Abstract
COVID‐19 pandemic caused by SARS‐CoV‐2 infection is a public health emergency. COVID‐19 typically exhibits respiratory illness. Unexpectedly, emerging clinical reports indicate that neurological symptoms continue to rise, suggesting detrimental effects of SARS‐CoV‐2 on the central nervous system (CNS). Here, we show that a Düsseldorf isolate of SARS‐CoV‐2 enters 3D human brain organoids within 2 days of exposure. We identified that SARS‐CoV‐2 preferably targets neurons of brain organoids. Imaging neurons of organoids reveal that SARS‐CoV‐2 exposure is associated with altered distribution of Tau from axons to soma, hyperphosphorylation, and apparent neuronal death. Our studies, therefore, provide initial insights into the potential neurotoxic effect of SARS‐CoV‐2 and emphasize that brain organoids could model CNS pathologies of COVID‐19.
Keywords: brain organoids, cell death, neurons, SARS‐CoV-2, Tau pathology
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Molecular Biology of Disease; Neuroscience
In vitro exposure of cerebral organoids to coronavirus results in preferential infection and cell death of neuronal cells.

Introduction
The novel coronavirus disease 2019 (COVID‐19) caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is spreading worldwide, and the outbreak continues to rise, posing a severe emergency (Worl Health Organization, 2020) . Understanding the biology of the current COVID‐19 pandemic is a high priority for combatting it efficiently. Thus, it is essential to gain initial insights into the infection mechanisms of SARS‐CoV‐2, including its target cell types and tropism, to contain its short‐ and long‐term effects on human health. Furthermore, it is vital to establish an experimental system that could allow designing measures on how to stop viral replication and protect human health rapidly. However, practical problems associated with the isolation and handling of highly infective viral strains and lack of reliable in vitro human model systems that can efficiently model COVID‐19 hamper these efforts.
Clinical symptoms of COVID‐19 patients include upper respiratory tract infection with fever, dry cough, and dyspnea, indicating that the respiratory tract is the first target (Yang et al, 2020b). However, emerging case reports showed that patients infected with SARS‐CoV‐2 suffered a sudden and complete loss of the olfactory function, stroke, and other severe neurological symptoms (Chen et al, 2020; Helms et al, 2020; Poyiadji et al, 2020; Sedaghat & Karimi, 2020; Virani et al, 2020). All of these indicate that SARS‐CoV‐2 could infect the central nervous system (CNS) and is therefore neurotropic (Baig et al, 2020; Conde Cardona et al, 2020; De Felice et al, 2020). Earlier studies show that SARS‐CoV target the brains of mice, and since the different coronaviruses share a similar structure, it is likely that SARS‐CoV‐2 exhibits the same infection mechanism and possibly invades into the brain (McCray et al, 2007). Indeed, a clinical report detected the presence of viral RNA in autopsy of brain samples (Puelles et al, 2020). Furthermore, a postmortem brain MRI analysis has identified the presence of hemorrhagic and encephalopathy syndromes suggesting that SARS‐CoV‐2 infection could cause neuronal stress and inflammations (Coolen et al, 2020). Thus, at this point, it is of utmost priority to test whether SARS‐CoV‐2 directly infects human neurons and productively replicates in the CNS.
To investigate the potential neurotropism of SARS‐CoV‐2, it is essential to employ a suitable in vitro human model system that recapitulates the physiological effects of SARS‐CoV‐2 infection. In this regard, the recently emerged human brain organoids that closely parallel the complex neural epithelium exhibiting a wide diversity of cell types could serve as a suitable model system to test the neurotoxic effects of SARS‐CoV‐2. Induced pluripotent stem cells (iPSCs)‐derived human brain organoids have revealed useful insights into human brain development and helped to model a variety of neurological disorders(Lancaster et al, 2013; Gabriel et al, 2016; Birey et al, 2017; Gabriel & Gopalakrishnan, 2017; Xiang et al, 2017; Goranci‐Buzhala et al, 2020). Notably, others and our work using brain organoids have revealed unprecedented insights into infection mechanisms, target cell types, and the toxicity effects of the Zika virus (ZIKV) during the recent ZIKV epidemic (Cugola et al, 2016; Qian et al, 2016; Gabriel et al, 2017). These studies validate organoids as a tool for studying not only genetic but also environmental hazards to the human brain.
Here, we report that SARS‐CoV‐2 readily targets neurons of 3D human brain organoids. Neurons invaded with SARS‐CoV‐2 at the cortical area display altered distribution of Tau, Tau hyperphosphorylation, and apparent neuronal death. Moreover, we show that although SARS‐CoV‐2 can readily target brain organoids, SARS‐CoV‐2 does not appear to efficiently replicate, suggesting that the CNS may not support the active replication of SARS‐CoV‐2.
Results
Isolation of an infectious SARS‐CoV‐2 virus
We isolated SARS‐CoV‐2 (SARS‐CoV‐2 NRW‐42) from a nasopharyngeal and oropharyngeal swab specimen of an infected patient admitted to our university hospital, University of Düsseldorf (see Materials and Methods section for culturing and propagation). To investigate whether SARS‐CoV‐2 replicates in inoculated African green monkey kidney cells (Vero CCL‐81), we performed real‐time quantitative polymerase chain reaction (qPCR) analysis with cell culture supernatant. The amount of SARS‐CoV‐2 RNA drastically increased from 0‐dpi until 3‐dpi (Appendix Fig S1A). Next, we analyzed the infectivity of generated SARS‐CoV‐2 particles by propagating virus‐containing supernatant to yet uninfected Vero cells. We confirmed the infection of new Vero cells by the emergence of virus‐induced cytopathic effects (CPEs) and an increase in SARS‐CoV‐2 RNA over 4‐dpi. The sequence (access number PRJNA627229 at the European Nucleotide Archive and the Sample accession number for NRW‐42 which is SRS6522060) showed only eight nucleotide exchanges compared to SARS‐CoV‐2 Wuhan‐Hu‐1 isolate.
Isolation and validation of COVID‐19 convalescent serum to detect SARS‐CoV‐2 infection
As of April 1, 2020, we could not procure commercial antibodies that can specifically determine SARS‐CoV‐2 infection. Therefore, we isolated COVID‐19 convalescent serum and tested if they can specifically recognize SARS‐CoV‐2 infections in our experiments. We obtained blood samples of four independent individuals who recently recovered from COVID‐19 (AB1, AB2, AB3, and AB4). Testing them in an enzyme‐linked immunosorbent assay (ELISA) that used the SARS‐CoV‐2 S1 domain of the spike protein as an antigen revealed that, except for AB2, the rest of the convalescent serum contained SARS‐CoV‐2‐specific IgG (Appendix Fig S1B). We then affinity‐purified the convalescent serum against the full length ORF of SARS‐CoV‐2‐N (see Materials and Methods section). In Western blots, which used extracts of brain organoids and Vero cells exposed to SARS‐CoV‐2, the antibodies affinity‐purified from convalescent serum specifically recognized a signal similar to the size of the nucleoprotein of SARS‐CoV‐2. The recombinant SARS‐CoV‐2‐N serves as a positive control in this experiment (Appendix Fig S1C). The convalescent serum AB4 also specifically recognized SARS‐CoV‐2‐infected Vero cells. To further validate the specificity of the AB4, we performed co‐immunostaining with a mouse monoclonal anti‐SARS‐CoV‐2 S and a polyclonal anti‐SARS‐CoV‐2 NP. As expected, all of these antibodies recognized only the SARS‐CoV‐2‐infected Vero cells (Appendix Fig S2A). Similarly, AB4 could specifically recognize somas of SARS‐CoV‐2‐positive cells in SARS‐CoV‐2 exposed brain organoids which were further labeled by the monoclonal anti‐SARS‐CoV‐2 S antibody (Appendix Fig S2B). In Western blots that used SARS‐CoV‐2‐exposed organoid extracts, both AB4 and mouse monoclonal antibodies recognized protein bands around 50 and 180 kDs, sizes similar to the nucleoprotein and uncleaved spike proteins Together, these experiments validate that AB4 detects SARS‐CoV‐2 infection (Appendix Fig S2C).
SARS‐CoV‐2 targets neurons of human brain organoids
Before we infected our 3D human brain organoids with the new SARS‐CoV‐2 NRW‐42 isolate, we first tested if our experimental conditions are suitable to infect the well‐studied ciliated human respiratory epithelial cells (hRECs), an apparent target for the SARS‐CoV‐2 (Lamers et al, 2020). We noticed that SARS‐CoV‐2 readily targets hRECs within 2 days of virus exposure (Fig 1A). We then tested if SARS‐CoV‐2 could infect 3D human brain organoids. To do this, we adapted our previously described protocol and differentiated brain organoids from two different iPSC lines (Donor 1, IMR90 and Donor 2, Crx‐iPS; Gabriel et al, 2017). In brief, we started with 10,000 iPSCs and induced differentiation into neural epithelium directly using SB431542 and dorsomorphin, the TGF beta and BMP4 inhibitors, respectively. Our differentiation condition did not also include an exogenous addition of retinoic acid, which could activate retinoic acid receptors (RAR) and induce an aberrant neuronal differentiation (Janesick et al, 2015; Gabriel et al, 2016, 2017; Gabriel & Gopalakrishnan, 2017). As this method skips embryoid bodies formation, it reduces the heterogeneity in organoid sizes simultaneously avoiding the formation of mesoderm and endoderm, which are not required for ectodermal differentiation at early stages of differentiation (Streit et al, 2000). As described before, organoids exhibit their specific neuronal cell types, which are spatially restricted. The ventricular zone (VZ) harbors proliferating neural progenitors cells (NPCs) that display typically elongated nuclei which align to form a lumen, a neural tube‐like structure. Cortical neurons are positioned basally to the VZ, forming a cortical plate (Fig 1B) (Lancaster & Knoblich, 2014; Giandomenico & Lancaster, 2017; Gopalakrishnan, 2019).
