Abstract
COVID‐19, caused by SARS‐CoV‐2, has resulted in severe and unprecedented economic and social disruptions in the world. Nucleocapsid (N) protein, which is the major structural component of the virion and is involved in viral replication, assembly and immune regulation, plays key roles in the viral life cycle. Here, we solved the crystal structures of the N‐ and C‐terminal domains (N‐NTD and N‐CTD) of SARS‐CoV‐2 N protein, at 1.8 and 1.5 Å resolution, respectively. Both structures show conserved features from other CoV N proteins. The binding sites targeted by small molecules against HCoV‐OC43 and MERS‐CoV, which inhibit viral infection by blocking the RNA‐binding activity or normal oligomerization of N protein, are relatively conserved in our structure, indicating N protein is a promising drug target. In addition, certain areas of N‐NTD and N‐CTD display distinct charge distribution patterns in SARS‐CoV‐2, which may alter the RNA‐binding modes. The specific antigenic characteristics are critical for developing specific immune‐based rapid diagnostic tests. Our structural information can aid in the discovery and development of antiviral inhibitors against SARS‐CoV‐2 in the future.
Keywords: 2019‐nCoV, antivirals, dimerization, nucleocapsid, RNA binding
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Structural Biology
High‐resolution structures of the N‐ and C‐terminal domains of SARS‐CoV‐2 N protein reveal conserved as well as unique features compared to other coronavirus N proteins.
Introduction
In December 2019, a new coronavirus (2019‐nCoV) was detected due to emerging viral pneumonia cases in Wuhan, China (Li et al, 2020; Tan et al, 2020; Wang et al, 2020a; Wu et al, 2020a; Zhou et al, 2020b; Zhu et al, 2020). The World Health Organization named the infectious disease “COVID‐19”, and it declared a global pandemic on 11 March 2020 (https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020).
2019‐nCoV, which is the seventh coronavirus capable of infecting humans, was later named “SARS‐CoV‐2” by the International Committee on Taxonomy of Viruses (ICTV) (Gorbalenya et al, 2020), although with some controversies (Jiang et al, 2020). The other six coronaviruses are the low‐pathogenicity members HCoV‐OC43, HCoV‐HKU1, HCoV‐NL63 and HCoV‐229E and the highly pathogenic SARS‐CoV and MERS‐CoV (Lu et al, 2020). Sequence comparison has shown that SARS‐CoV‐2 has the closest relationship (96.2%) with the bat SARS‐like coronavirus RaTG13 (Lu et al, 2020; Zhou et al, 2020a,b), but the origin of this virus is more than likely the bat; what remains to be identified is a possible intermediate host. Although many small‐molecule drugs targeting the viral polymerase and protease, such as remdesivir, lipinavir, ritonavir and hydroxychloroquine, have been shown to be promising at the beginning (Wang et al, 2020c; Zhang et al, 2020a), no therapeutics have yet been proven effective for the treatment of severe illness in recent clinical trials (Beigel et al, 2020; Cao et al, 2020), and hydroxychloroquine has been found no benefit to help patients with COVID‐19 recover (Cavalcanti et al, 2020; Hoffmann et al, 2020; Maisonnasse et al, 2020), indicating more effective antiviral drugs are yet to be developed, and combinational therapeutic approaches are necessary to improve patient outcomes in COVID‐19. Recently, several protective neutralization antibodies blocking viral entry were rapidly developed (Shi et al, 2020; Wu et al, 2020b,c), and diverse types of vaccine candidates are undergoing clinical evaluation (Wang et al, 2020b). However, there is still no clinically approved specific drug or vaccine available to treat the disease. Therefore, it is urgent to develop specific drugs/inhibitors against distinct targets and infection processes.
Among positive‐sense RNA viruses, the family Coronaviridae, of which SARS‐CoV‐2 is a member, has the largest genome (~ 30,000 bases). It contains two large overlapping open reading frames (ORF1a and ORF1b) and encodes four structural proteins, i.e. spike, envelope, membrane and nucleocapsid (N) proteins, and nine accessory proteins. ORF1a and ORF1b are further processed to generate 16 nonstructural proteins (Nsp1 to 16). Among the viral proteins, the N protein is the central component of virions. It binds to viral genomic RNA to package the RNA into a ribonucleoprotein (RNP) complex. Besides assembly, N proteins also possess other functions, including roles in viral mRNA transcription and replication (Zuniga et al, 2010; Cong et al, 2020), cytoskeleton organization and immune regulation (Surjit et al, 2004, 2006; Lu et al, 2011). Especially, SARS‐CoV‐2 N protein has been found to function as a viral suppressor of RNA silencing (VSR) through its double‐stranded RNA‐binding activity to combat host RNAi‐mediated antiviral responses (Mu et al, 2020). In addition, N protein could induce both humoral and cellular immune responses after infection (Ni et al, 2020; Xiang et al, 2020), making it a key target for diagnosis and vaccine development.
Coronavirus N proteins have two conserved and independently folded structural domains, called the N‐terminal domain (NTD) and C‐terminal domain (CTD) (Chang et al, 2014). The two domains are connected by an intrinsically disordered region (IDR) called the central linker region (LKR). The LKR includes a Ser/Arg (SR)‐rich region that contains putative phosphorylation sites (Peng et al, 2008). In addition, the N‐NTD and N‐CTD of coronaviruses are usually flanked by two IDRs, which are called N‐arm and C‐tail. Previous studies have revealed that N‐NTD is responsible for RNA binding, N‐CTD for both RNA binding and dimerization, and the IDR for modulating the RNA‐binding activity of N‐NTD and N‐CTD and oligomerization (Chang et al, 2014). The structures of both N‐NTD and N‐CTD from several human‐infecting coronaviruses have been solved (Yu et al, 2006; Chen et al, 2007; Saikatendu et al, 2007; Lin et al, 2014; Papageorgiou et al, 2016; Szelazek et al, 2017; Nguyen et al, 2019), but the structure of the full‐length N protein remains a mystery due to the disordered IDR and oligomeric characteristics (Cong et al, 2017). The importance of N protein in viral assembly, replication and host immune response regulation makes it an attractive target for developing broad‐spectrum antiviral inhibitors. Previous studies have shown that small‐molecule drugs can be developed by targeting the RNA‐binding activity and the oligomerization of the N protein (Lin et al, 2014; Chang et al, 2016; Zhang et al, 2020b).
Here, we determined the crystal structures of SARS‐CoV‐2 N‐NTD at a resolution of 1.8 Å and N‐CTD at a resolution of 1.5 Å, and we compared these with other human‐infecting coronavirus N‐NTD and N‐CTD structures to identify suitable target sites for anti‐COVID‐19 drug discovery and elucidate their structural drug‐binding features. These studies provide important structural information to guide the development of antiviral compounds against SARS‐CoV‐2 and targeting the N protein.
