Abstract
Objective
Glucagon is well known to regulate blood glucose but may be equally important for amino acid metabolism. Plasma levels of amino acids are regulated by glucagon-dependent mechanism(s), while amino acids stimulate glucagon secretion from alpha cells, completing the recently described liver-alpha cell axis. The mechanisms underlying the cycle and the possible impact of hepatic steatosis are unclear.
Methods
We assessed amino acid clearance in vivo in mice treated with a glucagon receptor antagonist (GRA), transgenic mice with 95% reduction in alpha cells, and mice with hepatic steatosis. In addition, we evaluated urea formation in primary hepatocytes from ob/ob mice and humans, and we studied acute metabolic effects of glucagon in perfused rat livers. We also performed RNA sequencing on livers from glucagon receptor knock-out mice and mice with hepatic steatosis. Finally, we measured individual plasma amino acids and glucagon in healthy controls and in two independent cohorts of patients with biopsy-verified non-alcoholic fatty liver disease (NAFLD).
Results
Amino acid clearance was reduced in mice treated with GRA and mice lacking endogenous glucagon (loss of alpha cells) concomitantly with reduced production of urea. Glucagon administration markedly changed the secretion of rat liver metabolites and within minutes increased urea formation in mice, in perfused rat liver, and in primary human hepatocytes. Transcriptomic analyses revealed that three genes responsible for amino acid catabolism (Cps1, Slc7a2, and Slc38a2) were downregulated both in mice with hepatic steatosis and in mice with deletion of the glucagon receptor. Cultured ob/ob hepatocytes produced less urea upon stimulation with mixed amino acids, and amino acid clearance was lower in mice with hepatic steatosis. Glucagon-induced ureagenesis was impaired in perfused rat livers with hepatic steatosis. Patients with NAFLD had hyperglucagonemia and increased levels of glucagonotropic amino acids, including alanine in particular. Both glucagon and alanine levels were reduced after diet-induced reduction in Homeostatic Model Assessment for Insulin Resistance (HOMA-IR, a marker of hepatic steatosis).
Conclusions
Glucagon regulates amino acid metabolism both non-transcriptionally and transcriptionally. Hepatic steatosis may impair glucagon-dependent enhancement of amino acid catabolism.
Keywords: Amino acids, Glucagon, Liver-alpha cell axis, Non-alcoholic fatty liver disease
Graphical abstract

Highlights
-
•
Glucagon increases amino acid-induced formation of urea within a few minutes.
-
•
Hepatic steatosis impairs glucagon-induced ureagenesis.
-
•
Fatty liver disease associates with hyperglucagonemia.
-
•
The liver-α cell axis is disrupted in animals and humans with fatty liver disease.
1. Introduction
Glucagon was discovered in 1923 as a hyperglycemic substance from the pancreas [1] and is known to increase hepatic glucose production. Glucagon also regulates amino acid metabolism in the liver via ureagenesis [2,3]. In a feedback manner, circulating amino acids stimulate glucagon secretion from the alpha cells [4], thereby completing the recently described liver-alpha cell axis [[5], [6], [7], [8], [9], [10]]. Expression of the urea cycle enzymes in the liver is regulated mainly by glucagon and glucocorticoids, and complete disruption of glucagon receptor signaling, as observed in mice [11,12] and humans [13] with non-functioning glucagon receptors, leads to marked hyperaminoacidemia. Chronic hyperaminoacidemia causes hyperglucagonemia and, in turn, alpha cell hypertrophy and hyperplasia [5]. Conversely, patients suffering from glucagon-producing tumors (glucagonomas) have dramatic hypoaminoacidemia, leading to a characteristic skin cell necrolysis [14] as well as increased urea synthesis in response to amino acid infusion [15]. In addition to these long-term consequences of disruption of the liver-alpha cell axis, it is known that a transient rise in circulating amino acids acutely stimulates glucagon secretion [16]. However, it is unclear whether glucagon is also involved in a corresponding minute-to-minute regulation of hepatic amino acid turnover and ureagenesis, which is relevant during an amino acid challenge, i.e., after intake of protein rich meals.
It has recently been reported that patients with non-alcoholic fatty liver disease (NAFLD) have elevated fasting levels of glucagon and amino acids [[17], [18], [19]]. We hypothesized that steatosis results in disturbed amino acid clearance and decreased ureagenesis in response to amino acids and that this may be related to impaired hepatic actions of glucagon.
Here, we examined both acute and long-term effects of glucagon receptor signaling on amino acid turnover and urea formation in rats and in mice using genetic and pharmacological approaches. Furthermore, we studied the effects of hepatic steatosis on amino acid metabolism in mice, rats, and humans.
2. Results
2.1. Glucagon receptor antagonist reduces amino acid-induced urea formation in mice
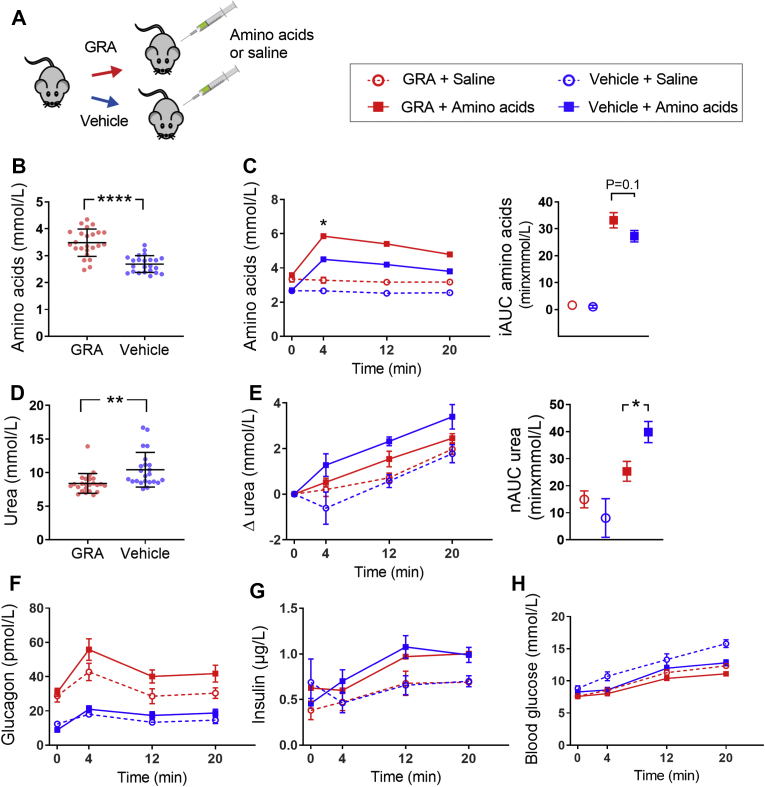
To investigate whether short-term blockage of the glucagon receptor alters amino acid metabolism, we treated mice with a glucagon receptor antagonist (GRA) (previously pharmacologically characterized [20]) (Figure 1A). We analyzed plasma sampled before (the basal state) and after intraperitoneal administration of mixed amino acids (Vamin, composition is found in Table S1). Baseline plasma levels of amino acids were increased in GRA-treated compared to vehicle-treated mice (3.5 ± 0.1 vs. 2.7 ± 0.1 mmol/L, P < 0.0001) (Figure 1B). Clearance of plasma amino acids, reflected by the incremental area under the curve (iAUC), tended to be lower in GRA-treated compared to vehicle-treated mice (33.1 ± 2.8 vs. 27.2 ± 2.1 min × mmol/L, P = 0.1) (Figure 1C). Basal urea levels were significantly lower in GRA-treated mice compared to vehicle-treated mice (8.5 ± 0.2 vs. 10.4 ± 0.3 mmol/L, P < 0.0001) (Figure 1D). Administration of mixed amino acids led to an increase in plasma urea levels, and mice receiving GRA + amino acids showed a diminished increase in plasma urea levels compared to vehicle + amino acids (netAUC: 25.3 ± 3.7 vs. 39.8 ± 3.9 min × mmol/L, P < 0.05) (Figure 1E), supporting the hypothesis that reduced amino acid clearance was due to decreased hepatic metabolism, and not caused by changes in peripheral uptake/elimination. In an attempt to correct for urea excretion in the urine, we ligated the kidneys of anesthetized mice; however, the surgical procedure resulted in lower levels of plasma urea compared to that observed in sham-operated mice independent of administration of amino acids or saline (Fig. S1).
Figure 1.
Pharmacological inhibition of glucagon receptor signaling alters amino acid metabolism and ureagenesis in mice. (A) Female C57BL/6JRj mice received a glucagon receptor antagonist (GRA, 100 mg/kg) or vehicle three hours prior to intraperitoneal administration of amino acids or saline. (B) Baseline plasma levels of total l-amino acids after treatment with GRA or vehicle (n = 23–24). (C) Plasma levels of total l-amino acids upon administration of saline (open circles) or amino acids (Vamin, 3.5 μmol/g body weight, closed squares) in mice treated with GRA (red symbols and lines) or vehicle (blue symbols and lines). Data are shown as incremental area under the curve (iAUC) in the right panel (n = 9–14). (D) Baseline plasma levels of urea after three hours of treatment with GRA or vehicle (n = 23–24). (E) Plasma levels of urea (baseline corrected) upon administration of saline or amino acids in mice treated with GRA or vehicle. Data are shown as net area under the curve (nAUC, positive and negative peaks were included) in the right panel (n = 9–14). Levels of (F) plasma glucagon, (G) plasma insulin, and (H) blood glucose in response to administration of amino acids or saline in mice treated with GRA or vehicle (n = 23–24 except for insulin: n = 15–17). (B + D) Error bars indicate SD, and data are analyzed by unpaired t-tests. (C, E, F, G, H) Data are shown as mean ± SEM. AUCs are analyzed by one-way ANOVA and time-resolved data by multiple t-tests on delta values and corrected for multiple testing using the Holm-Sidak algorithm; ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗∗P < 0.0001.
GRA treatment increased plasma glucagon levels 3-fold in the basal state (P < 0.0001) and 2-fold in response to administration of amino acids compared to vehicle treatment (P = 0.05) (Figure 1F). Plasma levels of insulin did not differ between GRA and vehicle-treated mice, neither at baseline nor upon administration of amino acids or saline (Figure 1G). The GRA used in our study has been shown to decrease hyperglycemia in ob/ob mice [21], and we likewise observed a small decrease in basal blood glucose levels in GRA-treated compared to vehicle-treated mice (7.7 ± 0.2 vs. 8.6 ± 0.2 mmol/L, P < 0.001). During the experiments, there was a slight increase in blood glucose, but the iAUC of blood glucose upon administration of saline in vehicle-treated mice (73.5 ± 11.3 min × mmol/L) was higher compared to mice receiving GRA + saline (53.9 ± 7.6 min × mmol/L, P = 0.07), vehicle + amino acids (46.0 ± 5.0 min × mmol/L, P = 0.04), and GRA + amino acids (38.4 ± 4.5 min × mmol/L, P < 0.01) (Figure 1H).
2.2. Exogenous glucagon restores amino acid metabolism in alpha cell-deficient mice
To further explore the acute effects of glucagon on amino acids metabolism, we investigated whether reductions in endogenous glucagon results in impaired amino acid-induced ureagenesis and if reintroduction of glucagon would restore amino acid turnover and ureagenesis. To address this, we injected diphtheria toxin (DT) to transgenic mice expressing the human diphtheria toxin receptor under control of the Gcg promoter [22], leading to acute loss of alpha cells and intestinal L cells. Another group of transgenic mice was injected with vehicle, and wild-type (WT) littermates received DT to control for any unspecific toxic effects of the DT treatment (Fig. S2). Glucagon-positive cells were clearly reduced in mice treated with DT compared to vehicle (Figure 2A), and the amount of extractable glucagon from pancreatic tissue was reduced by 95% (0.5 ± 0.1 vs. 9.0 ± 0.9 pmol/mg total protein, P < 0.0001) (Figure 2B). Consistently, DT-treated mice had a significant reduction in baseline plasma glucagon levels compared to vehicle-treated mice (5.0 ± 0.3 vs. 12.7 ± 1.3 pmol/L, P < 0.0001) and only a minor glucagon response to intraperitoneal administration of mixed amino acids (iAUC0–4min: 7.1 ± 1.0 vs. 19.4 ± 3.5 min × pmol/L, P < 0.01) (Figure 2C).