Figure 1. SARS‐CoV‐2 targets the cortical region of human brain organoids.
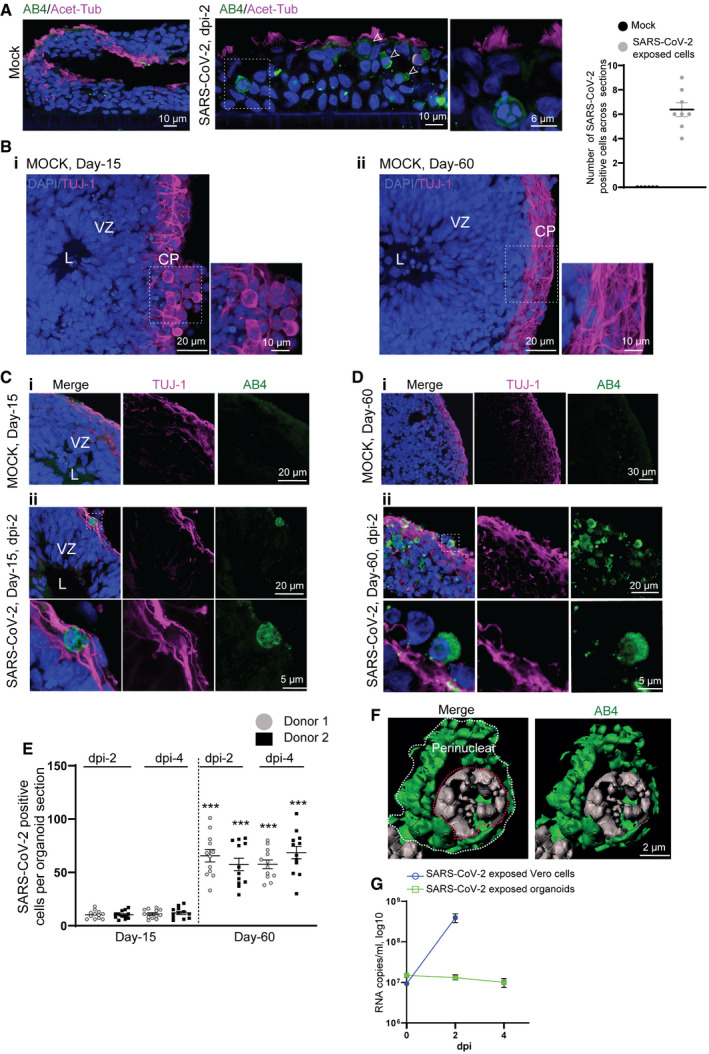
-
AA positive control experiment. SARS‐CoV-2 readily targets ciliated human respiratory epithelial cells (hRECs). Acetylated α‐tubulin labels cilia. Arrows point SARS‐CoV-2‐positive cells labeled by AB4 (green). Figures display scale bars. Bar diagram at right quantifies frequencies of SARS‐CoV-2‐positive cells in hRECs. At least six hREC sections from three (n = 3) independent samples were examined. Data presented as mean ± SEM.
-
BMock organoids of two age groups Day‐15 (i) and‐60 (ii) display typical cytoarchitecture of brain organoids. L, lumen, VZ, ventricular Zone is containing compact and palisade‐like elongated nuclei of neural progenitor cells (NPCs, blue) and CP, a cortical plate containing TUJ‐1-positive neurons (magenta). Note a distinct difference TUJ‐1 labeling pattern between younger (Day‐15) and older (Day‐60) brain organoid. Figures display scale bars. Representative images from eight organoids cultured in at least three independent batches (n = 3) derived from donor‐1 (IMR90) iPSC line.
-
CCompared to mock organoids (i), SARS‐CoV-2‐exposed Day‐15 organoids display SARS‐CoV-2‐positive cells (AB4, green) in their outer periphery, a region of the cortical plate (ii) that is specified by TUJ-1‐positive neurons (magenta). L, the lumen of a VZ, the inner area of an organoid where NPCs are located, is free from SARS‐CoV-2‐positive cells. Magnified region (dotted while box) is given below. At least 10 organoids from five different batches (n = 5) are tested. Figures display scale bars.
-
DSARS-CoV‐2-exposed Day‐60 organoids. Compared to Day 15 organoids and mock (i), Day‐60 organoids display an increased number of SARS‐CoV-2‐positive cells (AB4, green) in their cortical plate that is specified by TUJ-1‐positive neurons (magenta) (ii). Magnified region (dotted while box) is given below, showing the perinuclear location of SARS‐CoV-2 in cortical neurons. At least 10 organoids from five different batches (n = 5) are tested. Figures display scale bars.
-
EThe bar diagram quantifies frequencies of SARS‐CoV-2‐positive cells in different brain organoid sections derived from two donor iPSC lines (IMR90 and Crx‐iPS, see Materials and Methods). Please note that each point represents one organoid section. SARS‐CoV-2 shows an enhanced tropism for Day‐60 organoids. Note, comparative statistics are shown between different age groups and respective days post‐infection (dpi) of organoids, and the significance is given as Asterisks in Day‐60 groups. There is no significant difference in SARS‐CoV-2‐positive cells between 2‐ and 4‐dpi within each age groups. At least twelve organoids sections from four (n = 4) independent batches, from each donor and day post‐infections (dpi), were analyzed. One‐way ANOVA, followed by Tukey's multiple comparisons test, ***P < 0.001. Data presented as mean ± SEM.
-
FSubcellular localization SARS‐CoV-2 in cortical neurons. High‐resolution imaging and deconvolution show perinuclear localization of SARS‐CoV-2. SARS‐CoV-2 (AB4, green) and nucleus (gray). Figures display scale bars. Representative images from at least 200 cells are examined. White line surrounds perinuclear border, and red line encircles the nucleus.
-
GDetermination of viral progeny. Supernatants of SARS‐CoV-2 exposed Vero cells, and brain organoids were analyzed for viral RNA assessed by qRT–PCR. While an increase in viral RNA was detected in the supernatants of Vero cells, no apparent increase was identified in brain organoid supernatants. Data are obtained from five technical replicates from four (n = 4) independent batches of organoids. Data presented as mean ± SEM.
We exposed at least two different age groups of organoids (Day‐15 and Day‐60) to SARS‐CoV‐2 (TCID50/ml of 50 which is equivalent to 17.5 PFU/organoid, see Materials and Methods section for details) and analyzed after 2 and 4 days post‐infection (dpi). First, we began analyzing Day‐15 organoids, a developmental stage used to study ZIKV infections (Gabriel et al, 2017). At this developmental stage, organoids mostly constitute actively proliferating NPCs at the VZs and a primitive cortical plate containing fewer early neurons (Fig 1B). Testing the target cell types of SARS‐CoV‐2 in these organoids revealed that SARS‐CoV‐2 could mostly target the cortical plate specified by pan‐neuronal marker TUJ‐1 that is spatially distinct from the VZ (Fig 1C and Appendix Fig S2D). To exclude the possibility that the virus may have a limited capacity of diffusion to target NPCs at the inner part of the intact 3D organoids, we directly exposed NPCs’ 2D cultures to SARS‐CoV‐2. Compared to 2D cortical neuronal cultures, NPCs cultures displayed only fewer cells positive for SARS‐CoV‐2. These findings indicate that SARS‐CoV‐2 has a preferred tropism to neurons, as reported recently (preprint: Mesci et al, 2020; preprint: Song et al, 2020; Yang et al, 2020a) (Appendix Fig S3A). This is indeed in striking contrast to ZIKV, which directly targets NPCs present at the inner region of brain organoids and triggers them to prematurely differentiate into neurons leading to congenital microcephaly (Cugola et al, 2016; Qian et al, 2016; Gabriel et al, 2017).
Analyzing the cortical regions of Day‐60 organoids revealed that the number of SARS‐CoV‐2‐positive cells was significantly higher than in Day‐15 organoids. This suggests that SARS‐CoV‐2 prefers relatively mature neuronal cell types present in older organoids (Fig. 1D and E). Day‐60 organoids indeed displayed signs of maturation as judged by more MAP2‐positive neurons, S100β‐positive astrocytes, and fewer Iba‐1‐positive microglial cells (Appendix Fig S3B–D). Importantly, the perinuclear localization of SARS‐CoV‐2 in somas of cortical neurons is similar to the virus's localization pattern in Vero cells, indicating that SARS‐CoV‐2 can enter into neuronal cells of brain organoids (Fig 1F). Turning our analysis to the later time point of infection (dpi‐4 and dpi‐6) revealed no apparent increase in SARS‐CoV‐2‐positive cells although dpi‐6 organoids exhibited a slightly compromised integrity (Fig 1E and Appendix Fig S4A). Corroborating to this, we could not detect an increase in viral RNA in the supernatants between 2‐ and 4‐dpi (Fig 1G). In contrast to brain organoids, SARS‐CoV‐2 productively infects vascular, kidney, and gut organoids (Lamers et al, 2020; Monteil et al, 2020; Zhou et al, 2020). Notably, angiotensin‐converting enzyme 2 (ACE‐2), an entry receptor of SARS‐CoV‐2, is highly expressed in these organoid types. Testing the ACE‐2 expression at the mRNA level via a qRT–PCR revealed that both iPSCs‐derived brain organoids and neurons exhibited ~12.5‐ and 50‐fold lesser than human respiratory epithelial cells (hREC), which served as a positive control (Appendix Fig S4B). Our Western blots using anti‐ACE2 antibodies recognized ACE2 in organoid extracts only at higher exposure conditions (Appendix Fig S4C).
