Abstract
Retinal degeneration is a main class of ocular diseases. So far, retinal progenitor cell (RPC) transplantation has been the most potential therapy for it, in which promoting RPCs neuronal differentiation remains an unmet challenge. To address this issue, innovatively designed L/d-phenylalanine based chiral nanofibers (LPG and DPG) are employed and it finds that chirality of fibers can efficiently regulate RPCs differentiation. qPCR, western blot, and immunofluorescence analysis show that right-handed helical DPG nanofibers significantly promote RPCs neuronal differentiation, whereas left-handed LPG nanofibers decrease this effect. These effects are mainly ascribed to the stereoselective interaction between chiral helical nanofibers and retinol-binding protein 4 (RBP4, a key protein in the retinoic acid (RA) metabolic pathway). The findings of chirality-dependent neuronal differentiation provide new strategies for treatment of neurodegenerative diseases via optimizing differentiation of transplanted stem cells on chiral nanofibers.
Keywords: Chiral structures, Self-assembly, Nanofibers, Retinal progenitor cell, Neuronal differentiation
Graphical abstract
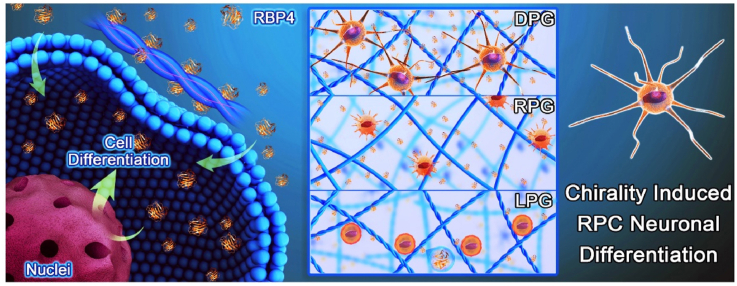
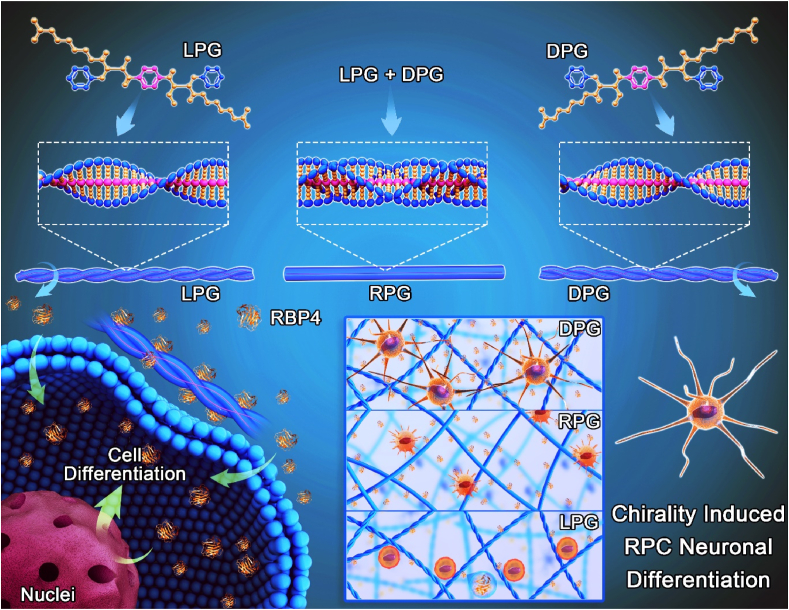
L/d-phenylalanine-based enantiomers are employed as building blocks to fabricate chiral nanofibers. Right-handed helical nanofibers significantly promote RPC neuronal differentiation, migration and synapse formation, which are mainly ascribed to activation of the RA metabolic pathway. This provides a feasible strategy to address retinal degeneration if chiral structure of extracellular microenvironment is considered.
Highlights
-
•
This is the first attempt to utilize chiral structure to induce retinal progenitor cell neuronal differentiation.
-
•
Right-handed nanofibers promote cell neuronal differentiation, whereas left-handed fibers decrease this effect.
-
•
The different cell differentiations are attributed to the stereoselective interaction between chiral fibers and proteins.
-
•
It provides new strategies for treatment of neurodegenerative diseases via optimizing chiral structures of biomaterials.
1. Introduction
Retinal degeneration is a main class of ocular diseases, which is mainly caused by the loss and death of retinal neuronal cells, leading to a decline in visual function or even blindness [1]. So far, transplantation of retinal progenitor cells (RPCs) has been one of the most potential therapy for such disease [2], in which promoting RPCs neuronal differentiation remains an unmet challenge since RPCs prefer to differentiate into glial cells rather than retinal neuronal cells [3]. Actually, stem/progenitor cells differentiation are closely associated with the characteristics of extracellular microenvironment, including chemical species [4], structures [5], mechanical [6] and chiral properties [7,8]. Among these, chiral structures as basic structural units of living organisms are particularly important for regulating cell differentiation [9,10]. Therefore, it's highly expected that chiral structure can guide RPCs to directionally differentiate into retinal neuronal cells, since native extracellular microenvironment of RPCs is full of chiral components (e.g. collagen with right-handed triple helix), however, this strategy has been rarely explored until now.
To regulate RPCs differentiation, biomimetic extracellular microenvironment is commonly fabricated via biochemical or biophysical cues. For example, considering biochemical effect on cell behaviors, matrix metalloproteinase 2 (MMP2) was loaded into poly (lactic-co-glycolic acid) (PLGA) microspheres to enhance cellular integration and thereby induce RPCs differentiate into mature retinal neuronal cells [11]. Besides, biophysical factors (e.g. morphology, stiffness, strength) have also been used to guide RPCs neuronal growth [12,13]. However, as one of most important biophysical factors, chiral structure and its potential role in RPCs differentiation have yet to be fully studied and remian poorly understood. It is desirable to explore influence of chirality on RPCs differentiation, since helical structure is one of the most important characters of extracellular fibrous matrix and involved in a wide diversity of physiological functions, which is necessary and important for revealing the underlying mechanism of chirality induced RPCs differentiation.
