Abstract
Ovarian tumour domain-containing protein 3 (OTUD3), a key OTU (ovarian tumour protease) family deubiquitylase, plays context-dependent roles in cancers. It suppresses tumorigenesis in breast, colon, liver and cervical cancer through stabilizing PTEN (phosphatase and tension homologue deleted on chromosome 10) while promotes lung tumorigenesis through stabilizing GRP78 (The glucose-regulated protein 78 kDa). The regulation especially post-translational modification of OTUD3 remains unclear. Here, we report that the carboxyl terminus of Hsc70-interacting protein (CHIP) is a ubiquitin ligase for OTUD3. CHIP interacts with, polyubiquitylates OTUD3 and promotes OTUD3 degradation. Knockdown of CHIP stabilizes OTUD3 which leads to elevated GRP78 levels in lung cancer cells. CHIP-knockdown lung cancer cells exhibit increased invasion in OTUD3 and GRP78 dependent manner. Further study demonstrates that CHIP-knockdown lung cancer cells are more prone to metastasize to mice lung when injected intravenously or subcutaneously. Moreover, the expression of CHIP is low in human lung cancer tissues and inversely correlates with OTUD3 expression and GRP78 expression. Furthermore, we identified CHIP mutations in human lung cancers, which reduce CHIP catalytic activity. These findings demonstrate that CHIP is a negative regulator of OTUD3 and CHIP suppresses lung cancer metastasis through inhibiting OTUD3-GRP78 signaling axis.
Subject terms: Tumour-suppressor proteins, Ubiquitylation
Introduction
The ubiquitin-proteasome system (UPS) which is primarily consisted of ubiquitin, ubiquitylation enzymes and the 26S proteasome modulates protein homoeostasis in eukaryotic organisms [1]. The 76-amino-acid ubiquitin protein is activated by a ubiquitin-activating enzyme (E1), then transferred to a ubiquitin-conjugating enzyme (E2) through the formation of a thioester bond. The ubiquitin ligase enzyme (E3) recognizes specific target proteins and facilitates transference of ubiquitin conjugated with E2 to the target proteins [2, 3]. The enzymatic cascades results in proteasomal degradation of proteins or non-proteolytic outcomes such as altered subcellular localization or activity [4]. Ubiquitylation of many proteins can be reversed by deubiquitylases (DUBs) [5]. Currently, about 100 DUBs have been identified, which are categorized into six families, including ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumour proteases (OTUs), motif interacting with ubiquitin-containing novel DUB family, Josephins (also termed MJDs) and Jad1/Pad/MPN-domain-containing metalloenzymes [6]. DUBs function as oncogenes or tumour suppressors in different types of cancers [7]. For example, USP7 exhibits oncogenic roles in various cancer tissues such as lung cancer, multiple myeloma, liver cancer and cervical cancer [8–11]; BAP1 plays tumour suppressive roles in cancers such as malignant mesothelioma and melanomas [12–14]; OTUD7B plays context-dependent roles in different cancer types [15, 16].
The OTU family DUBs are special as most of them are ubiquitin chain linkage specific [17]. Previously we found the OTU family member ovarian tumour domain-containing protein 3 (OTUD3) could deubiquitylate and stabilize the tumour suppressor PTEN [18]. In concert with this, OTUD3 transgenic mice display increased PTEN protein level and decreased susceptibility to breast tumorigenesis [18], while OTUD3 knockout (KO) mice exhibit decreased PTEN protein level and increased susceptibility to breast cancer [19]. Further studies reveal OTUD3 level is downregulated, concomitant with decreased PTEN level, in breast cancer as well as in hepatocellular cancer, colon cancer and cervical cancer [18, 19]. In addition, OTUD3 loss-of-function mutations further support a tumour suppressive role of OTUD3 [18]. However, we recently observed OTUD3 transgenic mice are more susceptible to KrasG12D driven lung cancer while OTUD3 KO mice are less susceptible [19], implicating an oncogenic role of OTUD3 in lung cancer. Furthermore, OTUD3 is overexpressed in lung cancer and the increased expression of OTUD3 correlates with poor survival in lung cancer patients [19]. Mechanistically, OTUD3 promotes lung adenocarcinoma progression through deubiquitylation and stabilization of GRP78 [19]. GRP78 is a major endoplasmic reticulum chaperone, which facilitates protein folding and participates in unfolded protein response [20, 21]. GRP78 is overexpressed in multiple cancers and involved in promoting tumour growth and metastasis [22–27]. Targeting of cell surface-localized GRP78 in cancers inhibits tumour growth and metastasis [28–30]. Thus, OTUD3 plays context-dependent roles in different types of cancers. However, the regulation especially post-translational modification of OTUD3 is still elusive.
The carboxyl terminus of Hsc70-interacting protein (CHIP) is a U-box subfamily ubiquitin ligase which interacts with Hsp70 and Hsp90 chaperones and promotes protein degradation [31–33]. CHIP contains three tandem tetratricopeptide-repeat (TPR) domains which mediate the interaction with chaperone and a U-box domain which recruits ubiquitin-conjugating enzymes (E2s) [32]. CHIP is involved in multiple physiological and pathological processes such as protein quality control, apoptosis, aging, immunity, neurological disorders, cardiac disease and cancer [34]. PTEN has been reported as a substate of CHIP [35]. CHIP interacts with PTEN and promotes PTEN ubiquitylation and proteasomal degradation [35]. CHIP also degrades other substrates such as c-Myc, p53, HIF-1-α and RUNX2 [36–39]. CHIP has been linked to several kinds of tumours. A tumour suppressor function has been reported for CHIP in cancers such as breast cancer, gastric cancer and ovarian cancer [40–45], whereas several studies suggest an oncogenic role of CHIP in prostate cancer and gallbladder cancer [46, 47]. Thus, CHIP may play context-dependent roles in tumorigenesis through specific oncogenes or tumour suppressors. However, what role CHIP plays in lung cancer is not elucidated.
In this study, we identified CHIP as a ubiquitin ligase for OTUD3. Loss of CHIP in lung cancer cell lines stabilizes OTUD3, which elevates GRP78 levels. The increased GRP78 facilitates lung cancer cell invasion. Moreover, we found that CHIP deficient lung cancer cells are more prone to metastasize to mice lung. Further studies reveal that CHIP level is downregulated in human lung cancer tissues and CHIP mutations in human lung cancers reduce CHIP catalytic activity. Overall, our data indicate that CHIP inhibits OTUD3-GRP78 signaling axis and suppresses tumour metastasis in lung cancer.
Results
CHIP downregulates OTUD3 levels
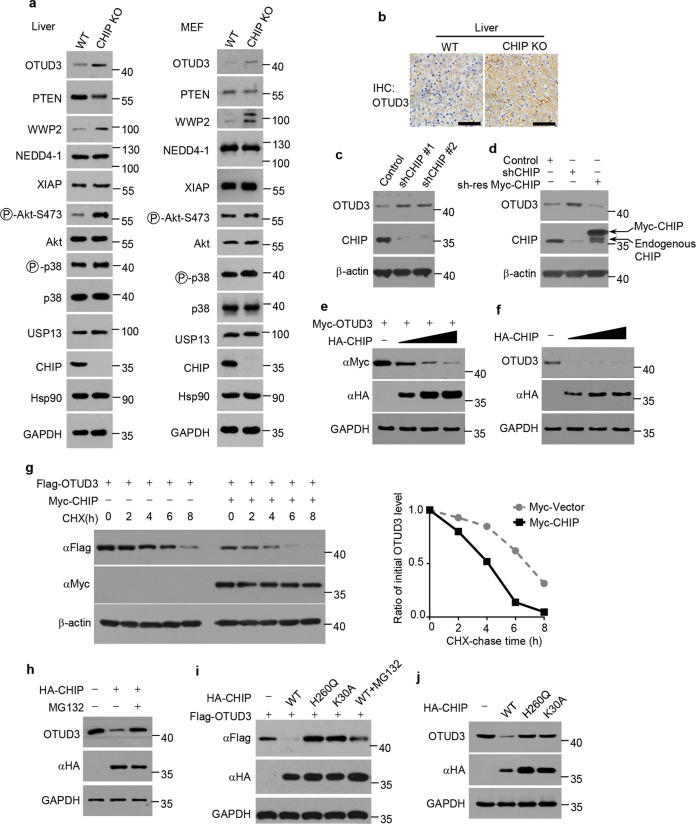
PTEN plays a central role in suppressing various human cancers [48]. Recently we took advantage of specific gene KO mice to elucidate the physiological ubiquitin ligase for PTEN. We found PTEN protein levels are upregulated in WWP2 KO mouse embryonic fibroblasts (MEFs) and several tissues [49]. Consistently, ubiquitylation of PTEN is decreased in WWP2 KO MEFs [49]. However, the PTEN protein levels have no changes in CHIP KO MEFs and tissues when compared with wildtype (WT) MEFs and tissues [49]. PTEN ubiquitylation is also similar between WT and CHIP KO MEFs [49]. We speculated CHIP KO might induce increased negative regulator for PTEN levels, which led to unchanged PTEN levels. Indeed, we found WWP2 levels were upregulated in CHIP KO liver and MEFs (Fig. 1a). Surprisingly, the OTUD3 levels were also elevated in CHIP KO liver and MEFs (Fig. 1a), which was confirmed by immunostaining OTUD3 in mice liver (Fig. 1b). The protein levels of other E3 ligases and DUBs for PTEN, such as NEDD4-1, XIAP and USP13, had no changes in CHIP KO MEFs (Fig. 1a). Thus, we speculated CHIP might be a negative regulator for OTUD3 levels.
Fig. 1. CHIP downregulates OTUD3 levels.
a Livers from newborn CHIP WT and KO littermates (P0) and MEFs from CHIP WT and KO embryos (E13.5) were collected and the indicated proteins were detected by western blotting. b Representative images of OTUD3 immunohistochemical staining in liver tissues from newborn CHIP WT and KO littermates (P0). Scale bars: 50 μm. c Control or CHIP shRNAs (shCHIP #1, shCHIP #2) were packaged into lentiviral particles and transduced into H1299 cells. The positive clones were selected by puromycin (2 µg/ml). The indicated proteins were detected by western blotting. d H1299 cells stably expressing CHIP shRNA (shCHIP #1) were transfected with or without shRNA-resistant (sh-res) Myc-tagged CHIP plasmids for 48 h. The indicated proteins were detected by western blotting. e 293T cells were transfected for 48 h with Myc-tagged OTUD3 and increasing amounts of HA tagged CHIP plasmids. The indicated proteins were detected by western blotting. f 293T cells transfected with increasing amounts of HA tagged CHIP plasmids were collected 48 h post-transfection. The indicated proteins were detected by western blotting. g 293T cells were transfected for 40 h with Flag-tagged OTUD3 alone or together with Myc-tagged CHIP plasmids, and then treated with cycloheximide (CHX) (100 µg/ml) for indicated time period. The indicated proteins were detected by western blotting. The graph shows quantification of relative OTUD3 levels. h 293T cells were transfected for 40 h with or without HA tagged CHIP plasmids, and then treated with or without MG132 (20 µM) for 8 h. The indicated proteins were detected by western blotting. i 293T cells were transfected for 40 h with Flag-tagged OTUD3 alone or together with HA tagged WT CHIP, H260Q mutant CHIP or K30A mutant CHIP plasmids, and then treated with or without MG132 (20 µM) for 8 h. The indicated proteins were detected by western blotting. j 293T cells transfected with or without HA tagged WT CHIP, H260Q mutant CHIP or K30A mutant CHIP plasmids were collected 48 h post-transfection. The indicated proteins were detected by western blotting.
