Abstract
Objective
Obesity due to overnutrition causes adipose tissue dysfunction, which is a critical pathological step on the road to type 2 diabetes (T2D) and other metabolic disorders. In this study, we conducted an unbiased investigation into the fundamental molecular mechanisms by which adipocytes transition to an unhealthy state during obesity.
Methods
We used nuclear tagging and translating ribosome affinity purification (NuTRAP) reporter mice crossed with Adipoq-Cre mice to determine adipocyte-specific 1) transcriptional profiles (RNA-seq), 2) promoter and enhancer activity (H3K27ac ChIP-seq), 3) and PPARγ cistrome (ChIP-seq) profiles in mice fed chow or a high-fat diet (HFD) for 10 weeks. We also assessed the impact of the PPARγ agonist rosiglitazone (Rosi) on gene expression and cellular state of adipocytes from the HFD-fed mice. We integrated these data to determine the transcription factors underlying adipocyte responses to HFD and conducted functional studies using shRNA-mediated loss-of-function approaches in 3T3-L1 adipocytes.
Results
Adipocytes from the HFD-fed mice exhibited reduced expression of adipocyte markers and metabolic genes and enhanced expression of myofibroblast marker genes involved in cytoskeletal organization, accompanied by the formation of actin filament structures within the cell. PPARγ binding was globally reduced in adipocytes after HFD feeding, and Rosi restored the molecular and cellular phenotypes of adipocytes associated with HFD feeding. We identified the TGFβ1 effector protein SMAD to be enriched at HFD-induced promoters and enhancers and associated with myofibroblast signature genes. TGFβ1 treatment of mature 3T3-L1 adipocytes induced gene expression and cellular changes similar to those seen after HFD in vivo, and knockdown of Smad3 blunted the effects of TGFβ1.
Conclusions
Our data demonstrate that adipocytes fail to maintain cellular identity after HFD feeding, acquiring characteristics of a myofibroblast-like cell type through reduced PPARγ activity and elevated TGFβ-SMAD signaling. This cellular identity crisis may be a fundamental mechanism that drives functional decline of adipose tissues during obesity.
Keywords: Adipose tissue, Obesity, Cellular identity, NuTRAP, PPARγ, TGFβ-SMAD
Abbreviations: T2D, Type 2 diabetes; HFD, high-fat diet; TZDs, thiazolidinediones; Rosi, rosiglitazone; PPARγ, peroxisome proliferator-activated receptor γ; TGFβ, Transforming growth factor beta; eWAT, epididymal white adipose tissue; iWAT, inguinal white adipose tissue; BAT, brown adipose tissue; NuTRAP, nuclear tagging and translating ribosome affinity purification; ChIP-seq, chromatin immunoprecipitation sequencing.
Highlights
-
•
Adipocytes after HFD intake exhibit defects in cellular identity maintenance.
-
•
Adipocytes develop actin filament networks in obesity.
-
•
Altered PPARγ activity mediates defective adipocyte identity phenotypes.
-
•
TGFβ-SMAD pathways promote HFD-induced aberrant phenotype of adipocytes.
1. Introduction
Obesity is a widespread and growing global health problem. In 2016, more than 1.9 billion adults were overweight, and over 650 million were obese, and these numbers are expected to continue to rise over the next decade [1]. Obesity causes a range of metabolic disorders, including cardiovascular disease, hepatic steatosis, and type 2 diabetes (T2D) [2]. Expansion of adipose tissue is the sine qua non of obesity and is accompanied by adipose tissue dysfunction characterized by impaired lipid handling, hormone resistance, and defective adipokine secretion. These changes are in turn believed to have systemic consequences, including hepatic steatosis and insulin resistance [3]. Several interrelated cellular pathways have been proposed to account for adipocyte dysfunction in obesity, including inflammation, fibrosis, hypoxia, oxidative stress, and ER stress [3]. However, the transcriptional mechanisms that underlie the transition from a healthy adipocyte to an unhealthy adipocyte are not completely understood.
Peroxisome proliferator-activated receptor γ (PPARγ) is a ligand-dependent transcription factor and master regulator of adipose biology [4]. PPARγ is essential for adipocyte differentiation and directly induces the expression of genes involved in insulin action and lipid transport, synthesis, and storage [4,5]. In addition, PPARγ is crucial for mature adipocyte maintenance and survival [6] as shown by lipodystrophy in adipocyte-specific PPARγ knockout mice [7]. High-fat feeding has been shown to alter the binding profile of PPARγ in whole adipose tissue [8]. Thiazolidinediones (TZDs) are PPARγ agonists that act as potent antidiabetic drugs, effectively lowering blood glucose, improving insulin sensitivity, and ameliorating hepatic steatosis [9]. The beneficial effects of TZDs have been shown to be mediated by activation of PPARγ in many different tissues and cell types, including macrophages, adipocytes, and endothelial cells [9], all of which are found within adipose tissue. It is not yet clear how either obesity or TZDs alter PPARγ binding in mature adipocytes.
One issue that generally confounds the study of adipocyte-specific processes is the heterogeneity of adipose tissue. Despite its bland appearance, adipose tissue is complex, consisting of a diverse array of cells, including fibroblasts, pericytes, endothelial cells, and various immune cells (for example, macrophages, neutrophils, and T cells). These non-adipocytes account for 40–70% of all cells in lean animals depending on depot and diet [10,11]. Under HFD conditions, the immune compartment increases in both numbers and inflammatory potential [12], presumably diluting the adipocyte fraction even further. Many studies of adipocytes have circumvented this issue by using the Rodbell protocol [13], which takes advantage of the propensity of mature lipid-laden adipocytes to float following cell dissociation by collagenase and low-speed centrifugation. Although this is a workhorse technique in the field of adipose biology, there are at least two important limitations. First, there is often less than optimal purity, as some non-adipocytes can adhere to floating adipocytes even after collagenase digestion [14]. Second, the method itself induces changes in the adipocytes because of activation of stress and TNF-mediated immune pathways induced by collagenase digestion [15]. To overcome these issues, we generated a transgenic mouse line called NuTRAP (nuclear tagging and translating ribosome affinity purification), which makes it possible to profile the transcriptome and epigenome of a specific cell type within heterogeneous tissue in vivo without cell dissociation [11]. Using the NuTRAP line allowed us to characterize the transcriptional and epigenomic features of pure adipocyte populations in vivo and define the unique cellular plasticity of temperature-responsive beige adipocytes [16].
We used NuTRAP mice to study how the transcriptional and chromatin states of adipocytes are altered by HFD and TZD treatment. Pathway analysis of the epigenomic and transcriptional changes induced by conditions of overnutrition suggest a partial loss of mature adipocyte cellular identity, including both increased myofibroblast and reduced adipocyte marker gene expression. These changes are accompanied by the formation of aberrant actin filament structures. Furthermore, we identify reduced PPARγ binding and enhanced TGFβ-SMAD3 pathway activation as key drivers of these state changes.
2. Material and methods
2.1. Animal studies
All of the animal experiments were conducted according to procedures approved by the Beth Israel Deaconess Medical Center (BIDMC) Institutional Animal Care and Use Committee (IACUC). The mice were housed under a 12-h light/dark cycle at 22 °C with free access to food and water. To label mature adipocytes, NuTRAP mice [11] (Jackson Laboratory, 029899) were crossed with Adipoq-Cre mice [17] (Jackson Laboratory, 010803). The mice were placed on a chow diet or high-fat diet (HFD, 60% kcal from fat, Research Diets, D12492i) at 6 weeks of age for 10 weeks.
2.2. Glucose homeostasis tests
For glucose tolerance tests, the mice were fasted for 12 h and injected intraperitoneally (IP) with glucose (1 g/kg body weight). Blood was collected at 0, 15, 30, 45, 60, and 120 min after glucose injection to measure glucose levels with a portable glucometer. For insulin-tolerance tests, the mice were fasted for 6 h and injected IP with insulin (0.75 U/kg body weight). Blood glucose levels were determined at 0, 15, 30,45, 60, and 120 min after insulin injection. For fasting plasma insulin levels, blood samples were collected after 6 f of fasting in EDTA-coated blood collection tubes and analyzed using an ELISA kit from Crystal Chem following the manufacturer's instructions.
2.3. Tamoxifen injection
Tamoxifen (Sigma, T5648) was dissolved in sunflower seed oil (Sigma, S5007) at a concentration of 20 mg/ml by rotating at 37 °C overnight. Tamoxifen was then injected intraperitoneally into the mice at a dose of 100 mg/kg for 3 consecutive days. After a 1-week washout period following the last injection, HFD feeding was started.
2.4. Rosiglitazone injection
Rosiglitazone (Rosi, Cayman, 71740) stock solution was prepared by dissolving in dimethyl sulfoxide (DMSO) at a concentration of 50 mg/ml. The stock solution was freshly diluted on each day prior to injection in saline containing 2% Tween 80 at a concentration of 1.6 mg/ml of Rosi (3.2% of the stock) and IP injected into mice at a dose of 8 mg/kg for 10 consecutive days. On the next day after the last injection, ITT was conducted and the mice were terminated 5 days later.
2.5. Tissue histology and immunofluorescence staining
For histology, fresh adipose tissues were collected and fixed in 10% formalin for 1 day, washed in phosphate-buffered saline (PBS), stored in 70% ethanol, and processed by the BIDMC histology core for paraffin sectioning and hematoxylin and eosin (H&E) staining. For whole mount immunofluorescence staining, fresh adipose tissues were cut into small pieces (3 mm × 3 mm), fixed in 10% formalin for 1 day, and washed in PBS. Fixed tissues were blocked and permeabilized in PBS containing 1% Triton X-100, 2% fetal bovine serum (FBS), and 1% bovine serum albumin (BSA) for 1 day and incubated with anti-PLIN1 (Abcam ab61682) at 1/1000 dilution in a 48-well plate for 1 day. After washing in PBS/Triton X-100/FBS/BSA, tissues incubated with phalloidin (Thermo Fisher Scientific, A12380, 1 U/200 μl), Alexa Fluor conjugated antibodies (1/2000), and Hoechst 33342 (2 μg/ml) for 1 day. The tissues were washed in PBS with 1% Triton X-100, placed in mounting solution on slides, and covered with coverslips. The slides were visualized using a Zeiss LSM 510 Meta confocal microscope. For adipocyte phalloidin staining, floating adipocytes were fixed with 4% PFA for 10 min in 1.5 ml tubes, permeabilized in PBS with 0.1% Triton X-100 for 15 min, and washed twice in PBS. The adipocytes were then stained with phalloidin and Hoechst for 20 min, washed twice, mounted on 1% agarose pads on slides, and visualized.