Since SARS‐CoV‐2 appears to preferably target neurons, we wondered if SARS‐CoV‐2 could productively replicate when exposed to an abundant number of mature neurons. To test this, we cultured organotypic slices of 60‐day‐old organoids, an alternative organoid culturing method that enhances neuronal maturation and viability. These cultures exhibit neuronal outgrowths as long‐range axonal fibers expressing mature neuronal markers of MAP2, Tau, synapsin‐1, and PSD95 (Gabriel et al, 2016; Giandomenico et al, 2019; Goranci‐Buzhala et al, 2020). After directly exposing these slices to SARS‐CoV‐2, we detected the virus localized at the cell bodies of the neurons which are labeled by MAP2 and Tau (Appendix Fig S5A and B). We noticed only a slight increase in SARS‐CoV‐2 RNA within 2 days of viral exposure (Appendix Fig S5C). These experiments demonstrate that SARS‐CoV‐2 enters neurons of brain organoids but does not actively replicate.
SARS‐CoV‐2‐positive neurons reveal aberrant Tau localization
Next, we identified that the SARS‐CoV‐2‐positive region of the cortical plate is further substantiated by Tau, a microtubule‐associated protein that stabilizes neuronal microtubules and promotes axonal growth (Fig 2) (Wang & Mandelkow, 2016). Tau dysfunction is implicated in Alzheimer's disease (AD) and other Tauopathies. Post‐translational modifications in Tau, in particular, phosphorylations, modulate the ability of Tau to bind and assemble microtubules. In Tauopathies, Tau is aberrantly phosphorylated (hyperphosphorylation; Cho & Johnson, 2004; Cohen et al, 2011; Castellani & Perry, 2019). A recent report showed that herpes simplex virus type 1 can induce AD‐like effects, including hyperphosphorylation of Tau in 3D human brain‐like tissue model (Cairns et al, 2020). This prompted us to investigate if SARS‐CoV‐2 has a consequence upon its entry into neurons.
Figure 2. SARS‐CoV‐2 deregulates of Tau in cortical neurons.
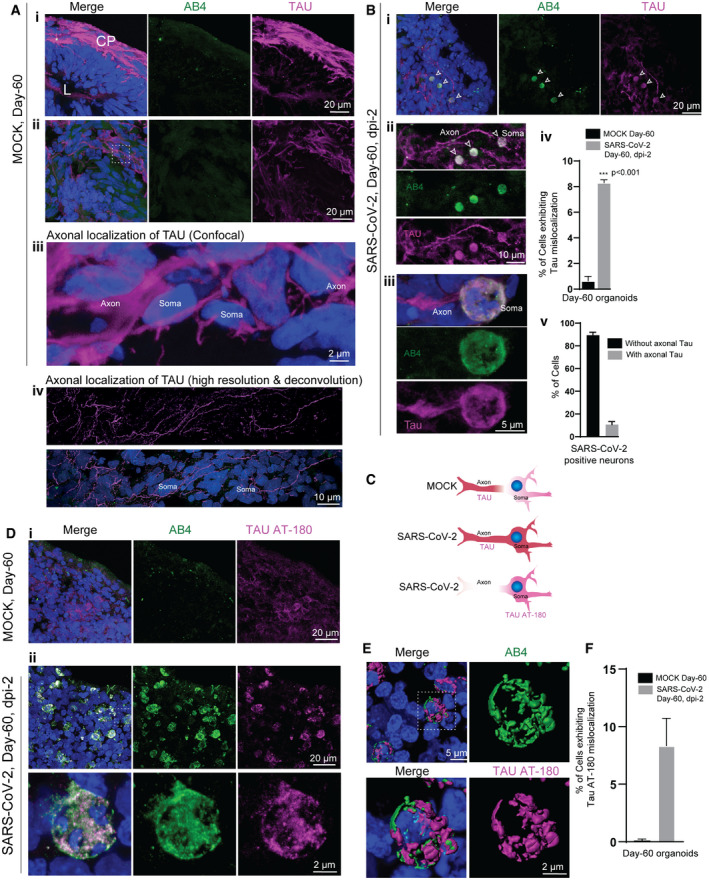
-
ATau immunoreactivity (magenta) specifies the cortical plate (CP) surrounding the lumen (L) (i). Selected optic sections at high magnification (ii and iii) and high‐resolution imaging (iv) show Tau localization only in axons of cortical neurons. Note the somas of neurons are free from Tau protein. At least eight organoids from four different batches (n = 4) are tested. Figures display scale bars.
-
BTau localization in SARS‐CoV-2‐positive neurons (AB4, green) in selected optic sections (i). Note, in contrast, to control groups, SARS‐CoV-2‐exposed organoids display mislocalized Tau (magenta) majorly into the somas of neurons (arrowheads). Selected confocal slices are shown to distinguish Tau mislocalization into neuronal soma (arrowheads). At high magnification, neuronal soma is further specified by the perinuclear localization of SARS‐CoV-2 (green) (ii and iii). Bar diagrams at right quantifies the percentage of neurons (Mock and SARS‐CoV-2 exposed) exhibiting mislocalized Tau (iv) and the fraction of SARS‐CoV-2‐positive neurons exhibiting Tau‐positive axons spanning different cortical areas (v). For statistics, at least 300 cells from six organoids from four different batches (n = 4) were tested. Figures display scale bars. Unpaired t‐test with Welch's correction, ***P < 0.001. Data presented as mean ± SD.
-
CSchematic cartoon of differential Tau distribution in mock compared to SARS‐CoV-2‐positive neurons. In mock, Tau is sorted mainly to axons. In SARS‐CoV-2‐positive neurons, Tau is missorted to the soma (determined by Pan‐Tau antibody). Furthermore, phosphorylated Tau (at T231) majorly localizes in the soma (bottom panel, determined by Tau AT‐180 antibody, see below).
-
DIn contrast to controls (i), Tau AT180 antibody (magenta) that specifically recognizes the phosphorylated Threonine 231 of Tau protein distinctly localizes at the somas of SARS‐CoV-2‐positive neurons (AB4, green) (ii). At least four organoids from two different batches (n = 2) are tested. Figures display scale bars.
-
ECo‐localization of SARS‐CoV-2 (AB4, green) and phosphorylated Tau protein (magenta) at somas of cortical neurons revealed by high‐resolution imaging and deconvolution. Representative images from at least 300 cells examined. Figures display scale bars.
-
FThe bar diagram quantifies the fraction of Tau AT180‐positive neurons that co‐localize with SARS‐CoV-2‐positive neurons. For statistics, at least 250 cells from four organoids and two independent batches (n = 2) were examined. Unpaired t‐test with Welch's correction. Data presented as mean ± SEM.
Under physiological conditions, Tau is mainly an axonal protein that localizes at the axons of mature neurons (Fig 2Ai–iv). Applying high‐resolution imaging followed by deconvolution, we could visualize Tau's localization (as probed by a Pan‐Tau antibody Tau5A6) exclusively in axons of the cortical neurons (Fig 2Av). The term Tau “missorting” is used when Tau protein is mislocalized into a cell soma and is observed at the early stages of Tau pathology (Zempel & Mandelkow, 2014).
Compared to control organoids where Tau normally localizes in axons, SARS‐CoV‐2‐positive neurons exhibited an altered Tau localization pattern, although it was challenging to visualize mislocalization of Tau in 3D tissues. Nevertheless, using selected confocal sections, we could image an altered Tau localization in SARS‐CoV‐2‐positive neurons. In particular, we identified an enhanced level of Tau into the somas of the SARS‐CoV‐2‐positive neurons. Importantly, we could visualize fractions of these neurons still contained Tau and TUJ‐1 in their axons, indicating that these neurons are still viable (Fig 2B and C and Appendix Fig S6A).
During the pathogenesis of AD and other Tauopathies, Tau also gets hyperphosphorylated at multiple sites. Sequential phosphorylation at different sites ultimately leads to hyperphosphorylation of Tau (Castellani & Perry, 2019). Phosphorylation of Threonine 231 (T231) is one of the first events in the cascade of phosphorylation, and it regulates the microtubule binding. Still, it is also implicated in disease progression such as detachment of Tau from axonal microtubules (Sengupta et al, 1998; Augustinack et al, 2002a,b; Luna‐Munoz et al, 2007; Alonso et al, 2010; Frost et al, 2015). More precisely, we found that compared to control organoids, early Tau phosphorylation marker AT180 recognizes pT231Tau localized at the soma of the SARS‐CoV‐2‐positive neurons (Fig 2D–F). Imaging the neurons for additional phosphorylated Tau using AT8 antibodies (specific for S202 and T205 of Tau) and p396 (specific for S396 of Tau) revealed that unlike pT231Tau, these phospho‐species were restricted to the axons and did not mislocalize to the soma of SARS‐CoV‐2‐positive neurons (Appendix Fig S6B–E). In summary, these results demonstrate the aberrant localization of Tau pT231Tau in SARS‐CoV‐2‐positive neurons suggesting the potential neuronal stress reactions upon virus entry.
