Abstract
The role of environmental condition on the infection by the novel pathogenic SARS-CoV-2 virus remains uncertain. In here, exploiting a large panel of publicly available genome-wide data, we investigated whether the human receptor ACE2 and human proteases TMPRSS2, FURIN and CATHEPSINs (B, L and V), which are involved in SARS-CoV-2 cell entry, are transcriptionally regulated by environmental cues. We report that more than 50 chemicals modulate the expression of ACE2 or human proteases important for SARS-CoV-2 cell entry. We further demonstrate that transcription factor AhR, which is commonly activated by pollutants, binds to the promoter of TMPRSS2 and enhancers and/or promoters of Cathepsin B, L and V encoding genes. Our exploratory study documents an influence of environmental exposures on the expression of genes involved in SARS-CoV-2 cell entry. These results could be conceptually and medically relevant to our understanding of the COVID-19 disease, and should be further explored in laboratory and epidemiologic studies.
Keywords: Coronavirus, SARS-CoV-2, COVID-19, ACE2, TMPRSS2, FURIN, Cathepsin genes, Environmental exposure
1. Introduction
Coronavirus disease 2019 (or COVID-19) is caused by a coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Oberfeld et al., 2020; Zheng, 2020). While a majority of infected individuals have minimal symptoms around 15–20% need to be hospitalised with severe respiratory complications (Cevik et al., 2020; Del Sole et al., 2020; Wolff et al., 2020). Based on first reports, several factors such as sex, age and underlying diseases (e.g. diabetes, auto-immune diseases or congenital heart diseases) were documented to cause a more severe course of the disease, and increase likelihood of death (Gallo Marin et al., 2020; Kopel et al., 2020; Wolff et al., 2020). Similar features and comorbidities were already observed for other respiratory diseases caused by coronaviruses such as the human severe acute respiratory syndrome (SARS-CoV) and the Middle East respiratory syndrome (MERS-CoV) (da Costa et al., 2020; Petrosillo et al., 2020).
It is also observed that some environmental factors cause a higher risk of infection by SARS-CoV-2 as well as to develop more severe symptoms of COVID-19. For instance, cigarette smoke increases the risk to develop severe symptoms of the disease, while, in contrast, epidemiologic reports show that smokers are under-represented in COVID-19 patients, possibly due to the protective role of nicotine against SARS-CoV-2 infection (Brake et al., 2020; Cai et al., 2020; Cai, 2020; Leung et al., 2020; Lippi and Henry, 2020; Olds and Kabbani, 2020). Several studies also report that atmospheric pollution, including particulate matters (PM2.5 and PM10) and high levels of nitrogen dioxide in the air are correlated with a higher incidence of COVID-19 and more severe forms of the disease (Bianconi et al., 2020; Bolaño-Ortiz et al., 2020; Borro et al., 2020; Comunian et al., 2020; Conticini et al., 2020; Copat et al., 2020; Ogen, 2020; Yao et al., 2020; Zoran et al., 2020). These data raised the issue of environmental exposures as a confounding factor in SARS-CoV-2 infection and disease course.
It has been demonstrated that SARS-CoV-2 uses the human angiotensin-converting enzyme-2 (ACE2) receptor for infection, as already observed for other coronaviruses, including SARS-CoV and HCoV-NL63 (Hoffmann et al., 2020b; Lan et al., 2020; Letko et al., 2020; Shang et al, 2020a, 2020b; Walls et al., 2020; Yan et al., 2020). In vitro, soluble ACE2 is sufficient to block SARS-CoV-2 infection in reconstituted human tissues (Monteil et al., 2020). Conversely, expression of human ACE2 in mouse tissues and its over-expression in human cells refractory to SARS-CoV-2 infection confers SARS-CoV-2 susceptibility (Bao et al., 2020; Ou et al., 2020; Winkler et al., 2020). It was further demonstrated that the SARS-CoV-2 spike (S) glycoprotein directly binds the receptor ACE2 (Hoffmann et al., 2020b; Lan et al., 2020; Shang et al., 2020b; Yan et al., 2020). ACE2 is expressed in many tissues including lung, liver, colon, oesophagus, intestine, heart and kidney tissues which is coherent with the variety of cell types that can be infected and might explain the variety of symptoms reported in COVID-19 patients (Cevik et al., 2020; Gkogkou et al., 2020; Hikmet et al., 2020; Hoffmann et al., 2020b; Li et al., 2020; Lukassen et al., 2020; Oberfeld et al., 2020; Qi et al., 2020; Zou et al., 2020).
The cellular entry of SARS-CoV-2 also depends on the priming (i.e. cleavage) of its transmembrane spike (S) glycoprotein by host cell proteases to promote virus-cell entry. Different proteases are important for SARS-CoV-2 cell entry, notably the transmembrane serine protease-2 (TMPRSS2) (Hoffmann et al., 2020b; Wrapp et al., 2020). It was observed that human cells over-expressing TMPRSS2 are more susceptible to coronaviruses infection than control cells, and that SARS-CoV-2 infection can be partially blocked by treatment with inhibitors of TMPRSS2 protease activity (Hoffmann et al., 2020b; Matsuyama et al., 2020; Ou et al., 2020; Shang et al., 2020a). It was also reported that Cathepsins lysosomal cysteine proteases have a major role in the virus cell entry. Chemical inhibition of Cathepsin L endosomal cysteine proteases activity (encoded by CTSL and CTSL2 also known as CTSV), and to a lesser extent inhibition of Cathepsin B (CTSB) activity, impairs SARS-CoV-2 cell entry (Hoffmann et al., 2020b; Ou et al., 2020; Shang et al., 2020a; Zhou et al., 2016). In addition, it was also shown that the subtilisin-like endoprotease FURIN cleaves the multibasic cleavage site (Arg-Arg-Ala-Arg) at the junction of the two subunits of the SARS-CoV-2 spike protein, favouring virus entry (Coutard et al., 2020; Hoffmann et al., 2020a; Jaimes et al., 2020; Shang et al., 2020a; Walls et al., 2020). This list is probably not exhaustive, as numerous investigations are still ongoing to properly catalogue the full spectrum of host cell proteases and pathways that have an effect on SARS-CoV-2 cell entry in different cell types.
We postulated that identifying and reporting environmental factors which possibly altered the expression of ACE2 and human proteases involved in the proteolytic processing of the S protein of SARS-CoV-2, might help to better understand the susceptibility to viral infection. We thus exploited large publicly available and freely re-useable gene expression and genomic datasets to support the hypothesis of a potential impact of environmental exposures on the expression of the genes ACE2, TMPRSS2, FURIN, CTSL, CTSV (also known as CTSL2) and CTSB, that in turns could influence virus entry into host cells.
2. Materials and methods
2.1. Genomic and tissue expression resources
Genomic and interactions data were analysed using the UCSC Genome Browser (Lee et al., 2020) and the GeneHancer database (Fishilevich et al., 2017). Genomic data were analysed on the human reference genome GRCh37/hg19. Interactions between GeneHancer regulatory elements (promoters and enhancers) and genes reported in the GeneHancer database were collected from different sources, including expression quantitative trait loci (eQTL) from the genotype-tissue expression (GTEX) project (version v6p), chromosome conformation capture (capture Hi-C) long range interactions, FANTOM eRNA-gene expression correlation or distance based associations (Fishilevich et al., 2017).
2.2. Gene expression datasets and analysis
Gene expression datasets were downloaded through the Gene Expression Omnibus (GEO) database (Barrett et al., 2013). The datasets downloaded and re-analysed are the following: 1) Brushings from intra-pulmonary airways (the right upper lobe carina), from normal smoking and non-smoking volunteers (including 34 current smokers, 23 nonsmokers and 18 former smokers) (GSE994) (Spira et al., 2004); 2) Bronchial epithelial cells from 21 former smokers and 9 current smokers (GSE112073) (Corbett et al., 2019); 3) Bronchial epithelium of fiberoptic bronchoscopy from 21 never smokers, 52 current smokers and 31 former smokers (GSE7895) (Beane et al., 2007); 4) Small airway epithelium from 38 nonsmokers and 45 smokers obtained by fiberoptic bronchoscopy (GSE11952) (Hübner et al., 2009); 5) Bronchial epithelial cells exposed to cigarette smoke from two brands of cigarettes (n = 4) (GSE10700) (Jorgensen et al., 2008).
