Abstract
TP53 wild-type breast tumors rarely undergo a complete pathological response after chemotherapy treatment. These patients have an extremely poor survival rate and studies show these tumors preferentially undergo senescence instead of apoptosis. These senescent cells persist after chemotherapy and secrete cytokines and chemokines comprising the senescence associated secretory phenotype, which promotes survival, proliferation, and metastasis. We hypothesized that eliminating senescent tumor cells would improve chemotherapy response and extend survival. Previous studies have shown “senolytic” agents selectively kill senescent normal cells, but their efficacy in killing chemotherapy-induced senescent cancer cells is unknown. We show that ABT-263, a BH3 mimetic that targets antiapoptotic proteins BCL2/BCL-XL/BCL-W, had no effect on proliferating cells, but rapidly and selectively induced apoptosis in a subset of chemotherapy-treated cancer cells, though sensitivity required days to develop. Low NOXA expression conferred resistance to ABT-263 in some cells, necessitating additional MCL1 inhibition. Gene editing confirmed breast cancer cells relied on BCL-XL or BCL-XL/MCL1 for survival in senescence. In a mouse model of breast cancer, ABT-263 treatment following chemotherapy led to apoptosis, greater tumor regression, and longer survival. Our results reveal cancer cells that have survived chemotherapy by entering senescence can be eliminated using BH3 mimetic drugs that target BCL-XL or BCL-XL/MCL1. These drugs could help minimize residual disease and extend survival in breast cancer patients that otherwise have a poor prognosis and are most in need of improved therapies.
Subject terms: Cancer models, Cell biology, Cancer genetics
Introduction
The tumor suppressor TP53 is mutated in half of all cancers [1]. When wild type, p53 will respond to cell stress, such as that caused by chemotherapy, by turning on transcription of genes involved in apoptosis, cell cycle arrest, and senescence [2, 3]. Thus, wild-type p53 activity is thought to be favorable for prognosis in cancer. TP53 is mutant in 30% of breast cancers [4], and accumulating evidence shows that for patients treated with chemotherapy, it is much more favorable to have a mutant TP53 tumor than wild type. TP53-mutant tumors are much more likely to undergo a pathological complete response (pCR) after neoadjuvant chemotherapy than TP53 wild-type tumors [5–10]. Our recent analysis showed patients with wild-type TP53 tumors have remarkably poor survival after chemotherapy [11]. For example, triple negative breast cancer (TNBC) patients that received chemotherapy had a median survival of ~45 months if their tumor was TP53 wild type, compared with patients harboring TP53-mutant tumors that had a median survival of ~260 months [11]. When modeled in mice, p53 wild-type mammary tumors underwent senescence instead of apoptosis in response to chemotherapy treatment, resulting in early relapse and poor survival [12]. In contrast, p53 mutant tumors underwent mitotic catastrophe and apoptosis resulting in greater tumor regression and longer time to relapse [12]. TP53 wild type, chemotherapy-treated human breast cancers also undergo senescence [13].
Because senescent cells are ostensibly permanently arrested, it is generally thought they would pose no threat to contribute to relapse. It has been thoroughly demonstrated, however, that many tumor cells made senescent by chemotherapy treatment produce cytokines and chemokines of the senescence associated secretory phenotype (SASP) in their dormant state [12, 14–19]. These factors promote tumorigenic properties such as proliferation, survival, angiogenesis [20–24], an increase in the cancer stem cell population [16, 17, 25, 26], and suppression of the immune system [27, 28]. Because patients with TP53 wild-type breast tumors respond so poorly to chemotherapy, eliminating the senescent cells that persist and promote relapse is critical for improving patient survival. Notably, ~70% of all breast cancers are TP53 wild type, and every molecular subtype contains tumors with wild-type TP53, including TNBC, for which there is an unmet need for better treatment options [4, 29, 30].
Normal, diploid, nontransformed, senescent cells accumulate in aged organisms as a tumor suppressive mechanism [31–35] and contribute to inflammation and aging phenotypes [28, 36]. Thus, the discovery of senolytic agents, or drugs that selectively kill senescent cells, has been a priority [37]. Agents identified include those that target the rapid metabolism of senescent cells [38] and several proapoptotic BH3 mimetic drugs, originally developed as cancer therapies [39]. These drugs could eliminate senescent, normal diploid fibroblasts in culture, or senescent cells in aged mice [37, 40–42]. They include the BCL2 inhibitor ABT-199 (venetoclax), which is FDA approved for treatment of chronic lymphocytic leukemia [43], and ABT-263 (navitoclax), which inhibits BCL2, BCL-XL, and BCL-W and has activity in hematopoietic cancers [43]. Lamentably, these drugs have relatively minimal activity in solid tumors [44], including minimal or mixed responses as single agents in human breast cancer cell lines and xenografts [45–48].
