Abstract
This study investigated genomic differences in Australian and Indian Pseudomonas aeruginosa isolates from keratitis (infection of the cornea). Overall, the Indian isolates were resistant to more antibiotics, with some of those isolates being multi-drug resistant. Acquired genes were related to resistance to fluoroquinolones, aminoglycosides, beta-lactams, macrolides, sulphonamides, and tetracycline and were more frequent in Indian (96%) than in Australian (35%) isolates (p = 0.02). Indian isolates had large numbers of gene variations (median 50,006, IQR = 26,967–50,600) compared to Australian isolates (median 26,317, IQR = 25,681–33,780). There were a larger number of mutations in the mutL and uvrD genes associated with the mismatch repair (MMR) system in Indian isolates, which may result in strains losing their efficacy for DNA repair. The number of gene variations were greater in isolates carrying MMR system genes or exoU. In the phylogenetic division, the number of core genes were similar in both groups, but Indian isolates had larger numbers of pan genes (median 6518, IQR = 6040–6935). Clones related to three different sequence types—ST308, ST316, and ST491—were found among Indian isolates. Only one clone, ST233, containing two strains was present in Australian isolates. The most striking differences between Australian and Indian isolates were carriage of exoU (that encodes a cytolytic phospholipase) in Indian isolates and exoS (that encodes for GTPase activator activity) in Australian isolates, large number of acquired resistance genes, greater changes to MMR genes, and a larger pan genome as well as increased overall genetic variation in the Indian isolates.
Keywords: antibiotic susceptibility, WGS, phylogenetic analysis, DNA mismatch repair system
1. Introduction
Pseudomonas aeruginosa is a ubiquitous bacterium which can cause opportunistic or nosocomial infections in immuno-compromised patients [1]. P. aeruginosa commonly causes corneal (keratitis) [2], respiratory, burn and wound infections, and infections related to medical or surgical devices including ventilator-associated pneumonia [3,4]. P. aeruginosa corneal infections are usually related to contact lens wear, but other risk factors for keratitis in non-contact lens wearers include ocular trauma, ocular surgery, and prior ocular surface disease [5,6,7,8].
The prevalence of multi-drug resistant (MDR) or extensively drug resistant strains of P. aeruginosa reduces treatment options, significantly increasing morbidity rates [9]. P. aeruginosa is naturally resistant to some antibiotics due to the possession of specific resistance genes such as catB that confers chloramphenicol resistance and an inducible ampC which encodes for a β-lactamase that hydrolyses cephalothin and ampicillin, conferring resistance to β-lactams [10]. Additionally, the regulation of efflux pumps also contributes towards an elevated resistance to antibiotics [11]. For example, expression of the efflux pump MexAB-OprM contributes towards intrinsic resistance to a broad spectrum of antibiotics [12], whereas the efflux pump MexXY-OprM is involved in the adaptive resistance to aminoglycosides [13]. Other resistance mechanisms in P. aeruginosa include the acquisition of transferrable resistance determinants, including those associated with transposons and integrons [14]. Antibiotic resistance of P. aeruginosa varies according to the region where the strains have been isolated [15,16] presumably due to the prescribing practices, availability of antibiotics, and perhaps their use in animal husbandry. Various epidemiological studies have identified MDR P. aeruginosa from different infections and these isolates have acquired different resistance characteristics. For example, aminoglycoside resistance [17] and ciprofloxacin persistence [18] are found in cystic fibrosis isolates of P. aeruginosa. Some of these MDR strains are clonal and such clonal strains are often the predominant global clinical MDR isolates [19] which spread resistance characteristics into the wider population which enables clonal lineages to expand with time.
ExoU has been associated with virulence of P. aeruginosa at the ocular surface. ExoU is a phospholipase that causes mammalian cell death [20] and exoU possession is common in strains isolated from ocular infections [21]. There is a correlation between carriage of exoU and elevated resistance to fluoroquinolones and aminoglycosides [22]. ExoU is carried by strains on a genomic island that also contains resistance genes for a range of antibiotics [23].
In addition to the acquisition of resistance genes, bacteria can develop resistance through mutation of genes so that antibiotic targets are modified. Mutation rates are elevated in strains that carry mutations in DNA mismatch repair (MMR) systems and hence such mutator strains will normally carry more mutations than non-mutator strains [24]. In P. aeruginosa, the MMR system is composed of mutS, mutL, and UvrD genes [25]. Strains of P. aeruginosa isolated from the lungs of cystic fibrosis patients have alterations in the DNA MMR system and this has been correlated with multiple antimicrobial resistance [23].
In Australia, there is a tight regulation of prescribing antibiotics, and antibiotics can only be obtained legally with a prescription from a qualified healthcare professional according to the Therapeutic Goods Act 1989. In India, on the other hand, whilst branded antibiotics exist, other forms such as counterfeit, substandard, and ‘spurious’ antibiotics have been reported [26], making surveillance and regulation difficult [27]. While the antibiotic consumption per person in Australia and India in 2010 was approximately similar, there was a more rapid increase between 2000 and 2010 in India [28]. These differences may affect antibiotic resistance development.
The aim of the current study was to compare the phenotypic resistance and genetic characteristics associated with resistance between strains isolated from Australia and India to better understand the underlying factors that may lead to an increased resistance in P. aeruginosa strains associated with ocular infection.
2. Results
2.1. Antibiotic Susceptibility
The minimum inhibitory concentrations (MICs) and minimum bactericidal concentrations (MBCs) of the P. aeruginosa isolates were determined (Table 1). Strains showing intermediate resistance (I) as well as full resistance to antibiotics were categorized as resistant (R) for subsequent analyses. Based on the Centers for Disease Control and Prevention’s (CDC, Atlanta, GA, USA) definition of multi-drug resistance as “an isolate that is resistant to at least one antibiotic in three or more drug classes”, isolates 198, 202, 216, 217, 218, 219, 220, and 221 were deemed to be multi-drug resistant. Australian isolates 223, 224, 225, 227, 233, and 235 were also resistant to three antibiotics but these antibiotics were not of different classes. Isolates 176, 193, and 206 were sensitive to all antibiotics, but all other isolates were resistant to at least one antibiotic. Overall, Indian isolates were more resistant to antibiotics compared to Australian isolates. Among Australian isolates (n = 14), resistance was 78% for imipenem, 57% for ceftazidime, 50% for ciprofloxacin, 21% for piperacillin, 14% for levofloxacin, 7% for tobramycin, and no isolates were resistant to gentamicin or polymyxin. In contrast, resistance in Indian isolates (n = 12) was 75% for ciprofloxacin, 58% for imipenem, 50% for levofloxacin, tobramycin, and ceftazidime, 41% for piperacillin, 40% for gentamicin, and 25% for polymyxin.
Table 1.
Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of antibiotics to Pseudomonas aeruginosa keratitis isolates.