To test our hypothesis, we knocked down CHIP in H1299 cells and detected OTUD3 levels. Compared with control cells, OTUD3 levels were increased in CHIP-knockdown cells (Fig. 1c). The increased OTUD3 levels were reversed by overexpression of shRNA-resistant CHIP (Fig. 1d). Furthermore, ectopic expression of CHIP downregulated exogenous and endogenous OTUD3 levels in a dose-dependent manner (Fig. 1e, f). These results further confirmed CHIP negatively regulates OTUD3 levels. We next determined whether CHIP accelerated OTUD3 degradation. We treated cells with the protein synthesis inhibitor cycloheximide (CHX). The half-life of OTUD3 was shortened in cells overexpressing CHIP (Fig. 1g), demonstrating CHIP accelerates OTUD3 degradation to downregulate OTUD3 levels. As CHIP is a U-box subfamily ubiquitin ligase, we analyzed whether the accelerated degradation of OTUD3 is proteasome dependent. Indeed, proteasome inhibitor MG132 prevented the downregulation of OTUD3 levels caused by CHIP (Fig. 1h). Previous studies reveal that CHIP H260Q mutant lacks E3 ligase activity while CHIP K30A mutant shows defective binding to Hsp70 [50, 51]. We overexpressed WT CHIP, H260Q mutant CHIP or K30A mutant CHIP and detected exogenous or endogenous OTUD3 levels. Overexpression of WT CHIP decreased exogenous and endogenous OTUD3 levels while H260Q or K30A mutant CHIP lost this effect (Fig. 1i, j), indicating CHIP downregulates OTUD3 in E3 ligase activity and HSP70 dependent manner.
CHIP interacts with OTUD3
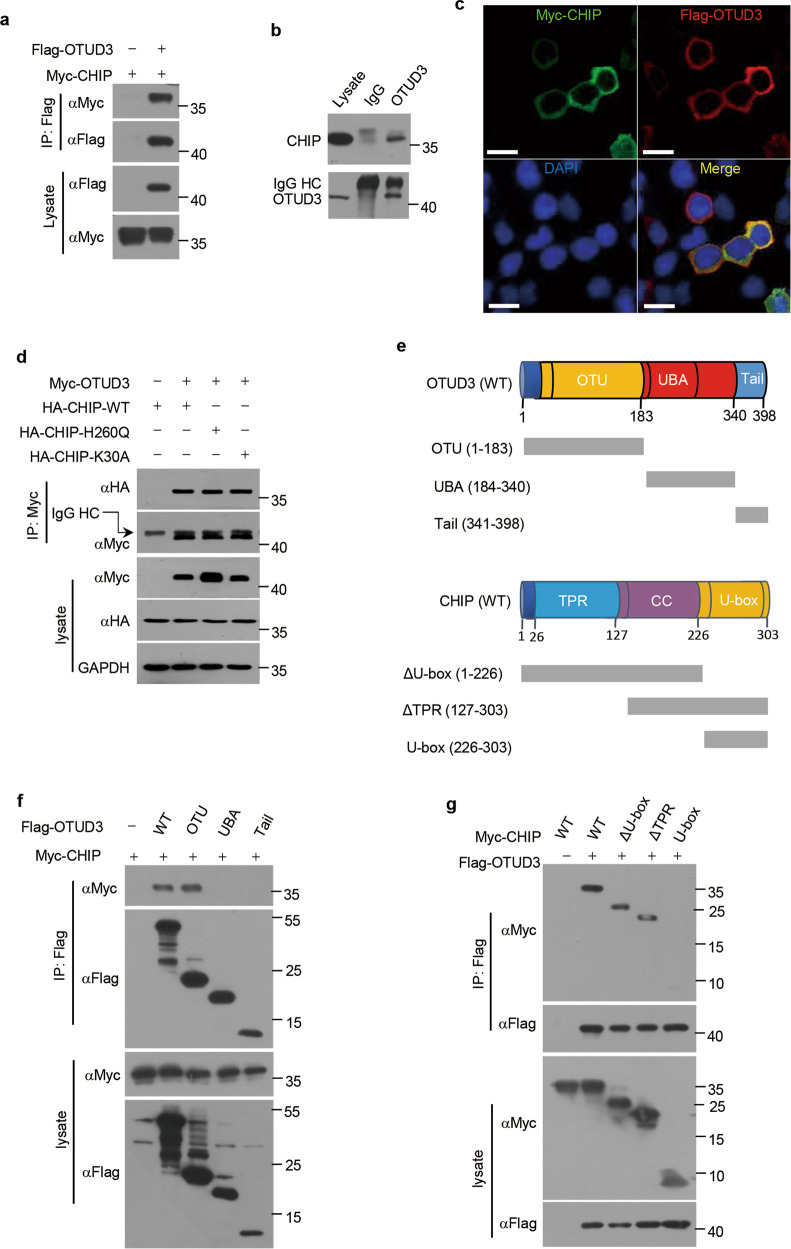
As CHIP reduces the steady-state level of OTUD3, we further examined whether CHIP interacts with OTUD3. We carried out co-immunoprecipitation assays following transfection of plasmids encoding Myc-tagged CHIP alone or together with plasmids encoding Flag-tagged OTUD3. Exogenous CHIP and OTUD3 proteins were co-immunoprecipitated (Fig. 2a). The interaction between endogenous CHIP and endogenous OTUD3 was further confirmed in H1299 cells (Fig. 2b). Immunostaining showed that CHIP and OTUD3 were colocalized in the cytoplasm (Fig. 2c). OTUD3 also bound CHIP H260Q mutant or CHIP K30A mutant, indicating the interaction between CHIP and OTUD3 is E3 ligase activity and Hsp70 independent (Fig. 2d). CHIP contains a TPR domain essential for chaperone binding, a coiled-coil (CC) domain and a U-box domain responsible for ubiquitin ligase activity. OTUD3 contains a OTU domain, a ubiquitin associated (UBA) domain and a C-terminal tail. To determine the interaction domain(s) of CHIP and OTUD3, we constructed several deletion mutants of CHIP and OTUD3 (Fig. 2e). Co-immunoprecipitation assays demonstrated that OTUD3 deletion mutants OTU interacted with CHIP while UBA and tail did not bind to CHIP (Fig. 2f), demonstrating OTU domain of OTUD3 mediated the interaction with CHIP. In addition, both ΔU-box deletion mutant and ΔTPR deletion mutant of CHIP interacted with OTUD3, while U-box did not, indicating CC domain of CHIP mediated the physical interaction with OTUD3 (Fig. 2g). Taken together, these results indicate that the interaction between OTUD3 and CHIP is mediated by OTU domain of OTUD3 and CC domain of CHIP.
Fig. 2. CHIP interacts with OTUD3.
a 293T cells were transfected for 36 h with Myc-tagged CHIP alone or together with Flag-tagged OTUD3 plasmids, and then immunoprecipitated with Flag antibody, followed by western blotting with indicated antibodies. b H1299 whole-cell lysates were immunoprecipitated with control IgG or OTUD3 antibody, followed by western blotting with indicated antibodies. c H1299 cells transfected with Myc-tagged CHIP and Flag-tagged OTUD3 were fixed 36 h post-transfection, and then incubated with primary antibodies. FITC-conjugated secondary antibody was used to detect CHIP (green) while TRITC-conjugated secondary antibody was used to detect OTUD3 (red). The nuclei were stained by DAPI. Scale bars: 40 μm. d 293T cells were transfected for 36 h with HA tagged WT CHIP, H260Q mutant CHIP or K30A mutant CHIP alone or together with Myc-tagged OTUD3 plasmids, and then immunoprecipitated with Myc antibody, followed by western blotting with indicated antibodies. e Overview of WT OTUD3 and its deletion mutants OTU (1-183), UBA (184-340) and Tail (341-398); WT CHIP and its deletion mutants ΔU-box (1-226), ΔTPR (127-303) and U-box (226-303). f 293T cells were transfected for 36 h with Myc-tagged CHIP alone or together with Flag-tagged WT OTUD3 or its deletion mutants, and then immunoprecipitated with Flag antibody, followed by western blotting with indicated antibodies. g 293T cells were transfected for 36 h with Myc-tagged WT CHIP or its deletion mutants alone or together with Flag-tagged OTUD3, and then immunoprecipitated with Flag antibody, followed by western blotting with indicated antibodies.