Herein, enantiomeric L/d-phenylalanine gelators (LPG, DPG) and their racemic mixture (RPG) are employed to mimic chiral extracellular microenvironment (Fig. 1a). LPG and DPG molecules can self-assemble into left-handed and right-handed helical nanofibers, respectively. Subsequent biological assays demonstrated that right-handed DPG nanofibers significantly enhance RPCs neuronal differentiation, especially, β3-tubulin (a retinal neuron marker) expression in RPCs cultured on DPG nanofibers is almost 3.0 times higher than those on LPG ones. The different phenomena of RPCs differentiation are mainly attributed to the stereoselective interaction between chiral helical nanofibers and retinol-binding protein 4 (RBP4, a key protein in the retinoic acid (RA) metabolic pathway) as confirmed by western blot analysis, enzyme-linked immunosorbent assay (ELISA), microarray gene analysis, and classic molecular dynamic (MD) simulation (Fig. 1b). The promoted RPCs neuronal differentiation on chiral cell growth nanofibers may provide new insight into retinal repair and have potential application in the treatment of retinal degeneration.
Fig. 1.
a) A schematic representation of left-handed helical LPG nanofibers, right-handed helical DPG nanofibers, and RPG nanofibers without helical chiral structure, respectively. (b) A schematic representation of chirality induced RPCs neuronal differentiation. Promoted neuronal differentiation of RPCs is achieved on right-handed DPG nanofibers.
2. Materials and methods
2.1. Materials synthesis and characterization
2.1.1. Materials synthesis
L/d-phenylalanine methyl ester hydrochloride, 1,4-benzenedicarbonyl dichloride and diglycol were purchased from Shanghai Titian Scientific Co., Ltd and used without further purification L(D)PG molecule (Fig. S1) was synthesized according to previous reference [14]. 1H NMR (400 M Hz, DMSO‑d6, δ): δ = 3.1 (m, 4H, CH2), 3.4 (m, 16H, CH2), 4.2 (q, 2H, OH), 4.7 (q, 2H, CH), 7.3 (m, 10H, Ar H), 7.8 (s, 4H, Ar H), 8.9 (d, 2H, NH) ppm. EI-MS for L(D)PFEG calcd. 636.71; found 637.28 [M + H]+.
2.1.2. Nanofiber films, non-assembled molecules and hydrogel preparation
LPG (or DPG or LPG + DPG) molecules were suspended in deionized water (3 mg/mL). Next, the solution was heated to 95 °C, and a clear solution was formed. After that, 400 μL, 200 μL, 100 μL, and 30 μL clear solutions were used to coat the 6-well, 12-well, 24-well, and 96-well plates, respectively. The hydrogel was observed in plate wells when the solution was cooled to room temperature within several minutes. After the plates were placed in 37 °C oven (Thermo) for 12 h, so that the hydrogel could form a thin film on each well. In addition, the non-assembled molecules were prepared by dissolving them in methanol (3 mg/mL). In animal experiments, the hydrogel was to prepared by dissolving the molecules in DMSO (3 mg/mL).
2.1.3. Scanning electron microscopy (SEM) study
SEM measurement was performed on a FEI QUANTA 250 Microscope. All tested samples (0.5 mg/mL) were prepared by depositing solutions on silicon slices and drying at room temperature. All measurements were performed with an operation voltage of 10 kV.
2.1.4. Circular dichroism (CD) and vibrational circular dichroism (VCD) spectroscopy
The test concentration of all samples for CD measurements were 0.5 mg/mL. For VCD measurements, the samples were firstly dissolved in hot water (above 90 °C). After cooling, 100 μL liquid samples were deposited on quartz wafers, and dried at room temperature. CD spectra were obtained using JACSO J-815 CD spectrometer with bandwidth of 0.5 nm. VCD spectra were measured at BioTools, Inc., Jupiter, FL, using a ChiralIR-2X Fourier Transform VCD (FT-VCD) spectrometer equipped with an MCT detector and the Dual PEM option for enhanced VCD baseline stability. Every VCD spectrum was the result of averaging a minimum of 8000 scans at a spectral resolution of 4 cm−1.
2.1.5. Classic molecular dynamic (MD) simulation
GROMACS 2016.4 was used as the MD simulation package with AMBER99SB-ILDM force field [15]. Periodic boundary was employed in simulation, where protein was immersed in a cubic box with a length of 70 Å. Simplified point charge model TIP3P [16] was used to represent water molecules. We first used steepest descent approach to minimize the energy of the system with a convergence criterion of 1000 kJ mol−1 nm−1 and a step of 0.01 nm. Energy-minimization was followed by 10 ns of NPT MD. Finally, equilibrated structure of system was obtained after 1 ns of NPT MD. The Berendsen thermostat [17] was applied to maintain a constant temperature of 335.15 K in the system with a coupling constant λ = 0.2 ps.
2.2. Cell experimental section
2.2.1. Separation and culture of RPCs
Retinal progenitor cells (RPCs) were obtained from fresh retinal tissue of postnatal day 1 C57BL/6 mice. Next, the isolated RPCs were placed in a T25 flask and cultured using a proliferation medium containing advanced DMEM/F12 (Invitrogen, Carlsbad, CA, USA), 2 mM l-glutamine (Invitrogen), 1% N2 neural supplement (Invitrogen), 20 ng/mL of epidermal growth factor (EGF, Invitrogen) and 100 U/mL of penicillin-streptomycin (Invitrogen). During the differentiation study, RPCs were incubated in differentiation medium with 10% fetal bovine serum (FBS, Invitrogen) without EGF.
All the animals were handled according to the Association for Research in Vision and Ophthalmology (ARVO)'s animal usage standards following authorization by the Animal Care and Use Committee of the Schepens Eye Research Institute, where the original source of cells was derived.
2.2.2. Live-dead assay
The live-dead assay was carried out as described previously [13]. Briefly, each well (5 × 104 cells per well) was coated with LPG, DPG or RPG (100 μL) films in 24-well plates. Next, according to the recommended measurements, each well was incubated with the live/dead kit reagents (Invitrogen). A fluorescence microscope (Olympus BX51, Japan) was used to acquire images after staining for 30 min. The images were processed and analyzed by ImageJ software.