Figure 2.
Glucagon acutely increases amino acid turnover in mice with a 95% reduction in pancreatic glucagon content. (A) Representative histological pictures of endocrine islets stained for glucagon in female transgenic (TgN(GCG.DTR)) mice treated with vehicle (left panel) or diphtheria toxin (DT, 10 ng/g, right panel). Images are shown at ×200 magnification. The scale bar indicates 50 μm. (B) Extracted pancreatic glucagon content in a separate group of transgenic (TG) mice treated with vehicle or DT. Error bars indicate SD (n = 6–8). (C) Plasma levels of glucagon in response to saline (open circles), amino acids (Vamin, 3.5 μmol/g body weight, closed squares), or amino acids + glucagon (200 ng/g body weight, triangles) in mice treated with DT (red symbols) or vehicle (blue symbols and lines). (D) Plasma levels of total l-amino acids (left panel) and incremental area under the curve (iAUC, right panel), and (E) plasma levels of urea (baseline corrected, left panel) and netAUC (nAUC, positive and negative peaks were included, right panel) in response to saline, amino acids, or amino acids + glucagon in TG mice treated with vehicle or DT. (F) Plasma insulin and (G) blood glucose levels in response to saline, amino acids, or amino acids + glucagon in TG mice treated with vehicle or DT. n = 9–15. Data in (C–G) are shown as mean ± SEM and AUCs were analyzed by one-way ANOVA corrected for multiple testing using the Holm-Sidak algorithm. ∗P < 0.05 and ∗∗P < 0.01. See also Fig. S2.
Similar to the previous experiment with mice treated with GRA, mice received an intraperitoneal injection with saline or mixed amino acids. The amino acid clearance, estimated from the iAUC, tended to be decreased in mice treated with DT compared to vehicle (46.9 ± 4.2 vs. 39.4 ± 5.0 min × mmol/L, P = 0.1). Exogenous glucagon administered together with amino acids increased the amino acid clearance rate in mice treated with DT compared to administration of amino acids alone (iAUC: 30.9 ± 2.2 vs. 46.9 ± 4.1 min × mmol/L, P < 0.01) (Figure 2D). Plasma levels of urea in response to mixed amino acids were lower in mice treated with DT compared to vehicle (netAUC: 23.6 ± 5.4 vs. 37.5 ± 2.5 min × mmol/L, P < 0.05), consistent with the observed effect of pharmacological inhibition of glucagon actions (Figure 1). The administration of exogenous glucagon together with amino acids restored the urea response to levels similar to those of mice with intact alpha cells (netAUC: 30.3 ± 3.0 vs. 37.5 ± 2.5 min × mmol/L, P = 0.4) (Figure 2E), demonstrating that glucagon acutely controls ureagenesis.
Upon administration of amino acids and glucagon, plasma insulin levels increased to levels significantly higher than those in all other groups when analyzed by iAUC (P < 0.0001) (Figure 2F). Blood glucose levels did not differ upon stimulation with saline, amino acids, or amino acids and glucagon or between mice treated with vehicle or DT (Figure 2G). Because the seven-day recovery period after DT treatment allows the intestinal glucagon-like peptide 1 (GLP-1)-producing L cells to grow back [22] (Fig. S2), differences in GLP-1 secretion were unlikely to be responsible for any differences in insulin secretion.
2.3. Glucagon regulates the secretion of metabolites from the perfused rat liver within minutes
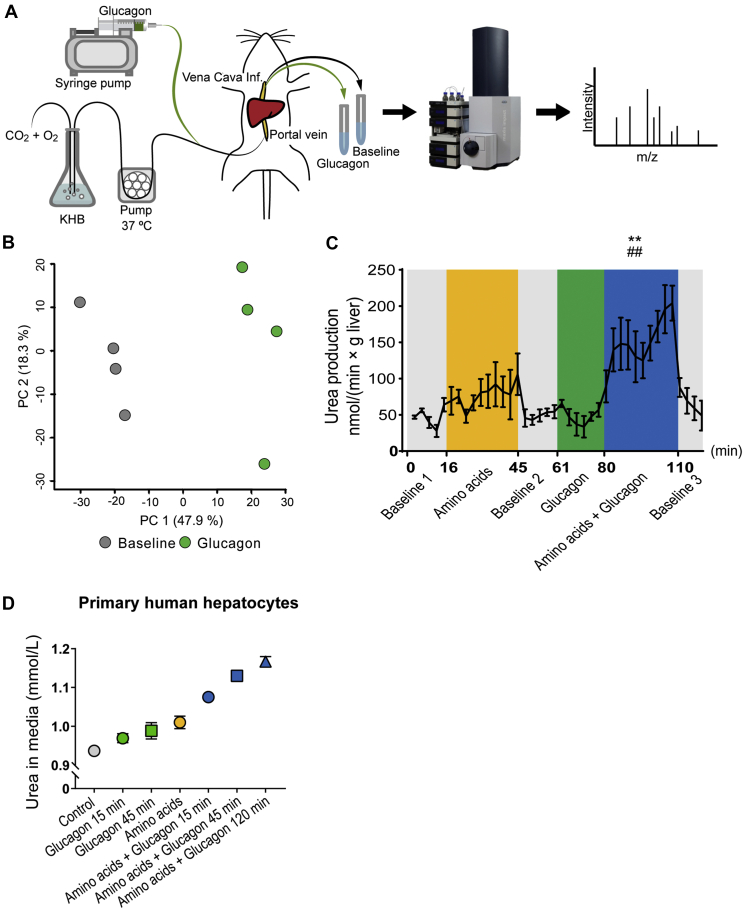
We perfused rat livers to further investigate the acute effects of glucagon on liver metabolism reflected by secreted metabolites (Figure 3A). The functionality of the perfused liver was assessed by measuring glucose production, oxygen uptake, carbon dioxide production, and changes in pH (Fig. S3). Untargeted metabolomics showed a large separation between glucagon stimulation and baseline samples in a principal component analysis (PCA) (Figure 3B and Fig. S3). Liquid chromatography tandem mass spectrometry (LC-MS/MS) identification of the significantly regulated metabolic features revealed 4 and 6 metabolites to be upregulated and downregulated, respectively, by glucagon stimulation (Fig. S3 and Table S2). The metabolite that increased the most upon glucagon stimulation was cyclic AMP (cAMP), consistent with the well-described increase in circulating cAMP upon hepatic glucagon stimulation in both rats and humans [23]. The downregulated metabolites were all amino acids (or amino acid derivatives) consistent with glucagon's stimulating effect on hepatic amino acid uptake and metabolism (Fig. S3).
Figure 3.
Glucagon acutely impacts the release of metabolites from perfused rat livers and potentiates amino acid-induced urea production in perfused rat livers and primary human hepatocytes. (A) Experimental design of the perfused rat liver using Krebs-Henseleit buffer (KHB). n = 4 male rats. (B) Principal component analysis based on both positive and negative mode LC-MS metabolomics of perfusate collected from perfused rat livers during baseline 2 (grey) and glucagon stimulation (10 nM, green) (n = 4). (C) Formation of urea (normalized to liver weight) in perfusate during baseline (gray), amino acids (Vamin, 1 mM, yellow), glucagon (10 nM, green), and amino acids + glucagon (blue) stimulations. Data are shown as mean ± SEM. Statistical analyses were performed on mean output: ##P < 0.01 compared to amino acid stimulation, ∗∗P < 0.001 compared to baseline. (D) Concentration of urea in media of primary human hepatocytes incubated with glucagon (200 nM, green), amino acids (Vamin, 175 mM, yellow), or both glucagon and amino acids (blue) at indicated time periods. Error bars indicate SDs of two technical replicates. See also Fig. S3 and S4.
2.4. Glucagon acutely increases ureagenesis in response to amino acids in situ and in vitro
Next, we aimed to determine whether the rapid increase in amino acid-induced ureagenesis observed in our in vivo mouse studies (Figure 2E) could be ascribed to direct effects of glucagon on hepatic ureagenesis. Production of urea from perfused livers tended to increase in response to stimulation with 1 mmol/L of mixed amino acids compared to baseline 1 (output: 75.3 ± 20.8 vs. 48.9 ± 3.4 nmol/(min × g liver), P = 0.4). Administration of glucagon alone did not change urea output, but simultaneous stimulation with mixed amino acids and glucagon led to a markedly higher urea response compared to the preceding baseline (output: 151.0 ± 24.3 vs. 51.2 ± 6.2 nmol/(min × g liver), P < 0.01) and compared to stimulation with amino acids alone (P < 0.01) (Figure 3C).
We tested the acute effects of glucagon in two commonly used immortal hepatocyte cell lines HepG2 and HepaRG®. HepG2 cells are derived from a well-differentiated hepatocellular carcinoma and display certain hepatocyte-like features, while HepaRG® cells, derived from a human hepatoma, can be treated with dimethyl sulfoxide (DMSO) to differentiate into hepatocyte-like cells. Glucagon increased the cell number in HepaRG® cells (Fig. S4), irrespectively of whether the culture medium contained full serum demonstrating some degree of preserved glucagon receptor signaling. However, neither HepG2 nor HepaRG® cells produced urea in response to glucagon and/or amino acids (Fig. S4). The lack of urea production can be explained by HepG2 cells not expressing two of the five urea cycle enzymes, ornithine transcarbamylase (OTC) and arginase (ARG1) [24]. Differentiation of HepaRG® cells by DMSO induced the expression of OTC but downregulated expression of carbamoylphosphate synthetase (CPS1) while argininosuccinate synthetase (ASS) and ARG1 transcripts were present but not regulated. In our work, HepaRG® did not express argininosuccinate lyase (ASL) (H.C. Bisgaard, unpublished); therefore, neither of these cell lines appears to be useful for studying the urea cycle.
Primary human hepatocytes are, on the other hand, capable of producing urea [25]. Using suspension cultures of primary human hepatocytes, levels of urea in the media were found to increase slightly, and time dependently, upon stimulation with glucagon (Figure 3D). Incubation with 175 mmol/L of mixed amino acids increased urea levels further, and, finally, co-incubation with amino acids and glucagon time-dependently increased urea production even further (Figure 3D). Together these data support that glucagon increases the amino acid-induced urea formation within minutes by directly acting on the hepatocytes.
2.5. Transcriptomic profiling of glucagon receptor knock-out mice reveals altered hepatic amino acid metabolism
In addition to the proposed acute regulation, glucagon is known to regulate ureagenesis at the transcriptional level [26]. We analyzed transcriptomic profiles of livers from global glucagon receptor knockout (Gcgr−/−) mice and wild-type littermates using paired-end RNA sequencing of extracted RNA depleted for ribosomal RNA (Figure 4A) and identified 890 genes showing differential expression in the livers of Gcgr−/− compared to wild-type littermates. Gene clustering of the 50 genes with the highest variance across samples revealed a clear separation between the two mouse genotypes (Figure 4B). Gcgr−/− mice had a 3-fold increase in plasma levels of amino acids (P < 0.0001) [10], and to investigate to what degree amino acid metabolism was altered at the transcriptional level, we mapped genes significantly regulated by glucagon using the gene ontology biological processes (GOBP). An enrichment analysis showed that most of the downregulated genes in Gcgr−/− mice (fold-change > 2) were involved in amino acid metabolism. As an example, 13% of the significantly changed genes mapped to alpha-amino acid catabolic processes compared to a background frequency of 0.4% (Figure 4C and Table S3). The urea cycle includes the five enzymes CPS1, OTC, ASS1, ASL, and ARG1, of which the first two are located in the mitochondria. mRNA levels of all of these, except for Otc, were significantly downregulated in Gcgr−/− mice (Table 1). Furthermore, transcription of N-acetylglutamate synthase (Nags) and the mitochondrial ornithine transporter 1 (Slc25a15), both essential for the urea cycle [26,27], were downregulated in Gcgr−/− mice. Several other amino acid transporters were downregulated, a number of which have also been identified by others upon inhibition of glucagon signaling [6,7,28]. Furthermore, expression of the liver-specific glutaminase (Gls2), an important provider of ammonia to the urea cycle, was reduced (Table 1).
