Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19), a newly emerging human infectious disease. Because no specific antiviral drugs or vaccines are available to treat COVID-19, early diagnostics, isolation, and prevention are crucial for containing the outbreak. Molecular diagnostics using reverse transcription polymerase chain reaction (RT-PCR) are the current gold standard for detection. However, viral RNAs are much less stable during transport and storage than proteins such as antigens and antibodies. Consequently, false-negative RT-PCR results can occur due to inadequate collection of clinical specimens or poor handling of a specimen during testing. Although antigen immunoassays are stable diagnostics for detection of past infection, infection progress, and transmission dynamics, no matched antibody pair for immunoassay of SARS-CoV-2 antigens has yet been reported. In this study, we designed and developed a novel rapid detection method for SARS-CoV-2 spike 1 (S1) protein using the SARS-CoV-2 receptor ACE2, which can form matched pairs with commercially available antibodies. ACE2 and S1-mAb were paired with each other for capture and detection in a lateral flow immunoassay (LFIA) that did not cross-react with SARS-CoV Spike 1 or MERS-CoV Spike 1 protein. The SARS-CoV-2 S1 (<5 ng of recombinant proteins/reaction) was detected by the ACE2-based LFIA. The limit of detection of our ACE2-LFIA was 1.86 × 105 copies/mL in the clinical specimen of COVID-19 Patients without no cross-reactivity for nasal swabs from healthy subjects. This is the first study to detect SARS-CoV-2 S1 antigen using an LFIA with matched pair consisting of ACE2 and antibody. Our findings will be helpful to detect the S1 antigen of SARS-CoV-2 from COVID-19 patients.
Keywords: COVID-19, SARS-CoV-2, Spike antigens, Human ACE2, Lateral flow immunoassay
Highlights
-
•
The human ACE2 and commercial antibody were paired with each other as capture and detection probes in a lateral flow immunoassay that was not cross-reactive with SARS-CoV S1 and MERS-CoV S1 proteins.
-
•
The limit of detection of our ACE2-LFIA was 1.86 × 105 copies/mL in the clinical specimen of COVID-19 patients without no cross-reactivity for nasal swabs from healthy subjects.
-
•
This is the first study to detect SARS-CoV-2 S1 antigen using an LFIA with matched pair consisting of ACE2 and antibody.
1. Introduction
Coronavirus disease 2019 (COVID-19) is an ongoing pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which was first reported in Wuhan in the Hubei province of China in December 2019 (Lan et al., 2020; Wu et al., 2020; Zhou et al., 2020; Zhu et al., 2020). The majority of COVID-19 patients develop pneumonia with symptoms such as fever, fatigue, anosmia, and cough (Huang et al., 2020a; Liu et al., 2020; Wang et al., 2020a). The virus has spread rapidly from the region of the initial outbreak to more than 200 countries, and the World Health Organization (WHO) recently reported more than 4.9 million confirmed cases and 375,656 deaths (WHO 2020). Because no specific antiviral drugs or vaccines are available to treat COVID-19, early diagnostics, isolation, and prevention are crucial for containing the outbreak. Currently, molecular diagnostics using reverse transcription polymerase chain reaction (RT-PCR) are the current gold standard for detection. Since the viral genome sequence was reported in January 2020 (GenBank: MN908947.3), this method has served as a powerful test for the detection of SARS-CoV-2 from COVID-19 patients.
Coronaviruses (CoVs) cause mild-to-moderate upper respiratory tract illnesses in human and animals, including bats, camels, pigs, cats, and mice. Over the past two decades, severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) caused severe epidemics which are still ongoing in the Middle East (Ge et al., 2013; Walls et al., 2020). SARS-CoV-2, a strain of SARS-related CoV (SARSr-CoV), has a single-positive strand RNA genome (29.8 kb) encoding four structural proteins: spike (S), envelope (E), matrix (M), and nucleocapsid (N). The entry of SARS-CoV-2 into host cells is facilitated by an interaction between the receptor-binding domain (RBD) of S protein and the peptidase domain of angiotensin-converting enzyme 2 (ACE2), previously identified as the entry receptor of SARS-CoV (Ge et al., 2013; Li et al. 2003, 2005; Song et al., 2018; Walls et al., 2020). Surprisingly, the binding affinity of SARS-CoV-2 S1 to ACE2 is higher than that of SARS-CoV S1 (Wrapp et al., 2020). Some variations may strengthen (Gln493/Asn479 and Lys417/Val404 in the SARS-CoV-2 RBD/SARS-CoV RBD) the interaction between SARS-CoV-2 RBD and ACE2. SARS-CoV-2 RBD Gln 493 strongly interacts with several residues (Lys31, His34 and Glu35) in ACE2 than SARS-CoV RBD (Lan et al., 2020; Yan et al., 2020).
Several COVID-19 immunoassays had been developed to detect specific antigens of, or antibodies against, SARS-CoV-2 (Huang et al., 2020b; Mavrikou et al., 2020). The CRISPR-powered COVID-19 testing of the target genes of SARS-CoV-2 (Morales-Narváez and Dincer 2020) has been reported. Because viral RNAs are less stable during transport and storage than proteins such as antigens and antibodies (Barr and Fearns 2016; Relova et al., 2018), false-negative RT-PCR results can arise due to improper collection of clinical specimens or poor handing of specimens during testing (Li et al., 2020; Xie et al., 2020). For that reason, immunoassays represent reliable diagnostics for the detection of past infection, infection progress, and transmission dynamics. In the case of antigen detection using ELISA (enzyme-linked immunosorbent assay) and RDTs (rapid diagnostic tests), binding of matched antibody pairs to virus-specific antigens is essential. To date, however, no study has reported a matched antibody pair for immunoassay of SARS-CoV-2 antigens such as the S and N proteins.
In this study, we designed and developed a novel rapid detection method for SARS-CoV-2 S protein using the SARS-CoV-2 receptor ACE2, which can form a matched pair with commercially available antibodies. The interaction between SARS-CoV-2 S1 protein and ACE2 (or antibodies) was examined by Western blot and ELISA, respectively. Efforts to identify a matched pair for the rapid detection of SARS-CoV-2 spike protein focused mainly on ACE2 and antibodies such as CR3022, F26G19, and S1-mAb. Ultimately, ACE2 and S1-mAb were paired with each other for capture and detection in a lateral flow-based immunoassay without cross-reactivity for SARS-CoV Spike 1 or MERS-CoV Spike 1 protein. Our ACE2-based lateral flow immunoassay (LFIA) for SARS-CoV-2 S1 was more sensitive (1 ng/reaction) to the RBD than the S1 protein (5 ng/reaction) of SARS-CoV-2. This is the first study to detect SARS-CoV-2 S1 protein using lateral flow immunoassay with a matched pair consisting of ACE2 and an antibody.
