Abstract
Studies have reported the genetic gives rise to male infertility. The aim of the present meta-analysis was to evaluate the association between PRM1 (rs737008 and rs2301365) and PRM2 (rs1646022 and rs2070923) polymorphisms and susceptibility to male infertility. The association between PRM1 and PRM2 polymorphisms and the risk of male infertility was evaluated using specific search terms in the Web of Science, Cochrane Library, PubMed, and Scopus databases without language restriction until January 28, 2020. The association was determined by odds ratio (OR) and 95% confidence interval (CI) on five genetic models using Review Manager 5.3 software. The funnel plot analysis and sensitivity analysis were done by the Comprehensive Meta-analysis 2.0 software. Out of 261 records retrieved from the databases, 17 studies were analyzed in the meta-analysis, including the four PRM polymorphisms. The pooled results as OR (P-value) showed 0.96 (0.44), 1.04 (0.70), 0.94 (0.51), 0.94 (0.48), and 1.03 (0.72) for PRM1 rs737008 polymorphism and 1.67 (0.0007), 1.73 (0.06), 1.50 (0.007), 1.56 (0.004), and 1.62 (0.33) for PRM1 rs2301365 polymorphism in allele, homozygous, heterozygous, recessive, and dominant models, respectively. Moreover, the pooled results as OR (P-value) showed 1.19 (0.004), 1.15 (0.26), 1.08 (0.70), 1.05 (0.76), and 0.98 (0.82) for PRM2 rs1646022 and 0.88 (0.04), 0.84 (0.10), 1.05 (0.81), 0.90 (0.24), and 0.80 (0.02) for PRM2 rs2070923 in allele, homozygous, heterozygous, recessive, and dominant models, respectively. The results showed PRM1 rs2301365 and PRM2 rs1646022 polymorphisms were associated with an elevated risk of male infertility and PRM2 rs2070923 polymorphism had a protective role in infertile men.
Subject terms: Genetics, Molecular medicine
Introduction
Infertility is defined as couples' inability to have a baby after one year of regular unprotected intercourse1. Male factor infertility affects up to 50% of couples' infertility and accounts for only 20% of total infertility2. Recently, however, the male factor infertility incidence has increased3,4. Male infertility is currently assessed through routine analysis according to sperm concentration/number, motility, and sperm morphology. However, there is a significant integration of semen characteristics between fertile and infertile males. In fact, around 15% of patients with male factor infertility according to WHO guidelines5 have normal semen parameters6. Thus, there are several limitations to routine conventional semen analysis in assessing male infertility, indicating that conventional semen parameters are poor predictors of reproductive outcome and that definitive diagnosis of male infertility cannot be made by routine analysis alone7. These limitations have led to the development of advanced methods for the study of sperm function, oxidative stress, fragmentation and DNA packing8. Non-obstructive azoospermia and severe oligozoospermia are two of the dominant phenotypes associated with severe spermatogenesis9. However, many factors relate to male infertility, like to reproductive tract disorders, chemical exposure, and infection9. Genetic factors account for 50% or more of all male infertility etiology, and approximately 7% of men worldwide suffer from infertility10. In order to indicate the underlying causes, extensive research has been done on the genetic reasons of male infertility in recent years.
There are two types of protamines (PRMNs), PRMN1 and PRMN2, which are encoded by two genes, PMN1 and PMN2, located on chromosome 16. In human sperm cells, 85% of histones are replaced by PRMN and from DNA in Protect against harmful agents. Altered ratio of histones to proteins has been shown to increase chromatin deficiency in sperm, increasing the risk of DNA damage and male infertility. In addition, an adequate ratio of PRMN1 and PRMN2 (normal 0.8–1.2) is needed for normal sperm function11. The expression of these two proteins in the sperm nucleus is approximately equal12. The complete translation of PRM1 and PRM2 mRNA happens throughout the elongated spermatids development, occurring in the production of positively charged PRMNs as a result of the high arginine content and this allows for strong binding to negatively charged DNA13. It was noticed a significantly diminished level of PRM1 mRNAs in spermatozoa isolated from crossbred Frieswal bulls with poor semen parameters, mostly featured by low progressive motility, in comparison to a group with good semen features14 and decreased PRM2 levels have been reported in various studies in infertile patients15. PRMs are believed to play a significant role in chromatin aggregation, transcriptional repression, haploid male genome conservation, sperm formation, and offspring production16. There were two previous meta-analyses reporting an association between PRM polymorphisms and the risk of male infertility including 8 studies17 and checking one PRM polymorphism and another9 included 13 studies with six PRM polymorphisms. Therefore, in the present meta-analysis including a meta-regression analysis of 17 studies, we investigated 13 PRM polymorphisms and then focused on the association between four functional PRM1 (rs737008 and rs2301365) and PRM2 (rs1646022 and PRM2 rs2070923) polymorphisms and male infertility susceptibility in case–control studies.
Materials and methods
The meta-analysis was done based on PRISMA statement, and the study question was formulated based on the PICOS framework18,19.
Participants (P): Men with infertility
Interventions (I): Prevalence of PRM1 and PRM2 polymorphisms
Comparisons (C): Male healthy controls
Outcomes (O): Risk of PRM1 and PRM2 polymorphisms
Study design (S): Case–control studies
Literature search
To search the association of PRM1 and PRM2 polymorphisms with the risk of male infertility, one author used the search terms ("male infertility") and (“PRM1” or “PRM2" or “Protamine 1” or “Protamine 2”) and (“gene*” or “variant*” or “polymorphism*” or “single-nucleotide polymorphism”) in the Web of Science, Cochrane Library, PubMed, and Scopus databases without language restriction until January 28, 2020. Another author checked the titles and abstracts to exclude the duplicates and irrelevant records and checked the full-texts of eligible studies. The databases were searched manually by crosschecking the references of original papers, review papers, and previous meta-analyses related to our topic in this meta-analysis to find the possibly missed studies. In addition, among studies retrieved, two previous meta-analyses had reported an association between PRM polymorphisms and the risk of male infertility9,17. One of them17 included 8 studies checking PRM1 rs2301365 polymorphism and showed an association between this polymorphism and the risk of male infertility just in Caucasians. Another9 included 13 studies (11 studies on PRM1 and 7 studies on PRM2 polymorphisms) with six PRM polymorphisms and showed an association between PRM1 rs737008, PRM1 rs2301365, and PRM2 rs1646022 polymorphisms and the risk of male infertility.
Inclusion and exclusion criteria
The inclusion criteria included (1) study focus on PRM1 polymorphisms rs35576928, rs737008, rs35262993, rs2301365, rs140477029, and rs193922261 and also PRM2 polymorphisms of rs1646022, rs779337774, rs545828790, rs201933708, rs115686767, rs200072135, and rs2070923 with male infertility susceptibility; (2) case–control studies on human beings that the cases were infertile patients with idiopathic infertility and including all subtypes (mainly azoospermia, cryptozoospermia, and oligozoospermia) and the controls were fertile; (3) including the details of genotype or allele frequency of cases and controls; (4) studies with complete full-text, and (5) studies with every language, (6) studies with or without deviation from the Hardy–Weinberg equilibrium (HWE) in controls. The exclusion criteria included (1) studies not concerning the association between PRM polymorphisms mentioned above and male infertility susceptibility; (2) animal articles, review studies, meta-analyses, and conference papers or editorial articles; (3) duplicate studies; and (4) studies with irrelevant data.
Data extraction and verification
The information retrieved from each study is mentioned in Tables 1, 2, and 3, including: (I) the first author’s name, (II) publication year, (III) region of origin and ethnicity, (IV) genotyping methods, (V) number of both cases and controls, (VI) HWE in the controls, (VII) control sources, and (VIII) prevalence of genotypes and alleles. Two authors independently extracted all the data of the studies included in the meta-analysis. In the case of disagreement between the two authors, another author resolved the disagreement by review and discussion.
Table 1.
Main characteristics of all studies entered to the meta-analysis.
| First author, publication year | Country | Ethnicity | No. of patients to controls | Method | Control source |
|---|---|---|---|---|---|
| Tanaka, 200324 | Japan | Asian | 226/270 | PCR sequence | PB |
| Aoki, 200625 | USA | Mixed | 192/96 | PCR sequence | HB |
| Ravel, 200726 | France | Caucasian | 281/111 | PCR–RFLP and sequence | PB |
| Gazquez, 200827 | Spain | Caucasian | 220/101 | PCR–RFLP and sequence | PB |
| Imken, 200928 | Morocco | Caucasian | 135/160 | PCR sequence | PB |
| Tuttelmann, 201029 | Germany | Caucasian | 171/77 | PCR sequence | PB |
| Jodar, 201123 | Spain and Sweden | Caucasian | 156/102 and 53/50 | PCR sequence | HB |
| Venkatesh, 201130 | India | Caucasian | 100/100 | PCR sequence | PB |
| Grassetti, 201231 | Italy | Caucasian | 110/53 | PCR sequence | HB |
| He, 201232 | China | Asian | 304/369 | Mass ARRAY | HB |
| Siasi, 201233 | Iran | Caucasian | 96/100 | PCR–RFLP, PCR–SSCP and PCR sequencing | HB |
| Yu, 201234 | China | Asian | 157/37 | Mass ARRAY | HB |
| Jamali, 201635 | Iran | Caucasian | 130/130 | PCR–RFLP | PB |
| Jiang, 201736 | China | Asian | 636/442 | Mass ARRAY | HB |
| Aydos, 201837 | Turkey | Caucasian | 100/100 | PCR | HB |
| Nabi, 201838 | Iran | Caucasian | 100/100 | PCR sequence | HB |
| Dehghanpour, 201939 | Iran | Caucasian | 65/65 | PCR sequence | HB |
PCR Polymerase chain reaction, RFLP restriction fragment length polymorphism, SSCP single-strand conformation polymorphism, HB hospital-based, PB population-based.
Table 2.