CHIP promotes ubiquitylation of OTUD3
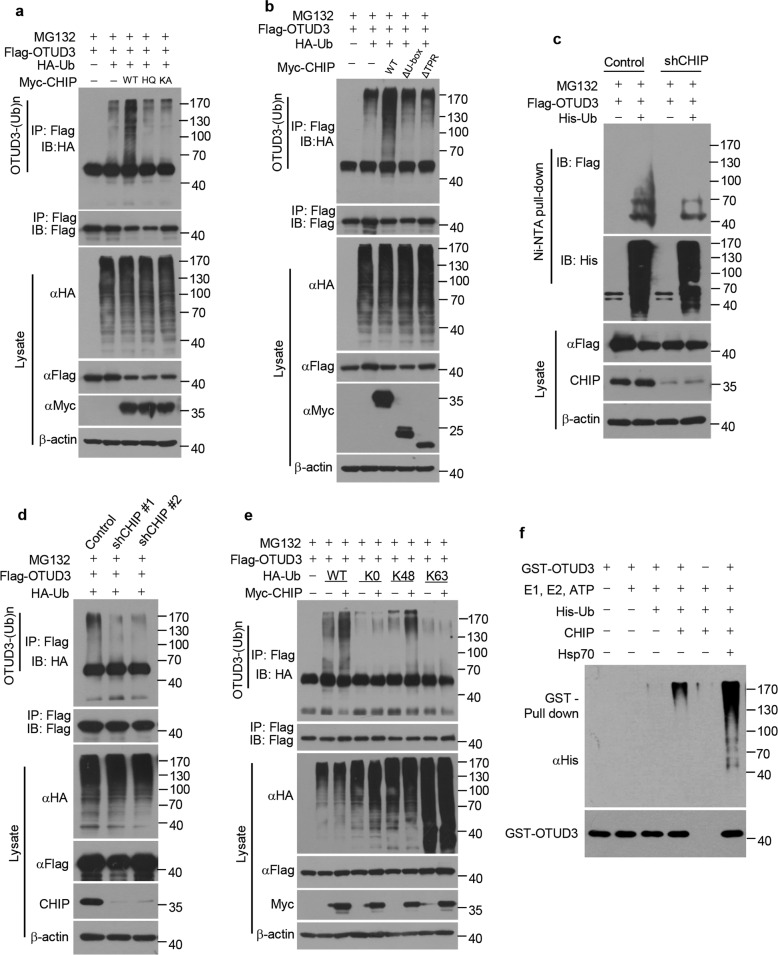
As CHIP is a ubiquitin ligase, we went on to examine whether CHIP promotes OTUD3 ubiquitylation. Ectopic expression of WT, but not H260Q and K30A, CHIP, promoted OTUD3 ubiquitylation in cells (Fig. 3a). Moreover, ΔU-box deletion mutant and ΔTPR deletion mutant also lost the ability to ubiquitylate OTUD3 (Fig. 3b), indicating CHIP promotes OTUD3 ubiquitylation in E3 ligase activity and Hsp70 dependent manner. Conversely, knockdown of CHIP decreased OTUD3 ubiquitylation in cells (Fig. 3c, d). We next examined which type of ubiquitin chain of OTUD3 was affected by CHIP. CHIP promoted Lys 48-linked ubiquitylation, but not monoubiquitylation nor the non-degradative Lys 63-linked ubiquitylation of OTUD3 (Fig. 3e). In vitro ubiquitylation assays showed that CHIP directly ubiquitylated OTUD3, especially in the presence of Hsp70 (Fig. 3f). Taken together, CHIP is a specific ubiquitin E3 ligase for OTUD3, which ubiquitylates and stabilizes OTUD3.
Fig. 3. CHIP promotes ubiquitylation of OTUD3.
a 293T cells were transfected for 40 h with Flag-tagged OTUD3, HA tagged Ub, Myc-tagged WT CHIP, H260Q (HQ) mutant CHIP or K30A (KA) mutant CHIP alone or in combination, and then treated for 8 h with MG132 (20 µM). Cell lysates were immunoprecipitated with Flag antibody, followed by western blotting with indicated antibodies. b 293T cells were transfected for 40 h with Flag-tagged OTUD3, HA tagged Ub, Myc-tagged WT CHIP, ΔU-box or ΔTPR alone or in combination, and then treated for 8 h with MG132 (20 µM). Cell lysates were immunoprecipitated with Flag antibody, followed by western blotting with indicated antibodies. c H1299 cells stably expressing control shRNA or CHIP shRNA were transfected for 40 h with Flag-tagged OTUD3 alone or together with His tagged Ub, and then treated for 8 h with MG132 (20 µM). Cell lysates were subjected to pull-down with Ni2+ beads, followed by western blotting with indicated antibodies. d H1299 cells stably expressing control shRNA or CHIP shRNAs (shCHIP #1, shCHIP #2) were transfected for 40 h with Flag-tagged OTUD3 and HA tagged Ub, and then treated for 8 h with MG132 (20 µM). Cell lysates were immunoprecipitated with Flag antibody, followed by western blotting with indicated antibodies. e 293T cells were transfected for 40 h with Flag-tagged OTUD3, Myc-tagged CHIP, HA tagged WT Ub or mutant Ub (K0, no Lys; K48, Lys 48-only; K63, Lys 63-only) alone or in combination, and then treated for 8 h with MG132 (20 µM). Cell lysates were immunoprecipitated with Flag antibody, followed by western blotting with indicated antibodies. f The bacteria-expressed and purified GST-tagged OTUD3 proteins were incubated with or without commercial E1, UbE2D3 (E2), ATP, CHIP (E3), Hsp70 and His tagged Ub for 2 h at 37 °C. The mixtures were subjected to GST pull-down and western blotting with indicated antibodies.
CHIP inhibits OTUD3-GRP78 signaling
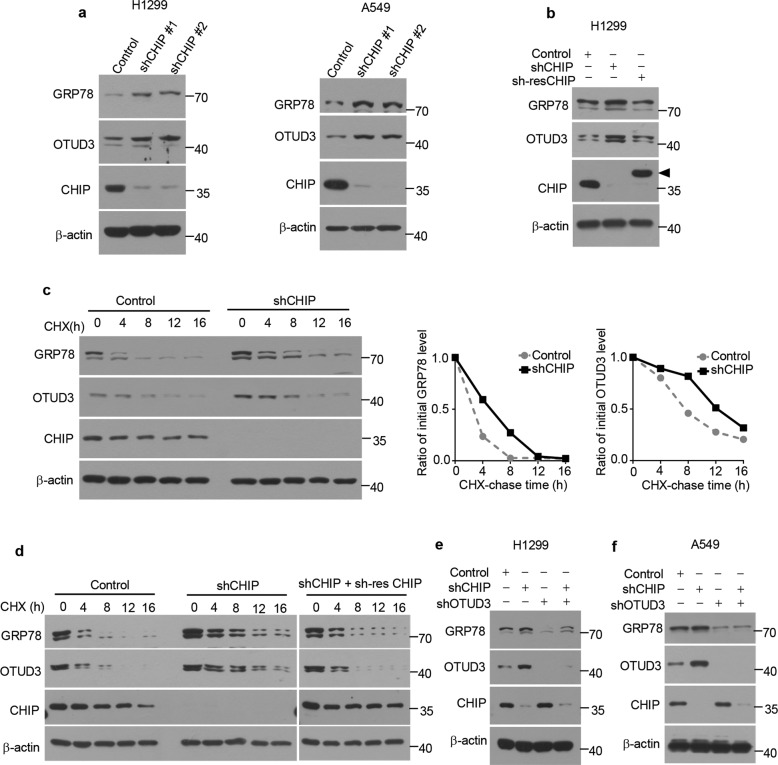
Previous studies showed that OTUD3 is a deubiquitylase of GRP78 [19]. To determine whether CHIP is involved in the regulation of GRP78 level, we knocked down CHIP in H1299 and A549 cells. Knockdown of CHIP increased OTUD3 and GRP78 levels (Fig. 4a). The increase of OTUD3 and GRP78 was reversed by overexpression of an shRNA-resistant CHIP (Fig. 4b). H1299 cells were treated with the protein synthesis inhibitor CHX. The half-life of OTUD3 and GRP78 was prolonged in CHIP-knockdown cells (Fig. 4c). The prolonged half-life of OTUD3 and GRP78 was reversed by overexpression of an shRNA-resistant CHIP (Fig. 4d). To further explore whether upregulated GRP78 level was OTUD3 dependent, we knocked down OTUD3 in CHIP-knockdown H1299 cells. OTUD3 knockdown dramatically decreased CHIP-knockdown-induced GRP78 level (Fig. 4e). Similar results were detected in A549 cells (Fig. 4f). Together, these data indicate that CHIP depletion contributes to the upregulation of GRP78 levels partially through OTUD3.
Fig. 4. CHIP inhibits OTUD3-GRP78 signaling.
a Control or CHIP shRNAs (shCHIP #1, shCHIP #2) were packaged into lentiviral particles and transduced into H1299 or A549 cells. The positive clones were selected by puromycin (2 µg/ml) and indicated proteins were detected by western blotting. b H1299 cells stably expressing CHIP shRNA (shCHIP #1) were infected with or without lentivirus expressing shRNA-resistant (sh-res) CHIP. The indicated proteins were detected by western blotting. Triangle: The shRNA-resistant (sh-res) CHIP was cloned into pCDH-MCS-T2A-copGFP-MSCV vector (System Biosciences, CD523A-1), which generated a high-molecular-weight CHIP as several amino acids were linked to CHIP C-terminal after T2A peptide self-cleavage. c H1299 cells stably expressing control or CHIP shRNA were treated with cycloheximide (CHX) (100 µg/ml) for the indicated time period. The indicated proteins were detected by western blotting. Quantification of relative GRP78 levels and OTUD3 levels is shown. d H1299 cells stably expressing control shRNA, CHIP shRNA together with or without shRNA-resistant CHIP were treated with cycloheximide (CHX) (100 µg/ml) for the indicated time period. The indicated proteins were detected by western blotting. Control shRNA, CHIP shRNA or OTUD3 shRNA alone or in combination were packaged into lentiviral particles and transduced into H1299 (e) or A549 (f) cells. The positive clones were selected by puromycin (2 µg/ml) and indicated proteins were detected by western blotting.
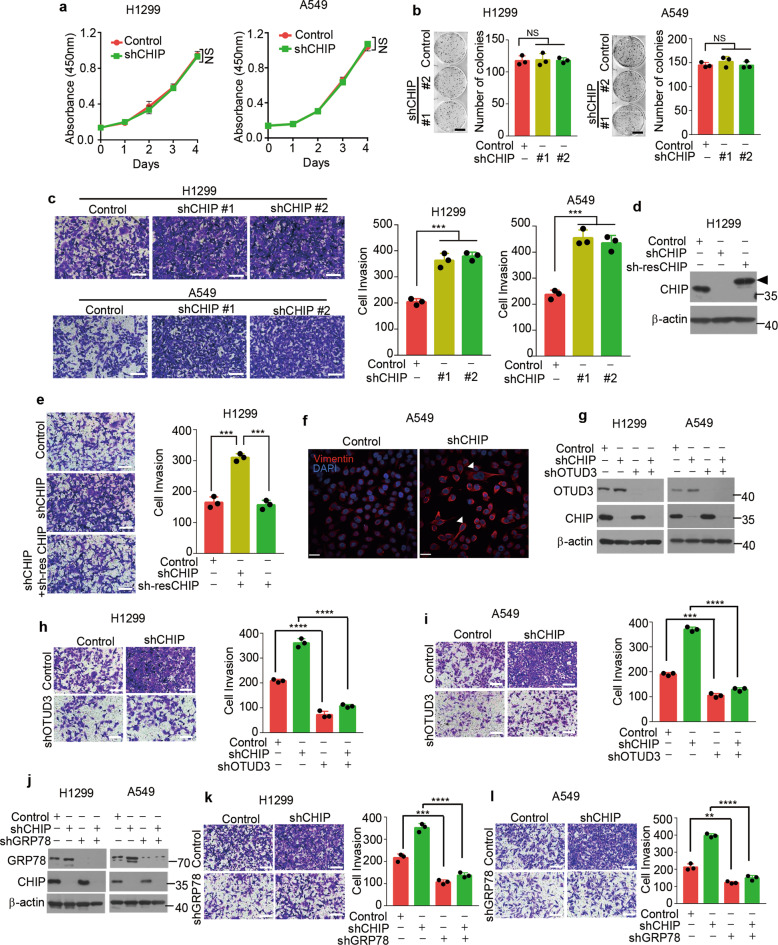
CHIP suppresses invasion of lung cancer cells
As CHIP negatively regulates OTUD3 and GRP78 levels, we propose that CHIP might function as a tumour suppressor in lung cancer cells. To test this hypothesis, we investigated the proliferation rates of control and CHIP-knockdown lung cancer cells. CHIP knockdown had no effect on cell proliferation in H1299 and A549 cells (Fig. 5a, b). As invasiveness of cancer cells are hallmarks of tumour progression, we further examined the invasive potential of these cells by invasion assays. CHIP knockdown increased the invasiveness of H1299 and A549 cells (Fig. 5c). Reconstitution of CHIP reversed the increased invasiveness of H1299 cells (Fig. 5d, e). Furthermore, CHIP deficient A549 cells were more spindle-liked and expressed increased vimentin, implicating increased invasive ability (Fig. 5f). To investigate whether CHIP suppresses invasion through regulating OTUD3 and GRP78 levels, we depleted endogenous OTUD3 or GRP78 in CHIP-knockdown H1299 and A549 cells (Fig. 5g, j). Depletion of OTUD3 or GRP78 attenuated cell invasion in CHIP-knockdown lung cancer cells (Fig. 5h, i, k, l). Collectively, these results demonstrate that CHIP suppresses cell invasion partially through OTUD3 and GRP78.
