17(3):3967–3971. doi: 10.3892/mmr.2017.8312. [DOI] [PubMed] [Google Scholar]
- 57.Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Research. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 58.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aiken C.E., Ozanne S.E. Sex differences in developmental programming models. Reproduction. 2013;145(1):R1–R13. doi: 10.1530/REP-11-0489. [DOI] [PubMed] [Google Scholar]
- 60.Gabory A., Attig L., Junien C. Developmental programming and epigenetics. American Journal of Clinical Nutrition. 2011;94(6 Suppl):1943S–1952S. doi: 10.3945/ajcn.110.000927. [DOI] [PubMed] [Google Scholar]
- 61.Tarrade A., Panchenko P., Junien C., Gabory A. Placental contribution to nutritional programming of health and diseases: epigenetics and sexual dimorphism. Journal of Experimental Biology. 2015;218(Pt 1):50–58. doi: 10.1242/jeb.110320. [DOI] [PubMed] [Google Scholar]
- 62.Chen P.Y., Ganguly A., Rubbi L., Orozco L.D., Morselli M., Ashraf D. Intrauterine calorie restriction affects placental DNA methylation and gene expression. Physiological Genomics. 2013;45(14):565–576. doi: 10.1152/physiolgenomics.00034.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Higashijima A., Miura K., Mishima H., Kinoshita A., Jo O., Abe S. Characterization of placenta-specific microRNAs in fetal growth restriction pregnancy. Prenatal Diagnosis. 2013;33(3):214–222. doi: 10.1002/pd.4045. [DOI] [PubMed] [Google Scholar]
- 64.Chiofalo B., Lagana A.S., Vaiarelli A., La Rosa V.L., Rossetti D., Palmara V. Do miRNAs play a role in fetal growth restriction? A fresh look to a busy corner. BioMed Research International. 2017;2017:6073167. doi: 10.1155/2017/6073167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vitale S.G., Lagana A.S., Nigro A., La Rosa V.L., Rossetti P., Rapisarda A.M. Peroxisome proliferator-activated receptor modulation during metabolic diseases and cancers: master and minions. PPAR Research. 2016;2016:6517313. doi: 10.1155/2016/6517313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berger J.P., Akiyama T.E., Meinke P.T. PPARs: therapeutic targets for metabolic disease. Trends in Pharmacological Sciences. 2005;26(5):244–251. doi: 10.1016/j.tips.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 67.Wu H.M., Ni X.X., Xu Q.Y., Wang Q., Li X.Y., Hua J. Regulation of lipid-induced macrophage polarization through modulating peroxisome proliferator-activated receptor-gamma activity affects hepatic lipid metabolism via a Toll-like receptor 4/NF-kappaB signaling pathway. Journal of Gastroenterology and Hepatology. 2020 doi: 10.1111/jgh.15025. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 68.Chen Z., He P., Ding X., Huang Y., Gu H., Ni X. PPARgamma stimulates expression of L-type amino acid and taurine transporters in human placentas: the evidence of PPARgamma regulating fetal growth. Scientific Reports. 2015;5:12650. doi: 10.1038/srep12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lendvai A., Deutsch M.J., Plosch T., Ensenauer R. The peroxisome proliferator-activated receptors under epigenetic control in placental metabolism and fetal development. American Journal of Physiology. Endocrinology and Metabolism. 2016;310(10):E797–E810. doi: 10.1152/ajpendo.00372.2015. [DOI] [PubMed] [Google Scholar]
- 70.Weber J.A., Baxter D.H., Zhang S., Huang D.Y., Huang K.H., Lee M.J. The microRNA spectrum in 12 body fluids. Clinical Chemistry. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ranade A.R., Cherba D., Sridhar S., Richardson P., Webb C., Paripati A. MicroRNA 92a-2∗: a biomarker predictive for chemoresistance and prognostic for survival in patients with small cell lung cancer. Journal of Thoracic Oncology. 2010;5(8):1273–1278. doi: 10.1097/JTO.0b013e3181dea6be. [DOI] [PubMed] [Google Scholar]
- 72.Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Research. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 73.Creemers E.E., Tijsen A.J., Pinto Y.M. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circulation Research. 2012;110(3):483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 74.Lopez-Riera M., Conde I., Quintas G., Pedrola L., Zaragoza A., Perez-Rojas J. Non-invasive prediction of NAFLD severity: a comprehensive, independent validation of previously postulated serum microRNA biomarkers. Scientific Reports. 2018;8(1):10606. doi: 10.1038/s41598-018-28854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed in this study are available from the figshare website repository (https://figshare.com) (DOI under embargo: 10.6084/m9.figshare.8015906; private link for reviewers: https://doi.org/10.6084/m9.figshare.8015906). The ChIPSeq data supporting this study's findings were deposited in the NCBI's Sequence Read Archive with the BioProject ID: PRJNA533940.