Prevalence of genotypes and alleles of PRM1 and PRM2 polymorphisms.
| First author, publication year | PRM1 polymorphism | Case | Control | Case | Control | HWE* | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CA | AA | CC | CA | AA | C | A | C | A | |||
| Tanaka, 200324 | rs737008 | 125 | 86 | 15 | 129 | 117 | 24 | 336 | 116 | 375 | 165 | 0.728 |
| Aoki, 200625 | rs737008 | 32 | 79 | 81 | 12 | 43 | 41 | 143 | 241 | 67 | 125 | 0.889 |
| Ravel, 200726 | rs737008 | 38 | 131 | 112 | 14 | 51 | 46 | 207 | 355 | 79 | 143 | 0.981 |
| Imken, 200928 | rs737008 | 16 | 55 | 64 | 16 | 74 | 70 | 87 | 183 | 106 | 214 | 0.578 |
| Tuttelmann, 201029 | rs737008 | 23 | 63 | 85 | 8 | 28 | 41 | 109 | 233 | 44 | 110 | 0.338 |
| Jodar, 2011a23 | rs737008 | 12 | 64 | 80 | 14 | 41 | 47 | 88 | 224 | 69 | 135 | 0.302 |
| Jodar, 2011b23 | rs737008 | 2 | 28 | 30 | 4 | 20 | 26 | 32 | 74 | 28 | 72 | 0.955 |
| Venkatesh, 201130 | rs737008 | 56 | 20 | 24 | 48 | 24 | 28 | 132 | 68 | 120 | 80 | < 0.001 |
| Grassetti, 201231 | rs737008 | 15 | 55 | 40 | 4 | 29 | 20 | 85 | 135 | 37 | 69 | 0.137 |
| He, 201232 | rs737008 | 161 | 112 | 31 | 209 | 142 | 25 | 434 | 174 | 560 | 192 | 0.894 |
| Siasi, 201233 | rs737008 | 22 | 32 | 42 | 24 | 29 | 47 | 76 | 116 | 77 | 123 | < 0.001 |
| Nabi, 201834 | rs737008 | 33 | 47 | 12 | 21 | 51 | 15 | 123 | 61 | 93 | 81 | 0.096 |
| Dehghanpour, 201935 | rs737008 | 0 | 62 | 3 | 17 | 37 | 11 | 62 | 68 | 71 | 59 | 0.232 |
| Ravel, 200726 | rs2301365 | 184 | 87 | 10 | 71 | 36 | 4 | 455 | 287 | 178 | 44 | 0.829 |
| Gazquez, 200827 | rs2301365 | 114 | 90 | 16 | 68 | 30 | 3 | 318 | 122 | 166 | 36 | 0.887 |
| Imken, 200928 | rs2301365 | 85 | 45 | 5 | 113 | 42 | 5 | 215 | 55 | 268 | 52 | 0.652 |
| Jodar, 2011a23 | rs2301365 | 88 | 55 | 13 | 60 | 38 | 4 | 231 | 81 | 158 | 46 | 0.501 |
| Jodar, 2011b23 | rs2301365 | 25 | 27 | 1 | 26 | 17 | 7 | 77 | 29 | 69 | 31 | 0.176 |
| He, 201232 | rs2301365 | 100 | 17 | 241 | 112 | 16 | 474 | 134 | 594 | 144 | 0.517 | |
| Yu, 201234 | rs2301365 | 61 | 70 | 26 | 17 | 19 | 1 | 192 | 122 | 53 | 21 | 0.109 |
| Jamali, 201635 | rs2301365 | 80 | 39 | 11 | 109 | 20 | 1 | 199 | 61 | 238 | 22 | 0.937 |
| Jiang, 201736 | rs2301365 | 378 | 229 | 29 | 277 | 144 | 21 | 985 | 287 | 698 | 187 | 0.681 |
| Aydos, 201837 | rs2301365 | 58 | 38 | 4 | 92 | 8 | 0 | 154 | 46 | 192 | 8 | 0.676 |
| First author, publication year | PRM2 polymorphism | GG | GC | CC | GG | GC | CC | G | C | G | C | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tanaka, 200324 | rs1646022 | 127 | 80 | 19 | 127 | 118 | 24 | 224 | 118 | 372 | 166 | 0.645 |
| Aoki, 200625 | rs1646022 | 77 | 30 | 85 | 39 | 13 | 44 | 184 | 200 | 91 | 101 | < 0.001 |
| Tuttelmann, 201028 | rs1646022 | 57 | 66 | 36 | 22 | 28 | 23 | 180 | 138 | 74 | 72 | 0.046 |
| Venkatesh, 201130 | rs1646022 | 100 | 0 | 0 | 98 | 0 | 2 | 200 | 0 | 196 | 4 | < 0.001 |
| Grassetti, 201231 | rs1646022 | 30 | 62 | 18 | 18 | 26 | 9 | 122 | 98 | 62 | 44 | 0.940 |
| Jamali, 201635 | rs1646022 | 4 | 39 | 7 | 93 | 31 | 6 | 207 | 53 | 217 | 43 | 0.120 |
| Jiang, 201736 | rs1646022 | 35 | 266 | 335 | 47 | 162 | 233 | 336 | 936 | 256 | 478 | 0.021 |
| Nabi, 201838 | rs1646022 | 31 | 59 | 10 | 36 | 56 | 8 | 121 | 79 | 128 | 72 | 0.031 |
| Dehghanpour, 201939 | rs1646022 | 29 | 25 | 11 | 20 | 41 | 4 | 83 | 47 | 81 | 49 | 0.005 |
| First author, publication year | PRM2 polymorphism | CC | CA | AA | CC | CA | AA | C | A | C | A | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tanaka, 200324 | rs2070923 | 125 | 82 | 19 | 127 | 118 | 25 | 332 | 120 | 372 | 168 | 0.747 |
| Aoki, 200625 | rs2070923 | 93 | 27 | 72 | 40 | 12 | 44 | 213 | 171 | 81 | 100 | < 0.001 |
| Tuttelmann, 201029 | rs2070923 | 78 | 55 | 26 | 38 | 26 | 9 | 211 | 107 | 102 | 44 | 0.187 |
| Venkatesh, 201130 | rs2070923 | 55 | 20 | 25 | 60 | 0 | 40 | 130 | 70 | 120 | 80 | < 0.001 |
| Grassetti, 201231 | rs2070923 | 42 | 54 | 14 | 23 | 25 | 5 | 138 | 82 | 71 | 35 | 0.628 |
| He, 201232 | rs2070923 | 87 | 57 | 162 | 99 | 73 | 204 | 231 | 381 | 271 | 481 | < 0.001 |
| Nabi, 201838 | rs2070923 | 15 | 57 | 28 | 23 | 34 | 43 | 87 | 113 | 80 | 120 | 0.003 |
| Dehghanpour, 201939 | rs2070923 | 21 | 22 | 22 | 11 | 26 | 28 | 64 | 66 | 48 | 56 | 0.254 |
HWE Hardy–Weinberg equilibrium.
*P-values of HWE for control group. The study of Jodar et al.17 included two studies.
Table 3.
Prevalence of genotypes and alleles of other PRM1 and PRM2 polymorphisms.
| First author, publication year | PRM1 polymorphism | Case | Control | Case | Control | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GG | GA | AA | GG | GA | AA | G | A | G | A | ||
| Aoki, 200625 | rs35262993 | 189 | 3 | 0 | 94 | 2 | 0 | 381 | 3 | 190 | 2 |
| Ravel, 200726 | rs35262993 | 111 | 0 | 0 | 281 | 0 | 0 | 222 | 0 | 562 | 0 |
| Imken, 200928 | rs35262993 | 133 | 2 | 0 | 155 | 5 | 0 | 315 | 5 | 271 | 2 |
| Tuttelmann, 201029 | rs35262993 | 167 | 4 | 0 | 75 | 2 | 0 | 338 | 4 | 152 | 2 |
| Grassetti, 201231 | rs35262993 | 109 | 1 | 0 | 53 | 0 | 0 | 106 | 1 | 119 | 0 |
| He, 201232 | rs35262993 | 292 | 1 | 0 | 373 | 1 | 0 | 585 | 1 | 747 | 1 |
| First author, publication year | PRM1 polymorphism | CC | CT | TT | CC | CT | TT | C | T | C | T |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Jodar, 2011a23 | rs140477029 | 155 | 1 | 0 | 102 | 0 | 0 | 311 | 1 | 204 | 0 |
| Dehghanpour, 201939 | rs140477029 | 65 | 0 | 0 | 65 | 0 | 0 | 130 | 0 | 130 | 0 |
| First author, publication year | PRM1 polymorphism | GG | GT | TT | GG | GT | TT | G | T | G | T |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Aoki, 200625 | rs35576928 | 189 | 3 | 0 | 94 | 2 | 0 | 381 | 3 | 190 | 2 |
| Ravel, 200726 | rs35576928 | 111 | 0 | 0 | 278 | 3 | 0 | 222 | 0 | 559 | 3 |
| Tuttelmann, 201029 | rs35576928 | 167 | 4 | 0 | 75 | 2 | 0 | 338 | 4 | 152 | 2 |
| Jodar, 2011a23 | rs35576928 | 155 | 1 | 0 | 102 | 0 | 0 | 311 | 1 | 204 | 0 |
| Jodar, 2011b23 | rs35576928 | 52 | 1 | 0 | 49 | 1 | 0 | 104 | 1 | 99 | 1 |
| Grassetti, 201231 | rs35576928 | 110 | 0 | 0 | 52 | 1 | 0 | 220 | 0 | 105 | 1 |
| He, 201232 | rs35576928 | 328 | 45 | 0 | 256 | 47 | 0 | 701 | 45 | 559 | 47 |
| Aydos, 201837 | rs35576928 | 100 | 0 | 0 | 100 | 0 | 0 | 200 | 0 | 200 | 0 |
| Nabi, 201838 | rs35576928 | 92 | 0 | 0 | 87 | 0 | 0 | 182 | 0 | 174 | 0 |
| Zeyadi, 201922 | rs35576928 | 9 | 6 | 0 | 9 | 1 | 0 | 24 | 0 | 19 | 1 |
| Dehghanpour, 201939 | rs35576928 | 65 | 0 | 0 | 65 | 0 | 0 | 130 | 0 | 130 | 0 |
| First author, publication year | PRM1 polymorphism | GG | GC | CC | GG | GC | CC | G | C | G | C |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ravel, 200726 | rs193922261 | 111 | 0 | 0 | 281 | 0 | 0 | 222 | 0 | 562 | 0 |
| Imken, 200928 | rs193922261 | 134 | 1 | 0 | 160 | 0 | 0 | 269 | 1 | 320 | 0 |
| First author, publication year | PRM2 polymorphism | CC | CT | TT | CC | CT | TT | C | T | C | T |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Siasi, 201233 | rs779337774 | 100 | 0 | 0 | 100 | 0 | 0 | 200 | 0 | 200 | 0 |
| Aydos, 201837 | rs779337774 | 98 | 2 | 0 | 100 | 0 | 0 | 198 | 2 | 200 | 0 |
| Nabi, 201838 | rs779337774 | 92 | 0 | 0 | 87 | 0 | 0 | 184 | 0 | 174 | 0 |
| Zeyadi, 201922 | rs779337774 | 33 | 3 | 4 | 9 | 1 | 0 | 69 | 11 | 19 | 1 |
| First author, publication year | PRM2 polymorphism | GG | GA | AA | GG | GA | AA | G | A | G | A |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nabi, 201838 | rs545828790 | 92 | 0 | 0 | 84 | 3 | 0 | 184 | 0 | 171 | 3 |
| Dehghanpour, 201939 | rs545828790 | 65 | 0 | 0 | 65 | 0 | 0 | 130 | 0 | 130 | 0 |
| First author, publication year | PRM2 polymorphism | GG | GC | CC | GG | GC | CC | G | C | G | C |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Grassetti, 201231 | rs201933708 | 110 | 0 | 0 | 52 | 1 | 0 | 220 | 0 | 105 | 1 |
| Nabi, 201838 | rs201933708 | 92 | 0 | 0 | 85 | 2 | 0 | 184 | 0 | 172 | 2 |
| Dehghanpour, 201939 | rs201933708 | 65 | 0 | 0 | 61 | 4 | 0 | 130 | 0 | 126 | 4 |
| First author, publication year | PRM2 polymorphism | CC | CT | TT | CC | CT | TT | C | T | C | T |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nabi, 201838 | rs115686767 | 92 | 0 | 0 | 85 | 2 | 0 | 184 | 0 | 172 | 2 |
| Dehghanpour, 201939 | rs115686767 | 65 | 0 | 0 | 61 | 4 | 0 | 130 | 0 | 126 | 4 |
| Aoki, 200625 | rs200072135 | 191 | 1 | 0 | 95 | 1 | 0 | 383 | 1 | 191 | 1 |
| Imken, 200928 | rs200072135 | 135 | 0 | 0 | 159 | 1 | 0 | 170 | 0 | 319 | 1 |
| Jodar, 2011a23 | rs200072135 | 111 | 0 | 0 | 49 | 1 | 0 | 222 | 0 | 99 | 1 |
The study of Jodar et al.17 included two studies.