SARS‐CoV‐2 induces neuronal cell death
Phosphorylation of Tau at T231 allows for isomerization of the following proline residue into distinct cis‐ and trans‐conformations by the propyl‐isomerase PIN1 (Lu et al, 1999). Cis‐pT231Tau is acutely produced by neurons after traumatic brain injury, leading to disruption of the axonal microtubule network and apoptosis (Nakamura et al, 2012; Kondo et al, 2015). Analyzing the nuclei of SARS‐CoV‐2‐positive cells (Fig 3A), we realized that they are highly condensed or fragmented exhibiting a strong reaction to 4′,6‐diamidino‐2‐phenylindole (DAPI) that labels nuclei, a feature quite frequently observed in dead cells. To test neuronal cell death as a consequence of SARS‐CoV‐2 infection, we stained the SARS‐CoV‐2‐exposed samples with terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) that detects fragmented DNA in dead cells (Darzynkiewicz et al, 2008). Compared to un‐exposed control organoids, we identified an overall increase in TUNEL‐positive cells in SARS‐CoV‐2‐exposed organoids suggesting that virus exposure has caused cell death within 2‐dpi (Fig 3B). Staining for SARS‐CoV‐2‐positive cells revealed that most of the virus‐positive cells were TUNEL‐positive. Besides, we also noticed that some SARS‐CoV‐2‐positive cells were also positive for caspase‐3, a protease that specifies programmed cell death (Fig 3C). Interestingly, a fraction of caspase‐positive cells displayed pT231Tau localization at the cell soma. Furthermore, TUNEL‐positive cells in un‐exposed control organoids (which could be after programmed cell death) did not contain pT231Tau suggesting that this different Tau phosphorylation pattern is associated with SARS‐CoV‐2 entry (Fig 3Ciii). Thus, it appears that Tau is aberrantly phosphorylated in response to the viral‐induced stress, which may elicit further cell death programs that remains to be elucidated.
Figure 3. SARS‐CoV‐2 induces of neuronal death.
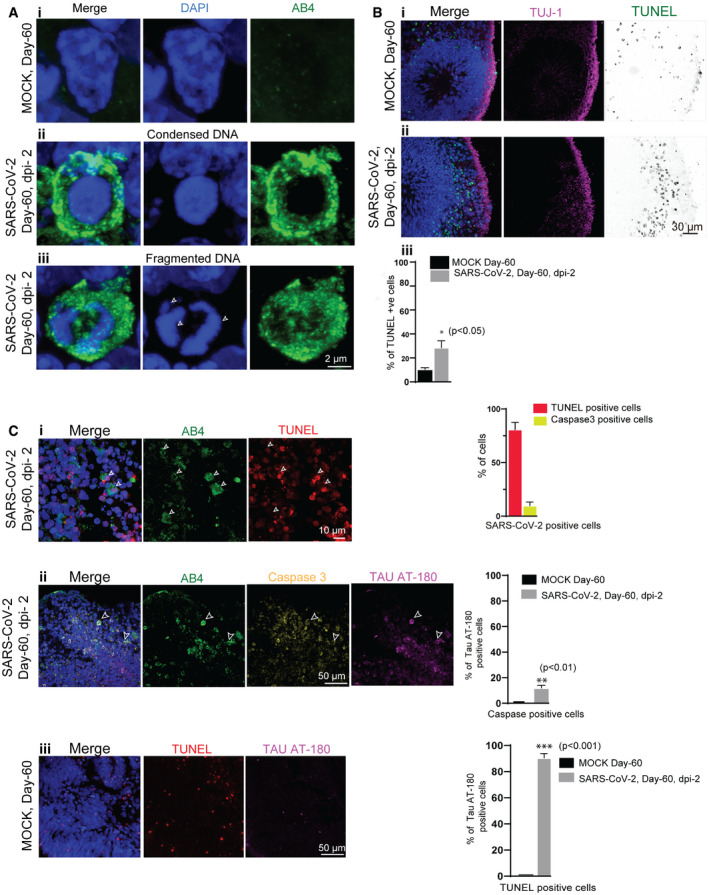
-
ACells from mock organoids display a healthy nucleus labeled by DAPI (blue) (i). SARS‐CoV-2‐positive cells (green) display condensed (middle panel, ii) and fragmented DNA (bottom panel, iii, arrows). At least 75 cells from two (n = 2) independent batches of organoids were examined. Figures display scale bars.
-
BCompared to mock organoids, (i) SARS‐CoV-2‐exposed organoids (ii) display increased TUNEL‐positive cells (displayed as inverted LUT) at the cortical plate that is specified by TUJ‐1 (magenta). At least four organoids from two (n = 2) independent batches of organoids were examined. Figures display scale bars. The bar diagram below quantifies the frequencies of TUNEL‐positive cells between mock and SARS‐CoV-2‐exposed organoids. Four organoids from two (n = 2) independent batches were examined. Unpaired t‐test, *P < 0.05. Data presented as mean ± SD.
-
CMost of the SARS‐CoV-2‐positive cells (AB4, green) are TUNEL‐positive (i) and some of the SARS‐CoV-2‐positive cells are caspase‐positive (ii). Caspase‐positive cells in SARS‐CoV-2‐exposed organoids display pT231Tau localization at the cell soma specified by AT‐180 (ii), which are not observed in mock organoids (iii). Arrowheads point SARS‐CoV-2‐positive cells (AB4, green) that are also positive for TUNNEL (red), caspase 3 (yellow), and Tau AT‐180 (magenta). Figures display scale bars. Bar diagrams at right quantifies proportions of TUNEL and caspase‐positive cells among SARS‐CoV-2‐positive cells. The second graph below quantifies the proportions of pT231Tau‐positive cells among caspase‐positive cells between control and virus exposed groups. At least 400 cells from four organoids and two independent (n = 2) batches were examined. Unpaired t‐test, **P < 0.01. Data presented as mean ± SEM. TUNEL‐positive cells in control un‐exposed organoids do not contain pT231Tau (iii). Figures display scale bars. Bar diagrams at right quantifies proportions of pT231Tau‐positive cells among TUNNEL‐positive cells between control and virus exposed groups. At least 350 cells from 4 organoids and two independent (n = 2) batches were examined. Unpaired t‐test, ***P < 0.001. Data presented as mean ± SEM.
Discussion
So far, the possible direct effect of SARS‐CoV‐2 on the CNS has been debated but not experimentally demonstrated (Baig et al, 2020; Conde Cardona et al, 2020:718; Coolen et al, 2020; Helms et al, 2020; Poyiadji et al, 2020). Thus, it was essential to examine whether SARS‐CoV‐2 can directly target human neurons and whether this leads to productive infection. In contrast to vascular, kidney, and intestinal organoids (Lamers et al, 2020; Monteil et al, 2020; Zhou et al, 2020), brain organoids does not appear to strongly support the active replication of SARS‐CoV‐2 at least until 6‐dpi. There are several reasons for this. Firstly, the developmental stages of brain organoids used in this work may not contain the full complement of SARS‐CoV‐2's host cell replication factors. As an example, efficient replication of SARS‐CoV requires ACE‐2 (Li et al, 2003) whose expression appears to be relatively low in brain organoids (Appendix Fig S4B). Next, in brain organoids, post‐mitotic neurons seem to be susceptible for SARS‐CoV‐2 that may not be permissive (Fig 1, Appendix Figs S3 and S5). Finally, brain organoids are simplified reductionist models and lack an additional cell type that can influence viral replication such as blood–brain barrier, vasculature, and mature glial cells, including microglia. To our surprise, we did notice the appearance of fewer Iba‐1 and S100β‐positive cells in our organoids pointing toward the need of further engineering of our differentiation conditions which could lead to the differentiation of mature microglia and astrocytes (Appendix Fig S3C and D). Thus, future experiments using aged organoids and bioengineered organoids with SARS‐CoV‐2 replication factors are required to conclude if brain organoids can support productive infection of SARS‐CoV‐2.
ACE‐2 is an entry receptor for SARS‐CoV and efficient replication of SARS‐CoV (SARS outbreak in the year 2003) and also depends on the expression level of ACE‐2 (Li et al, 2003; Hoffmann et al, 2020). Curiously, SARS‐CoV could only infect the brain of transgenic mice expressing an elevated level of human ACE‐2 but not non‐transgenic mice. This key finding suggests that the neurotropism of SARS‐CoV, to some extent, depends on the expression level of human ACE‐2 in the brain (McCray et al, 2007). Using our 3D human brain organoid system, we unexpectedly find that although these organoids express low level of ACE‐2, the human neurons are indeed a target for SARS‐CoV‐2. This finding offers a couple of possibilities. First, even a basal level of ACE2 expression is sufficient for viral entry into the neurons. Second, the presence of yet unknown neuron‐specific viral entry factors has to be elucidated. It is indeed intriguing that even a low level of ACE‐2 is sufficient for the viral entry, and this could explain why SARS‐CoV‐2 has a broad spectrum of target organs and cell types (Puelles et al, 2020).
Detection of Tau phosphorylation at T231 in SARS‐CoV‐2‐positive neurons is remarkable as it could trigger a cascade of downstream effects that finally could initiate neuronal stress and toxicity. Intriguingly, there is growing evidence that viral infections, particularly herpes simplex virus type I (HSV‐1), is a potential causative agent leading to Alzheimer's disease (AD). Indeed a recent work demonstrated infection of 3D human brain‐like tissue model with HSV‐1 and showed that the HSV‐1 infection is sufficient to elicit AD‐like effects, including hyperphosphorylation of Tau. Early Tau phosphorylation at T231 could be reversible (Castellani & Perry, 2019). However, phosphorylation events observed in conjunction with apparent neuronal cell death suggest that SARS‐CoV‐2 has potential detrimental effects on neurons at least in our organoid test system (Fig 3). Future biochemical experiments dissecting the ratio of soluble and sarkosyl‐stable Tau extracted from SARS‐CoV‐2‐positive neurons are required to obtain insights into the cause and effect of potential Tau pathology and neuronal death. Although we observe Tau abnormalities in SARS‐CoV‐2‐positive neurons, we could not conclude whether the observed effect is directly caused by the virus or an effect due to neuronal stress, which warrants future investigations.