2. Materials and methods
2.1. Enzyme-linked immunosorbent assay
MaxiSorp immunoplates (ThermoFisher SCIENTIFIC, MA, USA) were coated overnight at 4 °C with varying amounts (200, 50, 12.5, 3.13, 0.78, 0.2, 0.05, and 0 ng/mL) of SARS-CoV-2 S1 protein (S1 subunit, Cat. No. 40591-V08H; Sino Biological, Beijing, China) in 100 μL coating buffer per well. The immunoplates were blocked for 1 h with blocking buffer (Cat. No. DS98200; Invitrogen, CA, USA), and then hACE2, CR3022, F26G19, or S1-mAb in 100 μL blocking buffer was added to each well and incubated for 1 h. After washing with wash buffer (Cat. No. WB01; Invitrogen), the bound receptor and antibodies were incubated with horseradish peroxidase (HRP)-conjugated anti-mouse IgG (1:2000; Cat. No. 31437; Invitrogen), anti-human IgG (1:2000; Cat. No. 31413; Invitrogen), or anti-rabbit IgG (1:2000; Cat. No. 31463; Invitrogen) detection antibody, as appropriate, for 1 h. After extensive washing with wash buffer, stabilized chromogen (TMB solution, Cat. No. SB01; Invitrogen) was added. The immunoplates were allowed to react for 10 min, and the reaction was stopped by addition of 1 M HCl. The optical density (OD) value was measured at 450 nm using a microplate reader (BioTek, VT, USA).
2.2. Biolayer interferometry
Fc-tag tagged human ACE2 (ACE2, Cat. No. 10108-H05H), SARS-CoV-2 spike monoclonal antibody (S1-mAb, Cat. No. 40150-R007), SARS-CoV-2 spike protein (S1 subunit, Cat. No. 40591-V08H), and SARS-CoV-2 RBD (Cat. No. 40592-V08B) were purchased from Sino Biological. CR3022 and F26G19 were purchased from Abclon (Seoul, Korea). Plasmids encoding heavy and light chains of each antibody at a 1:6 ratio were transiently cotransfected into 293-F cells using PEI reagent (PolyScience, PA, USA). Six days after transfection, the supernatant was collected, and CR3022 and F26G19 were purified on Protein A columns (GE Healthcare, IL, USA). Binding affinities between SARS-CoV-2 spike antigens (S1 and RBD) and four different antibodies (ACE2, CR3022, F26G19, and S1-mAb) were analyzed by biolayer interferometry (BLI) on a BLITz instrument (ForteBio, CA, USA). After hydration of the biosensor tip for 10 min, BLI analysis began, followed by five steps: initial baseline (30 s), sample loading (300 s), baseline (120 s), binding (300 s), and dissociation (300 s). All of the initial baseline, baseline, and dissociation processes were performed using sample diluent buffer (0.02% Tween 20, 150 mM NaCl, and 1 mg/mL BSA in 10 mM PBS with 0.05% sodium azide, pH 7.4). In the sample loading step, each antibody (100 μg/mL) was immobilized on a biosensor tip coated with Protein A through the specific interaction between Protein A and the antibody Fc domain. After the baseline step, four different antigen concentrates were incubated with the antibody-conjugated biosensor tip to measure the association between antigen and antibody. From the four different binding curves obtained from each antigen concentrate, binding constants were calculated based on the 1:1 binding model.
2.3. Preparation of antibody-conjugated CNBs
A 1% stock solution of red cellulose nanobeads (CNBs) was purchased from Asahi Kasei Fibers Corporation (NanoAct, Cat. No. RE2AA; Miyazaki, Japan); the beads have an average diameter of 345 nm. A CNB conjugation kit, containing conjugation buffer, blocking buffer, and wash buffer, was purchased from DCN Diagnostics (CA, USA). Conjugation of the SARS-CoV-2 spike antigen-specific antibodies (CR3022, F26G19, S1mAb) and binding protein (ACE2) to the surface of CNBs was performed according to the manufacturer's instructions. Briefly, 0.12 mL of 0.5 mg/mL antibody was mixed with 0.060 mL of 1% stock CNB and 0.12 mL conjugation buffer, and the mixture was incubated for 2 h at 37 °C. Then, 7.2 mL blocking buffer was added to the mixture, which was incubated for an additional hour at 37 °C. The solution was centrifuged (14,400×g, 20 min, 4 °C), and the pellet was resuspended in 7.2 mL wash buffer. After centrifugation (14,400×g, 20 min, 4 °C), the washed pellet was resuspended in 0.5 mL wash buffer. The final concentration of antibody-conjugated CNB was approximately 0.1%. The exact concentration of the CNB was calculated by measuring the maximum absorbance at 554 nm by UV–vis spectrophotometry (Synergy H1; BioTek).
2.4. Preparation of LFIA strips
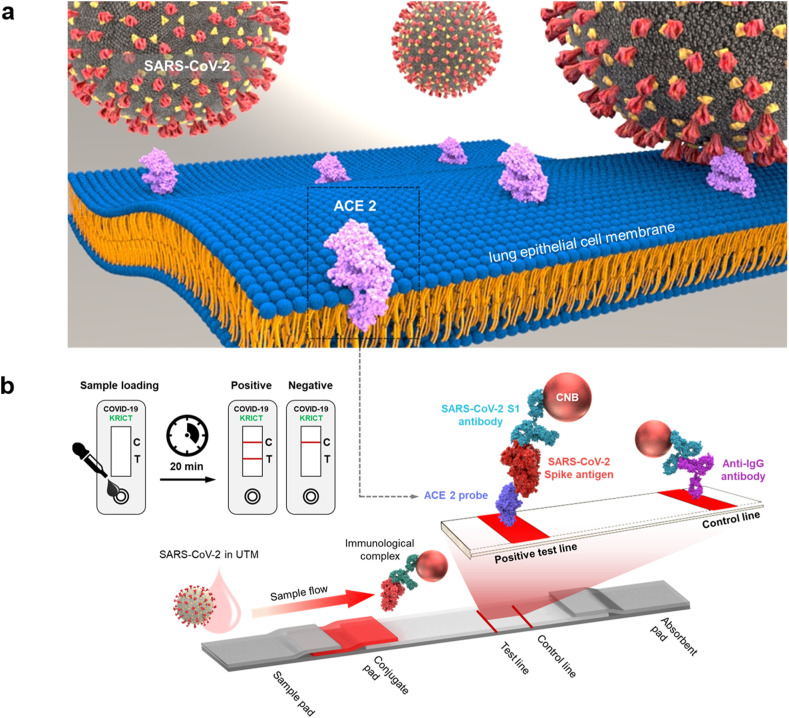
LFIA strips consisted of a sample pad (Ahlstrom, Helsinki, Finland), a conjugate pad (Ahlstrom, Helsinki, Finland), a nitrocellulose membrane (Advanced Microdevices, Haryana, India), and an absorbent pad (Ahlstrom, Helsinki, Finland) as shown in Fig. 1 b. The conjugate pad was treated with 0.1% Triton X-100 (Cat. No. T8787; Sigma-Aldrich, MO, USA) before CNB fixation. After complete drying, a 0.05% solution of antibody-conjugated CNB in stabilizing buffer (10 mM 2-amino-2-methyl-1-propanol (pH 9.0), 0.5% BSA, 0.5% β-Lactose, 0.05% Triton X-100, and 0.05% sodium azide) was sprayed on the conjugate pad, followed by incubation for 1 h at 37 °C in a vacuum oven (FDU-1200, EYELA, Tokyo, Japan; JSVO-30T, JSR, Gongju, Korea). A test line containing the capture probe and a control line were dispensed onto the nitrocellulose membrane using a line dispenser (BTM Inc., Uiwang, Korea) under the following conditions: dispensing speed, 50 mm/s; dispensing rate, 1 μL/cm. ACE2 (1 mg/mL) and 0.5 mg/mL anti-IgG antibody mixture [1:1:1 (v/v/v) anti-human IgG antibody (Cat. No. I2136, Sigma-Aldrich)/anti-rabbit IgG antibody (Cat. No. R5506, Sigma-Aldrich)/anti-mouse IgG antibody (Cat. No. A4416, Sigma-Aldrich)] in sample diluent buffer (Cat. No. ab154873; Abcam, Cambridge, UK) were used for the formation of the test line and control line, respectively. After line dispensing, the nitrocellulose membrane was dried for 1 h at 37 °C. To decrease the non-specific interaction between capture probes in test lines and detection probes, the nitrocellulose membrane was treated with the blocking solution (10 mM 2-amino-2-methyl-1-propanol (pH 9.0), 0.5% BSA, 0.5% β-Lactose, 0.05% Triton X-100, 0.05% sodium azide) for 1 h in a vacuum oven (37 °C). Each component of the LFIA strip was precisely assembled and cut to a 38 mm width, followed by integration into a housing for a single LFIA test.