Differentially expressed genes were determined using the GEO2R software using p-value correction with false-discovery rate correction (Benjamini & Hochberg) (Barrett et al., 2013). q-values (FDR corrected p-values) were calculated from a genome-wide analysis and fold change between treated and matching control condition (i.e. reported as log2 of the Fold Change). These values were used to prepare the different panels. A q-value < 0.05 was considered statistically significant.
2.3. Comparative toxicogenomics database search and analysis
The Comparative Toxicogenomics Database (CTD) (http://ctdbase.org/) reports manually curated chemical–gene, chemical–phenotype and chemical–exposure interactions for 15,681 chemicals and 46,689 genes (Davis et al., 2019). The database was interrogated on April 14, 2020 using the search function with gene name (i.e. ACE2, CTSB, CTSL, CTSV, FES, FURIN, IQGAP1, MX2, PIR, TMPRSS2 or UNC45) and human genome (Taxon:9606) as queries. The list of chemical-gene interactions obtained was then manually curated to remove studies involving mixture of chemicals, functional relationships and analyses of protein levels (Davis et al., 2019). Chemicals associated with ACE2, CTSB, CTSL, CTSV, FES, FURIN, IQGAP1, MX2, PIR, TMPRSS2 and UNC45 gene expression up-regulation or down-regulation were further analysed and intersected. Chemicals associated with the regulation of AhR, either at mRNA or protein level, were also retrieved from the database on April 14, 2020.
2.4. Identification of AhR consensus binding motif
Coordinates of promoters were retrieved from the GeneHancer database (Fishilevich et al., 2017). Coordinates were uploaded on the Galaxy interface (https://usegalaxy.org/) and DNA sequences corresponding to these coordinates were retrieved in batch in FASTA format using the function “Extract genomic DNA”. Motif search on these DNA sequences was then performed using the EMBOSS Fuzznuc function with the following criteria: no mismatch allowed to the searched pattern and a search on the Watson and Crick strand. We investigated the AhR/ARNT core sequence 5′-GCGTG-3’ (Jaspar MOTIF Ahr:Arnt_MA0006.1) embedded in the xenobiotic responsive elements (XREs) consisting of T/CNGCGTGA/CG/CA (N = any nucleotide) or a divergent AhR element 5′-CATG(N6)CT/ATG-3’ (Dere et al., 2011; Jackson et al., 2015; Murray et al., 2014).
2.5. ChIP-sequencing analysis of AhR binding sites in human cells
AhR ChIP-sequencing data were retrieved from the GEO database: GEO GSE90550 and GEO GSE116632 (Neavin et al., 2019; Yang et al., 2018).
GSE90550 reports AhR ChIP-sequencing experiments performed using an AhR antibody (Santa Cruz, H-211) in triplicate, starting from MCF7 breast cancer cells treated with 10 nM TCDD or vehicle for 45 min (Yang et al., 2018).
GSE116632 reports AhR ChIP-sequencing experiments performed using an AhR antibody (Cell Signalling Technology, D5S6H) in duplicate, starting from a lymphoblastoid cell line (GM17212) treated with 1 μM 3-methylcholanthrene or vehicle for 24 h (Neavin et al., 2019).
3. Results
3.1. Genomic and 3D organisation of the loci encoding the human receptor and proteases involved in SARS-CoV-2 cell entry
We thought that expression levels of ACE2, TMPRSS2, FURIN, CTSB, CTSL and CTSV might be impacted by the expression level of other genes they physically interact with within the nucleus. To get a better understanding of these potential regulatory mechanisms we analysed genomic and chromosome conformation capture data in human cells and listed genes and regulatory sequences that interact with ACE2, TMPRSS2, FURIN, CTSB, CTSL and CTSV.
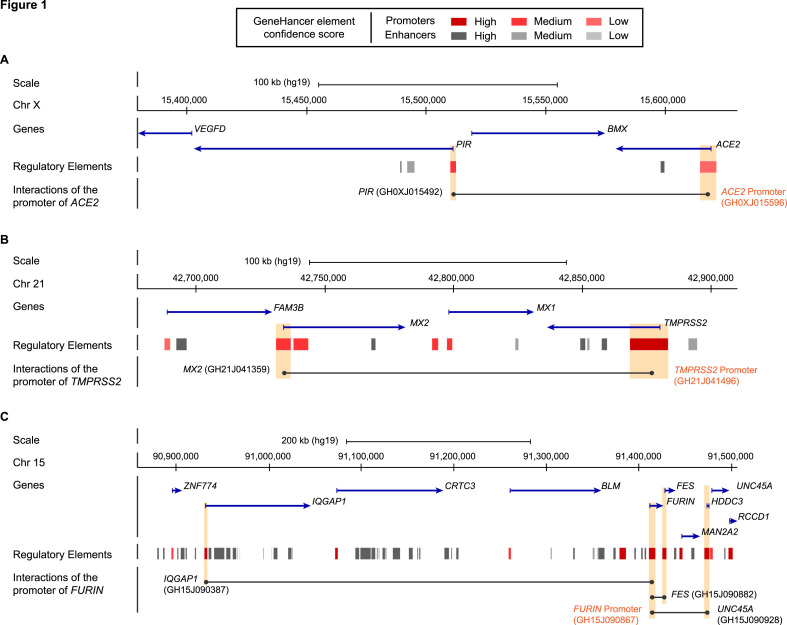
ACE2 is located on the short arm of chromosome X and it escapes X inactivation in female cells. In the GeneHancer database (Fishilevich et al., 2017), it is reported that the promoter of ACE2 is involved in long-distance interactions with the promoter of PIR, a transcriptional coregulator of the NF-kappa-B pathway (Liu et al., 2013), and an enhancer located in intron 9 of ACE2 (Fig. 1 A and Supplementary Table S1). Importantly, higher expression of PIR is recurrently detected in lung biopsies from smokers compared to nonsmokers indicating a regulation of this gene by cigarette smoke. It was also shown that PIR up-regulation may be associated with lung epithelial cell damages and eventually cell death (Gelbman et al., 2007).
Fig. 1.
Genomic organisation of ACE2, TMPRSS2 and FURIN regions. Schematic representation from the UCSC Genome browser (reference human genome GRC37/hg19) highlighting the regions of (A) ACE2 (chrX:15,380,000–15,630,000), (B) TMPRSS2 (chr21:42,678,000–42,910,000), (C) FURIN (chr15:90,860,000–91,507,000). The panels show from the top to bottom: the genomic coordinates on chromosome, the position of the genes and their orientations retrieved from the NCBI RefSeq Select database (one representative transcript per protein-coding gene), the position of the regulatory elements with enhancers represented in grey and promoters represented in red from GeneHancer Double Elite database, the interactions of the promoter of gene of interest with regulatory elements in its region, highlighted by orange boxes. Full list of chromosome contacts for each region analysed in panel A–C is reported in Supplementary Table S1. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
TMPRSS2 is located on chromosome 21 in the vicinity of MX Dynamin Like GTPase 1 (MX1) and MX Dynamin Like GTPase 2 (MX2) (Fig. 1B and Supplementary Table S1). In the GeneHancer database, TMPRSS2 promoter interacts with the promoter region of MX2. MX2 encodes an interferon-alpha inducible GTPase with antiviral activity against immunodeficiency virus type 1 (HIV1) and vesicular stomatitis virus (Haller et al., 2015; Kane et al, 2013, 2018; Melén et al., 1996; Pavlovic et al., 1990). An alternative promoter of TMPRSS2, upstream of the transcription start site (TSS), mediates interactions with enhancers located in intronic regions of TMPRSS2 (introns 6, 7 and 8) as well as with its canonical promoter (Supplementary Table S1 ).
FURIN (also known as FES upstream region) is located on chromosome 15. In the GeneHancer database, we found that the promoter of FURIN interacts with promoters of the FES Proto-Oncogene Tyrosine Kinase (FES), the Ras GTPase-activating-like protein IQGAP1 (IQGAP1) and Unc-45 Myosin Chaperone A (UNC45A) (Fig. 1C and Supplementary Table S1). FES, IQGAP1 and UNC45A have been described for their role in cell cycle progression and cell migration, and their expression altered in several cancers (Eisa et al., 2019; Johnson et al., 2009; Sodroski et al., 1984).