Results
Domain organization of the SARS‐CoV‐2 N protein
According to the sequence alignment analysis of SARS‐CoV‐2 and other coronaviruses, SARS‐CoV‐2 has a genome organization similar to that of SARS‐CoV (Fig 1A), and the SARS‐CoV‐2 N protein also shares the same modular organization with the SARS‐CoV N protein. It is composed of three IDRs (N‐arm, LKR and C‐tail) and two structural domains (NTD and CTD) (Fig 1B). SARS‐CoV‐2 has a close relationship with SARS‐CoV, and both belong to lineage B beta‐coronaviruses (Fig 1C). Previous studies have shown that coronavirus N‐NTD is the RNA‐binding domain, while N‐CTD is the dimerization domain and also has RNA‐binding activity. To further study the biochemical properties of SARS‐CoV‐2 N, we produced SARS‐CoV‐2 N‐NTD and N‐CTD in Escherichia coli, and we obtained the soluble protein domains by metal affinity chromatography and gel filtration chromatography (Fig EV1A and B). Sedimentation velocity analytical ultracentrifugation analyses determined that SARS‐CoV‐2 N‐NTD exists as a monomer (~ 15.4 kDa) (Fig 1D), while N‐CTD exists as a dimer (~ 28.7 kDa) (Fig 1E) in solution. These results further confirm that SARS‐CoV‐2 N protein shares the same modular organization with other CoV N proteins.
Figure 1. Domain organization of the SARS‐CoV‐2 nucleocapsid protein.
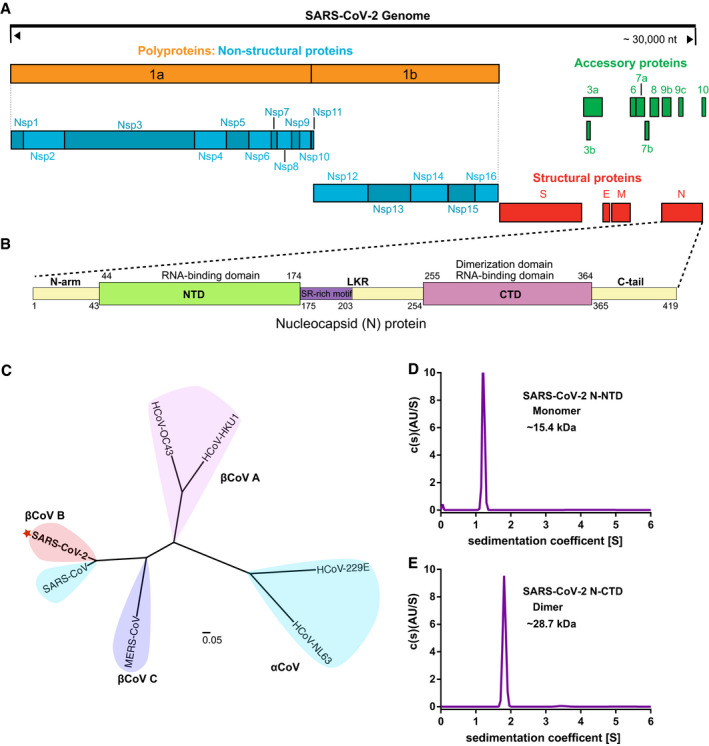
- Genome organization of SARS‐CoV-2.
- Schematic representation of SARS‐CoV-2 N protein domains. Three intrinsically disordered regions, i.e. N‐arm, linker region (LKR) and C‐tail, and the N‐terminal domain (NTD) and C‐terminal domain (CTD) are illustrated. The charged Ser/Arg (SR)‐rich motif (coloured purple) is shown.
- Phylogenetic analysis of N proteins from seven human‐infecting coronaviruses. SARS‐CoV-2 is highlighted by a red asterisk.
- Analytical ultracentrifugation absorbance data analysis of N‐NTD. The clear peak at ˜ 15.4 kDa molecular weight (MW) corresponds to the N‐NTD monomer.
- Analytical ultracentrifugation absorbance data analysis of N‐CTD. The clear peak at ˜ 28.7 kDa MW corresponds to the N‐CTD dimer.
Figure EV1. Biophysical characterization of SARS‐CoV‐2 N‐NTD and N‐CTD .
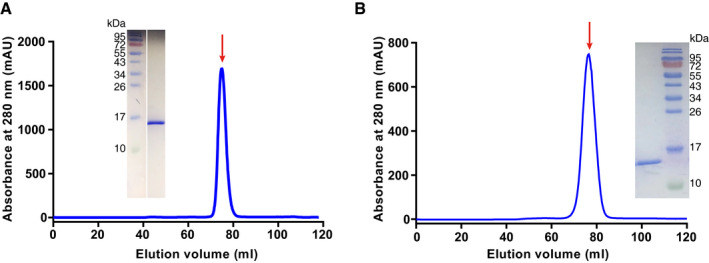
- Analytical gel filtration of SARS‐CoV-2 N‐NTD.
- Analytical gel filtration of SARS‐CoV-2 N‐CTD.
Structure of SARS‐CoV‐2 N‐NTD
The crystal structure of SARS‐CoV‐2 N‐NTD was solved by molecular replacement to a resolution of 1.8 Å, with R work and R free values of 19.7 and 22.7%, respectively (Table 1). Each asymmetric unit contains four N‐NTD molecules (Fig EV2A) with an overall root‐mean‐square difference (RMSD) of 0.16–0.28 Å between each protomer (Fig EV2B). Each N‐NTD molecule presents a right‐handed fist shape and consists of a four‐stranded antiparallel β‐sheet core subdomain, which is sandwiched between loops or short 310 helices and a protruding β‐hairpin region formed by β2 and β3 strands out of the core (Fig 2A and B). Based on the surface charge distribution, the protruding loops are positively charged, providing a putative site for RNA binding (Fig 2C). In addition, according to the complex structures of the previously solved HCoV‐NL63 N‐NTD with ribonucleotide 5′‐mono‐phosphates (AMP/GMP/UMP/CMP) (Lin et al, 2014), the core domain also contains an RNA‐binding site, which is conserved among different human‐infecting coronaviruses (Fig 2D).
Table 1.
