Abstract
Most patients with multiple myeloma (MM) die from progressive disease after relapse. To advance our understanding of MM evolution mechanisms, we performed whole-genome sequencing of 80 IGH-translocated tumour-normal newly diagnosed pairs and 24 matched relapsed tumours from the Myeloma XI trial. We identify multiple events as potentially important for survival and therapy-resistance at relapse including driver point mutations (e.g., TET2), translocations (MAP3K14), lengthened telomeres, and increased genomic instability (e.g., 17p deletions). Despite heterogeneous mutational processes contributing to relapsed mutations across MM subtypes, increased AID/APOBEC activity is particularly associated with shorter progression time to relapse, and contributes to higher mutational burden at relapse. In addition, we identify three enhanced major clonal evolution patterns of MM relapse, independent of treatment strategies and molecular karyotypes, questioning the viability of “evolutionary herding” approach in treating drug-resistant MM. Our data show that MM relapse is associated with acquisition of new mutations and clonal selection, and suggest APOBEC enzymes among potential targets for therapy-resistant MM.
Subject terms: Myeloma, Cancer genomics
Introduction
Multiple myeloma (MM) is caused by the expansion of clonal plasma cells in the bone marrow1. Over half of MM tumours have chromosomal translocations involving the immunoglobulin heavy chain locus, which leads to overexpression of oncogenes (CCND1, CCND3, MAF, MAFB, WHSC1/MMSET, and FGFR3) as an initiating event1. Despite recent advances, MM is essentially an incurable malignancy, and most patients die from progressive disease after multiple relapses irrespective of treatment. Our limited knowledge of the molecular changes associated with relapse is a barrier to developing new therapeutic strategies to overcome drug resistance.
To advance our understanding of the evolution of MM tumours and the mutational mechanisms that shape their history, we performed whole-genome sequencing (WGS) of 80 newly diagnosed MM tumour-normal pairs, 24 also had matched relapsed tumours2. WGS allowed us to examine the impact of noncoding mutations, complex structural rearrangements, and telomere structure on MM tumourigenesis analyses not possible in previous studies, which have been based on whole-exome sequencing (WES)3,4. Integrating information from multiple types of genomic alterations has allowed us to infer the order of mutational events, and show that relapse is associated with acquisition of new mutations and clonal selection.
Materials and methods
Samples
Bone marrow aspirates and blood samples were obtained from 80 patients with newly diagnosed MM being treated according to the UK National Cancer Research Institute Myeloma XI trial protocol2. Matched relapsed tumour DNAs were available for 24/80 primary patients. Tumour DNAs were extracted from plasma cells selected and sorted using CD138 microbeads as described, previously5. In all cases tumour purity was in excess of 30%. Germline DNA was derived from matched blood samples. Tumour IGH-translocation status was determined using multiplexed real-time PCR6. Hyperdiploid MM was defined as gain of at least two chromosomes as defined previously5. An entire chromosome was considered amplified if at least 90% of the chromosome overlapped with an amplification7. Clinical data and informed consent was obtained from all patients. Ethical approval for the study was obtained by the Oxfordshire Research Ethics Committee (MREC 17/09/09, ISRCTN49407852).
Whole genome sequencing
Sequencing libraries were prepared using Illumina SeqLab specific TruSeq Nano High Throughput library preparation kit (Illumina Inc, San Diego, CA 92122 USA), and paired end sequencing was conducted using Illumina HiSeqX technology. Raw WGS sequencing data were quality checked using FastQC (v.0.11.4) and aligned using the Burrows-Wheeler Alignment tool8 (BWA v0.7.13) to the human genome hg38 assembly using default parameters. Matching of tumour, normal, and relapsed samples was confirmed using NGSCheckMate9. Single nucleotide variants (SNVs) and indels were called using MuTect2 (v4.0.3.0)10 according to best practices, using The Genome Aggregation Database (gnomAD)11 file in GRCh38 provided as part of the GATK resource. Variants were filtered for cross-sample contamination, oxidation artefacts10, quality score7, and using a panel of normals generated from 80 germline samples. Variants with a germline population allele frequency >0.1% in gnomAD or in repetitive regions defined by University California Santa Cruz (UCSC) were excluded. Somatic indels were excluded if they were supported by <20% of tumour sample reads overlapping the position12 or were located within ten base pairs of a germline indel catalogued by gnomAD.
Reconstruction of clonal and subclonal copy number alterations (CNAs) for primary and relapsed tumours was conducted using Battenberg13. Since copy-neutral loss of heterozygosity (nLOH) is intrinsically more problematic to identify accurately14, these segments called by Battenberg were inspected manually against CNA calls overlapping within 10 Mb of two other CNA callers Sequenza15 and FACETS16. The copy number status of an nLOH segment was corrected and only reported if it was supported by at least two of the three CNA callers, and was excluded from downstream analysis if all methods were discordant. Tumour purity estimated by Battenberg was compared against and corrected using Ccube17. Somatic structural variants (SVs) were identified taking a consensus approach, as implemented by The Pancancer Analysis of Whole Genomes18, considering only variants identified by at least two of MANTA (v1.2.0)19, LUMPY (v0.2.13)20, or DELLY (v0.7.9)21. Chromothripsis regions were identified using ShatterSeek, adopting the criteria of at least four adjacent segments oscillating copy number states and at least six interleaved SVs22. All candidate chromothripsis regions were manually curated as previously advocated22. Chromoplexy was detected using ChainFinder (v1.0.1) with default parameters23 and hg38 UCSC cytoband definitions (http://hgdownload.cse.ucsc.edu/goldenpath/hg38/database/). As previously advocated22, chromoplexy was only called when at least three chromosomes were involved in a chain of SVs. Telomere length was estimated using Telomerecat24 with default parameters. Kataegis foci were identified using the KataegisPortal with default parameters (https://github.com/MeichunCai/KataegisPortal), and defined as having six or more consecutive mutations with an average mutational distance ≤1 Kb, excluding immune hypermutated regions25.
Identifying driver mutations
Coding drivers were identified using dNdScv with default parameters26. Nonsilent mutations in a curated list of 82 established coding drivers7,27 and all coding genes were compared in matched primary and relapsed tumours. To identify noncoding drivers we analysed promoter and cis-regulatory regions (CREs) as described previously7. Briefly, promoters were defined as intervals spanning 400 bp upstream and 250 bp downstream of transcription start site from GENCODE (release 25)28. CREs were defined using promoter capture Hi-C data generated on naïve B-cells29. Raw sequencing reads from European Genome-Phenome Archive (EGA; accession code EGAS00001001911) were aligned to hg38 using HiCUP (v0.6.1)30 and promoter–CRE interactions were called with CHiCAGO (v1.8)31. Only interactions with linear distance ≤1 Mb and CHiCAGO score ≥5 were considered7.