Fig. 1.
Cellular receptor (ACE2)-based LFIA. a) Schematic of ACE2 receptor recognition by SARS-CoV-2. ACE2, a type 1 membrane protein expressed in the lung, heart, kidney, and intestine, is the cellular receptor for SARS-CoV-2. b) Schematic of an ACE2-based LFIA consisting of a sample pad, conjugate pad, nitrocellulose membrane, and absorbent pad. The test line placed on the nitrocellulose membrane contains ACE2 for detection of the SARS-CoV-2 spike antigen. Anti-IgG antibody is used in the control line. The proposed LFIA can achieve sensitive and selective detection of SARS-CoV-2 spike antigen within 20 min.
2.5. Dot-blot assay for the discovery of sandwich pairs
To discover the optimum pair for detection of the SARS-CoV-2 spike antigen, a total of 12 capture probe–detection probe pairs (Supplementary Table 1) were tested. The affinity of the four different antibodies for SARS-CoV-2 S1 and RBD was confirmed through ELISA, Western blot, and BLI. Capture probe (1 mg/mL) and 0.5 mg/mL anti-IgG antibody mixture [1:1:1 (v/v/v) anti-human IgG antibody/anti-rabbit IgG antibody/anti-mouse IgG antibody] were used to form test dots and control dots, respectively. Dots were immobilized onto a nitrocellulose membrane (Advanced Microdevices) by loading 0.5 μL of the test or control solution. The nitrocellulose membrane was incubated with blocking solution (10 mM 2-amino-2-methyl-1-propanol (pH 9.0), 0.5% BSA, 0.5% β-lactose, 0.05% Triton X-100, 0.05% sodium azide) in a vacuum oven (1 h, 37 °C). The LFIA strip for the dot-blot assay was constructed as described in the previous section. For comparative analysis between the 12 pairs, 50 ng target antigen (SARS-CoV-2 Spike S1) was incubated with each detection probe in running buffer [10 mM adenosine monophosphate (AMP, pH 9.0), 5 mM EDTA, 200 mM Urea, 1% Triton X-100, 0.5% Tween 20, 500 mM NaCl, 1% PEG (MW 200)] for 10 min at RT, followed by loading of the mixed sample onto the LFIA strip. After 20 min, red signals indicating antigen detection were confirmed by the naked eye and smartphone camera. In addition, all dots were quantitatively analyzed using an image analyzer (Sapphire Biomolecular Imager, Azure Biosystems, CA, USA), and the relative intensity of each dot was calculated based on the average intensity of the dot minus the background level (Supplementary Table 1).
2.6. Sensitivity and specificity analysis of LFIA
For the development of highly sensitive and specific LFIA for the detection of SARS-CoV-2, the concentration of CNB (Supplementary Fig. 1), the concentration of immobilized ACE2 (Supplementary Fig. 2), and the composition of the running buffer (Supplementary Fig. 3) were optimized. Finally, 0.05% of CNB for the detection probe, 1 mg/mL of ACE2 for the immobilized capture probes, and running buffer [10 mM AMP, (pH 9.0), 5 mM EDTA, 200 mM urea, 1% Triton X-100, 0.5% Tween 20, 500 mM NaCl, 1% PEG (MW 200)] were selected. The SARS-CoV-2 S1 and SARS-CoV-2 RBD antigens were serially diluted in sample diluent buffer (Cat. No. ab154873; Abcam), and diluted samples were mixed with running buffer at a ratio of 1:9 (v/v). The final concentrations of the diluted samples ranged from 500 to 5 ng/mL. One hundred microliters of running buffer containing each antigen concentrate were added to the inlet of the LFIA device. In this system, the sample flows along with the LFIA strip by capillary force and first encounters the antibody (S1-mAb)-conjugated CNB. The antigen in the sample is captured by the S1-mAb conjugated CNB, and this antigen-probe complex is detected by a pre-immobilized capture reporter (ACE2) on the nitrocellulose membrane. After 20 min of sample loading, the appearance of red color in the test and control lines is confirmed and analyzed on a Sapphire Biomolecular Imager.
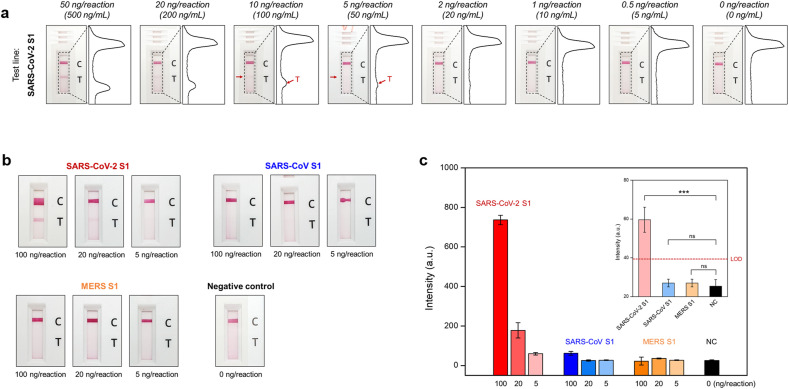
For specificity testing, two different corona-related spike antigens (i.e., SARS-CoV S1, and MERS S1 antigens) were prepared at three different concentrations (100, 20, and 5 ng/mL). Three concentrates of each antigen were loaded to the LFIA device, and the line intensities were quantified with a portable line analyzer (Light-G; WellsBio, Seoul, Korea). A positive intensity at the test line is more than 50 by the manufacturer's recommendation of the portable analyzer. All experiments were performed three times, and the limit of detection in Fig. 5 c was calculated as the mean value of the negative control plus three times the standard deviation.