Data collection and refinement statistics
| SARS‐CoV‐2 N‐NTD | SARS‐CoV‐2 N‐CTD | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.97919 | 0.97919 |
| Space group | P 1 21 1 | P 1 |
| Cell dimensions | ||
| a, b, c (Å) | 59.27, 55.43, 85.68 | 43.73, 50.31, 69.13 |
| α, β, γ (°) | 90.00, 95.38, 90.00 | 73.88, 89.94, 82.55 |
| Resolution (Å) | 50.00–1.80 (1.86–1.80)a | 50.00–1.39 (1.41–1.39) |
| R p.i.m. b | 0.023 (0.330) | 0.020 (0.321) |
| I/σI | 32.724 (2.538) | 30.265 (2.035) |
| CC1/2 | 0.998 (0.963) | 0.996 (0.808) |
| Completeness (%) | 98.4 (97.6) | 91.7 (67.2) |
| Redundancy | 6.6 (6.6) | 3.4 (3.0) |
| Refinement | ||
| Resolution (Å) | 29.50–1.80 | 26.83–1.50 |
| No. reflections | 46,709 | 81,130 |
| R work/R free c | 0.197/0.227 | 0.177/0.190 |
| No. atoms | ||
| Protein | 3,956 | 3,534 |
| Ligand/ion | 0 | 0 |
| Water | 611 | 740 |
| B‐factors | ||
| Protein | 29.6 | 17.2 |
| Ligand/ion | – | – |
| Water | 30.5 | 28.3 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.005 | 0.003 |
| Bond angles (°) | 0.808 | 0.651 |
| Ramachandran plot | ||
| Favoured (%) | 99.40 | 99.77 |
| Allowed (%) | 0.60 | 0.23 |
| Outliers (%) | 0.00 | 0.00 |
Values in parentheses are for highest‐resolution shell.
R p.i.m. = Σhkl[1/(N − 1)]1/2 Σi |I i − <I>|/ΣhklΣi I i, where I i is the observed intensity and <I> is the average intensity from multiple measurements.
R work = Σ | |F o| − |F c| |/Σ |F o|, where F o and F c are the structure‐factor amplitudes from the data and the model, respectively. R free is the R factor for a subset (5%) of reflections that was selected prior to refinement calculations and was not included in the refinement.
Figure EV2. Overall structure of four SARS‐CoV‐2 N‐NTD molecules in one asymmetric unit.
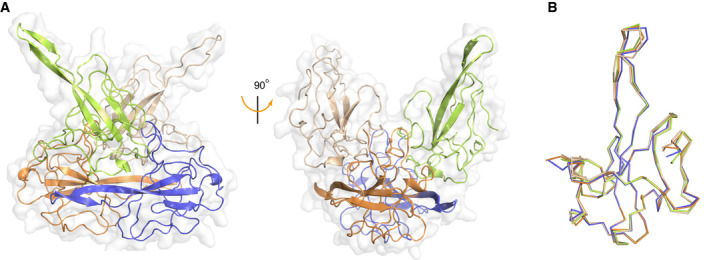
- Cartoon presentation of the four molecules in one asymmetric unit. Four protomers are differently coloured. Each protomer interacts with adjacent protomers with buried areas of 345–617 Å2. The surface is shown in transparent white.
- Superimposition of these four molecules shown as ribbon representation.
Figure 2. Structure of SARS‐CoV‐2 N‐NTD .
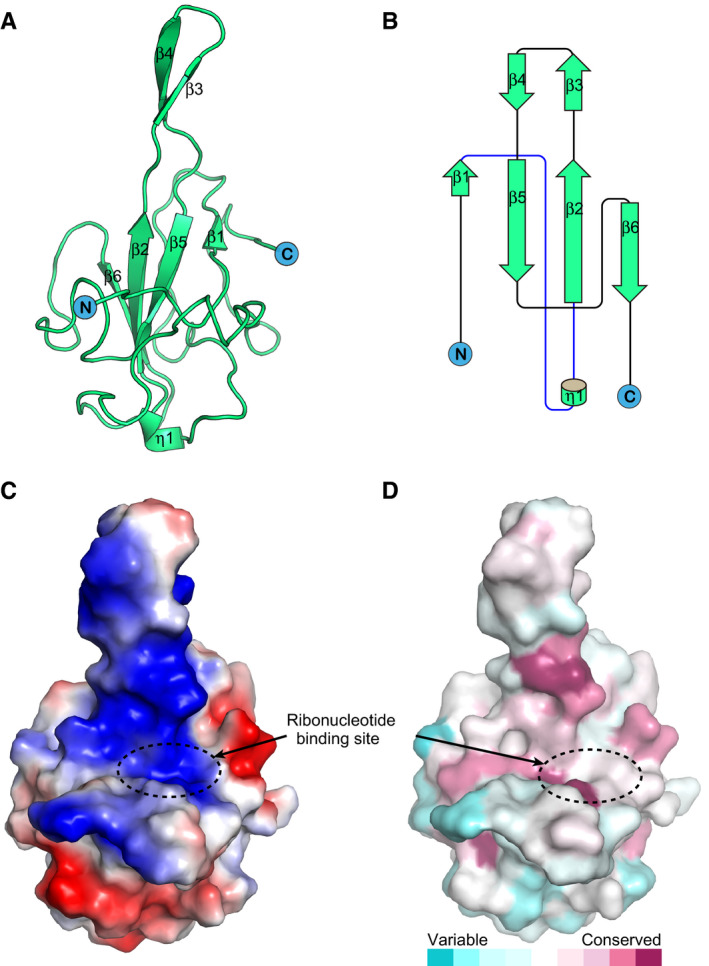
- Cartoon representation of SARS‐CoV-2 N‐NTD.
- Topology diagram for SARS‐CoV-2 N‐NTD; η represents the 310 helix and β represents the β‐sheet.
- The electrostatic surface potential of SARS‐CoV-2 N‐NTD. Red and blue colours indicate negative and positive potential, respectively. The RNA‐binding sites are highlighted in dotted circles and labelled.
- Surface representation of SARS‐CoV-2 N‐NTD coloured according to sequence conservation based the alignment of N protein sequences from seven human‐infecting CoVs using the ConSurf server (Ashkenazy et al, 2016); dark magenta indicates the most conserved, dark cyan indicates the most divergent.