CTSB is located on chromosome 8 while CTSL and CTSV are located on chromosome 9. In the GeneHancer database, there is no information regarding potential 3D conformational interactions between the promoters of cathepsins and promoters of other genes (Supplementary Table S1).
Our analysis indicates that promoters of ACE2 and TMPRSS2 interact through long range interactions with promoters of PIR and MX2 genes, which are regulated by environmental cues, such as cigarette smoke and viral infection. We also observed that FURIN promoter interacts with many genes important for cell architecture and cancer signalling. We thus considered the entire list of genes to document a possible relationship between environmental cues and the expression of human genes encoding the receptor and proteases of SARS-CoV-2 spike S glycoprotein.
3.2. A data mining approach shows that expression of genes encoding receptor and proteases involved in SARS-CoV-2 cell entry is modulated by a variety of chemicals
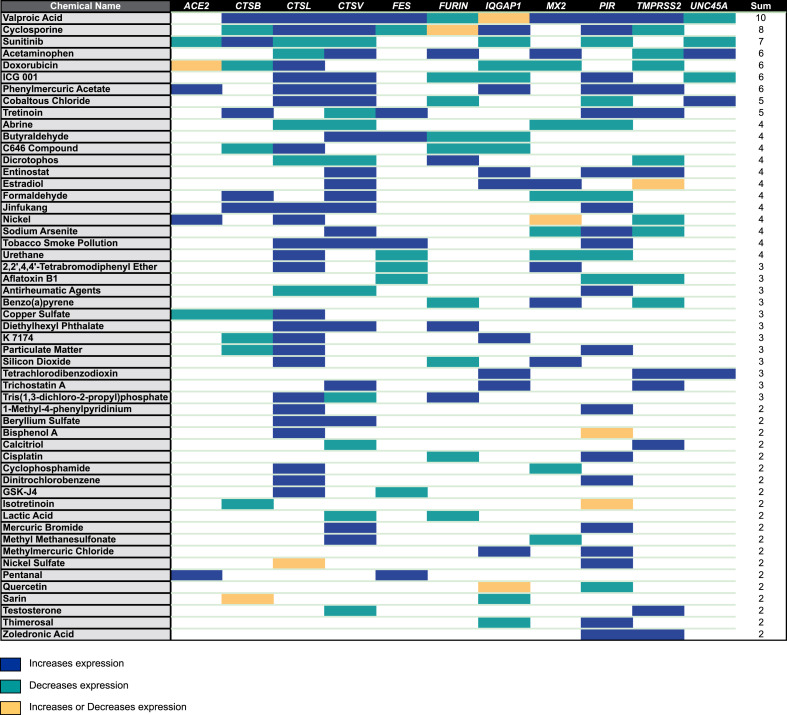
We queried the comparative toxicogenomics database (CTD) to identify studies reporting changes in expression levels of human genes encoding receptor and proteases important for SARS-CoV-2 cell entry upon chemical exposure (Davis et al., 2019). We selected for our analysis any study reporting gene expression in human tissues (or cells) and with negative or positive regulation of mRNA expression. We considered chemical components found in the environment, as well as chemicals used in medicine or industry. Using these criteria, we identified several chemical exposures regulating the expression of receptor ACE2 and human proteases TMPRSS2, FURIN and Cathepsins genes in human cells and tissues (Fig. 2 ; Supplementary Table S2). Notably, this approach confirms that PIR expression is enhanced by tobacco smoke pollution (Supplementary Table S2).
Fig. 2.
Data mining identification of compounds potentially involved in the regulation of human receptor ACE2 and proteases of SARS-CoV-2 S protein at mRNA level in human tissues. Data were retrieved from the Comparative Toxicogenomics Database (CTD). Colour code: blue square, the chemical increases gene expression; green square, the chemical decreases gene expression; yellow square, the chemical increases or decreases gene expression depending on studies; white, no regulation reported in the database. The full list of compounds and relationships is reported in Supplementary Table S2. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We unveiled several chemicals found in the environment modulating the expression of at least two genes analysed in our study (Fig. 2). Three pollutants can increase ACE2 expression as well as human proteases of SARS-CoV-2 S protein: phenylmercuric acetate (widely used as a fungicide and a disinfectant), nickel (a metal used in industry) and pentanal (an alkyl aldehyde used in industry). Phenylmercuric acetate is the only chemical reported in the database that enhances the expression of ACE2 and TMPRSS2, the main SARS-CoV-2 receptor/protease couple identified in in vitro assays, suggesting a potential interference with SARS-CoV-2 infection. We also noticed that several compounds, including bisphenol A, cyclophosphamide, tobacco smoke and particulate matter increase the expression of CTSL, a key regulator of SARS-CoV-2 cell entry by endocytosis. Finally, we also noted that Benzo(a)pyrene, a prototypical polycyclic aromatic hydrocarbon compound, modulates FURIN and TMPRSS2 expression (Fig. 2).
We also identified several chemicals used in medicine as modulator of ACE2 and human proteases of SARS-CoV-2 expression (Fig. 2). Quite noticeable is the impact of valproic acid, a chemical used for the treatment of mood disorders (Dong et al., 2008) on the expression of 10 out of the 11 genes analysed (Fig. 2). Valproic acid is a histone deacetylase inhibitor that also acts on DNA methylation patterns and hence affects a large number of gene targets, including most gene analysed here. Also intriguing, is the down-regulation of ACE2, CTSL and CTSV expression in cells treated with the anti-cancer tyrosine-kinase inhibitor sunitinib (Fig. 2) (Cabebe and Wakelee, 2006). We also observed that cyclosporine, a modulator of the immune response utilised for the treatment of several auto-immune diseases (e.g. psoriasis, atopic dermatitis) and in organ transplant protocols (Talal, 1988), impacts on the expression level of all the known human proteases involved in the processing of the SARS-CoV-2 spike glycoprotein, CSTB, CTSL, CTSV, FURIN and TMPRSS2 (Fig. 2).
In sum, our analysis reveals that >50 chemicals can potentially affect the expression of human receptor ACE2 and human proteases involved in SARS-CoV-2 cell entry, which may modulate the susceptibility to SARS-CoV-2 infection.
3.3. Cigarette smoke does not modulate the expression of human receptor and proteases of SARS-CoV-2 S protein in the respiratory tract
Cigarette smoke is a confounding factor in many lung pathologies and many studies suggest it may impact on COVID-19 prognosis (Leung et al., 2020). We thus further investigated whether cigarette smoke may regulate the expression of the receptor ACE2 and proteases involved in SARS-CoV-2 cell entry that may modulate the susceptibility to infection.