Fig. 5.
Sensitivity and specificity of the ACE2-based LFA a) Results of ACE2-based LFA for the detection sensitivity of SARS-CoV-2 S1 antigen. Serially diluted antigen concentrates (concentration range: 500 ng/mL to 5 ng/mL) were tested by ACE2-based LFA. After 20 min, the LFA strips were photographed with a smartphone. Moreover, the intensity of the test and control lines was converted to a peak histogram by an image analyzer. b) Results of the comparative analysis for the detection selectivity: positive control – SARS-CoV-2 S1, negative control – SARS-CoV S1, MERS S1, and buffer solution. Using three different concentrates (1 μg/mL, 200 ng/mL, and 50 ng/mL) of the antigen sample, the detection performance of ACE2-based LFA was demonstrated. c) Bar graph of peak intensities for test lines. After 20 min for the sample flow, the intensity of the test lines was measured by a portable line analyzer. Inset) the detection intensity for the 5 ng antigen per reaction of each control. Limit of detection (LOD) was determined by the mean value of negative controls plus three times the standard deviation. P-values: ns > 0.05, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
2.7. Purification and quantification of copy number of human coronavirus-OC43 (HCoV-OC43) and SARS-CoV-2
Purification of viral RNA from HCoV-OC43, γ-irradiated SARS-CoV-2 (Cat. No. NR-52287, BEI Resources, VA, USA) was performed using a QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Serial dilution of viral RNA is targeted for nucleocapsid (N) gene of HCoV-OC43 (1.16 × 1012-100 copies/μL), or envelope (Env) gene of SARS-CoV-2 (7.7 × 106-100 copies/μL) served as standards, and purified viral RNA was synthesized to complementary DNA, consistent with performing quantitative reverse transcription PCR (RT-qPCR) using the Luna® Universal Probe One-Step RT-qPCR Kit (New England BioLabs, MA, USA) according to manufacturer's protocol by following primers for HCov-OC43 gene: Forward 5′-AGC AAC CAG GCT GAT GTC AAT ACC Reverse 5′-AGC AGA CCT TCC TGA GCC TTC AAT, SARS-CoV-2 Env gene: Forward 5′-ACA GGT ACG TTA ATA GTT AAT AGC GT, Reverse 5′-ATA TTG CAG CAG TAC GCA CAC A, and specific probe (ACA CTA GCC ATC CTT ACT GCG CTT CG) labeled with FAM (6-carboxyfluorescein) and BHQ-1 (Back Hole Quencher-1). The single-step RT-qPCR was set for reverse transcription under 55 °C for 10 min, and amplification of 45 cycles under 95 °C for 10s, 60 °C for the 30s. The reaction was analyzed using the Bio-Rad CFX 96 Touch Real-Time PCR System (CA, USA).
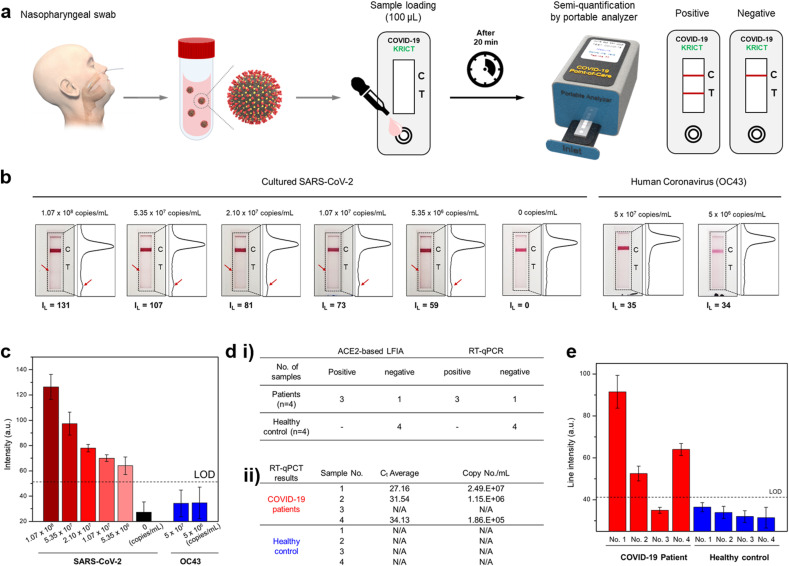
2.8. Assessment of the clinical performance of ACE2 based LFIA
By serial dilution of γ-radiated SARS-CoV-2 in running buffer, 1.07 × 108 copies/mL to 5.35 × 106 copies/mL of cultured SARS-CoV-2 samples were prepared. One hundred microliters of running buffer containing each viral concentrate were added to the LFIA device. After twenty minutes, the appearance of red color in the test lines was confirmed and quantified by a LightG portable analyzer (IL: line intensity). In addition, the intensity of the test and control lines were converted to peak histograms using a Sapphire Biomolecular Imager. Furthermore, human coronavirus-OC43 (HCoV-OC43), which was previously confirmed viral load by RT-qPCR, was also tested as a negative control. Two different concentrates (5 × 107 copies/mL, and 5 × 106 copies/mL) were loaded into the LFIA device. Twenty minutes after sample loading, non-specific interaction in the test line was evaluated by the same analytical methods. To evaluate the clinical relevance of ACE2-based LFIA, nasopharyngeal swab samples from COVID-19 patients (n = 4), and healthy subjects (n = 4) were applied to ACE2-based LFIA. Nasopharyngeal swab samples in universal transport media (UTM) from COVID-19 patients were kindly provided by Chonbuk National University Hospital (Korea). Moreover, nasopharyngeal swab samples from healthy subjects were purchased from LEE Biosolutions (Cat. No. 991-31-NC, MO, USA). Healthy nasopharyngeal swabs were suspended in UTM (Cat. No. UTNFS-3B-1, Noble Bio, Korea) and used for LFIA testing. Detail information for COVID-19 patients and Healthy subjects is presented in Supplementary Table 4. 50 μL of UTM obtained from nasopharyngeal swab samples from COVID-19 patients and healthy subjects were mixed with running buffer in a 1:1 (v/v) ratio and loaded into the LFIA device. After 20 min, the intensities of the test line were measured with the LightG portable analyzer. The detection limit was determined as the mean value of the healthy control group plus three times the standard deviation. RT-qPCR was also demonstrated to compare diagnostic performance using clinical samples. To detect the SARS-CoV-2 specific envelope gene (Env gene), the primer-probe set, which was previously confirmed (Jung et al., 2020), was used as follows: Forward 5′-ACA GGT ACG TTA ATA GTT AAT AGC GT, Reverse 5′-ATA TTG CAG CAG TAC GCA CAC A, and specific probe (ACA CTA GCC ATC CTT ACT GCG CTT CG) labeled with FAM (6-carboxyfluorescein) and BHQ-1 (Back Hole Quencher-1). The single-step RT-qPCR was set for reverse transcription under 55 °C for 10 min, and amplification of 45 cycles under 95 °C for 10s, 60 °C for the 30s. The reaction was analyzed using the Bio-Rad CFX 96 Touch Real-Time PCR System (CA, USA).