The overall structure of SARS‐CoV‐2 N‐NTD strongly resembles other CoV N‐NTD structures, with an RMSD of 0.33–0.76 Å (Fig 3A). We compared the SARS‐CoV‐2 N‐NTD structure with all of the other available human‐infecting CoV N‐NTD structures, i.e. SARS‐CoV, MERS‐CoV, HCoV‐OC43 and HCoV‐NL63. Of note, among these structures, two regions show the greatest structural differences. First, the orientations of the N‐terminal regions are distinct. The N‐terminal loops of SARS‐CoV‐2, SARS‐CoV and MERS‐CoV N‐NTD stretch outward, while the loops from HCoV‐OC43 and HCoV‐NL63 N‐NTD rotate towards the core subdomain. Second, the protruding β‐hairpin regions are flexible; particularly, the connecting flexible loops are invisible in HCoV‐OC43 and HCoV‐NL63 N‐NTD structures (Fig 3A). The ribonucleotide‐binding site is located at the core subdomain in the HCoV‐NL63 N‐NTD‐AMP complex structure. We then compared the residues involved in AMP binding in different coronaviruses (Fig 3B and C). The highly conserved residues S64, Y126 and R164 (HCoV‐OC43 N‐NTD numbering), located near the base of the pocket, bind to AMP via hydrogen‐bonding interactions, while the residues on the side wall of the pockets, including Y124 and G68, further stabilize the binding. However, this region is not conserved in all human‐infecting coronaviruses, and it could be further divided into two groups. The SARS‐CoV‐2, SARS‐CoV and MERS‐CoV NTD N‐terminal regions are similar to that of HCoV‐OC43, containing conserved residues Y124 and G68 (or Ala). The phenolic hydroxyl group of Y124 interacts with the AMP adenine ring via a hydrogen bond, and the backbone of G68 forms hydrogen bonds with the monophosphate group of AMP (Fig 3B). However, for HCoV‐NL63, residues H77 and P24 are present at equivalent positions, and both residues lack the capability of hydrogen bond formation, and they may further occlude AMP binding (Fig 3C). Therefore, the RNA‐binding modes must differ between these two groups. Whether this is a divergent characteristic between beta‐coronavirus (HCoV‐OC43, SARS‐CoV‐2, SARS‐CoV and MERS‐CoV) and alpha‐coronaviruses (HCoV‐NL63) needs further investigation (Fig 1C). The electrostatic potential surfaces also show different charge distribution patterns in different coronavirus N‐NTD structures, especially the N‐terminal loop, the top tip of the protruding region and the bottom of core subdomain (Fig 3D–H). However, all of the structures display an extended positively charged groove from the AMP‐binding site to the protruding region, indicating this patch is responsible for RNA binding in different coronaviruses.
Figure 3. Comparison of the structure of SARS‐CoV‐2 N‐NTD with other human‐infecting CoV N‐NTD structures.
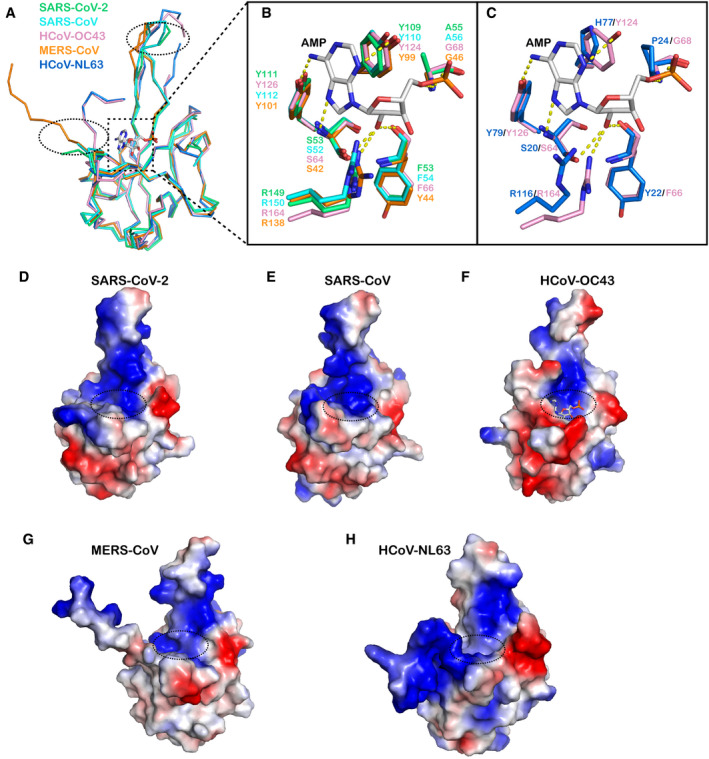
-
ASuperposition of the SARS‐CoV-2 N‐NTD structure with other human‐infecting CoV N‐NTD structures. The ribbon representation of each structure is coloured separately (SARS‐CoV, PDB: 2OFZ, cyan; HCoV‐OC43, PDB: 4LI4, pink; MERS‐CoV, PDB: 4UD1, orange; HCoV‐NL63, PDB: 5N4K, blue) (Saikatendu et al, 2007; Lin et al, 2014; Papageorgiou et al, 2016; Szelazek et al, 2017). The loops that show significant differences are highlighted by dotted circles. The AMP ligand in the HCoV‐OC43 N‐NTD-AMP complex is shown as a stick structure.
-
BStructural superimposition of SARS‐CoV-2, SARS‐CoV and MERS‐CoV N‐NTDs with that of HCoV‐OC43 bound to AMP to show the detailed interactions involved in AMP binding.
-
CStructural superimposition of HCoV‐NL63 N‐NTD with HCoV‐OC43 N‐NTD bound to AMP to show the detailed interactions involved in AMP binding.
-
D–HComparison of the electrostatic surfaces with other human‐infecting CoV N‐NTD structures. The electrostatic surface of N‐NTD from (D) SARS‐CoV-2, (E) SARS‐CoV, (F) HCoV‐OC43 bound to AMP, (G) MERS‐CoV and (H) HCoV‐NL63 are shown, and the AMP‐binding sites are highlighted in dotted circles. Red and blue colours indicate negative and positive potential, respectively.
Structure of SARS‐CoV‐2 N‐CTD
The crystal structure of SARS‐CoV‐2 N‐CTD was solved by molecular replacement to a resolution of 1.5 Å, with R work and R free values of 17.7 and 19.0%, respectively (Table 1). SARS‐CoV‐2 N‐CTD forms a tight homodimer and displays an overall rectangular slab shape (Fig 4A). Each protomer has the shape of the letter C (Fig 4B) and is comprised of five α‐helices, two β‐strands, and two 310 helices (Fig 4C). The β‐hairpin from one protomer is inserted into the cavity of the other protomer, resulting in the formation of the four‐stranded, antiparallel β‐sheet at the dimer interface. The β‐sheet forms one face of the slab dimer, while on the opposite face of the dimer, the surface is formed by α‐helices and loops. The dimer interface buries a surface area of 2,590 Å2. Extensive hydrogen bond interactions between the two hairpins and hydrophobic interactions between the β‐sheet and the alpha helices make the dimeric structure highly stable. This is consistent with our previous biochemical characterization that N‐CTD exists as a dimer in solution (Fig 1E).
Figure 4. Structure of SARS‐CoV‐2 N‐CTD .
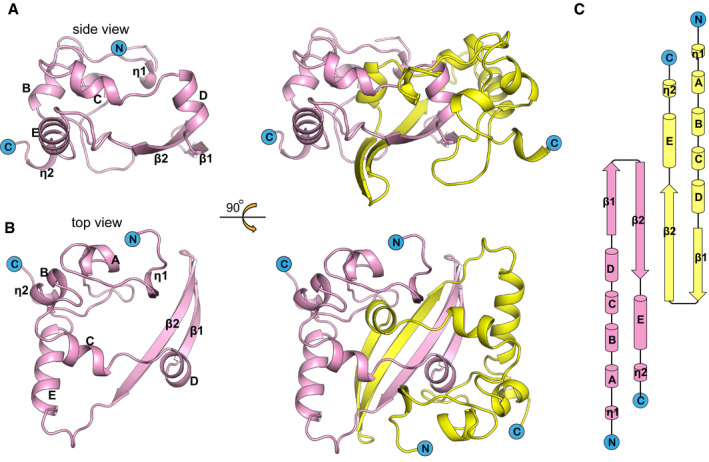
- Cartoon representation of SARS‐CoV-2 N‐CTD. Ribbon representation of SARS‐CoV-2 N‐CTD showing the separated monomer and dimer. One monomer is coloured in pink, and the other is coloured in yellow.
