Abstract
Innovations in organoid-based models of human tissues have made them an exciting experimental platform for studying development and disease. However, these models require systematic benchmarking against primary tissue to establish their value. We discuss key parameters that impact the utility of organoid models, primarily focusing on cerebral organoids as examples.
Cerebral Organoids Are a Window into Early Brain Development
Development of the mammalian brain takes place largely during prenatal stages and varies substantially across species. Although animal models have provided critical insights into the basic mechanisms of brain development, they fall short of replicating human-specific biology. Studies utilizing human embryonic or induced pluripotent stem cells have leveraged lessons from developmental biology to differentiate brain-specific progenitor cells. These progenitors can be cultured as three dimensional aggregates to harness the properties of self-assembly of embryonic brain tissue to recapitulate early features of the in vivo developing brain (Kadoshima et al., 2013; Lancaster et al., 2013). These three-dimensional cultures are often referred to as organoids and have become a valuable experimental model of early development, as organoidshave the potential to provide important mechanistic insights into developmental and disease processes occurring in tissues that do not harbor resident stem cells, including the brain.
Pluripotent stem cells are especially versatile because they enable simultaneous differentiation of multiple lineages, can be easily shared between laboratories to cross-validate biological insights, and can be derived from a diverse pool of genetic backgrounds. These innovations create an unprecedented opportunity to study the impact of genetic variants on cell type specification and organ development and function, including those related to human disease biology.
Given the ease of implementation, as well as experimental and genetic tractability of organoids as a model of developing human tissue, it is sometimes assumed that organoids obviate the need for primary tissue or animal model research and are poised to fuel drug discovery for human disease. However, to embrace cerebral organoids for research purposes, it is important to understand and critically evaluate not only their strengths, but also their potential short-comings. Practical utility of any experimental model depends on the fidelity and robustness of cell fate specification, the precision of self-assembly into higher-order tissues, and the accuracy and dynamics of functional processes in vitro and in vivo (Figure 1).