Figure 4.
Expression of amino acid metabolism genes is downregulated in glucagon receptor knockout mice and mice with hepatic steatosis. (A) Work flow of RNA sequencing of liver biopsies from global glucagon receptor knock-out mice (Gcgr−/−) and wild-type (WT) littermates. (B) Gene clustering of the 50 most differentially expressed genes between Gcgr−/− (pink) and WT (blue). Scale represents normalized counts using the variance stabilizing transformation (VST). (C) Gene ontology (GO) biological processes enriched for up- and downregulated genes in Gcgr−/− mice compared to WT littermates. Only genes downregulated by more than 2 (log2(fold-change)) are included. FDR<0.1 was applied to correct for multiple testing. Gcgr−/− mice: n = 4 male mice, 10 weeks of age; WT littermates: n = 5 male mice, 10 weeks of age. (D) Design of the study with female C57BL/6JRj mice (start weight: 21.7 ± 0.1 g) fed high-fat diet and 15% fructose water (HFD + FW) for 8–10 weeks with representative histological pictures of a liver section from a mouse fed HFD + FW (top) and control diet (bottom). Scale bar = 50 μm. (E) Gene expression of the same 50 genes shown in (B) in mice fed high-fat diet and 15% fructose water (HFD + FW) or control diet and water. Scale represents normalized counts using the VST. HFD + FW: n = 3 female mice; control diet: n = 3 female mice. (F) Venn diagram analysis showing the number of genes significantly (FDR<0.1) regulated in Gcgr−/− mice (blue) and mice fed HFD + FW (red) and overlapping genes. See also Tables S3 and S4.
Table 1.
Significantly altered expression of genes related to urea cycle and amino acid transport in the liver. Genes are ranked according to p-value. Genes overlapping between Gcgr−/− mice and mice fed high-fat diet and 15% fructose water (HFD + FW) are written in bold.
|
Gene symbol |
Gene name |
Gcgr−/−/WT |
HFD + FW/control |
||
|---|---|---|---|---|---|
| Fold change | Adjusted P-value |
Fold change | Adjusted P-value |
||
| Urea cycle | |||||
| Gls2 | Glutaminase liver isoform | 0.23 | 7.10E-50 | ||
| Ass1 | Argininosuccinate synthetase | 0.43 | 3.93E-12 | ||
| Arg1 | Arginase 1 | 0.70 | 1.21E-05 | ||
| Asl | Argininosuccinate lyase | 0.65 | 2.64E-05 | ||
| Cps1 | Carbamoyl phosphate synthetase I | 0.62 | 7.10E-04 | 0.56 | 5.13E-02 |
| Nags | N-Acetylglutamate synthase | 0.76 | 2.58E-02 | ||
| Glud1 | Glutamate dehydrogenase | 0.83 | 3.96E-02 | ||
| Amino acid transport | |||||
| Slc3a1 | Neutral and basic amino acid transport protein rBAT | 0.22 | 2.20E-53 | ||
| Slc7a2 | Cationic Amino Acid Transporter 2 | 0.36 | 1.23E-16 | 0.46 | 4.01E-06 |
| Slc25a15 | Mitochondrial ornithine transporter 1 | 0.71 | 2.46E-07 | ||
| Slc38a3 | Sodium-coupled neutral amino acid transporter 3/SNAT3 | 0.59 | 7.49E-06 | ||
| Slc43a1 | Large neutral amino acids transporter small subunit 3 | 0.29 | 8.31E-06 | ||
| Slc1a2 | Excitatory amino acid transporter 2 | 0.66 | 1.12E-03 | ||
| Slc38a4 | Sodium-coupled neutral amino acid transporter 4/SNAT4 | 0.68 | 9.31E-03 | ||
| Slc25a22 | Mitochondrial glutamate carrier 1 | 0.76 | 4.2E-02 | ||
| Slc38a2 | Sodium-coupled neutral amino acid transporter 2/SNAT2 | 0.72 | 8.39E-02 | 0.54 | 2.48E-02 |
2.6. Genes involved in amino acid metabolism are commonly downregulated in mice with hepatic steatosis and glucagon receptor knockout mice
It was recently shown that patients with non-alcoholic fatty liver disease (NAFLD) have reduced capacity for ureagenesis [29], and that gene expression of urea cycle-related enzymes is downregulated in NAFLD patients [30,31]. To test whether this is also the case in mice, animals were fed a high-fat diet and 15% fructose water (HFD + FW) for 8–10 weeks to induce a physiologically relevant model of hepatic steatosis (Figure 4D). Mice on HFD + FW gained significantly more weight than mice fed the control diet (5.2 ± 2.4 vs. 0.01 ± 0.2 g, P < 0.001) (Fig. S5) and had increased levels of hepatic fat content in histological preparations (Figure 4D) and increased levels of triglyceride in liver extracts (Fig. S5).
RNA sequencing revealed 168 significantly regulated genes in the livers of mice on HDF + FW of which 27 genes overlapped with significantly regulated genes in livers from Gcgr−/− mice (Figure 4F and Table S4). Five of the genes with no known association to amino acid metabolism overlapped, but in different directions (Table S4). Of the urea cycle enzymes downregulated in Gcgr−/− mice, Cps1 was also downregulated in mice on HFD + FW (Table 1). Furthermore, the amino acid transporters Slc7a2 and Slc38a2 were significantly downregulated (Table 1 and S4). Thus, at the transcriptional level, amino acid metabolism seems to be disturbed in mice with mild hepatic steatosis and in mice lacking glucagon signaling.
2.7. Increased liver fat results in impaired amino acid metabolism in mice
We next tested whether amino acid metabolism was disturbed in vivo in mice with hepatic steatosis similar to what we observed in mice with disrupted glucagon signaling (Figure 5A). Plasma levels of glucagon were increased in mice fed HFD + FW at baseline and increased significantly more at 4 min after administration of amino acids compared to controls (43.5 ± 2.8 vs. 32.0 ± 3.4 pmol/L, P < 0.05) (Figure 5B). Amino acid clearance, estimated as iAUC, tended to be lower in mice fed HFD + FW (6.8 ± 0.7 vs. 5.5 ± 0.3 min × mmol/L, P = 0.1) and plasma levels of amino acids were significantly higher 12 min after administration of amino acids compared to control mice (8.0 ± 0.6 vs. 6.8 ± 0.2 mmol/L, P < 0.05) (Figure 5C), indicating delayed uptake/metabolism. Four min after administration of amino acids, plasma levels of urea tended to be lower in mice fed HFD + FW compared to controls (P = 0.1) (Figure 5D). As expected, plasma levels of insulin were increased in mice fed HFD + FW at baseline (P < 0.01) as well as at each time point following administration of amino acids (P < 0.001) (Figure 5E). Because blood glucose levels did not differ between the two groups of mice, neither in the basal state nor upon administration of amino acids (Figure 5F), the elevated insulin levels may be indicative of insulin resistance.
Figure 5.
Increased liver fat impairs the liver-alpha cell axis in mice. (A) Design of the study with female C57BL/6JRj mice fed high-fat diet and 15% fructose water (HFD + FW) for 8–10 weeks. Plasma levels of (B) glucagon, (C) amino acids, (D) urea (baseline subtracted), (E) insulin, and (F) blood glucose levels in response to amino acids (Vamin, 3.5 μmol/g body weight) in mice fed HFD + FW (red) or control diet and water (blue) (n = 9–12). Data are shown as mean ± SEM and analyzed by multiple t-tests corrected for multiple testing using the Holm-Sidak algorithm ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. (G) Urea in the culture media of primary hepatocytes from male ob/ob and control mice upon stimulation with mixed amino acids (Vamin, 175 mM) or vehicle (n = 3 mice). Data are shown as mean ± SEM and data are analyzed by AUC using one-way ANOVA. ∗P < 0.05. (H) Formation of urea (normalized to liver weight) in perfusate during baseline (grey), amino acids (Vamin, 10 mM, yellow), glucagon (10 nM, green), and amino acids + glucagon (blue) stimulations in male Zucker lean (black, n = 5) and Zucker fatty rats (red, n = 5). Data are shown as mean ± SEM. See also Fig. S5 and S6.
Severely obese ob/ob mice similarly showed hyperglucagonemia, hyperaminoacidemia, and hyperinsulinemia in the basal state and had increased liver triglyceride levels (Fig. S6). Amino acid clearance in response to administration of mixed amino acids in vivo was reduced (Fig. S6). We furthermore measured formation of urea in primary hepatocytes isolated from ob/ob and control mice. Upon stimulation with a mixture of amino acids, there was a reduced production of urea in ob/ob hepatocytes compared to control hepatocytes (AUC: 35.5 ± 4.0 vs. 51.5 ± 2.8 nmol/μg protein × min, P < 0.05) (Figure 5G). Fat accumulation in the hepatocytes remained after hepatocyte isolation (Fig. S6).
2.8. Hepatic steatosis impairs glucagon-induced ureagenesis in rats
Our results indicate that hepatic steatosis may disrupt amino acid metabolism in a manner similar to that observed after reduced glucagon signaling at both transcriptional and functional levels. To directly test for impairment of glucagon-induced amino acid catabolism, we measured production of urea in perfused rat livers of Zucker fatty and lean rats in response to amino acids and glucagon. Zucker fatty rats had increased liver triglyceride (32.3 ± 3.4 vs. 12.6 ± 1.1 μmol/g liver, P < 0.01). The experimental setup was identical to the previously described liver perfusion experiment (Figure 3A), except for the perfusate concentration of amino acids being increased to 10 mmol/L. In the lean rats, administration of glucagon concomitantly with amino acids increased production of urea compared to amino acid stimulation alone (mean output: 123.5 ± 25.2 vs. 79.9 ± 14.3 mmol/min × g liver, P < 0.05) (Figure 5H). In the fatty rats, the production of urea did not increase significantly upon co-administration of glucagon compared to amino acids alone (mean output: 77.6 ± 13.7 vs. 66.9 ± 5.6 mmol/min × g liver, P = 0.4) (Figure 5H). Release of cAMP upon glucagon stimulation was not significantly different between lean and fatty rats (P = 0.6) (Fig. S6), suggesting that hepatic steatosis does not result in impaired glucagon signaling at the level of cAMP, but specifically reduces glucagon-induced ureagenesis.
2.9. Elevated plasma levels of glucagon and alanine as an indicator of reduced amino acid metabolism in patients with NAFLD
It was recently shown that patients with NAFLD have increased plasma levels of certain amino acids [18,32] and increased secretion of glucagon [17,18], potentially as a response to the increased levels of amino acids. We measured plasma levels of individual amino acids in a cohort of 25 patients with biopsy-proven NAFLD (12 with simple steatosis and 13 with non-alcoholic steatohepatitis (NASH)) and 10 age and sex-matched controls [29] (Figure 6A). Alanine and glutamic acid showed the largest increase in patients with NAFLD with a mean fold change of 1.4 and 1.7, respectively. In addition, the branched-chain amino acids, valine, leucine, and isoleucine, were increased in patients with simple steatosis and NASH compared to controls (Figure 6B). Glutamine levels were significantly lower in patients with simple steatosis and NASH (Figure 6B).
Figure 6.
Increased liver fat distorts the liver-alpha cell axis in humans. (A) Overview of the three cohorts included in the study. Discovery cohort: patients with biopsy-verified steatosis (light blue, n = 12) and non-alcoholic steatohepatitis (NASH, dark blue, n = 13), and controls (grey, n = 10). Validation cohort: patients with biopsy-verified steatosis (light blue, n = 18), NASH (dark blue, n = 17), and healthy controls (grey, n = 9). Weight loss study cohort: obese individuals were subjected to eight weeks of calorie restriction (800 kcal/day, n = 52) and thereafter a 52-week weight maintenance program (n = 39). (B) Fasting plasma levels of individual amino acids in individuals in the discovery cohort. Amino acids are ranked according to mean plasma levels in the control group. Bars indicate median and interquartile range. ∗P < 0.05 between control and both steatosis and NASH, †P < 0.05 between control and steatosis, #P < 0.05 between control and NASH. (C) Glucagon-alanine index (the product of fasting levels of plasma glucagon (pmol/L) and alanine (μmol/L)) in the discovery cohort and (D) validation cohort. Bars indicate median and interquartile range. The data were analyzed by the Kruskal–Wallis test and compared to the healthy control group using Dunn's post hoc test to correct for multiple testing. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. (E) Glucagon-alanine index in individuals before weight loss (week −8), after weight loss (week 0), and after 1-year weight loss maintenance (week 52). Statistical significance was tested by paired t-tests corrected for multiple testing using the Holm-Sidak algorithm. ∗P < 0.05. See also Figure S7.