Statistical analysis
The evaluation of the strength of association between PRM1 and PRM2 polymorphisms and male infertility risk was performed by odds ratio (OR) and 95% confidence interval (CI). Review Manager 5.3 software was applied to calculate the summary ORs based on five genetic models (allele, heterozygous, homozygous, recessive, and dominant). In this state, the statistical significance of pooled results was illustrated with the Z-test. P-value < 0.05 was considered statistically significant. In addition, heterogeneity across the studies was estimated by the Chi-square-based Q test20. If the Ph or Pheterogeneity was > 0.10 and heterogeneity or I2 < 50%, showing lack of heterogeneity between studies, we should use the fixed-effects model, but conversely, we used the random-effects model21.
The thirteen polymorphisms were assessed for the association with susceptibility to male infertility based on five genetic models. Among them, four polymorphisms were included in the meta-analysis: PRM1 (rs737008 and rs2301365) and PRM2 (rs1646022 and rs2070923). The prevalence rates of CC (wild-type homozygote), CA (heterozygote), and AA genotype (rare homozygote) were calculated for PRM1 rs737008, PRM1 rs2301365, and PRM2 rs2070923 polymorphisms. Further, the GG (wild-type homozygote), GC (heterozygote), and CC (rare homozygote) were calculated for PRM2 rs1646022 polymorphism. Subgroup analyses were further performed based on ethnicity, method, and control source. A sensitivity analysis was conducted in which the studies with deviation from HWE in the controls were deleted. A meta-regression analysis was performed to detect the confounding factors affecting the pooled results by IBM SPSS 22.0 software. In addition, sensitivity analyses, including “one remove study” and “cumulative analysis”, were conducted each time on previous analyses to determine the stability of the pooled results. Funnel plots and Egger’s liner regression test were used to examine the publication bias. The funnel plot analysis and sensitivity analysis were done by Comprehensive Meta-analysis 2.0 software.
Results
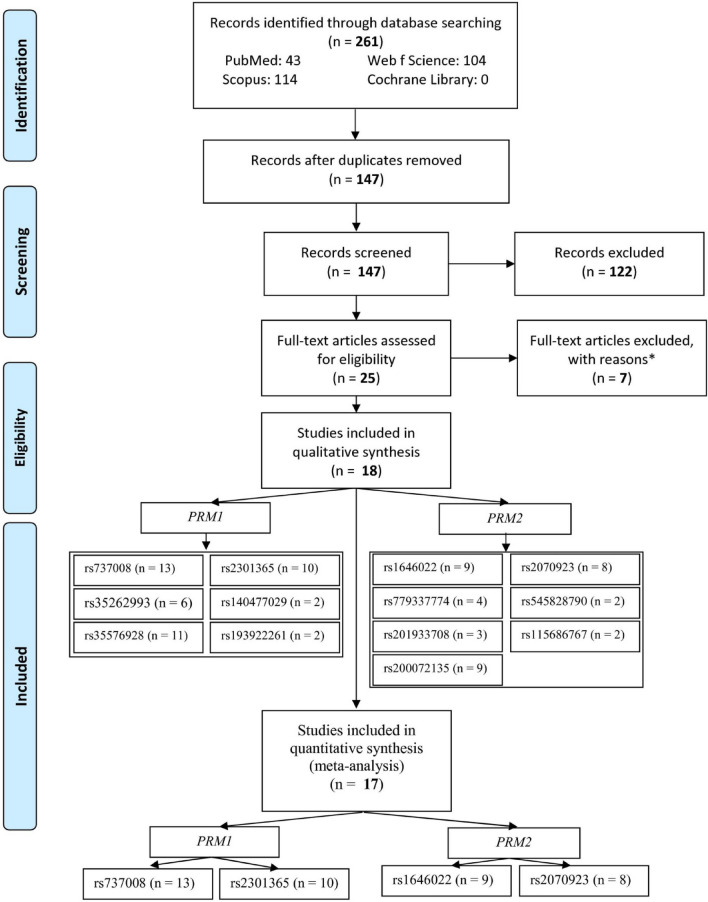
Out of 261 records retrieved in the databases, 25 articles including full-texts were evaluated for eligibility after excluding the duplicates and irrelevant records (Fig. 1). Among these full-texts, 7 of them were excluded with reasons (2 meta-analyses, 2 reviews, 1 animal study, and 2 studies with no control groups). Therefore, 18 studies were included in the systematic review, from which one study22 was excluded because it did not include four eligible polymorphisms. Finally, 17 studies including four polymorphisms of PRM1 rs737008, PRM1 rs2301365, PRM2 rs1646022, and PRM2 rs2070923 were analyzed in the meta-analysis. One study23 checked the rs737008 and rs2301365 polymorphisms in two different populations (13 for polymorphism of PRM1 rs737008, 10 for PRM1 rs2301365, 9 for PRM2 rs1646022, and 8 for PRM2 rs2070923).
Figure 1.
Flow-chart of the study selection. One of articles23 included two studies.
Table 1 presentations the features of studies entered to the meta-analysis. The studies23–39 were published from 2003 to 2019. Twelve studies23,26–31,33,35,37–39 were reported in Caucasian, four studies24,32,34,36 in Asian, and one25 in mixed ethnicities. The genotyping method was PCR-based in fourteen studies23–31,33,35,37–39 and Mass ARRAY in three studies32,34,36. The source of controls was hospital-based in ten studies25,31–33,33,34,36–39 and population-based in seven studies24,26–30,35.
Tables 2 and 3 show the prevalence of the genotypes and alleles of PRM1 and PRM2 polymorphisms. We included four polymorphisms (PRM1 rs737008, PRM1 rs2301365, PRM2 rs1646022, and PRM2 rs2070923) in the meta-analysis mentioned in Table 2. The other polymorphisms mentioned (PRM1 rs35262993, rs140477029, rs35576928, and rs193922261 polymorphisms and PRM2 rs779337774, rs545828790, rs201933708, rs115686767, and rs200072135 polymorphisms) in Table 3 were excluded from the meta-analysis because a lot of studies had no mutation or the percentage of mutation was very low. The P-values of HWE were less than 0.05 for the controls of PRM1 rs737008 polymorphism in two studies30,33, PRM2 rs1646022 in six studies25,29,30,36,38,39, and PRM2 rs2070923 in four studies25,30,32,38.
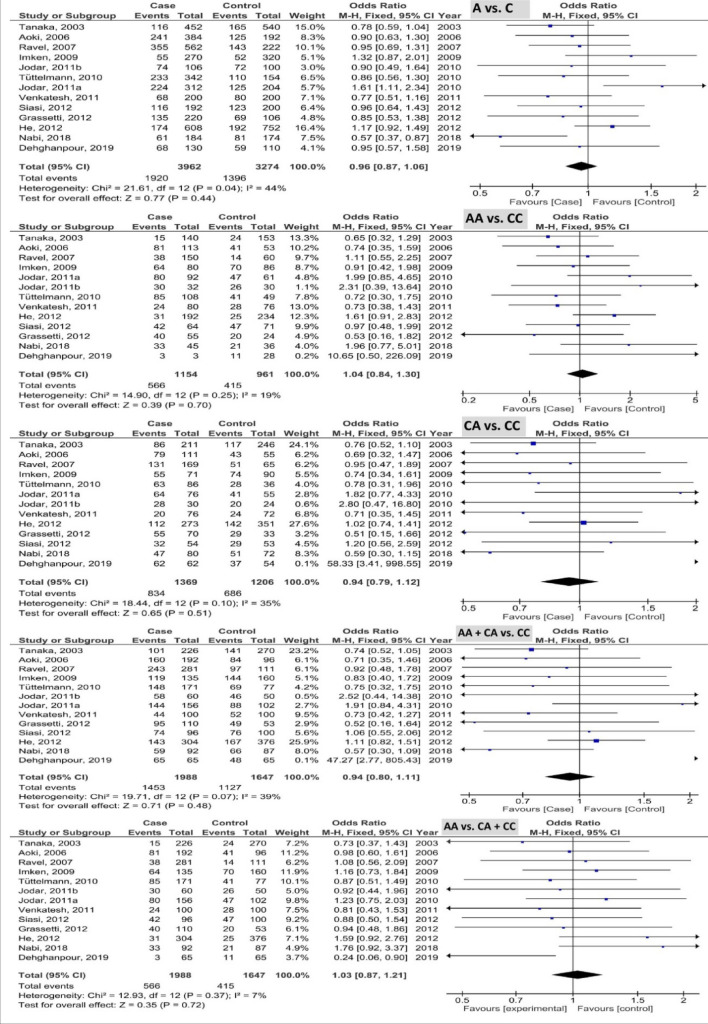
The pooled results of PRM1 rs737008 polymorphism based on five genetic models are illustrated in Fig. 2. The pooled results as OR (985%CI; P-value) showed 0.96 (0.87, 1.06; 0.44) with I2 = 44% (Pheterogeneity or Ph = 0.04), 1.04 (0.84, 1.30; 0.70) with I2 = 19% (Ph = 0.25), 0.94 (0.79, 1.12; 0.51) with I2 = 35% (Ph = 0.10), 0.94 (0.80, 1.11; 0.48) with I2 = 39% (Ph = 0.07), and 1.03 (0.87, 1.21; 0.72) with I2 = 7% (Ph = 0.37) in the allele, homozygous, heterozygous, recessive, and dominant models, respectively. Based on the results, this polymorphism was not associated with the male infertility susceptibility.