3. Results and discussion
3.1. Interaction between SARS-CoV-2 S1 antigens and human cellular receptor (ACE2)
The surface spike (S) glycoprotein of SARS-CoV is recognized by the ACE2 receptor, leading to cell entry (Yuan et al., 2020). The S proteins of SARS-CoV-2 and SARS-CoV are very similar (amino-acid sequence identity: ~77%) (Zhou et al., 2020). Several studies have shown that the S protein of SARS-CoV-2 binds to ACE2 with greater affinity than that of SARS-CoV (Park et al., 2020). Strong affinity between target antigen and antibodies (or receptor) is a prerequisite for development of not only therapeutics and vaccines, but also sensitive and accurate diagnostic platforms for antigen detection. We measured the interaction between SARS-CoV-2 S1 protein and ACE2 by Western blot analysis, indirect ELISA, and BLI to detect SARS-CoV-2 S protein.
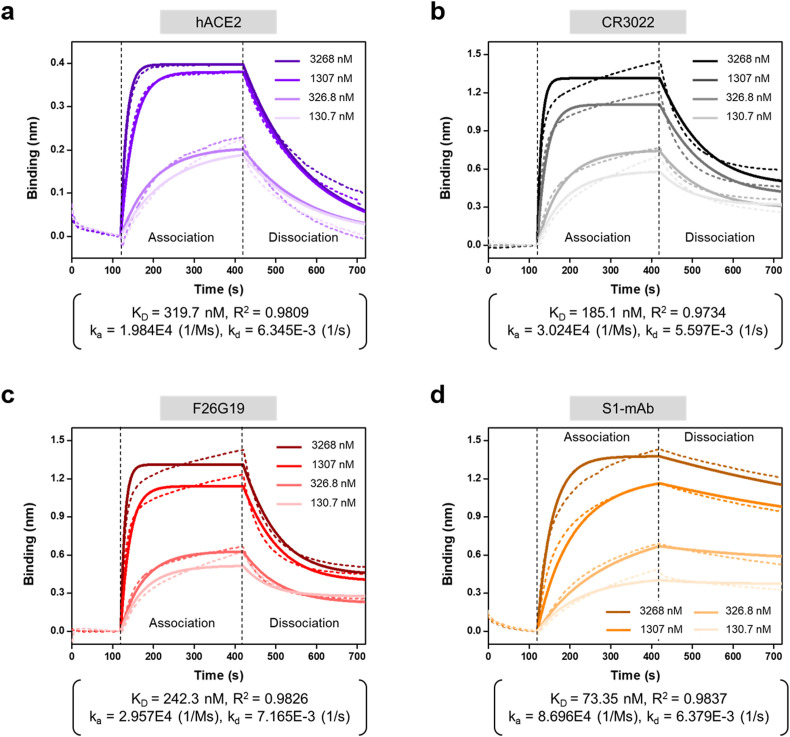
The interaction between receptor (or antibodies) and SARS-CoV-2 S1 protein was characterized by Western blot (Supplementary Fig. 4). The results revealed that anti-SARS-CoV-2 antibodies (CR3022, F26G19, S1-mAb) and human Fc tagged ACE2 receptor (ACE2-Fc) detect SARS-CoV-2 S1 protein. These interactions were also confirmed by ELISA showing that ACE2 receptor bound the SARS-CoV-2 S1 protein more strongly than the SARS-CoV S1 protein, but it did not bind the MERS-CoV S1 protein (Fig. 2a). By contrast, the commercial anti-SARS-CoV-2 antibodies bound to the SARS-CoV S1 protein and SARS-CoV-2 S1 protein with similar affinities (Fig. 2b–d). In the ELISA, the detection limits of the ACE2 receptor, CR3022, F26G19, and S1-mAb against SARS-CoV-2 S1 protein were approximately 125, 3.13, 3.13, and 0.78 ng/mL, respectively. In addition, the detection limits of the ACE2 receptor, CR3022, F26G19, and S1-mAb against SARS-CoV-2 RBD were approximately 3.13, 125, 0.05, and 0.05 ng/mL (Supplementary Fig. 5). To confirm the detailed kinetics of binding between ACE2 and the SARS-CoV-2 S1 protein, we performed BLI to measure the affinity of ACE2 for two different variants of the SARS-CoV-2 spike (S1 and RBD) protein. In addition, we used three different commercial antibodies (CR3022, F26G19, and S1-mAb) that bind SARS-CoV-2 S1 in the BLI experiments. BLI is a label-free technology for measuring biomolecular interactions based on changes in the interference pattern before and after binding events. The end of the BLI biosensor tip was coated with Protein A, which enables the efficient capture of the target antibody. Three commercial antibodies (CR3022, F26G19, and S1-mAb) and ACE2-Fc were immobilized onto the surface of the BLI biosensor through the specific interaction with protein A. We then examined the interactions of these antibodies with SARS-CoV-2 S1 and SARS-CoV-2 RBD. Representative real-time binding sensorgrams (dotted lines) and their fitting curves (solid line) for receptor (or antibody)-antigen are shown in Fig. 3 and Supplementary Fig. 6. Four different concentrates of antigens were used for the analysis of binding kinetics, and the kinetic constants were calculated from these four curves based on a 1:1 binding model (Fig. 3 and Supplementary Table 2). The KD values of ACE2 for S1 and RBD were 319.7 and 13.18 nM, respectively. The RBD of S1 is mainly in charge of engagement with a host cell receptor, ACE2 (Wang et al., 2020b). The previous study demonstrated that each monomer of the trimeric S protein of SARS-CoV binds to ACE2 (Pak et al., 2009). The proteolytically activated S protein of SARS-CoV-2 by proprotein convertase furin has had a higher binding affinity to ACE2 through its RBD (Shang et al., 2020). Therefore, we assume that the KD value of ACE2 is lower for RBD binding than for S1 binding because RBD can access ACE2 more efficiently than S1. This also implies that targeting monomeric RBD of SARS-CoV-2 S1 antigens to detect antigens using ACE2 and antibody pair may be more sensitive than targeting S1 protein in the present study although we have not checked out the detection of RBD itself in complex biological sample yet.