Because residual disease that persists following chemotherapy treatment is composed of tumor cells that have entered senescence [12, 13, 19], we tested if drugs that kill senescent normal cells could kill these persisting cancer cells that cause relapse and shorten survival.
Here, we show that ABT-263 induced extensive, rapid apoptosis in a subset of breast cancer cells made senescent by doxorubicin treatment, while some cell lines with low NOXA levels required a second drug targeting MCL1. In a p53 wild-type mouse mammary tumor model that undergoes senescence after chemotherapy treatment [12, 19], administration of ABT-263 following chemotherapy resulted in apoptosis and significantly greater tumor regression, resulting in longer time to relapse. ABT-263 could be used to eliminate the cancer cells that persist in a senescent state, reduce residual disease, and improve survival in patients with TP53 wild-type tumors who otherwise respond poorly to chemotherapy and have a dismal prognosis.
Results
ABT-263 induces apoptosis specifically in chemotherapy-treated cells
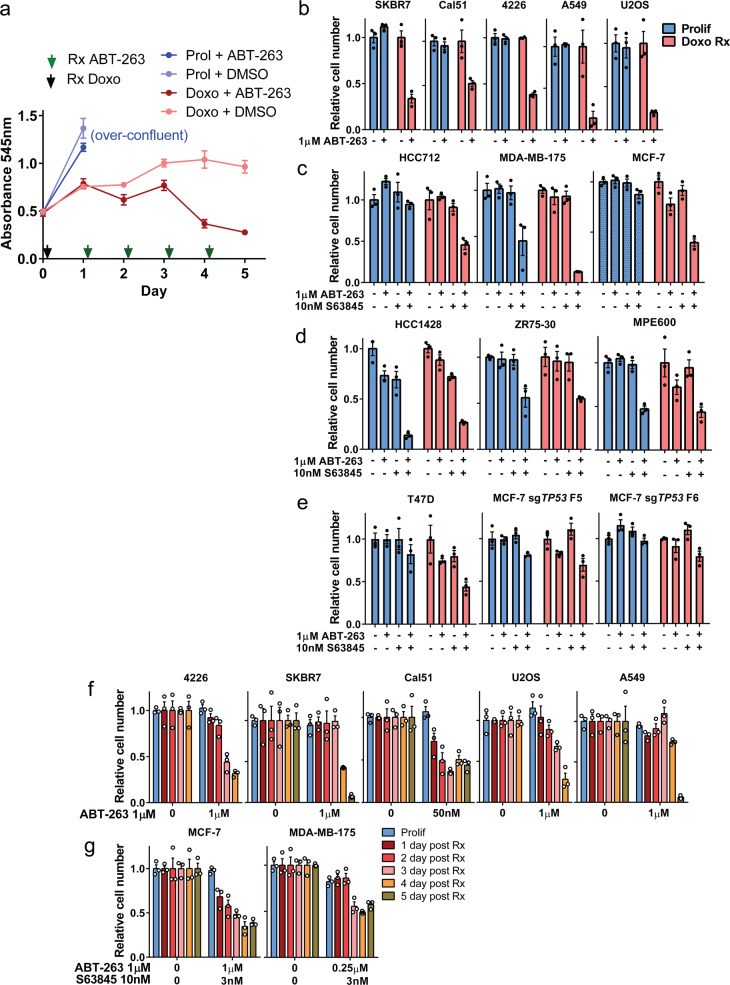
We examined the effects of a BH3 mimetic drug that has senolytic activity in normal cells [41, 42] on cancer cells made senescent by chemotherapy. Cell lines treated with doxorubicin (0.25 or 1 μM) adopted a senescent morphology and stained positive for senescence associated beta galactosidase (SAβGal) activity 7 days later (Supplementary Fig. 1) consistent with previous findings [12, 49, 50]. We next determined sensitivity to ABT-263 in the p53 wild-type mammary tumor cell line 4226 (created from an MMTV-Wnt1 tumor [50]) that was untreated and proliferating, or treated with doxorubicin for 1–4 days. Cells not treated with doxorubicin increased in cell number as they proliferated, before becoming overconfluent and lifting off. Treatment with ABT-263 had no effect on proliferating cells in this or subsequent experiments (Fig. 1a). Cell number increased only slightly 1 day after doxorubicin treatment, and remained relatively steady over the next 4 days, consistent with induction of cell cycle arrest and senescence over time. Addition of ABT-263 to cells 1 or 2 days following doxorubicin treatment had little effect on cell number measured 24 h later (Fig. 1a). Addition of a single dose of ABT-263 to cells 3 or more days after doxorubicin treatment resulted in significant reduction in cell number, with maximal effect observed at 5 days (Fig. 1a), a time point when cells express multiple senescent markers [12, 50, 51]. We next examined ABT-263 sensitivity in more TP53 wild-type cell lines. Because of the predictable cytostatic effects on cell number, doxorubicin-treated cells given DMSO were normalized to 1.0 and only compared with doxorubicin-treated cells given ABT-263, and likewise for proliferating cells. Nonnormalized data are shown for each proliferation assay in Supplementary Figs. 2–3.