Figure 2.
Forest plot of analysis of PRM1 rs737008 polymorphism based on five genetic models.
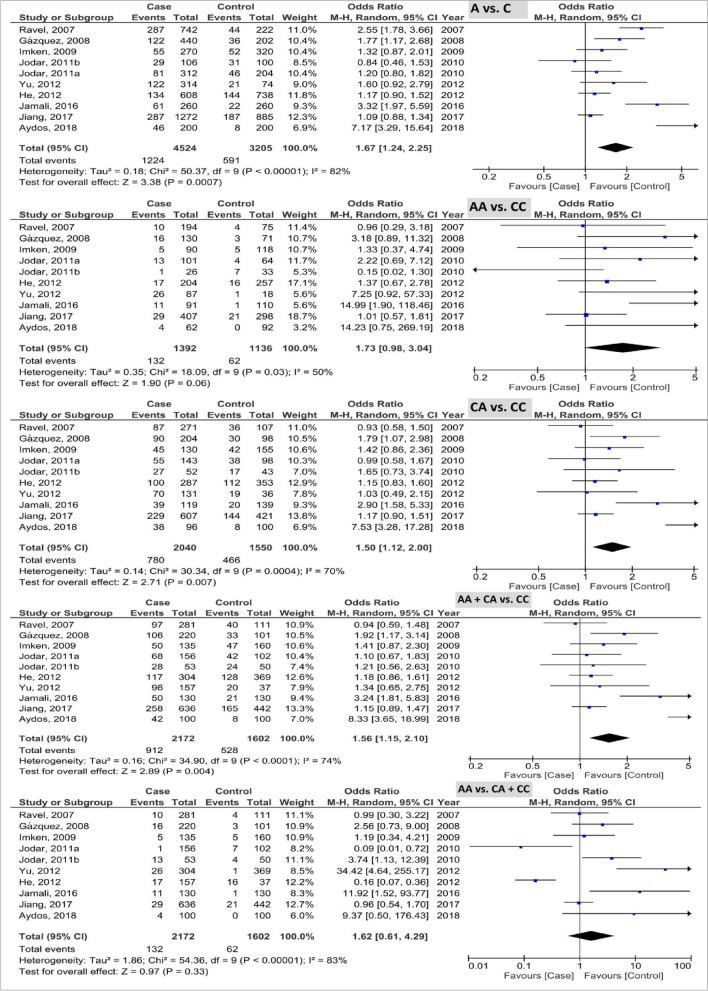
The pooled results of PRM1 rs2301365 polymorphism based on five genetic models are indicated in Fig. 3. The pooled results as OR (95% CI; P-value) showed the 1.67 (1.24, 2.25; 0.0007) with I2 = 82% (Ph < 0.00001), 1.73 (0.98, 3.04; 0.06) with I2 = 50% (Ph = 0.03), 1.50 (1.12, 2.00; 0.007) with I2 = 70% (Ph = 0.0004), 1.56 (1.15, 2.10; 0.004) with I2 = 74% (Ph < 0.0001), and 1.62 (0.61, 4.29; 0.33) with I2 = 83% (Ph < 0.00001) in the allele, homozygous, heterozygous, recessive, and dominant models, respectively. Based on the results, C allele and CA genotype of PRM1 rs2301365 polymorphism were associated with the elevated risk of male infertility.
Figure 3.
Forest plot of analysis of PRM1 rs2301365 polymorphism based on five genetic models.
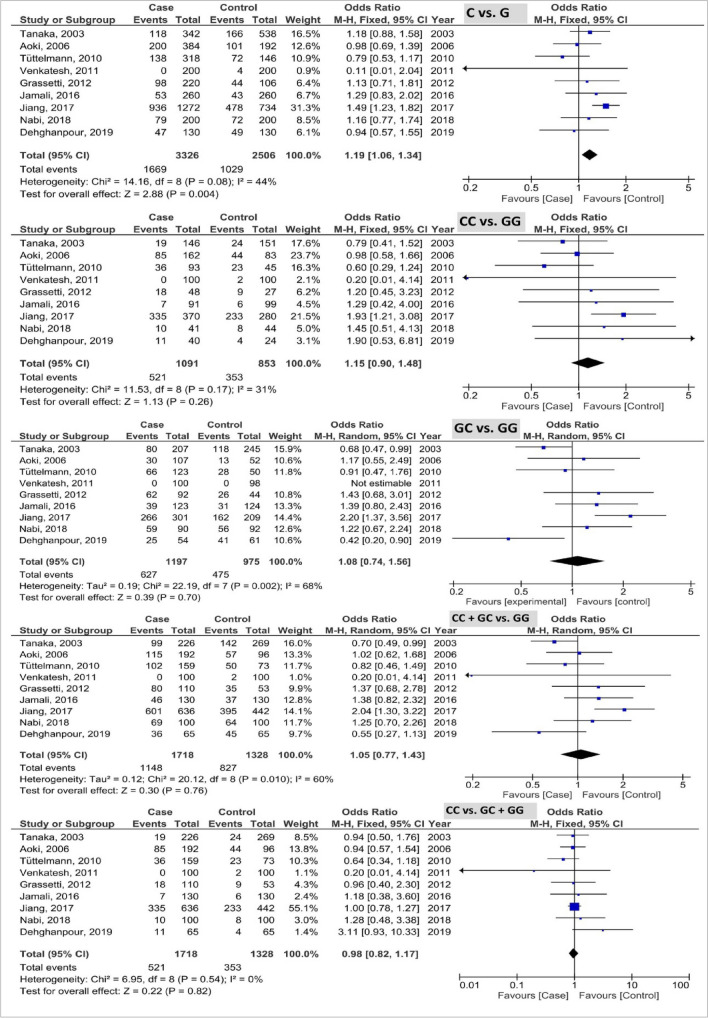
The pooled results of PRM2 rs1646022 polymorphism based on five genetic models are shown in Fig. 4. The pooled results as OR (95% CI; P-value) showed the 1.19 (1.06, 1.34; 0.004) with I2 = 44% (Ph = 0.08), 1.15 (0.90, 1.48; 0.26) with I2 = 31% (Ph = 0.17), 1.08 (0.74, 1.56; 0.70) with I2 = 68% (Ph = 0.002), 1.05 (0.77, 1.43; 0.76) with I2 = 60% (Ph = 0.010), and 0.98 (0.82, 1.17; 0.82) with I2 = 0% (Ph = 0.54) in the allele, homozygous, heterozygous, recessive, and dominant models, respectively. Based on the results, the G allele of PRM2 rs1646022 polymorphism was associated with the elevated risk of male infertility.
Figure 4.
Forest plot of analysis of PRM2 rs1646022 polymorphism based on five genetic models.
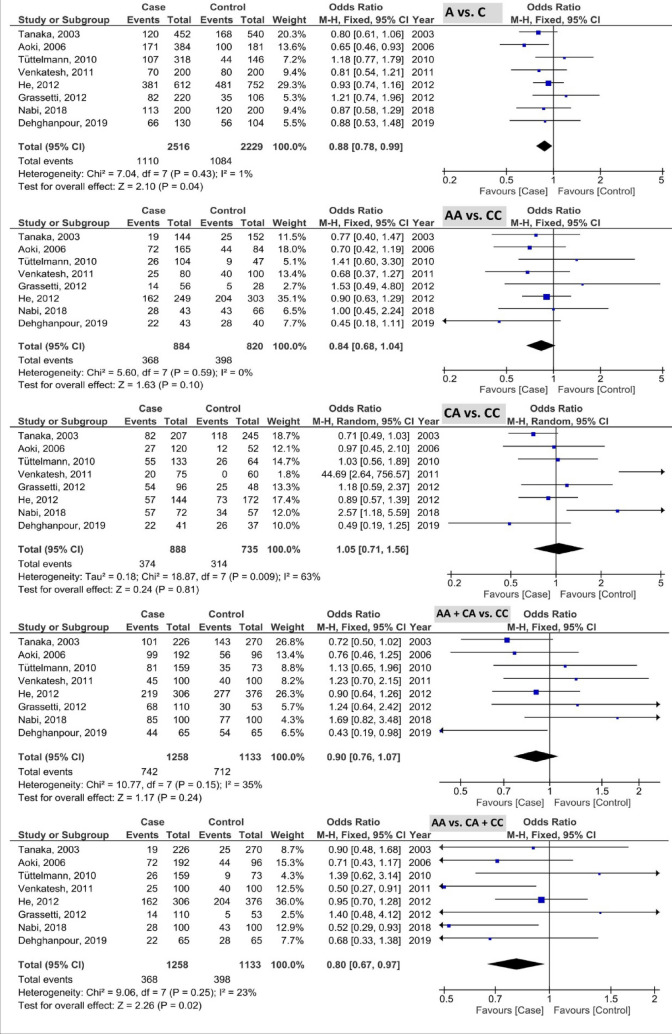
The pooled results of PRM2 rs2070923 polymorphism based on five genetic models are demonstrated in Fig. 5. The pooled results as OR (95% CI; P-value) showed the 0.88 (0.78, 0.99; 0.04) with I2 = 1% (Ph = 0.43), 0.84 (0.68, 1.04; 0.10) with I2 = 0% (Ph = 0.59), 1.05 (0.71, 1.56; 0.81) with I2 = 63% (Ph = 0.009), 0.90 (0.76, 1.07; 0.24) with I2 = 35% (Ph = 015), and 0.80 (0.67, 0.97; 0.02) with I2 = 23% (Ph = 0.25) in the allele, homozygous, heterozygous, recessive, and dominant models, respectively. Based on the results, the C allele and CC genotype of PRM2 rs2070923 polymorphism were associated with the reduced risk of male infertility.