Figure 1. Cellular Identity in Primary Tissue to Improve Organoid Models.
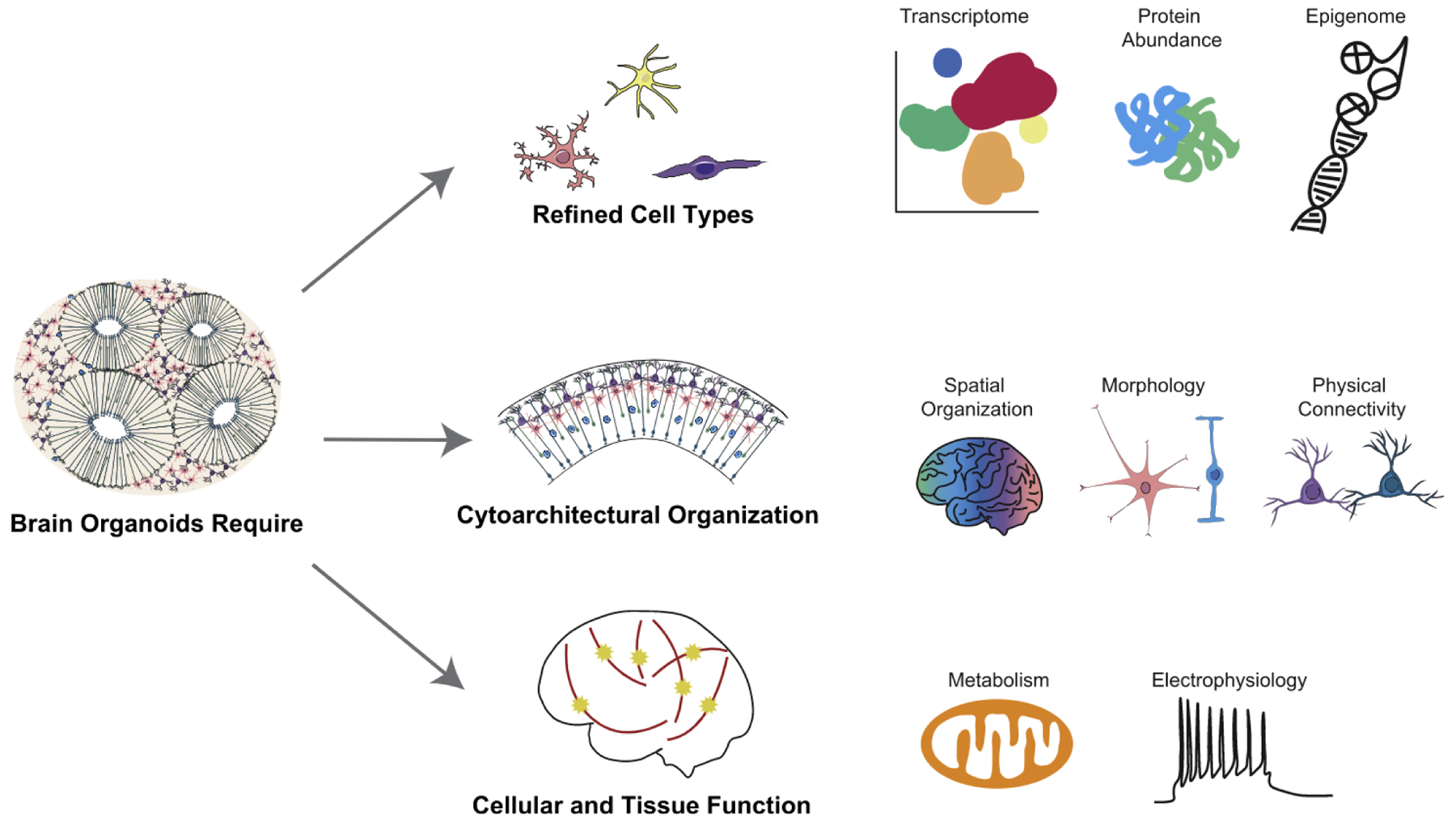
Many aspects of a cell within a primary tissue define its cell identity. Characterizing each of these properties in primary tissues is essential to measuring and optimizing them in organoid models. These datapoints include characterization of the cell type via methods including transcriptome, epigenome, and protein abundance; cytoarchitectural organization including parameters of spatial organization, morphology, and physical connectivity; and metrics of cellular and tissue function including metabolic state and electrophysiology.
Understanding Cell Type in Organoids
Most nervous system disorders are cell type specific, requiring reproducibility of accurate cell type identity in cerebral organoid models to gain meaningful insight into neurological and neuropsychiatric disorders. Cell types in organoid models were initially validated using staining for known marker genes, demonstrating that broadly defined cell classes emerge during in vitro differentiation (Kadoshima et al., 2013; Lancaster et al., 2013). However, the broadly defined cell classes, such as radial glia, intermediate progenitors, and upper and deep cortical layer excitatory neurons need to be more extensively characterized for it to be determined if they are truly reflective of the rich diversity of cell types found in the complex human cerebral cortex. Until recently, our understanding of cell identity has been very limited. However, the emergence of cellular resolution transcriptomics and epigenomics has revealed substantial heterogeneity within classically defined cell classes and under-scored the need for more nuanced understanding of human-specific cell types and features. Across most tissues, single-cell RNA-sequencing datasets are beginning to define a molecular taxonomy of cell types that will aid us in better understanding organ development, and these datasets can also be used to benchmark in-vitro-derived organoids (Velasco et al., 2019).