Fig. 1. ABT-263 treatment alone or in combination with S63845 can selectively eliminate chemotherapy-induced senescent cells.
a 4226 mammary tumor cell line was plated in 24 well plates in parallel, and 1 day later treated with 0.75 μM doxorubicin (Doxo, indicated by black arrow) or left untreated to continue proliferating (Prolif). For the following 4 days, one plate was treated with a single dose of 1 μM ABT-263 (as indicated by green arrows), and then cell number determined by MTT assay 24 h later. Absorbance was plotted for triplicate wells of each treatment group over time. Untreated cells grew to conflunce 2 days after plating and then lifted off the plate (noted as “over-confluent”). Error bars represent SEM. Indicated p53 wild type (b–d) or p53 mutant/edited (e) cancer cell lines were plated and the next day treated with doxorubicin (0.25–1 μM) for 24 h (Doxo Rx cells). Seven days after treatment, in parallel with untreated, proliferating cells (Prolif), ABT-263 (1 μM), S63845 (10 nM), or both were added as indicated and then MTT assay was performed 24 h later to determine cell number. f–g Indicated cell lines were plated and doxorubicin treated as in (b–e) and cell number determined by MTT assay after ABT-263 or ABT-263 + S63845 treatment each day following doxorubicin treatment. Bar graphs for (b–g) show mean ± SEM with individual data points plotted. Nonnormalized data for (b–g) are shown in Supplementary Fig. 2b–g. Statistics (complete analysis available in Supplementary Table 1) used were (b) Student’s t test comparing two groups: control vs. ABT-263 treated, for Prolif and Doxo Rx separately. (c–e) ANOVA with Tukey’s posttest comparing four groups: control, ABT-263, S63845, ABT-263 + S63845, for Prolif and Doxo Rx separately. f–g Student’s t test comparing two groups: control vs. ABT-263 or ABT-263/S63845 treated, for each day post Doxo Rx separately. In (b–g), the DMSO group (indicated by “−” or “0” signifying no ABT-263) was normalized to 1.0 for Prolif and 1.0 for each treatment group. Each treatment group was in triplicate and individual data points are shown. Error bars indicate SEM.
ABT-263 preferentially reduced cell number in senescent, TP53 wild-type breast cancer cell lines SKBR7, Cal51, 4226, and two other cell lines shown to undergo chemotherapy-induced senescence: U2OS (osteosarcoma) [52], and A549 (lung adenocarcinoma) [53]. The number of proliferating cells was unchanged by ABT-263 treatment (Fig. 1b). Interestingly, other TP53 wild-type breast cancer lines made senescent with doxorubicin were insensitive to ABT-263. Since MCL1 is a common predictor of resistance to ABT-263 [45, 54], we treated these cells with S63845, an MCL1 inhibitor [54]. While ABT-263 or S63845 treatment alone had little effect, treatment with both drugs resulted in a reduction in the number of doxorubicin-induced senescent HCC712, MDA-MB-175, and MCF-7 cells 24 h later (Fig. 1c). While some sensitivity was observed for proliferating MDA-MB-175, senescent cells were more sensitive (Fig. 1c). Interestingly, doxorubicin-induced senescent HCC1428, ZR75-30, and MPE-600 cells were either resistant to the treatments or both senescent and proliferating cells were similarly sensitive (Fig. 1d). These results suggest doxorubicin-induced senescent breast cancer cells can be grouped into different classes: ABT-263 sensitive (A-sensitive), ABT-263 plus S63845 sensitive (A/S sensitive), and insensitive or not preferentially sensitive to either or both treatments (insensitive). We next tested senolytic sensitivity in TP53-mutant cell lines that can undergo senescence in culture (Supplementary Fig. 1) [50, 55]. T47D and two MCF-7 clones with CRISPR-Cas9 edited TP53 showed some sensitivity to ABT-263/S63845 when made senescent by doxorubicin (Fig. 1e). Ruling out the possibility that senolytic drugs might interfere with the assay used to determine cell number (MTT), live-cell imaging analysis of cells expressing GFP or mCherry showed consistent results: greater sensitivity in doxorubicin-treated 4226 and MCF-7 (Supplementary Fig. 2a).