2.6. Cell culture
Murine 3T3-L1 preadipocytes (American Type Culture Collection) were maintained in Dulbecco's Modified Eagle Medium (DMEM) (Invitrogen) with 10% bovine calf serum (HyClone) and 1% penicillin/streptomycin (P/S, Gibco). Two days after confluency, the cells were induced to differentiate in DMEM containing 10% FBS (Peak Serum), 5 μg/ml insulin, 500 μM isobutylmethylxanthine, and 1 mμM dexamethasone for 2 days and then maintained in DMEM with FBS and insulin for another 2 days. Thereafter, the cells were maintained in DMEM containing 10% FBS. For lentivirus transduction experiments, viral supernatants were generated by co-transfection of lentiviral constructs with pM2D.G and psPAX2 plasmids in 293T cells and then collected after 48 h of transfection. Viral supernatants were incubated with 3T3-L1 adipocytes for 1 day and changed with fresh media, and adipocytes were used for experiments 4–5 days later. For TGFβ treatment, mouse TGFβ1 (R&D, 7666-MB-005) was dissolved at 50 μg/mg in 4 mM HCl containing 0.1% BSA for the stock solution and diluted in culture media at the indicated concentrations for treatment.
2.7. Protein isolation and Western blotting
Cell and tissue lysates were prepared using radioimmunoprecipitation assay (RIPA) lysis buffer (20 mM Tris pH 8, 140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, and 1 mM EDTA) supplemented with 1x Complete EDTA-free protease inhibitor (Roche). Protein concentrations were measured by a bicinchoninic acid (BCA) assay kit (Thermo Fisher Scientific, PI23228). Lysate samples were separated by 4%–15% gradient SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). Blots were incubated with primary and HRP-conjugated secondary antibodies and detected using Western Lightning ECL (PerkinElmer, NEL103001EA). Western blots were quantified using ImageJ software (NIH). The following antibodies were used: ACTA2 (Abcam, ab5694), Caveolin (BD Biosciences, 610059), PPARγ (Cell Signaling Technology, 2443S), GAPDH (Cell Signaling Technology, 2118S), Phospho-Akt (Ser473) (Cell Signaling Technology, 9271S), and Akt (Cell Signaling Technology, 9272S).
2.8. RNA isolation and quantitative real-time PCR
Total RNA was extracted from cells or tissues using TRIzol reagent (Invitrogen), and 500 ng of RNA was converted to cDNA using a High-Capacity cDNA Reverse Transcription kit following the manufacturer's instructions. Quantitative real-time PCR was conducted using SYBR Green PCR Master Mix (Applied Biosystems) in a QuantStudio5 system. Fold change was determined by comparing the target gene expression with reference gene 36B4. The primers used for qRT-PCR are provided in Supplemental Table 1.
2.9. TRAP and RNA-seq
TRAP was conducted as previously described [11,16] with minor modifications. Adipose tissue samples were Dounce homogenized in homogenization buffer (20 mM HEPES pH 7.5, 15 mM MgCl2, 150 mM KCl, 1% NP-40, 100 μg/ml cycloheximide, 1 mg/ml sodium heparin, 1 mM DTT, 0.2 U/μl RNasin, and 1x Roche Complete EDTA-free protease inhibitor) and incubated on ice for 15 min with vortexing every 5 min. After lysates were centrifuged at 13,000 rpm for 10 min, the top lipid layer was removed and the supernatant was collected in new tubes. Immunoprecipitation was conducted by rotating the lysates mixed with GFP antibody (Abcam, ab290) for 1 h at 4 °C and subsequently with Dynabeads Protein G (Thermo Fisher Scientific) for 1 h at 4 °C. After washing three times in wash buffer (20 mM HEPES pH 7.5, 15 mM MgCl2, 350 mM KCl, 1% NP-40, 100 μg/ml cycloheximide, and 1 mM DTT), immunoprecipitates were subjected to RNA extraction using a Qiagen Micro RNeasy kit according to the manufacturer's instructions. For RNA-seq library constructions, isolated RNA (100 ng) was processed by a Ribo-Zero rRNA removal kit (Epicentre) for ribosomal RNA removal and converted into cDNA by Maxima Reverse Transcriptase (Thermo Fisher Scientific). Second strands were generated by a NEBNext mRNA Second Strand Synthesis kit following the manufacturer's instructions. Sequencing libraries were generated by tagmentation (Nextera XT DNA Library Preparation kit), PCR amplification and sizes were selected using AMPure XP beads (Beckman Coulter). RNA-seq libraries were quantified by Qubit, analyzed by Agilent Bioanalyzer, and sequenced on an Illumina HiSeq2500.
2.10. Nuclei isolation, sorting, and ChIP-seq
Nuclear isolation, sorting, and ChIP-seq were conducted as previously described [11,16]. Adipose tissue samples were Dounce homogenized in nucleus preparation buffer (10 mM HEPES pH 7.5, 1.5 mM MgCl2, 10 mM KCl, 250 mM sucrose, 0.1% NP-40, and 0.2 mM DTT) and filtered through 100 μm cell strainers. Filtered homogenates were cross-linked by rotating with 1% paraformaldehyde (PFA) at room temperature for 4 min and subsequently quenched by 125 mM glycine for 10 min. The homogenates were centrifuged at 1000 g for 10 min, washed once, resuspended in nucleus-sorting buffer (10 mM Tris pH 7.5, 40 mM NaCl, 90 mM KCl, 2 mM EDTA, 0.5 mM EGTA, 0.1% NP-40, and 0.2 mM DTT), and filtered through 40 μm cell strainers. Nuclei were sorted using a BD FACS Aria II with gating on FSC, SSC, mCherry, and GFP fluorescence. For ChIP experiments, sorted adipocyte nuclei were pelleted by centrifugation at 1000g for 10 min and sonicated in nuclear lysis buffer (10 mM Tris pH 8, 1 mM EDTA, and 0.1% SDS) using a Covaris E220. Sheared chromatin was used for ChIP in ChIP dilution buffer (16.7 mM Tris pH 8, 1.2 mM EDTA, 167 mM NaCl, 1.1% Triton X-100, and 0.01% SDS) added with H3K27ac (Active Motif, 39133) or PPARγ antibody (Santa Cruz Biotechnology, sc-7196X). After overnight ChIP, the samples were washed in low-salt wash buffer (20 mM Tris pH 8, 1 mM EDTA, 140 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, and 0.1% SDS), high-salt wash buffer (20 mM Tris pH 8, 1 mM EDTA, 500 mM NaCl, 1% Triton X-100, 0.1% sodium deoxycholate, and 0.1% SDS), LiCl wash buffer (10 mM Tris pH 8, 1 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate, and 250 mM LiCl), and TE buffer (10 mM Tris pH 8 and 1 mM EDTA) twice in each step, eluted, and reverse cross-linked in elution buffer (10 mM Tris pH 8, 50 mM EDTA, 0.1% SDS, 300 mM NaCl, 0.8 mg/ml proteinase K, and 10 μg/ml RNase A). DNA was then extracted using AMPure XP beads. For ChIP-seq library construction, DNA was processed through end repair/phosphorylation using an End-It DNA End-Repair kit (Lucigen), A-tailing using the Klenow fragment (New England Biolabs, M0212), index adaptor ligation using Quick Ligase (NEB, M2200), PCR amplification using PfuUltra II Hotstart PCR Master Mix (Agilent Technologies, 600850), and size selection using gel electrophoresis and extraction. ChIP-seq libraries were quantified by Qubit, analyzed by Agilent Bioanalyzer, and sequenced on an Illumina NextSeq500.
2.11. Adipocyte isolation by flotation
Fresh adipose tissues were collected, minced, and digested in PBS containing Collagenase D (1.5 U/ml), Dispase II (2.4 U/ml), and CaCl2 (10 mM in PBS) while shaking for 40 min at 37 °C. After digestion, the tissues were resuspended in culture medium (DMEM/F12 containing 10% FBS and 1% P/S), filtered through 100 μm cell strainers, and centrifuged at 500 g for 5 min. Floating adipocytes were collected, washed in PBS twice, and subjected to TRIzol RNA isolation or phalloidin staining.
2.12. Bioinformatic analysis
Bioinformatic analysis was conducted as previously described [11,16]. For RNA-seq analysis, sequencing reads were aligned to the mm10 mouse genome using HISAT2 [18] and filtered by Picard (https://broadinstitute.github.io/picard) to remove PCR duplicates and low-quality reads. Filtered reads were assigned to transcriptome and quantified using featureCounts [19], and differential gene expression analysis was conducted using edgeR [20]. Lowly expressed genes (log2CPM < 2) were removed from the differential analysis, and we used a fold change (FC) cutoff of log2FC ≥ 0.5 and a false discovery rate (FDR) cutoff of FDR ≤ 0.05. Gene ontology (GO) analysis was conducted using Metascape [21]. Morpheus (https://software.broadinstitute.org/morpheus/) was used to visualize heatmaps, and the volcano plotter, an interactive R Shiny app that we developed for public use [16], was used to depict volcano and MA plots. For ChIP-seq analysis, sequencing reads were aligned to the mm10 mouse genome using Bowtie2 [22], filtered by Picard to remove duplicates, processed by Samtools [23], and subjected to peak-calling by MACS2 [24]. Peaks were quantified using bedtools [25], and normalization and differential analyses were conducted using edgeR. We used the cutoffs of log2CPM ≥2, log2FC ≥ 1, and FDR ≤0.05 for H3K27ac ChIP-seq and log2CPM ≥6, log2FC ≥ 1, and FDR ≤0.25 for PPARγ ChIP-seq. ChIP-seq signal tracks were visualized by the WashU Epigenome Browser [26] using BigWig files generated by bedtools [25] and bedGraphToBigWig [27]. ChIP-seq peak heatmaps were generated by deepTools2 [28] using BigWig files. Intersecting peaks were determined using bedtools, peaks were assigned to genes using HOMER [29], and GO analysis was conducted by Metascape. We used FIMO from the MEME package [30] with the JASPAR database [31] for motif enrichment analysis.