Recurrently mutated promoters and CREs were identified using a Poisson binomial model as previously described7,32, taking into account tumour ID, trinucleotide context, and replication timing. For CRE regions, mutations were excluded if they overlap with open reading frames, 5′-UTR, and 3′-UTR as defined by Ensembl7. For promoters, mutations overlapping with open reading frames were excluded. Replication timing was estimated as the average of two B-lymphocyte replicates33,34. For promoters and CREs mutated in ≥3 samples, the clustering of mutations was examined using a permutation approach considering the number of mutations occurring at the same nucleotide position as previously described7. For each promoter and CRE, a combined P-value from the mutational recurrence and clustering analyses were obtained using Fisher’s method7,35. The Benjamini–Hochberg false discovery rate (FDR) procedure was used to adjust for multiple testing with significant threshold at Q < 0.05. Promoters and CREs overlap with immune hypermutated regions were excluded to avoid false positives. We only report CREs and promoters mutated in at least three tumours.
Impact of cereblon and IMiD response pathway genes mutation on relapse
All patients we studied were treated with immunomodulatory drugs (IMiDs), either thalidomide or lenalidomide. Mutations in CRBN and associated genes have been proposed as being a mechanism of acquired drug resistance to IMiDs36,37. To examine this proposition, we specifically considered nonsynonymous mutations, CNAs, and SVs disrupting a curated list of 42 CRBN/IMiD genes—genes involved in the CRBN pathway regulation and IMiD response (Supplementary Table 1).
Chronology of mutational events
The chronological timing of SNVs and CNAs was estimated independently for the 80 primary tumours as previously described38. Briefly, for SNVs we considered only driver genes mutated in ≥4 samples to allow reliable estimation of relative timing. For CNAs we considered only large-scale autosomal events (≥3 Mb) present in ≥8 samples38. Cytobands were assigned based on UCSC hg38 definitions. One sample (8573) displayed hyperdiploid characteristics and was excluded from the analysis. Cancer cell fractions (CCFs) of each CNV event and SNV were estimated using Battenberg13. Each cytoband or driver gene was ordered by mean of CCF from highest to lowest. The Tukey’s range test and a stepwise approach were used to test for difference between the CCF means of consecutive cytobands or driver genes to define discrete clonality levels, as described previously38. As previously advocated38, 95% confidence intervals were calculated with basic bootstrap method with 1000 iterations using boot R package.
Analysis of copy number changes
Permutation was used to test the null hypothesis that the frequency of particular chromosome arm copy number events does not differ between primary and relapse MM. We first counted change in frequency of affected tumours at primary and relapse. We then randomly swapped condition labels for all matched primary and relapsed tumours 10,000 times, and recounted change in chromosome arm event frequency. Empirical P-values for each chromosome arm event were calculated as fraction of permutations with absolute net frequency change at least as great as the absolute net frequency change observed in the true primary/relapse labelling. We only considered chromosome arm events with net change in frequency in at least two tumours.
We employed a permutation-based approach to test the null hypothesis that additional relapse-associated CNA events occur by chance at pre-existing unstable genomic regions. For each autosomal chromosome arm, we counted the number of tumours with additional large-scale CNA on the considered chromosome arm at relapse. The tested chromosome arm in considered tumours with further CNA change were permutated 10,000 times among 44 possible chromosome arms loci (22 autosomal chromosomes with either p or q arm). The empirical P-values were calculated as the fraction of permutations with the number of additional CNA change were at least as great as the original tested chromosome arm.
Mapping evolutionary trajectories
Analysis of clonality was conducted using only SNVs in diploid regions, as miscalled copy number states can confound the analysis. Potential neutral tail mutations were identified using MOBSTER39 and excluded prior to clustering procedure to minimise calling false positive clones. For each primary and relapse tumour pair, we performed two-dimensional variant clustering using a Bayesian Dirichlet process implemented in DPclust3,13. Only those clusters with ≥1% of total mutations and ≥100 SNVs were considered. Muller plots were generated with Timescape R package version 1.10.0. For each cluster in primary tumour and matched relapse, the proportion of SNVs shared was calculated.
Mutational signatures
De novo extraction of signatures was performed on 80 primary and 24 relapsed genomes separately using non-negative matrix factorization40. We compared de novo mutational signatures with Catalogue of Somatic Mutations in Cancer (COSMIC) single base substitution (SBS) signatures version 3 by computing their cosine similarities41. A de novo mutational signature was assigned to a COSMIC signature if the cosine similarity was >0.75 as advocated12. We next performed signature fitting using deconstructSigs42 considering only those COSMIC signatures extracted de novo, as previously recommended43. In view of potential ambiguous assignment, we combined the contributions of the flat profile signatures 5, 8, and 4025,42,43, excluding signature 3 as this signature is unlikely to be active in MM43. As previously advocated, we compared mutational signature proportions in paired primary and relapsed samples using the chi-squared test13. Association between changes in mutational burden and AID/APOBEC mutational contribution for paired primary and relapsed tumours was calculated using Fisher’s exact test. Spearman correlation was performed to test the association between AID/APOBEC contribution of relapse-specific mutations and time to relapse.
Results
We carried out WGS on 80 newly diagnosed MM tumour-normal pairs from the Myeloma XI trial, and matched relapsed tumour from 24 patients. The 80 patients had either t(4;14) (n = 38), t(11;14) (n = 38), or t(14;16) (n = 4) MM, with one patient carrying both t(4;14) translocation and trisomy of chromosomes 9 and 15 (Table 1). Hyperdiploid (HD) and non-HD subtypes of MM have distinctive genomic landscapes and are a priori likely to have different evolutionary trajectories1. In this study, we restricted our analysis to IGH-translocated tumours to focus on examining evolutionary dynamics of non-HD myeloma. WGS resulted in a median of 38× coverage for normal samples (30–44×), 111× for primary tumours (82–155×), and 114× for the 24 relapsed tumours (102–154×) (Supplementary Table 2). 6 of the 80 patients have been the subject of a previous WES project4.
Table 1.