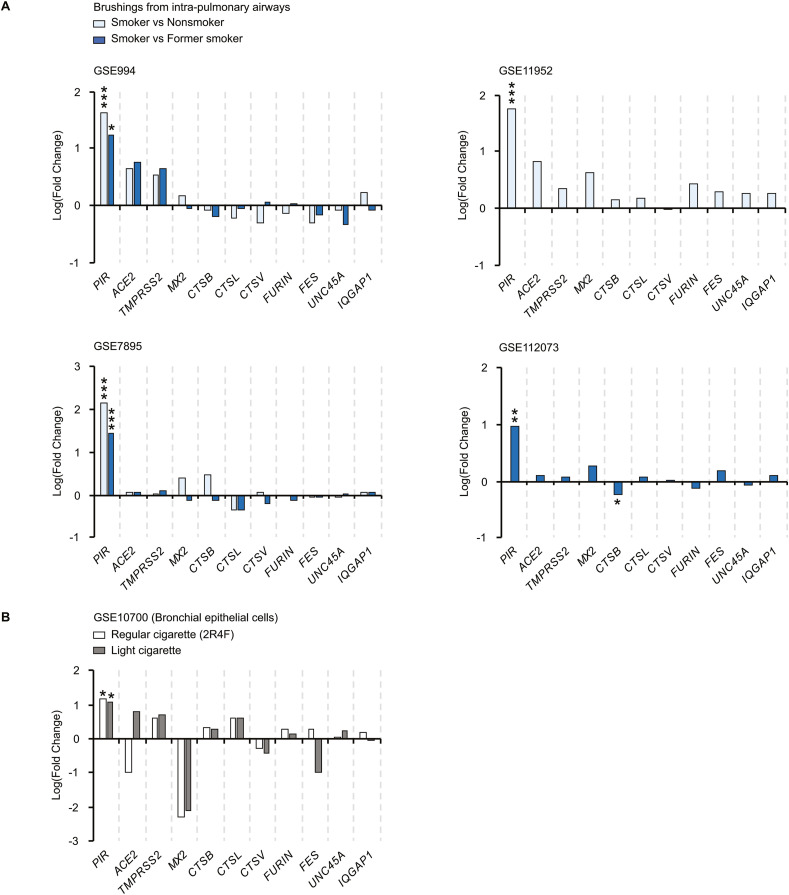
The analysis of the CTD database revealed that expression of CTSL and CTSV is increased upon cigarette smoke exposure in bronchial epithelial cells and monocytes and CTSV expression is also increased in 3D-bronchial tissues (Anthérieu et al., 2017; Ishikawa et al., 2019; Wright et al., 2012). We thus further investigated the level of expression of the different genes in the respiratory tract tissues of smokers, nonsmokers and former smokers. We first analysed gene expression from intra-pulmonary airways obtained from 34 current, 23 never and 18 former smokers (GSE994) (Spira et al., 2004). We observed significant higher levels of PIR expression in smokers’ biopsies compared to nonsmokers and former smokers biopsies, while other genes studied are expressed at similar levels (Fig. 3 A). In three additional cohorts (GSE11952, GSE7895 and GSE112073) we also observed similar results (Fig. 3A) (Beane et al., 2007; Corbett et al., 2019; Hübner et al., 2009). These data indicate that among the different genes studied, solely PIR is significantly regulated by cigarette smoke. To further assess these data, we re-analysed gene expression data from bronchial epithelial cells exposed in vitro to cigarette smoke (GSE10700) (Jorgensen et al., 2008). Importantly, in vitro, cigarette smoke also causes an up-regulation of PIR expression in bronchial epithelial cells after 24 h exposure, consistent with a direct effect of cigarette smoke on PIR expression (Fig. 3B). Other genes were not differentially expressed between smoke and control treated cells (Fig. 3B). Our analysis suggests that expression of human receptor and proteases of SARS-CoV-2 spike S glycoprotein are not directly regulated by cigarette smoke at the transcriptional level in lung cells, nor that PIR and ACE2 are co-regulated upon cigarette smoke exposure.
Fig. 3.
Expression level of genes encoding ACE2 and proteases of SARS-CoV-2 S protein in lung biopsies from smokers and non-smokers. (A) Gene expression in lung biopsies from smokers compared to nonsmokers and former smokers. *, p < 0.05; ***, p < 1e-5 (vs non or former smokers). (B) Gene expression in bronchial epithelial cells exposed to smoke from a brand of regular cigarette (2R4F) and a brand of typical “light” cigarette. *, p < 0.05 (vs mock treated).
3.4. Many aryl hydrocarbon receptor (AhR) agonists regulate the expression of ACE2 and human proteases of SARS-CoV-2 S protein
We thought to identify the molecular pathways that link chemicals with COVID-19 disease. Intriguingly, we identified several chemicals known as agonist of the aryl hydrocarbon receptor (AhR) (Murray et al., 2014). Among the 53 chemicals regulating the expression of ACE2 and proteases important for SARS-CoV-2 cell entry, 30 were associated with the function and/or regulation of AhR (P (X = 30) = 1.58e-23; enrichment: 9.54) based on the CTD nomenclature. For instance, exposure to particulate matters (PM), well-known agonists of AhR (Castañeda et al., 2018; Harmon et al., 2018), reduces CTSB expression in endothelial cells (Gong et al., 2007) and enhances CTSL expression in dermal fibroblasts (Park et al., 2018) and bronchial Beas-2B cells (Grilli et al., 2018). Pilling up evidences indicate that exposure to particulate matters, ranging from PM2.5 and PM10, is associated with greater COVID-19 disease severity and poor clinical outcome (Comunian et al., 2020; Yao et al., 2020; Zoran et al., 2020). We also identified, benzo(a)pyrene, a polycyclic aromatic hydrocarbon, that decreases FURIN expression in HepG2 liver cancer cells (Lizarraga et al., 2012) and TMPRSS2 expression in human prostate and liver cancer cells (Hrubá et al., 2011; Kreuzer et al., 2020). Tetrachlorobenzodioxin increases TMPRSS2 expression in MCF7 breast cancer cells (Spink et al., 2009). Calcitriol, the physiologically active form of vitamin D, increases TMPRSS2 expression in human breast cancer tissues (Sheng et al., 2016).
These data suggest that AhR signalling is an important player in the transcriptional regulation of human receptor and proteases of SARS-CoV-2 S protein, and that AhR signalling may impact on cell infection by SARS-CoV-2.
3.5. Analysis of aryl hydrocarbon receptor (AhR) binding sites in human cells suggests that TMPRSS2 is a direct transcriptional target of chemicals
We investigated whether AhR binds regulatory sequences involved in the regulation of human ACE2 and proteases of SARS-CoV-2 S protein. Upon binding of its ligand, AhR forms a heterodimer with the AhR nuclear translocator (ARNT), forming a DNA binding complex that translocates to the nucleus and binds and activates the transcription of target genes. AhR/ARNT binds the core sequence 5′-GCGTG-3’ (Jaspar MOTIF Ahr:Arnt_MA0006.1) embedded in the xenobiotic responsive elements (XREs) consisting of T/CNGCGTGA/CG/CA (N = any nucleotide) or a divergent AhR element 5′-CATG(N6)CT/ATG-3’ (Dere et al., 2011; Jackson et al., 2015; Murray et al., 2014). We thus performed a search for these AhR consensus binding motifs in the promoters of ACE2, CTSB, CTSL, FURIN and TMPRSS2 using coordinates from the GeneHancer database (since no promoter segment is reported in the database for CTSV we did not include this gene in our analysis). We identified consensus binding sites for AhR in the promoters of ACE2, CTSB, CTSL, FURIN and TMPRSS2 (Supplementary Table S3). These data suggest that AhR might directly regulate the expression of these genes. Nonetheless, the frequency of AhR motifs in the promoter of the genes of interest is not significantly higher than the number of expected AhR motif in promoters of the GeneHancer database (Supplementary Table S3).
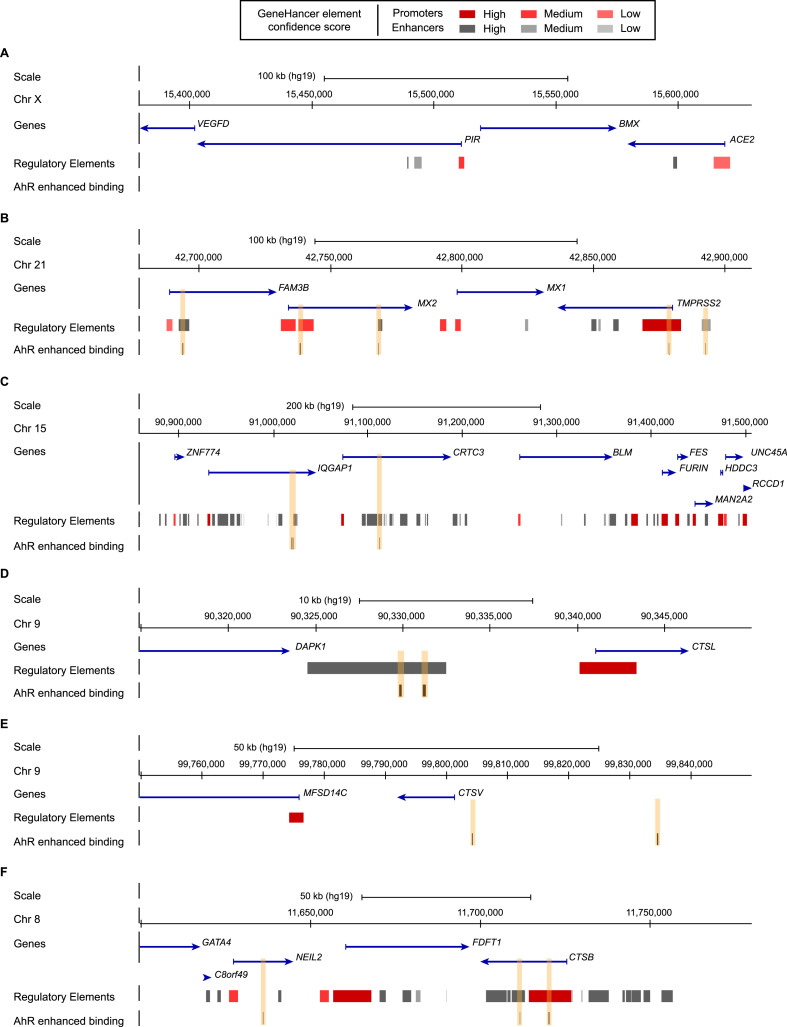
To support a role of AhR in the regulation of ACE2, CTSB, CTSL, FURIN and TMPRSS2 we thus searched for chromatin immunoprecipitation data for AhR in primary human cells or human tissues. Unfortunately, we could not find maps of AhR binding sites in “normal” cells, but identified two maps of AhR performed in MCF7 breast cancer cells 45 min after addition of the well known AhR pathway activator 2,3,7,8-tetrachlorodibenzo-p-dioxine (TCDD) and in lymphoblastoid cells 24 h after addition of AhR agonist 3-methylcholanthrene (Neavin et al., 2019; Yang et al., 2018).