2.13. Statistical analysis
We used two-tailed unpaired Student's t-test for pair-wise comparisons, and p < 0.05 was considered statistically significant unless otherwise specified. Differential analyses were conducted with the edgeR package using the exact test as described by the package's exactTest function.
2.14. Data availability
The raw and processed data reported in this paper are available at the Gene Expression Omnibus (GEO) repository with accession number GSE153120. Among the data, the RNA-seq data from the chow-fed NuTRAP mice were previously deposited (GSE92590) [11] and reanalyzed in this study.
3. Results
3.1. Altered gene expression signature of adipocytes after high-fat diet (HFD)
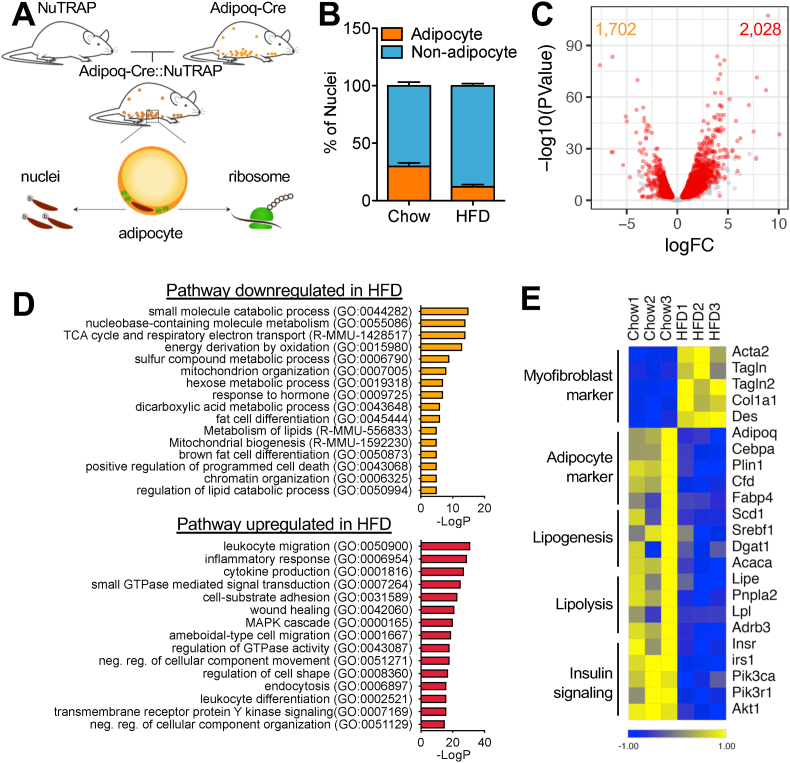
To identify molecular changes taking place specifically within mature adipocytes, we generated Adipoq-Cre::NuTRAP mice by crossing NuTRAP with adipocyte-specific Adipoq-Cre mice [17]. In the Adipoq-Cre::NuTRAP mice, adipocyte nuclei were biotinylated and labeled with mCherry protein, while adipocyte ribosomes were labeled with green fluorescent protein (GFP) (Figure 1A). The Adipoq-Cre::NuTRAP mice fed an HFD for 10 weeks exhibited elevated body weight and higher blood glucose and insulin levels compared to the fed chow mice concomitant with glucose intolerance, insulin resistance, and adipocyte hypertrophy with increased lipid accumulation in adipose tissues (Figure S1). To determine the fraction of adipocytes in the adipose tissues in the different diets, we isolated total nuclei from epididymal white adipose tissues (eWAT) of the Adipoq-Cre::NuTRAP mice and quantified the number of nuclei from adipocytes and non-adipocytes by flow cytometry. mCherry-positive adipocyte nuclei comprised approximately 30% of the total nuclei in eWAT on chow, and this percentage declined to ∼12% after 10 weeks of HFD feeding (Figure 1B). Because we had lower nuclei recovery rates with the HFD-fed than chow-fed mice, we were unable to directly compare the absolute numbers of nuclei. As the adipocyte number is known to increase on an HFD [32,33], the reduced adipocyte fraction on the HFD was likely due to increased numbers of non-adipocyte populations such as immune cells.
Figure 1.
NuTRAP/RNA-seq reveals aberrant alterations in molecular signature of adipocytes after HFD. (a) NuTRAP mice crossed with Adipoq-Cre mice to allow simultaneous labeling of adipocyte nuclei and ribosomes in adipocytes. (b) Adipocyte and non-adipocyte nuclei from eWAT of Adipoq-Cre::NuTRAP mice on chow and HFD were quantified by flow cytometry. Bars indicate mean ± SEM (n = 3–4 animals/group). (c) Volcano plot of the adipocyte-specific transcriptomes on chow and HFD by TRAP followed by RNA-seq (HFD/chow). Each dot indicates individual genes. Differentially regulated genes are shown in red (FDR ≤ 0.05, logFC ≥ 0.5, and logCPM ≥ 2). (d) GO pathway analysis with the differentially regulated genes. (e) Heatmap of myofibroblast and adipocyte markers and genes involved in adipocyte cellular metabolic processes. Columns represent biological replicates (n = 3 animals/group). Expression values (CPM) of each mRNA are represented by z scores.
To characterize the gene expression changes in the adipocytes, we conducted translating ribosome affinity purification (TRAP) [34] in the eWAT of the Adipoq-Cre::NuTRAP mice on chow and after 10 weeks of HFD feeding. The RNA-seq analysis identified 2,028 upregulated and 1,702 downregulated genes on the HFD (Figure 1C). The upregulated pathways included cytoskeleton organization-related processes, along with inflammatory pathways (Figure 1D). Intriguingly, we noted that several myofibroblast marker genes (for example, Acta2, Tagln, and Col1a1) become robustly induced in the adipocytes on the HFD. Downregulated pathways on the HFD included genes crucial for adipocyte identity and metabolic function, such as lipogenesis, lipolysis, and insulin signaling (Figure 1E). High-fat feeding is known to induce de novo adipogenesis [32], so it is possible that the altered gene expression profiles could reflect an influx of newly formed adipocytes. To test this possibility, we generated NuTRAP mice crossed with Adipoq-CreERT [35], which is activated in adipocytes upon tamoxifen injection. After tamoxifen injection, the mice were placed on chow or an HFD for 10 weeks; this model prevents newly differentiated adipocytes from being labeled and thus excluded them from our analysis. Using this approach, we again saw that myofibroblast markers were strongly induced, while adipocyte genes were suppressed (Figure S2). These results indicated that these pathways were altered in mature adipocytes and did not reflect changes due to new differentiation. Of note, we could not exclude some effects on newly differentiated adipocytes that formed under the stimulus of the HFD due to residual tamoxifen that may have persisted even after a long washout period [36].
3.2. Adipocytes undergo remodeling of cytoskeletal organizations on an HFD
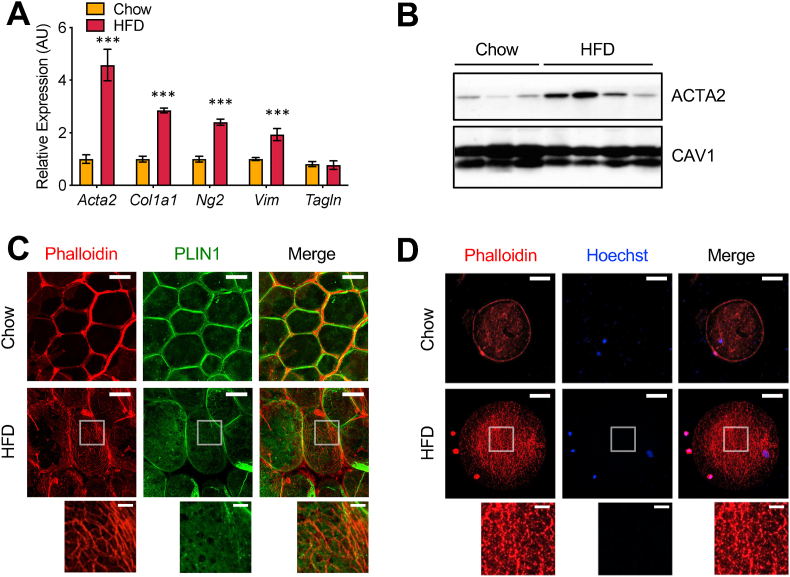
To confirm the myofibroblast-like gene expression signature of the adipocytes on the HFD obtained from the TRAP experiments, we used an independent method using a modified Rodbell protocol [13], often referred to as adipocyte flotation. Mature adipocytes were isolated after tissue digestion and centrifugation and used to conduct gene expression and protein analysis. Similar to the data from the TRAP experiments, a set of myofibroblast marker genes was highly induced in the adipocytes on the HFD (Figure 2A). ACTA2 (alpha smooth muscle actin; α-SMA) protein was also significantly increased in the adipocytes after HFD feeding (Figure 2B). To determine whether these molecular changes were accompanied by a cellular phenotype, we then assessed actin filament formation. Whole-mount tissue staining of the eWAT from the mice on chow or the HFD was conducted using phalloidin and an antibody against perilipin 1 (PLIN1) that stains lipid droplet membranes. In the chow-fed mice, phalloidin staining was restricted primarily to the interstitial regions, including blood vessels, with minimal staining of adipocytes. In contrast, after HFD feeding, we observed a mesh-like actin filament network structure in the cellular cortex region just underneath the plasma membrane of the hypertrophic adipocytes (Figure 2C). To ensure that the actin filament network structures were within the adipocytes and not in the extracellular matrix (ECM), we isolated epididymal adipocytes by flotation and stained them with phalloidin. We detected a small number of actin filament structures in the adipocytes from the chow-fed mice, while these structures became dramatically denser and more complex in the adipocytes from the HFD-fed mice (Figure 2D). These results suggested that the adipocytes on the HFD underwent substantial molecular and cellular remodeling and acquired some features of a myofibroblast-like cell.