Summary of demographic and treatment data.
| Sample ID | Karyotype | Gender | Age | Elapsed time (months) | Induction | Maintenance | Pathway |
|---|---|---|---|---|---|---|---|
| 1305 | 11;14 | Male | 51 | 38.34 | CTD | No | Intensive |
| 1334 | 11;14 | Female | 43 | 24.00 | CTD | Missing | Intensive |
| 5834 | 11;14 | Female | 69 | 29.93 | CTDa | No | Nonintensive |
| 6030 | 4;14 | Female | 36 | 19.75 | CTD | No | Intensive |
| 6178 | 11;14 | Female | 67 | 18.40 | RCD | Missing | Intensive |
| 6229 | 11;14 | Male | 74 | 9.23 | CTDa | Missing | Nonintensive |
| 6706 | 11;14 | Male | 59 | 25.43 | RCD | No | Intensive |
| 6988 | 11;14 | Male | 69 | 12.26 | RCDa | No | Nonintensive |
| 7020 | 4;14 | Female | 58 | 14.69 | CTD | Missing | Intensive |
| 7240 | 4;14 | Male | 55 | 11.30 | RCD | Lenalidomide | Intensive |
| 7801 | 14;16 | Female | 48 | 14.49 | CTD | Missing | Intensive |
| 7842 | 4;14 | Male | 66 | 17.64 | CTD | No | Intensive |
| 8237 | 4;14 | Female | 49 | 14.00 | CTD | No | Intensive |
| 9126 | 11;14 | Male | 64 | 16.23 | CTDa | Missing | Nonintensive |
| 9166 | 14;16 | Female | 68 | 27.24 | CCRD | No | Intensive |
| 9515 | 11;14 | Male | 68 | 26.15 | RCDa | Lenalidomide | Nonintensive |
| 9721 | 14;16 | Male | 64 | 29.44 | CTD | Lenalidomide | Intensive |
| 10,068 | 4;14 | Male | 71 | 13.77 | RCDa | Lenalidomide and Vorinostat | Nonintensive |
| 10,365 | 11;14 | Male | 76 | 9.33 | CTD | Missing | Intensive |
| 11,506 | 14;16 | Male | 77 | 11.83 | CTDa | Lenalidomide | Nonintensive |
| 11,668 | 4;14 | Male | 49 | 19.29 | RCDa | Missing | Nonintensive |
| 11,949 | 11;14 | Male | 76 | 14.65 | CTD | Missing | Intensive |
| 12,546 | 4;14 | Male | 77 | 30.59 | RCD | Missing | Intensive |
| 13,029 | 4;14 | Male | 62 | 6.90 | CTD | Missing | Intensive |
| 5695 | 11;14 | Male | 64 | NA | CTD | No | Intensive |
| 5699 | 11;14 | Female | 68 | NA | CTD | Missing | Intensive |
| 5836 | 11;14 | Male | 77 | NA | CTDa | No | Nonintensive |
| 5939 | 4;14 | Male | 65 | NA | CTD | Missing | Intensive |
| 6016 | 11;14 | Female | 55 | NA | RCD | Missing | Intensive |
| 6076 | 4;14 | Male | 72 | NA | RCDa | Lenalidomide | Nonintensive |
| 6163 | 4;14 | Male | 75 | NA | RCDa | Missing | Nonintensive |
| 6277 | 11;14 | Male | 56 | NA | RCD | Lenalidomide | Intensive |
| 6279 | 4;14 | Male | 62 | NA | RCD | Lenalidomide | Intensive |
| 6345 | 4;14 | Female | 72 | NA | CTDa | Missing | Nonintensive |
| 6415 | 11;14 | Female | 68 | NA | RCDa | Missing | Nonintensive |
| 6425 | 4;14 | Male | 67 | NA | RCD | Lenalidomide and Vorinostat | Intensive |
| 6501 | 11;14 | Female | 51 | NA | RCD | Missing | Intensive |
| 6702 | 4;14 | Female | 78 | NA | CTDa | Missing | Nonintensive |
| 7000 | 11;14 | Female | 78 | NA | CTDa | Missing | Nonintensive |
| 7005 | 4;14 | Male | 74 | NA | CTDa | Missing | Nonintensive |
| 7164 | 11;14 | Female | 80 | NA | RCDa | Missing | Nonintensive |
| 7348 | 4;14 | Male | 67 | NA | RCDa | No | Nonintensive |
| 7729 | 4;14 | Male | 65 | NA | RCD | Lenalidomide and Vorinostat | Intensive |
| 7794 | 4;14 | Female | 52 | NA | CTD | No | Intensive |
| 7880 | 4;14 | Female | 82 | NA | RCDa | Missing | Non-intensive |
| 7915 | 4;14 | Male | 59 | NA | CTD | Lenalidomide and Vorinostat | Intensive |
| 7925 | 4;14 | Male | 59 | NA | CTD | Missing | Intensive |
| 7950 | 4;14 | Male | 49 | NA | CTD | Lenalidomide and Vorinostat | Intensive |
| 7956 | 4;14 | Female | 56 | NA | CTD | Missing | Intensive |
| 8043 | 4;14 | Female | 81 | NA | CTDa | Missing | Non-intensive |
| 8245 | 11;14 | Female | 63 | NA | RCD | Lenalidomide | Intensive |
| 8567 | 11;14 | Female | 66 | NA | RCDa | Lenalidomide and Vorinostat | Nonintensive |
| 8573 | 4;14/HD | Female | 82 | NA | CTDa | Missing | Nonintensive |
| 8928 | 4;14 | Male | 52 | NA | CTD | Missing | Intensive |
| 8979 | 4;14 | Male | 76 | NA | CTDa | Missing | Nonintensive |
| 9069 | 11;14 | Male | 73 | NA | RCDa | Missing | Non-intensive |
| 9176 | 11;14 | Male | 78 | NA | RCDa | Missing | Nonintensive |
| 9210 | 11;14 | Male | 69 | NA | CTD | Missing | Intensive |
| 9249 | 11;14 | Male | 58 | NA | RCD | Lenalidomide | Intensive |
| 9289 | 11;14 | Male | 56 | NA | CTD | No | Intensive |
| 9292 | 4;14 | Female | 74 | NA | CTDa | Missing | Nonintensive |
| 9337 | 11;14 | Female | 71 | NA | CTDa | Missing | Nonintensive |
| 9376 | 4;14 | Female | 64 | NA | RCD | Missing | Intensive |
| 9409 | 11;14 | Male | 73 | NA | CTDa | Missing | Nonintensive |
| 9524 | 4;14 | Male | 51 | NA | RCDa | Lenalidomide | Nonintensive |
| 9544 | 11;14 | Male | 67 | NA | RCDa | No | Nonintensive |
| 9623 | 11;14 | Male | 58 | NA | RCD | Lenalidomide | Intensive |
| 9718 | 4;14 | Male | 66 | NA | RCDa | No | Nonintensive |
| 9917 | 11;14 | Male | 76 | NA | CTDa | Missing | Nonintensive |
| 9931 | 11;14 | Female | 55 | NA | RCD | Missing | Intensive |
| 10,085 | 11;14 | Female | 59 | NA | CCRD | Lenalidomide | Intensive |
| 10,212 | 11;14 | Female | 79 | NA | RCDa | Lenalidomide | Nonintensive |
| 10,597 | 4;14 | Male | 59 | NA | CCRD | No | Intensive |
| 10,772 | 4;14 | Female | 63 | NA | CCRD | Missing | Intensive |
| 10,801 | 11;14 | Male | 77 | NA | RCDa | Missing | Nonintensive |
| 11,029 | 4;14 | Female | 73 | NA | RCDa | Missing | Nonintensive |
| 11,897 | 4;14 | Male | 58 | NA | CCRD | Lenalidomide | Intensive |
| 12,101 | 4;14 | Male | 62 | NA | CCRD | Missing | Intensive |
| 12,227 | 11;14 | Male | 57 | NA | CCRD | No | Intensive |
| 12,541 | 11;14 | Male | 56 | NA | CTD | Missing | Intensive |
CTD cyclophosphamide, thalidomie, and dexamethasone, CTDa CTD with a reduced dose of dexamethasone and lower starting dose of thalidomide, RCD Lenalidomide (Revlimid), cyclophosphamide, and dexamethasone, RCDa RCD with a reduced dose of dexamethasone, CCRD carfilzomib, cyclophosphamide, lenalidomide, and dexamethasone. Intensive pathway: treatment with high dose melphalan after induction. NA: Matched relapsed data are not available.