| Strain Number | Fluoroquinolones * | Aminoglycosides | β-Lactams | Poly-Peptide | ||||
|---|---|---|---|---|---|---|---|---|
| 2nd Generation | 3rd Generation | Penicillin 4th Generation | Carba-Penem | Cephalosporin 3rd Generation | ||||
|
Cipro µg/mL ≤1, 2, ≥4 # |
Levo µg/mL ≤2, 4, ≥8 |
Genta µg/mL ≤4, 8, ≥16 |
Tobra µg/mL ≤4, 8, ≥16 |
Pipera µg/mL ≤16 |
Imi µg/mL ≤2, 4, ≥8 |
Ceftaz µg/mL ≤8, 16, ≥32 |
PMB µg/mL ≤2, 4, ≥8 |
|
| MIC/ MBC |
MIC/ MBC |
MIC/ MBC |
MIC/ MBC |
MIC/ MBC |
MIC/ MBC |
MIC/ MBC |
MIC/ MBC |
|
| 123 | 1/1 | 1/1 | 0.25/0.5 | 4/4 | 8/16 | 4(I)/8 | 2/2 | 1280(R)/1280 |
| 126 | 0.5/1 | 0.5/1 | 0.5/1 | 0.25/0.5 | 8/16 | 8(R)/16 | 128(R)/256 | 1/1 |
| 127 | 1/2 | 0.25/1 | 2/4 | 32(R)/128 | 4/16 | 4(I)/8 | 128(R)/256 | 0.5/1 |
| 162 | 0.5/1 | 0.5/1 | 0.25/0.5 | 0.25/1 | 8/8 | 4(I)/4 | 2/4 | 0.25/0.5 |
| 169 | 2(I)/4 | 0.25/0.5 | 0.25/0.5 | 0.25/0.5 | 4/8 | 2/4 | 1/2 | 0.25/0.25 |
| 176 | 0.5/1 | 0.25/0.5 | 0.25/0.5 | 0.25/0.5 | 4/8 | 2/8 | 2/4 | 0.25/0.5 |
| 181 | 1/4 | 0.25/0.5 | 0.25/0.5 | 0.25/0.5 | 32(R)/64 | 4(I)/8 | 16(I)/32 | 0.5/1 |
| 182 | 1/2 | 0.25/0.5 | 0.25/0.5 | 0.25/0.5 | 4/8 | 8(R)/16 | 1/2 | 0.25/0.5 |
| 223 | 64(R)/128 | 1/2 | 0.5/1 | 0.5/1 | 160(R)/320 | 1/2 | 16(I)/32 | 2/4 |
| 224 | 16(R)/32 | 1/2 | 0.25/0.5 | 0.25/0.5 | 8/16 | 64(R)/128 | 16(I)/32 | 1/2 |
| 225 | 64(R)/128 | 16(R)/32 | 0.5/2 | 1/2 | 16/32 | 64(R)/128 | 8/16 | 0.25/0.5 |
| 227 | 64(R)/128 | 64(R)/128 | 0.5/1 | 0.25/1 | 16/32 | 16(R)/32 | 16(I)/32 | 0.25/0.5 |
| 233 | 8(R)/16 | 1/2 | 1/2 | 105/1 | 16/32 | 4(I)/8 | 160(I)/320 | 0.5/1 |
| 235 | 16(R)/32 | 0.5/1 | 2/4 | 0.5/1 | 64(R)/128 | 4(I)/8 | 64(I)/128 | 0.25 |
| 188 | 2(I)/4 | 1/2 | 0.5/1 | 32(R)/64 | 16/65 | 0.5/1 | 4/8 | 2/4 |
| 189 | 0.25/1 | 1/2 | 0.25/0.5 | 16(R)/32 | 4/8 | 2/1 | 8/16 | 2/4 |
| 193 | 1/1 | 0.25/1 | 0.25/0.25 | 0.25/0.5 | 4/8 | 2/4 | 2/2 | 0.5/1 |
| 198 | 1280(R)/2560 | 320(R)/1280 | 2560(R)/5120 | 16(R)/16 | 8/8 | 1/2 | 8/8 | 4(I)/4 |
| 202 | 640(R)/1280 | 320(R)/640 | 8(I)/32 | 320(R)/640 | 16/64 | 8(R)/32 | 8/32 | 0.25/0.25 |
| 206 | 1/1 | 0.5/0.5 | 1/1 | 0.25/0.5 | 8/8 | 2/4 | 2/4 | 0.25/0.5 |
| 216 | 64(R)/128 | 4 (I)/8 | 1/2 | 0.5/2 | 160(R)/320 | 16(R)/32 | 64(R)/128 | 64(R)/128 |
| 217 | 64(R)/128 | 32(R)/64 | 1/2 | 1/2 | 64(R)/128 | 8(R)/16 | 32(R)/64 | 0.25/1 |
| 218 | 8(R)/16 | 1/2 | 0.5/1 | 0.5/1 | 160(R)/320 | 8(R)/16 | 64(R)/128 | 1/4 |
| 219 | ≥5120(R)/≥5120 | 640(R)/1280 | ≥5120(R)/≥5120 | 1280(R)/2560 | 2560(R)/5120 | 40(R)/80 | 16(I)/32 | 0.25/1 |
| 220 | 2(I)/4 | 0.25/0.5 | 0.5/1 | 0.5/1 | 0.25/0.5 | 8(R)/16 | 160(R)/320 | 8(R)/16 |
| 221 | 2560(R)/5120 | 2560(R)/5120 | 2560(R)/5120 | 2560(R)/5120 | 64(R)/128 | 16(R)/32 | 32(R)/64 | 0.25/1 |
Data for Australian isolates (shaded in gray). Data for 123–182 is from a previously published study [29]. Strains 188–221 were Indian keratitis isolates. R = resistant, I = intermediate resistance. * Cipro = Ciprofloxacin, Levo = Levofloxacin, Genta = Gentamicin, Tobra = Tobramycin, Pipera = Piperacillin, Imi = Imipenem, Ceftaz = Ceftazidime, PMB = Polymyxin B; # = Antibiotic breakpoints for sensitive, intermediate, resistant classifications.
2.2. General Features of the Genomes
The isolates after de novo assembly consisted of different numbers of contigs ranging from 50 for isolate 169 to 1917 for isolate 216. The average number of coding sequences was 6162 ± 359.2 for the Australian isolates and 6544 ± 889 for the Indian isolates. Isolates had an average of 66.1% G + C content. The tRNA copy number for the isolates ranged from 57 to 86 (which may vary between studies that use different assembly methods). The general features of the isolates are provided in Supplementary Table S1.
2.3. Acquired Resistance Genes
P. aeruginosa isolates were examined for horizontally acquired antibiotic resistance genes (Table 2) using the Resfinder database. Altogether, 33 different acquired antibiotic resistance genes for various classes of antibiotics including aminoglycosides, fluoroquinolones, beta-lactams were found in these isolates (Table 2).
Table 2.
Acquired resistance genes in P. aeruginosa isolates from India and Australia.
| Genes | Australian Isolates | Indian Isolates | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 123 | 126 | 127 | 162 | 169 | 176 | 181 | 182 | 223 | 224 | 225 | 227 | 233 | 235 | 188 | 189 | 193 | 198 | 202 | 206 | 216 | 217 | 218 | 219 | 220 | 221 | |
| Aminoglycoside resistance genes | ||||||||||||||||||||||||||
| aph(3’)-IIb | ||||||||||||||||||||||||||
| aph(6)-Id | l; | |||||||||||||||||||||||||
| rmtD2 | ||||||||||||||||||||||||||
| rmtB | ||||||||||||||||||||||||||
| aph(3’)-VI | ||||||||||||||||||||||||||
| aph(3’)-lIb | ||||||||||||||||||||||||||
| aph(3’’)-Ib | ||||||||||||||||||||||||||
| aac(6’)-Ib3 | ||||||||||||||||||||||||||
| aac(3)-IId | ||||||||||||||||||||||||||
| aadA1 | ||||||||||||||||||||||||||
| aac(6’)-Ib-cr | ||||||||||||||||||||||||||
| Fluoroquinolone resistance genes | ||||||||||||||||||||||||||
| crpP | ||||||||||||||||||||||||||
| qnrVC1 | ||||||||||||||||||||||||||
| Beta-lactamase resistance genes | ||||||||||||||||||||||||||
| blaPAO | ||||||||||||||||||||||||||
| blaLCR-1 | ||||||||||||||||||||||||||
| blaOXA-485 | ||||||||||||||||||||||||||
| blaOXA-486 | ||||||||||||||||||||||||||
| blaOXA-488 | ||||||||||||||||||||||||||
| blaOXA-396 | ||||||||||||||||||||||||||
| blaOXA-395 | ||||||||||||||||||||||||||
| blaOXA-50 | ||||||||||||||||||||||||||
| blaOXA-10 | ||||||||||||||||||||||||||
| blaTEM-1B | ||||||||||||||||||||||||||
| blaVIM-2 | ||||||||||||||||||||||||||
| blaPME-1 | ||||||||||||||||||||||||||
| blaPAU-1 | ||||||||||||||||||||||||||
| Sulphonamide, tetracycline, macrolide, fosfomycin, and chloramphenicol resistance genes | ||||||||||||||||||||||||||
| sul1 | ||||||||||||||||||||||||||
| tet(G) | ||||||||||||||||||||||||||
| mph(E) | ||||||||||||||||||||||||||
| mph(A) | ||||||||||||||||||||||||||
| msr(E)* | ||||||||||||||||||||||||||
| fosA | ||||||||||||||||||||||||||
| catB | ||||||||||||||||||||||||||
* msr(E) encodes macrolide and lincosamide resistance. Isolates shaded in grey indicate Australian strains. Black color represents gene presence.