Figure 5.
Forest plot of analysis of PRM2 rs2070923 polymorphism based on five genetic models.
Subgroup analysis
The results of subgroup analysis for PRM1 rs737008, PRM1 rs2301365, PRM2 rs2070923, and PRM2 rs1646022 polymorphisms are shown in Table 4. The AA + CA genotype in the studies with population-based controls was associated with the reduced risk of male infertility (OR 0.77; 95% CI 0.60, 0.98; P = 0.04) without heterogeneity. With regard to PRM1 rs2301365 polymorphism, the C allele and CA genotype in the Caucasian ethnicity were associated with the elevated risk of male infertility (OR 1.96; 95% CI 1.29, 2.97; P = 0.002 and OR 1.79; 95% CI 1.13, 2.83; P = 0.01, respectively). Also, the C allele (OR 1.59; 95% CI 1.15, 2.20; P = 0.005) and CC (OR 1.44; 95% CI 1.02, 2.03; P = 0.04) and CA (OR 1.39; 95% CI 1.01, 1.92; P = 0.04) genotypes in the studies with hospital-based controls were associated with the elevated risk of male infertility. For PRM1 rs2301365 polymorphism, the C allele and CA genotype in the studies with PCR-based method were associated with the elevated risk of male infertility (OR 1.96; 95% CI 1.29, 2.97; P = 0.002 and OR 1.79; 95% CI 1.13, 2.83; P = 0.01, respectively). About PRM2 rs2070923 polymorphism, the G allele had an elevated risk in male infertility compared to male fertility (OR 1.38; 95% CI 1.18, 1.63; P < 0.0001), which was similar to the G allele (OR 1.26; 95% CI 1.09, 1.46; P = 0.001) and GG genotype (OR 1.43; 95% CI 1.06, 1.94; P = 0.02) in the studies with hospital-based controls. With regard to mass ARRAY, the G allele (OR 1.49; 95% CI 1.23, 1.82; P < 0.0001) and GG (OR 1.93; 95% CI 1.21, 3.08; P = 0.006) and GC (OR 2.20; 95% CI 1.37, 3.56; P = 0.001) genotypes had an elevated risk in male infertility compared to male fertility. As for PRM2 rs1646022 polymorphism, the CC genotype was associated with a reduced risk of male infertility (OR 0.69; 95% CI 0.51, 0.94; P = 0.02) in the Caucasian ethnicity and C allele (OR 0.65; 95% CI 0.46, 0.93; P = 0.02) in the mixed ethnicity. Further, the C allele (OR 0.86; 95% CI 0.74, 0.99; P = 0.04) and CC genotype (OR 0.72; 95% CI 0.57, 0.92; P = 0.009) in the PCR-based method had a reduced risk of male infertility.
Table 4.
Subgroup analysis for PRM1 rs737008, PRM1 rs2301365, PRM2 rs2070923, and PRM2 rs1646022 polymorphisms.
| PRM1 rs737008 | A vs. C | AA vs. CC | CA vs. CC | AA + CA vs. CC | AA vs. CA + CC |
|---|---|---|---|---|---|
| OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | |
| Total (13) | 0.96 (0.87, 1.06), 0.44, 44, 0.04 | 1.05 (0.84, 1.31), 0.66, 19, 0.25 | 0.94 (0.79, 1.12), 0.51, 35, 0.10 | 0.94 (0.80, 1.11), 0.48, 39, 0.07 | 1.03 (0.87, 1.21), 0.72, 7, 0.37 |
| Ethnicity | |||||
| Asian (2) | 0.96, (0.65, 1.43), 0.86, 78, 0.03 | 1.04 (0.43,2.55), 0.93, 75, 0.04 | 0.90 (0.71, 1.15), 0.40, 30, 0.23 | 0.92 (0.61, 1.37), 0.67, 66, 0.09 | 1.10 (0.51, 2.38), 0.80, 68, 0.08 |
| Caucasian (10) | 0.96 (0.84,1.09), 0.50, 47, 0.05 | 1.08 (0.82, 1.42), 0.60, 10, 0.35 | 1.04 (0.80, 1.34), 0.79, 47, 0.05 | 0.98 (0.78, 1.25), 0.89, 46, 0.06 | 1.01 (0.84, 1.23), 0.90, 5, 0.40 |
| Mixed (1) | 0.92 (0.68, 1.23), 0.57 | 0.74 (0.35, 1.59), 0.44 | 0.69 (0.32, 1.47), 0.34 | 0.71 (0.35, 1.46), 0.36 | 0.98 (0.60, 1.61), 0.93 |
| Control source | |||||
| HB (8) | 0.97 (0.79, 1.20), 0.81, 54, 0.03 | 1.32 (0.97, 1.78), 0.07, 22, 0.25 | 1.06 (0.67, 1.66), 0.82, 57, 0.02 | 1.09 (0.60, 1.98), 0.78, 63, 0.01 | 1.09 (0.88, 1.35), 0.42, 32, 0.17 |
| PB (5) | 0.89 (0.76, 1.05), 0.16, 17, 0.31 | 0.81 (0.59, 1.12), 0.20, 0, 0.83 | 0.78 (0.60, 1.01), 0.06, 0, 0.98 | 0.77 (0.60, 0.98), 0.04, 0, 0.98 | 0.95 (0.73, 1.22), 0.67, 0, 0.77 |
| Method | |||||
| PCR-based (12) | 0.92 (0.82, 1.03), 0.15, 40, 0.07 | 0.97 (0.76, 1.24), 0.81, 10, 0.35 | 0.91 (0.74, 1.12), 0.39, 38, 0.09 | 0.88 (0.73, 1.07), 0.21, 36, 0.10 | 0.99 (0.83, 1.17), 0.88, 0, 0.50 |
| Mass ARRAY (1) | 1.17 (0.92, 1.49), 0.20 | 1.61 (0.91, 2.83), 0.10 | 1.02 (0.74, 1.41), 0.89 | 1.11 (0.82, 1.51), 0.49 | 1.59 (0.92, 2.76), 0.10 |
| PRM1 rs2301365 | A vs. C | AA vs. CC | CA vs. CC | AA + CA vs. CC | AA vs. CA + CC |
|---|---|---|---|---|---|
| OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | |
| Total (10) | 1.67 (1.24, 2.25), 0.0007, 82, < 0.00001 | 1.73 (0.98, 3.04), 0.06, 50, 0.03 | 1.50 (1.12, 2.00), 0.007, 70, 0.0004 | 1.56 (1.15, 2.10), 0.004, 74, < 0.0001 | 1.62 (0.61, 4.29), 0.33, 83, < 0.00001 |
| Ethnicity | |||||
| Asian (3) | 1.15 (0.98, 1.35), 0.08, 0, 0.43 | 1.34 (0.87, 2.05), .0.19, 42, 0.18 | 1.15 (0.94, 1.40), 0.16, 0, 0.95 | 1.17 (0.97, 1.41), 0.10, 0, 0.92 | 1.41 (0.15, 13.18), 0.76, 94, < 0.00001 |
| Caucasian (7) | 1.96 (1.29, 2.97), 0.002, 82, < 0.0001 | 1.96 (0.82, 4.70), 0.13, 55, 0.04 | 1.79 (1.13, 2.83), 0.01, 77, 0.0003 | 1.82 (1.13, 2.93), 0.01, 80, < 0.0001 | 1.82 (0.71, 4.68), 0.21, 61, 0.02 |
| Control source | |||||
| HB (8) | 1.59 (1.15,2.20), 0.005, 82, < 0.00001 | 1.44 (1.02, 2.03), 0.04, 45, 0.08 | 1.39 (1.01, 1.92), 0.04, 70, 0.001 | 1.44 (1.04, 1.98), 0.03, 73, 0.0006 | 1.38 (0.45, 4.23), 0.57, 85, < 0.00001 |
| PB (2) | 2.06 (0.83, 5.10), 0.12, 86, 0.007 | 3.91 (0.33, 45.85), 0.28, 76, 0.04 | 0.99 (0.99, 3.99), 0.05, 68, 0.08 | 2.11 (0.93, 4.75), 0.07, 78, 0.03 | 3.28 (0.32, 34.09), 0.32, 74, 0.05 |
| Method | |||||
| PCR-based (7) | 1.96 (1.29, 2.97), 0.002, 82, < 0.0001 | 1.96 (0.82, 4.70), 0.13, 55, 0.04 | 1.79 (1.13, 2.83), 0.01, 77, 0.0003 | 1.82 (1.13, 2.93), 0.01, 80, < 0.0001 | 1.82 (0.71, 4.68), 0.21, 61, 0.02 |
| Mass ARRAY (3) | 1.15 (0.98, 1.35), 0.08, 0, 0.43 | 1.34 (0.87, 2.05), 0.19, 42, 0.18 | 1.15 (0.94, 1.40), 0.16, 0, 0.95 | 1.17 (0.97, 1.41), 0.10, 0, 0.92 | 1.41 (0.15, 13.18), 0.76, 94, < 0.00001 |
| PRM2 rs1646022 | C vs. G | CC vs. GG | GC vs. GG | CC + GC vs. GG | CC vs. GC + GG |
|---|---|---|---|---|---|
| OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | |
| Total (9) | 1.19 (1.06, 1.34), 0.004, 44, 0.08 | 1.15 (0.90, 1.48), 0.26, 31, 0.17 | 1.08 (0.74, 1.56), 0.70, 68, 0.002 | 1.05 (0.77, 1.43), 0.76, 0.60, 0.01 | 0.98 (0.82, 1.17), 0.82, 0, 0.54 |
| Ethnicity | |||||
| Asian (2) | 1.38 (1.18, 1.63), < 0.0001, 42, 0.19 | 1.27 (0.53, 3.05), 0.59, 79, 0.03 | 1.21 (0.38, 3.85), 0.75, 93, 0.0001 | 1.18 (0.41, 3.39), 0.76, 92, 0.0003 | 0.99 (0.79, 1.24), 0.93, 0, 0.85 |