Mutational events in primary tumours
We began by surveying for important genetic alterations in the 80 primary MM tumours by considering the contribution of both protein-coding and noncoding SNVs and indels, as well as CNAs. As expected, significantly mutated genes (Q < 0.05) at presentation were DIS3, KRAS, NRAS, FGFR3, MAX, CCND1, TP53, IRF4, and PRKD2 (Fig. 1a and Supplementary Table 3). The promoters of 17 genes including BCL6, CXCR4, BIRC3, MYO1E, CRIP1, FLT3LG, and DPP9 were also significantly mutated as well as nine cis-regulatory elements (CREs) interacting with genes including PAX5, BCL6, ZCCHC7, and IFNGR1 (Supplementary Fig. 1 and Supplementary Tables 4, 5). The most frequent large-scale CNAs were deletion of 13q (73%), 22q (35%), and 1p (35%); and gain of 1q (45%). (Fig. 1a and Supplementary Fig. 2 and Supplementary Table 6). Aberrations of 13q was enriched in high-risk t(4;14) and t(14;16) MM (P = 3.5 × 10−5, odd ratio = 16.2, Fisher’s exact test).
Fig. 1. Frequency and chronology of coding drivers and major copy number events.
a Frequency of coding drivers and major copy number events (present in at least eight tumours) detected in 80 primary tumours; b, c Chronology of coding drivers and major copy number events, respectively. Red dots denote mean of cancer cell fractions (CCFs) for each event with blue lines indicating 95% confidence intervals of the relative timing. Bootstrap confidence intervals were estimated based on the cancer cell fractions of mutational events. X-axis is plotted as relative timing based on CCF contribution. Dotted red lines denote discrete clonality events. Frequency: number of tumours with each mutational event; Ins insertion, Del deletion, LOH loss of heterozygosity.
Chromothripsis was observed in 18/80 primary tumours (23%) with the most frequently affected chromosomes are 1 (4 tumours), 8, 11, and 22 (3 tumours) (Supplementary Fig. 3); whereas 3% (2/80) of primary tumours featured chromoplexy (Supplementary Fig. 4). The frequency of chromothripsis and chromoplexy identified is comparable to a previous report44. Chromoplexy resulted in the simultaneous disruption of multiple driver genes7,27 (KRAS, PRKD2, PTPN11, PTH2, BAX, CELA1, FTL, ARID2, and CDKN1B) in primary tumours. Overall across the 80 primary tumours, high-risk subtypes MM t(4;14) and t(14;16) were associated with a shorter telomeres (P = 9.2 × 10−5, Wilcoxon rank-sum test) (Supplementary Fig. 5).
By integrating somatic mutations and copy number profiles we inferred the relative timing of key driver alterations in MM (i.e., which events occur earlier relative to others). Mutations of CCND1, MAX, PRKD2, DIS3, and NRAS were identified as early events whereas mutations of KRAS, IRF4, FGFR3, TP53, and TET2 occurred as later events (Fig. 1b). Chronological timing of major CNAs (present in ≥10% of total samples)38 identified 21q gain and 13q deletion as being early events (Fig. 1c), consistent with a previous report that 13q deletions tend to be clonal45. 1p deletion and 1q gain, which has been linked to patient prognosis were identified as later events (Fig. 1c).
Mutational landscape of relapse
We next investigated the molecular features of MM relapse by analysis of the 24 primary-relapse pairs. Patients received cyclophosphamide and dexamethasone in combination with either thalidomide (CTD), lenalidomide (RCD), or both carfilzomib and lenalidomide (CCRD) as induction therapy. Fit and young patients received high-dose melphalan (intensive pathway). 9 of the 25 patients subsequently received lenalidomide maintenance therapy. Treatment histories of each patient are summarized in Table 1. None of the patients we studied had detectable CRBN mutations at relapse. We did, however observe increased IKZF3 mutation CCF and de novo mutations disrupting CRBN/IMiD genes in two patients at relapse—RBX1 mutation and copy number loss affecting UBE2A (Supplementary Table 7). Relapse was associated with a higher mutational burden than primary tumours (Supplementary Fig. 6a–b, P < 0.01, paired Wilcoxon rank-sum test). Varied proportions (9–63%) of SNVs and indels identified in primary tumours were not detectable at relapse (Supplementary Fig. 6c), suggesting eradication and heterogenous clonal dynamics of the respective clone. Despite the increased mutational burden, relapsed tumours did not exhibit significantly more kataegis (Supplementary Fig. 7 and Supplementary Table 8). Chromothripis and chromoplexy were each observed in only one additional relapsed tumour (7842 and 8237 respectively; Supplementary Figs. 8 and 9). Although both primary and relapsed tumours had shorter telomeres compared to plasma cells (P < 0.01, paired Wilcoxon rank-sum test), relapse was associated with longer telomeres (P = 5.3 × 10−3) (Supplementary Fig. 10).
A translocation bringing the IGH loci in proximity to MAP3K14 was gained at relapse in one tumour (Supplementary Fig. 11). Driver genes additionally mutated at relapse included FAM46C, TRAF2, LTB, FAM154B, NF1, XBP1, and IDH2 (Supplementary Fig. 12). Driver mutations most frequently acquired at relapse were those in KRAS and NRAS, detected in three and two tumours respectively. The increase in CCF of TET2 mutations implied selection of subclones (Supplementary Fig. 13). The promoters and CREs of an additional 16 genes were significantly mutated at relapse, including genes with established roles in the biology of MM or other B-cell malignancies such as XBP1, BCL7A, and BCL9 (Supplementary Tables 9 and 10).