An aminoglycoside resistance gene (aph(3’)-IIb), a beta-lactam resistance gene (blaPAO), a fosfomycin resistance gene (fosA), and a chloramphenicol resistance gene (catB7) were common to all isolates. The Australian isolates (123–182) had acquired only eight resistance genes, while the Indian isolates (188–221) had acquired 26 different resistance genes (Table 2). Five Indian isolates (198, 202, 217, 219, and 221, with large pan genomes) acquired the largest number of resistance genes. Of these five isolates, the pairs 198/219 and 202/221 had the most similar resistance gene profiles and each member of the pair were of the same sequence type, ST308 and ST316 respectively. As acquired resistance genes may be carried on integrons, the genomes of the P. aeruginosa isolates were analyzed for integrons using Integron Finder version 1.5.1. qnrCV1 was associated with a class 1 integron in isolates 202 and 221 and a Tn3 transposon in isolates 198 and 219.
Several types of non-synonymous variations were found in the core genome of the keratitis P. aeruginosa isolates when compared with the reference genome of PAO1 (Table 3). These non-synonymous mutations included single nucleotide polymorphisms (SNPs), multi-nucleotide polymorphisms (MNPs), deletions, insertions, and complex variations (where more than one change occurred at one specific location compared to the reference strain). The total variations in the isolates ranged from 76,080 in isolate 206 to 22,536 in isolate 181. There was a median of 26,317 (IQR = 25,681–33,780) variations in the genomes of Australian isolates and a median of 50,006 (IQR = 26,967–50,600) in the Indian isolates (p = 0.09). Based on the grouping of core genome phylogeny, isolates within group 2 (198, 202, 219, 220, 221, 233) had the most variations. Isolate 206, which had a unique sequence type and was placed in a separate group by pan genome analysis, had an exceptionally high number of variations (76,080) and SNPs (67,271).
Table 3.
Frequency of different types of variation in the genes of P. aeruginosa isolates.
| P. aeruginosa Isolates | Total Variants | Variant Complex | Variants Insertions | Variants Deletions | Variants MNP | Variant SNP |
|---|---|---|---|---|---|---|
| 123 | 28,279 | 1593 | 187 | 163 | 398 | 25,938 |
| 126 | 26,258 | 1416 | 164 | 159 | 355 | 24,164 |
| 127 | 25,760 | 1362 | 163 | 176 | 391 | 23,668 |
| 162 | 50,999 | 3481 | 281 | 257 | 951 | 46,029 |
| 169 | 50,283 | 3359 | 269 | 245 | 922 | 45,488 |
| 176 | 26,065 | 1372 | 168 | 161 | 342 | 24,022 |
| 181 | 22,536 | 1063 | 162 | 133 | 283 | 20,895 |
| 182 | 25,684 | 1359 | 172 | 180 | 368 | 23,605 |
| 223 | 25,672 | 1358 | 167 | 176 | 402 | 23,568 |
| 224 | 26,376 | 1435 | 163 | 165 | 353 | 24,260 |
| 225 | 28,070 | 1566 | 167 | 156 | 385 | 25,796 |
| 227 | 28,000 | 1560 | 162 | 154 | 370 | 25,754 |
| 233 | 52,392 | 3590 | 285 | 263 | 956 | 47,298 |
| 235 | 24,919 | 1349 | 162 | 171 | 354 | 22,883 |
| 188 | 25,833 | 1435 | 164 | 154 | 351 | 23,729 |
| 189 | 25,910 | 1458 | 165 | 155 | 365 | 23,767 |
| 193 | 26,567 | 1445 | 180 | 147 | 389 | 24,406 |
| 198 | 50,631 | 3503 | 280 | 236 | 945 | 45,667 |
| 202 | 49,981 | 3461 | 257 | 236 | 902 | 45,125 |
| 206 | 76,180 | 6449 | 336 | 371 | 1653 | 67,271 |
| 216 | 28,166 | 1548 | 183 | 164 | 433 | 25,838 |
| 217 | 51,119 | 3575 | 290 | 226 | 944 | 46,084 |
| 218 | 29,161 | 1676 | 182 | 181 | 430 | 26,692 |
| 219 | 50,507 | 3484 | 273 | 237 | 925 | 45,588 |
| 220 | 50,180 | 3452 | 267 | 234 | 894 | 45,332 |
| 221 | 50,030 | 3477 | 260 | 237 | 906 | 45,150 |
SNP = single nucleotide polymorphism; MNP = multi-nucleotide polymorphism. Isolate numbers highlighted in gray are from Australia.
Non-synonymous mutations were assessed in resistance genes of the P. aeruginosa isolates (Supplementary Table S2). There were no large differences in the mutations in resistance genes of any of the isolates except the antibiotic efflux-related genes opmH and rosC. opmH had ≥9 mutations in all isolates except 127, 162,169, 202, 218, 220, and 221 (mostly isolates of group 2 of core and pan genome phylogenies except 127 and 218). rosC had 20 non-synonymous mutations including insertions/deletions in isolate 206, 11 in 233, and ≥5 mutations in isolates 162, 169, 176, 202, 216, 217, 219, 220, and 221 (mostly isolates of group 2 of core and pan genome phylogenies except 176, 216), but ≤3 mutations in isolates 123, 126, 123, 181, 182, 188, 189, 193, 198, and 218 (mostly isolates of group 1 of core and pan genome phylogenies except 198). Mutations in efflux genes encoding efflux pumps were also found, including mexX, mexT, mexD, mexM, and mexY, although there was no significant difference between two groups in the possession of mutations in these genes. All other mutations in the genes were random without any association to sequence type, phylogeny, or susceptibility to antibiotics.
2.4. Possession of exoU and Mutations in the DNA Mismatch Repair System
ExoU was present in the genomes of all isolates in group 2 (core and pan genome phylogenetic group) as well as isolates 123 and 127. All other isolates possessed exoS with the exception of isolate 126 which possessed both exoU and exoS. To address differences in the numbers of sequence variants between the isolates, the genes involved in the DNA mismatch repair (MMR) system mutS (that encodes a protein which binds to errors in DNA), mutL (that encodes a protein that works in synergy with MutS and activates UvrD), and uvrD (a DNA helicase active in DNA replication) were examined. The mutations in the MMR system included SNPs, indels, and complex variants. The number of mutations in mutL ranged from 1 to 2 and mutations in mutS (which ranged between 0 and 2) were found in seven isolates (Table 4). In uvrD, the number of mutations ranged between 0 and 5 (Table 4). exoU containing isolates possessed a median of two (IQR = 1–3) mutations in mutL, zero (IQR = 0–2) mutations in mutS, and four (IQR = 2–5) mutations in uvrD, whereas exoS containing isolates possessed a median of zero (IQR = 0–1) mutations in mutL, zero (IQR = 0–1) mutations in mutS, and two (IQR = 0–2) median mutations in uvrD. There were significant differences in the number of mutL (p = 0.0021) and uvrD (p = 0.02) mutations in exoS and exoU isolates but not with mutS (p = 0.3). Isolate 206, an exoS strain and an outlier in the core genome analysis, had one mutation in mutL. Details of mutations occurring in nucleotide and respective proteins are provided in Supplementary Table S3.
Table 4.
Possession of exoU and exoS and number and type of non-synonymous mutations in the mismatch repair system genes in P. aeruginosa isolates.
| P. aeruginosa Isolates | Type III Secretion System Genes | mutL | mutS | uvrD |
|---|---|---|---|---|
| 123 | exoU | 1 SNP | 0 | 1 complex |
| 126 | exoU/exoS | 0 | 0 | 0 |
| 127 | exoU | 0 | 1 MNP | 1 MNP, 1 complex |
| 162 | exoU | 1 SNP | 0 | 2 SNP, 1 MNP, 2 complexes |
| 169 | exoU | 1 SNP | 1 complex | 2 SNP, 2 MNP, 1 complex |
| 176 | exoS | 1 SNP | 0 | 1 SNP |
| 181 | exoS | 0 | 0 | 0 |
| 182 | exoS | 0 | 0 | 1 MNP 1complex |
| 223 | exoS | 0 | 1 SNP | 1 MNP, 1 complex |
| 224 | exoS | 1 SNP | 0 | 1 MNP, 1 complex |
| 225 | exoS | 0 | 0 | 2 SNP, 2 MNP, 1 complex |
| 227 | exoS | 0 | 0 | 2 SNP, 2 MNP, 1 complex |
| 233 | exoU | 0 | 0 | 1 MNP, 1 complex |
| 235 | exoS | 0 | 0 | 0 |
| 188 | exoS | 0 | 0 | 1 MNP, 1 complex |
| 189 | exoS | 1 SNP | 0 | 1 MNP, 1 complex |
| 193 | exoS | 0 | 0 | 0 |
| 198 | exoU | 2 SNP | 0 | 1 SNP, 3 complexes |
| 202 | exoU | 1 SNP | 1 complex | 1 SNP, 2 MNP, 2 complexes |
| 206 | exoS | 1 MNP | 1 complex | 0 |
| 216 | exoS | 0 | 0 | 0 |
| 217 | exoU | 2 SNP | 1 complex | 1 SNP, 2 MNP, 1 complex |
| 218 | exoS | 0 | 0 | 0 |
| 219 | exoU | 2 SNP | 0 | 1 SNP, 1 MNP, 2 complexes |
| 220 | exoU | 1 SNP | 1 complex | 1 SNP, 2 MNP, 2 complexes |
| 221 | exoU | 1 SNP | 1 complex | 1 SNP, 2 MNP, 2 complexes |
SNP = single nucleotide polymorphism, MNP = multinucelotide polymorphism. Isolates shaded in grey indicate Australian strains.