Fig. 5. CHIP suppresses invasion of lung cancer cells.
a Cell proliferation assays in H1299 or A549 cells stably expressing control shRNA or CHIP shRNA. Relative cell number was determined by adding WST-8 and then measuring absorbance at 450 nm. n = 3 independent experiments. b Representative images of colony formation assays in H1299 or A549 cells stably expressing control or CHIP shRNAs (shCHIP #1, shCHIP #2) 2 weeks after seeding of 500 cells in six-well plate. Quantification of colonies >0.1 mm in diameter is shown. n = 3 independent experiments. Scale bar: 1 cm. c Representative images of invasion assays in H1299 or A549 cells stably expressing control or CHIP shRNAs (shCHIP #1, shCHIP #2) 24 h after seeding of 3 × 105 cells. Quantification of invaded cells stained by 0.1% crystal violet per field is shown. n = 3 independent experiments. Scale bar: 100 μm. d H1299 cells stably expressing CHIP shRNA (shCHIP #1) were infected with or without lentivirus expressing shRNA-resistant (sh-res) CHIP. The indicated proteins were detected by western blotting. Triangle: The shRNA-resistant (sh-res) CHIP was cloned into pCDH-MCS-T2A-copGFP-MSCV vector (System Biosciences, CD523A-1), which generated a high-molecular-weight CHIP as several amino acids were linked to CHIP C-terminal after T2A peptide self-cleavage. e Representative images of invasion assays in H1299 cells stably expressing control shRNA, CHIP shRNA together with or without shRNA-resistant CHIP 24 h after seeding of 3 × 105 cells. Quantification of invaded cells stained by 0.1% crystal violet per field is shown. n = 3 independent experiments. Scale bar: 100 μm. f Representative images for Vimentin staining (Red) of A549 cells stably expressing control shRNA or CHIP shRNA. Nuclei were stained with DAPI (blue). White triangles indicate spindle-liked A549 cells. Scale bar: 40 μm. g H1299 or A549 cells stably expressing control shRNA or CHIP shRNA were infected with or without lentivirus expressing OTUD3 shRNA and selected, the indicated proteins were detected by western blotting. Representative images of invasion assays in H1299 (h) or A549 (i) cells stably expressing the shRNAs described in g 24 h after seeding of 3 × 105 cells. Quantification of invaded cells stained by 0.1% crystal violet per field is shown. n = 3 independent experiments. Scale bar: 100 μm. j H1299 or A549 cells stably expressing control shRNA or CHIP shRNA were infected with or without lentivirus expressing GRP78 shRNA and selected, the indicated proteins were detected by western blotting. Representative images of invasion assays in H1299 (k) or A549 (l) cells stably expressing the shRNAs described in j 24 h after seeding of 3 × 105 cells. Quantification of invaded cells stained by 0.1% crystal violet per field is shown. n = 3 independent experiments. Scale bar: 100 μm. Data are shown as mean ± s.d. Statistical analyses in a were performed using two-way ANOVA tests, and in b, c, e, h, i, k, l were performed with Student’s t tests. **p < 0.01, ***p < 0.001, ****p < 0.0001; NS not significant.
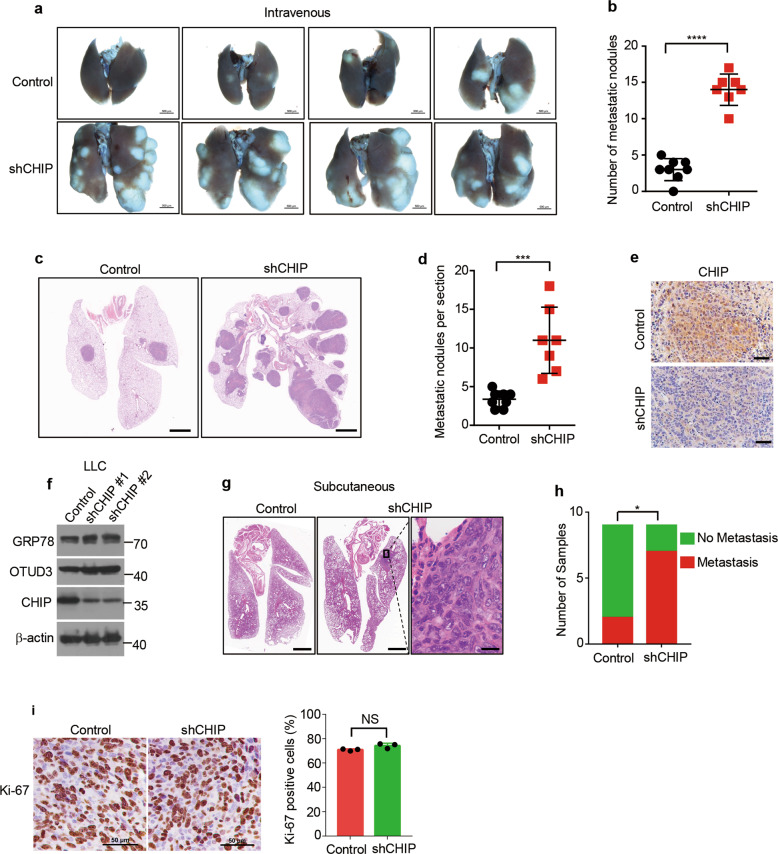
CHIP suppresses tumour metastasis
To investigate whether CHIP deficient lung cancer cells have greater metastatic potential, we introduced control and CHIP-knockdown A549 cells into the bloodstream of nude mice. CHIP-knockdown cells produced significantly more lung metastases (Fig. 6a, b). We further confirmed the observation by examining the metastatic nodules after HE staining (Fig. 6c, d). Immunostaining confirmed CHIP was deficient in metastatic nodules produced by CHIP-knockdown cells (Fig. 6e). Tumour metastasis needs several steps: detachment, invasion of blood vessels, immune escape, extravasation and colonization. As intravenous transplantation experiments lack the detachment and invasion process, we further performed subcutaneous transplantation experiments. We knocked down CHIP in mice lung cancer cell line LLC, which elevated OTUD3 and GRP78 levels (Fig. 6f). Control and CHIP-knockdown LLC cells were implanted subcutaneously into nude mice. Lung metastasis were observed by HE staining (Fig. 6g). We found that seven out of nine mice implanted with CHIP-knockdown cells produced lung metastasis, while two out of nine mice implanted with control cells produced metastasis (Fig. 6h). The proliferation rates of primary tumours had no difference (Fig. 6i). Taken together, these results demonstrate CHIP suppresses tumour metastasis in lung cancer.
Fig. 6. CHIP suppresses tumour metastasis.
a Representative images of the lungs of mice 4 weeks after intravenous injection with A549 cells expressing control shRNA or CHIP shRNA (2 × 106 cells per mice). Scale bar: 500 μm. b Quantification of metastatic nodules produced by A549 cells expressing control shRNA or CHIP shRNA described in a. n = 8 mice for control cells, n = 7 mice for shCHIP cells. c Representative images of the hematoxylin–eosin stained lungs of mice 4 weeks after intravenous injection with A549 cells expressing control shRNA or CHIP shRNA (2 × 106 cells per mice). Scale bar: 500 μm. d Quantification of metastatic nodules per section described in c. n = 8 mice for control cells, n = 7 mice for shCHIP cells. e Representative images of immunohistochemistry staining for CHIP in metastatic nodules produced by A549 cells expressing control shRNA or CHIP shRNA. Scale bar: 50 μm. f Control or CHIP shRNAs (shCHIP #1, shCHIP #2) were packaged into lentiviral particles and transduced into LLC cells. The positive clones were selected and the indicated proteins were detected by western blotting. g Representative images of the hematoxylin–eosin stained lungs of mice 4 weeks after subcutaneous implantation with LLC cells expressing control shRNA or CHIP shRNA (2 × 106 cells per mice). Scale bar of left two images: 500 µm, scale bar of right magnified image: 20 µm. h Quantification of mice with or without metastasis produced by LLC cells expressing control shRNA or CHIP shRNA. n = 9 mice for each cell line. i Representative images of immunohistochemistry staining for Ki-67 in subcutaneous tumours produced by LLC cells expressing control shRNA or CHIP shRNA. Quantification of percentage of Ki-67 positive cells is shown. n = 3 mice for each cell line. Scale bar: 50 μm. Data in b, d, i are shown as mean ± s.d. Statistical analyses in b, d, i were performed using Student’s t tests, and in h were performed with chi-square tests. *p < 0.05, ***p < 0.001, ****p < 0.0001. NS not significant.