Relapse was associated with additional CNAs, most frequently for 17p deletion (P < 2.2 × 10−6) (Fig. 2a, Supplementary Fig. 14, and Supplementary Table 11). We observed additional CNAs occurring at pre-existing unstable genomic regions, including the progression of copy-neutral loss of heterozygosity (nLOH) to LOH, LOH to complete deletion; as well as further copy number gains (Fig. 2b and Supplementary Fig. 15). Such trend was observed at a higher rate than expected by chance at 11q (P = 0.042) and 14q (P = 0.023) (Fig. 2c).
Fig. 2. Copy number alterations associated with relapse.
a Net change of CNA frequency in primary and matched relapse tumours; red and blue bars represent positive and negative changes respectively. Only significant events with changes in at least two tumours are shown. b Copy number profiles of patients 7842, 9166, and 9515. In 7842 copy number neutral loss of heterozygosity (nLOH) at chromosome 4 becomes LOH at relapse. In 9166 LOH at 13q progresses to complete loss of 13q. In 9515 copy number gain at chromosome 10 and 11 progresses to additional chromosome gain. Thick and thin lines represent clonal and subclonal copy number states, respectively. Yellow and blue lines denote total and minor copy number respectively (copy number states >5 not shown). c Patterns of copy number change across paired primary-relapse samples at 11q and 14q. Lines indicate relationship between primary and matched relapse tumours, with width being proportional to event frequency. Only chromosome arms with copy number alterations (CNAs) are plotted, with a copy number of 2 corresponding to nLOH.
Mutational processes active at relapse
At diagnosis, the major mutational signatures in tumours were those indicative of aging (SBS5), AID/APOBEC (SBS2, 9, and 13), and flat signatures (SBS5, 8, and 40) as previously observed7,25 (Supplementary Figs. 16 and 17). No additional mutational signatures potentially specific to treatment were extracted at relapse (Supplementary Fig. 18). Across all patients, we observed heterogeneous dynamic of mutational processes contributing to relapse (Supplementary Fig. 19). However, tumours with increased mutational burden at relapse were often associated with increased AID/APOBEC enzymes activity (P = 0.061, Fisher’s exact test). Despite the enrichment of APOBEC signatures in t(14;16) MM (P = 0.017, Wilcoxon rank-sum test) (Supplementary Fig. 17), we did not observe specific association of the signatures at relapse in this subtype (P = 0.20, Wilcoxon rank-sum test), consistent with previous finding46. Notably, patients with higher AID/APOBEC mutational contribution at relapse were associated with shorter refractory time (r = −0.43, P = 0.037, Spearman correlation) (Supplementary Figure 20). An increased C•G > G•C transversion rate in relapse-specific mutations was also observed (Q = 0.015, paired Wilcoxon rank-sum tests) (Supplementary Fig. 21), a feature previously reported in relapsed acute myeloid leukaemia47.
Evolutionary trajectories of relapse
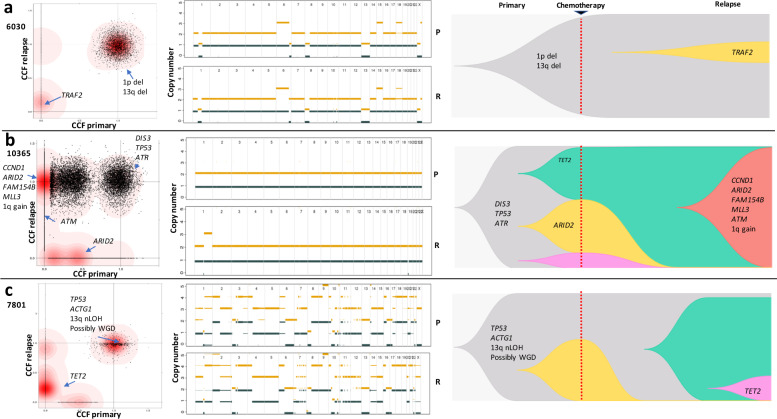
Three patterns of clonal evolution were apparent at relapse (Fig. 3). In Pattern 1 (3/24 patients), the dominant clone in primary survives treatment and gains additional mutations at relapse (Fig. 3a, Supplementary Fig. 22a). Tumours with Pattern 1 are characterised with no change in clonal composition of the dominant clones, suggesting that they were potentially unaffected by treatment. Pattern 2 (4/24 patients) is featured by subclonal expansion whereby a subclone in the primary survives treatment, and expands to become the dominant clone at relapse (Fig. 3b and Supplementary Fig. 22b). Tumours with Pattern 2 are also accompanied with “branching evolution” feature, where new clones emerge while others are lost. We suspect these clones might have mutations (e.g., TET2 and 6q deletion) giving them survival and selective advantage. Pattern 3 (17/24 patients) is characterised by the emergence of new clones at relapse, accompanied by the disappearance or decline of primary clones (Fig. 3c and Supplementary Fig. 22c). The three patterns of clonal evolution were not associated with therapy strategies (intensive versus nonintensive pathways) or molecular karyotypes (Fisher’s exact test). It was, however, of note that time to relapse was shorter with Pattern 2 (median 11.6 versus 19.3 months, P = 0.019, Wilcoxon rank-sum test).
Fig. 3. Evolutionary trajectories of relapse.
a Pattern 1 (3/24), dominant clone in primary survives treatment and gains additional mutations at relapse; b Pattern 2 (4/24), subclone in primary survives treatment and expands to become dominant clone at relapse; c Pattern 3 (17/24), eradication or decrease in frequency of one or more clones in primary and emergence of new clones not previously detected in primary. Left panels, two-dimensional density plots showing clustering of mutations by cancer cell fraction (CCF) in primary and relapse tumours. Darker red areas indicate location of a high posterior probability of a cluster. Clusters are annotated with coding driver mutations and major copy number alterations. Pattern 1: no disappearance of primary clusters on the horizontal axis accompanied by appearance of new clusters on the vertical axis. Pattern 2: existence of cluster positioned on the vertical top and horizontal centre. Pattern 3: disappearance of clusters on the horizontal axis accompanied by appearance of clusters on the vertical axis. Central panels, chromosomal copy-number profiles of primary (upper) and relapse (lower) tumours. Thick and thin lines represent clonal and sub-clonal copy number states respectively. Yellow and dark blue lines denote total and minor copy number alleles. Right panels, Muller plots of evolutionary trajectories. P primary, R relapse. WGD Whole genome duplication.