2.5. Sequence Type Analysis and Phylogenetics
All Australian isolates were of different sequence types (ST), except 225 and 227 which belonged to ST233. Among the 12 Indian isolates, one isolate was designated as belonging to a new sequence type, two isolates (198 and 219) belonged to ST308, two others (188 and 189) belonged to ST491, and three isolates (202, 220, and 221) belonged to ST316 (Table 5).
Table 5.
Sequence types of P. aeruginosa isolates.
| P. aeruginosa Isolates | Sequence Types | Core Genes | Shell Genes | Pan/Total Genes |
|---|---|---|---|---|
| 123 | ST218 | 5496 | 508 | 6004 |
| 126 | ST2726 | 5483 | 712 | 6195 |
| 127 | ST845 | 5483 | 938 | 6421 |
| 162 | ST298 | 5439 | 905 | 6344 |
| 169 | ST1027 | 5456 | 694 | 6150 |
| 176 | ST709 | 5547 | 1112 | 6659 |
| 181 | ST244 | 5588 | 1047 | 6662 |
| 182 | ST27 | 5486 | 1096 | 6582 |
| 223 | ST17 | 5471 | 1232 | 6703 |
| 224 | ST168 | 5483 | 607 | 6090 |
| 225 ¤ | ST233 | 5515 | 1338 | 6853 |
| 227 ¤ | ST233 | 5493 | 1304 | 6797 |
| 233 | NEWST | 5440 | 624 | 6064 |
| 235 | ST262 | 5470 | 540 | 6010 |
| 188 * | ST491 | 5490 | 535 | 6025 |
| 189 * | ST491 | 5492 | 531 | 6023 |
| 193 | ST760 | 5490 | 594 | 6084 |
| 198 † | ST308 | 5454 | 1428 | 6882 |
| 202 # | ST316 | 5425 | 1505 | 6930 |
| 206 | NEWST | 5331 | 1084 | 6415 |
| 216 | ST1527 | 5480 | 1488 | 6968 |
| 217 | ST1047 | 5448 | 1173 | 6621 |
| 218 | ST3083 | 5513 | 488 | 6001 |
| 219 † | ST308 | 5451 | 1796 | 7247 |
| 220 # | ST316 | 5430 | 948 | 6378 |
| 221 # | ST316 | 5425 | 1511 | 6936 |
| PA7 | ST1196 | 3599 | 4586 | 8185 |
| PA14 | ST253 | 5436 | 790 | 6226 |
Gray shading denotes Australian isolates. *, †, #, ¤ indicates strains belong to the same sequence types (STs).
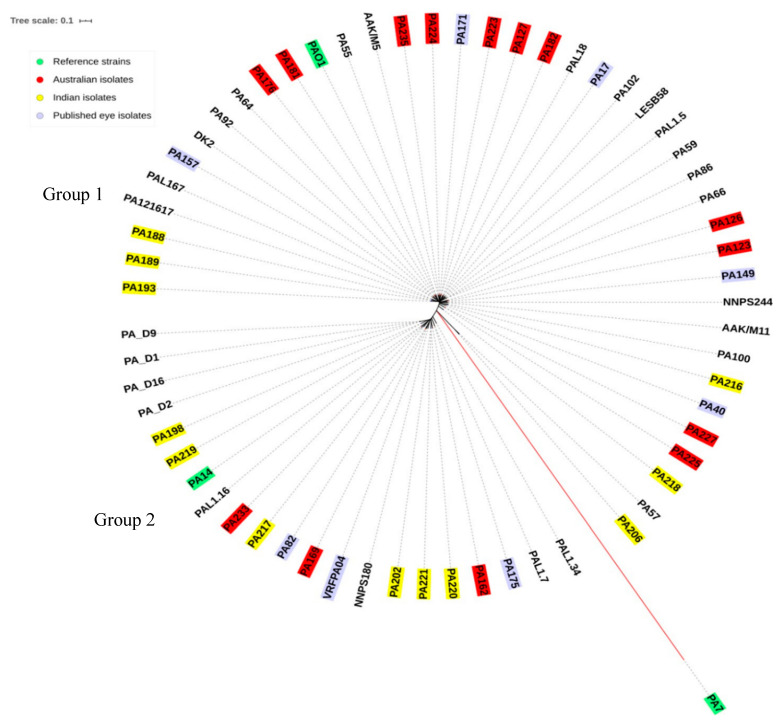
The number of core and total or pan (or total) genes were reported from the statistical summary of Roary v3.11.2. The core genomes of the isolates were aligned using PA7 (Accession number NC_009656.1), PA14 (Accession number NC_004863.1), and PAO1 (Accession number NC_002516.1) as reference strains. The eight published genomes of P. aeruginosa isolates from eye as well as strains from other sources were also included. The core genes of published isolates are provided in Supplementary Table S4. The isolates were sub-grouped based on the number of core genes; isolates with a similar number of core genes were closely aligned and isolates with the same sequence type were grouped together. The core genomes formed two groups in the phylogenetic tree (Figure 1). Isolates in group 1 tended to have a larger number of core genes than isolates in group 2. Isolate 206, PA57, and PA7 were outliers based on core genome phylogeny. The Australian and Indian isolates had a similar number of core genes, whereas the Indian isolates had a larger number of pan genes (10,889) due to the acquisition of shell genes (genes present in two or more strains) (Table 4).
Figure 1.
Core genome phylogeny of P. aeruginosa isolates using Parsnp. PAO1 was used as reference. PA7 and PA14 were also included.
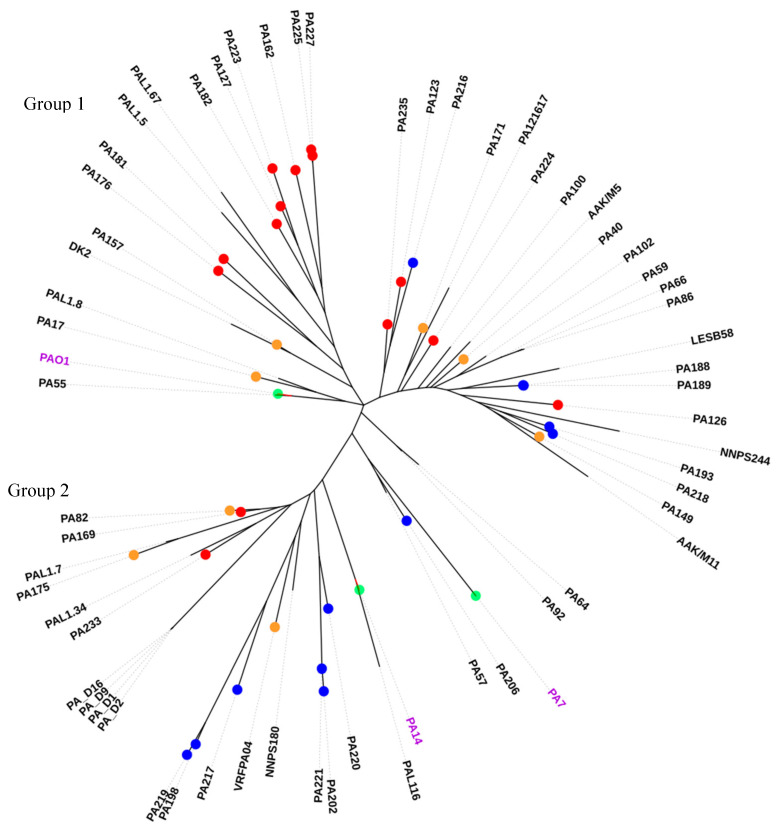
The phylogenetic relationships of these P. aeruginosa isolates were assessed by aligning their pan genome against PAO1 as a reference. The output generated using Roary showing the gene presence or absence in all isolates is provided in Supplementary Figure S1. This again divided the P. aeruginosa isolates into two major groups. Six multi-drug resistant Indian isolates (198, 202, 217, 219, 220, 221) and the VRFPA04 isolate (isolated from the cornea) were clustered in one group, which also contained the two Australian isolates 162 and 169. The Indian isolate 216 was categorized in a separate sub-group due to the large number of shell genes and possession of exoS.