Fueled by the availability of gene expression datasets from primary developing human tissues, algorithmic approaches have been applied to compare in vitro models to the blueprint of primary tissue, predominantly based on analyses of correlation or probabilistic models. However, comparing single-cell datasets across technology platforms, data generation centers, batches, and experimental models is far from simple. Current approaches to these comparisons have centered around identification of major marker genes that are almost always preserved across systems; mapping between datasets, such as canonical correlation analysis; or correlation between cluster marker genes. Each of these methods, based on correlation, is good at revealing coarse relationships between broad cell classes, but closer and more critical interrogation of cell-type-specific differential gene expression has already revealed important discrepancies between in vitro organoid models and their primary counterparts. For example, early single-cell transcriptomic comparisons between primary developing tissues and adherent in-vitro-derived cells of the human midbrain first raised the notion that in vitro systems may be recapitulating broad cell types, but not the more nuanced cell subtypes that can be identified by hierarchical clustering approaches (La Manno et al., 2016). This has been further highlighted by robust comparisons between primary datasets of the developing human cortex and multiple cortical and cerebral organoid datasets that have identified metabolic stress in the aggregate cultures as a roadblock to precise subtype specification (Bhaduri et al., 2020).
Because of the cell type heterogeneity that exists in the human brain, such purely correlational or differential expression-based approaches may or may not identify candidates that could effectively resolve current limitations of organoids. Thus, in addition to utilizing current gene expression comparison strategies, it may also prove useful to more comprehensively utilize gene network analysis (such as weighted gene co-expression network analysis) or trajectory methods (such as RNA velocity, scVelo, and Monocle) to highlight nuanced differences between primary and in vitro models and identify biological processes that could be targeted to improve the organoid systems. Because each of these analytical methods is noisy, especially in single-cell contexts, it is essential for researchers to focus on the disparities between datasets that emerge in order to identify unifying themes across analyses, rather than focusing solely on the similarities between systems. However, it is also essential that researchers interpret noisy data from the perspective of developmental biology and identify molecular processes or manipulations that will make biological sense rather than those that may reflect technical artifacts. This detail-oriented emphasis on differences between primary and organoid systems will require acknowledgment from the community at large that brain organoid models remain imperfect and a commitment to focus on data-driven limitations of cell type. As an example, comparison of numerous kidney organoid protocols to primary kidney samples with single-cell transcriptomics highlighted that off-target populations and impaired maturation were being driven by inappropriate activation of BDNF signaling (Wu et al., 2018).
In addition to impaired subtype specification, neural organoids also frequently contain off-target populations that are either outside of the anatomical region specified or represent a non-neural lineage; microglia and vascular cells can both be observed in cortical organoid single-cell datasets (Bhaduri et al., 2020). Limitations such as these are being widely ascribed to impaired maturation signatures in both progenitor and differentiated cell populations in retinal, cortical, and other organoid types. Comparisons across many PSC lines highlighted that limitations, including prevalent off-target populations or commitment to alternative lineages, are highly line dependent, suggesting that more work is required to understand why certain PSC lines are more effective than others. These studies are also a cautionary tale that current issues with reproducibility, imperfect cell type specification, and widespread lack of maturation may limit the over-arching utility of organoid systems to study cell-type-specific normal or disease processes.
New Approaches Required to Improve Tissue Organization
Building upon protocol improvements for reproducible generation of broad cell classes, another challenge for organoid research is the ability to organize cells into spatial patterns that support functional interactions between cell types. In brain organoids, rudimentary organization of progenitor zones is a common feature, in particular organization of radial glia-like cells into elongated rosette-like structures that mirror the organization of the ventricular zones of the developing cortex and formation of a secondary proliferative population away from the rosette, which mirrors the outer subventricular zone. Formation of the postmitotic neuronal mantle is more variable, and organization of the principle neurons into layers is not accurately recapitulated. Moreover, while several studies have been used to dissect the molecular and physical mechanisms of tangential expansion of the neuroepithelium, we currently lack models of cortical folding, a hallmark feature of the human cerebral cortex.