Discussion
Using high-depth WGS, we provide for an enhanced genetic model of the development and progression of MM. Our study expands upon previous findings, which have been based on WES/targeted sequencing3,4,36,46,48,49, low coverage WGS50, or fluorescence in situ hybridization and/or array technology46,51. While we have restricted our analysis to MM with an initiating translocation, our findings provide evidence for a common origin of tumour subpopulations with many tumours being composed of at least one subclone, reflecting the clonal heterogeneity present in both primary and relapse.
In addition to known coding drivers, we extend the number of potential non-coding drivers in MM, including those associated with CXCR4 and BIRC3. Somatic mutations in BCL6 promoters are common in MM52; however, since the gene is a common target of normal activation-induced deaminase (AID) in the germinal centre53, the relevance of these promoter mutations to MM biology is questionable. Noncoding regulatory regions additionally disrupted at relapse, included those targeting XBP1, RBX1, and SCML1. Common pathways affected by coding and noncoding mutations arising in MM relapse included those associated with WNT-signalling, MAPK-signalling, and NOTCH-signalling, base excision repair, cell cycle, telomere maintenance, and cellular senescence (Table 2). Notably, relapse was characterised by frequent additional CNAs, the most common being 17p deletion. Since the additional CNAs often occurred at unstable genomic regions such as 11q and 14q, it suggests increased chromosome instability are important means to escape therapy, analogous to that seen with chronic myeloid leukaemia in response to imatinib54. Our findings suggest that 21q gain, 13q deletion, and mutation of CCND1, MAX, PRKD2, DIS3, and NRAS are early events. The chronology of coding events identified from our study are broadly consistent with previous WES-based analyses1,55,56, any discrepancies are likely to be a consequence of sample size, representation of MM subtype, and number of coding drivers considered.
Table 2.
Summary of relapse-specific coding driver mutations, promoter mutations, CRE mutations, driver translocations, and large-scale genomic changes identified in 24 primary tumour-relapse pairs grouped by subtype.
| Subtype | Coding drivers | Promoters | CREs | Driver translocations | Frequent large-scale genomic changes |
|---|---|---|---|---|---|
| t(4;14) | KRAS; TP53; FGFR3; FAM46C; TRAF2; NF1; XBP1 | MTFRL1; FLT3LG; IL12A; POLG; XBP1; B3GALNT1; ALG10B | ABCA10; ABCA5 | MAP3K14 t(17,14)(q21,q32) |
17p deletion Further copy number changes at unstable genomic regions (11q and 14q) Increased telomere length |
| t(11;14) | PRDM1; LTB; IDH2; KRAS; NRAS; CCND1; ATM; FAM154B; MLL3 | RBX1; FAM81A; POLG; KCTD13; SCML1 | SCAF8 | ||
| t(14;16) | NRAS; TET2 | MYO1E; ALG10B; TMSB4X; KCTD13; SCML1 |
CRE cis-regulatory element.
Overall, the mutational load was higher in relapse MM and aberrations previously linked to MM resurfaced in both primary pretreatment and relapse tumours in our cohort, including mutations in RAS genes, DIS3, TP53, FGFR3, and PAX5 CRE mutations. As well as highlighting mutation of genes with established roles in MM, we identified a number of frequently acquired de novo coding mutations (e.g., FAM46C, TRAF2, NF1, and XBP1), de novo translocation (MAP3K14) and pre-existing mutations (e.g., TET2). Longer telomeres at relapse could be associated with treatment as observed in chronic myeloid leukemia57. Therapy targeting telomerase/telomeres should be further explored in MM as lengthened telomeres may provide a mechanism for treatment resistance58.
By performing high-depth WGS, we have been able to better refine the patterns of genomic evolution at relapse in MM compared to previous studies3,4. Notably, the “branching evolution” and “differential clonal response” models described by Bolli et al.3 often co-occurred as one single model (Pattern 2) in our analysis. Additionally, we did not find evidence for an association between t(11;14) MM with a “no change/linear” model3. The study by Jones et al. which included a small number of overlapping cases failed to identify Pattern 2 whereby a subclone survives treatment and expands at relapse4. Insights into tumour evolution has the potential to inform clinical decisions59. “Evolutionary herding”, in which clonal composition of tumours is tunnelled by a treatment to increase their sensitivity to another treatment, has been proposed as a strategy to combat treatment-resistance in tumours60. Despite a limited number of samples, we found little evidence that the evolutionary trajectory of MM is solely dictated by molecular karyotype or significantly influenced by current therapeutic strategies, questioning the viability of “evolutionary herding” in controlling drug resistance in MM. It was however noteworthy that Pattern 2 was associated with significant shorter time to relapse. Going forward, further strategies should be explored to accurately predict tumour dynamics and tailor patient therapy61.
Higher proportion of C•G>G•C at relapse is associated with DNA damage by oxidative stresses62, possibly due to oncogene activation and/or enhanced metabolism in relapsed MM63. AID/APOBEC activity contributes to increased mutational burden and associated with shorter time to relapse. APOBEC mutagenesis has been shown to promote survival and therapy escape in cancer through driving subclonal diversity, immune evasion, and genomic instability64. Collectively, these data suggest APOBEC family enzymes as potential therapeutic targets for treatment-resistance MM.
Inevitably, due to technical limitations, our ability to detect mutations in rare cells (mostly related to currently achievable levels of coverage with WGS) and spatial sampling constraints, our models potentially underestimate clonal heterogeneity in MM. We did however observe the loss of primary tumour clones at relapse in 21 of 24 cases, suggesting that some subclones are eradicated by therapy (Supplementary Fig. 22). Nevertheless, treatment failed to eradicate the founding clone in all cases. Our data also imply the acquisition of new mutations, which subsequently undergo selection and clonal expansion, potentially contributing to disease progression. It is likely that some mutations gained at relapse may alter the growth properties of MM cells, or confer resistance to additional chemotherapy.
Presently strategies to improve the poor cure rates of relapsing MM are limited. The forces shaping the evolutionary trajectory of MM have relevance to informing patient management. Williams et al. proposed that following a “big bang”, neutral evolution is a major feature of many cancers65. Application of same model to MM exome sequencing data suggested that neutral evolution is also a significant feature of MM66. Serious criticism has however been levelled at the assumptions on which the Williams et al. model is predicated67–70. In the light of such critique, as well as findings from our current WGS analysis and MM sequencing studies performed by other researchers71, it is apposite to reappraise the role of neutral evolution in MM. It seems highly unlikely neutral evolution is a dominant evolutionary force in MM and its evolutionary trajectory is essentially Darwinian-shaped by selection and subsequent expansion of diverse clones in patients.