In conclusion, COVID‐19 research has taken center stage in biomedical research. It is noteworthy that three coronavirus epidemics have occurred within the last two decades, and thus, the future zoonotic coronavirus outbreak is not unexpected. With the advent of emerging human organoid research, which did not exist 20 years ago, we should be able to model the current SARS‐CoV‐2 infections and sufficiently prepare us for the future. Recent works utilizing kidney, gut, and liver organoids have already revealed insights into the infection mechanisms (Lamers et al, 2020; Monteil et al, 2020; Yang et al, 2020a; Zhou et al, 2020). Adding to them is the current work that establishes brain organoids as a test system for SARS‐CoV‐2 infection and provides indications for potential neurotoxic effects of SARS‐CoV‐2. Since organoids are an experimentally tractable human in vitro system and convenient to culture as well as to infect, organoid systems may serve well as a test‐bed to screen for anti‐SARS‐CoV‐2 agents. The presented work only provides initial insights into primitive brain‐like tissues and requires further experiments to dissect viral replication mechanisms and whether there are ACE2 independent pathways for viral entry. It is important to note that although the virus seems to preferably target neurons, future experiments are required to test if the virus can have extended access across the entire organoids. Advanced experiments utilizing a mature state of brain organoids, bioengineered organoids, and orthogonal experiments with complementary in vivo experimental models are assured to dissect the neuropathology of SARS‐CoV‐2.
Materials and Methods
Clinical specimens
For the isolation of infectious SARS‐CoV‐2 particles, nasopharyngeal and oropharyngeal swab specimens from one individual with positive qRT–PCR results for SARS‐CoV‐2 infection were used. The swab specimen was transported in a viral cultivation medium and stored at 4°C overnight. Freezing at −20°C was found to interfere with the infectivity of viral particles. Before the inoculation of susceptible cells, 500 μl maintenance medium (Dulbecco's Modified Eagle Medium (Thermo Fisher), 2% fetal calf serum (PAN Biotech), 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco) were added to the swab specimen. To get rid of major impurities, samples were briefly centrifuged (3,000 g; 60 s) and the supernatant was transferred to new vials.
Human respiratory epithelial cells and culturing
To obtain respiratory epithelia, a MedScand Cytobrush Plus GT (Cooper Surgical, Trumbull, USA) with a gentle‐touch tip was rinsed with isotonic saline before use. Afterward, the brush was inserted into the inferior nasal meatus followed by rotatory and linear motions against the medial and superior side. Isolated cells were transferred into a 15‐ml centrifuge tube (Corning Incorporated, New York, USA) with 5 ml pre‐warmed RPMI 1640 medium containing 2% Antibiotic‐Antimycotic 100× (Gibco® Life Technology, Grand Island, USA). The brushes were vigorously shaken several times within the tube, and cells were pelletized by centrifugation at 900 rpm for 5 min at room temperature. hRECs were re‐suspended in Dulbecco's Modified Eagle Medium/Nutrient Mixture F‐12 (DMEM/F‐12, Gibco® Life Technology, New York, USA) supplemented with 2% UltroserTM G Serum Substitute (Pall Corporation, Port Washington, USA) and 2% Antibiotic‐Antimycotic 100 x and seeded on T‐25 or T‐75 rat‐tail collagen‐coated tissue flasks (Greiner Bio‐One, Kremsmünster, Austria), according to the pellet size, respectively, and incubated at 37°C, 5% CO2.
To reduce the risk of contamination, the medium was replaced after 24 h, and the flasks were then integrated into the regular feeding procedure (exchange of medium every 48–72 h). After 1 week, the concentration of Antibiotic‐Antimycotic was reduced to 1%. Reaching confluency of 90%, the collagen layer was digested by incubating with 200 U/ml collagenase type IV (Worthington Biochemical Company, New Jersey, USA) for 30–60 min, followed by several washing steps with DMEM/F‐12 supplemented with 1% Antibiotic‐Antimycotic. To reduce the number of fibroblasts, the pellet was re‐suspended in 7 ml DMEM/F12 supplemented with 2% UltroserTM G, seeded on tissue culture treated T‐25 flasks (Corning Incorporated, New York, USA) and incubated for 1 h at 37°C, 5% CO2. The cells were then separated by incubating with Trypsin‐EDTA 0.05% for 5 min before the reaction was stopped with FBS followed by centrifugation at 900 rpm for 5 min at room temperature.
After re‐suspending in PneumaCultTM‐Ex Medium (STEMCELLTM Technologies, Vancouver, Canada), 4 × 105 cells/ml were seeded on collagen‐coated 6.5 mm Transwell®, 0.4 μm pore Polyester membrane inserts (Corning Incorporated, New York, USA) with 250 μl medium on the apical side and 500 μl on the basolateral side, respectively. Before airlift, after 3–5 days, depending on cell confluency, PneumaCultTM‐Ex Medium was replaced every day at the apical and basolateral side. To perform airlift, the medium on the apical side was carefully removed, whereas the basolateral medium was exchanged with PneumaCultTM‐ALI Medium. The airlifted inserts were then integrated into the regular feeding procedure and incubated at 37°C, 5% CO2. A fully differentiated pseudostratified epithelium is expected 15–30 days after airlift and resembles human airway epithelium (in vivo) with respect to function and morphology.
Inoculation of Vero cells
In compliance with the German committee's decision on biological agents (ABAS) of the Federal Institute for Occupational Safety and Health, all experimental studies involving infectious SARS‐CoV‐2 were performed within the biosafety level 3 (P3) facility at the University Hospital Düsseldorf. To isolate SARS‐CoV‐2 from a clinical specimen, 2.5 × 105 Vero cells (ATCC‐CCL‐81, obtained from LGC Standards) were seeded into T‐25 cell culture flasks in maintenance medium and cultured at 37°C in a humidified cell culture incubator. The following day, SARS‐CoV‐2 inoculum was prepared by diluting 200 μl of a clinical specimen with 800 μl maintenance medium. The medium was removed from Vero cells, and 1 ml inoculum (1 ml of maintenance medium for control Vero cells) was added onto the Vero cell monolayer. Vero cells were incubated for 1 h on a laboratory shaker at 37°C in a humidified incubator. Afterward, 4 ml of maintenance medium was added. To monitor viral replication, 100 μl of supernatant was directly harvested as the first sample (0 h post‐inoculation) and every 24 h for 4 days post‐inoculation. Additionally, cells were imaged by light microscopy.
Real‐time qPCR analysis for quantification of SARS‐CoV‐2 RNA copies per ml
For extraction, 100 μl cell culture supernatant was incubated with 400 μl AVL buffer (viral lysis buffer used for purifying viral nucleic acids; cat No. 19073, Qiagen, Hilden Germany) for 10 min at RT and mixed with 400 μl 100% ethanol. RNA extraction was performed with 200 μl cell culture mix using the EZ1 Virus Mini Kit v2. (cat. no. 955134, Qiagen, Hilden, Germany) following the manufacturer's instructions. A total of 60 μl were eluted from the 200 μl starting material. 5 μl of the eluate was tested in qRT–PCR using the real‐time TaqMan®‐technique. A 113 base pair amplicon in the E‐gene of SARS‐Cov‐2 was amplified and detected, as described by Corman et al (2020) with minor modifications. The thermal protocol described has been shortened to 40 cycles of 95° C. We used the LightMix® Modular SARS and Wuhan CoV E‐gene (Cat.‐No. 53‐0776‐96) and the LightMix® Modular EAV RNA Extraction Control. We used the AgPath‐ID® One‐Step RT–PCR Kit (Applied Biosystems, Cat. No. 4387391). RT–PCR was performed with an ABI 7500 FAST sequence detector system (PE Applied Biosystems, Weiterstadt, Germany). As a DNA‐standard, a plasmid (pEX‐A128‐nCoV2019‐E‐gene) that encompasses the amplified region was created and serially diluted after purification. The software constructed a standard graph of the CT values obtained from serial dilutions of the standard. The CT values of the unknown samples are plotted on the standard curves, and the number of SARS‐CoV‐2 RNA copies was calculated.
For gene expression analysis of ACE2, quantitative RT–PCR analysis was performed by using qPCR MasterMix (PrimerDesign Ltd) and fluorescence emission was monitored by LightCycler 1.5 (Roche). For normalization, primers #5163 (5′ CCA CTC CTC CAC CTT TGA 3′) and #5164 (5′ ACC CTG TTG CTG TAG CCA 3′) were used monitoring cellular GAPDH expression. Expression was then calculated as .
Propagation of infectious SARS‐CoV‐2 particles
For propagation of infectious SARS‐CoV‐2 particles from Vero cell culture supernatant, 2.5 × 105 Vero cells were seeded into T‐25 cell culture flasks in maintenance medium and incubated at 37°C in a humidified cell culture incubator. The next day, the supernatant of inoculated Vero cells at day four post‐inoculation (see above) was diluted with maintenance medium (1:2, 1:10, 1:100, 1:1,000) in a total volume of 5 ml and added to the cells, which were incubated for 4 days at 37°C.
Determining SARS‐CoV‐2 viral titer by TCID50 assay in 96‐well plates with Vero cells
For determination of viral titer in TCID50/ml, 5 × 103 Vero cells were seeded in the first 10 columns of 96‐well plate in 100 μl maintenance medium and incubated at 37°C in a humidified cell culture incubator for 24 h. In a new 96‐well plate, 180 μl maintenance medium was added to all wells of the first 10 columns. For serial dilutions of the virus stock, 20 μl of the stock solution was added to the wells of the first column. Then, 20 μl of the first dilution was transferred to the wells of the next column to obtain 10‐fold serial dilutions up to 10−9. The tenth column of the 96‐well plate serves as a control. After exchanging the medium of the previously prepared Vero cell plate with 100 μl fresh maintenance medium, 100 μl of each virus dilution was transferred to the Vero cell plate. After incubation at 37°C for 4 days, microscopic inspection of the plate was used to monitor cytopathic effects (CPEs) in the form of detached cells. TCID50/ml was determined as:
D S = dilution factor of consecutive dilutions (10); N = total number of wells showing CPE; R = replicates per dilution (8); D 0 = dilution factor of the first dilution (10); V = volume per well in μl (200 μl).