Figure 2.
Adipocytes undergo remodeling of cytoskeletal organizations on HFD. (a) Gene expression analysis of myofibroblast markers in adipocytes on chow and HFD by qRT-PCR. RNA extracted from adipocytes isolated by the flotation method. Bars indicate mean ± SEM (n = 4–6 animals/group) (∗∗∗p < 0.005). (b) ACTA2 (smooth muscle α-2 actin) protein levels in adipocytes on chow and HFD by Western blotting. CAV1 (caveolin-1) was used as a loading control. (c) Whole mount staining showing actin filament formation in adipocytes on chow and HFD. Adipose tissues stained with anti-PLIN1 antibody for lipid droplets (green) and phalloidin for actin filaments (red) visualized by confocal microscopy. Scale bars: 50 μm and 10 μm (inset). (d) Isolated adipocytes showing actin filament formation in the cell on HFD. Actin filaments visualized by phalloidin staining (red) and nuclei visualized by Hoechst staining (blue). Scale bars: 50 μm and 10 μm (inset). Note that there are small non-adipocyte cells, seemingly macrophages, with strong phalloidin signals attached on the surface of adipocytes in HFD.
3.3. PPARγ activity is globally reduced in adipocytes on HFD
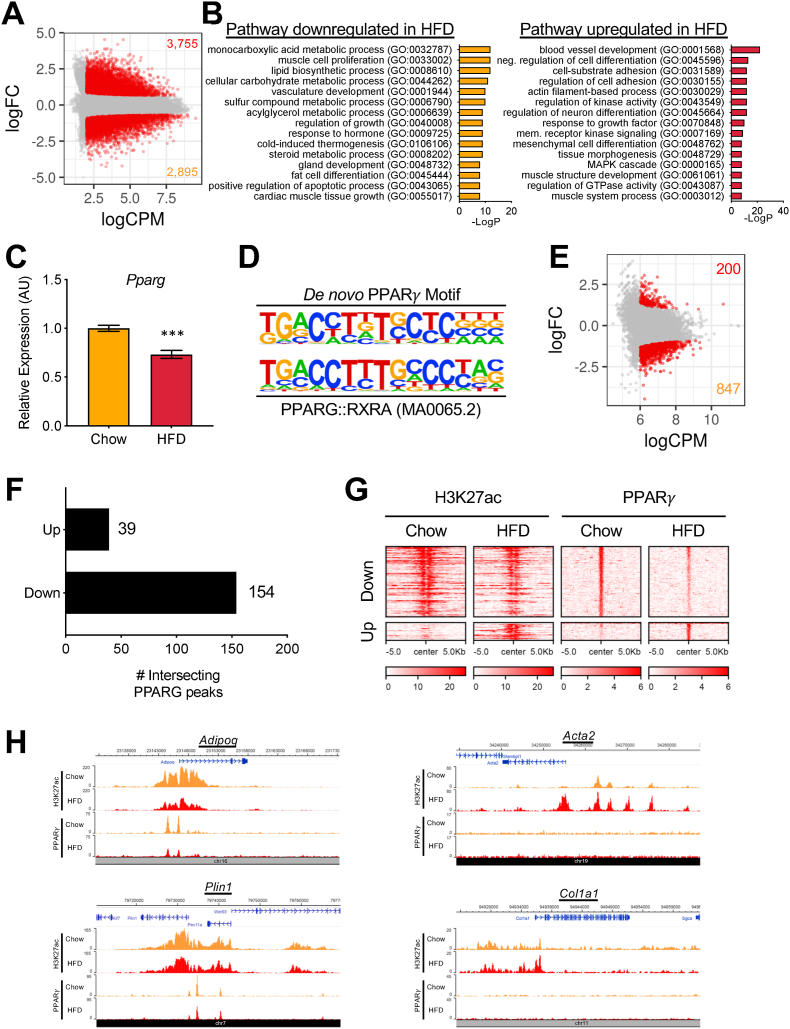
To understand the mechanisms driving altered gene expressions in adipocytes on HFD, we profiled the activity of gene regulatory elements in the adipocytes from the chow and HFD by conducting ChIP-seq for H3K27ac, a histone mark for active promoters and enhancers, with adipocyte nuclei isolated from the Adipoq-Cre::NuTRAP mice. Among the 41,933 H3K27ac peaks identified in the adipocytes, we found 2,895 downregulated and 3,755 upregulated peaks on the HFD (Figure 3A). To find biological processes involved with the differentially regulated peaks, we conducted pathway enrichment analyses using the genes that were associated with the peaks. Similar to the TRAP/RNA-seq data, we found that downregulated H3K27ac peaks were involved in lipid/nutrient metabolic processes, while upregulated H3K27ac peaks pointed to pathways related to cell adhesion and actin filament organization (Figure 3B).
Figure 3.
PPARγ binding is globally reduced in adipocytes on HFD. (a) MA plot of the H3K27ac ChIP-seq data (HFD/chow). Each dot indicates individual peaks. Differentially regulated peaks are shown in red (FDR ≤ 0.05, logFC ≥ 0.5, and logCPM ≥ 2). (b) GO pathway analysis with the genes associated with the differentially regulated H3K27ac peaks. (c) Pparg mRNA expression in isolated adipocytes on chow and HFD by qRT-PCR analysis. Bars indicate mean ± SEM (n = 4–6 animals/group). (∗∗∗p < 0.005). (d) Position weight matrices of the PPARγ motif built by de novo motif analysis with the PPARγ ChIP-seq peaks in comparison with the PPARG::RXRA motif (MA0065.2) from the Jaspar database. (e) MA plot of the PPARγ ChIP-seq data (HFD/chow). Each dot indicates individual peaks. Differentially regulated peaks are shown in red (FDR ≤ 0.25, logFC ≥ 1.0, and logCPM ≥ 6). (f) Numbers of the differentially regulated PPARγ peaks intersecting with H3K27ac in each direction of change on HFD. (g) Heatmap showing intersecting differentially regulated H3K27ac and PPARγ peaks of adipocytes on chow and HFD. Amplitude of each peak center (±5 kb) is represented by the z score and shown in red. (h) H3K27ac and PPARγ ChIP-seq signal tracks on chow and HFD at adipocyte markers Adipoq and Plin1 and myofibroblast markers Acta2 and Col1a1.
PPARγ is the master transcriptional regulator of adipose biology required for adipogenesis, adipocyte function, and survival [4,5]. We noted that many PPARγ-responsive genes in the mature adipocytes were downregulated by the high-fat diet. In fact, Pparg itself showed diminished expression after the HFD (Figure 3C). We next asked whether the in vivo genome-wide binding profile of PPARγ in adipocytes differed after HFD feeding. We used flow cytometry to sort and collect adipocyte nuclei from the eWAT of the Adipoq-Cre::NuTRAP mice on chow or HFD; these were subjected to ChIP-seq with an antibody specific for PPARγ. De novo motif analysis of the top 500 peaks found that the top over-represented DNA element corresponded to the PPARG::RXRA motif (MA0065.2) in the Jaspar database (Figure 3D), confirming the data's high quality. Among 7,665 moderate or strong PPARγ peaks (logCPM ≥ 6), we identified 1,047 differentially regulated peaks; more than 80% (847 peaks) were downregulated while only 200 peaks were upregulated (Figure 3E). Adipocyte-abundant genes, such as Adipoq, Plin1, and Lipe, showed strong PPARγ peaks significantly downregulated on the HFD (Figure S3A). The downregulated PPARγ peaks were associated with genes involved in adipocyte differentiation and cellular metabolism similar to the TRAP/RNA-seq and H3K27ac ChIP-seq data (Figure S3B), whereas the upregulated PPARγ peaks did not show any significantly associated biological pathways. These results indicate that PPARγ activity is globally reduced in adipocytes during obesity.
We next determined whether the changes in H3K27ac activity that occur with HFD correlate with changes in PPARγ activity. We quantified the number of intersecting peaks between the differentially regulated H3K27ac and PPARγ peaks in each direction. We found 193 intersecting peaks, in which the majority (79.8%; 154/193) were downregulated, while 20.2% (39/193) were upregulated (Figure 3F–G). Indeed, one of the key adipocyte marker genes, Adipoq, displayed PPARγ and H3K27ac peaks in the promoter region simultaneously decreased by the HFD (Figure 3H). In contrast, a major myofibroblast marker gene, Acta2, displayed strongly induced multiple H3K27ac peaks at its upstream genomic region but had no PPARγ peaks (Figure 3H). Taken together, these results suggest that PPARγ acts more importantly for HFD-repressed genes than HFD-induced genes.