2.2.3. Cell Counting Kit 8 assay
Cell Counting Kit 8 assay (CCK-8) (Yeasen) was utilized to evaluate cell proliferation. RPCs were seeded in 96-well plates at 1 × 104 cells per well under proliferation conditions. After 0 h, 24 h, 48 h, and 72 h of incubation, the CCK-8 reagent was incubated in the culture for 4 h. Then, a microplate reader (ELX800, Bio Tek, USA) was used to measure the optical density at 450 nm.
2.2.4. Total RNA Separation and quantitative polymerase chain reaction (qPCR)
Digested RPCs (2 × 105 cells per well) were seeded in 6-well plates coated with films (400 μL), then were cultured under differentiation conditions for 7 days. Total RNA was extracted from the cultured cells with TRIzol (Invitrogen) according to the manufacturer's instructions. The harvested total RNA concentrations were detected by spectrophotometry using NanoDrop 2000 software. In addition, the samples whose purity met the standard of OD260/280 nm ratios between 1.9 and 2.1 were used to synthesize the cDNA. cDNA was obtained from the reverse transcription of total RNA with a final reaction volume of 10 μL using a Primer Script RT reagent kit (Perfect Real Time; TaKaRa). The primer details are presented in Table 1. After normalization to the expression of β-actin, the data were expressed as a fold change for each treatment group relative to the control group.
2.2.5. Western blot analysis
RPCs (2 × 105 cells per well) were seeded on 6-well plates coated with films (400 μL) per well. Total proteins were obtained after treatment with differentiation culture for 7 days and quantitatively analyzed using a BCA kit (Pierce). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were used to separate total proteins. After the proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA), the membranes were incubated with various antibodies, including mouse monoclonal anti-Rhodopsin, anti-β3-tubulin, anti-GFAP (Millipore), anti-PKC-α (BD), anti-β-actin (Proteintech), anti-Mcp-1 (Proteintech), rabbit polyclonal anti-ALDH1A1 (ABclonal), anti-RBP4 (Proteintech), and anti-IL-6 (Absin) followed by incubation with secondary antibodies (Sigma-Aldrich). The ECL detection kit (Tanton) was used to detect protein expression.
2.2.6. Immunofluorescence
Cell suspensions (5 × 104 cells per well) were seeded on film-coated (100 μL) 24-well plates for 7 days (differentiation condition). Immunofluorescence was performed to evaluate the differentiation ability of RPCs. RPCs were fixed with 4% paraformaldehyde for 30 min, and then the cells were blocked with 10% normal goat serum for 1 h after treatment with 0.03% Triton X-100 in phosphate buffered saline buffer. Next, the samples were incubated with different primary antibodies (1: 200 dilution) including mouse monoclonal anti-β3-tubulin (Millipore), anti-glial fibrillary acidic protein (GFAP) (Millipore), anti-PKC-α (BD), and anti-Rhodopsin (Millipore), overnight at 4 °C. After that, the cells were incubated with secondary antibodies (Alexa Fluor546-goat anti-mouse/rabbit IgG, BD, 1:800) for 1 h in the dark. Hoechst dye (Invitrogen) were used to counterstain the cell nuclei. The stained cells were observed under a fluorescence microscope (Olympus BX51, Japan). Finally, the data were analyzed by ImageJ software.
2.2.7. Cell migration assays
First, 6-well plates were coated with various films (400 μL). RPCs were seeded on plates (4 × 105 cells per well) and cultured in differentiation medium to generate cell monolayers. When the cells reached 90–95% confluence, a 200 μL pipette tip was used to produce scratch wounds. After washing with PBS, the scratch wounds were recorded by fluorescence microscopy at 0 h, 48 h and 72 h. Next, the data were processed by ImageJ software. The closure rate of the wounds in different groups at each time point (0 h, 48 h and 72 h) was calculated using the following formula: would healing (closure %) = [(X0 – X1)/X0] × 100%. X0 represents the initial wound area and X1 represents the residual wound area at each time point (0 h, 48 h and 72 h).
2.2.8. ELISA
All the groups were treated as previously described in western blot. After 2,7 days of cell culture, the supernatant of the cell culture medium was collected for ELISA. Next, 150 μL of a standard solution of RBP4 (480 ng/mL) was added to an Eppendorf tube as the reference standard. To this solution was added 150 μl of the reference standard diluent, followed by vortexing for 30 s to obtain a 240-ng/mL stock solution of the reference standard. We then prepared 120-ng/mL, 60-ng/mL, 30-ng/mL, and 15-ng/mL stock solutions of the reference in Eppendorf tubes by performing serial dilutions in 150 μl of the reference standard diluent. Finally, the RBP4 concentration was measured by ELISA according to the manufacturer's instructions (Shanghai Lengton Bioscience Co., LTD).
2.2.9. Global gene expression
The RPCs (2 × 105 cells per well) were seeded on 6-well plates coated with films (400 μL) per well, then were cultured under differentiation conditions for 7 days. The RPCs were lysed in 1 mL of TRIzol (Invitrogen), and total RNA from each sample was quantified using a NanoDrop ND-1000 instrument (Thermo). 1 μg total RNA was used to prepare the sequencing library in the following steps: 1. Total RNA is enriched by oligo (dT) magnetic beads (rRNA removed); 2. RNA-seq library preparation using KAPA Stranded RNA-Seq Library Prep Kit (Illumina), which incorporates dUTP into the second cDNA strand and renders the RNA-seq library strand-specific. The completed libraries were qualified with Agilent 2100 Bioanalyzer and quantified by absolute quantification qPCR method. To sequence the libraries on the Illumina HiSeq 4000 instrument, the barcoded libraries were mixed, denatured to single stranded DNA in NaOH, captured on Illumina flow cell, amplified in situ, and subsequently sequenced for 150 cycles for both ends on Illumina HiSeq instrument. Image analysis and base calling were performed using Solexa pipeline v1.8 (Off-Line Base Caller software, v1.8). Sequence quality was examined using the FastQC software. The trimmed reads (trimmed 5′, 3′-adaptor bases using cutadapt) were aligned to reference genome using Hisat2 software. The transcript abundances for each sample was estimated with StringTie, and the FPKM value for gene and transcript level were calculated with R package Ballgown. The differentially expressed genes and transcripts were filtered using R package Ballgown. The novel genes and transcripts were predicted from assembled results by comparing to the reference annotation using StringTie and Ballgown, then use CPAT to assess the coding potential of those sequences. Then use rMATS to detecting alternative splicing events and plots. Principle Component Analysis (PCA) and correlation analysis were based on gene expression level, Hierarchical Clustering, Gene Ontology, Pathway analysis, Gene Ontology, Pathway analysis, scatter plots and volcano plots were performed with the differentially expressed genes in R, Python or shell environment for statistical computing and graphics.