Because increased plasma levels of alanine previously have been associated with increased liver fat [9] and alanine recently was reported to be an essential regulator of the liver-alpha cell axis in mice, and since glucagon levels were also increased in NAFLD patients in this cohort, we calculated a glucagon-alanine index (the product of fasting plasma levels of glucagon and alanine) [9] as a potential marker of impaired hepatic glucagon sensitivity with respect to amino acid metabolism. In this cohort (discovery cohort), the index was significantly increased in patients with simple steatosis, and numerically even higher in NASH patients (Figure 6C and Fig. S7). We followed up on these findings in a validation cohort, including patients from a different clinical center with biopsy-verified steatosis and NASH and matched controls (Figure 6A). The index was again significantly increased in patients with steatosis and NASH compared to controls (Figure 6D and Fig. S7).
To address whether the glucagon-alanine index may change as a result of a reduction of hepatic steatosis, we calculated the glucagon-alanine index in a cohort of healthy obese individuals at baseline (week −8), after eight weeks of weight loss (week 0), and one year after weight loss (week 52) (Figure 6A) [33]. On average, individuals lost 12% body weight during the weight loss phase (week −8 (mean ± SEM): 97.6 ± 1.6 kg; week 0: 85.6 ± 1.5 kg) and body weight remained stable during the weight maintenance program (week 52: 86.5 ± 1.9 kg). The weight loss resulted in a marked decrease in HOMA-IR (week −8: 9.0 ± 0.8; week 0: 4.9 ± 0.3; week 52: 5.3 ± 0.6), which is a commonly used index reflecting hepatic insulin resistance and is significantly associated with fat accumulation in the liver [34]. The glucagon-alanine index decreased following the initial weight loss and remained low after one year of weight loss maintenance, and the change in the index correlated with the size of weight loss (P < 0.01) (Figure 6E and Fig. S7). Collectively, these data suggest that the liver-alpha cell axis, in which alanine might be a key regulatory amino acid, seems to be disturbed (in a potentially reversible manner) in patients with NAFLD, and that the glucagon-alanine index might be used as a disease marker of NAFLD, particularly relevant for the evaluation of amino acid metabolism.
3. Discussion
Using various animal models, we demonstrated here that glucagon enhances amino acid metabolism by both non-transcriptional and transcriptional mechanism(s). We show that hepatic steatosis and pharmacological or genetic lack of glucagon receptor signaling in mice exhibit similar metabolic perturbations on plasma glucagon and amino acid levels and similar expression profiles of genes involved in hepatic amino acid catabolism. Collectively, our study provides evidence that glucagon, in addition to the known transcriptional mechanisms, also regulates ureagenesis through rapid-acting, non-transcriptional mechanisms, and that hepatic resistance against glucagon's effect on amino acid metabolism may be induced by steatosis. This finding has biological and clinical relevance because the disrupted amino acid metabolism results in hyperglucagonemia, which may contribute to the development of type 2 diabetes [35].
Transcriptomic analysis of livers of Gcgr−/− mice confirmed changes in hepatic amino acid metabolism as previously reported [[5], [6], [7]]. However, potential metabolic adaptations of knock-out mice make the results obtained with this model difficult to interpret, and we therefore turned to a more acute model of interrupted glucagon receptor signaling using a highly specific GRA. This approach resulted in a decrease in baseline blood glucose levels, consistent with previously reported studies in both rodents and humans [10,36]. GRA treatment caused hyperglucagonemia and hyperaminoacidemia and a concomitant decrease in plasma urea demonstrating the importance of glucagon receptor signaling in the liver-alpha cell axis.
To study ureagenesis in mice with an inducible lack of endogenous glucagon, we administered diphtheria toxin (DT) to transgenic mice expressing the human DT receptor under control of the proglucagon promoter, which resulted in an acute loss of pancreatic alpha cells and L cells, leaving only 5% of the pancreatic glucagon remaining in the islets and a much reduced glucagon response to amino acid stimulation. Baseline levels of amino acids were increased, whereas baseline urea and blood glucose levels did not differ, the latter observation being consistent with the notion that relative glucagon deficiency affects amino acid metabolism more than glucose metabolism, at least in this mouse model. Urea formation may be kept within the normal range due to increased substrate levels, or because the lower fasting glucagon plasma levels were nevertheless sufficient to maintain ureagenesis under non-challenged conditions.
Importantly, we found that exogenous glucagon could rescue the hampered amino acid clearance in mice lacking alpha cells and resulted in an accelerated amino acid disappearance rate. We also observed an increased secretion of insulin upon administration of glucagon and amino acids compared to amino acids alone, consistent with the recently emphasized importance of glucagon for the regulation of insulin secretion [37,38]. One of insulin's main functions is to suppress protein catabolism and enhance amino acid uptake and protein synthesis in insulin-sensitive tissues [39]. We recently investigated the effects of inhibition of the insulin receptor by the insulin receptor antagonist S961 [40] and found that treatment with S961 resulted in decreased uptake of amino acids after administration of a mixed amino acid [20]. We therefore cannot rule out that the increased levels of insulin in the DT-treated mice stimulated with glucagon and amino acids may have increased peripheral amino acid uptake, thereby contributing to the circulating pool of amino acids.
Several of our results point to an acute regulation by glucagon of ureagenesis: Firstly, when mice lacking endogenous glucagon received exogenous glucagon simultaneously with mixed amino acids, plasma levels of urea increased to a level equal to that of control mice. Secondly, in the perfused rat liver, exogenous glucagon markedly increased formation of urea in response to amino acids compared to stimulation with amino acids alone. Thirdly, amino acid-induced urea production by primary human hepatocytes was potentiated by addition of glucagon in a time-dependent manner. The small increase in urea production in response to glucagon alone is most likely due to the presence of amino acids in the culture medium. Taken together, our results indicate that glucagon signaling contributes to the minute-to-minute regulation of amino acid metabolism via the urea cycle. The rapid changes in urea production upon glucagon signaling cannot alone be explained by transcriptional changes. Thus, allosteric and/or covalent modifications might be involved, which may include N-acetyl glutamate-mediated activation of CPS1 [41], acetylation [42], deacetylation [43], phosphorylation [44], and regulated activity of amino acid transporters [45].
Patients with NAFLD have elevated fasting levels of glucagon [1,2], and it has been suggested that hyperglucagonemia seen in type 2 diabetes [[3], [4], [5]] may be related to impaired glucagon actions due to fatty liver rather than being related to impaired glucose regulation or disturbed alpha cell function [1]. We hypothesized that reduced glucagon sensitivity would result in reduced amino acid clearance and decreased formation of urea in response to stimulation with amino acids. Mice with hepatic steatosis (ob/ob mice and mice fed HFD + FW) showed hyperinsulinemia and hyperglucagonemia at baseline and in response to amino acids. Whether this was due to increased alpha cell stimulation or to an increased alpha cell mass was not determined. Basal levels of total amino acids were increased in ob/ob mice, but not significantly different in mice fed HFD + FW as would be expected if impairment of glucagon-induced ureagenesis had developed, perhaps because of the concomitant hyperinsulinemia [20]. However, the clearance of amino acids in response to an amino acid challenge decreased in both mouse models of steatosis in line with decreased formation of urea in hepatocytes from ob/ob mice compared to controls. This may, at least partly, be explained by reduced transcription of the rate-limiting urea cycle enzyme CPS1 in the mice fed HFD + FW. In support of this, hypermethylation of promoter regions of the urea cycle genes Otc and Cps1 has been reported both in animals models of NASH and in patients with NAFLD [30]. In addition to Cps1, two amino acid transporters, Slc7a2 and Slc38a2, were downregulated in mice on HFD + FW as well as in Gcgr−/- mice, which is in line with human data showing downregulation in patients with NAFLD [31,46]. SLC38A2 transports neutral amino acids (i.e., alanine), and, in support of its possible role in the liver-alpha cell axis, transcription of Slc38a2 was increased in rats by feeding a high-protein diet, by glucagon administration, and in hepatocytes incubated with forskolin (an activator of adenylyl cyclase) [47]. The three mentioned genes may be sufficient to drive the altered amino acid metabolism observed, but the relatively low number of mice included in the transcriptomic analyses may have resulted in a smaller number of genes being significantly regulated, and other genes could therefore be involved in the glucagon-dependent regulation of hepatic amino acid metabolism.
In addition to transcriptional alterations in hepatic steatosis, the acute glucagon-stimulated urea formation in perfused rat livers was abolished in rats with hepatic steatosis. Thus, hepatic steatosis seems to cause impairment of the glucagon-induced amino acid metabolism and ureagenesis (“glucagon resistance”). We similarly observed in a clinical study that plasma levels of amino acids in obese individuals did not change upon glucagon infusion compared to lean individuals [46]. Formation of urea in response to amino acids alone was not different between rats with hepatic steatosis and controls, which may be due to the relatively high concentration of amino acids (10 mmol/L). It may be speculated that glucose metabolism would also be affected by hepatic glucagon resistance, but we recently found that endogenous glucose production increased similarly in obese individuals compared to lean controls in response to increasing glucagon levels [46]. This is in line with the similar cAMP release upon glucagon stimulation in perfused livers from lean and fat rats and suggests that the regulation by glucagon of amino acid and glucose metabolism, respectively, are separate entities and function independently from each other. Because ureagenesis is initiated in the mitochondria, mitochondrial dysfunction could be involved as observed in patients with NAFLD [48]. HFD feeding has been reported to decrease glucagon receptor expression in rat hepatocytes [49,50], and NAFLD patients similarly have decreased expression of the glucagon receptor [31]. In our mouse model with mild steatosis, the expression of the glucagon receptor was not downregulated, but this might be due to the mild degree of steatosis observed. Nevertheless, because glucagon receptor antagonism leads to increased hepatic fat [51], it is important to further explore whether inhibited glucagon signaling may be related to the development of NAFLD.
Elevated plasma levels of glucagon and amino acids associated with NAFLD [9,18,52] could reflect a disturbed liver-alpha cell axis with reduced amino acid catabolism, again resulting in increasing plasma levels of amino acids, causing hypersecretion of glucagon. As evident from the individual amino acid measurements in NAFLD patients, not all amino acids are equally affected. Glutamine has been suggested to be specifically important in the liver-alpha cell axis in mice [7], which is in line with increased plasma levels in Gcgr−/− mice [7,10] and decreased expression of liver specific glutaminase (GLS2) in Gcgr−/− mice and in patients with steatosis and NASH [31], as well as increased expression of glutamine synthetase in the same patients [31]. In the current study, plasma glutamine levels were reduced in both steatosis and NASH patients, suggesting that glutamine may not be an important regulator of the liver-alpha cell axis.
Alanine, which is also elevated in plasma from Gcgr−/− mice and in humans with high HOMA-IR, was increased in patients with NAFLD. Therefore, we consider alanine as a key regulator of the liver-alpha cell axis. The glucagon-alanine index has been previously shown to be associated with HOMA-IR [9], which is in line with an increase in the glucagon-alanine index in patients with NAFLD compared to healthy controls and the decrease in the glucagon-alanine index upon weight loss, which reduces hepatic steatosis [53]. We did not include a normal-weight age- and gender-matched cohort to the obese individuals in the weight loss study because we found it more important to investigate the weight loss-induced changes in the glucagon-alanine index. There is large biological variation in the index, which may reflect the existence of other potential mediators in the liver-alpha cell axis. It is likely that other amino acids, in addition to alanine, are affected by impaired actions of glucagon. Indeed, all amino acids should be studied individually with regard to their dependency on glucagon for catabolism and their glucagonotropic effects. Measurements of individual amino acids were unfortunately not feasible in our mouse models of interrupted glucagon signaling due to volume restrictions.