The analysis in MCF7 cells indicates that AhR binding is enhanced at the promoter of TMPRSS2 and CTSB upon TCDD treatment (Fig. 4 A–F). This analysis also shows that AhR binding is enhanced upon TCDD exposure to the promoter of MX2 and enhancers of TMPRSS2 and CTSB, and in the vicinity of CTSV (Fig. 4A–F).
Fig. 4.
AhR binding in ACE2, TMPRSS2, FURIN, CTSL, CTSV and CTSB regions upon TCDD exposure. Schematic representation from the UCSC Genome browser (reference human genome GRC37/hg19) highlighting the regions of (A) ACE2 (chrX:15,380,000–15,630,000), (B) TMPRSS2 (chr21:42,678,000–42,910,000), (C) FURIN (chr15:90,860,000–91,507,000), (D) CTSL (chr9:90,315,000–90,350,000), (E) CTSV (chr9:99,750,000–99,850,000) and (F) CTSB (chr8:11,600,000–11,780,000). The panels show from the top to bottom: the genomic coordinates on chromosome, the position of the genes and their orientations retrieved from the NCBI RefSeq Select database (one representative transcript per protein-coding gene), the position of the regulatory elements with enhancers represented in grey and promoters represented in red from GeneHancer Double Elite database, the position of the AhR binding sites enhanced upon TCDD exposure highlighted by orange boxes. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
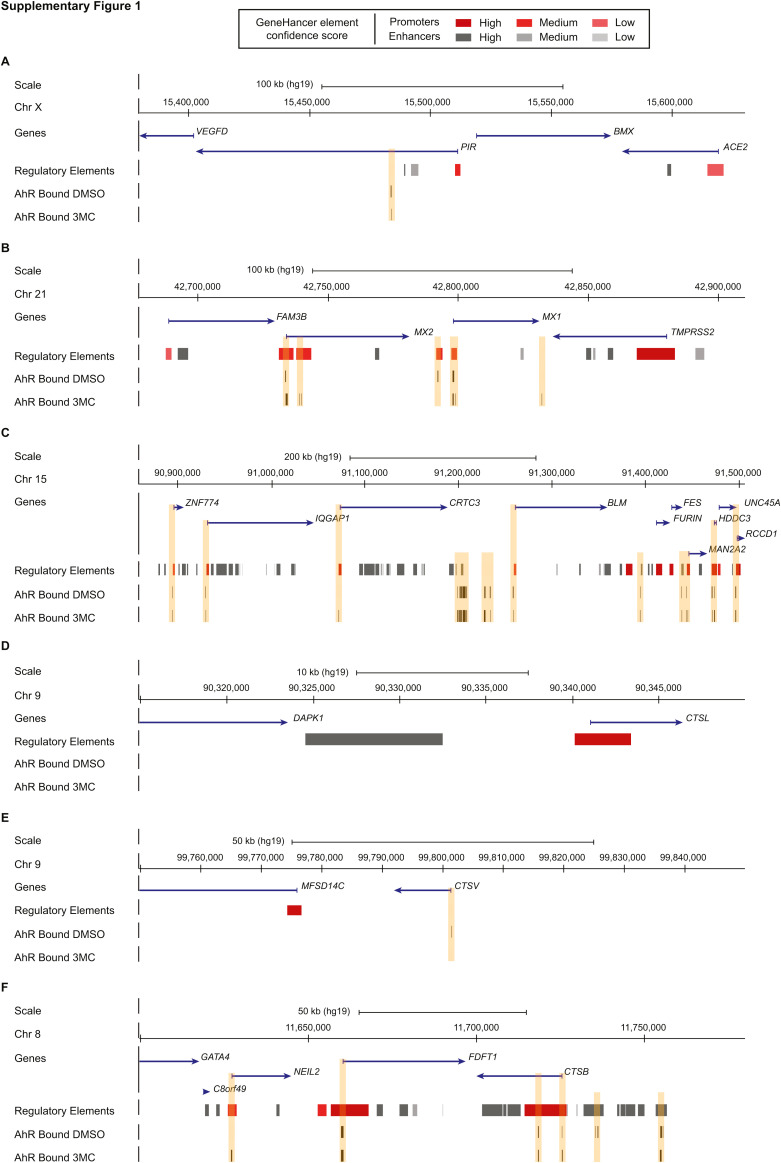
The analysis in lymphoblastoid cells indicates that AhR binds to the promoters, or enhancers, of CTSB, MX1, MX2, FURIN, IQGAP1, FES and UNC45A in untreated condition (Supplementary Figure 1). Upon 3-methylcholanthrene treatment, AhR binding is still detected at these sites (Supplementary Figure 1).Figure figs1
Our analysis revealed that TMPRSS2 and CTSB are direct targets of AhR in human cells upon AhR-signalling activation. These later analyses provide molecular support for a direct link between chemical exposure and SARS-CoV-2 infection.
4. Discussion
4.1. Chemicals as confounding factors in SARS-CoV-2 infection and disease
Refinement of the COVID-19 course is still ongoing and our report provides evidence that the mRNA expression of the human receptor ACE2 and proteases, which are important for SARS-CoV-2 entry into the cells, is modulated by environmental cues. This information may help better understand the molecular basis of the susceptibility to SARS-CoV-2 and other coronaviruses infection, the course of the disease and help design future epidemiologic studies by considering the exposition to chemicals revealed in this study. Proper molecular and cellular assays will nonetheless be needed to properly assess and validate the correlation described in this study in different cell types, reconstituted tissues, organoids and human samples.
We identify numerous chemicals, either pollutants from the environment or used for the treatment of chronic diseases, as potential regulators of ACE2 and human proteases of SARS-CoV-2 spike glycoprotein in human tissues. Higher levels of TMPRSS2 expression favour human cell infection in vitro, while expression of human ACE2 renders mice susceptible to SARS-CoV-2 infection (Bao et al., 2020; Matsuyama et al., 2020; Monteil et al., 2020; Winkler et al., 2020). These observations suggest that chemicals that promote higher expression levels of human receptor ACE2 and proteases in tissues might increase the risk of infection. This interpretation is obviously simplistic and detailed epidemiologic and clinical studies are needed to refine these data and address their robustness and reproducibility in the human population. Individuals treated for chronic pathologies and disorders, or chronically exposed to chemicals in their life, may present altered expression levels of ACE2 and proteases mediating SARS-CoV-2 cell entry compared to the rest of the population, which may account for the differences in the susceptibility to infection as well as the disease outcome between individuals. For instance, medical issues have emerged regarding the use of modulators of the immune system such as methotrexate, anti-TNF therapies or cyclosporine (Russell et al., 2020). Studies also proposed that particulate matters or high levels of nitrogen dioxide in the air may be favouring more severe forms of COVID-19 (Comunian et al., 2020; Conticini et al., 2020; Ogen, 2020; Yao et al., 2020). In support of these observations, our study identifies several compounds, frequently detected in the airaira, as potential regulators of ACE2 and proteases of SARS-CoV-2 S protein that might modulate the efficiency of viral infection. Importantly, we did not find a clear relationship between ACE2 expression levels and smoking habits in the cohorts we re-analysed. This is in contrast to other studies based on immunofluorescence assays (Brake et al., 2020; Cai et al., 2020) or showing that cigarette smoke is associated with higher levels of ACE2 expression in aged male (>65 years old) but not in female nor in younger male (Leung et al., 2020). Our analysis merely investigated based on publicly available data a potential relationship between environmental exposure (i.e. single exposure) and expression of receptor ACE2 and human proteases important for SARS-CoV-2 cell entry. Further work will be need to properly assess these cellular data in the human population, and in response to more complex “exposomes”.