2.2.10. RBP4 absorption
RBP4 protein (Novoprotein) was adhered to films of LPG-, DPG-, and RPG-coated cell culture plates infiltrated with 10 μg/mL of RBP4 solution for 1, 3 and 6 h at 37 °C. Next, Milli-Q water was used to wash away non-adhered RBP4 proteins. Rabbit anti-RBP4 (1:200, Proteintech) and Alexa Fluor546-goat anti-rabbit IgG (1:800, BD) were used as the primary antibody and secondary antibody, respectively. The stained samples were photographed using a fluorescence microscope (Olympus BX51, Japan).
2.3. In vivo toxicity
All 36 nude mice at approximately 4 weeks of age were divided into four groups. The control group was subcutaneously injected with 300 μL of saline on each side of the back area of every mouse. The other groups were treated similarly as the control group but treated with 300 μL of LPG, DPG, and RPG hydrogels. Three mice in each group were sacrificed at 14 and 28 days postinjection. The organs (heart, liver, spleen, lung, and kidney) were cut into portions for hematoxylin and eosin (H&E) staining at 14 and 28 days postinjection. Tissues at the site of injection were isolated to perform CD68 (Abcam, 1:100) immunohistochemical staining at 14 days postinjection. Photos were taken by laser scanning microscopy (Nikon).
2.4. Statistical analysis
The data were presented as means ± SD and were analyzed by Student's t-tests (GraphPad Prism 6 software). *P < 0.05 was considered significant for statistical tests.
3. Results and discussion
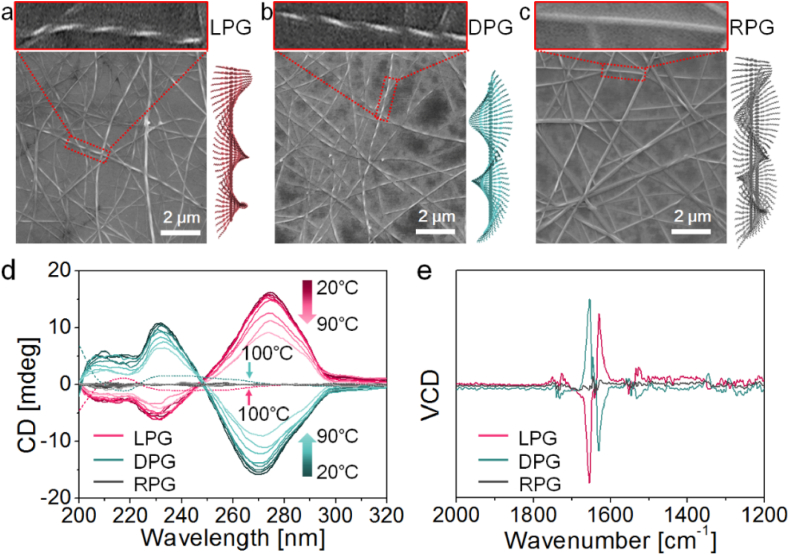
The scanning electronic microscopy (SEM) images demonstrated that LPG and DPG molecules self-assembled into left-handed and right-handed helical nanofibers, respectively (Fig. 2a and b). Both LPG and DPG nanofibers had similar diameters of 70–100 nm and helical pitches of 0.3–0.35 μm. Additionally, we found that the racemic mixture of LPG and DPG (abbreviated as RPG) co-assembled into nanofibers (diameter: 90–130 nm) without any chiral structure (Fig. 2c). The helicity of LPG and DPG nanofibers was further confirmed via circular dichroism (CD) and vibrational circular dichroism (VCD) measurements (Fig. 2d and e). At room temperature (20 °C), LPG and DPG assemblies presented two strong CD peaks at 230 nm and 270 nm, respectively. With increasing testing temperature, the intensity of the CD signals for both LPG and DPG gradually decreased due to disassembly triggered by heating. At 100 °C, the disappearance of CD signals at 230 nm and 270 nm suggested the disintegration of LPG and DPG self-assembled nanofibers, respectively. Two new weak CD peaks at 218 nm and 232 nm were ascribed to the molecular chirality from phenylalanine in LPG and DPG, respectively [14,18]. In the VCD spectra, the two groups of conversed peaks at 1655 cm−1 and 1630 cm−1 for LPG and DPG also demonstrated the inversion of the chiral structure from LPG to DPG nanofibers.
Fig. 2.
SEM images of (a) left-handed helical LPG nanofibers, (b) right-handed helical DPG nanofibers, and (c) non-helical RPG nanofibers. (d) CD spectra of LPG, DPG, and RPG assemblies. (e) VCD spectra of LPG, DPG, and RPG assemblies.