A general limitation of our in vivo studies is that the estimation of urea formation solely is based on quantifying plasma levels of urea. It is possible that small changes in urea production might not be measurable in plasma because of efficient renal urea excretion, a process that might even be stimulated by glucagon [54]. Quantitative urine collection is not easy in rodents, and therefore rats are usually nephrectomized before measurement of the urea synthesis rate [55]. We performed a similar procedure in mice anesthetized with isoflurane; however, the kidney ligation resulted in a decrease in plasma urea compared to sham operated mice, independent of stimulus with amino acids or saline indicating that the surgical procedure itself caused the differences (Fig. S1). Another limitation to our study is the mixed use of male and female animals. As isoflurane is known to inhibit glucagon secretion [56], we decided to perform our in vivo studies in conscious mice. Therefore, an element of stress may have influenced the results.
4. Conclusion
Our study demonstrates that lack of glucagon signaling as well as hepatic steatosis results in impaired amino acid metabolism, and, furthermore, that glucagon plays a role in the minute-to-minute regulation of amino acid turnover and urea formation, which is impaired in hepatic steatosis. Our results therefore support the existence of a liver-alpha cell axis [5,8] in which glucagon controls plasma levels of amino acids through activation of hepatic ureagenesis. Disruption of the axis, e.g., due to increased hepatic fat accumulation, results in hyperaminoacidemia and hyperglucagonemia, and given that the hyperglycemic effects of hyperglucagonemia seem to be preserved in this situation [46], these observations point toward a diabetogenic role of steatosis-induced disruption of the liver-alpha cell axis [35].
5. Materials and methods
5.1. Animal experiments
All animal experiments were performed in accordance with the Danish Animal Experiments Inspectorate, Ministry of Environment and Food of Denmark, permits 2013-15-2934-00833 and 2018-15-0201-01397, the EU directive 2010/63/EU, and the National Institutes of Health (publication No. 85-23). All studies were approved by the local ethical committee.
Female C57BL/6JRj mice (10–12 weeks of age), male ob/ob (10 weeks of age), and male Wistar rats (250 g) were obtained from Janvier Labs (Saint-Berthevin, France). Transgenic mice (TgN(GCG.DTR)) were previously described [22]. Male Zucker lean and fatty rats were obtained from Charles River (Italy). Female TgN(GCG.DTR), male Gcgr−/−, and wild-type littermates of both mouse strains were obtained from the local breeding facility (7–12 weeks of age). Mice were housed in groups of maximum eight and rats in groups of maximum four in individually ventilated cages. All animals followed a 12-h light cycle (lights on 6 AM to 6 PM) with ad libitum access to chow (catalog no. 1319, Altromin Spezialfutter, Lage, Germany) and water and were allowed to acclimatize for at least one week after arrival before use. During all studies with transgenic animals, the investigator was blinded to the genotype to avoid bias. On the experimental day, the mice were fasted from 8 AM to 12 PM with free access to water.
5.1.1. Amino acid metabolism upon GRA treatment
Forty-eight female C57BL/6JRj mice (12 week of age) received 0 or 100 mg/kg body weight of a glucagon receptor antagonist (GRA, 25–2648, a gift from Novo Nordisk A/S [21]) dissolved in 5% ethanol, 20% propyleneglycol, 10% 2-hydroxypropyl-β-cyclodextrin (vol./vol.), and phosphate buffer at pH 7.5–8.0 to a concentration of 10 mg/mL. GRA or vehicle was administered orally via gavage (100 μL) 3 h before the experiments as recommended in prior studies [10,21]. Blood samples were collected from the retrobulbar plexus (75 μL) at baseline immediately before administration of the test substance (t = 0 min) and at four subsequent time points (4, 12, and 20 min). Blood glucose was measured in tail blood at the same time points with a handheld plasma glucometer (Accu-Check mobile, Roche). At time zero, amino acids (3.5 μmol/g body weight, Vamin®; catalog no. B05ABA01; Fresenius Kabi, Copenhagen, Denmark, amino acid composition can be found in Table S1) diluted in saline (0.9% NaCl + 1% bovine serum albumin (BSA)) were administered intraperitoneally (200 μL). Blood samples were collected in ethylenediaminetetraacetic acid (EDTA)-coated capillary tubes (Vitrex Medical, Herlev, Denmark) and kept on ice until centrifugation (4,000 g, 10 min). Plasma was transferred into pre-chilled tubes and stored at −20 °C until analyzed for concentrations of total l-amino acids, urea, insulin, and glucagon. At the end of the study, mice were euthanized by cervical dislocation.
5.1.2. Amino acid metabolism upon alpha cell loss
Seventy-eight female TgN (GCG.DTR) mice and 32 wild-type littermates (8–12 weeks of age) received 0 or 10 ng/g body weight of diphtheria toxin (DT; catalog no. D0564; Sigma–Aldrich, Brøndby, DK) dissolved in phosphate-buffered saline (PBS) + 0.1% BSA (catalog no. A6003; Sigma–Aldrich, Brøndby, DK) to a concentration of 10 μg/mL. DT or vehicle was administered intraperitoneally (200–250 μL) every second day for 9 days (5 injections in total). Body weight and blood glucose levels were measured every day of DT administration, but treatment with DT did not affect either of these. Mice were allowed to recover for seven days before further experiments to allow intestinal L-cells to grow back [22]. Basal blood samples (75 μL) were collected from the retrobulbar plexus, and blood glucose levels were measured after tail tip puncture. Immediately thereafter, mixed amino acids (3.5 μmol/g body weight Vamin), amino acids + glucagon (200 ng/g body weight) or saline (0.9% NaCl + 1% BSA) was administered intraperitoneally (200 μL, all diluted in saline). Three additional blood samples (75 μL/time point) were collected, and blood glucose was measured 4, 12, and 20 min after the injection. Blood samples were handled as described above. Tissue samples from the pancreas and ileum were snap-frozen on dry ice and stored at −80 °C until protein extraction or submerged into 4% paraformaldehyde for histological analysis.
5.1.3. Amino acid metabolism in HFD fed mice and ob/ob mice
Sixteen female C57BL/6JRj mice (20 g) were fed a high-fat diet (HFD) for 8–10 weeks containing 31.7 g/100 g lard, 3.2 g/100 g soybean oil, and 0.028% cholesterol (LAD, D12492; Research Diets, New Brunswick, NJ, USA) supplemented with 15% fructose in the drinking water. Another 12 mice were fed a control chow diet (D12450J, Research Diets, New Brunswick, NJ, USA). Food and water were changed and weighed three times a week. Blood glucose levels and body weight of the mice were measured once a week. Several studies have shown great variability in the weight gain caused by high-fat diet feeding in C57BL/6 mice with reported differences between responders and non-responders in glucose metabolism, β-oxidation, and branched-chain amino acid catabolism [57,58]. Based on results from Boulange et al. [58], we defined responders and non-responders as mice with a final body weight above and below 27 g, respectively (9 out of 16 animals were responders and thus included in the following studies). As a model of severely obese mice, 5 male ob/ob and 4 C57BL/6JRj control mice (23 weeks of age) were also included. Basal blood samples (75 μL) were collected from the retrobulbar plexus immediately prior to administration of the test substance, and blood glucose levels were measured after tail tip puncture. At time zero, a mixture of amino acids (3.5 μmol/g body weight Vamin), glucagon (200 ng/g body weight), amino acids + glucagon, or saline (0.9% NaCl + 1% BSA) was administered intraperitoneally (200 μL). Three additional blood samples (75 μL/time point) were collected and blood glucose levels measured 4, 12, and 20 min after the injection. Blood samples were handled as described above. The liver was excised and weighed. A small piece of the right medial lobe was submerged into 4% paraformaldehyde (for histology) and another piece into RNAlater and stored at −20 °C (for RNA sequencing). The rest of the liver was snap-frozen in liquid nitrogen for lipid extraction.
5.1.4. Primary mouse hepatocyte isolation and culture
Male ob/ob and C57BL/6JRj mice, 11 weeks of age, were included. Primary hepatocytes were isolated as previously described [59]. Mice were anesthetized with Avertin (stock 1 g/ml tribromoethanol in T-amyl alcohol, dose 0.5 mg/g body weight intraperitoneally) and an incision in the abdomen was made to locate the inferior vena cava. In ob/ob mice, overlying fat was removed by careful strokes with a cotton swab to expose the vein. A 24G catheter was inserted into the vein, connected to a peristaltic pump and when perfusion started, the portal vein was cut. The liver was perfused with 50 ml of wash buffer (calcium and magnesium free Hanks Buffered Saline Solution (#14170112, Thermo Fisher Scientific), 10 mM of HEPES, 76 mg/L of EGTA) at 3.5 ml/min. Next, the liver was digested with 50 ml of digestion buffer (Williams E medium (#32551020, Thermo Fisher Scientific), 0.4 mg/ml of collagenase (#C5138, Sigma)). The liver was transferred to a Petri dish containing Williams E medium supplemented with 10% fetal bovine serum (FBS) and gently dissociated before being filtered through a 100 μm cell strainer. Hepatocytes were spun down for 2 min at 50 g, and the pellets were washed twice in plating medium (MEM-199 (#41150, Thermo Fisher Scientific), and 10% FBS, 1% penicillin/streptomycin (P/S)). Hepatocytes were resuspended in plating medium and filtered through a 70-μm cell strainer before cell number and viability were assessed using a NucleoCounter (NC-200, ChemoMetec). Viability was on average > 60% in both ob/ob and control mice even though slightly less viability was obtained from ob/ob mice. Hepatocytes were seeded at 130,000 living cells/cm2 in collagen-coated 12-well plates and allowed to attach for a minimum of 2 h in plating medium before being changed into culture medium (MEM-199, 0.5% FBS, 1% P/S, 1 μM dexamethasone, 1 nM insulin) overnight. Experiments were performed on the following day.
Cells were stimulated in fresh MEM-199 medium supplemented with vehicle (water adjusted to the pH of Vamin) or 25% (v/v) Vamin for 15, 45, and 120 min after which media were collected for urea quantification. Cells were rinsed in ice cold PBS, lysed in 200 μl of lysis buffer (10% glycerol, 1% IGEPAL, 50 mM of HEPES, 150 mM of NaCl, 10 mM of NaF, 1 mM of EDTA, 1 mM of EGTA, 20 mM of sodium pyrophosphate, 2 mM of sodium ortho-vanadate, 20 mM of β-glycerophosphate, 5 mM of nicotinamide, 4 μM of Thiamet G and protease inhibitors (SigmaFast, #S8820, Sigma), pH 7.4) and scraped off the well. Lysate was centrifuged for 20 min, 16,000 g, 4 °C.
5.1.5. Hepatic lipid extraction and quantification
The exact weight of each tissue sample was noted, and 1.8 mL of extraction buffer (3% triton X-100 (25% solution in ethanol) in sodium-acetate buffer (0.15M, pH 4.9)) was added. Samples were homogenized for 2 min (3030 Hz) using ice cold blocks, then placed in a 98 °C heat block for 2 min, and subsequently centrifuged at 9,000 g for 10 min. The supernatant was transferred to large COBAS vials for measurement of triglyceride content using standard methods in a COBAS analyzer.
5.1.6. Rat liver perfusion
Four non-fasted male Wistar rats (250 g), five Zucker lean rats (12 weeks, 290 g), and five Zucker fatty rats (12 weeks, 440 g) were anesthetized by a subcutaneous injection with Hypnorm/Midazolam (0.3 ml/100 g body weight; 0.079 mg fentanyl citrate, 2.5 mg fluanisone, 1.25 mg midazolam) and placed on a 37 °C heating table. The abdominal cavity was opened up to the diaphragm, and the large intestine was removed. A catheter was inserted into the portal vein and the liver was perfused (5 mL/min) with Krebs-Henseleit Buffer (118 mM of NaCl, 4.7 mM of KCl, 1.2 mM of MgSO4, 1.25 mM of CaCl2, 1.2 mM of KH2PO4, 25 mM of NaHCO3, 10 mM of glucose) gassed with 95% O2 and 5% CO2. The buffer was heated to 37 °C by passing it through a Uniper UP-100 perfusion system (Hugo Sachs; Harvard Apparatus, March-Hugstetten, Germany). Immediately after, the diaphragm and ribcage were dissected exposing the upper part of the inferior vena cava into which a catheter was inserted to collect the outflow (perfusion effluent). The flow was then increased to 30 mL/min for Wistar rats and 20 mL/min for Zucker rats. An amino acid mixture (Vamin) was added directly to the perfusion buffer in a final concentration of 1 mM for Wistar rats and 10 mM for Zucker rats. Glucagon was infused via a sidearm infusion pump (1 uM, flow rate 0.33 mL/min) for a final concentration of 10 nM reaching the liver. Perfusion samples were collected every 3 min throughout the protocol. After collection, samples were immediately chilled on ice and stored at −20 °C until analysis. For Wistar rats, samples of perfusion buffer were collected just before and after passing the perfused rat livers at times 1, 70, and 110 min. The samples were transferred to cassettes compatible with the blood gas analyzer ABL90-series (Radiometer, Brønshøj, Denmark) and measured immediately after collection.