To estimate MOI, we first calculated the viral titer as TCID50/ml of our generated SARS‐CoV‐2 by an end‐point dilution assay as previously described (Flint et al, 2015). In brief, based on induced cytopathic effects, we calculated the TCID50/ml using the above formula based on the Spearman‐Karber method (Ramakrishnan, 2016). To further confirm this calculation with respect to the novelty of this formula, we also applied the commonly used Reed and Muench method (Lei et al, 2020). Both of these methods resulted in a TCID50/ml of 5,000 that we then used to calculate the PFU/ml. Applying poisson distribution, we estimated that the amount of infectious viral particles per ml (PFU/ml) in our stock is 3,500 PFU/ml.
In the context of our infection experiments, we provided 5 μl virus stock per organoid. According to our calculation, the 5 μl volume of SARS‐CoV‐2 stock contains approximately 17.5 PFUs. Having then estimated the number of viable cells after disintegrating organoids (an average of 100,000 for Day 15 and 200,000 for day 60), we could determine the multiplicity of infection (MOI). Considering 17.5 PFUs, our estimated MOI is 1.8 × 10−4 and 8.8 × 10−5 for day 15 and day 60, respectively. Importantly, we found that such a low viral load is sufficient for our studies.
SARS‐CoV‐2 infection
All experiments including SARS‐CoV‐2 infections were performed in a P3 safety laboratory (see above). Neurons and brain organoids were tested and found free from mycoplasma contamination using the mycoplasma kit (Minevera, Cat. No. 11‐1050). For viral exposure, 15‐ and 60‐day‐old organoids were transferred from spinner flasks into low‐adherent 12 well plates. Each well contained one organoid in 2 ml differentiation medium and added with SARS‐CoV‐2 and was incubated as stationary suspension culture. To exclude that the observed effects were not induced by SARS‐CoV‐2, the control organoids (control, uninfected) were treated with supernatants of non‐infected Vero cells.
Generation of convalescent serum, ELISA validation, and affinity purification of SARS‐CoV‐2‐N specific antibodies
AB1 and AB2 were obtained 23 and 16 days after the diagnosis of SARS‐CoV‐2 infection. AB3 and AB4 were obtained 27 and 28 days after the diagnosis of SARS‐CoV‐2 infection (by PCR). Blood samples were drawn directly into serum collection tubes and spun for 15 min at 1,450 g. After centrifugation, the clear supernatant was aliquoted and stored at −80oC. ELISA was performed using semi‐quantitative SARS‐CoV‐2‐IgA and SARS‐CoV‐2‐IgG ELISAs that detect binding against the recombinant S1 domain of the SARS‐CoV‐2 spike protein (Euroimmun, Lübeck, Germany).
The full length ORF of SARS‐CoV‐2‐N was amplified from the vector pUC57‐2019‐nCoV‐N (GeneScript) with the primers: 5′‐aaaaaagtcgacatgtctgataatggacccc‐3′ and 5′‐aaaaaaggatccttaggcctgagttgagtc‐3′ and ligated into the expression vector pET15b via XhoI and BamHI allowing expression of SARS‐CoV‐2‐N with an N‐terminal His6‐tag. The correct sequence was validated by sequencing. pET15b‐SARS‐CoV‐2 was then heat‐shock transformed into BL21(DE3)‐Rosetta2‐pLysS bacteria, plated on LB plates containing 50 μg/ml carbenicillin and 34 μg/ml chloramphenicol. Following over night incubation at 37°C, a single colony was transferred into 20 ml of 2YT medium (1.6% Bacto tryptone, 1% Yeast extract, 0.5% NaCl) and bacteria were grown over night at 37°C. This preculture was used to inoculate a main culture of 1 l 2YT at the next morning. Expression of the nucleoprotein was induced at an OD600 of 0.8 with 1 mM IPTG and continued over night at 30°C. Bacteria were harvested by centrifugation at 4°C for 20 min at 5,000 g and the pellets were frozen at −80°C.
One pellet corresponding to 250 ml of culture was thawed and re‐suspended in 25 ml lysis buffer (PBS, 100 μg/ml lysozyme, 20 μl DNase, 20 mM MgSO4, 1 mM DTT) and incubated for 1 h at RT. Following a protocol from (Schlager et al, 2012), 1% of SDS was added to the lysate which was then transferred to 2 ml Eppendorf tubes and sonicated five times for 2 min in a cooled Misonix S‐4000 water bath sonicator, applying an amplitude of 100%. The samples were then placed on ice for 30 min in order to precipitate excess of SDS. The chilled lysates were centrifuged at 4°C for 20 min at 20,000 g and the supernatant was passed through a 0.45 μm syringe cellulose‐acetate filter (VWR).
For purification of SARS‐CoV‐2 N, the lysate was passed over a 3 ml Ni‐NTA agarose column (Qiagen) equilibrated with PBS, 0.1% Sarkosyl (w/v), 1 mM DTT by gravity flow. The column was then washed with 30 ml WB1 (PBS, 0.1% Sarkosyl (w/v), 5 mM imidazole, 1 mM DTT), and 30 ml of WB2 (PBS, 0.1% Sarkosyl (w/v), 20 mM imidazole, 1 mM DTT). Bound proteins were eluted with EB (PBS, 0.1% Sarkosyl (w/v), 200 mM imidazole, 1 mM DTT) into 1 ml aliquots. Samples were analyzed by SDS–PAGE regarding purity (by Coomassie stain) and identity (by Western Blot using the anti‐SARS‐CoV‐2‐nucleocapsid mouse IgG antibody clone #6F10 from BioVision (#A2060) at a dilution of 1:10,000). The total yield was around 100 mg/l culture, and purity was estimated to be > 95% (including < 5% of degradation products).
The eluate was then directly coupled to activated NHS‐sepharose (GE Healthcare) at 1 mg protein/1 ml of NHS‐beads. Incubation was carried out over night at 4°C. At the next day, the NHS‐sepharose was first blocked with 100 mM of TRIS–HCl pH 8 for 3 h at RT and then washed five times each with 100 mM TRIS pH8 and 100 mM NaAc pH 4.5, 150 mM NaCl. One microliter of SARS‐CoV‐2‐N‐NHS‐sepharose was then incubated with 2 ml of AB4 serum in a 5 ml tube (Sarstedt) at 4°C over night. At the next day, the beads were filled into a 3 ml column and washed with 10 ml of PBS. Bound antibodies were then diluted with 100 mM of glycine pH 2.5 in 100 μl steps. Eluates were immediately neutralized with 40 mM of unbuffered TRIS base and spectrometrically quantified by using a NanoDrop. The total yield was around 250 μg of SARS‐CoV‐2‐N specific antibodies from 2 ml of serum.
Generation of iPSCs‐derived cortical neurons
We differentiated iPSCs into NPCs using STEMdiff Neural Induction Medium (Stem cell technologies, USA). Five days later, the formed neurospheres were collected and cultured on poly‐l‐ornithine (PLO)‐/laminin coated dishes. Seven days later, using a neural rosette selection medium (Stem cell technologies, USA), we re‐cultured neural rosettes to generate NPCs. NPCs were differentiated into cortical neurons as described previously (37). Briefly, NPCs were seeded on poly‐l‐ornithine (PLO)‐/laminin coated coverslips. Forty‐eight hours later, NPCs were switched to cortical neuronal differentiation medium consisting of BrainPhys basal medium(38) supplemented with 1× B27 (without vitamin A, Thermo Scientific, USA), 1× N2 (Thermo Scientific, USA), 20 ng/ml BDNF (PeproTech, USA), 20 ng/ml GDNF (PeproTech, USA), 20 ng/ml NT3, 1 μM cAMP (Sigma, USA), and 0.2 μM ascorbic acid (Sigma, USA). Fresh medium was added every 2–3 days.
Generation of iPSCs‐derived brain organoids and outgrowths
Organoids were generated from two different iPS cell lines, namely IMR90 (Donor 1, Miltenyi, 130‐096‐726) and Crx‐iPS (Donor 2) as described previously (Gagliardi et al, 2018). We adapted previously described protocol to differentiate iPSCs into brain organoids described earlier (Lancaster et al, 2013; Gabriel et al, 2016). Five‐day‐old neurospheres were harvested and embedded in matrigel (Corning, USA) drops. Differentiation medium mixture of DMEM/F12 and Neural Basal Medium (in 1:1 ratio), supplemented with 1:200 N2, 1:100 l‐glutamine, 1:100 B27 w/o vitamin A, 100 U/ml penicillin, 100 μg/ml streptomycin, 23 μM insulin (Sigma‐Aldrich), 0.05 mM MEM non‐essential amino acids (NAA), and 0.05 mM β‐mercaptoethanol (Life Technologies) was used to differentiate the matrigel embedded droplets in suspension culture. After 4 days of culturing, embedded neurospheres were transferred to spinner flasks (IBS, Integra biosciences) containing the same differentiation medium supplemented with 0.5 μmol dorsomorphin (Sigma‐Aldrich, USA).
Organoids with outgrowing neurons were generated as described previously using 60‐day‐old organoids (Gabriel et al, 2016). Organoids were sliced and plated onto coverslips previously coated with poly‐l‐ornithine and laminin. The slices were grown in organoid medium for 14 days until the extended neural structures were observed under a stereomicroscope.