Organoids are limited in their ability to grow beyond a certain size, and cellular organization remains rudimentary. Interestingly, some aspects of tissue organization and cell type composition are better recapitulated in organoid systems such as liver and intestine derived from primary adult resident stem cells, suggesting that important differences exist between the starting material of organoids. Innovations in culture technologies such as spinning bioreactors and orbital shakers improve oxygenation and nutrient availability to the organoid center and support overall tissue health. Additionally, endothelial cell transplantation methods have supported the formation of primitive vasculature to promote more endogenous-like tissue development. Recent innovations have included sectioning of organoids and organotypic slice-culture to preserve cytoarchitectural organization that can be lost over time in standard organoid culture. Organoid slicing may allow organoids to develop more complex cell layering and organized axon outgrowth that are visually reminiscent of that in the adult cortex (Giandomenico et al., 2019). These innovations and other bioengineering solutions are beginning to address current limitations to allow the growth of larger and more mature organoids. However, as has been noted in other fields such as cancer biology, organoids still suffer from variability in cytoarchitectural organization and size disparities between pluripotent cell lines, despite the optimization of reproducible cortical organoid protocols (Velasco et al., 2019). We can look forward to additional exciting engineering advances, but these should also continue to be benchmarked against the developmental sequence by which primary human cortex develops its cytoarchitectural features.
Metabolic and Physiological Processes in Organoids
Despite their unprecedented ability to resolve transcriptomic cell types and states and contribute to the improvement of in vitro approaches, gene expression studies have limitations in capturing the defining features of cellular identity. Cellular properties including morphology, epigenetic state, protein and small RNA abundance, connectivity, organelle composition and distribution, and functional parameters collectively refine cell identity beyond what is reflected by transcriptomics alone. Multimodal comparisons, including combinations of proteomics, epigenetic profiling, metabolomics, and electrophysiological characterization of cell types in primary tissues and organoid models will more effectively identify cellular composition and fuel hypothesis-driven optimization of organoid protocols. This framework will likely be informative across many organoid models of human tissues, not only the brain. Importantly, few comprehensive multivariate characterizations have been performed in organoids so far, and among those that have, it is often difficult to interpret their meaning. For example, it is difficult to evaluate the fidelity of studies of organoid physiology because similar datasets have not been generated from primary human samples.
Recognizing the discrepancies between organoids and in vivo tissues, a number of ongoing funding initiatives have been unrolled to promote development of new strategies to improve the fidelity of organoid models. For example, transcriptomic analysis of primary pancreatic islets identified strong nuclear receptor signals that were not present in organoid models. Addition of ERRy signaling to these in vitro culture systems enabled insulin production in response to glucose (Yoshihara et al., 2016), a metric of maturation that was previously only attainable with transplantation of organoids into mice or other animals. It is unclear if similar approaches can be applied to overcome more fundamental challenges associated with PSC cultures, such as genomic instability and spontaneous mutations.
Integrating Models to Increase Scientific Insights
Organoid cultures are often considered to be a preferred model for conducting experimental studies in human tissues, driving interest and stimulating innovation to develop improved culture systems. Functional differences between organoids and primary tissue could have important consequences for gaining physiologically relevant insights into disease mechanisms. These potentially important differences are often unexplored. For example, experiments using organoids to model Zika virus infection provided different insights with respect to pathology, cellular tropism, and the required entry factors than experiments using primary tissues (Ming et al., 2016). These discrepancies may come at a cost to patient health and safety if candidate pathways or mechanisms are not validated prior to the development of candidate therapeutics.
As a counterbalance to organoid models, primary human tissues and animal models continue to serve as the gold standard in biomedical research. These experimental platforms enable studies of higher-order features that organoids do not recapitulate, such as complex and stereotyped brain circuitry, or animal behavior and physiology in an in vivo context (Figure 2). Innovative technologies such as single-cell transcriptomics and spatially resolved cell mapping techniques applied in the context of comparative studies can provide important context for molecular homology across species, and they may even identify new organisms that recapitulate human cell types or circuits. Similarly, bioengineering innovations and culture improvements may enable protracted cultures of human tissues that would enable functional genetic experiments (Ting et al., 2018). Classical animal models, non-model organisms, and ex vivo cultured tissues can be used to conduct functional experiments harnessing innovations in gene editing, cell-type-specific viruses, and other molecular tools for accessing and perturbing specific genes, cell types, and brain circuits. These complementary approaches are particularly important and should not be replaced by organoid modeling. There is a risk that organoid-based experimentation may be promoted at the expense of primary tissue by restrictive policies limiting federal funding for research using primary prenatal tissue in the United States (Goldstein, 2019), and at the expense of animal models that are being systematically reduced or even stopped at institutions in the United Kingdom and Europe.