5.2. Human samples
-
1.
Discovery cohort: Fasting plasma samples were available from 25 patients with biopsy-proven NAFLD (12 with simple steatosis (58% men) and 13 with non-alcoholic steatohepatitis (NASH, 77% men)) and 10 age and sex matched healthy controls (70% men). Patients were recruited from the Department of Hepatology and Gastroenterology, Aarhus University Hospital. The study was approved by the Central Denmark Region Committees on Health Research Ethics (1-10-72-140-14). Results from the same cohort of patients and controls have previously been reported [29,31].
-
2.
Validation cohort: Ten healthy controls (60% women; median age 34.5 years, range: 26–54; median BMI 23.3, range: 20.7–26.4) and 35 patients (59% women; median age 54 years, range: 22–77; median BMI 33.3, range: 23.7–46.5) with histologically verified NAFLD (18 simple steatosis with fibrosis grade 0–1 and 17 NASH with fibrosis grade 2–3) were included. All patients were recruited from the Gastrounit, Copenhagen University Hospital Hvidovre (VEK H-17029039). Plasma samples were acquired in the fasting state.
-
3.
Weight loss study: Fasting plasma samples were obtained from 52 healthy obese individuals (85% women) that followed a very low-calorie diet (800 kcal/day) for eight weeks resulting in an average weight reduction of 13%. Participants then entered a 52-week weight maintenance program, during which 50% of the cohort received liragutide (1.2 mg/day), whereas the other 50% did not. Results from the same cohort of patients have previously been reported [60]. The study was approved by The Ethical Committee of The Capital Region of Denmark (reference number: H-4-2010-134).
5.3. Cell lines
Frozen human hepatocytes (catalog no. HMCPP5; Gibco, Thermo Fisher Scientific, Waltham, MA, USA) were thawed according to the manufacturers' recommendation. HepG2 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FBS. HeparG cells were maintained in William's E medium supplemented with 10% fetal bovine serum or serum free media, 100 U/ml of penicillin, 100 g/ml of streptomycin, 5 g/ml of insulin, and 5 × 10−5 M of hydrocortisone hemisuccinate.
5.3.1. Cell culture
Primary human hepatocytes were kept in suspension at 37 °C either in DMEM without fetal calf serum or DMEM with 25% (v/v) Vamin, and treated with 200 nM of glucagon for 15, 45, or 120 min (each step contained 500,000 hepatocytes). Hepatocytes kept in suspension in DMEM without glucagon served as a negative control. At the end of the experiment, the media was separated from the cells by centrifugation at 1,500 rpm. The cells were treated with radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris–HCl (pH 7.5), 150 mM of NaCl, 1 mM of Na2EDTA, 1% Triton X-100) for 20 min after 10 s sonication. The lysate was centrifuged at 15,000 rpm and the supernatant was stored at −20 °C. HepG2 cells were stimulated with glucagon in a final concentration of 20 nM. HepaRG cells were stimulated with 20 nM glucagon, or 20 nM glucagon +2% Vamin with or without previous 1.7% DMSO treatment. After the end of the stimulation period (3 h), cell media or cell lysate were obtained and centrifuged (1,500 g, 4 °C, 5 min) to remove any cells or debris and kept at −20 °C until analysis.
5.4. Protein extraction and hormone quantification
Snap-frozen tissue of pancreas and ileum from 6 to 7 TgN(GCG.DTR) mice treated with DT or vehicle, or wild-type mice treated with DT, were subject to peptide extraction using 1% trifluoroacetic acid as described previously [61]. The extracts were purified using pH resistant tc18 cartridges (catalog no. WAT036810; Waters), dried overnight under a gentle air stream, and reconstituted in TRIS buffer (100 mmol/l TRIS buffer (Sigma–Aldrich) supplemented with 0.1% human serum albumin (catalog no. 126658; Merck), 20 mmol/l EDTA and 0.6 mmol/l thimerosal (catalog no. T-5125, Sigma Chemical), pH 8.5). Levels of pancreatic glucagon and ileal total GLP-1 were quantified using in-house radioimmunoassays (RIA) (glucagon: codename 4305 [62]; GLP-1: codename 390 [63]).
5.5. Histology
Liver and pancreas specimens were fixed in 10% formalin and embedded in paraffin. All embedded tissue blocks were sectioned by microtome and placed on glass slides. The liver specimens were dewaxed, stained with hematoxylin and eosin, and coverslipped. The occurrence and localization of glucagon in the pancreas specimens were studied by immunohistochemistry using the polyclonal glucagon antibody 4304 (in-house from J.J. Holst lab [64]). In short, the sections were dewaxed and incubated overnight with the glucagon antibody and diluted 1:5,000. On day two, sections were incubated with biotin goat anti-rabbit antibody (catalog no. BA-1000, Vector Laboratories. Burlingame, CA, USA) 1:200, followed by incubation with in Vectastain ABC complex (catalog no. PK-4000, Vector Laboratories) and 3,3′-diaminobenzidine (DAB, catalog no. SK-4100, Vector Laboratories) and finally lightly counterstained with hematoxylin and coverslipped.
5.6. Biochemical measurements
Plasma amino acid levels were quantified using an l-Amino Acid Assay Kit (catalog no. ab65347; Abcam, Cambridge, UK) according to the manufacturer's protocol. Plasma levels of the individual amino acids were measured using liquid chromatography as previously described [18]. Plasma levels of glucagon were measured using a validated [56] two-site enzyme immunoassay (catalog no. 10-1281-01; Mercodia, Uppsala, Sweden) according to the manufacturer's protocol. Plasma levels of insulin were quantified using an enzyme-linked immunosorbent assay (ELISA, catalog no. 10-1247-01, Mercodia, Uppsala, Sweden). Levels of urea were quantified using QuantiChrom Urea Assay Kit (catalog no. DIUR-100; BioAssay Systems, Hayward, CA) following the 5-μL protocol for plasma and 50-μL protocol for perfusate and cell culture media. cAMP for supplementary data was measured using an ELISA (catalog no. ab65355; Abcam; Cambridge, UK).
5.7. RNA sequencing
Five male glucagon receptor knock-out mice and five male wild-type littermates (10 weeks of age) (bred in-house) were subject to a short-term fast of 4 h before anesthesia with 2.5% isoflurane. A liver biopsy was taken from the right medial lobe and stored in RNAlater at −20 °C until analysis. Liver biopsies from three female mice fed high-fat diet + fructose water or control were collected after euthanizing the mice immediately after the amino acid stimulating experiment described above. Total RNA was purified by an AllPrep DNA/RNA/protein extraction kit (QIAgen) according to manufacturer's instructions and qualified using a 2100 Bioanalyzer instrument (Agilent Genomics). RNA-sequencing was done using TruSeq Stranded Total RNA Library Prep Kit (Illumina) according to the manufacturer's protocol. In brief, ribosomal RNA was depleted using TruSeq Standed RNA Ribominus Gold (Illumina), and cDNA libraries were purified using AMPure® XP Beads (Beckman). Following end-repair and poly(A)-tailing, adaptor ligation was performed with Truseq RNA single indexes set A/B (Illumina). RNAseq libraries were paired-end sequenced (2 × 125 bp) on an Illumina HiSeq2500 instrument and raw sequencing data were processed using CASAVA v. 1.8 conversion software (Illumina).
The RAW files (FastQ) were processed at the Danish National Supercomputer for Life Sciences, and the read quality was evaluated using FastQC. Reads were mapped to the mouse genome (GRCm38). We normalized the data using the algorithm variance stabilizing transformation (VST) offered by the DESeq2 package [65] (version 2_1.20.0) in R (version 3.5.0), which ensures homoscedasticity and normalizes with respect to library size. We used the same R package to identify differentially expressed genes. Gene ontology (GO) enrichment was performed at the level of biological processes (BP) and the list of all identified genes was used as background [65,66].
5.8. Metabolomic analysis
Samples of perfusion buffer collected during baseline and glucagon stimulation of the perfused rat liver were subjected to small molecule liquid chromatography Mass Spectrometry (LC-MS) analysis. Samples from each stimulation period (5 samples from baseline and 6 samples from glucagon) for each animal (n = 4) were combined in equal volumes before being prepared for LC-MS. Samples were dried via speed vacuum overnight (>16 h) and extracted with 0.5 mL of ice-cold LC-MS grade acetonitrile/2-propanol/methanol (1:1:0.4). Samples were suspended in extraction solvent with vigorous pipetting and vortexing followed by shaking (1,400 rpm) for 10 min in a pre-chilled (4 °C) Eppendorf Thermomixer. Clarified supernatants (centrifuged at 20,817 g and 4 °C for 10 min) were transferred to fresh 1.5-mL centrifuge tubes and dried via speed vacuum. Samples were suspended in 50 μL of ice-cold 5% (v/v) LC-MS grade acetonitrile containing 0.1% (v/v) formic acid. Clarified supernatants (centrifuged same as above) were transferred to ice-cold autosampler vials. Samples were injected (10 μL) onto a Phenomenex Luna Omega Polar C18 (2.1 × 100 mm) column and separated using a gradient between mobile phase A (0.1% (v/v) formic acid) and B (0.1% (v/v) formic acid in acetonitrile). Samples were analyzed in both negative and positive ESI full scan (m/z: 50–1,000) modes using a Bruker Impact II QTOF. Data for the initial analysis was acquired in MS1 only. Raw files from the LC-MS analysis were processed in MetaboScape 3.0 (Bruker, Germany). Mass recalibration was performed with lock mass and sodium formate clusters. Intensity threshold for feature detection was 1,500 counts in positive mode and 500 in negative mode. The minimum peak length was set to 6 spectra or 4 spectra for ‘recursive feature extraction’. Chromatographic peaks were aligned and metabolic features extracted based on the H+, Na+ and K+ adducts for the positive mode, H− and Cl− for the negative mode and isotope clustering. Extracted metabolic features in CSV files were merged from positive and negative mode and analyzed using MetaboAnalyst 4.0 [67]. Data were filtered using IQR, normalized by sum, log transformed and Pareto scaled. PCA score plot and paired t-test analysis was performed. A metabolic feature was considered significant if the q value from FDR was <0.05. Significantly dysregulated features were identified by matching m/z ratios and MS/MS fragmentation patterns to Metlin [68] and/or HMDB [69].
5.9. Calculations and statistical analysis
A glucagon-alanine index was calculated as previously described [9] by multiplying fasting plasma levels of glucagon (pmol/L) and alanine (mmol/L). For mouse experiments, area under the curve (AUC) is presented as incremental AUC (iAUC, positive changes from baseline) except for plasma urea levels, where netAUC (the sum of positive and negative changes from baseline) was applied. Amino acid clearance was estimated as iAUC. For rat liver perfusions, output was calculated as mean peak value for stimulation periods and compared with analysis of variance (ANOVA) with repeated measurements. All data are presented as mean ± SEM unless otherwise stated. Unpaired t-tests were used to analyze data from two independent groups. To analyze more than two groups, a one-way ANOVA or multiple t-tests corrected for multiple testing using the Holm-Sidak algorithm were applied. A p value < 0.05 was considered significant. Calculations were performed using GraphPad Prism version 7.02. R version 3.5.0 was used to analyze RNA sequencing data.
5.10. Data availability
The metabolomics data generated are available at MetaboLights [70] under the accession number MTBLS1508 (www.ebi.ac.uk/metabolights/MTBLS1508). RNA sequencing data is available upon request to the Corresponding Author Nicolai J. Wewer Albrechtsen (hgk795@ku.dk).