MM cells routinely acquire a small number of additional mutations at relapse, and some of these mutations may contribute to clonal selection and therapy resistance. While mutations in CRBN and associated genes have been implicated as a mechanism of acquired drug resistance to IMiDs, our analysis suggests mutation per se is unlikely to be a universal basis of acquired IMiD resistance. This does not preclude epigenetic alterations, which are a feature of relapse influencing drug transport, escape from apoptosis, and dysregulated intracellular signalling pathways, all of which can contribute to resistance72.
Here, we have demonstrated that relapsed MM harbour significantly more mutations than primary tumours and clonal selection of mutations occurs at relapse, which are accompanied by subclonal heterogeneity. Theoretically, these data provide a rationale for identifying disease-causing mutations for MM, which may be amenable to targeted therapies to avoid the use of cytotoxic drugs, many of which are mutagens. However, it remains to be determined whether the current arsenal of therapies directed against downstream effectors of mutated genes will be effective given that the MM genome in an individual patient is likely to be continuously evolving. It is conceivable that in the near future, chemotherapy-based regimens may be relegated to fifth or sixth line treatment after patients have failed proteasome inhibitors, IMiDs and/or immunotherapy. Although speculative, however successful immunotherapy will be in an individual patient, Darwinian evolution of MM would imply that such therapy is unlikely to affect cure. It is therefore likely that eradication of the founding clone, as well as all of its subclones, will be required to effect complete cure.
Supplementary information
Acknowledgements
This work was supported by grants from Myeloma UK, Bloodwise and Cancer Research UK (C1298/A8362). We are grateful to the NCRI Haemato-oncology subgroup and to all investigators for recruiting patients to Myeloma XI. These data were generated as part of the Myeloma XI trial. M.K. is supported by a fellowship from the David Forbes-Nixon Foundation.
Author contributions
P.H.H., M.K., and R.S.H. conceived and designed the study; P.H.H., A.J.C., D.C., and B.K. performed bioinformatics; A.S. and S.K. generated data; G.J., G.J.M., and G.C. provided samples; P.H.H. and R.S.H. wrote the manuscript with contributions from A.J.C., M.K., and D.C. All authors reviewed the final manuscript.
Data availability
Raw promoter capture Hi-C data for naïve B-cells were obtained from European Genome-Phenome Archive (EGA; accession code EGAS00001001911). Replication timing data for B-lymphocytes was downloaded from Replication Domain Database34. Raw WGS data generated as part of this study can be accessed through EGA accession code EGAD00001005491.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Martin Kaiser, Email: Martin.Kaiser@icr.ac.uk.
Richard S. Houlston, Email: Richard.Houlston@icr.ac.uk
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41408-020-00367-2).
References
- 1.Manier S, et al. Genomic complexity of multiple myeloma and its clinical implications. Nat. Rev. Clin. Oncol. 2017;14:100–113. doi: 10.1038/nrclinonc.2016.122. [DOI] [PubMed] [Google Scholar]
- 2.Shah V, et al. Prediction of outcome in newly diagnosed myeloma: a meta-analysis of the molecular profiles of 1905 trial patients. Leukemia. 2018;32:102–110. doi: 10.1038/leu.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolli N, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones JR, et al. Clonal evolution in myeloma: the impact of maintenance lenalidomide and depth of response on the genetics and sub-clonal structure of relapsed disease in uniformly treated newly diagnosed patients. Haematologica. 2019;104:1440–1450. doi: 10.3324/haematol.2018.202200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker BA, et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010;116:e56–e65. doi: 10.1182/blood-2010-04-279596. [DOI] [PubMed] [Google Scholar]
- 6.Kaiser MF, et al. A TC classification-based predictor for multiple myeloma using multiplexed real-time quantitative PCR. Leukemia. 2013;27:1754–1757. doi: 10.1038/leu.2013.12. [DOI] [PubMed] [Google Scholar]
- 7.Hoang PH, et al. Whole-genome sequencing of multiple myeloma reveals oncogenic pathways are targeted somatically through multiple mechanisms. Leukemia. 2018;32:2459–2470. doi: 10.1038/s41375-018-0103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S, et al. NGSCheckMate: software for validating sample identity in next-generation sequencing studies within and across data types. Nucleic Acids Res. 2017;45:e103. doi: 10.1093/nar/gkx193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costello M, et al. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013;41:e67. doi: 10.1093/nar/gks1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Letouze E, et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat. Commun. 2017;8:1315. doi: 10.1038/s41467-017-01358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soong D, et al. CNV Radar: an improved method for somatic copy number alteration characterization in oncology. BMC Bioinform. 2020;21:98. doi: 10.1186/s12859-020-3397-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Favero F, et al. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol. 2015;26:64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan, K., Macintyre, G., Liu, W. & Markowetz, F. Ccube: a fast and robust method for estimating cancer cell fractions. bioRxiv, 484402, 10.1101/484402 (2018).
- 18.Wala, J. A. et al. Selective and mechanistic sources of recurrent rearrangements across the cancer genome. bioRxiv, 187609, 10.1101/187609 (2017).