In addition, it remains to be investigated how these pollutants interfere with additional steps of SARS-CoV-2 biology, including its replication, its dissemination in the organism and how these environmental and chemical cues influence the course and outcome of COVID-19. For instance, AhR signalling appears as a key player in response to mouse coronavirus infection and to Zika virus infection (Giovannoni et al., 2020; Grunewald et al., 2020). It is thus likely that AhR signalling activity might contribute to the pathology of SARS-CoV-2. Furthermore, in vitro chemical screens identified numerous compounds that impair SARS-CoV-2 replication in cellular models, at different stages of the virus life cycle (Bojkova et al., 2020; Sauvat et al., 2020; Touret et al., 2020).
4.2. AhR signalling might contribute to COVID-19 prognosis
Analysis of two different maps of AhR binding sites in the human genome indicates that TMPRSS2, FURIN and CTSB are direct transcriptional targets of AhR signalling. Upon TCDD exposure, our analysis reveals enhanced binding of AhR at the promoter of TMPRSS2, consistent with a direct relationship between chemical exposure and TMPRSS2 expression. We also observed that promoters of FURIN and CTSB are bound by AhR in lymphocytes, but that AhR agonist 3-MC does not enhance the binding. This suggests that different agonists trigger different levels of AhR signalling and/or that the binding of AhR to promoters of receptor ACE2 and proteases of SARS-CoV-2 S protein is differentially modulated by AhR agonists in different cell types.
Importantly, while ACE2 and TMPRS22 promoters interact with promoters of PIR and MX2, that are massively up-regulated upon smoking and viral infection respectively, we did not observe a co-regulation of these genes in our analyses. This co-regulation might nonetheless occur under conditions that need to be properly and exhaustively investigated such as mixture of compounds and more complex environmental signals.
In sum, our study provides preliminary evidences that receptor ACE2 and human proteases important for SARS-CoV-2 cell entry are regulated by many chemicals linked with AhR signalling, which should prompt further analysis of the relationship between “exposome” and SARS-CoV-2 pathology.
Author contribution
Manon Watzky, Maud de Dieuleveult, Anne Letessier, Claude Saint-Ruf and Benoit Miotto, Conceptualization, Data acquisition, Formal analysis, Methodology, Drafting the manuscript. Benoit Miotto, Funding acquisition.
Funding
Laboratory of B.M. is partner of Labex “Who Am I?” #ANR-11-LABX-0071 and the Université de Paris IdEx #ANR-18-IDEX-0001 funded by the French Government through its “Investments for the Future” program. This work was funded by Fondation pour la Recherche Médicale (AJE20151234749), Labex “Who am I?” (ANR-11-LABX-0071 and ANR-11-IDEX-005-02), CNRS, INSERM and University of Paris. MW is supported by a PhD fellowship from the French Government. MdD is supported by a post-doctoral fellowship from Labex “Who Am I?” #ANR-11-LABX-0071.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are grateful to Dr Morgane Bomsel (INSERM U1016, Paris, France) for discussions during the course of this study. We thank research laboratories and consortiums for making their studies publicly available and freely re-usable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envres.2020.110317.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
References
- Anthérieu S., Garat A., Beauval N., Soyez M., Allorge D., Garçon G., Lo-Guidice J.-M. Comparison of cellular and transcriptomic effects between electronic cigarette vapor and cigarette smoke in human bronchial epithelial cells. Toxicol. Vitro. 2017;45:417–425. doi: 10.1016/j.tiv.2016.12.015. [DOI] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., Qu Y., Li F., Lv Q., Wang W., Xue J., Gong S., Liu M., Wang G., Wang S., Song Z., Zhao Linna, Liu P., Zhao Li, Ye F., Wang H., Zhou W., Zhu N., Zhen W., Yu H., Zhang X., Guo L., Chen L., Wang C., Wang Y., Wang X., Xiao Y., Sun Q., Liu H., Zhu F., Ma C., Yan L., Yang M., Han J., Xu W., Tan W., Peng X., Jin Q., Wu G., Qin C. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., Yefanov A., Lee H., Zhang N., Robertson C.L., Serova N., Davis S., Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beane J., Sebastiani P., Liu G., Brody J.S., Lenburg M.E., Spira A. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol. 2007;8:R201. doi: 10.1186/gb-2007-8-9-r201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianconi V., Bronzo P., Banach M., Sahebkar A., Mannarino M.R., Pirro M. Particulate matter pollution and the COVID-19 outbreak: results from Italian regions and provinces. Arch. Med. Sci. 2020;16:985–992. doi: 10.5114/aoms.2020.95336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Münch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolaño-Ortiz T.R., Camargo-Caicedo Y., Puliafito S.E., Ruggeri M.F., Bolaño-Diaz S., Pascual-Flores R., Saturno J., Ibarra-Espinosa S., Mayol-Bracero O.L., Torres-Delgado E., Cereceda-Balic F. Spread of SARS-CoV-2 through Latin America and the Caribbean region: a look from its economic conditions, climate and air pollution indicators. Environ. Res. 2020;191:109938. doi: 10.1016/j.envres.2020.109938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borro M., Di Girolamo P., Gentile G., De Luca O., Preissner R., Marcolongo A., Ferracuti S., Simmaco M. Evidence-based considerations exploring relations between SARS-CoV-2 pandemic and air pollution: involvement of PM2.5-mediated up-regulation of the viral receptor ACE-2. Int. J. Environ. Res. Publ. Health. 2020;17 doi: 10.3390/ijerph17155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brake S.J., Barnsley K., Lu W., McAlinden K.D., Eapen M.S., Sohal S.S. Smoking upregulates angiotensin-converting enzyme-2 receptor: a potential adhesion site for novel coronavirus SARS-CoV-2 (Covid-19) J. Clin. Med. 2020;9 doi: 10.3390/jcm9030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabebe E., Wakelee H. Sunitinib: a newly approved small-molecule inhibitor of angiogenesis. Drugs Today. 2006;42:387–398. doi: 10.1358/dot.2006.42.6.985633. [DOI] [PubMed] [Google Scholar]
- Cai G., Bossé Y., Xiao F., Kheradmand F., Amos C.I. Tobacco smoking increases the lung gene expression of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020 doi: 10.1164/rccm.202003-0693LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H. Sex difference and smoking predisposition in patients with COVID-19. Lancet Respir Med. 2020;8 doi: 10.1016/S2213-2600(20)30117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda A.R., Pinkerton K.E., Bein K.J., Magaña-Méndez A., Yang H.T., Ashwood P., Vogel C.F.A. Ambient particulate matter activates the aryl hydrocarbon receptor in dendritic cells and enhances Th17 polarization. Toxicol. Lett. 2018;292:85–96. doi: 10.1016/j.toxlet.2018.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik M., Bamford C.G.G., Ho A. COVID-19 pandemic-a focused review for clinicians. Clin. Microbiol. Infect. 2020;26:842–847. doi: 10.1016/j.cmi.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comunian S., Dongo D., Milani C., Palestini P. Air pollution and covid-19: the role of particulate matter in the spread and increase of covid-19's morbidity and mortality. Int. J. Environ. Res. Publ. Health. 2020;17 doi: 10.3390/ijerph17124487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticini E., Frediani B., Caro D. Can atmospheric pollution be considered a co-factor in extremely high level of SARS-CoV-2 lethality in Northern Italy? Environ. Pollut. 2020;114465 doi: 10.1016/j.envpol.2020.114465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copat C., Cristaldi A., Fiore M., Grasso A., Zuccarello P., Signorelli S.S., Conti G.O., Ferrante M. The role of air pollution (PM and NO2) in COVID-19 spread and lethality: a systematic review. Environ. Res. 2020;191:110129. doi: 10.1016/j.envres.2020.110129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett S.E., Nitzberg M., Moses E., Kleerup E., Wang T., Perdomo Catalina, Perdomo Claudia, Liu G., Xiao X., Liu H., Elashoff D.A., Brooks D.R., O'Connor G.T., Dubinett S.M., Spira A., Lenburg M.E. Gene expression alterations in the bronchial epithelium of e-cigarette users. Chest. 2019;156:764–773. doi: 10.1016/j.chest.2019.