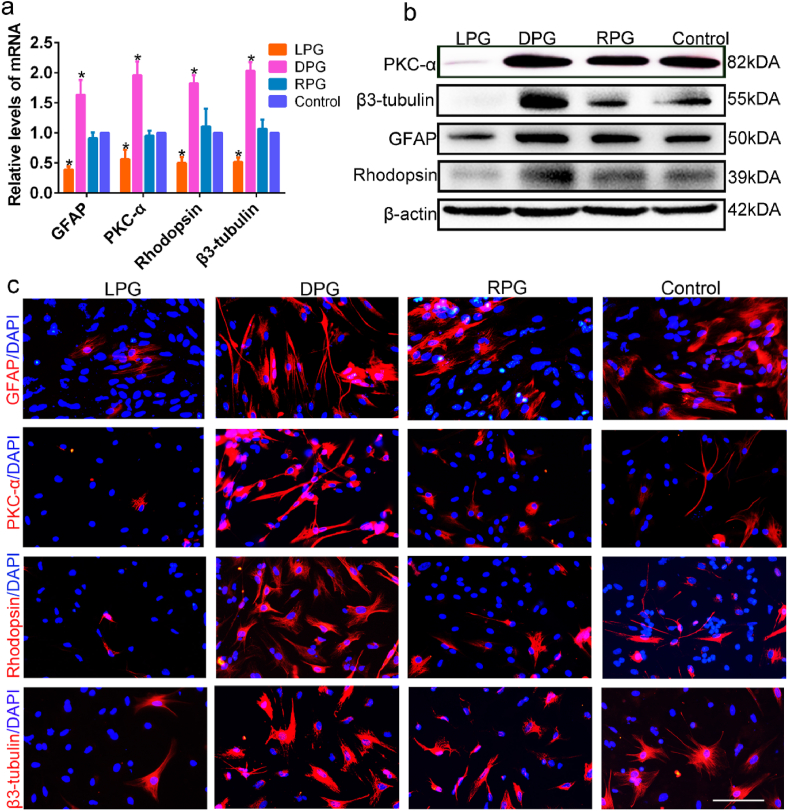
Before investigating RPCs differentiation on different chiral fibrous films, immunofluorescence was used to identify RPCs (Fig. S2). The live-dead staining (Fig. S3) and CCK-8 assays (Fig. S4) were performed to assess the cytocompatibility of chiral nanofibers. Our data revealed that RPCs were more than 80% viable on chiral nanofibers. The gene expression levels of inflammatory cytokines (Mcp-1 and IL-6) [19] were markedly decreased on the chiral nanofibers compared with the control group (Fig. S5a), a finding that was consistent with the results of western blot assays (Fig. S5b). Hematoxylin and eosin (H&E) staining assays (Figs. S6 and S7) revealed that the main organs showed no obvious cell invasion, toxicity, inflammatory response and pathological response compared with the control group (as evaluated by a pathologist). By day 14, no significant inflammatory response at the site of injection and no CD68-positive macrophages (indicating an inflammatory response) [20] were observed in the treated group (Fig. S8). These data implied that chiral nanofibers could reduce inflammation in vitro and in vivo. To explore the effect of chiral nanofibers on the formation of retinal neuronal networks, the differentiation capacity of RPCs was detected regarding morphology, migration behavior, differentiation-related gene and protein expression levels after culturing RPCs on nanofibrous films. Both the outgrowth of neurites and cell migration play crucial roles in the differentiation of stem cells into neuronal cells [21,22]. After 5 days of differentiation culture, faster and slender outgrowth of neurites (Fig. S9) and faster RPCs migration were observed on DPG fibers, suggesting that DPG nanofibers with right-handed helical structures enhanced RPCs differentiation compared with left-handed LPG nanofibers (Fig. S10). Because the coated nanofiber films were extremely thin, the process to produce scratch wounds also detached the nanofiber films in cell migration assays (Fig. S11). However, nanofiber films could still exist on both sides of the scratch, and migrating cells were still be affected by the chiral effect of the nanofiber films. qPCR data indicated that the neuron differentiation-related gene expression levels were notably enhanced on DPG group such as β3-tubulin (a general marker for neuronal cells), Rhodopsin (a marker for photoreceptors) and PKC-α (a marker for bipolar cells), while LPG repressed them (Fig. 3a). In addition, western blot (Fig. 3b) and immunofluorescence (Figs. 3c and S12) results demonstrated that the neuron-differentiation-related protein levels were obviously upregulated and the percentages of immunoreactive cells were significantly increased on DPG fibrous films. For example, β3-tubulin (64% vs 32.6%), Rhodopsin (68.6% vs 15%) and PKC-α (75.3% vs 18.6%), which were decreased on LPG films. In the DPG group, the high expression of GFAP may be due to the increase in the proportion of positive cells, but the number of positive cells did not increase significantly. Notably, DPG mainly promoted the differentiation of RPCs toward retinal neuronal cells such as photoreceptors (light-sensing cells), which are the most crucial cells to restore vision in many cases of blindness [23]. Additionally, qPCR results showed that non-assembled DPG (N-DPG) had a weaker impact on cell differentiation and chiral assembled structure of DPG significantly enhanced RPCs differentiation (Fig. S13). In this study, both non-assembled (N-DPG) and assembled molecules (DPG) were derived from d-amino acids. It has been reported that amplification of chirality from chiral molecules to chiral assemblies presented much more pronounced regulated cell behaviors [14]. Consistent with these studies, our data suggested that chiral assembled structure of DPG enhanced the effect of chiral amplification.
Fig. 3.
Chirality affected differentiation of RPCs. (a) The qPCR assays revealed that differentiation-related markers (β3-tubulin, GFAP, PKC-a, Rhodopsin) were increased in the DPG group but decreased in the LPG group. (b) Western blot analysis showed that DPG nanofibers obviously enhanced differentiation-related markers, however, LPG nanofibers exhibited an opposite effect in RPCs differentiation. (c) Immunofluorescence assays indicated that greater numbers of β3-tubulin, GFAP, PKC-a, Rhodopsin clusters in the DPG group than in the LPG, RPG and control. Scale bar: 50 μm *P < 0.05. The error bars show the standard deviation of the mean for n = 3 independent experiments.