The second group (group 2 of the pan genome analysis) included most of the Australian (123, 126, 127, 162, 176, 181, 182, 223, 224, 225, 227, 235) and Indian (188, 189, 193, 216 218) isolates along with reference strain PAO1 (Figure 2). Overall, the multi-drug resistant Indian isolates had a large pan genome (total of 10,889 genes obtained from the statistical summary in Roary v3.11.2). The pan genome grouping of isolates was broadly based on the number of pan (or total) genes and possession of either exoU or exoS in each group, except two Australian isolates 123 and 127 which were in group 2 but possessed exoU. The other exception to this grouping pattern was for isolates 181 and 182 which had large pan genomes and were clustered into group 1 but carried exoS.
Figure 2.
Pan genome phylogeny of P. aeruginosa isolates. Branches with no color representation indicate non-ocular isolates. Red color indicates Australian, blue color represents Indian, and orange color represents published eye isolates. Green color represents reference strains. Purple color represents reference strains.
The isolates of group 2 usually had a large number of pan genes and were exoU+. Isolates having similar numbers of pan genes were sub-grouped together. For example, isolate 193 (pan genes = 6084) and 218 (pan genes = 6001) were sub-grouped together. Isolate 218 had a similar number of pan genes to isolate 123 (pan genes = 6001), but isolate 218 possessed exoS, while 123 possessed exoU, and thus these were not grouped together. Isolates belonging to the same sequence type were also grouped together. The MDR isolates, the isolates with same STs, and isolates with large gene variations were clustered in one pan-group. The previously published isolates PA_D1, PA_D2, PA_D9, and PA_D16 with the same ST and those with large shell genes were grouped with the MDR isolates of the current study.
3. Discussion
This study investigated genomic differences in Australian and Indian P. aeruginosa isolates from keratitis. Phenotypically, more resistance was found in Indian isolates compared to Australian isolates as has been shown in previous studies [30,31]. Unregulated antibiotic use in India has been linked to increased antibiotic resistance [32]. Resistance to antibiotics is problematic even in the treatment of keratitis, where a topical application of antibiotics is used. Infection with antibiotic resistant strains results in prolonged infection [33], more severe keratitis [5], and an increase in the cost of treatment [34,35].
Indian P. aeruginosa strains harbored more resistance genes compared to Australian isolates, although aph(3’)-IIb, blaPAO1 (fosA), and catB7 were found in all isolates, which was consistent with previous studies [31,36]. qnrVC1 was found in four Indian isolates but no Australian isolates. This fluoroquinolone resistance gene has not been previously reported in P. aeruginosa ocular isolates [31], but it has been reported in burns isolates and has been identified as being carried on an integron [37]. Similarly, in the current study qnrVC1 was carried on a class 1 integron in isolates 202 and 221, but integrated into a Tn3 transposon in isolates 198 and 219. This gene has also been isolated from the high risk ST773 clone of P. aeruginosa from urine in Hungary [38]. High risk clones are isolates with high mutational rates in resistance genes and those that have acquired a large number of resistance genes. As previously described, resistance to fluoroquinolones in keratitis P. aeruginosa isolates was also due to mutations in the quinolone resistance determining regions of gyrA and parC [15]. Possession of qnrVC1 and mutations in gyrA and parC were associated with high levels of fluroquinolone resistance. The possession of large numbers of acquired resistance genes by Indian isolates likely contributed to the higher rates of resistance of these isolates. The Indian isolates 198, 202, 217, 219, 220, and 221 also had a high number of gene variations which is an independent mechanism of resistance.
The aminoglycoside resistance gene aph(6)-Id which encodes for streptomycin resistance was found in six Indian isolates, including the four that carried qnrVC1, but in no Australian isolates. Previously, aph(6)-Id was found in only one Indian ocular isolate from 1997 [31], but has been found in cystic fibrosis P. aeruginosa isolates [39] and has been associated with the transposon Tn5393 on a plasmid in one strain of P. aeruginosa [39]. As streptomycin is no longer used in clinical treatment [40], this resistance may not be clinically relevant but does suggest environmental selection for the persistence of this gene.
The total number of gene variants found in the Indian isolates 198, 202, 219, and 221 were greater than Australian isolates. However, there were a small number of SNPs found in the genes associated with resistance for these isolates. The Indian isolate 206 (NEWST) had a high number of SNPs in antibiotic resistance genes mexC, mexD, mexM, mexX, mexS, opmE, mexP, mexK, oprJ, ampC, rosC, and mprF. There was no difference in the mutations of other mex genes including mexX, mexT, mexD, mexM, and mexY between Australian and Indian isolates. Given that most isolates from both countries, whether they were sensitive or resistant, had a similar number of mutations in the resistance genes, it is likely that the resistance to antibiotics was related to the possession of acquired resistance genes rather than mutations in chromosomal genes.
In the Australian isolates, four out of the eight isolates (50%) carried exoU, while one isolate was both exoU+/exoS+ and three (38%) were exoS+. In Indian isolates, 50% carried exoU and 50% carried exoS. A previous study has also shown an equal ratio of both genes [41] in keratitis isolates. The possession of the exoU genotype in P. aeruginosa ocular isolates has been related to elevated resistance to disinfectants [42], fluoroquinolones [43], and multiple antibiotics [41]. Furthermore, one study reported worst clinical outcomes and more resistance by exoU carrying isolates [43]. The isolates of this study showed similar findings because the exoU+ isolates 198, 202, 217, 219, 220, 221, and 233 were also MDR.
The DNA mismatch repair system (MMR) in P. aeruginosa is based on the protein trimer MutS-MutL-UvrD and functions to correct errors and preserve the integrity of the genome [24,44,45]. The mutH component of MMR, which is important in other Gram negative bacteria, such as E. coli [46], has not been found previously in P. aeruginosa [25] and was not present in the isolates of the current study. Mutations in mutS, mutL and uvrD can reduce the ability of the bacterium to repair DNA lesions [46]. Strong mutator strains have defects in their MMR system and mutations in mutS predominate [47]. Mutations in the MMR can be a reason for the development of hypermutations in isolates. In cystic fibrosis, hypermutations were found to be a key factor in the development of MDR resistant P. aeruginosa strains [23]. Similar findings were found in this study where isolates 198, 202, 206, 219, and 221 had mutations in the MMR genes, and these isolates had an overall larger variation in their genomes. In the current study, isolates had more mutations in mutL and uvrD, suggesting the strains may not be strong mutators (which is usually associated with mutation in mutS), but nevertheless can undergo uncorrected genetic changes. Indeed, the P. aeruginosa isolates in the current study which had mutations in mutL and uvrD had greater numbers of SNPs, insertions and deletions, acquired genes, and had large pan genomes. Among these isolates, 198, 202, 219, and 221 possessed either the transposon Tn3 or class 1 integrons which carried the acquired genes. This also might be due to mutated MMR, as mutations in MMR genes increase the chances of horizontal gene transfer in mutator isolates [47]. The number of mutations in MMR was greater in exoU possessing isolates with large gene variations. exoU is carried on genomic islands [48,49] and these exoU carrying isolates had larger pan genomes with possession of mobile genetic elements. Therefore, the isolates with the mutated MMR systems may have a greater ability of strains to accumulate gene variations and the acquisition of exoU. Isolate 206, on the other hand, possessed exoS and was not MDR but possessed a large number of SNPs and a large pan genome with one mutation in each of mutS and mutL. Further in-depth studies are required to understand the influence of the MMR system on genomic changes in P. aeruginosa.
Analysis of the sequence types of the P. aeruginosa ocular isolates revealed the presence of three clones, two in the Indian and one in the Australian isolates. The isolates with the same STs had mostly the same phenotypic and genotypic features. The exception to this was isolate 220 that had acquired fewer resistance genes compared to the other two isolates 202 and 221 of ST316. Previously, five ocular P. aeruginosa strains from India isolated in 1997 were of sequence type ST308 [31]. The two isolates of ST308 in the current study, isolated in 2017 and 2018, had acquired more resistance genes compared to isolates from 1997 [31]. This indicates that the clonal isolates have continued to evolve over this time period, although the specific selection factors driving those changes are yet to be elucidated. None of the isolates were collected from the same patient. The majority of the isolates with the same STs grouped in the same phylogeny including previously published isolates (PA_D1, PA_D2, PA_D9, P_D16) with ST1971.