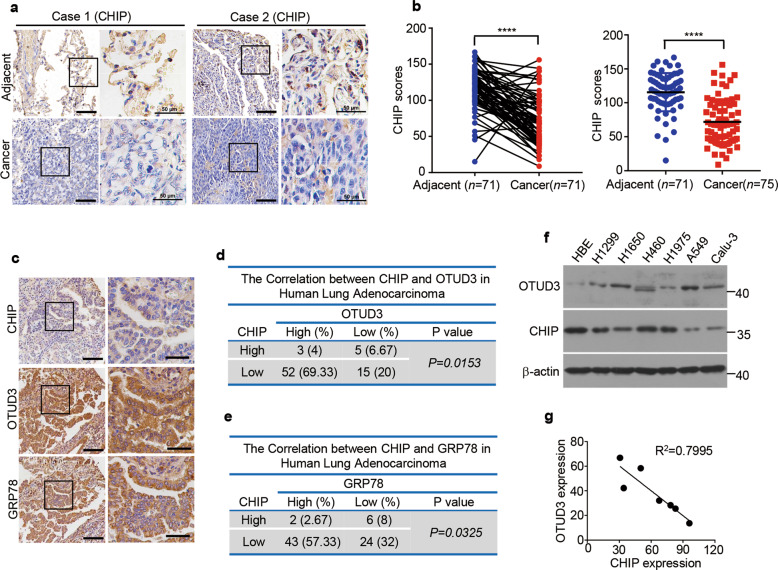
CHIP expression is downregulated in human lung adenocarcinoma
We further analyzed whether CHIP had the clinical evidence supporting its relevance to lung cancer. We detected the expression of CHIP in human lung adenocarcinoma (n = 75). We found that CHIP expression was significantly decreased in lung adenocarcinoma compared with the matched adjacent normal tissues (Fig. 7a, b). Moreover, the expression of CHIP was inversely correlated with OTUD3 and GRP78 expression (Fig. 7c–e). The negative correlation between CHIP and OTUD3 was further confirmed in human lung cancer cell lines (Fig. 7f, g). We also found that the expression levels of CHIP were lower in A549 cells and Calu-3 cells, which were more prone to metastasize (Fig. 7f, g). These results suggest that CHIP is decreased in lung adenocarcinoma, concomitant with OTUD3 and GRP78 upregulation to promote tumorigenesis.
Fig. 7. CHIP expression is downregulated in human lung adenocarcinoma.
a Representative images of CHIP immunohistochemical staining in lung cancer and matched adjacent tissues of two cases from tissue microarrays containing 75 pairs of human lung adenocarcinoma together with matched adjacent normal lung tissues. The outlined areas in the left images are magnified on the right. Scale bars: 100 µm (left), 50 µm (right). b CHIP expression H-scores were compared between adjacent tissues and lung cancer tissues. Data in the right panel are shown as mean ± s.d. Statistical analyses were performed using paired Student’s t tests (left) or Wilcoxon unpaired t test (right). ****p < 0.0001. c Representative images of immunohistochemical staining for CHIP, OTUD3 or GRP78 in human lung adenocarcinoma (n = 75). Scale bars: 100 µm (left), 50 µm (right). CHIP, OTUD3 and GRP78 expression in human lung adenocarcinoma (n = 75) was quantified using H-score and divided into high expression (CHIP score >120, OTUD3 score > 120, GRP78 score > 150) and low expression (CHIP score < 120, OTUD3 score < 120, GRP78 score < 150), the correlation of CHIP expression levels with OTUD3 expression levels (d) or with GRP78 expression levels (e) is analyzed. Chi-square test, p = 0.0153 (d), p = 0.0325 (e). f The expression levels of OTUD3 and CHIP in indicated lung cancer cell lines were analyzed by western blotting. g Regression analysis comparing CHIP and OTUD3 expression in lung cancer cell lines described in f. Pearson’s correlation test. p = 0.0066.
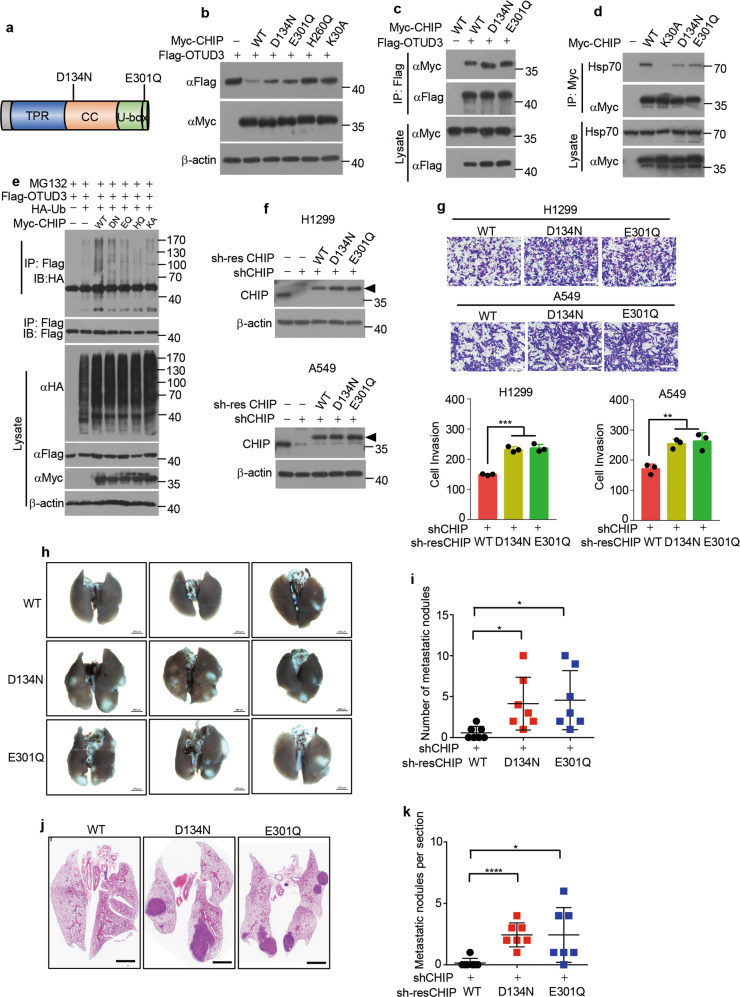
Identification of CHIP mutations in human lung cancer
As CHIP functions as a tumour suppressor in lung cancer, we further examined whether CHIP had mutations in lung cancer. We searched 5006 samples of non-small cell lung cancer in the cBioPortal [52, 53] database and found CHIP mutations D134N (sample ID: TCGA-75-5126-01) and E301Q (sample ID: CRUK0016-R1; CRUK0016-R2) (Fig. 8a). We next investigated whether these mutations abolished CHIP function. Myc-tagged D134N and E301Q plasmids were constructed and overexpressed with OTUD3 in cells. Indeed, CHIP D134N and E301Q mutants partially lost the ability to degrade OTUD3 protein (Fig. 8b). To investigate the mechanism, we tested the interaction between CHIP mutants and OTUD3. However, the interaction between CHIP mutants and OTUD3 was the same as that between WT CHIP and OTUD3 (Fig. 8c), suggesting that these mutations did not impair the binding to OTUD3. As a chaperone-dependent E3 ubiquitin ligase, CHIP exerts its ubiquitin ligase function when it binds to Hsp70 or Hsp90 chaperone [31]. Mouse CHIP Asp135 (equivalent to human CHIP Asp134) has been reported to form hydrogen bonding with Hsp90 [51]. Thus we detected whether CHIP mutants had decreased ability to interact with Hsp70. Indeed, the interaction between CHIP mutants and Hsp70 was weaker than that between WT CHIP and Hsp70 (Fig. 8d), which might account for the reduced catalytic activity of CHIP mutants. Subsequent ubiquitylation assay demonstrated that CHIP D134N and E301Q mutants partially lost their ability to ubiquitylate OTUD3 (Fig. 8e). We next investigated whether D134N and E301Q mutant CHIP had reduced capacity to suppress tumour metastasis. We stably overexpressed WT CHIP or mutant CHIP in CHIP-knockdown lung cancer cells (Fig. 8f). The number of invaded CHIP mutant lung cancer cells was more than that of invaded CHIP WT lung cancer cells (Fig. 8g), demonstrating mutant CHIP had reduced capacity to inhibit cell invasion. Furthermore, we injected CHIP WT or CHIP mutant A549 cells into tail vein of nude mice. The CHIP mutant cells produced more metastatic nodules in lung than CHIP WT cells (Fig. 8h, i), which was confirmed by HE staining (Fig. 8j, k). Taken together, These results demonstrate that CHIP D134N and E301Q mutations reduce CHIP catalytic activity and tumour suppressive function in lung cancer.
Fig. 8. CHIP D134N and E301Q mutations in lung cancer reduce E3 ligase function.
a Schematic illustration of CHIP D134N and E301Q mutations. b 293T cells were transfected for 48 h with Flag-tagged OTUD3 alone or together with Myc-tagged WT CHIP or mutant CHIP (D134N, E301Q, H260Q, K30A). The indicated proteins were detected by western blotting. c 293T cells were transfected for 36 h with Myc-tagged WT CHIP or mutant CHIP (D134N, E301Q) together with or without Flag-tagged OTUD3, and then immunoprecipitated with Flag antibody, followed by western blotting with indicated antibodies. d H1299 cells were transfected for 36 h with or without Myc-tagged WT CHIP or mutant CHIP (K30A, D134N, E301Q), and then immunoprecipitated with Myc antibody, followed by western blotting with indicated antibodies. e 293T cells were transfected for 40 h with Flag-tagged OTUD3, HA tagged Ub, Myc-tagged WT CHIP, D134N (DN) mutant CHIP, E301Q (EQ) mutant CHIP, H260Q (HQ) mutant CHIP or K30A (KA) mutant CHIP alone or in combination, and then treated for 8 h with MG132 (20 µM). Cell lysates were immunoprecipitated with Flag antibody, followed by western blotting with indicated antibodies. f H1299 or A549 cells stably expressing CHIP shRNA were infected with or without lentivirus expressing shRNA-resistant (sh-res) WT CHIP or mutant CHIP (D134N, E301Q). The indicated proteins were detected by western blotting. Triangle: The shRNA-resistant (sh-res) CHIP was cloned into pCDH-MCS-T2A-copGFP-MSCV vector (System Biosciences, CD523A-1), which generated a high-molecular-weight CHIP as several amino acids were linked to CHIP C-terminal after T2A peptide self-cleavage. g Representative images of invasion assays in H1299 or A549 cells stably expressing WT CHIP or mutant CHIP (D134N, E301Q) described in f 24 h after seeding of 3 × 105 cells. Quantification of invaded cells stained by 0.1% crystal violet per field is shown. n = 3 independent experiments. Scale bar: 100 μm. h Representative images of the lungs of mice 4 weeks after intravenous injection with A549 cells stably expressing WT CHIP or mutant CHIP (D134N, E301Q) (2 × 106 cells per mice). Scale bar: 500 μm. i Quantification of metastatic nodules described in h. n = 7 mice for each cell line. j Representative images of the hematoxylin–eosin stained lungs of mice 4 weeks after intravenous injection with A549 cells stably expressing WT CHIP or mutant CHIP (D134N, E301Q). Scale bar: 500 μm. k Quantification of metastatic nodules per section described in j. n = 7 mice for each cell line. Data are shown as mean ± s.d. Statistical analyses in g, i, k were performed using Student’s t tests. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Discussion
The DUBs regulate important processes in cells and post-translational modifications of DUBs are crucial to their functions [5]. Previously we identified deubiquitylase OTUD3 exhibits tumour suppressive roles through deubiquitylating and stabilizing PTEN in breast cancer, hepatocellular cancer, colon cancer and cervical cancer [18, 19]. OTUD3 transgenic mice are less susceptible to breast tumorigenesis, while OTUD3 KO mice exhibit increased susceptibility to breast cancer [18, 19]. Recently we found OTUD3 plays oncogenic roles in lung cancer [19]. OTUD3 transgenic mice are more prone to develop KrasG12D driven lung cancer while OTUD3 KO mice are less prone to [19]. Furthermore, OTUD3 expression levels are upregulated in lung cancer and elevated expression levels of OTUD3 correlates with poor survival in lung cancer patients [19]. However, the reason why OTUD3 is elevated in lung cancer and the post-translational modification of OTUD3 is unknown. In this study, we found OTUD3 can be ubiquitylated and identified CHIP as a E3 ligase mediates ubiquitylation of OTUD3 and promotes the proteasomal degradation of OTUD3.