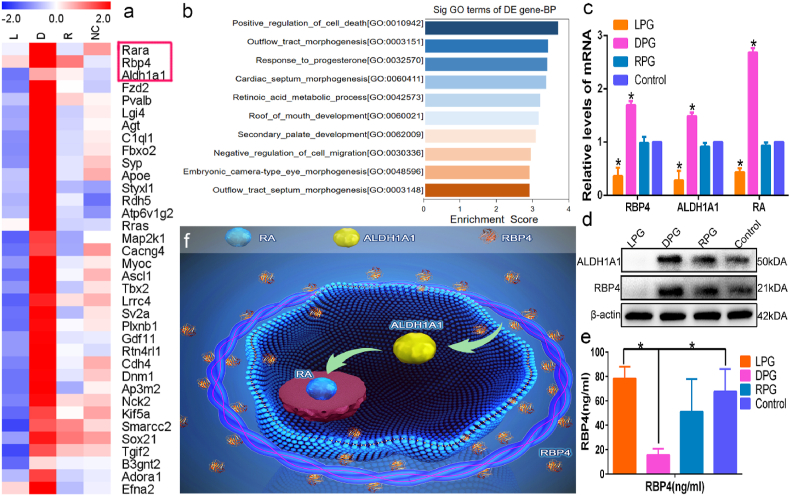
To gain deeper insight, a global microarray gene analysis was implemented. High-throughput sequencing analysis indicated that neural differentiation-related transcripts were downregulated in the LPG-treated group, markedly upregulated in the DPG-treated group, and not visibly changed in the RPG-treated group (Fig. 4a). These data implied that neural differentiation-related pathways might be activated in the DPG group. In-depth pathway analysis (Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis) revealed that the retinoic acid (RA) metabolism pathway was significantly activated in the DPG group (Fig. 4b). Notably, four different analyses (biological process, cell component, molecular function and neural-differentiation transcripts) all showed that retinaldehyde dehydrogenases 1A1 (ALDH1A1) molecule (a fatal role in RA pathway [24]) was markedly elevated (Figs. S14 and S15). qPCR and western blot results showed that molecules, involved in the RA pathway (RBP4, ALDH1A1, RA), were significantly upregulated at the gene and protein levels in DPG group (Fig. 4c and d), suggesting that the RA metabolism pathway (a vital factor for RPCs differentiation [25]) could be activated by right-handed DPG nanofibers. Additionally, our results showed that the gene and protein expression levels of RBP4, ALDH1A1 and RA were decreased in LPG group compared to the control group (Fig. 4c and d), which indicated that LPG may be weaker to stimulate the RA pathway. Therefore, LPG inhibited RPCs differentiation.
Fig. 4.
Chirality affected the process of retinoic acid metabolism. (a) Hierarchical clustering indicated that a variety of neural differentiation-related transcripts were increased in the DPG group but decreased in the other groups. Gene expression values are expressed using a red-blue color scheme. Red denotes higher-than-median expression values, white corresponds to median-valued expression values, and blue indicates lower-median-valued expression values. (b) Results of biological process (BP) analysis in GO enrichment analysis. (c) The qPCR results indicated upregulated levels of retinoic acid metabolic process related proteins (RBP4, ALDH1A1, RA) in the DPG compared with the LPG, RPG and control groups. (d) Western blot assays were consistent with the qPCR results. (e) ELISA showed the relationship between absorbance at 450 nm and RBP4 obtained from cell culture medium (7 days of differentiation). (f) A schematic representation of the RA metabolic pathway regulating RPCs differentiation. *P < 0.05. The error bars show the standard deviation of the mean for n = 3 independent experiments.
RBP4 is the sole specific transporter for retinoic acid precursorprecursors [[26], [27], [28]]. The transformation of the retinoic acid precursor into RA requires oxidation, which is performed by ALDH1A1 [29,30]. Subsequently, RA can trigger the differentiation of stem/progenitor cells such as hematopoietic stem cells [31], neural progenitor cells [32,33], and embryonic stem cells [34]). Herein, the first step in the RA pathway was further studied. The content of RBP4 (approximately 27 ng/mL) in the differentiation medium (10% fetal bovine serum) was tested by ELISA (Fig. S16). Additionally, the content of RBP4 in the supernatant was detected by ELISA under differentiation conditions for 2 days. The results showed that DPG could stimulate more RBP4 (approximately 36 ng/mL) secretion from RPCs (Fig. S17). On day 7, ELISA revealed the content of RBP4 in the supernatant was decreased in DPG group (Fig. 4e). This may be related to the pre-differentiation of the cells on day 2. RPCs had stronger differentiation ability and needed to consume more RBP4 to enhance differentiation ability on day 7. ELISA and protein absorption assays (Fig. S18) revealed significantly increased absorption of RBP4 in the DPG group than in the other groups. Although, the potential way that RBP4 binds to the DPG was unclear, it may be a non-covalent bond [35], which was considered to be the primary interaction. The non-covalent bonding facilitated a reversible capture/release of RBP4 from the supernatant. Increased RBP4 absorption could provide more retinoic acid precursors to synthesize retinoic acid. As shown in Fig. 4e and Fig. S18, the surface of LPG adsorbed less RBP4, which implied that LPG had negative effect on RA pathway and inhibited cells differentiation. Therefore, these data showed that chiral nanofibers affected RPCs differentiation through the RBP4-initiated RA metabolic pathway (Fig. 4f), laying the foundation for different cellular behaviors.
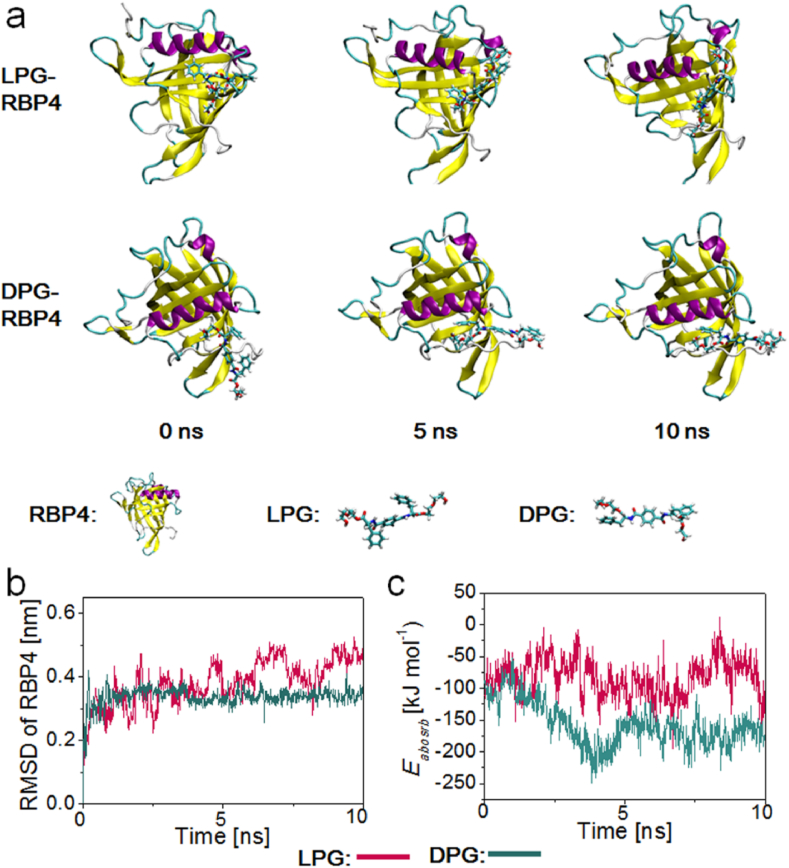
Classic molecular dynamics (MD) simulations were explored to reveal the steric interaction between the RBP4 protein (one of the main proteins that plays a key role in regulating retinal neuronal cells differentiation) and LPG/DPG molecule. The snapshots demonstrated that the binding of DPG and RBP4 could reach a stable stage within 5 ns, while the interaction between LPG and RBP4 could not reach an equilibrium stage until 10 ns (Fig. 5a; Movies S1 and S2, Supporting Information). This result suggested that RBP4 had a higher stereoaffinity for DPG than LPG. The smaller root-mean-square deviation (RMSD) value of RBP4 in the DPG-RBP4 complex (0.48 nm for the LPG-RBP4 complex and 0.35 nm for the DPG-RBP4 complex; Fig. 5b) and lower interaction energy between RBP4 and DPG (−105 kJ mol−1 for LPG-RBP4 complex, −181 kJ mol−1 for DPG-RBP4 complex, Fig. 5c) further confirmed stronger binding of RBP4 on DPG than on LPG. The MD simulation data confirmed a greater stereoaffinity of RBP4 to DPG than to LPG, ultimately leading to promoted neuronal differentiation of RPCs on DPG.