Core and pan genome phylogenies of the isolates produced two almost identical groups, which was in agreement with previously published studies [31,50]. Both phylogenies included isolates from either Australia or India, but those in group 2 tended to be the MDR Indian isolates and possessed higher numbers of antibiotic resistance genes. About 65% of all ocular isolates grouped together which indicated less diversity in the ocular P. aeruginosa isolates [31,51,52]. The grouping of MDR strains from this study with PA14 along a MDR ocular isolate VRFPA04 [36] in both core genome and pan genome analysis, and the grouping of the sensitive strains with PAO1 along the commonly studied cystic fibrosis isolates DK2 and LESB58, was similar to a previous study examining older isolates from India and Australia [31]. Isolate 206, which had the smallest number of core genes and was of a new sequence type, was an outlier in the core genome phylogeny similar to the taxonomic outlier PA7 [53]. However, isolate 206 was grouped together with other isolates in the pan genome because it had acquired a large number of genes. Acquired genes are part of the pan rather than the core genome [53] and the presence of larger pan genomes in MDR P. aeruginosa isolates points towards the acquisition of new genes [54]. Previously, a smaller core genome size of 4910 genes has been reported in ocular P. aeruginosa isolates [31]. However, the current study found a core genome size similar to P. aeruginosa from different sources, comprising 5316–5233 genes [55,56]. The core genome (which is almost 90% of total genome) refers to the conserved genes present in a species [57] which might differ in each individual strain within that species. Additionally, SNPs can be a result of poor sequencing quality and hence it is important to have a good sequencing depth at those positions to identify them as a mutation rather than sequencing error [58]. Grouping of all the isolates including ocular and non-ocular remained the same in both core and pan genome phylogeny.
4. Materials and Methods
4.1. P. aeruginosa Strains and Susceptibility Testing
Twenty-six P. aeruginosa keratitis isolates, eight isolated in Australia from 2004 to 2006, six from 2018 and 2019 (total 14 Australian isolates), and twelve isolated in India between 2017 and 2018, were included in this study. These isolates were selected from a larger collection of strains based on their antibiotic susceptibilities (those phenotypically resistant to multiple antibiotics, some resistant to one or multiple antibiotics, and some which were sensitive to all antibiotics). The susceptibilities of Australian strains (2004–2006) included in this study have been previously published [29]. Strains were selected after comparing their susceptibilities to antibiotics that are used to treat ocular infections. For genetic comparisons, the data of 34 P. aeruginosa isolates from eyes and other sources were also included. The general characteristics of these isolates are described in Supplementary Table S4. The genomes of these isolates were downloaded from the NCBI database and reannotated for this study using the same parameters as of the isolates of this study to avoid any bias in results.
The minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of various antibiotics which are commonly used to treat P. aeruginosa keratitis [16] were assessed for the isolates using the broth microdilution method in 96-well plates following the Clinical and Laboratory Standard Institute guidelines [59]. The antibiotics tested were ciprofloxacin, levofloxacin, gentamicin, ceftazidime (Sigma-Aldrich, St. Louis, MO, USA), polymyxin B (Sigma-Aldrich, Vandtårnsvej, Søborg, Denmark), tobramycin, piperacillin (Cayman Chemical Company, Ann Arbor, MI, USA), and imipenem (LKT Laboratories Inc., St. Paul, MN, USA). The susceptibility results were interpreted using the EUCAST v9 [60] and CLSI [61] 2017 breakpoints.
4.2. Genomic Sequencing
DNeasy Blood and Tissue Kits (Qiagen, Hilden, Germany) were used for DNA extraction as per the manufacturer’s recommendations. The Nextera XT DNA library preparation kit (Illumina, San Diego, CA, USA) was used to prepare paired-end libraries. All the libraries were multiplexed on one MiSeq run. FastQC version 0.117 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc) was used to assess the quality of sequenced genomes using raw reads. Version 0.38 of Trimmomatic [61] was used for trimming the adapters from the reads following de novo assembly using Spades v3.13.0 [62]. Genomes were annotated using Prokka v1.12 [63].
Sequence types were investigated using PubMLST https://pubmlst.org/. Pan genomes of the P. aeruginosa isolates were analyzed using Roary v3.11.2 [64] using PAO1 as a reference, while core genome phylogeny was constructed using Harvest Suite Parsnp v1.2 [65] with strains PAO1, PA7, and PA14 used as reference strains. The output file ‘genes_ presence_absence’ was used to compare the P. aeruginosa isolates. Acquired resistance genes were identified using the online database Resfinder v3.1 (Centre for Genomic Epidemiology, DTU, Denmark) [66]. Integron Finder v1.5.1 was used to identify any integrons present in the isolates. Mutations in the genes were detected using Snippy V2 [67]. Isolates with same sequence types were compared for nucleotide similarities using the MUMmer online web tool (http://jspecies.ribohost.com/jspeciesws/#analyse).
Using the Pseudomonas genome database (http://www.pseudomonas.com) and comprehensive antibiotic resistance database (https://card.mcmaster.ca), 76 genes related to P. aeruginosa resistance were selected to investigate the presence of single nucleotide polymorphisms. All isolates were analyzed for the presence of the type III secretion system associated virulence factors exoU and exoS using the BlastN database.
4.3. Statistical Analysis
The statistical analysis was performed using GraphPad Prism v8. Medians were calculated with the ‘descriptive statistics’ option during analysis of variance (ANOVA). P-values less than 0.05 were considered as significant. Fischer’s Exact test was used to find the difference between acquired genes. To analyze the significant difference in the DNA mismatch repair genes between exoU and exoS isolates and gene variations in the isolates, the Mann–Whitney test was used.
5. Conclusions
Indian isolates and Australian isolates were clearly distinct in carrying a type III secretion system related to exoU and exoS. There was an association in the isolates for carrying acquired resistance genes with a large number of pan genes. Indian isolates were more resistant to antibiotics compared to Australian isolates. Additionally, isolates of P. aeruginosa from ocular infection had a large number of genetic variations (mutations) and a mutated mismatch repair system. However, the isolates collected from the same region or time will give a clearer idea of these differences.
Acknowledgments
The authors would like to acknowledge the Singapore Centre for Environmental Life Sciences Engineering (SCELSE), whose research is supported by the National Research Foundation Singapore, Ministry of Education, Nanyang Technological University, and National University of Singapore, under its Research Centre of Excellence Programme. We would also like to acknowledge Engineer Ahsan Ullah Khan, Independent Monitoring Unit Haripur KPK, Pakistan, for his help in the computational analysis and Doctor Nicole Carnt, School of Optometry and Vision Science UNSW, Sydney, for her help in the isolates collection. We are also thankful to UNSW high performance computing facility KATANA for providing us with the cluster time for the data analysis.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-6382/9/9/600/s1, Table S1: Details of the Pseudomonas aeruginosa isolates used in the current study. Table S2: Gene variations of resistance genes in Pseudomonas aeruginosa isolates (the gray-shaded strains were isolated from Australia). Nucleotide accession: The nucleotide sequences are available in the GenBank under the Bio project accession number PRJNA590804. Table S3: Types of mutations in the mismatch repair system; Table S4: Genomics features of P. aeruginosa isolates; Figure S1: Pan-genome phylogenetic tree. The data on the right of the figure shows the presence and absence of genes. The tree was built using the genome of PAO1 as a reference.