Fig. 2.
Indirect ELISA results from spike antigens of three different coronaviruses (SARS-CoV S1, SARS-CoV-2 S1, and MERS S1). a) The interactions between these S1 antigens and ACE2 were examined with serially diluted samples (concentration range: 200 to 0.05 ng/mL). In addition, three different antibodies, CR3022 (black) (b); F26G19 (red) (c); and S1-mAb (orange) (d), were tested for their interaction with spike antigens, using the same concentration range. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
Biolayer interferometry (BLI) results of ACE2, CR3022, F26G19, and S1-mAb against the SARS-CoV-2 S1 antigen. Dotted lines represent the response curves of BLI measurement, and solid lines represent the fitting curves based on a 1:1 binding model. Binding kinetics were measured for four different concentrations of the S1 antigen.
On the other hand, the KD values of CR3022, F26G19, and S1-mAb were, respectively, 185.1, 242.3, and 73.35 nM for S1, and 21.52, 17.99, and 12.42 nM for RBD. Among these commercial antibodies, S1-mAb had the highest affinity for SARS-CoV-2 S1, consistent with previous ELISA results. CR3022 and F26G19 are neutralizing antibodies that target the RBD of SARS-CoV (Park et al., 2020). Although recent studies showed that these antibodies can also bind the RBD of SARS-CoV-2 (Yuan et al., 2020), their affinities for SARS-CoV-2 S1 and RBD were lower than that of S1-mAb, which was produced using SARS-CoV-2 S1 as an immunizing antigen. In regard to binding of SARS-CoV-2 S1, ACE2 had the highest KD value. However, the affinity of ACE2 for SARS-CoV-2 RBD was comparable to those of other commercial antibodies, raising the important possibility of substituting a commercial antibody for ACE2 in the diagnostic platform for antigen detection. The KD values in our experiments are in line with recently reported results, although the exact KD values vary depending on the specific approach used in each experiment (Tian et al., 2020; Wang et al., 2020b).
3.2. Identification of the sandwich pair for detection of the SARS-CoV-2 S1 antigen
Successful detection of the target antigen requires two types of probes, the capture probe and detection probe, which recognize different sites of a specific antigen. Under unpredictable circumstances like the current COVID-19 outbreak, however, the development of two different antibody pairs would be too time-consuming and expensive, and therefore could not be applied to rapid diagnosis in the field with the goal of preventing the spread of disease. To overcome this drawback of antibody production, we used the cellular receptor, human ACE2, whose binding affinity for target antigen is comparable to those of antibodies, instead of one of the antibodies.
We used a dot-blot assay to discover the pair for sandwich antigen detection. ACE2 and three commercially available antibodies (CR3022, F26G19, and S1-mAb) were immobilized on a nitrocellulose membrane (NC membrane) as capture probes. Then, the other proteins (antibodies or receptor) not used in the immobilization were conjugated with cellulose nanobeads (CNB) for signal generation as detection probes, and the detection performance was evaluated. As shown in Fig. 4a and b, a total of 12 pairs were used for detection of SARS-CoV-2 S1 antigen, and non-specific interactions between capture and detection probes were also assessed. Furthermore, the intensity of each dot was analyzed on an image scanner (Fig. 4c and Supplementary Table 1). When ACE2 was used as the capture probe, sandwich detection of the S1 antigen was successfully achieved with all three detection probes. The result implies that the ACE2 binding site of the SARS-CoV-2 S1 antigen does not overlap with the epitopes of antibodies. In addition, we observed no false-positive signals in any of the negative controls (i.e., without antigen).
Fig. 4.
Identification of the sandwich pair for detection of SARS-CoV-2 spike antigen. a) Schematic diagram of LFIA using ACE2 as the capture probe and sandwich analysis results obtained from paired antibodies (CR3022, F26G19, and S1mAb). SARS-CoV-2 S1 antigen (50 ng) was used as a positive control, and buffer containing no S1 antigen was used as a negative control. After 20 min, the strips were captured by a smartphone, and their peak intensities were analyzed. b) Schematic diagram of LFIA, using antibodies as the capture probe, and their sandwich analysis results. c) Peak intensities of capture probe (PC)–detection probe (PD) pairs. A total of 12 pairs of positive controls (50 ng S1 antigen) were tested, and their intensities were analyzed. Peak intensity was calculated by subtracting the background intensity of the strip from the average intensities of the dots.
On the other hand, when antibody was used as a capture probe, false-positive signals were observed due to a non-specific interaction between the capture and detection antibodies. Notably, non-specific interactions occurred between S1-mAb and CR3022 and between S1-mAb and F26G19 (the red circle of the negative control in Fig. 4b). In addition, the epitope of CR3022 appeared to overlap with the epitope of F26G19, resulting in a decrease in the signal of the test dot. The results of this experiment emphasize that discovery of suitable antibody pairs for the sandwich assay is complicated by several variables. The introduction of ACE2 as a replacement for antibodies could accelerate the development of antigen diagnostic kits and facilitate efficient management of outbreaks. Moreover, overall signals, in both test and control dots, were diminished in the case of ACE2-conjugated CNBs (Fig. 4b and Supplementary Table 1). Evaluation of 12 capture–detection probe pairs allowed selection of the most suitable pair for the sensitive detection of SARS-CoV-2 S1 antigen: capture probe, ACE2, and detection probe, S1-mAb.
3.3. Sensitivity and specificity of ACE2-based LFIA for SARS-CoV-2 S1
The LFIA sensor strip consists of a sample pad, a conjugation pad, a nitrocellulose membrane, and an absorbent pad (Fig. 1b). The sample containing the target antigen is introduced onto the sample pad and sequentially encounters the CNBs dried on the conjugate pad. The CNBs have already been coated with detection antibody (S1-mAb) through hydrophobic and/or electrostatic interactions. The detection antibody-coated CNB captures the target SARS-CoV-2 S1 antigen and transfers it to the nitrocellulose membrane. The test line containing the capture probe (ACE2) detects the SARS-CoV-2 S1 antigen that was previously captured by the detection probes (S1-mAb-conjugated CNB), allowing sandwich detection of the SARS-CoV-2 S1 antigen. Meanwhile, the control line serves to determine whether the sample has flowed through and the biomolecule on the conjugate pad is active. For this purpose, anti-IgG Ab was used to capture all antibodies that were already conjugated with the CNBs. The test and control lines were formed on the nitrocellulose membrane using a line dispenser. As the detection of the target antigen on the test lines of LFIA, the red signal from the CNBs makes it possible to visually confirm whether the sample contains the target antigen or not. To avoid non-specific interaction between capture probes in test lines and detection probes, the nitrocellulose membrane was adequately treated with blocking solution. Unbound detection probes pass through the nitrocellulose membrane and ultimately reach the absorbent pad located at the end of the strip, which serves to maintain the capillary force that drives sample flow.