3.4. Treatment with a PPARγ agonist reverses the molecular and cellular phenotypes associated with HFD feeding
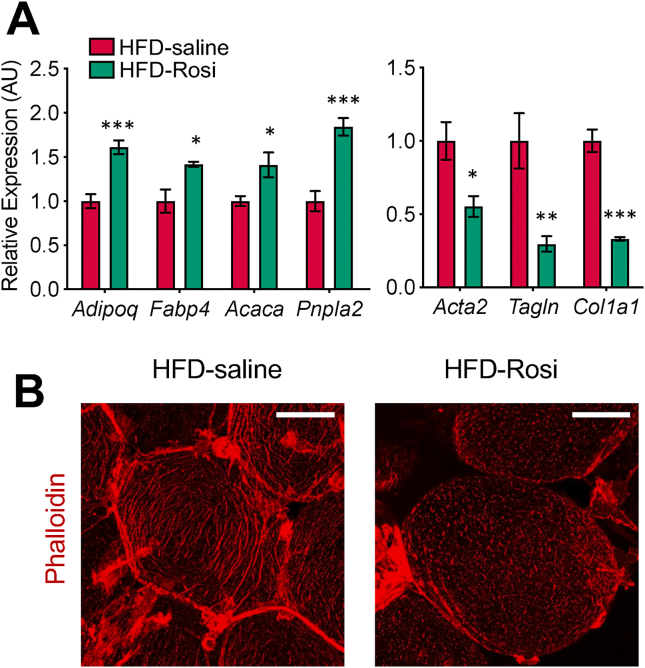
We next tested whether the PPARγ agonist rosiglitazone (Rosi) is able to reverse the aberrant molecular and cellular phenotypes of adipocytes on HFD. We injected Rosi intraperitoneally into the HFD-fed mice for 10 consecutive days, significantly increasing insulin sensitivity and body weight (Figure S4A and S4B). Gene expression analysis of whole eWAT samples from these mice found a significant induction of adipocyte marker/functional genes (for example, Adipoq, Fabp4, and Pnpla2) but no changes in myofibroblast marker genes (for example, Acta2, Tagln, and Col1a1) (Figures S4C-S4D). Adipocyte-specific gene expression analysis using TRAP ribosome-pulldown found not only similar induction of adipocyte marker/functional genes, but also significant repression of myofibroblast marker gene expression (Figure 4A).
Figure 4.
Rosi treatment reverses the molecular and cellular phenotypes in adipocytes associated with HFD. (a) Gene expression analysis of the adipocyte and myofibroblast markers in adipocytes using RNA extracted by TRAP on HFD and HFD with Rosi treatment (8 mg/kg IP for 10 days). Bars indicate mean ± SEM (n = 5 animals/group) (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.005). (b) Whole mount staining showing actin filament formation by phalloidin staining in adipocytes on HFD and HFD with Rosi treatment. Scale bar: 50 μm.
These results suggest that as fibrotic genes are substantially more highly expressed in non-adipocytes than adipocytes, changes in the myofibroblast marker gene expression in adipocytes by Rosi treatment are masked by the high background and undetectable by whole tissue analysis. Adipocyte-specific analysis showed that Rosi acts within adipocytes and can reverse molecular phenotypes of adipocytes. Using phalloidin staining, we again found that the adipocytes on the HFD displayed complex networks of actin filaments, which became fragmented and partially resolved with Rosi treatment (Figure 4B). Taken together, these results suggest that decreased PPARγ expression and binding is a key mechanism underlying much of the aberrant cellular morphology and molecular changes in adipocytes associated with overnutrition.
3.5. TGFβ-SMAD mediates HFD-induced aberrant cellular state changes in adipocytes
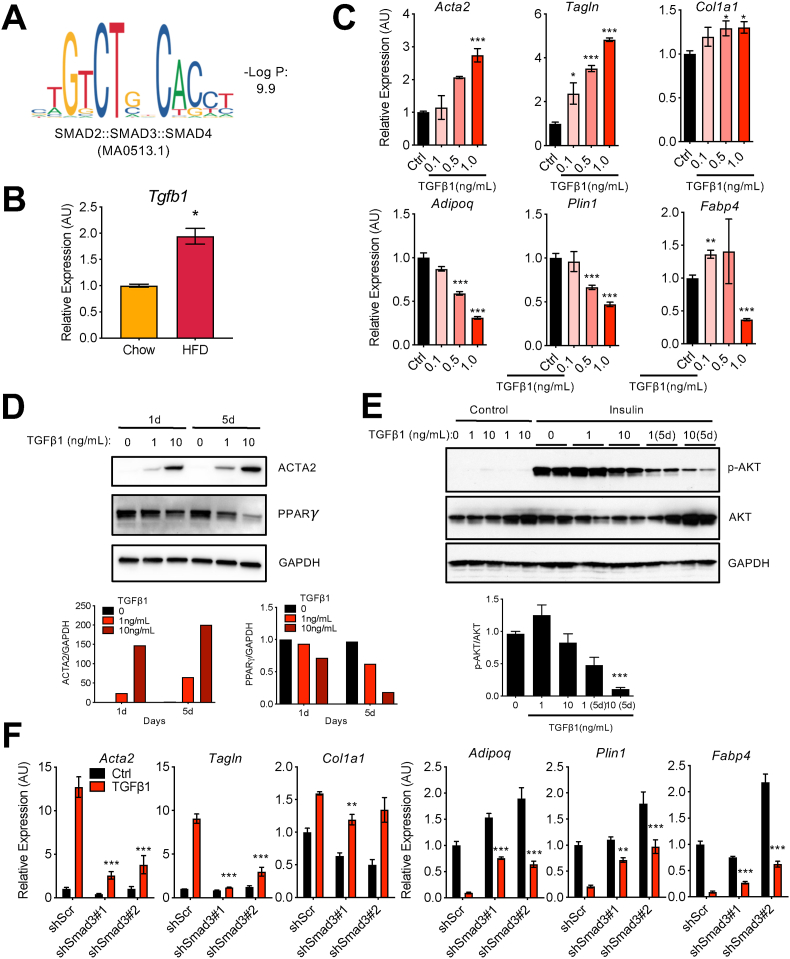
To identify potential transcription factors that account for enhanced gene expression in adipocytes during HFD (for example, Acta2 or Tagln), we looked for transcription factor binding motifs over-represented in H3K27ac peaks that were upregulated by HFD. We found that a motif for the SMAD transcription factor family (SMAD2::SMAD3::SMAD4), canonical transcriptional effectors of the TGFβ signaling pathway, was significantly enriched in HFD-induced H3K27ac peaks (Figure 5A). Consistent with this, the expression of Tgfb1, a ligand that drives the activity of SMADs, was significantly induced in the whole eWAT after HFD feeding [37] (Figure 5B). Together, these data provide a mechanism by which obesity-associated increases in tissue-level TGFβ drives the myofibroblast phenotype in adipocytes via activation of SMADs, which directly regulate a myofibroblast gene program.
Figure 5.
TGFβ-SMAD3 pathway promotes aberrant changes in adipocytes in vitro. (a) SMAD2::SMAD3::SMAD4 motif over-represented in the H3K27ac peaks induced in adipocytes on HFD. (b) Tgfb1 mRNA expression in whole eWAT samples on chow and HFD by qRT-PCR analysis. Bars indicate mean ± SEM (n = 4–6 animals/group) (∗p < 0.05). (c) Gene expression analysis of the adipocyte and myofibroblast markers by qRT-PCR in differentiated 3T3-L1 adipocytes treated with TGFβ1 (1 ng/ml) for 24 h at the indicated concentrations. Bars indicate mean ± SEM (n = 3 replicates). (control vs TGFβ1 treatments; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.005). (d) Protein levels of PPARγ and ACTA2 in differentiated 3T3-L1 adipocytes treated with TGFβ1 treatment for 24 h or 5 days at the indicated concentrations. GAPDH was used as a loading control. Bar graphs represent the quantification of ACTA2 and PPARγ protein levels relative to GAPDH. (e) Insulin signaling measured by analyzing AKT phosphorylation (p-AKT). Differentiated 3T3-L1 adipocytes were treated with TGFβ1 for 24 h or 5 days at the indicated concentrations, serum-starved for 4 h, and then stimulated with 100 nM insulin for 20 min. GAPDH was used as a loading control. Bar graph is the quantification of p-AKT/AKT by ImageJ (∗∗∗p < 0.005). (f) Effects of Smad3 knockdown on the expression of adipocyte and myofibroblast marker genes. Differentiated 3T3-L1 adipocytes were transduced with lentivirus expressing control scrambled shRNA or two different shSmad3 RNA for 4 days and treated with TGFβ1 (ng/mL) for 24 h. Bars indicate mean ± SEM (n = 3 replicates) (control with TGFβ1 vs shSmad3 with TGFβ1; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.005).
We tested this hypothesis in vitro by examining the myofibroblast gene expression program by qPCR in differentiated 3T3-L1 adipocytes exposed to TGFβ1. The “myofibroblast” genes Acta2, Tagln, and Col1a1 were induced by TGFβ1 in a dose-dependent manner, while adipocyte marker genes such as Adipoq, Plin1, and Fabp4 were repressed (Figure 5C). TGFβ1 treatment increased ACTA2 and reduced PPARγ protein levels (Figure 5D). TGFβ1 treatment also inhibited insulin signaling, as shown by reduced AKT phosphorylation upon insulin stimulation, in a dose-dependent manner (Figure 5E), indicating that TGFβ1 treatment caused aberrant changes in cultured in vitro adipocytes, which mimics the dysfunctional changes occurring in adipocytes in vivo.
We treated differentiated 3T3-L1 adipocytes with the TGFβ receptor inhibitor SB-431542 (SB) or Rosi and tested their impacts on gene expression. SB almost completely blunted the induction of myofibroblast marker gene expression (Acta2, Tagln, and Col1a1) and reverted adipocyte marker gene expression (Adipoq, Plin1, and Fabp4) in response to TGFβ1 treatment (Figure S5A and B). Rosi also significantly mitigated the gene expression changes by TGFβ1 (Figure S5A and B). We also knocked down Smad3 using two different small hairpin RNAs (shRNAs) by an ∼80% decrease in mRNA expression (Figure S5C) and found that the knockdown of Smad3 significantly inhibited the gene expression from turning toward myofibroblast-like states (Figure 5F). Taken together, these results suggest that the TGFβ-SMAD pathway drives aberrant cellular state changes in adipocytes during obesity.
4. Discussion
The pathological features of dysfunctional adipose tissue during obesity include aberrant lipid storage, insulin resistance, elevated inflammation, and tissue fibrosis [38]. Much of the attention in this area has focused on the altered cellular composition of the fat pad during overnutrition, primarily regarding the influx of pro-inflammatory immune cells. In fact, our own work corroborates this dramatically by showing that the relative percentage of adipocyte nuclei in the epididymal depot declines from 30% to 10% after HFD feeding. Other changes in cellular abundance have also been proposed to accompany obesity, including the recruitment of pro-fibrotic myofibroblasts from undifferentiated precursor cells, an event believed to drive the development of adipose fibrosis [39,40]. Less well understood, however, are the changes that occur within adipocytes themselves, specifically the transcriptional events that promote metabolic dysfunction. In this study, we used NuTRAP mice as a model to determine transcriptional and epigenomic events that occur within adipocytes under nutritional stress.