- Rotation of (A) along the x‐axis by 90° to show the top view.
- Topology diagram for SARS‐CoV-2 N‐CTD. The α‐helices are numbered A, B and C, η represents the 310 helix and β represents the β‐sheet, according to a previous SARS N‐CTD structural element numbering rule (Yu et al, 2006).
We then compared the SARS‐CoV‐2 N‐CTD structure with the available N‐CTD structures of SARS‐CoV, MERS‐CoV and HCoV‐NL63, and we found that the overall structure is highly similar, with an RMSD of 0.31–0.98 Å (Fig 5A). They all show a conserved positively charged groove in the helix face of the N‐CTD dimer, due to the distribution of several positively charged residues, including K256, K257, K261 and R262 in SARS‐CoV‐2 N‐CTD (Figs 5B and EV3). Interestingly, the electrostatic potential surfaces in the β‐sheet faces of these proteins show distinct features. The MERS‐CoV structure displays a positively charged central region, whereas SARS‐CoV‐2 and SARS‐CoV structures both display a negatively charged region. For NL63, the central region of its β‐sheet face shows a highly negatively charged region (Fig 5B). These differences may affect the binding modes of RNA recognition. Our structure shows SARS‐CoV‐2 N‐CTD functions as the dimerization domain and the RNA‐binding domain involved in RNP assembly.
Figure 5. Comparison of the structure of SARS‐CoV‐2 N‐CTD with other coronavirus N‐CTD structures.

- Electrostatic surface views of the four N‐CTD structures. Red and blue colours indicate negative and positive potential, respectively.
Figure EV3. Sequence alignment of N proteins from seven human‐infecting coronaviruses.

The secondary structure elements α‐helix, β‐sheet and turn are shown as spirals (α‐helix), arrows (β‐strand) and “TT” letters (turn), respectively. The residue numbers of each sequence are labelled on the left of the alignment, and black dots above the sequence alignment mark the residue number according to SARS‐CoV‐2 N at intervals of 10. Identical residues are highlighted in red background, and similar residues are labelled in red and boxed in blue lines.
Drug target sites in the N protein
As the RNA‐binding activity of the N protein is critical for viral RNP formation and genome replication, blocking RNA binding of N‐NTD has been proved to be a good strategy to develop antiviral drugs. The compound PJ34, which targets the ribonucleotide‐binding site in N‐NTD, displays potent inhibition of the RNA‐binding activity of the HCoV‐OC43 N protein and can inhibit viral replication of HCoV‐OC43 (Lin et al, 2014). PJ34 mimics the binding of AMP to N‐NTD, and it fits in the ribonucleotide‐binding pocket of N‐NTD with an extended trend to the N‐terminal loop. We have compared the corresponding binding site for PJ34 in our SARS‐CoV‐2 N‐NTD structure with that in the HCoV‐OC43 N‐NTD structure (Fig 6A), and we found that the key residues that are involved in the interactions, including S51, F53, Y109, Y111 and R149 (in SARS‐CoV‐2 N‐NTD numbering), are conserved (Fig 6B).
Figure 6. Conserved drug target sites in SARS‐CoV‐2 N protein.

-
A, BSuperimposition of the structure of SARS‐CoV-2 N‐NTD (light blue) with HCoV‐OC43 N‐NTD (light pink) bound to PJ34 (magenta) (PDB: 4KXJ) (Lin et al, 2014). The detailed interactions are shown in (B). The water molecule mediating hydrogen‐bonding interactions is shown as a red sphere.
-
C, DSuperimposition of the structure of SARS‐CoV-2 N‐NTD (light blue) with MERS‐CoV N‐NTD dimer (grey) or MERS‐CoV N‐NTD bound to P3 (cyan) (PDB: 6KL6) (Lin et al, 2020). The detailed interactions are shown in (D).
Another strategy is to block normal N protein oligomerization, thereby halting RNP formation or inducing abnormal aggregation. Recently, the novel inhibitor 5‐benzyloxygramine (P3) was identified by virtual screening (Lin et al, 2020). This compound could mediate MERS‐CoV N‐NTD non‐native dimerization and induce N protein aggregation. It was shown to have potent antiviral activity against MERS‐CoV. The complex structure of MERS‐CoV N‐NTD and P3 shows that P3 targets the non‐native interface of dimeric N‐NTD and binds to two hydrophobic pockets in two N‐NTD protomers. P3 occupies the N‐terminal vector‐fusion residue‐binding cavity of NTD promoter 1 in the ligand‐free MERS‐CoV N‐NTD structure (Fig 6C) and further stabilizes the dimeric status by massive hydrophobic interactions. We compared the binding cavity of P3 with the corresponding site in our SARS‐CoV‐2 N‐NTD structure, and we found that almost all of the residues involved in the interactions are conserved, except F135 in MERS‐CoV, which is replaced by I146 in SARS‐CoV‐2 (Fig 6D). Although both residues are nonpolar amino acids, the difference in hydrophobic force may affect the dimerization efficiency.
Discussion
In this study, we solved the crystal structures of both SARS‐CoV‐2 N‐NTD and N‐CTD. Both structures show conserved features with other coronaviruses (Yu et al, 2006; Chen et al, 2007; Saikatendu et al, 2007; Lin et al, 2014; Papageorgiou et al, 2016; Szelazek et al, 2017; Nguyen et al, 2019). N‐NTD has a right‐handed fist shape consisting of an antiparallel β‐sheet core subdomain and a protruding β‐hairpin region. N‐CTD is present as a tight intertwined homodimer and displays an overall rectangular slab shape. Based on the surface electronic distribution and superimposition with other coronavirus N‐NTD structures, we compared the ribonucleotide‐binding pocket in SARS‐CoV‐2 N‐NTD, and we found that the binding residues are highly conserved, which could be a verified target for antiviral drugs that block the RNA‐binding activity of the N protein.