| Caucasian (6) | 1.02 (0.84, 1.24), 0.86, 12, 0.34 | 0.99 (0.65, 1.50), 0.94, 0, 0.44 | 1.04 (0.78, 1.39), 0.78, 48, 0.10 | 1.03 (0.79, 1.35), 0.81, 27, 0.23 | 0.98 (0.67, 1.43), 0.90, 27, 0.23 |
| Mixed (1) | 0.98 (0.69, 1.39), 0.91 | 0.98 (0.58, 1.66), 0.94 | 1.17 (0.55, 2.49), 0.69 | 1.02 (0.62, 1.68), 0.93 | 0.94 (0.57, 1.54), 0.80 |
| Control source | |||||
| HB (5) | 1.26 (1.09, 1.46), 0.001, 39, 0.16 | 1.43 (1.06, 1.94), 0.02, 0, 0.43 | 1.18 (0.69, 2.01), 0.55, 70, 0.009 | 1.19 (0.79, 1.80), 0.41, 61, 0.04 | 1.03 (0.84, 1.27), 0.74, 0, 0.45 |
| PB (4) | 1.05 (0.86, 1.29), 0.62, 48, 0.12 | 0.74 (0.48, 1.14), 0.18, 0, 0.57 | 0.92 (0.59, 1.44), 0.71, 55, 0.11 | 0.84 (0.65, 1.09), 0.20, 44, 0.15 | 0.79 (0.53, 1.18), 0.25, 0, 0.56 |
| Method | |||||
| PCR-based (9) | 1.05 (0.91, 1.21), 0.52, 0, 0.48 | 0.94 (0.70, 1.26), 0.68, 0, 0.65 | 0.91 (0.73, 1.13), 0.39, 47, 0.08 | 0.91 (0.75, 1.11), 0.37, 31, 0.18 | 0.96 (0.73, 1.26), 0.75, 0, 0.44 |
| Mass ARRAY (1) | 1.49 (1.23, 1.82), < 0.0001 | 1.93 (1.21, 3.08), 0.006 | 2.20 (1.37, 3.56), 0.001 | 2.04 (1.30, 3.22), 0.002 | 1.00 (0.78, 1.27), 0.99 |
| PRM2 rs2070923 | A vs. C | AA vs. CC | CA vs. CC | AA + CA vs. CC | AA vs. CA + CC |
|---|---|---|---|---|---|
| OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | |
| Total (8) | 0.88 (0.78, 0.99), 0.04, 1, 0.43 | 0.84 (0.68, 1.04), 0.10, 0, 0.59 | 1.05 (0.71, 1.56), 0.81, 0.63, 0.009 |
0.90 (0.76, 1.07), 0.24, 35, 0.15 |
00.80 (0.67, 0.97), 0.02, 23, 0.25 |
| Ethnicity | |||||
| Asian (2) | 0.88 (0.74, 1.04), 0.13, 0, 0.41 | 0.87 (0.64, 1.19), 0.38, 0, 0.68 | 0.78 (0.58, 1.03), 0.08, 0, 0.44 | 0.81 (0.63, 1.03), 0.09, 0, 0.37 | 1.68 (0.44, 6.44), 0.45, 80, 0.03 |
| Caucasian (5) | 0.96 (0.79, 1.17), 0.71, 0, 0.60 | 0.86 (0.60, 1.23), 0.40, 19, 0.29 | 1.40 (0.66, 3.00), 0.38, 73, 0.005 | 1.11 (0.83, 1.47), 0.48, 40, 0.16 | 0.69 (0.51, 0.94), 0.02, 39, 0.16 |
| Mixed (1) | 0.65 (0.46, 0.93), 0.02 | 0.70 (0.42, 1.19), 0.19 | 0.97 (0.45, 2.10), 0.93 | 0.76 (0.46, 1.25), 0.28 | 0.71 (0.43, 1.17), 0.17 |
| Control source | |||||
| HB (5) | 0.88 (0.75, 1.02), 0.10, 13, 0.33 | 0.84 (0.65, 1.08), 0.17, 0, 0.45 | 1.06 (0.68, 1.67), 0.79, 52, 0.08 | 0.91 (0.72, 1.14), 0.41, 46, 0.12 | 0.81 (0.65, 1.01), 0.06, 17, 0.31 |
| PB (3) | 0.88 (0.72, 1.07), 0.19, 17, 0.30 | 0.84 (0.57, 1.24), 0.38, 0, 0.38 | 1.26 (0.46, 3.41), 0.65, 80, 0.006 | 0.89 (0.69, 1.16), 0.41, 41, 0.19 | 0.82 (0.46, 1.44), 0.48, 53, 0.12 |
| Method | |||||
| PCR-based (7) | 0.86 (0.74, 0.99), 0.04, 10, 0.35 | 0.80 (0.61, 1.05), 0.10, 0, 0.51 | 1.11 (0.68, 1.83), 0.67, 68, 0.004 | 0.90 (0.74, 1.10), 0.32, 44, 0.10 | 0.72 (0.57, 0.92), 0.009, 16, 0.31 |
| Mass ARRAY (1) | 0.93 (0.74, 1.16), 0.52 | 0.90 (0.63, 1.29), 0.58 | 0.89 (0.57, 1.39), 0.61 | 0.90 (0.64, 1.26), 0.54 | 0.95 (0.70, 1.28), 0.73 |
PCR Polymerase chain reaction, HB hospital-based, PB population-based. Bold numbers indicate statistically significant differences.
Meta-regression analysis
The results of meta-regression analysis for four polymorphisms based on publication year are shown in Table 5. The publication year could be a cofounding factor for PRM1 rs737008, PRM1 rs2301365, and PRM2 rs1646022 polymorphisms.
Table 5.
Meta-regression analysis for PRM1 rs737008, PRM1 rs2301365, PRM2 rs2070923, and PRM2 rs1646022 polymorphisms based on publication year.
| Polymorphism | Indexes | Allele | Homozygote | Heterozygous | Recessive | Dominant |
|---|---|---|---|---|---|---|
| PRM1 rs737008 | R | 0.152 | 0.639 | 0.573 | 0.572 | 0.066 |
| Adjusted R2 | − 0.66 | 0.354 | 0.267 | 0.266 | − 0.086 | |
| P-value | 0.620 | 0.019 | 0.041 | 0.041 | 0.831 | |
| PRM1 rs2301365 | R | 0.545 | 0.660 | 0.619 | 0.630 | 0.241 |
| Adjusted R2 | 0.209 | 0.365 | 0.306 | 0.322 | − 0.060 | |
| P-value | 0.104 | 0.038 | 0.057 | 0.051 | 0.503 | |
| PRM2 rs1646022 | R | 0.225 | 0.698 | 0.267 | 0.358 | 0.534 |
| Adjusted R2 | − 0.085 | 0.414 | − 0.083 | 0.004 | 0.183 | |
| P-value | 0.561 | 0.036 | 0.522 | 0.344 | 0.139 | |
| PRM2 rs2070923 | R | 0.234 | 0.059 | 0.012 | 0.249 | 0.251 |
| Adjusted R2 | − 0.103 | − 0.163 | − 0.166 | − 0.094 | − 0.093 | |
| P-value | 0.576 | 0.889 | 0.977 | 0.552 | 0.549 |
Allele: A vs. C, homozygous: AA vs. CC, heterozygous: AG vs. CC, recessive: AA + CA vs. CC, and dominant: AA vs. CA + CC, for PRM1 rs737008, PRM1 rs2301365, and PRM2 rs2070923 polymorphisms. Allele: C vs. G, homozygous: CC vs. GG, heterozygous: GC vs. GG, recessive: CC + GC vs. GG, and dominant: CC vs. GC + GG, for PRM2 rs1646022 polymorphism. Bold numbers indicate statistically significant differences.
Sensitivity analysis
We excluded the studies with a deviation of HWE in the controls, including two studies30,33 for polymorphism of PRM1 rs737008, six25,29,30,36,38,39 for PRM2 rs1646022, and four25,30,32,38 for PRM2 rs2070923. The results after excluding are presented in Table 6. Moreover, the sensitivity analysis based on “one study removed” and “cumulative analysis” on the previous analyses did not change the results and therefore confirmed the stability of the pooled data.
Table 6.
Sensitivity analysis at the studies without deviation of HWE in the controls.
| Polymorphism (number of studies) | Allele | Homozygote | Heterozygous | Recessive | Dominant |
|---|---|---|---|---|---|
| OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | OR (95% CI), P, I2, Ph | |
| PRM1 rs737008 (11) | 0.96 (0.82, 1.14), 0.66, 51, 0.03 | 1.11 (0.86, 1.42), 0.42, 27, 0.19 | 0.95 (0.79, 1.14), 0.57, 43, 0.06 | 0.96 (0.81, 1.14), 0.65, 47, 0.04 | 1.07 (0.89, 1.27), 0.48, 16, 0.29 |
| PRM2 rs1646022 (2) | 1.20 (0.96, 1.48), 0.10, 0, 0.92 | 0.96 (0.59, 1.56), 0.87, 0, 0.67 | 1.05 (0.61, 1.80), 0.87, 67, 0.05 | 1.04 (0.63, 1.73), 0.88, 66, 0.05 | 0.98 (0.62, 1.56), 0.93, 0, 0.94 |
| PRM2 rs2070923 (4) | 0.94 (0.77, 1.14), 0.53, 12, 0.33 | 0.88 (0.58, 1.31), 0.52, 31, 0.22 | 0.80 (0.61, 1.06), 0.12, 11, 0.34 | 0.82 (0.63, 1.06), 0.12, 47, 0.13 | 0.97 (0.67, 1.41), 0.87, 0, 0.52 |
Allele: A vs. C, homozygous: AA vs. CC, heterozygous: AG vs. CC, recessive: AA + CA vs. CC, and dominant: AA vs. CA + CC, for PRM1 rs737008, and PRM2 rs2070923 polymorphisms. Allele: C vs. G, homozygous: CC vs. GG, heterozygous: GC vs. GG, recessive: CC + GC vs. GG, and dominant: CC vs. GC + GG, for PRM2 rs1646022 polymorphism.