Our transcriptional analysis revealed many gene expression changes in mature adipocytes after high-fat feeding, which could be broadly categorized into two major categories. The first group of changes represent reduced expression of many of the genes that define mature adipocytes and underlie their primary functions of lipogenesis, lipolysis, and insulin signaling. The second category of gene expression change was exemplified by the upregulation of genes typically associated with myofibroblast identity and function, including Acta2, Tagln, and various collagen-encoding genes. Taken together, the overall pattern suggests a reprogramming of the cell state away from a metabolically competent mature adipocyte and toward a more pro-inflammatory, pro-fibrotic state more typically associated with myofibroblasts. This partial loss of identity is distinct from de-differentiation, as we do not see a reversion of the gene expression profile to a preadipocyte-like state. An obvious example of this is Lep (which encodes leptin), which is not expressed in preadipocytes but becomes highly induced in obesity.
However, as in de-differentiation, reductions in PPARγ activity in adipocytes occur under nutritional stress. PPARγ is a master regulator of adipose biology, essential not only for adipocyte differentiation, but also for survival of mature adipocytes [4,5]. Many studies have assessed PPARγ levels in adipose tissue during obesity, but the results have been confusing, with increased, decreased, and unchanged adipose PPARγ levels reported [[41], [42], [43], [44]]. These conflicting results likely reflect the fact that the cellular composition of adipose tissue changes in obesity, and so the relative expression of PPARγ in component cell types may dominate. Here we report reduced Pparg mRNA levels in adipocytes after high-fat feeding. Interestingly, the majority of the PPARγ cistrome was unchanged. Of the roughly 1,000 sites that changed after HFD feeding (out of 10,037 total sites), the vast majority were downregulated, and those sites were prominently located at critical genes associated with adipokine secretion, insulin action, and lipid handling. Whether this simply reflected reduced PPARγ protein levels or possibly altered ligand binding after HFD was not clear. Of note, Soccio et al. determined the PPARγ cistrome in whole adipose tissue from chow and HFD-fed mice; they noted that upregulated sites were 2.5 times more common than downregulated sites [8]. In that study, upregulated sites correlated well with known macrophage genes [8]. Overall, these data demonstrate the importance of cell type-specific cistrome determination.
There is a paucity of information regarding the transcriptional pathways that mediate abnormal metabolism in adipocytes from obese subjects. The glucocorticoid receptor (GR) was shown to mediate insulin resistance in cultured adipocytes [45], but surprisingly, mice bearing targeted ablation of GR in adipocytes were not protected from insulin resistance associated with high-fat feeding [38]. Inflammatory transcription factors are also candidates to play a role in adipocyte insulin resistance, although manipulation of NF-κB specifically within adipocytes has not yielded the expected results [46,47]. IRF3 is another pro-inflammatory transcription factor that may promote metabolic dysfunction within adipocytes in high-fat diet conditions [48]. Other transcription factors that drive pathways relevant to insulin sensitivity and resistance in adipocytes include VDR, HIF1α, ChREBP, and FoxO1 [45,[49], [50], [51]].
To identify novel transcriptional pathways in adipose biology, we have made extensive use of motif findings from epigenomic data, especially H3K27ac [16,45,52]. This approach has been used to discover the role of transcription factors in adipogenesis, insulin resistance, and whitening of beige adipocytes [16,45,52]. In the current study, we identified the Smad motif as over-represented in H3K27ac peaks that became induced after HFD. Smads are transcription factors activated by TGF-BMP signaling. Elevated TGFβ1 levels in adipose tissues and serum are associated with increased adiposity in humans and mice [53], while the TGFβ1-Smad3 pathway has negative effects on adipogenesis [54,55]. Smad3 global knockout mice are protected from HFD-induced obesity, hepatic steatosis, and insulin resistance by enhancing energy expenditure through increased thermogenic beige adipocyte formation in subcutaneous fat tissues [56,57], and the administration of TGFβ neutralizing antibodies similarly protects mice from metabolic defects due to HFD [56]. Smad3 and other genes in the TGFβ signaling pathway are also associated with metabolic syndrome, including T2D and cardiovascular disease, in genome-wide association studies [58,59]. However, the direct roles of the TGFβ1-Smad3 pathway in mature adipocytes remains elusive.
In our study, TGFβ1 treatment induced gene expression changes in cultured adipocytes that mimic those in adipocytes from HFD-fed mice. TGFβ1 also reduced insulin signaling in treated adipocytes. The TGFβ signaling pathway is known to promote fibrosis in many tissue types by promoting myofibroblast formation from fibroblasts and other progenitor cells [60]. Our results indicate that TGFβ1 signaling affects not only proliferative cells, but also post-mitotic mature cell types, such as adipocytes, which obtain features of myofibroblasts, leading to the disruption of cellular identity maintenance and functional failure. While treatments designed to inhibit TGFβ signaling have mostly been proposed as cancer or fibrosis therapies [61], adipocyte-specific anti-TGFβ treatment might have beneficial metabolic outcomes by preventing the development of diabetes in obesity through the inhibition of aberrant adipocyte cellular reprogramming. In conclusion, our study demonstrates that aberrant adipocyte cellular reprogramming is a key element of adipose tissue dysfunction during obesity that is driven by reduced PPARγ activity and elevated TGFβ-SMAD signaling.
Adipogenesis is accompanied by substantial morphological change, as spindle-shaped fibroblast-like preadipocytes transition to the typical rounded mature adipocytes. Although most focus has been on the accumulation of lipid droplets during adipogenesis, there is also extensive reorganization of the cytoskeleton, including decreased actin expression and rearrangements to cortical structures [62]. These cytoskeletal changes are not simply passive markers of differentiation, but can also modulate and maintain the developmental process. Thus, the inhibition of actin filament polymerization and actomyosin contraction using chemical inhibitors cytochalasin D and blebbistatin, respectively, induces the formation of a rounded morphology and subsequently promotes adipocyte differentiation [63,64]. Similarly, inhibition of Ras homolog family member A (RhoA) and Rho-associated protein kinases (ROCKs) disrupts actin cytoskeleton structures and leads to increased adipogenesis [64]. Of note, all of these studies used adipocytes cultured in vitro, in which the conditions imposed by monolayer growth can have profound effects on the cytoskeleton. In our study, we observed the actin filament structure in mature adipocytes in vivo to document changes in response to obesity. On the chow diet, minimal actin filament structures were detected in adipocytes by phalloidin staining, but high-fat feeding was accompanied by the appearance of highly complex filamentous structures. We observed this actin filament network structure throughout the adipose tissues and without obvious heterogeneity. Subcutaneous inguinal white adipose tissue (iWAT) displayed similar filamentous actin structures but to a lesser extent (data not shown). Since this whole mount staining method examines a small number of cells, more quantitative high-throughput techniques, such as single cell/nuclei RNA-seq and spatial transcriptomics, may reveal regional and depot-dependent differences, if any. Treatment with Rosi partially reversed these morphological changes. Our results are consistent with a recent study that demonstrated correlations between adipocyte size, actin cytoskeletal complexity, and insulin resistance [65]. These results differ from another study, however, in which it was reported that larger adipocytes with functional declines had reduced actin polymerization [66]. Interestingly, chemical compounds that interfere with actin cytoskeletal dynamics either by stabilizing actin polymerization or inhibiting polymerization have negative impacts on insulin-stimulated glucose uptake in adipocytes, apparently by disrupting glucose transporter type 4 (GLUT4) vesicular trafficking [67,68]. Whether the more extensive changes noted in our study have an impact on adipocyte function or are merely “passenger effects” without significant functional consequences remains to be determined.
Although adipocytes are a terminally differentiated post-mitotic cell type, recent studies indicated that they can alter their cellular identity in response to environmental and physiological cues. For example, beige adipocytes, a type of thermogenic adipocyte, interconvert between white and brown adipocyte identities in response to changes in temperature [16,69]. In the mammary gland, subcutaneous adipocytes de-differentiate and then re-differentiate during the cycle of lactation and involution [70]. This is consistent with the observation that mature adipocytes isolated from tissues can de-differentiate completely back to proliferating preadipocytes after isolation and in vitro ceiling culture [71]. Most relevant to our study, the conversion of dermal adipocytes into myofibroblasts has been noted to follow severe skin injury. Dermal adipocytes adjacent to a skin wound were shown to undergo extensive lipolysis and de-differentiation into myofibroblast-like cells that contribute to deposition of extracellular matrix [72]. A similar phenomenon was studied in a bleomycin exposure model of systemic sclerosis, a disease in which dermal fat is lost and replaced by fibrotic tissue. In those experiments, fate mapping of adiponectin + cells after bleomycin showed loss of adipocytes with the subsequent appearance of labeled myofibroblasts [73]. Interestingly, skin biopsies from patients with systemic sclerosis show evidence of enhanced nuclear corepressor (NCoR) activity, and mice lacking NCoR in adipocytes are protected from skin fibrosis [74]. NCoR is a repressor of many nuclear receptors, including PPARγ [75]. Other studies also indicated a high degree of plasticity among dermal adipocytes [76]. Our studies of visceral adipocytes do not fully recapitulate these observations in dermal adipocytes. Most importantly, we do not see evidence of de-differentiation in visceral adipocytes in obesity; the cytoskeletal changes that we observe occur in cells that are clearly mature adipocytes. The gene expression pattern includes reduced adipocyte marker genes, but they are attenuated, not ablated, and the overall profile differs from preadipocytes. Unlike dermal adipocytes, which undergo a pattern of de-differentiation and subsequent re-differentiation along a myofibroblast lineage, we propose that visceral adipocytes undergo a partial trans-differentiation to a myofibroblast-like state, involving conversion of cellular identity without intervening de-differentiation.