Figure 2. Integrating Organoids, Animal Models, and Primary Human Tissue Samples.
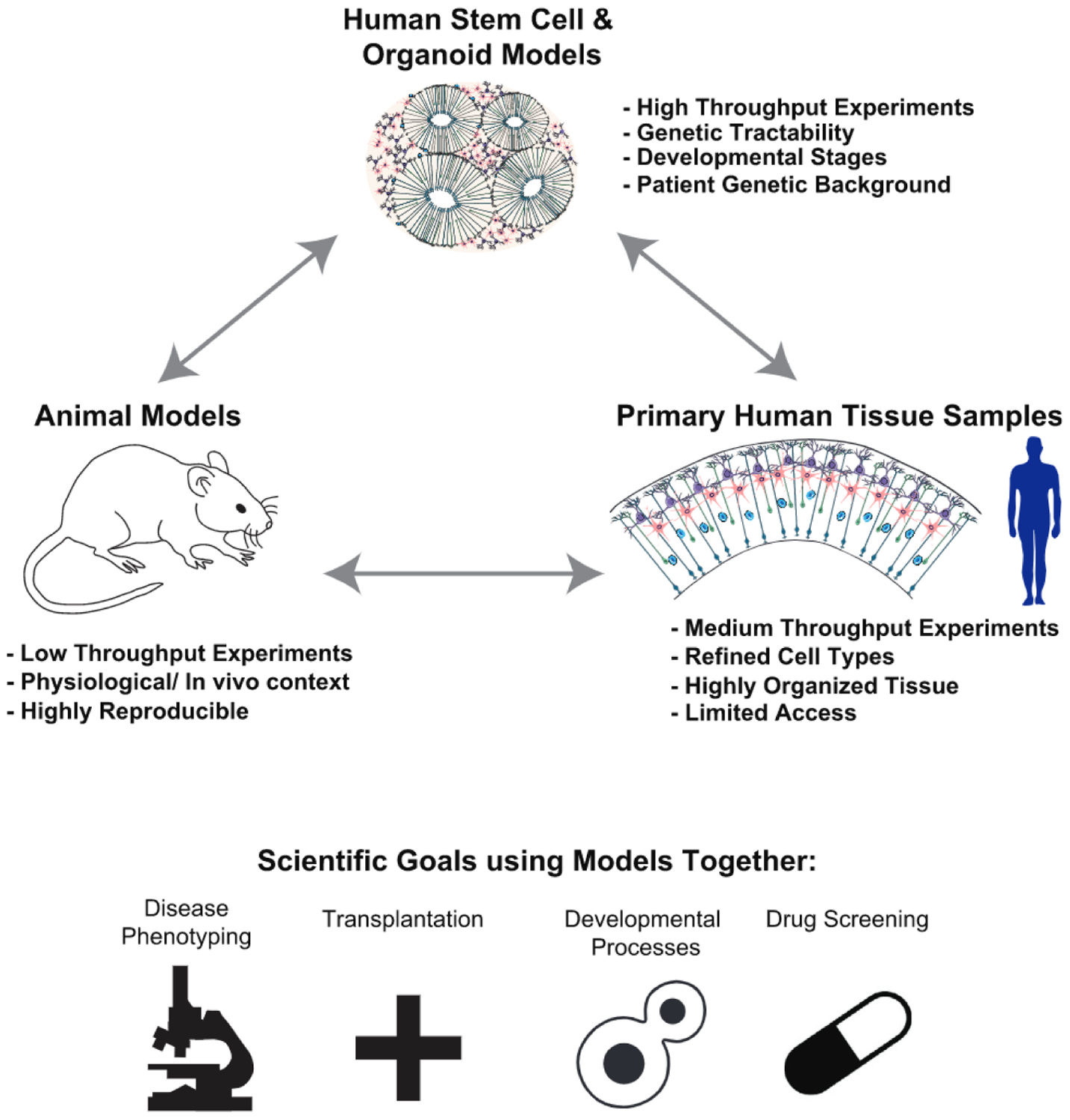
Each experimental system contains strengths and weaknesses in terms of ease of use, physiological relevance, and ability to inform human disease. Together, integration of multiple systems for biological questions regarding how normal development proceeds or how disease manifests can offer the most integrated understanding of biology rather than any one system on its own.