Author Contributions
Conceptualization, M.K., F.S., and M.D.P.W.; methodology, M.K., F.S., S.S., S.A.R., and M.D.P.W.; writing—original draft preparation, M.K.; writing—review and editing, F.S., S.A.R., and M.D.P.W.; supervision, F.S. and M.D.P.W.; funding acquisition, F.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Richards M.J., Edwards J.R., Culver D.H., Gaynes R.P. Nosocomial infections in medical intensive care units in the United States. National Nosocomial Infections Surveillance System. Crit. Care Med. 1999;27:887–892. doi: 10.1097/00003246-199905000-00020. [DOI] [PubMed] [Google Scholar]
- 2.Abjani F., Khan N.A., Jung S.Y., Siddiqui R. Status of the effectiveness of contact lens disinfectants in Malaysia against keratitis-causing pathogens. Exp. Parasitol. 2017;183:187–193. doi: 10.1016/j.exppara.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Stapleton F., Carnt N. Contact lens-related microbial keratitis: How have epidemiology and genetics helped us with pathogenesis and prophylaxis. Eye (Lond.) 2012;26:185–193. doi: 10.1038/eye.2011.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trouillet J.L., Vuagnat A., Combes A., Kassis N., Chastre J., Gibert C. Pseudomonas aeruginosa ventilator-associated pneumonia: Comparison of episodes due to piperacillin-resistant versus piperacillin-susceptible organisms. Clin. Infect. Dis. 2002;34:1047–1054. doi: 10.1086/339488. [DOI] [PubMed] [Google Scholar]
- 5.Green M., Apel A., Stapleton F. Risk factors and causative organisms in microbial keratitis. Cornea. 2008;27:22–27. doi: 10.1097/ICO.0b013e318156caf2. [DOI] [PubMed] [Google Scholar]
- 6.Hooi S.H., Hooi S.T. Culture-proven bacterial keratitis in a Malaysian general hospital. Med. J. Malays. 2005;60:614–623. [PubMed] [Google Scholar]
- 7.Parmar P., Salman A., Kalavathy C.M., Kaliamurthy J., Thomas P.A., Jesudasan C.A. Microbial keratitis at extremes of age. Cornea. 2006;25:153–158. doi: 10.1097/01.ico.0000167881.78513.d9. [DOI] [PubMed] [Google Scholar]
- 8.Sharma N., Sinha R., Singhvi A., Tandon R. Pseudomonas keratitis after laser in situ keratomileusis. J. Cataract. Refract. Surg. 2006;32:519–521. doi: 10.1016/j.jcrs.2005.12.061. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee S., Agrawal D. Multi-drug resistant Pseudomonas aeruginosa keratitis and its effective treatment with topical colistimethate. Indian J. Ophthalmol. 2016;64:153–157. doi: 10.4103/0301-4738.179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livermore D.M. beta-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 1995;8:557–584. doi: 10.1128/CMR.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X.-Z., Ma D., Livermore D.M., Nikaido H. Role of efflux pump (s) in intrinsic resistance of Pseudomonas aeruginosa: Active efflux as a contributing factor to beta-lactam resistance. Antimicrob. Agents Chemother. 1994;38:1742–1752. doi: 10.1128/AAC.38.8.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poole K. Pseudomonas aeruginosa: Resistance to the max. Front. Microbiol. 2011;2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hocquet D., Vogne C., El Garch F., Vejux A., Gotoh N., Lee A., Lomovskaya O., Plésiat P. MexXY-OprM efflux pump is necessary for adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 2003;47:1371–1375. doi: 10.1128/AAC.47.4.1371-1375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livermore D.M. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: Our worst nightmare? Clin. Infect. Dis. 2002;34:634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 15.Lomholt J.A., Kilian M. Ciprofloxacin susceptibility of Pseudomonas aeruginosa isolates from keratitis. Br. J. Ophthalmol. 2003;87:1238–1240. doi: 10.1136/bjo.87.10.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willcox M.D. Review of resistance of ocular isolates of Pseudomonas aeruginosa and staphylococci from keratitis to ciprofloxacin, gentamicin and cephalosporins. Clin. Exp. Optom. 2011;94:161–168. doi: 10.1111/j.1444-0938.2010.00536.x. [DOI] [PubMed] [Google Scholar]
- 17.Poonsuk K., Tribuddharat C., Chuanchuen R. Aminoglycoside resistance mechanisms in Pseudomonas aeruginosa isolates from non-cystic fibrosis patients in Thailand. Can. J. Microbiol. 2013;59:51–56. doi: 10.1139/cjm-2012-0465. [DOI] [PubMed] [Google Scholar]
- 18.Diver J.M., Schollaardt T., Rabin H.R., Thorson C., Bryan L.E. Persistence mechanisms in Pseudomonas aeruginosa from cystic fibrosis patients undergoing ciprofloxacin therapy. Antimicrob. Agents Chemother. 1991;35:1538–1546. doi: 10.1128/AAC.35.8.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzvinec M., Izdebski R., Butic I., Jelic M., Abram M., Koscak I., Baraniak A., Hryniewicz W., Gniadkowski M., Andrasevic A.T. Sequence types 235, 111, and 132 predominate among multidrug-resistant Pseudomonas aeruginosa clinical isolates in Croatia. Antimicrob. Agents Chemother. 2014;58:6277–6283. doi: 10.1128/AAC.03116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips R.M., Six D.A., Dennis E.A., Ghosh P. In vivo phospholipase activity of the Pseudomonas aeruginosa cytotoxin ExoU and protection of mammalian cells with phospholipase A2 inhibitors. J. Biol. Chem. 2003;278:41326–41332. doi: 10.1074/jbc.M302472200. [DOI] [PubMed] [Google Scholar]
- 21.Finck-Barbançon V., Goranson J., Zhu L., Sawa T., Wiener-Kronish J.P., Fleiszig S.M.J., Wu C., Mende-Mueller L., Frank D.W. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 22.Subedi D., Vijay A.K., Willcox M. Overview of mechanisms of antibiotic resistance in Pseudomonas aeruginosa: An ocular perspective. Clin. Exp. Optom. 2018;101:162–171. doi: 10.1111/cxo.12621. [DOI] [PubMed] [Google Scholar]
- 23.Maciá M.D., Blanquer D., Togores B., Sauleda J., Pérez J.L., Oliver A. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob. Agents Chemother. 2005;49:3382–3386. doi: 10.1128/AAC.49.8.3382-3386.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Modrich P. Mechanisms and biological effects of mismatch repair. Annu. Rev. Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 25.Oliver A., Baquero F., Blazquez J. The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: Molecular characterization of naturally occurring mutants. Mol. Microbiol. 2002;43:1641–1650. doi: 10.1046/j.1365-2958.2002.02855.x. [DOI] [PubMed] [Google Scholar]
- 26.Alsan M., Schoemaker L., Eggleston K., Kammili N., Kolli P., Bhattacharya J. Out-of-pocket health expenditures and antimicrobial resistance in low-income and middle-income countries: An economic analysis. Lancet Infect. Dis. 2015;15:1203–1210. doi: 10.1016/S1473-3099(15)00149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sánchez M., Sivaraman S. News Media Reporting of Antimicrobial Resistance in Latin America and India. In: Sosa A.d.J., Byarugaba D.K., Amábile-Cuevas C.F., Hsueh P.-R., Kariuki S., Okeke I.N., editors. Antimicrobial Resistance in Developing Countries. Springer; New York, NY, USA: 2010. pp. 525–537. [Google Scholar]
- 28.Van Boeckel T.P., Gandra S., Ashok A., Caudron Q., Grenfell B.T., Levin S.A., Laxminarayan R. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014;14:742–750. doi: 10.1016/S1473-3099(14)70780-7. [DOI] [PubMed] [Google Scholar]
- 29.Khan M., Stapleton F., Willcox M.D.P. Susceptibility of contact lens-related Pseudomonas aeruginosa keratitis isolates to multipurpose disinfecting solutions, disinfectants, and antibiotics. Transl. Vis. Sci. Technol. 2020;9:2. doi: 10.1167/tvst.9.5.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choy M.H., Stapleton F., Willcox M.D.P., Zhu H. Comparison of virulence factors in Pseudomonas aeruginosa strains isolated from contact lens- and non-contact lens-related keratitis. J. Med. Microbiol. 2008;57:1539–1546. doi: 10.1099/jmm.0.2008/003723-0. [DOI] [PubMed] [Google Scholar]
- 31.Subedi D., Vijay A.K., Kohli G.S., Rice S.A., Willcox M. Comparative genomics of clinical strains of Pseudomonas aeruginosa strains isolated from different geographic sites. Sci. Rep. 2018;8:15668. doi: 10.1038/s41598-018-34020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porter G., Grills N. Medication misuse in India: A major public health issue in India. J. Public Health (Oxf.) 2016;38:e150–e157. doi: 10.1093/pubmed/fdv072. [DOI] [PubMed] [Google Scholar]