Progression into the senescence program is necessary for ABT-263 sensitivity
Following doxorubicin treatment, p53 wild-type cells incur DNA strand breaks, arrest the cell cycle, and rapidly activate p53 target genes such as CDKN1A, BAX, BBC3, and PMAIP1 [3], but take days to acquire senescent phenotypes such as SAβGal staining and morphological changes [2, 49]. We treated cells with ABT-263 at different time points post-doxorubicin to determine the timing of sensitivity. Cell number was not reduced in A-sensitive cell lines 4226 and U2OS until ~3 days following doxorubicin treatment, while SKBR7 and A549 cells required 4 and 5 days (Fig. 1f). Cal51 showed some sensitivity to ABT-263 1 day after doxorubicin treatment, which increased over the next 2 days. A/S-sensitive cell lines MCF-7 and MDA-MB-175 also became maximally sensitive to ABT-263/S63845 treatment 3–4 days after doxorubicin treatment (Fig. 1g). These results suggest ABT-263 does not target merely arrested cells or cells with DNA damage. For the majority of cell lines, 3–5 days of progression into the senescence program was required for maximal senolytic sensitivity.
Other senescence-inducing treatments confer ABT-263 sensitivity
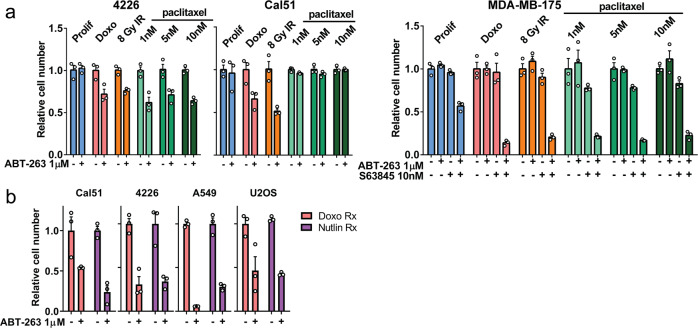
It is possible that sensitivity to ABT-263 occurs over days, specifically as a result of doxorubicin treatment, rather than induction of senescence per se, thus, we tested if other commonly used breast cancer treatments could render cancer cells sensitive to senolytic drugs. We found that A-sensitive and A/S-sensitive cell lines made senescent by exposure to IR (causes DNA strand breaks) or paclitaxel (a mitotic spindle poison) were sensitive to senolytic drugs. One exception, Cal51 exhibited primary resistance to paclitaxel and thus was not induced to become sensitive to ABT-263 (Fig. 2a). We next determined if cells could be made sensitive to senolytic drugs without DNA damage or mitotic stress. We treated cells with Nutlin-3a, a drug that potently activates p53 ectopically, in the absence of stress, by blocking MDM2–p53 interactions [56]. Nutlin induces characteristic senescent phenotypes [50, 57], and also conferred sensitivity to senolytic drugs after 6–7 days of treatment, in all four of the cell lines tested, to an extent equal to doxorubicin (Fig. 2b).