Although both N‐NTD and N‐CTD structures resemble those of other coronaviruses, especially SARS‐CoV, the electrostatic potential surfaces display different charge distribution patterns in some areas, including the N‐terminal loop, the top tip of the protruding region, the bottom of the core subdomain in N‐NTD and the β‐sheet face of N‐CTD. These divergent features may alter the RNA‐binding modes or binding efficiency. Of note, we found that the orientations of the N‐terminal loops of N‐NTD were distinct. The N‐terminal loops of SARS‐CoV‐2, SARS‐CoV and MERS‐CoV N‐NTD stretch outward, while the loops from HCoV‐OC43 and HCoV‐NL63 N‐NTD rotate towards the core subdomain. This result suggests that different N‐NTD may have group‐specific characteristics and that the distinctions may alter the locations of N‐arms and then lead to different structural and functional properties. In addition, as the N protein could interact with many host proteins during the infection process and cause a robust humoral immune response after infection, the divergent charge distribution surfaces could be critical to identify specific interacting cellular proteins and develop specific immune‐based rapid diagnostic test (Burbelo et al, 2020; Ni et al, 2020). Besides the serological assays for detection of SARS‐CoV‐2 antibodies (Hörber et al, 2020), the rapid detection of viral antigens based on specific antibodies can provide essential information for disease monitoring (Che et al, 2004). Therefore, to develop high‐affinity and specific anti‐N antibodies would be critical to enhance the rapid molecular diagnosis for COVID‐19. During our manuscript preparation, other groups also solved crystal structures of N‐NTD or N‐CTD, which show similar structural features (Kang et al, 2020; Ye et al, 2020).
Until now, there are no structures of the full‐length N proteins from coronaviruses reported, due to their dynamic and oligomeric characteristics. Based on our dimeric structure of N‐CTD and previous studies, the N protein forms a dimer by N‐CTD interaction, and the disordered LKRs serve as the two arms connecting the two N‐NTDs to the N‐CTD dimers (Chen et al, 2007; Nguyen et al, 2019). In the RNP complex, the N‐CTDs form the helical core. The RNA molecule twists around the helical groove and further interacts with the NTDs (Chang et al, 2014). According to previous cryo‐electron tomography studies of the vRNP complex isolated from murine hepatitis virus (MHV) and SARS‐CoV virions, each dimeric N unit packs in a long loose helical or a supercoiled organization (Neuman et al, 2006; Bárcena et al, 2009; Gui et al, 2017). Recently, preprint: Klein et al (2020) observed that SARS‐CoV‐2 RNPs organizing like string beads, indicating the high sterical flexibility of RNP assembly. However, a lack of high‐resolution structures of RNP limits our understanding of the precise RNP assembly mechanism.
Coronavirus N proteins have been attractive targets for antiviral drugs as they are critical for viral replication and assembly. Few inhibitors have been identified through various approaches based on HCoV‐OC43 or MERS‐CoV (Lin et al, 2014; Zhang et al, 2020b). We analysed the binding sites of the representative inhibitors in our crystal structures and found that the drug targeting sites are relatively conserved, indicating further structure‐based drug optimization for broad‐spectrum agent development is very promising. Our structural information of SARS‐CoV‐2 N‐NTD and C‐CTD can aid in the discovery and development of antiviral inhibitors against SARS‐CoV‐2 in the future.
Materials and Methods
Gene cloning protein production and purification
SARS‐CoV‐2 N‐NTD (amino acid residues 44–174, GISAID accession ID: EPI_ISL_402119) fused at its N‐terminus with a hexa‐histidine tag was cloned into the pET‐21a vector (Novagen) with NdeI and XhoI restriction sites (Li et al, 2019). Transformed E. coli strain BL21 (DE3) clones were grown in LB medium containing 100 μg/ml ampicillin to an OD600 of 0.6–0.8 at 37°C. Expression of the recombinant proteins was induced by the addition of 0.5 mM isopropyl‐β‐D‐1‐thiogalactopyranoside (IPTG), and incubation was continued for a further 16 h at 16°C. Cells were harvested by centrifugation at 7,000 g for 15 min at 4°C and then resuspended in lysis buffer [20 mM Tris–HCl (pH 8.0) and 150 mM NaCl] and further homogenized with a low‐temperature ultra‐high pressure cell disrupter (JNBIO, China). The lysate was clarified by centrifugation at 20,000 g for 60 min at 4°C. The supernatant was purified by metal affinity chromatography using a HisTrap HP 5 ml column (GE Healthcare). Proteins were eluted using lysis buffer supplemented with 300 mM imidazole. The proteins were further purified by gel filtration chromatography using a HiLoad 16/600 Superdex® 75 PG (GE Healthcare) with a running buffer of 20 mM Tris–HCl (pH 8.0) and 50 mM NaCl, and the collected protein fractions were concentrated to 15 mg/ml using a membrane concentrator with a molecular weight cut‐off of 10 kDa (Millipore).
SARS‐CoV‐2 N‐CTD (amino acid residues 255–364) was constructed and expressed in the same way as N‐NTD. During metal affinity chromatography, N‐CTDs were eluted using lysis buffer supplemented with 500 mM imidazole. The proteins were further purified by gel filtration chromatography using a HiLoad 16/600 Superdex® 75 PG (GE Healthcare) with a running buffer of 20 mM Tris–HCl (pH 8.0) and 150 mM NaCl, and the collected protein fractions were concentrated to 10 mg/ml using a membrane concentrator with a molecular weight cut‐off of 10 kDa (Millipore).
Crystallization, data collection and structure determination
Crystallization trials were set up with commercial crystallization kits (Hampton Research) using the sitting drop vapour diffusion method. The resultant drop was then sealed, equilibrating against 100 μl reservoir solution at 18°C. Diffractable crystals of SARS‐CoV‐2 N‐NTD were obtained in 0.1 M SPG (succinic acid, sodium phosphate monobasic monohydrate and glycine mix buffer pH 6.0) (Molecular Dimensions, MD2‐59) with 25% w/v polyethylene glycol 1,500 at 18°C. SARS‐CoV‐2 N‐CTD crystals were obtained in 4 M potassium formate, 0.1 M Bis‐Tris propane (pH 9.0) and 2% w/v polyethylene glycol monomethyl ether 2,000 at 18°C. Crystals were flash‐cooled in liquid nitrogen after a brief soaking in reservoir solution with the addition of 17% (v/v) glycerol. The X‐ray diffraction data were collected under cryogenic conditions (−173°C) at Shanghai Synchrotron Radiation Facility (SSRF) beam line BL19U1 and indexed, integrated and scaled with HKL3000 (Minor et al, 2006).
SARS‐CoV‐2 N‐NTD and N‐CTD structures were solved by the molecular replacement method using Phaser (Read, 2001) from the CCP4 programme suite (Winn et al, 2011), with the structures of SARS‐CoV N‐NTD (PDB: 2OFZ) (Saikatendu et al, 2007) and N‐CTD (PDB: 2CJR) (Chen et al, 2007) as the search models, respectively. Initial restrained rigid‐body refinement and manual model building were performed using REFMAC5 (Murshudov et al, 1997) and COOT (Emsley & Cowtan, 2004), respectively. Further refinement was performed using Phenix (Adams et al, 2010). Final statistics for data collection and structure refinement are presented in Table 1.