Western blot
The gel electrophoretic separation of proteins was performed under denaturing conditions in the presence of SDS in a non‐continuous gel system, which consisted of a 5% stacking gel and 10% resolving gel, which was then transferred to nitrocellulose membranes. Once the transfer was finished, the membrane was soaked into 5% milk in TRIS–HCl‐based buffer (TBST) for a minimum of 30 min at RT. After incubating with primary antibodies overnight at 4°C, the blots were treated with secondary antibodies at RT for 1 h. Super Signal West Pico or Femto Chemiluminescent substrates (Pierce) were used for detection. Antibody dilutions for Western blots: human convalescent serum AB4 (1:400), polyclonal rabbit SARS‐CoV‐2 (1:500, Biozol, GTX‐GTX632604), monoclonal mouse anti‐SARS‐CoV‐2 (1:500, GeneTex, GTX635679), and mouse anti‐GAPDH (1:20,000, Proteintech, 60004‐I‐Ig). Secondary antibodies goat anti‐mouse IgG (H+L) HRP (1:5,000, 31430, Thermo Fisher Scientific), goat anti‐rabbit IgG (H+L) HRP (1:5,000, 31466, Invitrogen), anti‐human secondary antibodies conjugated to HRP (1:5,000, Thermo Scientific).
Immunofluorescence and confocal microscopy
For light microscopy analysis, monolayer cells (Vero and aspics‐derived neurons) were fixed for 10 min. Brain organoids were fixed for 30 min. We used 4% paraformaldehyde/PBS as a fixative (Gabriel et al, 2016). Organoids were incubated in 30% sucrose overnight at 4°C, embedded in Tissue‐Tek O.C.T. compound (Sakura, Netherlands). Organoids were cryofrozen at −80°C before sectioning into 10–15 μm thin slices using Cryostat Leica CM3050 S. Thin sections and cells were permeabilized with a buffer containing 0.5% Triton X100 for 10 min. Specimens were blocked with 0.5% fish gelatin/PBS for 1 hr, both at room temperature. For SOX2 staining, antigen retrieval was required. For this, sections were treated with repeated heating (microwave) in odium citrate buffer (10 mM Sodium citrate, 0.05% Tween 20, pH 6.0) and were applied before permeabilization and blocking.
We used different antibodies as follows: human convalescent serum (AB1 to AB3 (1:40), AB4, 1:40 or 1:50 or 1:100), rabbit anti‐TUJ‐1 (1:400, Sigma‐Aldrich, T2200), monoclonal mouse anti‐phospho‐Tau AT180 (1:100, Thermo Scientific, MN1040), polyclonal rabbit anti‐phospho‐Tau (S396), (1:100, Thermo Fisher), monoclonal mouse anti‐SARS‐CoV‐2 (1:200, GeneTex, GTX635679), rabbit anti‐SARS‐CoV‐2 NP (1:200, Biozol, GTX‐GTX632604), rabbit anti S‐100 beta (1:100, Abcam, ab52642), rabbit anti Iba‐1 (1:100, Abcam, ab178846), rabbit anti‐MAP2 (1:100, Proteintech, Cat# 17490‐1‐AP), mouse anti‐Pan‐Tau (1:100, DSHB, 5A6). Specimens with primary antibodies were incubated overnight at 4°C. For secondary antibodies, donkey anti‐rabbit IgG (H+L) secondary antibody, Alexa Fluor 594 (Thermo Scientific, Cat# A21207), donkey anti‐rabbit IgG (H+L) secondary antibody, Alexa Fluor 647 (Thermo Scientific, Cat# A31573), donkey anti‐rabbit IgG (H+L) secondary antibody, HRP (Thermo Scientific, Cat# A16023), goat anti‐mouse IgG (H+L) secondary antibody, Alexa Fluor 488 (Thermo Scientific, Cat# A28175), Goat anti‐mouse IgG (H+L) secondary antibody, Alexa Fluor 594 (Thermo Scientific, Cat# A‐11032), Goat anti‐mouse IgG (H+L) secondary antibody, Alexa Fluor 647 (Thermo Scientific, Cat# A‐21236), Alexa Fluor Dyes conjugated either with goat/donkey anti‐mouse, anti‐human, or anti‐rabbit (1:500 or 1:1,000, molecular probes, Invitrogen) was used. For DNA staining, DAPI at a concentration of 1 μg/ml (Thermo Scientific, Cat# 32670) was used, and the coverslips were mounted using Mowiol (Carl Roth, Germany). The raw images were collected using a Leica SP8 confocal system (Leica microsystems, Germany) and processed with the help of Adobe Photoshop (Adobe Systems, USA). For deconvolution, the captured image files were processed using ZEN software (2.3, SP1, black, 64 bit, release version 14.0.0.0; ZEISS, Oberkochen, Germany) for 3D reconstruction and deconvolution. After deconvolution, files were imported into Fiji and further processed using Image J, Adobe Photoshop CC 2018, and Adobe Illustrator CC 2018. For 3D surface and volume rendering, raw image files were processed using Imaris (64× version 7.7.1).
TUNEL assay
Apoptotic cells were detected by using DeadEnd™ Fluorometric TUNEL System (Promega, G3250, USA) according to the manufacturer′s protocol.
Ethical approval and patient samples
Serum samples AB1 and AB2 were obtained under a protocol approved by the ethical committee, medical faculty, University Hospital Düsseldorf, Heinrich‐Heine‐University (study number 5350). Serum samples AB3 and AB4 were obtained under a protocol approved by the Institutional Review Board of the University of Cologne (protocol 16‐054). Human respiratory epithelial cells (hREC) were obtained by nasal brush biopsy from healthy control individuals. The study was endorsed by the local ethical committee at the University of Münster, and each patient gave written informed consent (Study number, 2015‐104‐f‐S, Flimmerepithel) and 2020‐274‐f‐S (COVID‐19). Trained physicians from the Department of General Pediatrics, University Hospital of Münster, performed biopsies.
Statistical analysis
The statistical analyses were performed using GraphPad Prism version 8. All experiments were performed at least in triplicates, and the statistical significance of each dataset was analyzed using Student's t‐test followed by Welche's correction and non‐parametric one‐way ANOVA followed by Tukey's post hoc test. For immunofluorescence‐based experiments, we randomized the samples to avoid any bias. The values are expressed as mean ± SD or SEM. Independent experiments have been represented by “n”.
Author contributions
AR and JG conceptualize the project. AR designed, coordinated and conducted experiments. LM and PNO performed virology works. HS supervised virology. EG, PA‐I, AM‐S and AM provided technical support. AM‐S and CK purified antibodies. OG provided iPS cells. HG, DW and FK provided convalescent serum. KW and HO provided airway cells. AW, MA, SH, TH, AD, OA and JT involved in sequencing and viral strains. JG and HS supervised the work. AR and JG wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Review Process File
Acknowledgements
We want to thank Dr. Boris Görg for offering generous support with their microscope facility. We want to thank Ms Gladiola Goranci and Nazlican Altnisk for their excellent technical assistance. This work was financially supported by a grant from Fritz‐Thyssen Foundation and Foundation and by the Jürgen Manchot Foundation. We would like to thank the diagnostics department of the Institute of Virology, University Hospital Düsseldorf.
The EMBO Journal (2020) 39: e106230
Contributor Information
Heiner Schaal, Email: schaal@uni-duesseldorf.de.
Jay Gopalakrishnan, Email: jay.gopalakrishnan@hhu.de.
Data availability
No amenable data sets.