To demonstrate the detection capability of the ACE2-based LFIA (ACE2-LFIA), we performed the sensitivity analysis using serially diluted samples of SARS-CoV-2 specific antigen (S1 and RBD, concentration range: 5–500 ng/mL) as shown in Fig. 5a and Supplementary Figs. 7 and 8. Twenty minutes after sample loading, the window of the LFIA device was photographed using a smartphone, and the intensity of the test and control lines was analyzed with an image scanner and analyzer that converts the line color intensity to signal peaks, as shown in Fig. 5a. In the case of S1 detection, the detection signal gradually decreased as the dilution factor increased, but even if only 5 ng antigen was present in the sample, the signal was still present. In addition, we confirmed that the detection signal was clearly higher for RBD than for S1 for all diluted samples, and even 1 ng RBD present in the sample could be successfully detected by our ACE2-LFIA. In addition, we observed no false-positive signals in the negative control (i.e., the absence of target antigen). Cellulose nanobeads (CNB), a colorimetric label used in LFIA, have been reported to have higher sensitivity than colloidal gold (Sakurai et al., 2014). Hence, we also compared the detection sensitivities of colloidal gold and CNBs (Supplementary Fig. 9). Colloidal gold-based LFIA detected RBD antigen at concentrations as low as 20 ng, and was thus significantly less sensitive than CNB-based LFIA. Because CNBs have higher color intensity and surface area than colloidal gold in the same volume, CNB-based LFIA was approximately ten times more sensitive than colloidal gold-based LFIA, consistent with recently reported results (Sakurai et al., 2014).
Next, we evaluated the specificity of ACE2-LFIA using S1 antigens from other coronaviruses (SARS-CoV and MERS-CoV). Three different concentrations (100, 20, and 5 ng/reaction) of each S1 antigen were introduced into the LFIA device, and the intensity of test lines was measured after 20 min using a portable line analyzer. It took less than 10 s to measure the line intensity of each LFIA device, enabling rapid and accurate diagnosis in point-of-care or laboratory settings. Real images of the LFIA device and portable line analyzer are shown in Supplementary Fig. 10. The SARS-CoV-2 S1 (<5 ng antigen) and RBD (<1 ng antigen) were successfully detected by the LFIA device, even at low antigen concentration (Fig. 5a and Supplementary Fig. 8). The sensitivity difference between SARS-CoV-2 S1 and RBD using ACE2-based LFIA might be associated with the KD value of ACE2 and S1-mAb were lower for RBD binding than for S1 binding (Supplementary Table 2). Meanwhile, the MERS-CoV S1 antigen was not detected even at relatively high concentrations (100 ng antigen); however, 100 ng antigen of SARS-CoV S1 was slightly detected, as shown in Fig. 5b and c, and Supplementary Table 3. This means that more than 100 ng antigen of SARS-CoV S1 might not be distinguished using the ACE2-LFIA. The intensity slightly increased at a high concentration of SARS-CoV S1; however, the proposed ACE2-LFIA has the potential to distinguish SARS-CoV-2 from other coronaviruses due to the significant difference in intensity. The inset graph in Fig. 5c shows the detection intensity for the S1 antigens from three different coronaviruses at an antigen concentration of 5 ng. The detection signal of SARS-CoV-2 S1 was clearly higher than the limit of detection (LOD, mean value of negative controls + 3 × standard deviation), whereas those of SARS-CoV S1 and MERS-CoV S1 were similar to that of the negative control. Thus, the proposed ACE2-LFIA has the ability to detect the SARS-CoV-2 antigen with high sensitivity and without significant cross-reactivity from other coronaviruses.
3.4. Laboratory confirmation using clinical samples
Cultured viral and clinical samples were used for laboratory confirmation of ACE2-LFIA (Fig. 6 a). Viral load of cultured SARS-CoV-2 and human CoV OC43 was measured by quantitative RT-PCR (RT-qPCR) analysis with the standard curve of the E and N gene, respectively (Supplementary Fig. 11). The limit of detection of ACE2-LFIA was 5.35 × 106 copies/mL of the cultured viral sample of SARS-CoV-2; however, there were no positive signals (>50) from ACE2-LFIA tests with cultured samples of human CoV OC43 (Fig. 6b).
Fig. 6.
Laboratory confirmation of ACE2-based LFIA using clinical samples a) Schematic diagram of COVID-19 test using ACE2-based LFIA. A nasopharyngeal swab from the COVID-19 patient is placed into the UTM. 50 μL of UTM containing the SARS-CoV-2 is mixed with running buffer in a 1:1 (v/v) ratio, and 100 μL of mixed solution is loaded into the LFIA device. After 20 min, the line intensity of the LFIA strip is semi-quantified by the portable analyzer. b) Results of ACE2-based LFA for the detection sensitivity of cultured SARS-CoV-2. Serially diluted virus concentrates (concentration range: 1.07 × 108 copies/mL to 5.35 × 106 copies/mL) were tested. After 20 min, the LFIA strip was taken with a smartphone and scanned with an image analyzer. The line intensities of the test and control lines were converted to peak histograms. Also, the intensity of the test lines was measured by a portable line analyzer (IL: line intensity of test line). Furthermore, human coronavirus (OC43) was tested as a negative control. c) Bar graph of intensities for test lines measured by the portable analyzer. The limit of detection (LOD) was determined by the mean value of negative controls (0 copies/mL of SARS-CoV-2) plus three times the standard deviation. d) Laboratory confirmation of ACE2-based LFIA compared to the RT-qPCR using clinical samples. i) Nasopharyngeal swab samples of COVID-19 patients (n = 4) and healthy subjects (n = 4) were tested both ACE2-based LFIA and RT-qPCR. Sensitivity was determined by the number of true positive samples divided by the number of positive samples tested. Moreover, specificity was determined by the number of true negative samples divide by the number of negative samples tested. ii) RT-qPCR results for the detection of the SARS-CoV-2 specific gene (Env gene). Ct value and their correspondent viral load in the clinical samples were evaluated. e) Results of ACE2-based LFIA on laboratory confirmation using clinical COVID-19 patient samples. Twenty minutes after sample loading, the test line intensities of the LFIA strips were measured with a portable line analyzer. The limit of detection (LOD) was determined by the mean value of negative controls (healthy control) plus three times the standard deviation.