Funding
This study was supported by the Novo Nordisk Foundation (NNF) Tandem Program (31526), NNF Project support in Endocrinology and Metabolism – Nordic Region (34250), and NNF Excellence Emerging Investigator Grant – Endocrinology and Metabolism (NNF19OC0055001).
Author contributions
MW-S, KDG, JP, JJH, and NJWA designed the study and experiments. MW-S, KDG, JAW, and JEH performed mouse in vivo experiments. MW-S, REK, and DBA designed and performed rat liver perfusion experiments. MWS, ASH, MD, and JTT performed studies with isolated mouse hepatocytes. SASK measured samples from in vivo and in vitro experiments. SLJ performed pancreas perfusion experiments. ASD analyzed data of mouse liver RNA-sequencing. SAJT and MPG performed the LC-MS experiments. SAJT, KS, and CGV analyzed the metabolomics data. CGV and KS took part in the MetaboScape 3.0 development in collaboration with Bruker GmbH. SST, LG, MPW, PLE, and HV provided patient material and clinical data. CØ performed histology. HCB provided cell lines, HepaRG RNA-sequencing data, and interpreted liver perfusion experiments. MW-S and NJWA drafted the manuscript. FKK revised the manuscript. NJWA and JJH designed and interpreted the study, supervised data analyses and revised the manuscript. All authors approved the final version of the manuscript.
Acknowledgments
We thank Maureen J. Charron, Departments of Biochemistry, Obstetrics and Gynecology and Women's Health, and Medicine, Albert Einstein College of Medicine, New York, New York for providing glucagon receptor knock-out mice. We also thank laboratory technician Ramya Kweder, Biotech Research & Innovation Center, University of Copenhagen, Denmark, for performing cell culture studies. The assistance from laboratory technicians Lene Albæk and Jeppe Bach, Department of Biomedical Sciences, University of Copenhagen, Denmark, is truly appreciated. We acknowledge the work implemented regarding the weight loss study by Julie R. Lundgren and Eva W. Lehmann, Department of Biomedical Sciences, University of Copenhagen, Denmark, and Sten Madsbad, Department of Endocrinology, Hvidovre University Hospital, Denmark. We would like to acknowledge Florian Meier, Department of Proteomics and Signal Transduction, Max Planck Institute of Biochemistry, for support of the collaboration with Bruker GmbH. We would also like to thank the Center for Genomic Medicine, Rigshospitalet, Denmark for performing RNA sequencing. The graphical abstract was created with BioRender. Novo Nordisk Foundation Center for Protein Research is supported financially by the Novo Nordisk Foundation (Grant agreement NNF14CC0001).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101080.
Contributor Information
Jens J. Holst, Email: jjholst@sund.ku.dk.
Nicolai J. Wewer Albrechtsen, Email: hgk795@ku.dk.
Conflict of interest
The authors declare no competing interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kimball C.P., Murlin J.R. Aqueous extracts of pancreas: III. Some precipitation reactions of insulin. Journal of Biological Chemistry. 1923;58:337–348. [Google Scholar]
- 2.Boden G., Rezvani I., Owen O.E. Effects of glucagon on plasma amino acids. Journal of Clinical Investigation. 1984;73(3):785–793. doi: 10.1172/JCI111272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamberg O., Vilstrup H. Regulation of urea synthesis by glucose and glucagon in normal man. Clinical Nutrition. 1994;13(3):183–191. doi: 10.1016/0261-5614(94)90099-x. [DOI] [PubMed] [Google Scholar]
- 4.Assan R., Attali J.R., Ballerio G., Boillot J., Girard J.R. Glucagon secretion induced by natural and artificial amino acids in the perfused rat pancreas. Diabetes. 1977;26(4):300–307. doi: 10.2337/diab.26.4.300. [DOI] [PubMed] [Google Scholar]
- 5.Solloway M.J., Madjidi A., Gu C., Eastham-Anderson J., Clarke H.J., Kljavin N. Glucagon couples hepatic amino acid catabolism to mTOR-dependent regulation of alpha-cell mass. Cell Reports. 2015;12(3):495–510. doi: 10.1016/j.celrep.2015.06.034. [DOI] [PubMed] [Google Scholar]
- 6.Kim J., Okamoto H., Huang Z., Anguiano G., Chen S., Liu Q. Amino acid transporter slc38a5 controls glucagon receptor inhibition-induced pancreatic alpha cell hyperplasia in mice. Cell Metabolism. 2017;25(6):1348–1361. doi: 10.1016/j.cmet.2017.05.006. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean E.D., Li M., Prasad N., Wisniewski S.N., Von Deylen A., Spaeth J. Interrupted glucagon signaling reveals hepatic alpha cell axis and role for L-glutamine in alpha cell proliferation. Cell Metabolism. 2017;25(6):1362–1373. doi: 10.1016/j.cmet.2017.05.011. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holst J.J., Wewer Albrechtsen N.J., Pedersen J., Knop F.K. Glucagon and amino acids are linked in a mutual feedback cycle: The liver-alpha-cell axis. Diabetes. 2017;66(2):235–240. doi: 10.2337/db16-0994. [DOI] [PubMed] [Google Scholar]
- 9.Wewer Albrechtsen N.J., Faerch K., Jensen T.M., Witte D.R., Pedersen J., Mahendran Y. Evidence of a liver-alpha cell axis in humans: hepatic insulin resistance attenuates relationship between fasting plasma glucagon and glucagonotropic amino acids. Diabetologia. 2018;61(3):671–680. doi: 10.1007/s00125-017-4535-5. [DOI] [PubMed] [Google Scholar]
- 10.Galsgaard K.D., Winther-Sørensen M., Ørskov C., Kissow H., Poulsen S.S., Vilstrup H. Disruption of glucagon receptor signaling causes hyperaminoacidemia exposing a possible liver-alpha-cell axis. American Journal of Physiology-Endocrinology and Metabolism. 2018;314(1):E93–E103. doi: 10.1152/ajpendo.00198.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelling R.W., Du X.Q., Dichmann D.S., Romer J., Huang H., Cui L. Lower blood glucose, hyperglucagonemia, and pancreatic alpha cell hyperplasia in glucagon receptor knockout mice. Proceedings of the National Academy of Sciences of the U S A. 2003;100(3):1438–1443. doi: 10.1073/pnas.0237106100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longuet C., Robledo A.M., Dean E.D., Dai C., Ali S., McGuinness I. Liver-specific disruption of the murine glucagon receptor produces alpha-cell hyperplasia: evidence for a circulating alpha-cell growth factor. Diabetes. 2013;62(4):1196–1205. doi: 10.2337/db11-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larger E., Wewer Albrechtsen N.J., Hansen L.H., Gelling R.W., Capeau J., Deacon C.F. Pancreatic α-cell hyperplasia and hyperglucagonemia due to a glucagon receptor splice mutation. Endocrinology, Diabetes & Metabolism Case Reports. 2016;2016:16–81. doi: 10.1530/EDM-16-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norton J.A., Kahn C.R., Schiebinger R., Gorschboth C., Brennan M.F. Amino acid deficiency and the skin rash associated with glucagonoma. Annals of Internal Medicine. 1979;91(2):213–215. doi: 10.7326/0003-4819-91-2-213. [DOI] [PubMed] [Google Scholar]
- 15.Almdal T.P., Heindorff H., Bardram L., Vilstrup H. Increased amino acid clearance and urea synthesis in a patient with glucagonoma. Gut. 1990;31(8):946–948. doi: 10.1136/gut.31.8.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rocha D.M., Faloona G.R., Unger R.H. Glucagon-stimulating activity of 20 amino acids in dogs. Journal of Clinical Investigation. 1972;51(9):2346–2351. doi: 10.1172/JCI107046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Junker A.E., Gluud L., Holst J.J., Knop F.K., Vilsboll T. Diabetic and nondiabetic patients with nonalcoholic fatty liver disease have an impaired incretin effect and fasting hyperglucagonaemia. Journal of Internal Medicine. 2016;279(5):485–493. doi: 10.1111/joim.12462. [DOI] [PubMed] [Google Scholar]
- 18.Wewer Albrechtsen N.J., Junker A.E., Christensen M., Haedersdal S., Wibrand F., Lund A.M. Hyperglucagonemia correlates with plasma levels of non-branched-chain amino acids in patients with liver disease independent of type 2 diabetes. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2018;314(1):G91–G96. doi: 10.1152/ajpgi.00216.2017. [DOI] [PubMed] [Google Scholar]
- 19.Pedersen J.S., Rygg M.O., Kristiansen V.B., Olsen B.H., Serizawa R.R., Holst J.J. Nonalcoholic fatty liver disease impairs the liver–alpha cell axis independent of hepatic inflammation and fibrosis. Hepatology Communications. 2020 doi: 10.1002/hep4.1562. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galsgaard K.D., Winther-Sorensen M., Pedersen J., Kjeldsen S.A.S., Rosenkilde M.M., Wewer Albrechtsen N.J. Glucose and amino acid metabolism in mice depend mutually on glucagon and insulin receptor signaling. American Journal of Physiology. Endocrinology and Metabolism. 2019;316(4):E660–E673. doi: 10.1152/ajpendo.00410.2018. [DOI] [PubMed] [Google Scholar]
- 21.Kodra J.T., Jørgensen A.S., Andersen B., Behrens C., Brand C.L., Christensen I.T. Novel glucagon receptor antagonists with improved selectivity over the glucose-dependent insulinotropic polypeptide receptor. Journal of Medicinal Chemistry. 2008;51(17):5387–5396. doi: 10.1021/jm7015599. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen J., Ugleholdt R.K., Jorgensen S.M., Windelov J.A., Grunddal K.V., Schwartz T.W. Glucose metabolism is altered after loss of L cells and alpha-cells but not influenced by loss of K cells. American Journal of Physiology. Endocrinology and Metabolism. 2013;304(1):E60–E73. doi: 10.1152/ajpendo.00547.2011. [DOI] [PubMed] [Google Scholar]
- 23.Strange R.C., Mjos O.D. The sources of plasma cyclic AMP: studies in the rat using isoprenaline, nicotinic acid and glucagon. European Journal of Clinical Investigation. 1975;5(2):147–152. doi: 10.1111/j.1365-2362.1975.tb00440.x. [DOI] [PubMed] [Google Scholar]
- 24.Mavri-Damelin D., Eaton S., Damelin L.H., Rees M., Hodgson H.J.F., Selden C. Ornithine transcarbamylase and arginase I deficiency are responsible for diminished urea cycle function in the human hepatoblastoma cell line HepG2. The International Journal of Biochemistry & Cell Biology. 2007;39(3):555–564. doi: 10.1016/j.biocel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Bhogal R.H., Hodson J., Bartlett D.C., Weston C.J., Curbishley S.M., Haughton E. Isolation of primary human hepatocytes from normal and diseased liver tissue: a one hundred liver experience. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0018222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris S.M., Jr. Regulation of enzymes of the urea cycle and arginine metabolism. Annual Review of Nutrition. 2002;22:87–105. doi: 10.1146/annurev.nutr.22.110801.140547. [DOI] [PubMed] [Google Scholar]
- 27.Camacho J.A., Obie C., Biery B., Goodman B.K., Hu C.A., Almashanu S. Hyperornithinaemia-hyperammonaemia-homocitrullinuria syndrome is caused by mutations in a gene encoding a mitochondrial ornithine transporter. Nature Genetics. 1999;22(2):151–158. doi: 10.1038/9658. [DOI] [PubMed] [Google Scholar]