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa V.G., Moreli M.L., Saivish M.V. The emergence of SARS, MERS and novel SARS-2 coronaviruses in the 21st century. Arch. Virol. 2020;165:1517–1526. doi: 10.1007/s00705-020-04628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., McMorran R., Wiegers J., Wiegers T.C., Mattingly C.J. The comparative toxicogenomics database: update 2019. Nucleic Acids Res. 2019;47:D948–D954. doi: 10.1093/nar/gky868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Sole F., Farcomeni A., Loffredo L., Carnevale R., Menichelli D., Vicario T., Pignatelli P., Pastori D. Features of severe COVID-19: a systematic review and meta-analysis. Eur. J. Clin. Invest. 2020 doi: 10.1111/eci.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E., Lo R., Celius T., Matthews J., Zacharewski T.R. Integration of genome-wide computation DRE search, AhR ChIP-chip and gene expression analyses of TCDD-elicited responses in the mouse liver. BMC Genom. 2011;12:365. doi: 10.1186/1471-2164-12-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Nelson M., Grayson D.R., Costa E., Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13614–13619. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisa N.H., Jilani Y., Kainth K., Redd P., Lu S., Bougrine O., Abdul Sater H., Patwardhan C.A., Shull A., Shi H., Liu K., Elsherbiny N.M., Eissa L.A., El-Shishtawy M.M., Horuzsko A., Bollag R., Maihle N., Roig J., Korkaya H., Cowell J.K., Chadli A. The co-chaperone UNC45A is essential for the expression of mitotic kinase NEK7 and tumorigenesis. J. Biol. Chem. 2019;294:5246–5260. doi: 10.1074/jbc.RA118.006597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishilevich S., Nudel R., Rappaport N., Hadar R., Plaschkes I., Iny Stein T., Rosen N., Kohn A., Twik M., Safran M., Lancet D., Cohen D. Database; Oxford: 2017. GeneHancer: Genome-wide Integration of Enhancers and Target Genes in GeneCards. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo Marin B., Aghagoli G., Lavine K., Yang L., Siff E.J., Chiang S.S., Salazar-Mather T.P., Dumenco L., Savaria M.C., Aung S.N., Flanigan T., Michelow I.C. Predictors of COVID-19 severity: a literature review. Rev. Med. Virol. 2020 doi: 10.1002/rmv.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbman B.D., Heguy A., O'Connor T.P., Zabner J., Crystal R.G. Upregulation of pirin expression by chronic cigarette smoking is associated with bronchial epithelial cell apoptosis. Respir. Res. 2007;8:10. doi: 10.1186/1465-9921-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni F., Bosch I., Polonio C.M., Torti M.F., Wheeler M.A., Li Z., Romorini L., Rodriguez Varela M.S., Rothhammer V., Barroso A., Tjon E.C., Sanmarco L.M., Takenaka M.C., Modaresi S.M.S., Gutiérrez-Vázquez C., Zanluqui N.G., Dos Santos N.B., Munhoz C.D., Wang Z., Damonte E.B., Sherr D., Gehrke L., Peron J.P.S., Garcia C.C., Quintana F.J. AHR is a Zika virus host factor and a candidate target for antiviral therapy. Nat. Neurosci. 2020;23:939–951. doi: 10.1038/s41593-020-0664-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkogkou E., Barnasas G., Vougas K., Trougakos I.P. Expression profiling meta-analysis of ACE2 and TMPRSS2, the putative anti-inflammatory receptor and priming protease of SARS-CoV-2 in human cells, and identification of putative modulators. Redox Biol. 2020;36:101615. doi: 10.1016/j.redox.2020.101615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong K.W., Zhao W., Li N., Barajas B., Kleinman M., Sioutas C., Horvath S., Lusis A.J., Nel A., Araujo J.A. Air-pollutant chemicals and oxidized lipids exhibit genome-wide synergistic effects on endothelial cells. Genome Biol. 2007;8:R149. doi: 10.1186/gb-2007-8-7-r149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grilli A., Bengalli R., Longhin E., Capasso L., Proverbio M.C., Forcato M., Bicciato S., Gualtieri M., Battaglia C., Camatini M. Transcriptional profiling of human bronchial epithelial cell BEAS-2B exposed to diesel and biomass ultrafine particles. BMC Genom. 2018;19:302. doi: 10.1186/s12864-018-4679-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald M.E., Shaban M.G., Mackin S.R., Fehr A.R., Perlman S. Murine coronavirus infection activates the aryl hydrocarbon receptor in an indoleamine 2,3-dioxygenase-independent manner, contributing to cytokine modulation and proviral TCDD-inducible-PARP expression. J. Virol. 2020;94 doi: 10.1128/JVI.01743-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O., Staeheli P., Schwemmle M., Kochs G. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol. 2015;23:154–163. doi: 10.1016/j.tim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Harmon A.C., Hebert V.Y., Cormier S.A., Subramanian B., Reed J.R., Backes W.L., Dugas T.R. Particulate matter containing environmentally persistent free radicals induces AhR-dependent cytokine and reactive oxygen species production in human bronchial epithelial cells. PloS One. 2018;13 doi: 10.1371/journal.pone.0205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikmet F., Méar L., Edvinsson Å., Micke P., Uhlén M., Lindskog C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020;16 doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrubá E., Vondráček J., Líbalová H., Topinka J., Bryja V., Souček K., Machala M. Gene expression changes in human prostate carcinoma cells exposed to genotoxic and nongenotoxic aryl hydrocarbon receptor ligands. Toxicol. Lett. 2011;206:178–188. doi: 10.1016/j.toxlet.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Hübner R.-H., Schwartz J.D., De Bishnu P., Ferris B., Omberg L., Mezey J.G., Hackett N.R., Crystal R.G. Coordinate control of expression of Nrf2-modulated genes in the human small airway epithelium is highly responsive to cigarette smoking. Mol. Med. 2009;15:203–219. doi: 10.2119/molmed.2008.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa S., Matsumura K., Kitamura N., Takanami Y., Ito S. Multi-omics analysis: repeated exposure of a 3D bronchial tissue culture to whole-cigarette smoke. Toxicol. Vitro. 2019;54:251–262. doi: 10.1016/j.tiv.2018.10.001. [DOI] [PubMed] [Google Scholar]
- Jackson D.P., Joshi A.D., Elferink C.J. Ah receptor pathway intricacies; signaling through diverse protein partners and DNA-motifs. Toxicol Res (Camb) 2015;4:1143–1158. doi: 10.1039/c4tx00236a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., Millet J.K., Whittaker G.R. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. iScience. 2020;23:101212. doi: 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M., Sharma M., Henderson B.R. IQGAP1 regulation and roles in cancer. Cell. Signal. 2009;21:1471–1478. doi: 10.1016/j.cellsig.2009.02.023. [DOI] [PubMed] [Google Scholar]
- Jorgensen E., Stinson A., Shan L., Yang J., Gietl D., Albino A.P. Cigarette smoke induces endoplasmic reticulum stress and the unfolded protein response in normal and malignant human lung cells. BMC Canc. 2008;8:229. doi: 10.1186/1471-2407-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M., Rebensburg S.V., Takata M.A., Zang T.M., Yamashita M., Kvaratskhelia M., Bieniasz P.D. Nuclear pore heterogeneity influences HIV-1 infection and the antiviral activity of MX2. Elife. 2018;7 doi: 10.7554/eLife.35738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M., Yadav S.S., Bitzegeio J., Kutluay S.B., Zang T., Wilson S.J., Schoggins J.W., Rice C.M., Yamashita M., Hatziioannou T., Bieniasz P.D. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature. 2013;502:563–566. doi: 10.1038/nature12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopel J., Perisetti A., Roghani A., Aziz M., Gajendran M., Goyal H. Racial and gender-based differences in COVID-19. Front Public Health. 2020;8:418. doi: 10.3389/fpubh.2020.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer K., Frenzel F., Lampen A., Braeuning A., Böhmert L. Transcriptomic effect marker patterns of genotoxins - a comparative study with literature data. J. Appl. Toxicol. 2020;40:448–457. doi: 10.1002/jat.3928. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lee C.M., Barber G.P., Casper J., Clawson H., Diekhans M., Gonzalez J.N., Hinrichs A.S., Lee B.T., Nassar L.R., Powell C.C., Raney B.J., Rosenbloom K.R., Schmelter D., Speir M.L., Zweig A.S., Haussler D., Haeussler M., Kuhn R.M., Kent W.J. UCSC Genome Browser enters 20th year. Nucleic Acids Res. 2020;48:D756–D761. doi: 10.1093/nar/gkz1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J.M., Yang C.X., Tam A., Shaipanich T., Hackett T.-L., Singhera G.K., Dorscheid D.R., Sin D.D. ACE-2 expression in the small airway epithelia of smokers and COPD patients: implications for COVID-19. Eur. Respir. J. 2020;55 doi: 10.1183/13993003.00688-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.-Y., Li L., Zhang Y., Wang X.-S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty. 2020;9:45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi G., Henry B.M. Active smoking is not associated with severity of coronavirus disease 2019 (COVID-19) Eur. J. Intern. Med. 2020 doi: 10.1016/j.ejim.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Rehmani I., Esaki S., Fu R., Chen L., de Serrano V., Liu A. Pirin is an iron-dependent redox regulator of NF-κB. Proc. Natl. Acad. Sci. U.S.A. 2013;110:9722–9727. doi: 10.1073/pnas.1221743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizarraga D., Gaj S., Brauers K.J., Timmermans L., Kleinjans J.C., van Delft J.H.M. Benzo[a]pyrene-induced changes in microRNA-mRNA networks. Chem. Res. Toxicol. 2012;25:838–849. doi: 10.1021/tx2003799. [DOI] [PubMed] [Google Scholar]
- Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W., Hennig B.P., Kreuter M., Conrad C., Eils R. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. e105114. 2020 doi: 10.15252/embj.2020105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U.S.A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melén K., Keskinen P., Ronni T., Sareneva T., Lounatmaa K., Julkunen I. Human MxB protein, an interferon-alpha-inducible GTPase, contains a nuclear targeting signal and is localized in the heterochromatin region beneath the nuclear envelope. J. Biol. Chem. 1996;271:23478–23486. doi: 10.1074/jbc.271.38.23478. [DOI] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., Romero J.P., Wirnsberger G., Zhang H., Slutsky A.S., Conder R., Montserrat N., Mirazimi A., Penninger J.M. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray I.A., Patterson A.D., Perdew G.H. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat. Rev. Canc. 2014;14:801–814. doi: 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neavin D.R., Lee J.-H., Liu D., Ye Z., Li H., Wang L., Ordog T., Weinshilboum R.M. Single nucleotide polymorphisms at a distance from aryl hydrocarbon receptor (AHR) binding sites influence AHR ligand-dependent gene expression. Drug Metab. Dispos. 2019;47:983–994. doi: 10.1124/dmd.119.087312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberfeld B., Achanta A., Carpenter K., Chen P., Gilette N.M., Langat P., Said J.T., Schiff A.E., Zhou A.S., Barczak A.K., Pillai S. SnapShot: COVID-19. Cell. 2020;181:954. doi: 10.1016/j.cell.2020.04.013. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogen Y. Assessing nitrogen dioxide (NO2) levels as a contributing factor to coronavirus (COVID-19) fatality. Sci. Total Environ. 2020;726:138605. doi: 10.1016/j.scitotenv.2020.138605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olds J.L., Kabbani N. Is nicotine exposure linked to cardiopulmonary vulnerability to COVID-19 in the general population? FEBS J. 2020 doi: 10.1111/febs.15303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-Y., Byun E.J., Lee J.D., Kim S., Kim H.S. Air pollution, autophagy, and skin aging: impact of particulate matter (PM10) on human dermal fibroblasts. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19092727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovic J., Zürcher T., Haller O., Staeheli P. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J. Virol. 1990;64:3370–3375. doi: 10.1128/jvi.64.7.3370-3375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020;26:729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020 doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell B., Moss C., George G., Santaolalla A., Cope A., Papa S., Van Hemelrijck M. Associations between immune-suppressive and stimulating drugs and novel COVID-19-a systematic review of current evidence. Ecancermedicalscience. 2020;14:1022. doi: 10.3332/ecancer.2020.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvat A., Ciccosanti F., Colavita F., Di Rienzo M., Castilletti C., Capobianchi M.R., Kepp O., Zitvogel L., Fimia G.M., Piacentini M., Kroemer G. On-target versus off-target effects of drugs inhibiting the replication of SARS-CoV-2. Cell Death Dis. 2020;11:656. doi: 10.1038/s41419-020-02842-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Proc. Natl. Acad. Sci. U.S.A; 2020. Cell Entry Mechanisms of SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng L., Anderson P.H., Turner A.G., Pishas K.I., Dhatrak D.J., Gill P.G., Morris H.A., Callen D.F. Identification of vitamin D3 target genes in human breast cancer tissue. J. Steroid Biochem. Mol. Biol. 2016;164:90–97. doi: 10.1016/j.jsbmb.2015.10.012. [DOI] [PubMed] [Google Scholar]
- Sodroski J.G., Goh W.C., Haseltine W.A. Transforming potential of a human protooncogene (c-fps/fes) locus. Proc. Natl. Acad. Sci. U.S.A. 1984;81:3039–3043. doi: 10.1073/pnas.81.10.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spink B.C., Bennett J.A., Pentecost B.T., Lostritto N., Englert N.A., Benn G.K., Goodenough A.K., Turesky R.J., Spink D.C. Long-term estrogen exposure promotes carcinogen bioactivation, induces persistent changes in gene expression, and enhances the tumorigenicity of MCF-7 human breast cancer cells. Toxicol. Appl. Pharmacol. 2009;240:355–366. doi: 10.1016/j.taap.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spira A., Beane J., Shah V., Liu G., Schembri F., Yang X., Palma J., Brody J.S. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10143–10148. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talal N. Cyclosporine as an immunosuppressive agent for autoimmune disease: theoretical concepts and therapeutic strategies. Transplant. Proc. 1988;20:11–15. [PubMed] [Google Scholar]
- Touret F., Gilles M., Barral K., Nougairède A., van Helden J., Decroly E., de Lamballerie X., Coutard B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 2020;10:13093. doi: 10.1038/s41598-020-70143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., Ritter J.H., Kang L.-I., Dort S., Robichaud A., Head R., Holtzman M.J., Diamond M.S. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020 doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff D., Nee S., Hickey N.S., Marschollek M. Risk factors for Covid-19 severity and fatality: a structured literature review. Infection. 2020 doi: 10.1007/s15010-020-01509-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright W.R., Parzych K., Crawford D., Mein C., Mitchell J.A., Paul-Clark M.J. Inflammatory transcriptome profiling of human monocytes exposed acutely to cigarette smoke. PloS One. 2012;7 doi: 10.1371/journal.pone.0030120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.Y., Ahmed S., Satheesh S.V., Matthews J. Genome-wide mapping and analysis of aryl hydrocarbon receptor (AHR)- and aryl hydrocarbon receptor repressor (AHRR)-binding sites in human breast cancer cells. Arch. Toxicol. 2018;92:225–240. doi: 10.1007/s00204-017-2022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Pan J., Wang Weidong, Liu Z., Kan H., Qiu Y., Meng X., Wang Weibing. Association of particulate matter pollution and case fatality rate of COVID-19 in 49 Chinese cities. Sci. Total Environ. 2020;741:140396. doi: 10.1016/j.scitotenv.2020.140396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16:1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Pan T., Zhang Junsong, Li Q., Zhang X., Bai C., Huang F., Peng T., Zhang Jianhua, Liu C., Tao L., Zhang H. Glycopeptide antibiotics potently inhibit cathepsin L in the late endosome/lysosome and block the entry of ebola virus, Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV) J. Biol. Chem. 2016;291:9218–9232. doi: 10.1074/jbc.M116.716100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoran M.A., Savastru R.S., Savastru D.M., Tautan M.N. Assessing the relationship between surface levels of PM2.5 and PM10 particulate matter impact on COVID-19 in Milan. Italy. Sci. Total Environ. 2020;738:139825. doi: 10.1016/j.scitotenv.2020.139825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020 doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.