Fig. 5.
MD simulations proved the stereoselective interaction between RBP4 and LPG/DPG molecule. (a) Snapshots, (b) RMSD, and (c) binding energy of RBP4 adsorbing on LPG and DPG during MD simulations.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bioactmat.2020.09.027.
The following are the supplementary data related to this article:
LPG recognized RBP4 and reached an equilibrium state within 10 ns.
DPG recognized RBP4 and reached an equilibrium state within 10 ns.
4. Conclusions
In conclusion, chiral assembled nanofibers constructed by enantiomeric L/d-phenylalanine derivatives are employed to study chirality induced RPCs differentiation. Compared with left-handed LPG nanofibers, right-handed DPG nanofibers obviously promote neuronal differentiation, migration and synapse formation of RPCs, which can be attributed to the RA pathway activated by increased adhesion of RBP4 on DPG nanofibers. This is the first attempt to utilize the chiral structure of the extracellular microenvironment to induce RPCs neuronal differentiation. This study provides novel insights to control RPCs differentiation, although the bioeffects of chiral nanofibers in vivo remain to be explored. The achievement has profound significance for the treatment of refractory ocular diseases caused by retinal degeneration and other neurodegenerative diseases (e.g. Alzheimer's disease and Parkinson's disease).
Ethical approval and consent to participate
All the animals were not only treated according to ARVO standards but were also in experimental protocols approved by the Ethics Committee of the Ninth People's Hospital affiliated with Shanghai Jiao Tong University School of Medicine.
CRediT authorship contribution statement
Na Sun: Conceptualization, Methodology, Writing - original draft. Xiaoqiu Dou: Conceptualization, Methodology, Writing - original draft. Zhimin Tang: Investigation, Data curation. Dandan Zhang: Investigation, Data curation. Ni Ni: Investigation, Data curation. Jiajing Wang: Resources, Supervision, Validation, Resources, Investigation. Huiqin Gao: Resources, Supervision, Validation. Yahan Ju: Resources, Supervision, Validation. Xiaochan Dai: Resources, Supervision, Validation. Changli Zhao: Resources, Supervision, Validation. Ping Gu: Resources, Supervision, Validation. Jing Ji: Writing - review & editing. Chuanliang Feng: Writing - review & editing.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
The author thanks Dr. Xia Ronghui for his evaluation of the HE images. This research was funded by the National Nature Science Foundation of China (NSFC 51833006), the National Key R&D Program of China (2018YFC1106100, 2018YFC1106101), SJTU Trans-med Awards Research (WF540162603), the Innovation Program of Shanghai Municipal Education Commission (201701070002E00061), the Shanghai Municipal Commission of Health and Family Planning (201840073) and the Science and Technology Commission of Shanghai (17DZ2260100).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2020.09.027.
Contributor Information
Xiaoqiu Dou, Email: douxiaoqiu@sjtu.edu.cn.
Ping Gu, Email: guping2009@sjtu.edu.cn.
Jing Ji, Email: flowerrainday@sina.com.