Fig. 2. ABT-263 treatment alone or in combination with S63845 can selectively eliminate cells treated with senescence-inducing therapies.
a Indicated cell lines were plated, and then exposed to 8 Gy gamma irradiation, 1, 5, or 10 nM paclitaxel, or doxorubicin, and media changed 24 h later. Five days later, cells were treated with indicated senolytic drug, and MTT assay was performed 24 h later to determine cell number. b Indicated cell lines were plated, and then treated with Nutlin-3a every other day (2.5–15 μM; total of four doses). One day after fourth dose of Nutlin-3a, cells were treated with ABT-263 (1 μM), and MTT assay was performed 24 h later to determine cell number. Each treatment group was in triplicate and individual data points are shown. Error bars indicate SEM. Nonnormalized data are shown in Supplementary Fig. 3a–b. Statistics (complete analysis available in Supplementary Table 2) used were (a, 4226 and Cal51) Student’s t test comparing two groups: control vs. ABT-263 treated, for Prolif, Doxo, IR, 1 nM, 5 nM, 10 nM paclitaxel Rx separately. (a, MDA-MB-175) ANOVA with Tukey’s posttest comparing four groups: control, ABT-263, S63845, ABT-263 + S63845, for Prolif and the indicated treatment groups separately. b Student’s t test comparing two groups: Doxo + DMSO vs. Doxo + ABT-263 treated and Nutlin-3a + DMSO vs. Nutlin-3a + ABT-263.
ABT-263 induces apoptosis in doxorubicin-induced senescent cells
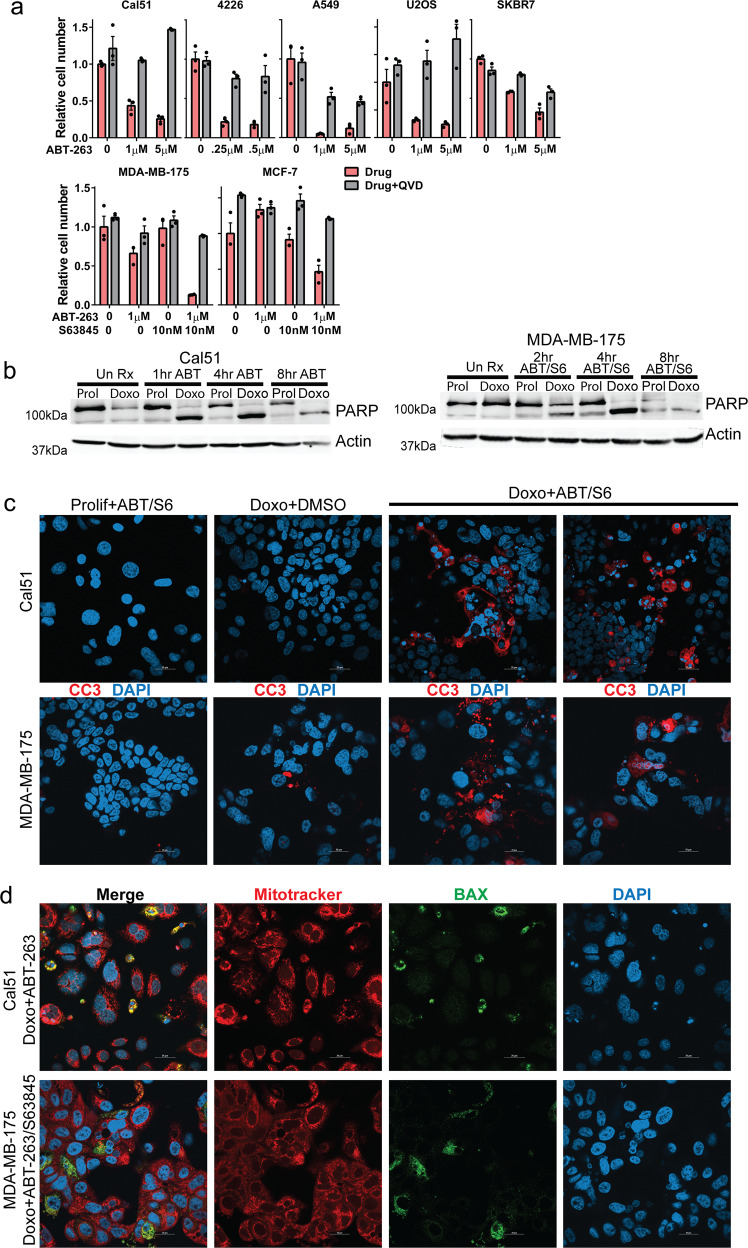
To determine the cause of reduced cell number in ABT-263 treated cells (Fig. 1), doxorubicin-induced senescent cells were treated with ABT-263 in the presence or absence of Q-VD-OPh, a pan-caspase inhibitor. Addition of Q-VD-OPh rescued the reduction in cell number caused by ABT-263 treatment, suggesting that ABT-263 induced apoptosis in A-sensitive cells (Fig. 3a, top). Q-VD-OPh also rescued A/S-sensitive MCF-7 and MDA-MB-175 senescent cells from death induced by combined treatment with ABT-263 and S63845 (Fig. 3a, lower). Treatment of doxorubicin-induced senescent Cal51 and MDA-MB-175 cells with the appropriate senolytic drugs (ABT-263 and ABT-263/S63845, respectively), resulted in the complete conversion of PARP to its cleaved form within 2–4 h (Fig. 3b). No PARP cleavage was observed in doxorubicin-induced senescent cells that were not treated with ABT-263, suggesting these senescent cells are resistant to apoptosis, consistent with results from other studies [58]. Senolytic drugs did not induce PARP cleavage in the untreated, proliferating Cal51 cells and had only a minimal effect on proliferating MDA-MB-175, consistent with no change or little change in cell number of proliferating cells treated with senolytic (Fig. 1) and previous studies showing minimal activity of these drugs in solid