Adipocytes are not the only cells that undergo loss of identity in response to nutritional stress. Some pancreatic β cells, for example, partially de-differentiate to a progenitor-like state during T2D, with reduced insulin expression and gain of other hormone (for example, glucagon) expression [77]. Cell plasticity is also implicated in non-nutritional physiological responses. During chronic periportal liver injury, hepatocytes de-differentiate into liver progenitor-like cells simultaneously expressing markers for hepatocytes and biliary epithelial cells and regenerate hepatocytes and duct-like cells [78]. These examples suggest that alterations in cellular identity may play an important role in physiological and pathological responses implicated in a broad range of human diseases.
Author contributions
The experiments were designed by H.C.R., L.T.-Y.T., and E.D.R. and executed by H.C.R., M.K., and D.T. The computational analysis was conducted by H.C.R., A.L., and C.J. The manuscript was written by H.C.R. and E.D.R. with input from all of the other authors.
Acknowledgments
We thank Jessica Hall for help with the rosiglitazone injection experiments and all members of the Rosen lab for their helpful advice and discussion. We are grateful to the Flow Cytometry Core, Histology Core, and Molecular Medicine Core at Beth Israel Deaconess Medical Center as well as the Functional Genomics and Bioinformatics Core at the Boston Nutrition and Obesity Research Center (BNORC). This study was supported by an American Diabetes Association Postdoctoral Fellowship 1-17-PDF-133, BNORC Pilot and Feasibility grant (P30DK046200), Boston Area Diabetes Endocrinology Research Center (P30DK057521) Pilot and Feasibility grant to H.C.R., a Department of Defense grant W81XWH-14-PRMRP-DA to L.T.Y.T., and NIH R01 DK102173, DK102170, DK113669, and DK085171 to E.D.R.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101086.
Contributor Information
Hyun Cheol Roh, Email: hyunroh@iu.edu.
Evan D. Rosen, Email: erosen@bidmc.harvard.edu.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.World Health Organization Obesity and overweight. https://www.who.int/en/news-room/fact-sheets/detail/obesity-and-overweight n.d.
- 2.Pi-Sunyer F.X. The medical risks of obesity. Obesity Surgery. 2002;12(1):S6–S11. doi: 10.1007/BF03342140. [DOI] [PubMed] [Google Scholar]
- 3.Kusminski C.M., Bickel P.E., Scherer P.E. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nature Reviews Drug Discovery. 2016;15(9):639–660. doi: 10.1038/nrd.2016.75. [DOI] [PubMed] [Google Scholar]
- 4.Tontonoz P., Spiegelman B.M. Fat and beyond: the diverse biology of PPARgamma. Annual Review of Biochemistry. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 5.Siersbæk R., Nielsen R., Mandrup S. PPARγ in adipocyte differentiation and metabolism - novel insights from genome-wide studies. FEBS Letters. 2010;584(15):3242–3249. doi: 10.1016/j.febslet.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Tamori Y., Masugi J., Nishino N., Kasuga M. Role of peroxisome proliferator-activated receptor-γ in maintenance of the characteristics of mature 3T3-L1 adipocytes. Diabetes. 2002;51(7):2045–2055. doi: 10.2337/diabetes.51.7.2045. [DOI] [PubMed] [Google Scholar]
- 7.Wang F., Mullican S.E., DiSpirito J.R., Peed L.C., Lazar M.A. Lipoatrophy and severe metabolic disturbance in mice with fat-specific deletion of PPARγ. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(46):18656–18661. doi: 10.1073/pnas.1314863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soccio R.E., Li Z., Chen E.R., Foong Y.H., Benson K.K., Dispirito J.R. Targeting PPARγ in the epigenome rescues genetic metabolic defects in mice. Journal of Clinical Investigation. 2017;127(4):1–12. doi: 10.1172/JCI91211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soccio R.E., Chen E.R., Lazar M.A. Thiazolidinediones and the promise of insulin sensitization in type 2 diabetes. Cell Metabolism. 2014;20(4):573–591. doi: 10.1016/j.cmet.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynes M.D., Tseng Y.H. Deciphering adipose tissue heterogeneity. Annals of the New York Academy of Sciences. 2018;1411(1):5–20. doi: 10.1111/nyas.13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roh H.C., Tsai L.T.Y., Lyubetskaya A., Tenen D., Kumari M., Rosen E.D. Simultaneous transcriptional and epigenomic profiling from specific cell types within heterogeneous tissues in vivo. Cell Reports. 2017;18(4):1048–1061. doi: 10.1016/j.celrep.2016.12.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reilly S.M., Saltiel A.R. Adapting to obesity with adipose tissue inflammation. Nature Reviews Endocrinology. 2017;13(11):633–643. doi: 10.1038/nrendo.2017.90. [DOI] [PubMed] [Google Scholar]
- 13.Rodbell M. Metabolism of isolated fat cells. I. Effects of hormones on glucose metabolism and lipolysis. Journal of Biological Chemistry. 1964;239:375–380. [PubMed] [Google Scholar]
- 14.Ebke L.A., Nestor-Kalinoski A.L., Slotterbeck B.D., Al-Dieri A.G., Ghosh-Lester S., Russo L. Tight association between macrophages and adipocytes in obesity: implications for adipocyte preparation. Obesity. 2014;22(5):1246–1255. doi: 10.1002/oby.20634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruan H., Zarnowski M.J., Cushman S.W., Lodish H.F. Standard isolation of primary adipose cells from mouse epididymal fat pads induces inflammatory mediators and down-regulates adipocyte genes. Journal of Biological Chemistry. 2003;278(48):47585–47593. doi: 10.1074/jbc.M305257200. [DOI] [PubMed] [Google Scholar]
- 16.Roh H.C., Tsai L.T.Y., Shao M., Tenen D., Shen Y., Kumari M. Warming induces significant reprogramming of beige, but not Brown, adipocyte cellular identity. Cell Metabolism. 2018;27(5):1121–1137. doi: 10.1016/j.cmet.2018.03.005. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eguchi J., Wang X., Yu S., Kershaw E.E., Chiu P.C., Dushay J. Transcriptional control of adipose lipid handling by IRF4. Cell Metabolism. 2011;13(3):249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao Y., Smyth G.K., Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 20.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications. 2019;10(1) doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E. Model-based analysis of ChIP-seq (MACS) Genome Biology. 2008;9(9) doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou X., Wang T. Using the wash U epigenome browser to examine genome-wide sequencing data. Current Protocols in Bioinformatics. 2012;40(1):10.10.1–10.10.14. doi: 10.1002/0471250953.bi1010s40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26(17):2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Research. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grant C.E., Bailey T.L., Noble W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Research. 2014;42(D1):142–147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Q.a., Tao C., Gupta R.K., Scherer P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nature Medicine. 2013;19(10):1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun K., Kusminski C.M., Scherer P.E. Adipose tissue remodeling and obesity. Journal of Clinical Investigation. 2011;121(6):2094–2101. doi: 10.1172/JCI45887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heiman M., Schaefer A., Gong S., Peterson J.D., Day M., Ramsey K.E. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135(4):738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wernstedt Asterholm I., Tao C., Morley T.S., Wang Q.A., Delgado-Lopez F., Wang Z.V. Adipocyte inflammation is essential for healthy adipose tissue expansion and remodeling. Cell Metabolism. 2014;20(1):103–118. doi: 10.1016/j.cmet.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye R., Wang Q.A., Tao C., Vishvanath L., Shao M., McDonald J.G. Impact of tamoxifen on adipocyte lineage tracing: inducer of adipogenesis and prolonged nuclear translocation of Cre recombinase. Molecular Metabolism. 2015;4(11):771–778. doi: 10.1016/j.molmet.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samad F., Yamamoto K., Pandey M., Loskutoff D.J. Elevated expression of transforming growth factor-beta in adipose tissue from obese mice. Molecular Medicine (Cambridge, Mass.) 1997;3(1):37–48. [PMC free article] [PubMed] [Google Scholar]
- 38.Shen Y., Roh H.C., Kumari M., Rosen E.D. Adipocyte glucocorticoid receptor is important in lipolysis and insulin resistance due to exogenous steroids, but not insulin resistance caused by high fat feeding. Molecular Metabolism. 2017 doi: 10.1016/j.molmet.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marcelin G., Ferreira A., Liu Y., Atlan M., Aron-Wisnewsky J., Pelloux V. A PDGFRα-mediated switch toward CD9high adipocyte progenitors controls obesity-induced adipose tissue fibrosis. Cell Metabolism. 2017;25(3):673–685. doi: 10.1016/j.cmet.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 40.Datta R., Podolsky M.J., Atabai K., Datta R., Podolsky M.J., Atabai K. Fat fibrosis: friend or foe? Find the latest version. JCI Insight. 2018;3(19) doi: 10.1172/jci.insight.122289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vidal-Puig A.J., Considine R.V., Jimenez-Liñan M., Werman A., Pories W.J., Caro J.F. Peroxisome proliferator-activated receptor gene expression in human tissues. Effects of obesity, weight loss, and regulation by insulin and glucocorticoids. Journal of Clinical Investigation. 1997;99(10):2416–2422. doi: 10.1172/JCI119424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Auboeuf D., Rieusset J., Fajas L., Vallier P., Frering V., Riou J.P. Tissue distribution and quantification of the expression of mRNAs of peroxisome proliferator–activated receptors and liver X receptor-α in humans: No alteration in adipose tissue of obese and NIDDM patients. Diabetes. 1997;46(8):1319–1327. doi: 10.2337/diab.46.8.1319. [DOI] [PubMed] [Google Scholar]