Publication bias
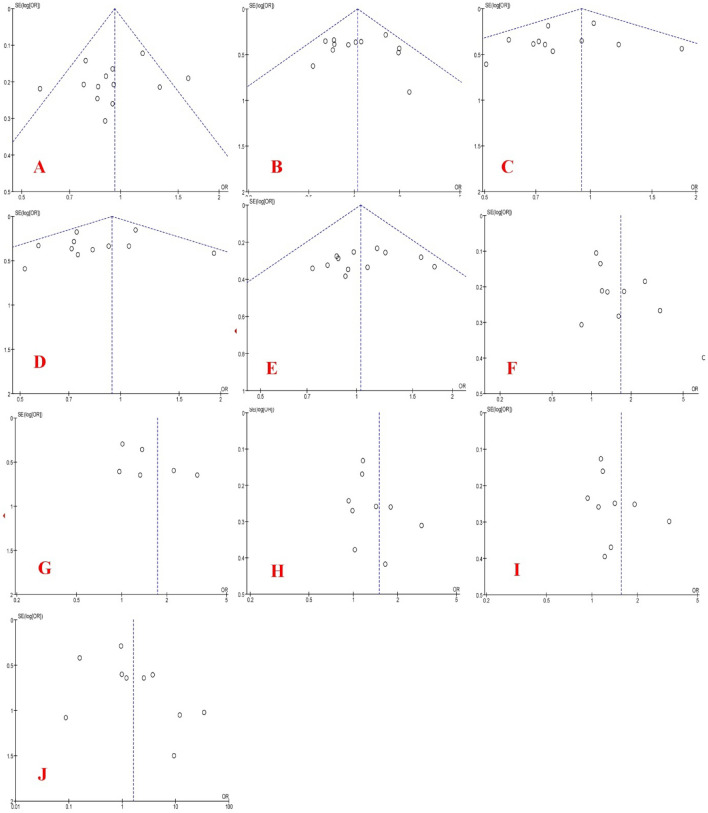
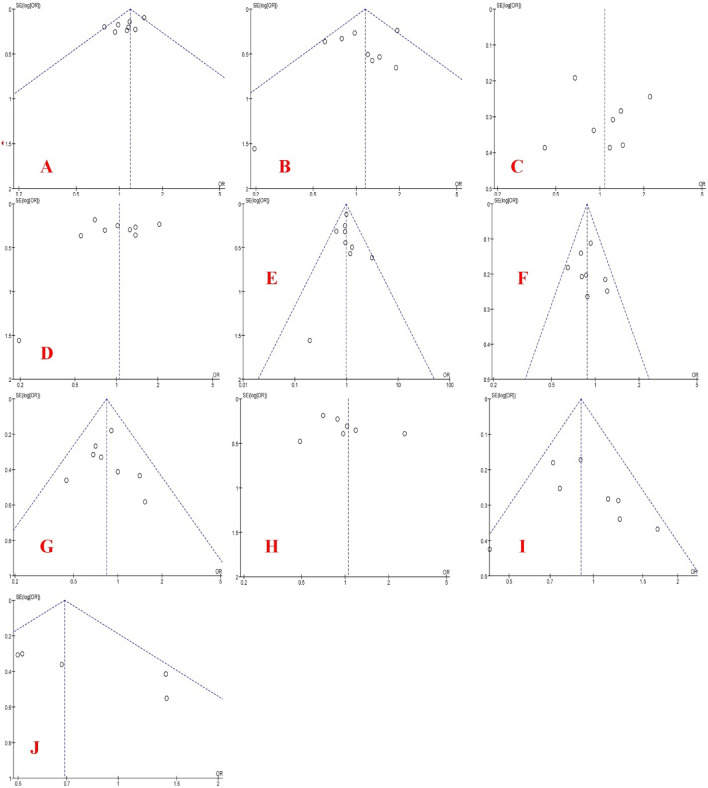
The funnel plots of PRM1 and PRM2 polymorphisms based on five genetic models are shown in Figs. 6 and 7, respectively. As the results showed, Egger’s test revealed the publication bias for AA + CA vs. CC (P < 0.001) and AA vs. CA + CC models (P = 0.04) in PRM1 rs737008 polymorphism and C vs. G model (P = 0.016) in PRM2 rs1646022 polymorphism. In addition, Begg’s test revealed the publication bias for AA + CA vs. CC (P = 0.001) model in PRM1 rs737008 polymorphism, CA vs. CC (P = 0.025) and AA + CA vs. CC models (P = 0.039) in PRM1 rs2301365 polymorphism.
Figure 6.
Funnel plots of PRM1 polymorphism based on five genetic models (allelic, homozygote, heterozygote, recessive, and dominant models, respectively): (A–E) for rs737008 and (F–J) for rs2301365.
Figure 7.
Funnel plots of PRM2 polymorphism based on five genetic models (allele, homozygote, heterozygote, recessive, and dominant models, respectively): (A–E) for rs1646022 and (F–J) for rs2070923.
Discussion
There is considerable empirical evidence to suggest that PRMs are necessary for male infertility and that PRM1 and PRM2 have a fundamental role in sperm chromatin density and spermatogenesis40,41 Any single nucleotide polymorphism in the coding and non-coding areas of PRM1 and PRM2 genes may cause significant abnormalities in their expression9. The changes in one set of genes and expression patterns impact the spermatogenesis process and its products, resulting in spermatogenesis dysfunction and leading to male infertility42. Nowadays, the findings on the association of PRM genes with male infertility are not fully convincing, and there are not sufficient studies on this topic32. A research confirmed that the expression of PRMs is uniquely related to the transcription/translation factors43. The present meta-analysis showed that PRM1 rs737008 polymorphism was not associated with the risk of male infertility. PRM1 rs2301365 and PRM2 rs1646022 polymorphisms were associated with an elevated risk of male infertility and PRM2 rs2070923 polymorphism had a protective role in infertile men. In addition, the subgroup analysis showed the effect of ethnicity, control source, and genotyping method on the association of PRM polymorphisms with the risk of male infertility. The results of meta-regression showed that publication year was a cofounding factor involved in the association between PRM1 rs737008, PRM1 rs2301365, and PRM2 rs1646022 polymorphisms and susceptibility to male infertility. Although single nucleotide polymorphism of G197T that lead to arginine to serine conversion was required in highly protected clusters of arginine for normal DNA binding has been found in 10% of unrelated infertile cases whose sperms were phenotypically same as those from mice with PRMN deficiency44.
It has been shown that PRM1 and PRM2 variants are related to male infertility in both humans and animals25,26. In the animal model, reduction of PRM causes sperm morphology defects due to decreased motility and infertility as a result of haploid germ deficiency45–47. Using gene–gene interaction analysis, Jiang et al.36 examined twelve combined genotypes of PRM polymorphisms. Their results showed a significant association between the combined genotypes and male infertility. One study reported that sperm concentration, motility, and morphology significantly decreased in patients with an aberrant PRM ratio48. PRM protection is very important in mammals and minor alternations in the coding and non-coding regions of PRM genes may cause important abnormalities in the expression or maintenance of gene expression stability9. PRMs may act as a checkpoint for spermatogenesis, where abnormal PRM expression causes the induction of an apoptotic process that may explain the decrease in sperm production12. In addition, studies have shown that abnormal PRM expression is related to defective spermatogenesis12. There is some evidence that PRM mutations or polymorphisms may induce alternations at the protein level and their composition in sperm chromatin, resulting in sperm deficiency46,47. Semen quality decreases with age and characteristic molecular changes occur during aging (increased damage of sperm DNA, sperm infection changes, and plasma miRNA profile changes). In addition, the logistic regression models have illustrated an association between age and semen parameters49.
As the present meta-analysis demonstrated, ethnicity, control source, and genotyping method of PRM polymorphisms are important and may contribute to the difference in susceptibility to male infertility. A meta-analysis17 reported an association between PRM1 rs2301365 polymorphism and the risk of male infertility in the Caucasians, not in the Asians. As in our meta-analysis, there was an elevated risk of male infertility for PRM1 rs2301365 polymorphism only in Caucasians and for PRM2 rs1646022 polymorphism only in Asians. In addition, there was significantly a decreased risk of PRM1 rs737008 in population-based controls, elevated risk of PRM1 rs2301365 and PRM2 rs1646022 in hospital-based controls. Also, with regards to method, an elevated risk of PRM1 rs2301365 and a decreased risk of PRM2 rs2070923 in PCR-based method and an elevated risk of PRM2 rs1646022 in Mass ARRAY method. It is noteworthy that the expression of genes, environmental factors, and spermatogenesis disorder can play an important role in male sterility9. Another possible reason for these inconsistent findings can be a particular selection of the clinical subtypes of male infertility and PRM1 and PRM2 variations in different populations examined9. Therefore, existence of heterogeneity among studies may be due to the differences genotyping method, clinical subtypes of male infertility, ethnicity, publication year, control source, and even number of recruited patients38.
This meta-analysis had two significant limitations. First, the clinical data such as age, abstinence time, serum hormone index, and semen quality and parameters were not analyzed due to lack of information. Second, the meta-analysis did not evaluate the gene–gene and gene-environment interactions due to lack of information in the published studies.
Conclusions
The present meta-analysis evaluated four PRM polymorphisms (PRM1 rs737008, PRM1 rs2301365, PRM2 rs1646022, and PRM2 rs2070923). The results showed PRM1 rs2301365 and PRM2 rs1646022 polymorphisms were associated with an elevated risk of male infertility and PRM2 rs2070923 polymorphism had a protective role in infertile men. In addition, ethnicity, control source, and genotyping method impacted the PRM polymorphisms and susceptibility to male infertility. Based on the results, the future studies need to evaluate these polymorphisms in a large number of participants in various areas, with an emphasis on environmental factors, interactions, age, method, and selection of controls (deviation of HWE and source).
Acknowledgements
This meta-analysis was supported by Kermanshah University of Medical Sciences, Kermanshah, Iran (Project code: 990339).