tumor cell lines [44–48]. Treatment with appropriate senolytic drugs induced strong activation of caspase 3, as detected by immunofluorescence staining (Fig. 3c) and immunoblot (Supplementary Fig. 4a), but only in senescent cells. We also found the apoptotic mediator BAX was localized to mitochondria of senolytic treated senescent cells, but not control cells (Fig. 3d, Supplementary Fig. 5). Time course microscopy confirmed doxorubicin-induced senescent 4226 and MDA-MB-175 cells underwent rapid and near-synchronous cell death when treated with ABT-263 or ABT-263/S63845. Pan-caspase inhibitor Q-VD-OPh rescued the cell death (Supplementary Fig. 4b).
Fig. 3. ABT-263 alone or in combination with S63845 induces apoptosis in doxorubicin-induced senescent cells.
a Indicated cell lines were plated, treated with doxorubicin for 24 h, and then 7 days later treated with DMSO, ABT-263, S63845, or ABT-263 + S63845 with or without the pan-caspase inhibitor Q-VD-OPh (20 μM) as indicated in the figure. MTT assay was performed 24 h after drug treatments. b Parallel plates of proliferating (Prolif) and doxorubicin-induced senescent (Doxo) Cal51 and MDA-MB-175 cells were treated with ABT-263 (Cal51) or ABT-263 + S63845 (MDA-MB-175) as indicated and harvested 1, 2, 4, and 8 h after treatment as indicated. Immunoblots were performed for PARP and actin. c Indicated cell lines were plated on glass coverslips and treated as indicated, followed by IF staining for cleaved-caspase 3 and DAPI. Shown are representative images captured by confocal microscopy. Three fields of view for each treatment group were quantified for cleaved-caspase 3 staining, and ANOVA with Tukey’s posttest was performed (Cal51: Prolif + ABT vs. Doxo + ABT p < 0.0001; Doxo + DMSO vs. Doxo + ABT p < 0.0001; MDA-MB-175: Prolif + ABT/S6 vs. Doxo + ABT/S6 p = 0.0003; Doxo + DMSO vs. Doxo + ABT/S6 p = 0.0003). d Indicated cell lines were plated on glass coverslips and treated as indicated, followed by staining for MitoTracker (red), DAPI (blue), and IF staining for BAX (green). Shown are representative images captured by confocal microscopy. Three fields of view for each treatment group were quantified for BAX and MitoTracker costaining, and ANOVA with Tukey’s posttest was performed (Cal51: Prolif + ABT vs. Doxo + ABT p = 0.0359; Doxo+DMSO vs. Doxo + ABT p = 0.0052; MDA-MB-175: Prolif + ABT/S6 vs. Doxo + ABT/S6 p = 0.0004; Doxo + DMSO vs. Doxo + ABT/S6 p = 0.0006). Statistics (complete analysis available in Supplementary Table 3) used for (a) were ANOVA with Tukey’s posttest for all comparisons within a cell line. Control minus Q-VD-OPh (indicated by “0” representing DMSO treatment) was normalized to 1.0. Each treatment group was in triplicate and individual data points are shown. Error bars indicate SEM.