Several lines of evidence support the conclusion that CHIP is a true ubiquitin ligase for OTUD3. First, CHIP interacts with OTUD3. Co-immunoprecipitation assays showed that endogenous CHIP bound endogenous OTUD3. Furthermore, CHIP and OTUD3 were colocalized in the cytoplasm. Second, CHIP negatively regulates OTUD3 levels. OTUD3 levels were elevated in CHIP KO mice. Overexpression of CHIP downregulated OTUD3 levels in E3 ligase activity and Hsp70 dependent manner. CHIP expression levels were reduced in lung adenocarcinoma and were negatively correlated with OTUD3 levels. Third, CHIP promotes OTUD3 ubiquitylation. Overexpression of CHIP promoted OTUD3 ubiquitylation in E3 ligase activity and Hsp70 dependent manner, which was confirmed by in vitro ubiquitylation assay. We also found CHIP promoted Lys 48-linked ubiquitylation, but not non-degradative Lys 63-linked ubiquitylation of OTUD3. However, whether CHIP promotes other types of OTUD3 ubiquitylation needs further investigation. Although we analyzed the regulation of OTUD3 by CHIP in the context of lung cancer, we do not exclude that this regulation exists in other cell types or tissues. To our knowledge, CHIP is the first ubiquitin ligase identified to promote OTUD3 ubiquitylation and degradation.
OTUD3 contains 28 lysine residues, among which OTU contains 10 lysine residues, UBA domain contains 7 lysine residues and tail domain contains 11 lysine residues. These lysine residues are potential ubiquitylation sites as well as potential neddylation, SUMOylation and acetylation sites. In addition, OTUD3 also contains 23 serine residues and 11 threonine residues, which are potential phosphorylation sites. These potential post-translational modifications are worthy of further investigation in the future.
GRP78 as a major endoplasmic reticulum chaperone is overexpressed in cancer [21]. Induced GRP78 in cancer regulates a variety of processes that are essential for tumourigenesis such as proliferation, apoptosis, angiogenesis, invasion and metastasis [54]. Several studies revealed that GRP78 expression is increased in lung cancer and is higher in advanced cancer tissues [22, 55]. Overexpression of GRP78 also predicts poor prognosis in lung adenocarcinoma [56]. Thus, GRP78 exhibits tumour promoting roles in lung cancer. Previously we found OTUD3 is a deubiquitylase for GRP78 [19]. Depletion of OTUD3 in mice reduced GRP78 levels and suppresses KrasG12D-driven lung adenocarcinoma progression and markedly increases survival [19]. Here we found deficiency of CHIP upregulates GRP78 expression in lung cancer in an OTUD3 dependent manner. Cell invasion assays demonstrated that GRP78 mediated the increased invasion of CHIP deficient lung cancer cells. What’s more, CHIP expression negatively correlated with GRP78 levels in lung adenocarcinoma. We uncovered a new regulatory mechanism of GRP78 expression in lung cancer.
CHIP maintains the protein homoeostasis and participates in multiple cellular processes [34]. Several evidence shows that CHIP plays tumour suppressive roles as it promotes ubiquitylation and degradation of oncogenic proteins, such as c-ErbB2/neu, mutant p53, HIF-1α, pyruvate kinase isoenzyme M2 (PKM2), c-Myc and OCT4 [36–38, 42, 45, 57]. CHIP also displays oncogenic roles through degrading tumour suppressors such as forkhead box protein O1, apoptosis inducing factor and interferon regulatory factor-1 [58–60]. Here, we present several lines of evidence to support the concept that CHIP exhibits a tumour suppressive role in lung cancer. First, the expression levels of CHIP were significantly reduced in lung cancer tissues as compared with adjacent tissues. CHIP expression negatively correlated with OTUD3 and GRP78 protein levels in lung cancer. Second, we identified CHIP mutations D134N and E301Q in lung adenocarcinoma. These two mutations reduce CHIP E3 ligase activity and lung cancer cells expressing these mutations were more prone to metastasize to mice lung. Third, CHIP functions as a suppressor of tumour metastasis. Depletion of CHIP in lung cancer cells significantly increased invasive ability and CHIP deficient lung cancer cells were more prone to colonize in mice lung when implanted intravenously or subcutaneously. To further investigate the tumour suppressive functions of CHIP in vivo, CHIP KO mice should be utilized. However, approximately one third of CHIP KO mice die at the embryonic stage, the other CHIP KO mice die within 2 months after birth [49]. Thus, it is practically impossible to observe tumorigenesis or metastasis in CHIP KO mice. Generation of CHIP conditional KO mice in the future may in favour of studying the relation between CHIP and lung cancer.
Tissue microarray analysis showed that CHIP expression levels were reduced in human lung cancer. The downregulated expression levels of CHIP have also been found in breast cancer, colorectal cancer and gastric cancer [40, 43, 44, 61]. The hypermethylated CHIP promoter may account for the reduced CHIP expression level in colorectal cancer and gastric cancer [44, 61]. Whether CHIP promoter is hypermethylated in lung cancer is worthy of investigation. CHIP expression levels are also regulated by microRNA. The microRNA miR-764-5p has been reported to inhibit CHIP mRNA translation in MC3T3-E1 cells [62]. Whether this regulation exists and is disturbed in lung cancer also needs further tests. In addition, other mechanisms rendering downregulation of CHIP expression in lung cancer may also exist. As CHIP plays important roles in suppressing lung cancer, elevating CHIP levels in lung cancer might be an effective approach to inhibit lung cancer metastasis.
Collectively, our study provides a new insight into the post-translational modification of OTUD3 in lung cancer and uncovers that CHIP is a E3 ligase for OTUD3 polyubiquitination. Functional study reveals that loss of CHIP expression facilitates tumour metastasis in vitro and in vivo. In conclusion, our results demonstrate that CHIP is an essential regulator of OTUD3 and that the CHIP-OTUD3-GRP78 signaling axis plays a critical role in lung cancer metastasis.
Materials and methods
Cell culture and reagents
A549, HEK293T, NCI-H1299 (H1299), HBE, NCI-H1650 (H1650), NCI-H460 (H460), NCI-H1975 (H1975) and Calu-3 cell lines were obtained from the American Type Culture Collection. HEK293T, HBE and Calu-3 cells were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS). H1299, H1650, H460 and H1975 cells were cultured in RPMI-1640 (Gibco) supplemented with 10% FBS. A549 cells were cultured in Ham’s F12K (Gibco) supplemented with 10% FBS. CHIP WT and KO MEF cells were isolated from E13.5 embryos of mice and cultured in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin (Gibco). MG132 (MCE), Puromycin (Mediatech), CHX (CST) and Polybrene (Sigma).
Plasmids and antibodies
HA tagged CHIP WT, CHIP H260Q and CHIP K30A were kindly provided by ZC. Full-length OTUD3 WT, full-length OTUD3 C76A, OTU, UBA, tail truncations, full-length CHIP WT, full-length CHIP H260Q, full-length CHIP K30A, full-length CHIP D134N, full-length CHIP E301Q, ΔU-box, ΔTPR, U-box truncations, were cloned into the pCMV-Myc, pFlag-CMV-2 vectors as indicated. GST-tagged OTUD3 were cloned into the pGEX-4T-2 vector.
The following antibodies were used for western blotting. Anti-CHIP (A1169, 1:4000) and anti-β-actin (Ac026, 1:40,000) antibodies were purchased from ABclonal Technology. Anti-OTUD3 (HPA028543, 1:500) antibody was purchased from Sigma. Anti-PTEN (9188, 1:2000), anti-AKT (9272, 1:2000), anti-pAKT-S473 (4060, 1:2000), anti-p38 (8690, 1:1000), anti-p-p38 (4511, 1:1000) and anti-USP13 (12577, 1:1000) antibodies were purchased from Cell Signaling Technology. Anti-WWP2 (ab103527, 1:500) antibody was purchased from Abcam. Anti-GRP78 (11587-1-AP, 1:1000), anti-NEDD4-1 (21698-1-AP, 1:1000), anti-Hsp90 (13171-1-AP, 1:1000) antibodies were purchased from Proteintech. Anti-Myc (M047-3, 1:1000), anti-HA (M180-3, 1:1000), anti-Flag (M185-3, 1:1000) and anti-His (M089-3, 1:2000) antibodies were purchased from MBL. Anti-GAPDH (sc-25778, 1:1000), anti-GST (sc-374171, A-6, 1:1000) and anti-XIAP (sc-55550, 1:1000) antibodies were purchased from Santa Cruz Biotechnology.
For immunoprecipitation, anti-OTUD3 (HPA028543, Sigma, 1:100), anti-Flag (M185-3, MBL, 1:200), anti-Myc (M047-3, MBL, 1:500) and Normal IgG (sc-2003, Santa Cruz Biotechnology, 1:500) antibodies were used. For immunofluorescence and immunohistochemistry, anti-Myc (M047-3, MBL, 1:1000), anti-Flag (M185-3, MBL, 1:1000), anti-vimentin (ab92547, Abcam, 1:200), anti-CHIP (A1169, ABclonal Technology, 1:400), anti-OTUD3 (HPA028543, Sigma, 1:200), anti-GRP78 (11587-1-AP, Proteintech, 1:200) and anti-Ki-67 (12202, CST, 1:200) were used.
Lentivirus packaging and infection
Lentiviral vectors (GV248) carrying CHIP shRNA were from GeneChem. The OTUD3 and GRP78 shRNA sequences were cloned into the pLKO-puro vector. The shRNA-resistant CHIP WT, shRNA-resistant CHIP D134N and shRNA-resistant CHIP E301Q were cloned into pCDH-MCS-T2A-copGFP-MSCV vector (System Biosciences, CD523A-1). The indicated vector was transfected into HEK293T cells together with pSPAX.2 and pMD.2G for 24 h, and cell culture media were collected and filtered. The viruses were used to infect cells in the presence of polybrene (4 μg/ml). Forty-eight hours later, A549, H1299 or HEK293T cells were cultured in medium containing puromycin (2 μg/ml) for the selection of stable clones. The positive clones were identified and verified by western blotting. The shRNA sequences are below. CHIP no. 1, 5′-AGAGGAAGAAGCGAGACAT-3′; no. 2, 5′-AGTCTGTGAAGGCGCACTT-3′; OTUD3 5′-TGGAAATCAGGGCTTAAAT-3′; GRP78 5′-AACCATCCCGTGGCATAAA-3′.