- 33.Wilhelmus K.R., Abshire R.L., Schlech B.A. Influence of fluoroquinolone susceptibility on the therapeutic response of fluoroquinolone-treated bacterial keratitis. Arch. Ophthalmol. 2003;121:1229–1233. doi: 10.1001/archopht.121.9.1229. [DOI] [PubMed] [Google Scholar]
- 34.Keay L., Edwards K., Naduvilath T., Taylor H.R., Snibson G.R., Forde K., Stapleton F. Microbial keratitis predisposing factors and morbidity. Ophthalmology. 2006;113:109–116. doi: 10.1016/j.ophtha.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Keay L., Edwards K., Dart J., Stapleton F. Grading contact lens-related microbial keratitis: Relevance to disease burden. Optom. Vis. Sci. 2008;85:531–537. doi: 10.1097/OPX.0b013e31817dba2e. [DOI] [PubMed] [Google Scholar]
- 36.Murugan N., Malathi J., Umashankar V., Madhavan H.N. Unraveling genomic and phenotypic nature of multidrug-resistant (MDR) Pseudomonas aeruginosa VRFPA04 isolated from keratitis patient. Microbiol. Res. 2016;193:140–149. doi: 10.1016/j.micres.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Belotti P.T., Thabet L., Laffargue A., Andre C., Coulange-Mayonnove L., Arpin C., Messadi A., M’zali F., Quentin C., Dubois V. Description of an original integron encompassing blaVIM-2, qnrVC1 and genes encoding bacterial group II intron proteins in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2015;70:2237–2240. doi: 10.1093/jac/dkv103. [DOI] [PubMed] [Google Scholar]
- 38.Kocsis B., Toth A., Gulyas D., Ligeti B., Katona K., Rokusz L., Szabo D. Acquired qnrVC1 and blaNDM-1 resistance markers in an international high-risk Pseudomonas aeruginosa ST773 clone. J. Med. Microbiol. 2019;68:336–338. doi: 10.1099/jmm.0.000927. [DOI] [PubMed] [Google Scholar]
- 39.Tauch A., Schluter A., Bischoff N., Goesmann A., Meyer F., Puhler A. The 79,370-bp conjugative plasmid pB4 consists of an IncP-1beta backbone loaded with a chromate resistance transposon, the strA-strB streptomycin resistance gene pair, the oxacillinase gene bla(NPS-1), and a tripartite antibiotic efflux system of the resistance-nodulation-division family. Mol. Genet. Genom. 2003;268:570–584. doi: 10.1007/s00438-002-0785-z. [DOI] [PubMed] [Google Scholar]
- 40.Sundin G.W., Bender C.L. Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol. Ecol. 1996;5:133–143. doi: 10.1111/j.1365-294X.1996.tb00299.x. [DOI] [PubMed] [Google Scholar]
- 41.Zhu H., Conibear T.C., Bandara R., Aliwarga Y., Stapleton F., Willcox M.D. Type III secretion system-associated toxins, proteases, serotypes, and antibiotic resistance of Pseudomonas aeruginosa isolates associated with keratitis. Curr. Eye Res. 2006;31:297–306. doi: 10.1080/02713680500536746. [DOI] [PubMed] [Google Scholar]
- 42.Lakkis C., Fleiszig S.M. Resistance of Pseudomonas aeruginosa isolates to hydrogel contact lens disinfection correlates with cytotoxic activity. J. Clin. Microbiol. 2001;39:1477–1486. doi: 10.1128/JCM.39.4.1477-1486.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borkar D.S., Acharya N.R., Leong C., Lalitha P., Srinivasan M., Oldenburg C.E., Cevallos V., Lietman T.M., Evans D.J., Fleiszig S.M. Cytotoxic clinical isolates of Pseudomonas aeruginosa identified during the Steroids for Corneal Ulcers Trial show elevated resistance to fluoroquinolones. BMC Ophthalmol. 2014;14:54. doi: 10.1186/1471-2415-14-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tiraby J.G., Fox M.S. Marker discrimination in transformation and mutation of Pneumococcus. Proc. Natl. Acad. Sci. USA. 1973;70:3541–3545. doi: 10.1073/pnas.70.12.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Acharya S., Foster P.L., Brooks P., Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell. 2003;12:233–246. doi: 10.1016/S1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 46.Rayssiguier C., Thaler D.S., Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 47.Chopra I., O’Neill A.J., Miller K. The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist. Updates. 2003;6:137–145. doi: 10.1016/S1368-7646(03)00041-4. [DOI] [PubMed] [Google Scholar]
- 48.Sato H., Frank D.W., Hillard C.J., Feix J.B., Pankhaniya R.R., Moriyama K., Finck-Barbançon V., Buchaklian A., Lei M., Long R.M., et al. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 2003;22:2959–2969. doi: 10.1093/emboj/cdg290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kulasekara B.R., Kulasekara H.D., Wolfgang M.C., Stevens L., Frank D.W., Lory S. Acquisition and evolution of the exoU locus in Pseudomonas aeruginosa. J. Bacteriol. 2006;188:4037–4050. doi: 10.1128/JB.02000-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Freschi L., Jeukens J., Kukavica-Ibrulj I., Boyle B., Dupont M.J., Laroche J., Larose S., Maaroufi H., Fothergill J.L., Moore M., et al. Clinical utilization of genomics data produced by the international Pseudomonas aeruginosa consortium. Front. Microbiol. 2015;6:1036. doi: 10.3389/fmicb.2015.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winstanley C., Langille M.G., Fothergill J.L., Kukavica-Ibrulj I., Paradis-Bleau C., Sanschagrin F., Thomson N.R., Winsor G.L., Quail M.A., Lennard N., et al. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool Epidemic Strain of Pseudomonas aeruginosa. Genome Res. 2009;19:12–23. doi: 10.1101/gr.086082.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klockgether J., Cramer N., Wiehlmann L., Davenport C.F., Tummler B. Pseudomonas aeruginosa genomic structure and diversity. Front. Microbiol. 2011;2:150. doi: 10.3389/fmicb.2011.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roy P.H., Tetu S.G., Larouche A., Elbourne L., Tremblay S., Ren Q., Dodson R., Harkins D., Shay R., Watkins K., et al. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS ONE. 2010;5:e8842. doi: 10.1371/journal.pone.0008842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freschi L., Vincent A.T., Jeukens J., Emond-Rheault J.G., Kukavica-Ibrulj I., Dupont M.J., Charette S.J., Boyle B., Levesque R.C. The Pseudomonas aeruginosa pan-genome provides new insights on its population structure, horizontal gene transfer, and pathogenicity. Genome Biol. Evol. 2019;11:109–120. doi: 10.1093/gbe/evy259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ozer E.A., Allen J.P., Hauser A.R. Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genom. 2014;15:737. doi: 10.1186/1471-2164-15-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valot B., Guyeux C., Rolland J.Y., Mazouzi K., Bertrand X., Hocquet D. What it takes to be a Pseudomonas aeruginosa? The core genome of the opportunistic pathogen updated. PLoS ONE. 2015;10:e0126468. doi: 10.1371/journal.pone.0126468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolfgang M.C., Kulasekara B.R., Liang X., Boyd D., Wu K., Yang Q., Miyada C.G., Lory S. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA. 2003;100:8484–8489. doi: 10.1073/pnas.0832438100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smits T.H.M. The importance of genome sequence quality to microbial comparative genomics. BMC Genom. 2019;20:662. doi: 10.1186/s12864-019-6014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.EUCAST . Breakpoint Tables for Interpretation of MICs and Zone Diameters. The European Committee on Antimicrobial Susceptibility Testing; Växjö, Sweden: 2019. Version 6.0. [Google Scholar]
- 61.Patel J.B., Cockerill F.R., Alder J., Bradford P.A., Eliopoulos G.M., Hardy D.J., Hindler J.A., Jenkins S.G., Lewis J.S., Miller L.A., et al. Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2014. Twenty-Fourth Informational Supplement. [Google Scholar]
- 62.Nurk S., Bankevich A., Antipov D., Gurevich A., Korobeynikov A., Lapidus A., Prjibelsky A., Pyshkin A., Sirotkin A., Sirotkin Y., et al. Assembling Genomes and Mini-metagenomes from Highly Chimeric Reads. In: Deng M., Jiang R., Sun F., Zhang X., editors. Research in Computational Molecular Biology RECOMB 2013. Springer; Berlin/Heidelberg, Germany: 2013. Lecture Notes in Computer Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics (Oxf. Engl.) 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 64.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T., Fookes M., Falush D., Keane J.A., Parkhill J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Treangen T.J., Ondov B.D., Koren S., Phillippy A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seeman T. Snippy: Fast Bacterial Variant Calling from NGS Reads. GitHub Australia; Melbourne, Australia: 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.