References
- Alonso AD, Di Clerico J, Li B, Corbo CP, Alaniz ME, Grundke‐Iqbal I, Iqbal K (2010) Phosphorylation of tau at Thr212, Thr231, and Ser262 combined causes neurodegeneration. J Biol Chem 285: 30851–30860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustinack JC, Sanders JL, Tsai LH, Hyman BT (2002a) Colocalization and fluorescence resonance energy transfer between cdk5 and AT8 suggests a close association in pre‐neurofibrillary tangles and neurofibrillary tangles. J Neuropathol Exp Neurol 61: 557–564 [DOI] [PubMed] [Google Scholar]
- Augustinack JC, Schneider A, Mandelkow EM, Hyman BT (2002b) Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer's disease. Acta Neuropathol 103: 26–35 [DOI] [PubMed] [Google Scholar]
- Baig AM, Khaleeq A, Ali U, Syeda H (2020) Evidence of the COVID‐19 virus targeting the CNS: tissue distribution, host‐virus interaction, and proposed neurotropic mechanisms. ACS Chem Neurosci 11: 995–998 [DOI] [PubMed] [Google Scholar]
- Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KRC, Panagiotakos G, Thom N et al (2017) Assembly of functionally integrated human forebrain spheroids. Nature 545: 54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns DM, Rouleau N, Parker RN, Walsh KG, Gehrke L, Kaplan DL (2020) A 3D human brain‐like tissue model of herpes‐induced Alzheimer's disease. Sci Adv 6: eaay8828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani RJ, Perry G (2019) Tau biology, tauopathy, traumatic brain injury, and diagnostic challenges. J Alzheimers Dis 67: 447–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, Ma K, Xu D, Yu H, Wang H et al (2020) Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ 368: m1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JH, Johnson GV (2004) Primed phosphorylation of tau at Thr231 by glycogen synthase kinase 3beta (GSK3beta) plays a critical role in regulating tau's ability to bind and stabilize microtubules. J Neurochem 88: 349–358 [DOI] [PubMed] [Google Scholar]
- Cohen TJ, Guo JL, Hurtado DE, Kwong LK, Mills IP, Trojanowski JQ, Lee VM (2011) The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat Commun 2: 252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde Cardona G, Quintana Pajaro LD, Quintero Marzola ID, Ramos Villegas Y, Moscote Salazar LR (2020) Neurotropism of SARS‐CoV 2: mechanisms and manifestations. J Neurol Sci 412: 116824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen T, Lolli V, Sadeghi N, Rovai A, Trotta N, Taccone FS, Creteur J, Henrard S, Goffard JC, De Witte O et al (2020) Early postmortem brain MRI findings in COVID‐19 non‐survivors. Neurology 10.1212/WNL.0000000000010116 [DOI] [PubMed] [Google Scholar]
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brunink S, Schneider J, Schmidt ML et al (2020) Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill 25: 2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimaraes KP, Benazzato C, Almeida N, Pignatari GC, Romero S et al (2016) The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534: 267–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Galkowski D, Zhao H (2008) Analysis of apoptosis by cytometry using TUNEL assay. Methods 44: 250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice FG, Tovar‐Moll F, Moll J, Munoz DP, Ferreira ST (2020) Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and the central nervous system. Trends Neurosci 43: 355–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint SJ, Racaniello VR, Rall GF, Skalka AM, Enquist LW (2015) Principles of virology. Washington, DC: ASM Press; [Google Scholar]
- Frost B, Gotz J, Feany MB (2015) Connecting the dots between tau dysfunction and neurodegeneration. Trends Cell Biol 25: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel E, Wason A, Ramani A, Gooi LM, Keller P, Pozniakovsky A, Poser I, Noack F, Telugu NS, Calegari F et al (2016) CPAP promotes timely cilium disassembly to maintain neural progenitor pool. EMBO J 35: 803–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel E, Gopalakrishnan J (2017) Generation of iPSC‐derived human brain organoids to model early neurodevelopmental disorders. J Vis Exp 122: 55372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci‐Buzhala G, Krut O, Peters F, Nikolic M et al (2017) Recent zika virus isolates induce premature differentiation of neural progenitors in human brain organoids. Cell Stem Cell 20: 397–406. [DOI] [PubMed] [Google Scholar]
- Gagliardi G, Ben M'Barek K, Chaffiol A, Slembrouck‐Brec A, Conart JB, Nanteau C, Rabesandratana O, Sahel JA, Duebel J, Orieux G et al (2018) Characterization and transplantation of CD73‐positive photoreceptors isolated from human iPSC‐derived retinal organoids. Stem Cell Reports 11: 665–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giandomenico SL, Lancaster MA (2017) Probing human brain evolution and development in organoids. Curr Opin Cell Biol 44: 36–43 [DOI] [PubMed] [Google Scholar]
- Giandomenico SL, Mierau SB, Gibbons GM, Wenger LMD, Masullo L, Sit T, Sutcliffe M, Boulanger J, Tripodi M, Derivery E et al (2019) Cerebral organoids at the air‐liquid interface generate diverse nerve tracts with functional output. Nat Neurosci 22: 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan J (2019) The emergence of stem cell‐based brain organoids: trends and challenges phosphorylates. BioEssays 41: e1900011 [DOI] [PubMed] [Google Scholar]
- Goranci‐Buzhala G, Mariappan A, Gabriel E, Ramani A, Ricci‐Vitiani L, Buccarelli M, D'Alessandris QG, Pallini R, Gopalakrishnan J (2020) Rapid and efficient invasion assay of glioblastoma in human brain organoids. Cell Rep 31: 107738 [DOI] [PubMed] [Google Scholar]
- Helms J, Kremer S, Merdji H, Clere‐Jehl R, Schenck M, Kummerlen C, Collange O, Boulay C, Fafi‐Kremer S, Ohana M et al (2020) Neurologic features in severe SARS‐CoV‐2 infection. N Engl J Med 382: 2268–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine‐Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A et al (2020) SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271–280.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janesick A, Wu SC, Blumberg B (2015) Retinoic acid signaling and neuronal differentiation. Cell Mol Life Sci 72: 1559–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo A, Shahpasand K, Mannix R, Qiu J, Moncaster J, Chen CH, Yao Y, Lin YM, Driver JA, Sun Y et al (2015) Antibody against early driver of neurodegeneration cis P‐tau blocks brain injury and tauopathy. Nature 523: 431–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers MM, Beumer J, van dVJ, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, Paul vSJ, Mykytyn AZ, Duimel HQ et al (2020) SARS‐CoV‐2 productively infects human gut enterocytes. Science 369: 50–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA (2013) Cerebral organoids model human brain development and microcephaly. Nature 501: 373–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA (2014) Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345: 1247125 [DOI] [PubMed] [Google Scholar]
- Lei C, Yang J, Hu J, Sun X (2020) On the calculation of TCID50 for quantitation of virus infectivity. Virol Sin. 10.1007/s12250-020-00230-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC et al (2003) Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426: 450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP (1999) The prolyl isomerase Pin1 restores the function of Alzheimer‐associated phosphorylated tau protein. Nature 399: 784–788 [DOI] [PubMed] [Google Scholar]
- Luna‐Munoz J, Chavez‐Macias L, Garcia‐Sierra F, Mena R (2007) Earliest stages of tau conformational changes are related to the appearance of a sequence of specific phospho‐dependent tau epitopes in Alzheimer's disease. J Alzheimers Dis 12: 365–375 [DOI] [PubMed] [Google Scholar]
- McCray PB Jr, Pewe L, Wohlford‐Lenane C, Hickey M, Manzel L, Shi L, Netland J, Jia HP, Halabi C, Sigmund CD et al (2007) Lethal infection of K18‐hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol 81: 813–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesci P, Macia A, Saleh A, Martin‐Sancho L, Yin X, Snethlage C, Avansini S, Chanda SK, Muotri A (2020) Sofosbuvir protects human brain organoids against SARS‐CoV‐2. BioRxiv 10.1101/2020.05.30.125856 [PREPRINT] [DOI] [Google Scholar]
- Monteil V, Kwon H, Prado P, Hagelkruys A, Wimmer RA, Stahl M, Leopoldi A, Garreta E, Hurtado Del Pozo C, Prosper F et al (2020) Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ACE2. Cell 181: 905–913.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Greenwood A, Binder L, Bigio EH, Denial S, Nicholson L, Zhou XZ, Lu KP (2012) Proline isomer‐specific antibodies reveal the early pathogenic tau conformation in Alzheimer's disease. Cell 149: 232–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyiadji N, Shahin G, Noujaim D, Stone M, Patel S, Griffith B (2020) COVID‐19‐associated acute hemorrhagic necrotizing encephalopathy: CT and MRI features. Radiology 296: E119–E120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles VG, Lutgehetmann M, Lindenmeyer MT, Sperhake JP, Wong MN, Allweiss L, Chilla S, Heinemann A, Wanner N, Liu S et al (2020) Multiorgan and renal tropism of SARS‐CoV‐2. N Engl J Med 383: 590–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C et al (2016) Brain‐region‐specific organoids using mini‐bioreactors for modeling ZIKV exposure. Cell 165: 1238–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan MA (2016) Determination of 50% endpoint titer using a simple formula. World J Virol 5: 85–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlager B, Straessle A, Hafen E (2012) Use of anionic denaturing detergents to purify insoluble proteins after overexpression. BMC Biotechnol 12: 95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedaghat Z, Karimi N (2020) Guillain Barre syndrome associated with COVID‐19 infection: a case report. J Clin Neurosci 76: 233–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A, Kabat J, Novak M, Wu Q, Grundke‐Iqbal I, Iqbal K (1998) Phosphorylation of tau at both Thr 231 and Ser 262 is required for maximal inhibition of its binding to microtubules. Arch Biochem Biophys 357: 299–309 [DOI] [PubMed] [Google Scholar]
- Song E, Zhang C, Israelow B, Lu P, Weizman OE, Liu F, Dai Y, Szigeti‐Buck K, Yasumoto Y, Wang G et al (2020) Neuroinvasive potential of SARS‐CoV‐2 revealed in a human brain organoid model. BioRxiv 10.1101/2020.06.25.169946 [PREPRINT] [DOI] [Google Scholar]
- Streit A, Berliner AJ, Papanayotou C, Sirulnik A, Stern CD (2000) Initiation of neural induction by FGF signalling before gastrulation. Nature 406: 74–78 [DOI] [PubMed] [Google Scholar]
- Virani A, Rabold E, Hanson T, Haag A, Elrufay R, Cheema T, Balaan M, Bhanot N (2020) Guillain‐Barre Syndrome associated with SARS‐CoV‐2 infection. IDCases 20: e00771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Mandelkow E (2016) Tau in physiology and pathology. Nat Rev Neurosci 17: 5–21 [DOI] [PubMed] [Google Scholar]
- Worl Health Organization (2020) Naming the coronavirus disease (COVID‐19) and the virus that causes it. https://wwwwhoint/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it
- Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, Cakir B, Kim KY, Lombroso AP, Hwang SM et al (2017) Fusion of regionally specified hPSC‐derived organoids models human brain development and interneuron migration. Cell Stem Cell 21: 383–398.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Han Y, Nilsson‐Payant BE, Gupta V, Wang P, Duan X, Tang X, Zhu J, Zhao Z, Jaffre F et al (2020a) A human pluripotent stem cell‐based platform to study SARS‐CoV‐2 tropism and model virus infection in human cells and organoids. Cell Stem Cell 27: 125–136.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, Wu Y, Zhang L, Yu Z, Fang M et al (2020b) Clinical course and outcomes of critically ill patients with SARS‐CoV‐2 pneumonia in Wuhan, China: a single‐centered, retrospective, observational study. Lancet Respir Med 8: 475–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zempel H, Mandelkow E (2014) Lost after translation: missorting of Tau protein and consequences for Alzheimer disease. Trends Neurosci 37: 721–32 [DOI] [PubMed] [Google Scholar]
- Zhou J, Li C, Liu X, Chiu MC, Zhao X, Wang D, Wei Y, Lee A, Zhang AJ, Chu H et al (2020) Infection of bat and human intestinal organoids by SARS‐CoV‐2. Nat Med 26: 1077–1083 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Review Process File
Data Availability Statement
No amenable data sets.