Senescent cells are preferentially targeted by senolytic drugs
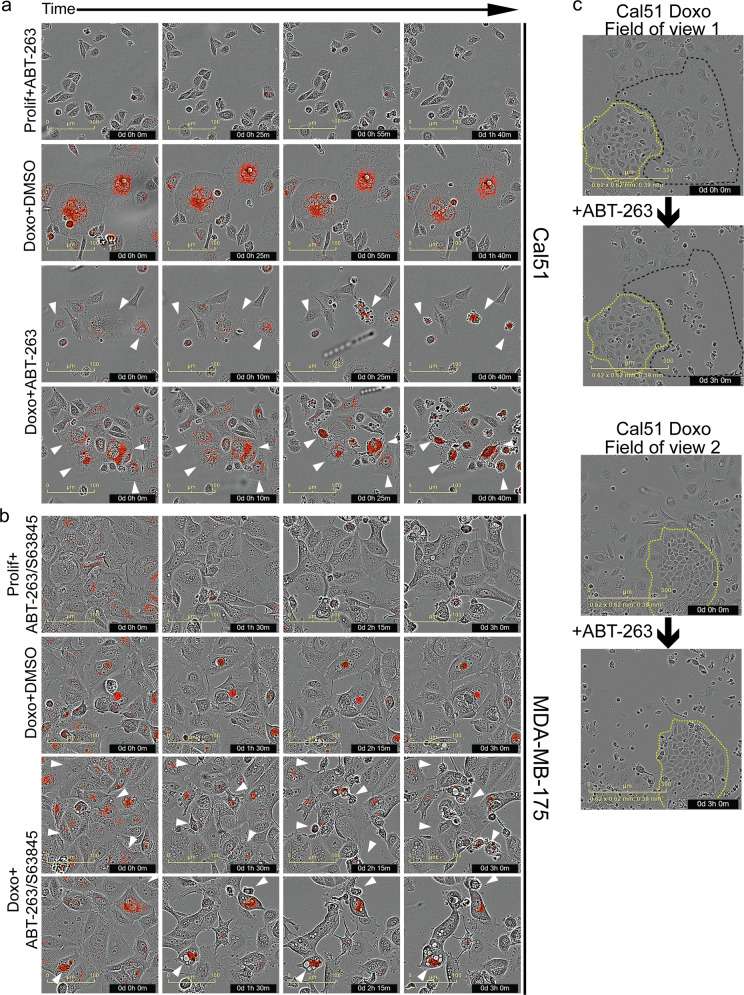
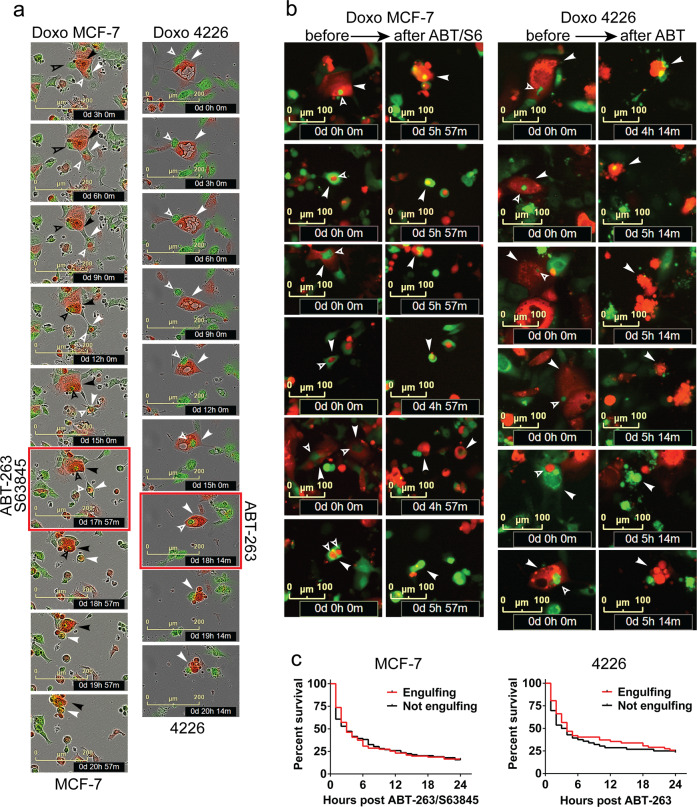
Senescent cells have an expanded lysosome compartment that is detected by SAβGal [59–61] and intense LysoTracker dye staining [50]. We further verified this correlation by performing sequential staining of the same field of view in three different cell lines. Doxorubicin-treated cells were highly positive for LysoTracker and SAβGal, while untreated, proliferating cells generally stained weakly for both (Supplementary Fig. 6). Using LysoTracker as a real time, live-cell marker of senescence, time course imaging showed that doxorubicin-treated A/S- or A-sensitive cell lines that were positive for LysoTracker underwent cell death within hours after treatment with senolytic drugs. Proliferating cells that were LysoTracker negative/low, or senescent cells that were LysoTracker positive and treated with DMSO, did not die (Fig. 4a, b). To test if doxorubicin primed cells for senolytic sensitivity independent of the senescent phenotype, we took advantage of Cal51, a cell line we observed presenting outgrowths of colonies ~1 week after doxorubicin treatment. Time-lapse imaging of doxorubicin-treated Cal51 showed cells with senescent morphology were highly sensitive to ABT-263, but the proliferating colonies in the same field of view were entirely resistant (Fig. 4c). While these data do not strictly prove that senolytic sensitivity is an integral part of senescence, taken together, our findings suggest it is the tumor cells that survive treatment by entering senescence that are sensitive to ABT-263/S63845, and this sensitivity is not specific to doxorubicin, DNA damage, or even cell stress.