- 43.Rieusset J., Andreelli F., Auboeuf D., Roques M., Vallier P., Riou J.P. Insulin acutely regulates the expression of the peroxisome proliferator-activated receptor-gamma in human adipocytes. Diabetes. 1999;48(4):699–705. doi: 10.2337/diabetes.48.4.699. [DOI] [PubMed] [Google Scholar]
- 44.Giusti V., Verdumo C., Suter M., Gaillard R.C., Burckhardt P., Pralong F. Expression of peroxisome proliferator – activated tissue of obese women. Diabetes. 2003;52(July):1673–1676. doi: 10.2337/diabetes.52.7.1673. [DOI] [PubMed] [Google Scholar]
- 45.Kang S., Tsai L.T., Zhou Y., Evertts A., Xu S., Griffin M.J. Identification of nuclear hormone receptor pathways causing insulin resistance by transcriptional and epigenomic analysis. Nature Cell Biology. 2015;17(1) doi: 10.1038/ncb3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang T., Zhang J., Yin J., Staszkiewicz J., Gawronska-Kozak B., Jung D.Y. Uncoupling of inflammation and insulin resistance by NF-κB in transgenic mice through elevated energy expenditure. Journal of Biological Chemistry. 2010;285(7):4637–4644. doi: 10.1074/jbc.M109.068007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao Z., Zhang J., Henagan T.M., Lee J.H., Ye X., Wang H. P65 inactivation in adipocytes and macrophages attenuates adipose inflammatory response in lean but not in obese mice. American Journal of Physiology - Endocrinology and Metabolism. 2015;308(6):E496–E505. doi: 10.1152/ajpendo.00532.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumari M., Wang X., Lantier L., Lyubetskaya A., Eguchi J., Kang S. IRF3 promotes adipose inflammation and insulin resistance and represses browning. Journal of Clinical Investigation. 2016;126(8):2839–2854. doi: 10.1172/JCI86080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakae J., Cao Y., Oki M., Orba Y., Sawa H., Kiyonari H. Forkhead transcription factor FoxO1 in adipose tissue regulates energy storage and expenditure. Diabetes. 2008;57(3):563–576. doi: 10.2337/db07-0698. [DOI] [PubMed] [Google Scholar]
- 50.Lee Y.S., Kim J.W., Osborne O., Oh D.Y., Sasik R., Schenk S. Increased adipocyte O2 consumption triggers HIF-1α, causing inflammation and insulin resistance in obesity. Cell. 2014;157(6):1339–1352. doi: 10.1016/j.cell.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vijayakumar A., Aryal P., Wen J., Syed I., Vazirani R.P., Moraes-Vieira P.M. Absence of carbohydrate response element binding protein in adipocytes causes systemic insulin resistance and impairs glucose transport. Cell Reports. 2017;21(4):1021–1035. doi: 10.1016/j.celrep.2017.09.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mikkelsen T.S., Xu Z., Zhang X., Wang L., Gimble J.M., Lander E.S. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143(1):156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alessi M.C., Bastelica D., Morange P., Berthet B., Leduc I., Verdier M. Plasminogen activator inhibitor 1, transforming growth factor-β1, and BMI are closely associated in human adipose tissue during morbid obesity. Diabetes. 2000;49(8):1374–1380. doi: 10.2337/diabetes.49.8.1374. [DOI] [PubMed] [Google Scholar]
- 54.Tsurutani Y., Fujimoto M., Takemoto M., Irisuna H., Koshizaka M., Onishi S. The roles of transforming growth factor-β and Smad3 signaling in adipocyte differentiation and obesity. Biochemical and Biophysical Research Communications. 2011;407(1):68–73. doi: 10.1016/j.bbrc.2011.02.106. [DOI] [PubMed] [Google Scholar]
- 55.Choy L., Derynck R. Transforming growth factor-β inhibits adipocyte differentiation by Smad3 interacting with CCAAT/enhancer-binding protein (C/EBP) and repressing C/EBP transactivation function. Journal of Biological Chemistry. 2003;278(11):9609–9619. doi: 10.1074/jbc.M212259200. [DOI] [PubMed] [Google Scholar]
- 56.Yadav H., Quijano C., Kamaraju A.K., Gavrilova O., Malek R., Chen W. Protection from obesity and diabetes by blockade of TGF-??/Smad3 signaling. Cell Metabolism. 2011;14(1):67–79. doi: 10.1016/j.cmet.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan C.K., Leuenberger N., Tan M.J., Yan Y.W., Chen Y., Kambadur R. Smad3 deficiency in mice protects against insulin resistance and obesity induced by a high-fat diet. Diabetes. 2011;60(2):464–476. doi: 10.2337/db10-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin E., Kuo P.H., Liu Y.L., Yang A.C., Tsai S.J. Transforming growth factor-β signaling pathway-associated genes SMAD2 and TGFBR2 are implicated in metabolic syndrome in a Taiwanese population. Scientific Reports. 2017;7(1):1–8. doi: 10.1038/s41598-017-14025-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perry J.R.B., McCarthy M.I., Hattersley A.T., Zeggini E., Weedon M.N., Frayling T.M. Interrogating type 2 diabetes genome-wide association data using a biological pathway-based approach. Diabetes. 2009;58(6):1463–1467. doi: 10.2337/db08-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evans R.A., Tian Y.C., Steadman R., Phillips A.O. TGF-β1-mediated fibroblast–myofibroblast terminal differentiation—the role of smad proteins. Experimental Cell Research. 2003;282(2):90–100. doi: 10.1016/S0014-4827(02)00015-0. [DOI] [PubMed] [Google Scholar]
- 61.Akhurst R.J., Hata A. Targeting the TGFβ signalling pathway in disease. Nature Reviews Drug Discovery. 2012;11(10):790–811. doi: 10.1038/nrd3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang W., Thein S., Wang X., Bi X., Ericksen R.E., Xu F. BSCL2/seipin regulates adipogenesis through actin cytoskeleton. Remodelling. 2014;23(2):502–513. doi: 10.1093/hmg/ddt444. [DOI] [PubMed] [Google Scholar]
- 63.Schiller Z.A., Schiele N.R., Sims J.K., Lee K., Kuo C.K. Adipogenesis of adipose-derived stem cells may be regulated via the cytoskeleton at physiological oxygen levels in vitro. Stem Cell Research & Therapy. 2013;4(4) doi: 10.1186/scrt230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McBeath R., Pirone D.M., Nelson C.M., Bhadriraju K., Chen C.S. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Developmental Cell. 2004;6(4):483–495. doi: 10.1016/S1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 65.Hansson B., Morén B., Fryklund C., Vliex L., Wasserstrom S., Albinsson S. Adipose cell size changes are associated with a drastic actin remodeling. Scientific Reports. 2019;9(1):1–14. doi: 10.1038/s41598-019-49418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim J.I., Park J., Ji Y., Jo K., Han S.M., Sohn J.H. During adipocyte remodeling, lipid droplet configurations regulate insulin sensitivity through F-actin and G-actin reorganization. Molecular and Cellular Biology. 2019;39(20):e00210–e00219. doi: 10.1128/MCB.00210-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yonashiro R., Sugiura A., Miyachi M., Fukuda T., Matsushita N., Inatome R. Mutant SOD1 and attenuates mutant SOD1-induced reactive oxygen species generation. Molecular Biology of the Cell. 2009;20:4524–4530. doi: 10.1091/mbc.E09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kanzaki M., Pessin J.E. Insulin-stimulated GLUT4 translocation in adipocytes is dependent upon cortical actin remodeling. Journal of Biological Chemistry. 2001;276(45):42436–42444. doi: 10.1074/jbc.M108297200. [DOI] [PubMed] [Google Scholar]
- 69.Rosenwald M., Perdikari A., Rülicke T., Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nature Cell Biology. 2013;15(5):1–11. doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- 70.Wang Q.A., Song A., Chen W., Schwalie P.C., Zhang F., Vishvanath L. Reversible de-differentiation of mature white adipocytes into preadipocyte-like precursors during lactation. Cell Metabolism. 2018:1–7. doi: 10.1016/j.cmet.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsumoto T., Kano K., Kondo D., Fukuda N., Iribe Y., Tanaka N. Mature adipocyte-derived de-differentiated fat cells exhibit multilineage potential. Journal of Cellular Physiology. 2008;215(1):210–222. doi: 10.1002/jcp.21304. [DOI] [PubMed] [Google Scholar]
- 72.Shook B.A., Wasko R.R., Mano O., Rutenberg-Schoenberg M., Rudolph M.C., Zirak B. Dermal adipocyte lipolysis and myofibroblast conversion are required for efficient skin repair. Cell Stem Cell. 2020:1–16. doi: 10.1016/j.stem.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marangoni R.G., Korman B.D., Wei J., Wood T.A., Graham L.V., Whitfield M.L. Myofibroblasts in murine cutaneous fibrosis originate from adiponectin-positive intradermal progenitors. Arthritis and Rheumatology. 2015;67(4):1062–1073. doi: 10.1002/art.38990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Korman B., Marangoni R.G., Lord G., Olefsky J., Tourtellotte W., Varga J. Adipocyte-specific repression of PPAR-gamma by NCoR contributes to scleroderma skin fibrosis. Arthritis Research and Therapy. 2018;20(1):1–11. doi: 10.1186/s13075-018-1630-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li P., Fan W., Xu J., Lu M., Yamamoto H., Auwerx J. Adipocyte NCoR knockout decreases PPARγ phosphorylation and enhances PPARγ activity and insulin sensitivity. Cell. 2011;147(4):815–826. doi: 10.1016/j.cell.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Z., Gupta R.K., Scherer P.E. Dermal adipose tissue has high plasticity and undergoes reversible de-differentiation in mice. Journal of Clinical Investigation. 2019;129(12):5327–5342. doi: 10.1172/JCI130239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Talchai C., Xuan S., Lin H.V., Sussel L., Accili D. Pancreatic β cell de-differentiation as a mechanism of diabetic β cell failure. Cell. 2012;150(6):1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li W., Li L., Hui L. Cell plasticity in liver regeneration. Trends in Cell Biology. 2020;30(4):329–338. doi: 10.1016/j.tcb.2020.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and processed data reported in this paper are available at the Gene Expression Omnibus (GEO) repository with accession number GSE153120. Among the data, the RNA-seq data from the chow-fed NuTRAP mice were previously deposited (GSE92590) [11] and reanalyzed in this study.