Chuanliang Feng, Email: clfeng@sjtu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lorach H., Goetz G., Smith R., Lei X., Mandel Y., Kamins T., Mathieson K., Huie P., Harris J., Sher A., Palanker D. Nat. Med. 2015;21:476–482. doi: 10.1038/nm.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alves C.H., Pellissier L.P., Wijnholds J. Prog. Retin. Eye Res. 2014;40:35–52. doi: 10.1016/j.preteyeres.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Ji J., Zhang D.D., Wei W., Shen B.Q., Zhang Y., Wang Y.Y., Tang Z.M., Ni N., Sun H., Liu J.Q., Fan X.Q., Gu P. Cytotherapy. 2018;20:74–86. doi: 10.1016/j.jcyt.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Smith Q., Gerecht S. Cell Stem Cell. 2016;19:289–290. doi: 10.1016/j.stem.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Liu X., Carbonell C., Braunschweig A.B. Chem. Soc. Rev. 2016;45:6289–6310. doi: 10.1039/c6cs00349d. [DOI] [PubMed] [Google Scholar]
- 6.Mehlenbacher R.D., Kolbl R., Lay A., Dionne J.A. Nat. Rev. Mater. 2017;3:17080. [Google Scholar]
- 7.Yao X., Hu Y.W., Cao B., Peng R., Ding J.D. Biomaterials. 2013;34:9001–9009. doi: 10.1016/j.biomaterials.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 8.Zhao X.L., Xu L.G., Sun M.Z., Ma W., Wu X.L., Xu C.L., Kuang H. Nat. Commun. 2017;8:2007. doi: 10.1038/s41467-017-02268-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dou X.Q., Mehwish N., Zhao C.L., Liu J.Y., Xing C., Feng C.L. Acc. Chem. Res. 2020;53:852–862. doi: 10.1021/acs.accounts.0c00012. [DOI] [PubMed] [Google Scholar]
- 10.Sun T., Han D., Riehemann K., Chi L., Fuchs H. J. Am. Chem. Soc. 2007;129:1496–1497. doi: 10.1021/ja0686155. [DOI] [PubMed] [Google Scholar]
- 11.Tucker B.A., Redenti S.M., Jiang C.H., Swift J.S., Klassen H.J., Smith M.E., Wnek G.E., Young M.J. Biomaterials. 2010;31:9–19. doi: 10.1016/j.biomaterials.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Pritchard C.D., Arnér K.M., Neal R.A., Neeley W.L., Bojo P., Bachelder E., Holz J., Watson N., Botchwey E.A., Langer R.S., Ghosh F.K. Biomaterials. 2010;31:2153–2162. doi: 10.1016/j.biomaterials.2009.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Z.M., Jiang F., Zhang Y.H., Zhang Y., Yang Y., Huang X.L., Wang Y.Y., Zhang D.D., Ni N., Liu F., Luo M., Fan X.Q., Zhang W.A., Gu P. Biomaterials. 2019;194:57–72. doi: 10.1016/j.biomaterials.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 14.Dou X.Q., Wu B.B., Liu J.Y., Zhao C.L., Qin M.G., Wang Z.M., Schönherr H., Feng C.L. ACS Appl. Mater. Interfaces. 2019;11:38568–38577. doi: 10.1021/acsami.9b15710. [DOI] [PubMed] [Google Scholar]
- 15.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 17.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 18.Dou X.Q., Zhang J., Feng C.L. ACS Appl. Mater. Interfaces. 2015;7:20786–20792. doi: 10.1021/acsami.5b05828. [DOI] [PubMed] [Google Scholar]
- 19.Navarro-González J.F., Mora-Fernández C., Fuentes M.M., García-Pérez J. Nat. Rev. Nephrol. 2011;7:327–340. doi: 10.1038/nrneph.2011.51. [DOI] [PubMed] [Google Scholar]
- 20.Sendler M., Weiss F.U., Golchert J., Homuth G., van den Brandt C., Mahajan U.M., Partecke L.I., Döring P., Gukovsky I., Gukovskaya A.S., Wagh P.R., Lerch M.M., Mayerle J. Gastroenterology. 2018;154:704–718. doi: 10.1053/j.gastro.2017.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu T., Xue J., Xia Y. Angew. Chem. Int. Ed. Engl. 2020;59:15626–15632. doi: 10.1002/anie.202002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge W., He F., Kim K.J., Blanchi B., Coskun V., Nguyen L., Wu X., Zhao J., Heng J.I., Martinowich K., Tao J., Wu H., Castro D., Sobeih M.M., Corfas G., Gleeson J.G., Greenberg M.E., Guillemot F., Sun Y.E. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1319–1324. doi: 10.1073/pnas.0510419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franke K., Vlasits A. Science. 2020;368:1057–1058. doi: 10.1126/science.abc2294. [DOI] [PubMed] [Google Scholar]
- 24.Blaner W.S. Pharmacol. Therapeut. 2019;197:153–178. doi: 10.1016/j.pharmthera.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y.Y., Zhang D.D., Tang Z.M., Zhang Y., Gao H.Q., Ni N., Shen B.Q., Sun H., Gu P. Cell Death Dis. 2018;9:444. doi: 10.1038/s41419-018-0473-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chou C.M., Nelson C., Tarlé S.A., Pribila J.T., Bardakjian T., Woods S., Schneider A., Glaser T. Cell. 2015;161:634–646. doi: 10.1016/j.cell.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiser P.D., Golczak M., Palczewski K. Chem. Rev. 2014;114:194–232. doi: 10.1021/cr400107q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y., Clarke O.B., Kim J., Stowe S., Kim Y.K., Assur Z., Cavalier M., Godoy-Ruiz R., von Alpen D.C., Manzini C., Blaner W.S., Frank J., Quadro L., Weber D.J., Shapiro L., Hendrickson W.A., Mancia F. Science. 2016;353:aad8266. doi: 10.1126/science.aad8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowles J., Feng C.W., Miles K., Ineson J., Spiller C., Koopman P. Nat. Commun. 2016;7:10845. doi: 10.1038/ncomms10845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato S., Matsukawa T., Koriyama Y., Sugitani K., Ogai K. Prog. Retin. Eye Res. 2013;37:13–30. doi: 10.1016/j.preteyeres.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Cabezas-Wallscheid N., Buettner F., Sommerkamp P., Klimmeck D., Ladel L., Thalheimer F.B., Pastor-Flores D., Roma L.P., Renders S., Zeisberger P., Przybylla A., Schönberger K., Scognamiglio R., Altamura S., Florian C.M., Fawaz M., Vonficht D., Tesio M., Collier P., Pavlinic D., Geiger H., Schroeder T., Benes V., Dick T.P., Rieger M.A., Stegle O., Trumpp A. Cell. 2017;169:807–823. doi: 10.1016/j.cell.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 32.Yu D., Ma M., Liu Z.W., Pi Z.F., Du X.B., Ren J.S., Qu X.G. Biomaterials. 2020;255:120160. doi: 10.1016/j.biomaterials.2020.120160. [DOI] [PubMed] [Google Scholar]
- 33.Zhang R., Li Y., Hu B.B., Lu Z.G., Zhang J.C., Zhang X. Adv. Mater. 2016;28:6345–6352. doi: 10.1002/adma.201600554. [DOI] [PubMed] [Google Scholar]
- 34.Su G.G., Guo D.H., Chen J., Liu M., Zheng J., Wang W.B., Zhao X.Y., Yin Q.Q., Zhang L., Zhao Z.F., Shi J.D., Lu W.G. Nucleic Acids Res. 2019;47:6737–6752. doi: 10.1093/nar/gkz482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baranes K., Moshe H., Alon N., Schwartz S., Shefi O. ACS Chem. Neurosci. 2014;5:370–376. doi: 10.1021/cn500015s. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LPG recognized RBP4 and reached an equilibrium state within 10 ns.
DPG recognized RBP4 and reached an equilibrium state within 10 ns.