Cell transfections, immunoprecipitation and immunoblotting
Cells were transfected with indicated plasmids using Lipofectamine 2000 (Invitrogen) reagent according to the manufacturer’s protocol. For immunoprecipitation assays, cells were lysed with HEPES lysis buffer (20 mM HEPES, pH 7.2, 50 mM NaCl, 0.5% Triton X-100, 1 mM NaF and 1 mM dithiothreitol) supplemented with protease-inhibitor cocktail (Roche). Immunoprecipitations were performed using the indicated primary antibody and protein A/G agarose beads (Santa Cruz) at 4 °C. The immunocomplexes were then washed with HEPES lysis buffer four times. Both lysates and immunoprecipitates were examined using the indicated primary antibodies followed by detection with the related secondary antibody and the SuperSignal west pico chemiluminescence substrate (Thermo).
Protein half-life assay
For the exogenous OTUD3 half-life assay, Plasmids encoding OTUD3 and CHIP were transfected into HEK293T cells. Twenty-four hours later, the cells were treated with the protein synthesis inhibitor CHX (CST, 100 µg/ml) for the indicated durations before collection. For the endogenous OTUD3 and GRP78 half-life assay, the H1299 cells stably expressing CHIP shRNA were treated with CHX (CST, 100 µg/ml) for the indicated durations before collection. Endogenous OTUD3 and GRP78 levels were detected by western blotting.
Fluorescence microscopy
For detection of subcellular localization by immunofluorescence, after fixation with 4% paraformaldehyde and permeabilization in 0.2% Triton X-100 (PBS), cells were incubated with the indicated Myc (M047-3, MBL, 1:1000), Flag (M185-3, MBL,1:1000) or vimentin (ab92547, Abcam, 1:200) antibodies for 8 h at 4 °C, followed by incubation with TRITC-conjugated or FITC-conjugated secondary antibody for 1 h at room temperature. The nuclei were stained with DAPI (Sigma), and images were visualized with a Zeiss LSM 510 Meta inverted confocal microscope.
In vivo ubiquitylation assay
For in vivo ubiquitylation assays, Myc-CHIP, Flag-OTUD3 and HA-ubiquitin were transfected into H1299 or HEK293T cells with Lipofectamine 2000. Forty hours later, the cells were treated with 20 µM of the proteasome inhibitor MG132 for 8 h. The cells were washed with PBS, pelleted and lysed in HEPES buffer (20 mM HEPES, pH 7.2, 50 mM NaCl, 1 mM NaF, 0.5% Triton X-100) plus 0.1% SDS, 20 µM MG132 and protease-inhibitor cocktail. The lysates were centrifuged to obtain cytosolic proteins and incubated with anti-Flag antibody for 3 h and protein A/G agarose beads (Santa Cruz) for a further 8 h at 4 °C. Then the beads were washed three times with HEPES buffer. The proteins were released from the beads by boiling in SDS–PAGE sample buffer and analyzed by immunoblotting with anti-HA antibody.
In vitro ubiquitylation assay
The reaction was carried out at 37 °C for 2 h in 20 µl reaction buffer (20 mM Tris-HCl, pH 7.2, 5 mM MgCl2, 50 mM NaCl, 1 mM 2-mercaptoethanol, 10% glycerol) containing the following components: 50 µM of ubiquitin, 100 nM human E1, 2 µM UbE2D3, 2 µM CHIP, 2 µM Hsp70, 2 mM ATP (all from Ubiquitin-Proteasome Biotechnologies, Cat. no. J5110), 5 µM GST–OTUD3. The reaction was terminated by adding 0.4 ml pull-down buffer (20 mM Tris-HCl, pH 7.5, 500 mM NaCl, 1% Triton X-100, 0.02% BSA, and 5 mM β-mercaptoethanol). After addition of 40 µl of glutathione–Sepharose, the samples were rotated at 4 °C for 6 h. The beads were washed with 1 ml of pull-down buffer three times. The proteins bound to beads were released by boiling in 50 µl of 2× SDS–PAGE sample buffer for 10 min. The samples were then resolved by 8% SDS–PAGE followed by immunoblot analysis using a monoclonal anti-His antibody.
Cell proliferation assay
The A549 or H1299 cells stably knocked down for control or CHIP were plated in 96-well plates (100 µl cell suspensions, 1 × 104 cells/ml). Twenty-four hours later, 10 µl WST-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt] was added to each well and incubated at 37 °C for 1 h followed by absorbance measurement at 450 nm.
Colony formation assays
For colony formation assay, H1299 and A549 cells expressing the indicated genes were diluted to the single-cell suspension and 500 or 1000 cells were cultured in every well of six-well plate at 37 °C with 5% CO2 incubator for 2 weeks. Then the colonies were stained with 0.1% crystal violet and counted.
Invasion assay
Invasion assay was performed in a 24-well plate with 8.0 µm polycarbonate membrane inserts (Corning) coated by 60 µl Matrigel matrix (Corning). In brief, 3 × 105 H1299 or A549 cells resuspended by serum-free medium were plated in each insert. The inserts were placed in the well containing 10% FBS (Hyclone) culture medium. After 24 h, non-invaded cells were removed and the inserts were washed in PBS, fixed in 4% formaldehyde for 10 min and stained with 0.1% crystal violet for 5 min. The wells were photographed and the stained cells were counted.
Cohort and immunohistochemistry
Tumour tissue microarrays, purchased from Shanghai Outdo Biotech Company, contain 75 pairs of lung adenocarcinoma together with matched adjacent normal lung tissue, respectively. Immunohistochemistry was performed by using the avidin-biotin complex method (Vector Laboratories), including heat-induced antigen-retrieval procedures. Incubation with antibodies against CHIP (1:400; A1169, ABclonal Technology), OTUD3 (1:200; HPA028543, Sigma), GRP78 (1:200, 11587-1-AP, Proteintech) and Ki-67 (1:200, 12202, CST) was carried out at 4 °C for 12 h. All staining was assessed by a quantitative imaging method; the percentage of immunostaining and the staining intensity were recorded. An H-score was calculated using the following formula: H-Score = ∑ (PI × I) = (percentage of cells of weak intensity × 1) + (percentage of cells of moderate intensity × 2) + (percentage of cells of strong intensity × 3). PI indicates the percentage of positive cells vs. all cells, and I represents the staining intensity.
Tumour metastasis assay
BALB/c nude mice (4–6 weeks old, 18.0 ± 2.0 g, male) were randomly divided into indicated groups; the mice in the groups were injected intravenously or subcutaneously with the indicated cells stably expressing the indicated shRNAs or constructs (2 × 106 cells in a volume of 100 µl PBS). The mice were dissected to observe the metastatic organs 4 weeks after injection. For the subcutaneously injected mice, the subcutaneous tumours were removed 2 weeks after injection. Hematoxylin and eosin staining was reviewed to identify the cancer tissue and normal tissue.
Statistical analysis
The statistical significance of differences between two groups was calculated with the two-tailed Student’s t test, and error bars represent standard deviation of the mean (s.d.). Statistical comparisons of the means of multiple groups were calculated using two-way ANOVA. Correlations between two variables were performed using chi-square test or Pearson correlation test. Statistical analyses, unless otherwise indicated, were performed using GraphPad Prism 5 or 7. Data are shown as mean ± s.d. P < 0.05 was considered statistically significant.
Acknowledgements
This work was supported by National Key R&D Program of China (2017YFA0505602).
Funding
This work was supported by National Key R&D Program of China (2017YFA0505602).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All experimental procedures in mice were approved by the Laboratory Animal Center of Chinese Academy of Military Medical Sciences and complied with all relevant ethical regulations.
Footnotes
Edited by S. Fulda
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chun-Ping Cui, Email: cui_chunping2000@aliyun.com.
Lingqiang Zhang, Email: zhanglq@nic.bmi.ac.cn.