- 19.Chen X, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 20.Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortés-Ciriano I, et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat. Genet. 2020;52:331–341. doi: 10.1038/s41588-019-0576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farmery JHR, Smith ML, Diseases NB-R, Lynch AG. Telomerecat: a ploidy-agnostic method for estimating telomere length from whole genome sequencing data. Sci. Rep. 2018;8:1300. doi: 10.1038/s41598-017-14403-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoang PH, Cornish AJ, Dobbins SE, Kaiser M, Houlston RS. Mutational processes contributing to the development of multiple myeloma. Blood Cancer J. 2019;9:60. doi: 10.1038/s41408-019-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martincorena I, et al. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171:1029–1041. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker BA, et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood. 2018;132:587–597. doi: 10.1182/blood-2018-03-840132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Javierre BM, et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167:1369–1384. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wingett S, et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res. 2015;4:1310. doi: 10.12688/f1000research.7334.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cairns J, et al. CHiCAGO: robust detection of DNA looping interactions in capture Hi-C data. Genome Biol. 2016;17:127. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Melton C, Reuter JA, Spacek DV, Snyder M. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat. Genet. 2015;47:710–716. doi: 10.1038/ng.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weddington N, et al. ReplicationDomain: a visualization tool and comparative database for genome-wide replication timing data. BMC Bioinform. 2008;9:530. doi: 10.1186/1471-2105-9-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sima J, et al. Identifying cis elements for spatiotemporal control of mammalian DNA replication. Cell. 2019;176:816–830. doi: 10.1016/j.cell.2018.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rheinbay E, et al. Recurrent and functional regulatory mutations in breast cancer. Nature. 2017;547:55–60. doi: 10.1038/nature22992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kortum KM, et al. Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes. Blood. 2016;128:1226–1233. doi: 10.1182/blood-2016-02-698092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gooding, S. et al. Multiple cereblon genetic changes associate with acquired resistance to lenalidomide or pomalidomide in multiple myeloma. Blood10.1182/blood.2020007081 (2020). Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 38.Aktas Samur A, et al. Deciphering the chronology of copy number alterations in multiple myeloma. Blood Cancer J. 2019;9:39. doi: 10.1038/s41408-019-0199-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caravagna G, et al. Subclonal reconstruction of tumors by using machine learning and population genetics. Nature Genetics. 2020;52:898–907. doi: 10.1038/s41588-020-0675-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinde J, et al. Palimpsest: an R package for studying mutational and structural variant signatures along clonal evolution in cancer. Bioinformatics. 2018;34:3380–3381. doi: 10.1093/bioinformatics/bty388. [DOI] [PubMed] [Google Scholar]
- 41.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 2016;17:31. doi: 10.1186/s13059-016-0893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maura F, et al. A practical guide for mutational signature analysis in hematological malignancies. Nat. Commun. 2019;10:2969. doi: 10.1038/s41467-019-11037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maura F, et al. Genomic landscape and chronological reconstruction of driver events in multiple myeloma. Nat. Commun. 2019;10:3835. doi: 10.1038/s41467-019-11680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaufmann H, et al. Both IGH translocations and chromosome 13q deletions are early events in monoclonal gammopathy of undetermined significance and do not evolve during transition to multiple myeloma. Leukemia. 2004;18:1879–1882. doi: 10.1038/sj.leu.2403518. [DOI] [PubMed] [Google Scholar]
- 46.Weinhold N, et al. Clonal selection and double-hit events involving tumor suppressor genes underlie relapse in myeloma. Blood. 2016;128:1735–1744. doi: 10.1182/blood-2016-06-723007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker BA, et al. Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood. 2012;120:1077–1086. doi: 10.1182/blood-2012-03-412981. [DOI] [PubMed] [Google Scholar]
- 49.Corre J, et al. Multiple myeloma clonal evolution in homogeneously treated patients. Leukemia. 2018;32:2636–2647. doi: 10.1038/s41375-018-0153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Egan JB, et al. Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood. 2012;120:1060–1066. doi: 10.1182/blood-2012-01-405977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keats JJ, et al. Clonal competition with alternating dominance in multiple myeloma. Blood. 2012;120:1067–1076. doi: 10.1182/blood-2012-01-405985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alvarez-Prado AF, et al. A broad atlas of somatic hypermutation allows prediction of activation-induced deaminase targets. J. Exp. Med. 2018;215:761–771. doi: 10.1084/jem.20171738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hochhaus A, et al. Molecular and chromosomal mechanisms of resistance to imatinib (STI571) therapy. Leukemia. 2002;16:2190–2196. doi: 10.1038/sj.leu.2402741. [DOI] [PubMed] [Google Scholar]
- 55.Lohr JG, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25:91–101. doi: 10.1016/j.ccr.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maura F, et al. Role of AID in the temporal pattern of acquisition of driver mutations in multiple myeloma. Leukemia. 2019;13:1–5. doi: 10.1038/s41375-019-0689-0. [DOI] [PubMed] [Google Scholar]
- 57.Brümmendorf TH, et al. Normalization of previously shortened telomere length under treatment with imatinib argues against a preexisting telomere length deficit in normal hematopoietic stem cells from patients with chronic myeloid leukemia. Ann. N. Y. Acad. Sci. 2003;996:26–38. doi: 10.1111/j.1749-6632.2003.tb03229.x. [DOI] [PubMed] [Google Scholar]
- 58.Lipinska N, et al. Telomerase and drug resistance in cancer. Cell Mol. Life Sci. 2017;74:4121–4132. doi: 10.1007/s00018-017-2573-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fittall MW, Van Loo P. Translating insights into tumor evolution to clinical practice: promises and challenges. Genome Med. 2019;11:20. doi: 10.1186/s13073-019-0632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Acar, A. et al. Exploiting evolutionary herding to control drug resistance in cancer. bioRxiv, 566950, 10.1101/566950 (2019).
- 61.Lipinski KA, et al. Cancer evolution and the limits of predictability in precision cancer medicine. Trends Cancer. 2016;2:49–63. doi: 10.1016/j.trecan.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kino K, Sugiyama H. UVR-induced G-C to C-G transversions from oxidative DNA damage. Mutat. Res. 2005;571:33–42. doi: 10.1016/j.mrfmmm.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 63.Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic. Res. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venkatesan S, et al. Perspective: APOBEC mutagenesis in drug resistance and immune escape in HIV and cancer evolution. Ann. Oncol. 2018;29:563–572. doi: 10.1093/annonc/mdy003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. Identification of neutral tumor evolution across cancer types. Nat. Genet. 2016;48:238–244. doi: 10.1038/ng.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson DC, et al. Neutral tumor evolution in myeloma is associated with poor prognosis. Blood. 2017;130:1639–1643. doi: 10.1182/blood-2016-11-750612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tarabichi M, et al. Neutral tumor evolution? Nat. Genet. 2018;50:1630–1633. doi: 10.1038/s41588-018-0258-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McDonald TO, Chakrabarti S, Michor F. Currently available bulk sequencing data do not necessarily support a model of neutral tumor evolution. Nat. Genet. 2018;50:1620–1623. doi: 10.1038/s41588-018-0217-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balaparya A, De S. Revisiting signatures of neutral tumor evolution in the light of complexity of cancer genomic data. Nat. Genet. 2018;50:1626–1628. doi: 10.1038/s41588-018-0219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang H-Y, et al. Is the evolution in tumors Darwinian or non-Darwinian? Natl Sci. Rev. 2017;5:15–17. doi: 10.1093/nsr/nwx076. [DOI] [Google Scholar]
- 71.Bahlis NJ. Darwinian evolution and tiding clones in multiple myeloma. Blood. 2012;120:927–928. doi: 10.1182/blood-2012-06-430645. [DOI] [PubMed] [Google Scholar]
- 72.Pinto V, et al. Multiple myeloma: available therapies and causes of drug resistance. Cancers. 2020;12:407. doi: 10.3390/cancers12020407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw promoter capture Hi-C data for naïve B-cells were obtained from European Genome-Phenome Archive (EGA; accession code EGAS00001001911). Replication timing data for B-lymphocytes was downloaded from Replication Domain Database34. Raw WGS data generated as part of this study can be accessed through EGA accession code EGAD00001005491.