Biochemical characterization of N proteins
The purified protein was analysed with an analytical gel filtration assay with a calibrated Superdex® 75 10/300 GL column (GE Healthcare). The sample was further analysed with SDS–PAGE.
The analytical ultracentrifugation assay was performed according to a previously reported method (Wang et al, 2017). The proteins were prepared in 20 mM Tris (pH 8.0) and 150 mM NaCl at a concentration of 0.8 mg/ml. The assay was performed on an optimal ProteomeLab XL‐I analytical ultracentrifuge (Beckman Coulter) at a speed of 228,000 g. The molecular mass analysis was performed with XL‐I data analysis software.
Author contributions
GFG and HS designed and supervised the study. YP, ND, YL and SD conducted the experiments. JQ collected the data sets and solved the structures. HS, TL and GFG analysed the data and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank the staff of BL19U1 beamlines at Shanghai Synchrotron Radiation Facility. We thank Qian Wang (Institute of Microbiology CAS) for help with analytical ultracentrifugation experiment. This work was supported by the Ministry of Science and Technology of the People's Republic of China (2020YFC0845900), the National Science and Technology Major Project (2018ZX10733403), and the National Natural Science Foundation of China (NSFC) (81702015). H.S. is supported by the Youth Innovation Promotion Association CAS (2017117). G.F.G. is supported partly by the COVID‐19 Emergency Project of CAS and the External Cooperation Program of CAS (153211KYSB20160001).
The EMBO Journal (2020) 39: e105938
Contributor Information
George F Gao, Email: gaof@im.ac.cn.
Hao Song, Email: songhao@im.ac.cn.
Data availability
Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 7CDZ (https://www.rcsb.org/structure/7CDZ) and 7CE0 (https://www.rcsb.org/structure/7CE0).
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW et al (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben‐Tal N (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44: W344–W350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bárcena M, Oostergetel GT, Bartelink W, Faas FG, Verkleij A, Rottier PJ, Koster AJ, Bosch BJ (2009) Cryo‐electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc Natl Acad Sci USA 106: 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S et al (2020) Remdesivir for the treatment of Covid‐19 – preliminary report. N Engl J Med 10.1056/NEJMoa2007764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo PD, Riedo FX, Morishima C, Rawlings S, Smith D, Das S, Strich JR, Chertow DS, Davey RT, Cohen JI (2020) Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome Coronavirus 2 in patients with coronavirus disease 2019. J Infect Dis 222: 206–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, Ruan L, Song B, Cai Y, Wei M et al (2020) A trial of lopinavir‐ritonavir in adults hospitalized with severe Covid‐19. N Engl J Med 382: 1787–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcanti AB, Zampieri FG, Rosa RG, Azevedo LCP, Veiga VC, Avezum A, Damiani LP, Marcadenti A, Kawano‐Dourado L, Lisboa T et al (2020) Hydroxychloroquine with or without Azithromycin in mild‐to‐moderate Covid‐19. N Engl J Med 10.1056/NEJMoa2019014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CK, Hou MH, Chang CF, Hsiao CD, Huang TH (2014) The SARS coronavirus nucleocapsid protein–forms and functions. Antiviral Res 103: 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CK, Jeyachandran S, Hu NJ, Liu CL, Lin SY, Wang YS, Chang YM, Hou MH (2016) Structure‐based virtual screening and experimental validation of the discovery of inhibitors targeted towards the human coronavirus nucleocapsid protein. Mol BioSyst 12: 59–66 [DOI] [PubMed] [Google Scholar]
- Che XY, Qiu LW, Pan YX, Wen K, Hao W, Zhang LY, Wang YD, Liao ZY, Hua X, Cheng VC et al (2004) Sensitive and specific monoclonal antibody‐based capture enzyme immunoassay for detection of nucleocapsid antigen in sera from patients with severe acute respiratory syndrome. J Clin Microbiol 42: 2629–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Chang CK, Chang YW, Sue SC, Bai HI, Riang L, Hsiao CD, Huang TH (2007) Structure of the SARS coronavirus nucleocapsid protein RNA‐binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J Mol Biol 368: 1075–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Kriegenburg F, de Haan CAM, Reggiori F (2017) Coronavirus nucleocapsid proteins assemble constitutively in high molecular oligomers. Sci Rep 7: 5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Ulasli M, Schepers H, Mauthe M, V'Kovski P, Kriegenburg F, Thiel V, de Haan CAM, Reggiori F (2020) Nucleocapsid protein recruitment to replication‐transcription complexes plays a crucial role in coronaviral life cycle. J Virol 94: e01925‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW et al (2020) The species severe acute respiratory syndrome‐related coronavirus: classifying 2019‐nCoV and naming it SARS‐CoV‐2. Nat Microbiol 5: 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M, Liu X, Guo D, Zhang Z, Yin CC, Chen Y, Xiang Y (2017) Electron microscopy studies of the coronavirus ribonucleoprotein complex. Protein Cell 8: 219–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Mosbauer K, Hofmann‐Winkler H, Kaul A, Kleine‐Weber H, Kruger N, Gassen NC, Muller MA, Drosten C, Pohlmann S (2020) Chloroquine does not inhibit infection of human lung cells with SARS‐CoV‐2. Nature 10.1038/s41586-020-2575-3 [DOI] [PubMed] [Google Scholar]
- Hörber S, Soldo J, Relker L, Jürgens S, Guther J, Peter S, Lehmann R, Peter A (2020) Evaluation of three fully‐automated SARS‐CoV‐2 antibody assays. Clin Chem Lab Med 10.1515/cclm-2020-0975 [DOI] [PubMed] [Google Scholar]
- Jiang S, Shi Z, Shu Y, Song J, Gao GF, Tan W, Guo D (2020) A distinct name is needed for the new coronavirus. Lancet 395: 949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Yang M, Hong Z, Zhang L, Huang Z, Chen X, He S, Zhou Z, Zhou Z, Chen Q et al (2020) Crystal structure of SARS‐CoV‐2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B 10: 1228–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Cortese M, Winter SL, Wachsmuth‐Melm M, Neufeldt CJ, Cerikan B, Stanifer ML, Boulant S, Bartenschlager R, Chlanda P (2020) SARS‐CoV‐2 structure and replication characterized by in situ cryo‐electron tomography. bioRxiv 10.1101/2020.06.23.167064 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Chai Y, Song H, Weng C, Qi J, Sun Y, Gao GF (2019) Crystal structure of african swine fever virus dUTPase reveals a potential drug target. MBio 10: e02483‐19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY et al (2020) Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. N Engl J Med 382: 1199–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Liu CL, Chang YM, Zhao J, Perlman S, Hou MH (2014) Structural basis for the identification of the N‐terminal domain of coronavirus nucleocapsid protein as an antiviral target. J Med Chem 57: 2247–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SM, Lin SC, Hsu JN, Chang CK, Chien CM, Wang YS, Wu HY, Jeng US, Kehn‐Hall K, Hou MH (2020) Structure‐based stabilization of non‐native protein‐protein interactions of coronavirus nucleocapsid proteins in antiviral drug design. J Med Chem 63: 3131–3141 [DOI] [PubMed] [Google Scholar]