To measure viral load (copy/mL) in clinical samples, RT-qPCR analysis was performed with nasopharyngeal (NP) and nasal swabs from COVID-19 patient (n = 4) and healthy subjects (n = 4), respectively. Viral loads in NP swab of COVID-19 patients were investigated with the standard curve of the E gene of SARS-CoV-2: 2.49 × 107 copies/mL in Patient 1, 1.15 × 106 copies/mL in Patient 2, and 1.86 × 105 copies/mL in Patient 4. Unfortunately, SARS-CoV-2 RNA from Patient 3 was not detected by RT-qPCR analysis (Fig. 6d and Supplementary Fig. 12). This result was also confirmed by other independent RT-qPCR analysis, and might be associated with the degradation of viral RNAs in UTM. Laboratory confirmation of ACE2-LFIA with clinical specimens of COVID-19 patients showed three positive results from only confirmed clinical specimens (n = 3) by our RT-qPCR analysis (Fig. 6d and e). The limit of detection of our ACE2-LFIA was 1.86 × 105 copies/mL in the clinical specimen of COVID-19 Patient 4; however, there were no positive signals from ACE2-LFIA tests with nasal swabs from healthy subjects (n = 4). Therefore, ACE2-based LFIA tests will be helpful to detect the S1 antigen of SARS-CoV-2 from COVID-19 patients. Further developments of ACE2-based LFIA with more specific antibodies, aptamers, affimers, or nanobodies will be needed for more sensitive and specific detection of the SARS-CoV-2 S1 antigen.
4. Conclusions
A matched pair of antibodies that bind virus-specific antigens is essential for sandwich immunoassays such as ELISA and RDT. To date, no study has reported a matched antibody pair for immunoassay of SARS-CoV-2 antigens, such as the S and N proteins. Wrapp et al. reported that human ACE2 has a higher affinity for SARS-CoV-2 S1 than SARS-CoV S1, suggesting the possibility of an ACE2-based immunoassay for SARS-CoV-2. In this study, we examined the interaction between SARS-CoV-2 S1 and ACE2 (or antibodies) by Western blot and ELISA. The possibility of the matched pair between ACE2 and antibodies for the rapid detection of the SARS-CoV-2 S1 antigen was mainly investigated in the present study. Ultimately, ACE2 and S1-mAb were paired with each other as capture and detection probes in a lateral flow immunoassay (LFIA) that was not cross-reactive with SARS-CoV S1 or MERS-CoV S1 proteins. The SARS-CoV-2 S1 (<5 ng of recombinant proteins/reaction) was detected by the ACE2-based LFIA. The limit of detection of our ACE2-LFIA was 1.86 × 105 copies/mL in the clinical specimen of COVID-19 patients without no cross-reactivity for nasal swabs from healthy subjects. This is the first study to detect SARS-CoV-2 S1 antigen using an LFIA with matched pair consisting of ACE2 and antibody. Our findings will be helpful to detect the S1 antigen of SARS-CoV-2 from COVID-19 patients. Further studies of ACE2-based LFIA with more specific antibodies, aptamers, affimers, or nanobodies will be needed for more sensitive and specific antigen diagnostic of clinical specimens of COVID-19 patients.
CRediT authorship contribution statement
Jong-Hwan Lee: Conceptualization, Methodology, Investigation, Data curation, Writing. Minsuk Choi: Methodology, Investigation, Data curation, Writing. Yujin Jung: Methodology, Investigation. Sung Kyun Lee: Methodology, Investigation. Chang-Seop Lee: Methodology, Investigation. Jung Kim: Investigation. Jongwoo Kim: Investigation. Nam Hoon Kim: Investigation. Bum-Tae Kim: Funding acquisition, Supervision. Hong Gi Kim: Project administration, Funding acquisition, Supervision, Conceptualization, Investigation, Writing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the National Culture Collection for Pathogens of the Korean CDC for providing clinical SARS-CoV-2 isolates. This work was supported by the National Research Council of Science and Technology (NST) and National Research Foundation (NRF) funded by the Ministry of Science and ICT, Republic of Korea (grant number: CRC-16-01-KRICT and NRF-2020M3E9A1043749) and Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant numbers: HI20C0033 & HI20C0363).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112715.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Barr J.N., Fearns R. Genetic instability of RNA viruses. Genome Stability. 2016:21–35. [Google Scholar]
- Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.J., Luo C.M., Tan B., Wang N., Zhu Y., Crameri G., Zhang S.Y., Wang L.F., Daszak P., Shi Z.L. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wen T., Shi F.-J., Zeng X.-Y., Jiao Y.-J. Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega. 2020;5(21):12550–12556. doi: 10.1021/acsomega.0c01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y., Park G.-S., Moon J.H., Ku K., Beak S.-H., Lee C.-S., Kim S., Park E.C., Park D., Lee J.-H., Byeon C.W., Lee J.J., Maeng J.-S., Kim S.-J., Kim S.I., Kim B.-T., Lee M.J., Kim H.G. Comparative analysis of primer–probe sets for RT-qPCR of COVID-19 causative virus (SARS-CoV-2) ACS Infect. Dis. 2020;6(9):2513–2523. doi: 10.1021/acsinfecdis.0c00464. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li D., Wang D., Dong J., Wang N., Huang H., Xu H., Xia C. False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-based CT diagnosis and insights from two cases. Korean J. Radiol. 2020;21(4):505–508. doi: 10.3348/kjr.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Fang Y.Y., Deng Y., Liu W., Wang M.F., Ma J.P., Xiao W., Wang Y.N., Zhong M.H., Li C.H., Li G.C., Liu H.G. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin. Med. J. 2020;133(9):1025–1031. doi: 10.1097/CM9.0000000000000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrikou S., Moschopoulou G., Tsekouras V., Kintzios S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sensors. 2020;20(11):3121. doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Narváez E., Dincer C. The impact of biosensing in a pandemic outbreak: COVID-19. Biosens. Bioelectron. 2020;163:112274. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J.E., Sharon C., Satkunarajah M., Auperin T.C., Cameron C.M., Kelvin D.J., Seetharaman J., Cochrane A., Plummer F.A., Berry J.D., Rini J.M. Structural insights into immune recognition of the severe acute respiratory syndrome coronavirus S protein receptor binding domain. J. Mol. Biol. 2009;388(4):815–823. doi: 10.1016/j.jmb.2009.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T., Lee S.-Y., Kim S., Kim M.J., Kim H.G., Jun S., Kim S.I., Kim B.T., Park E.C., Park D. 2020. Spike Protein Binding Prediction with Neutralizing Antibodies of SARS-CoV-2. bioRxiv. [Google Scholar]
- Relova D., Rios L., Acevedo A.M., Coronado L., Perera C.L., Perez L.J. Impact of RNA degradation on viral diagnosis: an understated but essential step for the successful establishment of a diagnosis network. Vet Sci. 2018;5(1) doi: 10.3390/vetsci5010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai A., Takayama K., Nomura N., Yamamoto N., Sakoda Y., Kobayashi Y., Kida H., Shibasaki F. Multi-colored immunochromatography using nanobeads for rapid and sensitive typing of seasonal influenza viruses. J. Virol. Methods. 2014;209:62–68. doi: 10.1016/j.jviromet.2014.08.025. [DOI] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117(21):11727. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. Plos Pathog. 2018;14(8) doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y., Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microb. Infect. 2020;9(1):382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. J. Am. Med. Assoc. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904. doi: 10.1016/j.cell.2020.03.045. e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Coronavirus disease (COVID-19)_case report. [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.