Conclusion
Cerebral organoids have emerged as attractive, and easy to implement, in vitro models of the human brain that can be reproducibly produced from pluripotent cell lines. However, differences between organoid and primary cell types could obscure insights into endogenous brain function and may provide misleading assumptions concerning neuropathology with the potential to misinform therapeutics. Studies have begun to benchmark organoid cells against native counterparts, but a more comprehensive and rigorous comparison is required. The in vitro-generated human cells need to be referenced against the blueprint of primary datasets evaluating cellular and tissue efficacy using genetic, morphological, and biochemical measures. A practical roadmap for evaluating the fidelity and robustness of organoid models of human tissues rests upon continued access to primary tissue from developing and adult human material, investments made to generate high-quality reference cellular resolution datasets, and development of new algorithmic approaches that could highlight similarities as well as differences between these models.
REFERENCES
- Bhaduri A, Andrews MG, Mancia Leon W, Jung D, Shin D, Allen D, Jung D, Schmunk G, Haeussler M, Salma J, et al. (2020). Cell stress in cortical organoids impairs molecular subtype specification. Nature 578, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giandomenico SL, Mierau SB, Gibbons GM, Wenger LMD, Masullo L, Sit T, Sutcliffe M, Boulanger J, Tripodi M, Derivery E, et al. (2019). Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat. Neurosci 22, 669–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A (2019). NIH issues strict new requirements for fetal tissue research funding. The Washington Post, July 26, 2019. https://www.washingtonpost.com/health/nih-issues-strict-new-requirements-for-fetal-tissue-research-funding/2019/07/26/e9f7e6f8-afdf-11e9-bc5c-e73b603e7f38_story.html. [Google Scholar]
- Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, and Sasai Y (2013). Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA 770, 20284–20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, et al. (2016). Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, and Knoblich JA (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, Tang H, and Song H (2016). Advances in Zika Virus Research: Stem Cell Models, Challenges, and Opportunities. Cell Stem Cell 19, 690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JT, Kalmbach B, Chong P, de Frates R, Keene CD, Gwinn RP, Cobbs C, Ko AL, Ojemann JG, Ellenbogen RG, et al. (2018). A robust ex vivo experimental platform for molecular-genetic dissection of adult human neocortical cell types and circuits. Sci. Rep 8, 8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, Paulsen B, Nguyen L, Adiconis X, Regev A, et al. (2019). Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Uchimura K, Donnelly EL, Kirita Y, Morris SA, and Humphreys BD (2018). Comparative Analysis and Refinement of Human PSC-Derived Kidney Organoid Differentiation with Single-Cell Transcriptomics. Cell Stem Cell 23, 869–881 e868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara E, Wei Z, Lin CS, Fang S, Ahmadian M, Kida Y, Tseng T, Dai Y, Yu RT, Liddle C, et al. (2016). ERRy Is Required for the Metabolic Maturation of Therapeutically Functional Glucose-Responsive (3 Cells. Cell Metab. 23, 622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]