Author contributions
H.N. designed the study. M.S. analyzed the data and wrote the manuscript. M.N. and M.M. critically revised the work. All authors have approved the final content of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vander Borght M, Wyns C. Fertility and infertility: Definition and epidemiology. Clin. Biochem. 2018;62:2–10. doi: 10.1016/j.clinbiochem.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Thonneau P, et al. Incidence and main causes of infertility in a resident population (1,850,000) of three French regions (1988–1989) Hum. Reprod. 1991;6:811–816. doi: 10.1093/oxfordjournals.humrep.a137433. [DOI] [PubMed] [Google Scholar]
- 3.Turner KA, et al. Male infertility is a women's health issue-research and clinical evaluation of male infertility is needed. Cells. 2020;9:990. doi: 10.3390/cells9040990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zandieh Z, et al. Comparing reactive oxygen species and DNA fragmentation in semen samples of unexplained infertile and healthy fertile men. Ir. J. Med. Sci. 2018;187:657–662. doi: 10.1007/s11845-017-1708-7. [DOI] [PubMed] [Google Scholar]
- 5.WHO . WHO Laboratory Manual for the Examination and Processing of Human Semen. Geneva: WHO Press; 2010. [PubMed] [Google Scholar]
- 6.Garolla A, et al. Sperm selected by both birefringence and motile sperm organelle morphology examination have reduced deoxyribonucleic acid fragmentation. Fertil. Steril. 2014;101:647–652. doi: 10.1016/j.fertnstert.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 7.Majzoub A, Agarwal A, Esteves SC. Clinical utility of sperm DNA damage in male infertility. Panminerva Med. 2019;61:118. doi: 10.23736/S0031-0808.18.03530-9. [DOI] [PubMed] [Google Scholar]
- 8.Esteves SC, Sharma RK, Gosálvez J, Agarwal A. A translational medicine appraisal of specialized andrology testing in unexplained male infertility. Int. Urol. Nephrol. 2014;46:1037–1052. doi: 10.1007/s11255-014-0715-0. [DOI] [PubMed] [Google Scholar]
- 9.Jiang W, et al. Polymorphisms in Protamine 1 and Protamine 2 predict the risk of male infertility: A meta-analysis. Sci. Rep. 2015;5:15300. doi: 10.1038/srep15300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yatsenko AN, et al. Association of mutations in the zona pellucida binding protein 1 (ZPBP1) gene with abnormal sperm head morphology in infertile men. Mol. Hum. Reprod. 2012;18:14–21. doi: 10.1093/molehr/gar057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamidian S, et al. The effect of vitamin C on the gene expression profile of sperm protamines in the male partners of couples with recurrent pregnancy loss: A randomized clinical trial. Clin. Exp. Reprod. Med. 2020;47:68. doi: 10.5653/cerm.2019.03188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrell DT, Emery BR, Hammoud S. Altered protamine expression and diminished spermatogenesis: What is the link? Hum. Reprod. Update. 2007;13:313–327. doi: 10.1093/humupd/dml057. [DOI] [PubMed] [Google Scholar]
- 13.Hamad MF. Quantification of histones and protamines mRNA transcripts in sperms of infertile couples and their impact on sperm’s quality and chromatin integrity. Reprod. Biol. 2019;19:6–13. doi: 10.1016/j.repbio.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Ganguly I, et al. Differential expression of protamine 1 and 2 genes in mature spermatozoa of normal and motility impaired semen producing crossbred Frieswal (HF× Sahiwal) bulls. Res. Vet. Sci. 2013;94:56–62. doi: 10.1016/j.rvsc.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Belokopytova IA, Kostyleva EI, Tomilin AN, Vorob'ev VI. Human male infertility may be due to a decrease of the protamine P2 content in sperm chromatin. Mol. Reprod. Dev. 1993;34:53–57. doi: 10.1002/mrd.1080340109. [DOI] [PubMed] [Google Scholar]
- 16.Cho C, et al. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat. Genet. 2001;28:82–86. doi: 10.1038/ng0501-82. [DOI] [PubMed] [Google Scholar]
- 17.He Q, Deng L, Deng S, Jin T. Association of protamine1 gene c.-190C>A polymorphism with male infertility risk: A meta-analysis. Int. J. Clin. Exp. Med. 2019;12:3047–3055. [Google Scholar]
- 18.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group Preferred reporting items for systematic reviews and meta-anal-yses: The PRISMA statement. J. Clin. Epidemiol. 2009;62:1006–1012. doi: 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Liberati A, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. J. Clin. Epidemiol. 2009;62:e1–34. doi: 10.1016/j.jclinepi.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Midgette AS, et al. Cost-effectiueness of streptokinase for acute myocardial infarction: A combined meta-analysis and decision analysis of the effects of infarct location and of likelihood of infarction. Med. Decis. Making. 1994;14:108–117. doi: 10.1177/0272989X9401400203. [DOI] [PubMed] [Google Scholar]
- 22.Zeyadi M, Alaauldeen SM, Al-Sallami ASM, Albaldawy MT. Single Nucleotide Polymorphism in Protamine 1 and Protamine 2 genes in fertile and infertile for men of Al-Najaf City. J. Phys. Conf. Ser. 2019;1234:012081. doi: 10.1088/1742-6596/1234/1/012081. [DOI] [Google Scholar]
- 23.Jodar M, et al. Polymorphisms, haplotypes and mutations in the protamine 1 and 2 genes. Int. J. Androl. 2011;34:470–485. doi: 10.1111/j.1365-2605.2010.01115.x. [DOI] [PubMed] [Google Scholar]
- 24.Tanaka H, Miyagawa Y, Tsujimura A, Matsumiya K, Okuyama A, Nishimune Y. Single nucleotide polymorphisms in the protamine-1 and -2 genes of fertile and infertile human male populations. Mol. Hum. Reprod. 2003;9:69–73. doi: 10.1093/molehr/gag010. [DOI] [PubMed] [Google Scholar]
- 25.Aoki VW, Christensen GL, Atkins JF, Carrell DT. Identification of novel polymorphisms in the nuclear protein genes and their relationship with human sperm protamine deficiency and severe male infertility. Fertil. Steril. 2006;86:1416–1422. doi: 10.1016/j.fertnstert.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 26.Ravel C, et al. Mutations in the protamine 1 gene associated with male infertility. Mol. Hum. Reprod. 2007;13:461–464. doi: 10.1093/molehr/gam031. [DOI] [PubMed] [Google Scholar]
- 27.Gazquez C, Oriola J, de Mateo S, Vidal-Taboada JM, Ballescà JL, Oliva R. A common Protamine 1 promoter polymorphism (−190 C→A) correlates with abnormal sperm morphology and increased protamine P1/P2 ratio in infertile patients. J. Androl. 2008;29:540–548. doi: 10.2164/jandrol.107.004390. [DOI] [PubMed] [Google Scholar]
- 28.Imken L, et al. Mutations in the protamine locus: Association with spermatogenic failure? Mol. Hum. Reprod. 2009;15:733–738. doi: 10.1093/molehr/gap056. [DOI] [PubMed] [Google Scholar]
- 29.Tuttelmann F, et al. A common haplotype of protamine 1 and 2 genes is associated with higher sperm counts. Int. J. Androl. 2010;33:e240–248. doi: 10.1111/j.1365-2605.2009.01003.x. [DOI] [PubMed] [Google Scholar]
- 30.Venkatesh S, Kumar R, Deka D, Deecaraman M, Dada R. Analysis of sperm nuclear protein gene polymorphisms and DNA integrity in infertile men. Syst. Biol. Reprod. Med. 2011;57:124–132. doi: 10.3109/19396368.2011.562960. [DOI] [PubMed] [Google Scholar]
- 31.Grassetti D, et al. Protamine-1 and -2 polymorphisms and gene expression in male infertility: An Italian study. J. Endocrinol. Investig. 2012;35:882–888. doi: 10.3275/8111. [DOI] [PubMed] [Google Scholar]
- 32.He XJ, et al. PRM1 variant rs35576928 (Arg>Ser) is associated with defective spermatogenesis in the Chinese Han population. Reprod. Biomed. Online. 2012;25:627–634. doi: 10.1016/j.rbmo.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Siasi E, Aleyasin A, Mowla J, Sahebkashaf H. Association study of six SNPs in PRM1, PRM2 and TNP2 genes in Iranian infertile men with idiopathic azoospermia. Iran J. Reprod. Med. 2012;10:329–336. [PMC free article] [PubMed] [Google Scholar]
- 34.Yu QF, et al. Association of PRM1–190C-> A polymorphism with teratozoospermia. Zhonghua Nan Ke Xue. 2012;18:314–317. [PubMed] [Google Scholar]
- 35.Jamali S, Karimian M, Nikzad H, Aftabi Y. The c.-190 C>A transversion in promoter region of protamine1 gene as a genetic risk factor for idiopathic oligozoospermia. Mol. Biol. Rep. 2016;43:795–802. doi: 10.1007/s11033-016-4017-8. [DOI] [PubMed] [Google Scholar]
- 36.Jiang W, et al. Polymorphisms of protamine genes contribute to male infertility susceptibility in the Chinese Han population. Oncotarget. 2017;8:61637–61645. doi: 10.18632/oncotarget.18660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aydos OSE, et al. Genetic polymorphisms in PRM1, PRM2, and YBX2 genes are associated with male factor infertility. Genet. Test Mol. Biomark. 2018;22:55–61. doi: 10.1089/gtmb.2017.0040. [DOI] [PubMed] [Google Scholar]
- 38.Nabi A, Khalili MA, Moshrefi M, Sheikhha MH, Zare Mehrjardi E, Ashrafzadeh HR. Polymorphisms in protamine 1 and 2 genes in asthenozoospermic men: A case-control study. Int. J. Reprod. Biomed. (Yazd) 2018;16:379–386. doi: 10.29252/ijrm.16.6.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dehghanpour F, Fesahat F, Miresmaeili SM, Zare Mehrjardi E, Honarju A, Talebi AR. Analysis of PRM1 and PRM2 polymorphisms in Iranian infertile men with idiopathic teratozoospermia. Int. J. Fertil. Steril. 2019;13:77–82. doi: 10.22074/ijfs.2019.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanippayoor RL, Alpern JH, Moehring AJ. Protamines and spermatogenesis in drosophila and homo sapiens: A comparative analysis. Spermatogenesis. 2013;3:e24376. doi: 10.4161/spmg.24376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Depa-Martynow M, Kempisty B, Jagodzinski PP, Pawelczyk L, Jedrzejczak P. Impact of protamine transcripts and their proteins on the quality and fertilization ability of sperm and the development of preimplantation embryos. Reprod. Biol. 2012;12:57–72. doi: 10.1016/S1642-431X(12)60077-1. [DOI] [PubMed] [Google Scholar]
- 42.Wu W, et al. Idiopathic male infertility is strongly associated with aberrant promoter methylation of methylenetetrahydrofolate reductase (MTHFR) PLoS ONE. 2010;5:e13884. doi: 10.1371/journal.pone.0013884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- 44.Iguchi N, Yang S, Lamb DJ, Hecht NB. An SNP in protamine 1: A possible genetic cause of male infertility? J. Med. Genet. 2006;43:382–384. doi: 10.1136/jmg.2005.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seki Y, et al. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134:2627–2638. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- 46.Takeda N, et al. Viable offspring obtained from Prm1-deficient sperm in mice. Sci. Rep. 2016;6:27409. doi: 10.1038/srep27409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho C, et al. Protamine 2 deficiency leads to sperm DNA damage and embryo death in mice. Biol. Reprod. 2003;69:211–217. doi: 10.1095/biolreprod.102.015115. [DOI] [PubMed] [Google Scholar]
- 48.Aoki VW, Liu L, Carrell DT. Identification and evaluation of a novel sperm protamine abnormality in a population of infertile males. Hum. Reprod. 2005;20:1298–1306. doi: 10.1093/humrep/deh798. [DOI] [PubMed] [Google Scholar]
- 49.Paoli D, et al. Cytological and molecular aspects of the ageing sperm. Hum. Reprod. 2019;34:218–227. doi: 10.1093/humrep/dey357. [DOI] [PubMed] [Google Scholar]