- Lu X, Pan J, Tao J, Guo D (2011) SARS‐CoV nucleocapsid protein antagonizes IFN‐β response by targeting initial step of IFN‐β induction pathway, and its C‐terminal region is critical for the antagonism. Virus Genes 42: 37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N et al (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonnasse P, Guedj J, Contreras V, Behillil S, Solas C, Marlin R, Naninck T, Pizzorno A, Lemaitre J, Goncalves A et al (2020) Hydroxychloroquine use against SARS‐CoV‐2 infection in non‐human primates. Nature 10.1038/s41586-020-2558-4 [DOI] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M (2006) HKL‐3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62: 859–866 [DOI] [PubMed] [Google Scholar]
- Mu J, Xu J, Zhang L, Shu T, Wu D, Huang M, Ren Y, Li X, Geng Q, Xu Y et al (2020) SARS‐CoV‐2‐encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci China Life Sci 63: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum‐likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Neuman BW, Adair BD, Yoshioka C, Quispe JD, Orca G, Kuhn P, Milligan RA, Yeager M, Buchmeier MJ (2006) Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J Virol 80: 7918–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen THV, Lichiere J, Canard B, Papageorgiou N, Attoumani S, Ferron F, Coutard B (2019) Structure and oligomerization state of the C‐terminal region of the Middle East respiratory syndrome coronavirus nucleoprotein. Acta Crystallogr D Struct Biol 75: 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Ye F, Cheng ML, Feng Y, Deng YQ, Zhao H, Wei P, Ge J, Gou M, Li X et al (2020) Detection of SARS‐CoV‐2‐specific humoral and cellular immunity in COVID‐19 convalescent individuals. Immunity 52: 971–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papageorgiou N, Lichiere J, Baklouti A, Ferron F, Sevajol M, Canard B, Coutard B (2016) Structural characterization of the N‐terminal part of the MERS‐CoV nucleocapsid by X‐ray diffraction and small‐angle X‐ray scattering. Acta Crystallogr D Struct Biol 72: 192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng TY, Lee KR, Tarn WY (2008) Phosphorylation of the arginine/serine dipeptide‐rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J 275: 4152–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RJ (2001) Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr 57: 1373–1382 [DOI] [PubMed] [Google Scholar]
- Saikatendu KS, Joseph JS, Subramanian V, Neuman BW, Buchmeier MJ, Stevens RC, Kuhn P (2007) Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N‐terminal domain of N protein. J Virol 81: 3913–3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, Song T, Bi X, Han C, Wu L et al (2020) A human neutralizing antibody targets the receptor‐binding site of SARS‐CoV‐2. Nature 584: 120–124 [DOI] [PubMed] [Google Scholar]
- Surjit M, Liu B, Jameel S, Chow VT, Lal SK (2004) The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS‐1 cells in the absence of growth factors. Biochem J 383: 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M, Liu B, Chow VT, Lal SK (2006) The nucleocapsid protein of severe acute respiratory syndrome‐coronavirus inhibits the activity of cyclin‐cyclin‐dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem 281: 10669–10681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szelazek B, Kabala W, Kus K, Zdzalik M, Twarda‐Clapa A, Golik P, Burmistrz M, Florek D, Wladyka B, Pyrc K et al (2017) Structural characterization of human coronavirus NL63 N protein. J Virol 91: e02503‐16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W, Zhao X, Ma X, Wang W, Niu P, Xu W, Gao GF, Wu G (2020) A novel coronavirus genome identified in a cluster of pneumonia cases ‐ Wuhan, China 2019–2020. China CDC Wkly 2: 61–62 [PMC free article] [PubMed] [Google Scholar]
- Wang H, Han M, Qi J, Hilgenfeld R, Luo T, Shi Y, Gao GF, Song H (2017) Crystal structure of the C‐terminal fragment of NS1 protein from yellow fever virus. Sci China Life Sci 60: 1403–1406 [DOI] [PubMed] [Google Scholar]
- Wang C, Horby PW, Hayden FG, Gao GF (2020a) A novel coronavirus outbreak of global health concern. Lancet 395: 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Peng Y, Xu H, Cui Z, Williams RO (2020b) The COVID‐19 vaccine race: challenges and opportunities in vaccine formulation. AAPS PharmSciTech 21: 225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G (2020c) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019‐nCoV) in vitro . Cell Res 30: 269–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A et al (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY et al (2020a) A new coronavirus associated with human respiratory disease in China. Nature 579: 265–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Li C, Xia S, Tian X, Kong Y, Wang Z, Gu C, Zhang R, Tu C, Xie Y et al (2020b) Identification of human single‐domain antibodies against SARS‐CoV‐2. Cell Host Microbe 27: 891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Wang F, Shen C, Peng W, Li D, Zhao C, Li Z, Li S, Bi Y, Yang Y et al (2020c) A noncompeting pair of human neutralizing antibodies block COVID‐19 virus binding to its receptor ACE2. Science 368: 1274–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, Zhou Q, Ye H, Ma Y, Li H et al (2020) Antibody detection and dynamic characteristics in patients with COVID‐19. Clin Infect Dis 10.1093/cid/ciaa461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, West AMV, Silletti S, Corbett KD (2020) Architecture and self‐assembly of the SARS‐CoV‐2 nucleocapsid protein. Protein Sci 10.1002/pro.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu IM, Oldham ML, Zhang J, Chen J (2006) Crystal structure of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between corona‐ and arteriviridae. J Biol Chem 281: 17134–17139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R (2020a) Crystal structure of SARS‐CoV‐2 main protease provides a basis for design of improved alpha‐ketoamide inhibitors. Science 368: 409–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Tan Y, Ling Y, Lu G, Liu F, Yi Z, Jia X, Wu M, Shi B, Xu S et al (2020b) Viral and host factors related to the clinical outcome of COVID‐19. Nature 583: 437–440 [DOI] [PubMed] [Google Scholar]
- Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, Wang P, Liu D, Yang J, Holmes EC et al (2020a) A novel bat coronavirus closely related to SARS‐CoV‐2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol 30: 2196–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL et al (2020b) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R et al (2020) A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382: 727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga S, Cruz JL, Sola I, Mateos‐Gomez PA, Palacio L, Enjuanes L (2010) Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J Virol 84: 2169–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Review Process File
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 7CDZ (https://www.rcsb.org/structure/7CDZ) and 7CE0 (https://www.rcsb.org/structure/7CE0).