- 28.Yang J., MacDougall M.L., McDowell M.T., Xi L., Wei R., Zavadoski W.J. Polyomic profiling reveals significant hepatic metabolic alterations in glucagon-receptor (GCGR) knockout mice: implications on anti-glucagon therapies for diabetes. BMC Genomics. 2011;12:281. doi: 10.1186/1471-2164-12-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eriksen P.L., Sørensen M., Grønbæk H., Hamilton-Dutoit S., Vilstrup H., Thomsen K.L. Non-alcoholic fatty liver disease causes dissociated changes in metabolic liver functions. Clinics and Research in Hepatology and Gastroenterology. 2019;43(5):551–560. doi: 10.1016/j.clinre.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 30.De Chiara F., Heebøll S., Marrone G., Montoliu C., Hamilton-Dutoit S., Ferrandez A. Urea cycle dysregulation in non-alcoholic fatty liver disease. Journal of Hepatology. 2018;69(4):905–915. doi: 10.1016/j.jhep.2018.06.023. [DOI] [PubMed] [Google Scholar]
- 31.Eriksen P.L., Vilstrup H., Rigbolt K., Suppli M.P., Sorensen M., Heeboll S. Non-alcoholic fatty liver disease alters expression of genes governing hepatic nitrogen conversion. Liver International. 2019;39(11):2094–2101. doi: 10.1111/liv.14205. [DOI] [PubMed] [Google Scholar]
- 32.Jin R., Banton S., Tran V.T., Konomi J.V., Li S., Jones D.P. Amino acid metabolism is altered in adolescents with nonalcoholic fatty liver disease-an untargeted, high resolution metabolomics study. The Journal of Pediatrics. 2016;172:14–19. doi: 10.1016/j.jpeds.2016.01.026. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iepsen E.W., Lundgren J., Dirksen C., Jensen J.E., Pedersen O., Hansen T. Treatment with a GLP-1 receptor agonist diminishes the decrease in free plasma leptin during maintenance of weight loss. International Journal of Obesity. 2015;39(5):834–841. doi: 10.1038/ijo.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isokuortti E., Zhou Y., Peltonen M., Bugianesi E., Clement K., Bonnefont-Rousselot D. Use of HOMA-IR to diagnose non-alcoholic fatty liver disease: a population-based and inter-laboratory study. Diabetologia. 2017;60(10):1873–1882. doi: 10.1007/s00125-017-4340-1. [DOI] [PubMed] [Google Scholar]
- 35.Wewer Albrechtsen N.J., Galsgaard K.D., Janah L., Winther-Sørensen M., Holst J.J., Pedersen J. The liver-alpha cell axis and type 2 diabetes. Endocrine Reviews. 2019;40(5):1353–1366. doi: 10.1210/er.2018-00251. [DOI] [PubMed] [Google Scholar]
- 36.Kazda C.M., Ding Y., Kelly R.P., Garhyan P., Shi C., Lim C.N. Evaluation of efficacy and safety of the glucagon receptor antagonist LY2409021 in patients with type 2 diabetes: 12- and 24-week phase 2 studies. Diabetes Care. 2016;39(7):1241–1249. doi: 10.2337/dc15-1643. [DOI] [PubMed] [Google Scholar]
- 37.Svendsen B., Larsen O., Gabe M.B.N., Christiansen C.B., Rosenkilde M.M., Drucker D.J. Insulin Secretion Depends on Intra-islet Glucagon Signaling. Cell Reports. 2018;25(5):1127–1134. doi: 10.1016/j.celrep.2018.10.018. e2. [DOI] [PubMed] [Google Scholar]
- 38.Hjorth S.A., Adelhorst K., Pedersen B.B., Kirk O., Schwartz T.W. Glucagon and glucagon-like peptide 1: selective receptor recognition via distinct peptide epitopes. Journal of Biological Chemistry. 1994;269(48):30121–30124. [PubMed] [Google Scholar]
- 39.Biolo G., Wolfe R.R. Insulin action on protein metabolism. Baillière's Clinical Endocrinology and Metabolism. 1993;7(4):989–1005. doi: 10.1016/s0950-351x(05)80242-3. [DOI] [PubMed] [Google Scholar]
- 40.Schaffer L., Brand C.L., Hansen B.F., Ribel U., Shaw A.C., Slaaby R. A novel high-affinity peptide antagonist to the insulin receptor. Biochemical and Biophysical Research Communications. 2008;376(2):380–383. doi: 10.1016/j.bbrc.2008.08.151. [DOI] [PubMed] [Google Scholar]
- 41.Hensgens H.E., Verhoeven A.J., Meijer A.J. The relationship between intramitochondrial N-acetylglutamate and activity of carbamoyl-phosphate synthetase (ammonia). The effect of glucagon. European Journal of Biochemistry. 1980;107(1):197–205. doi: 10.1111/j.1432-1033.1980.tb04640.x. [DOI] [PubMed] [Google Scholar]
- 42.Xiong Y., Guan K.L. Mechanistic insights into the regulation of metabolic enzymes by acetylation. The Journal of cell biology. 2012;198(2):155–164. doi: 10.1083/jcb.201202056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nogueiras R., Habegger K.M., Chaudhary N., Finan B., Banks A.S., Dietrich M.O. Sirtuin 1 and sirtuin 3: physiological modulators of metabolism. Physiol Rev. 2012;92(3):1479–1514. doi: 10.1152/physrev.00022.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Avruch J., Witters L.A., Alexander M.C., Bush M.A. Effects of glucagon and insulin on cytoplasmic protein phosphorylation in hepatocytes. Journal of Biological Chemistry. 1978;253(13):4754–4761. [PubMed] [Google Scholar]
- 45.Gebhardt R., Kleemann E. Hormonal regulation of amino acid transport system N in primary cultures of rat hepatocytes. European Journal of Biochemistry. 1987;166(2):339–344. doi: 10.1111/j.1432-1033.1987.tb13520.x. [DOI] [PubMed] [Google Scholar]
- 46.Suppli M.P., Bagger J.I., Lund A., Demant M., van Hall G., Strandberg C. Glucagon resistance at the level of amino acid turnover in obese subjects with hepatic steatosis. Diabetes. 2020;69(6):1090. doi: 10.2337/db19-0715. [DOI] [PubMed] [Google Scholar]
- 47.Ortiz V., Alemán G., Escamilla-Del-Arenal M., Recillas-Targa F., Torres N., Tovar A.R. Promoter characterization and role of CRE in the basal transcription of the rat SNAT2 gene. American Journal of Physiology-Endocrinology and Metabolism. 2011;300(6):E1092–E1102. doi: 10.1152/ajpendo.00459.2010. [DOI] [PubMed] [Google Scholar]
- 48.Nassir F., Ibdah J.A. Role of mitochondria in nonalcoholic fatty liver disease. International Journal of Molecular Sciences. 2014;15(5):8713–8742. doi: 10.3390/ijms15058713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Charbonneau A., Melancon A., Lavoie C., Lavoie J.-M. Alterations in hepatic glucagon receptor density and in Gsα and Giα2 protein content with diet-induced hepatic steatosis: effects of acute exercise. American Journal of Physiology-Endocrinology and Metabolism. 2005;289(1):E8–E14. doi: 10.1152/ajpendo.00570.2004. [DOI] [PubMed] [Google Scholar]
- 50.Charbonneau A., Unson C.G., Lavoie J.M. High-fat diet-induced hepatic steatosis reduces glucagon receptor content in rat hepatocytes: potential interaction with acute exercise. The Journal of physiology. 2007;579(Pt 1):255–267. doi: 10.1113/jphysiol.2006.121954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guzman C.B., Zhang X.M., Liu R., Regev A., Shankar S., Garhyan P. Treatment with LY2409021, a glucagon receptor antagonist, increases liver fat in patients with type 2 diabetes. Diabetes, Obesity and Metabolism. 2017;19(11):1521–1528. doi: 10.1111/dom.12958. [DOI] [PubMed] [Google Scholar]
- 52.Gaggini M., Carli F., Rosso C., Buzzigoli E., Marietti M., Della Latta V. Altered amino acid concentrations in NAFLD: Impact of obesity and insulin resistance. Hepatology. 2018;67(1):145–158. doi: 10.1002/hep.29465. [DOI] [PubMed] [Google Scholar]
- 53.Hsu C.C., Ness E., Kowdley K.V. Nutritional approaches to achieve weight loss in nonalcoholic fatty liver disease. Advances in Nutrition. 2017;8(2):253–265. doi: 10.3945/an.116.013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bankir L., Roussel R., Bouby N. Protein- and diabetes-induced glomerular hyperfiltration: role of glucagon, vasopressin, and urea. American Journal of Physiology-Renal Physiology. 2015;309(1):F2–F23. doi: 10.1152/ajprenal.00614.2014. [DOI] [PubMed] [Google Scholar]
- 55.Hansen B.A., Vilstrup H. A method for determination of the capacity of urea synthesis in the rat. Scandinavian Journal of Clinical and Laboratory Investigation. 1985;45(4):315–320. doi: 10.3109/00365518509161013. [DOI] [PubMed] [Google Scholar]
- 56.Wewer Albrechtsen N.J., Kuhre R.E., Windelov J.A., Orgaard A., Deacon C.F., Kissow H. Dynamics of glucagon secretion in mice and rats revealed using a validated sandwich ELISA for small sample volumes. American Journal of Physiology. Endocrinology and Metabolism. 2016;311(2):E302–E309. doi: 10.1152/ajpendo.00119.2016. [DOI] [PubMed] [Google Scholar]
- 57.Burcelin R., Crivelli V., Dacosta A., Roy-Tirelli A., Thorens B. Heterogeneous metabolic adaptation of C57BL/6J mice to high-fat diet. American Journal of Physiology-Endocrinology and Metabolism. 2002;282(4):E834–E842. doi: 10.1152/ajpendo.00332.2001. [DOI] [PubMed] [Google Scholar]
- 58.Boulange C.L., Claus S.P., Chou C.J., Collino S., Montoliu I., Kochhar S. Early metabolic adaptation in C57BL/6 mice resistant to high fat diet induced weight gain involves an activation of mitochondrial oxidative pathways. Journal of Proteome Research. 2013;12(4):1956–1968. doi: 10.1021/pr400051s. [DOI] [PubMed] [Google Scholar]
- 59.Dall M., Trammell S.A.J., Asping M., Hassing A.S., Agerholm M., Vienberg S.G. Mitochondrial function in liver cells is resistant to perturbations in NAD+ salvage capacity. Journal of Biological Chemistry. 2019;294(36):13304–13326. doi: 10.1074/jbc.RA118.006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iepsen E.W., Lundgren J., Holst J.J., Madsbad S., Torekov S.S. Successful weight loss maintenance includes long-term increased meal responses of GLP-1 and PYY3–36. European Journal of Endocrinology. 2016;174(6):775–784. doi: 10.1530/EJE-15-1116. [DOI] [PubMed] [Google Scholar]
- 61.Wewer Albrechtsen N.J., Kuhre R.E., Toräng S., Holst J.J. The intestinal distribution pattern of appetite- and glucose regulatory peptides in mice, rats and pigs. BMC Research Notes. 2016;9(1):60. doi: 10.1186/s13104-016-1872-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holst J.J. Evidence that glicentin contains the entire sequence of glucagon. Biochemical Journal. 1980;187(2):337–343. doi: 10.1042/bj1870337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ørskov C., Rabenhoj L., Wettergren A., Kofod H., Holst J.J. Tissue and plasma concentrations of amidated and glycine-extended glucagon-like peptide I in humans. Diabetes. 1994;43(4):535–539. doi: 10.2337/diab.43.4.535. [DOI] [PubMed] [Google Scholar]
- 64.Ørskov C., Holst J.J., Poulsen S.S., Kirkegaard P. Pancreatic and intestinal processing of proglucagon in man. Diabetologia. 1987;30(11):874–881. doi: 10.1007/BF00274797. [DOI] [PubMed] [Google Scholar]
- 65.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Research. 2018;46(W1):W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith C.A., O'Maille G., Want E.J., Qin C., Trauger S.A., Brandon T.R. METLIN: a metabolite mass spectral database. Therapeutic Drug Monitoring. 2005;27(6):747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 69.Wishart D.S., Jewison T., Guo A.C., Wilson M., Knox C., Liu Y. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Research. 2013;41(Database issue):D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haug K., Cochrane K., Nainala V.C., Williams M., Chang J., Jayaseelan K.V. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Research. 2019;48(D1):D440–D444. doi: 10.1093/nar/gkz1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metabolomics data generated are available at MetaboLights [70] under the accession number MTBLS1508 (www.ebi.ac.uk/metabolights/MTBLS1508). RNA sequencing data is available upon request to the Corresponding Author Nicolai J. Wewer Albrechtsen (hgk795@ku.dk).