References
- 1.Cheng J, North BJ, Zhang T, Dai X, Tao K, Guo J, et al. The emerging roles of protein homeostasis-governing pathways in Alzheimer’s disease. Aging Cell. 2018;17:e12801. doi: 10.1111/acel.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciechanover A. The unravelling of the ubiquitin system. Nat Rev Mol Cell Biol. 2015;16:322–4. doi: 10.1038/nrm3982. [DOI] [PubMed] [Google Scholar]
- 3.Kwon YT, Ciechanover A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem Sci. 2017;42:873–86. doi: 10.1016/j.tibs.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Gilberto S, Peter M. Dynamic ubiquitin signaling in cell cycle regulation. J Cell Biol. 2017;216:2259–71. doi: 10.1083/jcb.201703170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 6.Mevissen TET, Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem. 2017;86:159–92. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 7.Cheng J, Guo J, North BJ, Wang B, Cui CP, Li H, et al. Functional analysis of deubiquitylating enzymes in tumorigenesis and development. Biochim Biophys Acta Rev Cancer. 2019;1872:188312. doi: 10.1016/j.bbcan.2019.188312. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C, Lu J, Zhang QW, Zhao W, Guo JH, Liu SL, et al. USP7 promotes cell proliferation through the stabilization of Ki-67 protein in non-small cell lung cancer cells. Int J Biochem Cell Biol. 2016;79:209–21. doi: 10.1016/j.biocel.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 9.Chauhan D, Tian Z, Nicholson B, Kumar KG, Zhou B, Carrasco R, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22:345–58. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai JB, Shi GM, Dong ZR, Ke AW, Ma HH, Gao Q, et al. Ubiquitin-specific protease 7 accelerates p14(ARF) degradation by deubiquitinating thyroid hormone receptor-interacting protein 12 and promotes hepatocellular carcinoma progression. Hepatology. 2015;61:1603–14. doi: 10.1002/hep.27682. [DOI] [PubMed] [Google Scholar]
- 11.Su D, Ma S, Shan L, Wang Y, Wang Y, Cao C, et al. Ubiquitin-specific protease 7 sustains DNA damage response and promotes cervical carcinogenesis. J Clin Investig. 2018;128:4280–96. doi: 10.1172/JCI120518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Testa JR, Cheung M, Pei J, Below JE, Tan Y, Sementino E, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–5. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiesner T, Obenauf AC, Murali R, Fried I, Griewank KG, Ulz P, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–21. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang B, Jie Z, Joo D, Ordureau A, Liu P, Gan W, et al. TRAF2 and OTUD7B govern a ubiquitin-dependent switch that regulates mTORC2 signalling. Nature. 2017;545:365–9. doi: 10.1038/nature22344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang JH, Wei W, Guo ZX, Shi M, Guo RP. Decreased Cezanne expression is associated with the progression and poor prognosis in hepatocellular carcinoma. J Transl Med. 2015;13:41. doi: 10.1186/s12967-015-0396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–84. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan L, Lv Y, Li H, Gao H, Song S, Zhang Y, et al. Deubiquitylase OTUD3 regulates PTEN stability and suppresses tumorigenesis. Nat Cell Biol. 2015;17:1169–81. doi: 10.1038/ncb3218. [DOI] [PubMed] [Google Scholar]
- 19.Du T, Li H, Fan Y, Yuan L, Guo X, Zhu Q, et al. The deubiquitylase OTUD3 stabilizes GRP78 and promotes lung tumorigenesis. Nat Commun. 2019;10:2914. doi: 10.1038/s41467-019-10824-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munro S, Pelham HR. An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- 21.Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67:3496–9. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, He Z, Zhang J, Wang Y, Wang T, Tong S, et al. Overexpression of endoplasmic reticulum molecular chaperone GRP94 and GRP78 in human lung cancer tissues and its significance. Cancer Detect Prev. 2005;29:544–51. doi: 10.1016/j.cdp.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Jiang Y, Jia Z, Li Q, Gong W, Wang L, et al. Association of elevated GRP78 expression with increased lymph node metastasis and poor prognosis in patients with gastric cancer. Clin Exp Metastasis. 2006;23:401–10. doi: 10.1007/s10585-006-9051-9. [DOI] [PubMed] [Google Scholar]
- 24.Pootrakul L, Datar RH, Shi SR, Cai J, Hawes D, Groshen SG, et al. Expression of stress response protein Grp78 is associated with the development of castration-resistant prostate cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2006;12:5987–93. doi: 10.1158/1078-0432.CCR-06-0133. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez-Gronow M, Cuchacovich M, Llanos C, Urzua C, Gawdi G, Pizzo SV. Prostate cancer cell proliferation in vitro is modulated by antibodies against glucose-regulated protein 78 isolated from patient serum. Cancer Res. 2006;66:11424–31. doi: 10.1158/0008-5472.CAN-06-1721. [DOI] [PubMed] [Google Scholar]
- 26.Dong D, Ni M, Li J, Xiong S, Ye W, Virrey JJ, et al. Critical role of the stress chaperone GRP78/BiP in tumor proliferation, survival, and tumor angiogenesis in transgene-induced mammary tumor development. Cancer Res. 2008;68:498–505. doi: 10.1158/0008-5472.CAN-07-2950. [DOI] [PubMed] [Google Scholar]
- 27.Dong D, Stapleton C, Luo B, Xiong S, Ye W, Zhang Y, et al. A critical role for GRP78/BiP in the tumor microenvironment for neovascularization during tumor growth and metastasis. Cancer Res. 2011;71:2848–57. doi: 10.1158/0008-5472.CAN-10-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rauschert N, Brandlein S, Holzinger E, Hensel F, Muller-Hermelink HK, Vollmers HP. A new tumor-specific variant of GRP78 as target for antibody-based therapy. Lab Investig. 2008;88:375–86. doi: 10.1038/labinvest.2008.2. [DOI] [PubMed] [Google Scholar]
- 29.Liu R, Li X, Gao W, Zhou Y, Wey S, Mitra SK, et al. Monoclonal antibody against cell surface GRP78 as a novel agent in suppressing PI3K/AKT signaling, tumor growth, and metastasis. Clin Cancer Res Off J Am Assoc Cancer Res. 2013;19:6802–11. doi: 10.1158/1078-0432.CCR-13-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasche L, Menoret E, Dubljevic V, Menu E, Vanderkerken K, Lapa C, et al. A GRP78-directed monoclonal antibody recaptures response in refractory multiple myeloma with extramedullary involvement. Clin Cancer Res Off J Am Assoc Cancer Res. 2016;22:4341–9. doi: 10.1158/1078-0432.CCR-15-3111. [DOI] [PubMed] [Google Scholar]
- 31.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19:4535–45. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, et al. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276:42938–44. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 33.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 34.Paul I, Ghosh MK. A CHIPotle in physiology and disease. Int J Biochem Cell Biol. 2015;58:37–52. doi: 10.1016/j.biocel.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 35.Ahmed SF, Deb S, Paul I, Chatterjee A, Mandal T, Chatterjee U, et al. The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. J Biol Chem. 2012;287:15996–6006. doi: 10.1074/jbc.M111.321083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paul I, Ahmed SF, Bhowmik A, Deb S, Ghosh MK. The ubiquitin ligase CHIP regulates c-Myc stability and transcriptional activity. Oncogene. 2013;32:1284–95. doi: 10.1038/onc.2012.144. [DOI] [PubMed] [Google Scholar]
- 37.Muller P, Hrstka R, Coomber D, Lane DP, Vojtesek B. Chaperone-dependent stabilization and degradation of p53 mutants. Oncogene. 2008;27:3371–83. doi: 10.1038/sj.onc.1211010. [DOI] [PubMed] [Google Scholar]
- 38.Luo W, Zhong J, Chang R, Hu H, Pandey A, Semenza GL. Hsp70 and CHIP selectively mediate ubiquitination and degradation of hypoxia-inducible factor (HIF)-1alpha but Not HIF-2alpha. J Biol Chem. 2010;285:3651–63. doi: 10.1074/jbc.M109.068577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Huang M, Zheng H, Wang Y, Ren F, Shang Y, et al. CHIP promotes Runx2 degradation and negatively regulates osteoblast differentiation. J Cell Biol. 2008;181:959–72. doi: 10.1083/jcb.200711044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kajiro M, Hirota R, Nakajima Y, Kawanowa K, So-ma K, Ito I, et al. The ubiquitin ligase CHIP acts as an upstream regulator of oncogenic pathways. Nat Cell Biol. 2009;11:312–9. doi: 10.1038/ncb1839. [DOI] [PubMed] [Google Scholar]
- 41.Luan H, Mohapatra B, Bielecki TA, Mushtaq I, Mirza S, Jennings TA, et al. Loss of the nuclear pool of ubiquitin ligase CHIP/STUB1 in breast cancer unleashes the MZF1-cathepsin pro-oncogenic program. Cancer Res. 2018;78:2524–35. doi: 10.1158/0008-5472.CAN-16-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho Y, Kang HG, Kim SJ, Lee S, Jee S, Ahn SG, et al. Post-translational modification of OCT4 in breast cancer tumorigenesis. Cell Death Differ. 2018;25:1781–95. doi: 10.1038/s41418-018-0079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gan L, Liu DB, Lu HF, Long GX, Mei Q, Hu GY, et al. Decreased expression of the carboxyl terminus of heat shock cognate 70 interacting protein in human gastric cancer and its clinical significance. Oncol Rep. 2012;28:1392–8. doi: 10.3892/or.2012.1957. [DOI] [PubMed] [Google Scholar]
- 44.Wang S, Wu X, Zhang J, Chen Y, Xu J, Xia X, et al. CHIP functions as a novel suppressor of tumour angiogenesis with prognostic significance in human gastric cancer. Gut. 2013;62:496–508. doi: 10.1136/gutjnl-2011-301522. [DOI] [PubMed] [Google Scholar]
- 45.Shang Y, He J, Wang Y, Feng Q, Zhang Y, Guo J, et al. CHIP/Stub1 regulates the Warburg effect by promoting degradation of PKM2 in ovarian carcinoma. Oncogene. 2017;36:4191–4200. doi: 10.1038/onc.2017.31. [DOI] [PubMed] [Google Scholar]
- 46.Liu C, Lou W, Yang JC, Liu L, Armstrong CM, Lombard AP, et al. Proteostasis by STUB1/HSP70 complex controls sensitivity to androgen receptor targeted therapy in advanced prostate cancer. Nat Commun. 2018;9:4700. doi: 10.1038/s41467-018-07178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang ZL, Kim M, Huang SM, Lee HJ, Kim JM. Expression of carboxyl terminus of Hsp70-interacting protein (CHIP) indicates poor prognosis in human gallbladder carcinoma. Oncol Lett. 2013;5:813–8. doi: 10.3892/ol.2013.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee YR, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol. 2018;19:547–62. doi: 10.1038/s41580-018-0015-0. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Zhang P, Zhang Q, Li C, Zou W, Chang Z, et al. WWP2 is a physiological ubiquitin ligase for phosphatase and tensin homolog (PTEN) in mice. J Biol Chem. 2018;293:8886–99. doi: 10.1074/jbc.RA117.001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem. 2001;276:33111–20. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 51.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, et al. Chaperoned ubiquitylation-crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–38. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 52.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee AS. Glucose-regulated proteins in cancer: molecular mechanisms and therapeutic potential. Nat Rev Cancer. 2014;14:263–76. doi: 10.1038/nrc3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun Q, Hua J, Wang Q, Xu W, Zhang J, Zhang J, et al. Expressions of GRP78 and Bax associate with differentiation, metastasis, and apoptosis in non-small cell lung cancer. Mol Biol Rep. 2012;39:6753–61. doi: 10.1007/s11033-012-1500-8. [DOI] [PubMed] [Google Scholar]
- 56.Kwon D, Koh J, Kim S, Go H, Min HS, Kim YA, et al. Overexpression of endoplasmic reticulum stress-related proteins, XBP1s and GRP78, predicts poor prognosis in pulmonary adenocarcinoma. Lung Cancer. 2018;122:131–7. doi: 10.1016/j.lungcan.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 57.Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci USA. 2002;99:12847–52. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li F, Xie P, Fan Y, Zhang H, Zheng L, Gu D, et al. C terminus of Hsc70-interacting protein promotes smooth muscle cell proliferation and survival through ubiquitin-mediated degradation of FoxO1. J Biol Chem. 2009;284:20090–8. doi: 10.1074/jbc.M109.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oh KH, Yang SW, Park JM, Seol JH, Iemura S, Natsume T, et al. Control of AIF-mediated cell death by antagonistic functions of CHIP ubiquitin E3 ligase and USP2 deubiquitinating enzyme. Cell Death Differ. 2011;18:1326–36. doi: 10.1038/cdd.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narayan V, Pion E, Landre V, Muller P, Ball KL. Docking-dependent ubiquitination of the interferon regulatory factor-1 tumor suppressor protein by the ubiquitin ligase CHIP. J Biol Chem. 2011;286:607–19. doi: 10.1074/jbc.M110.153122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Ren F, Wang Y, Feng Y, Wang D, Jia B, et al. CHIP/Stub1 functions as a tumor suppressor and represses NF-kappaB-mediated signaling in colorectal cancer. Carcinogenesis. 2014;35:983–91. doi: 10.1093/carcin/bgt393. [DOI] [PubMed] [Google Scholar]
- 62.Guo J, Ren F, Wang Y, Li S, Gao Z, Wang X, et al. miR-764-5p promotes osteoblast differentiation through inhibition of CHIP/STUB1 expression. J Bone Miner Res Off J Am Soc Bone Miner Res. 2012;27:1607–18. doi: 10.1002/jbmr.1597. [DOI] [PubMed] [Google Scholar]