Fig. 4. Doxorubicin-treated cells with senescent phenotypes are sensitive to senolytic drugs.
Cal51 (a) or MDA-MB-175 (b) cells were plated and the next day treated with doxorubicin (0.25 μM) for 24 h (Doxo Rx cells) or left untreated and proliferating (Prolif). Seven days after treatment, cells were stained with LysoTracker (75 nM), treated with DMSO or indicated senolytic drug(s), and then placed on an IncuCyte and imaged over 4 h. Shown are representative time-lapse images for the different treatment groups. Arrowheads indicate LysoTracker positive cells that undergo cell death in the time lapse. Scale bar represents 100 μM. c Cal51 cells were plated and the next day treated with doxorubicin (0.25 μM) for 24 h. Cells were then placed on IncuCyte for live-cell imaging. Six days later, cells were treated with ABT-263 (1 μM), cell death of proliferating and senescent cells was determined visually via live-cell imaging and two representive fields of view are shown, pre and 3 h post addition of ABT-263. Scale bar represents 300 μM.
Chemotherapy-induced senescent cells cannibalize neighboring cells to prolong their survival [50]. We next determined if senescent cells that cannibalize other cells are resistant to treatment with senolytic drugs. A/S-sensitive MCF-7 cells and A-sensitive 4226 cells both derive a survival benefit from frequent engulfments [50]. For the indicated cell lines, we mixed GFP and mCherry expressing cells, treated with doxorubicin to induce senescence and the engulfment phenotype, and then treated with appropriate senolytic drugs. Time-lapse microscopy showed active engulfment by both cell lines (Fig. 5a), followed by rapid cell death after exposure to senolytic drugs (Fig. 5a, b). Senolytic drug treatment was equally effective at killing cells whether they had engulfed another cell or not (Fig. 5c).
Fig. 5. Cell cannibalism phenotype does not protect doxorubicin treated, senescent cells from senolytic drugs.
a GFP and mCherry expressing MCF-7 or 4226 cells were plated together, and the next day treated with doxorubicin (0.25 μM for MCF-7; 0.75 μM for 4226) for 24 h (Doxo). On day 3, cells were placed on an IncuCyte and imaged every 3 h. On day 7, frequent cell engulfment was observed, and cells were treated with indicated senolytic at the image time point with the red outline. Cells were continually imaged every hour. Time-lapse images show representative predator cells (solid arrowheads) engulfing prey cells (open arrowheads), and subsequent cell death induced by senolytic treatment. b Representative images of doxorubicin-treated senescent, engulfing MCF-7 or 4226 cells captured before (left) and after (right) senolytic treatment. Solid arrowheads indicate engulfing predator cells, and open arrowheads indicate engulfed prey cells. Time stamps are noted in the image. Scale represents 100 μM. c Single cells from the MCF-7 and 4226 imaged in (b) were followed in time to determine if they had engulfed or not, and then were scored for the time point when they adopted characteristic features of apoptosis and cell death following senolytic treatment. Survival data from two independent experiments were plotted on a Kaplan–Meier curve with >30 cells in each group in each experiment for both cell lines. Differences in survival for both cell lines were not significant by Log-rank (Mantel–Cox) test.
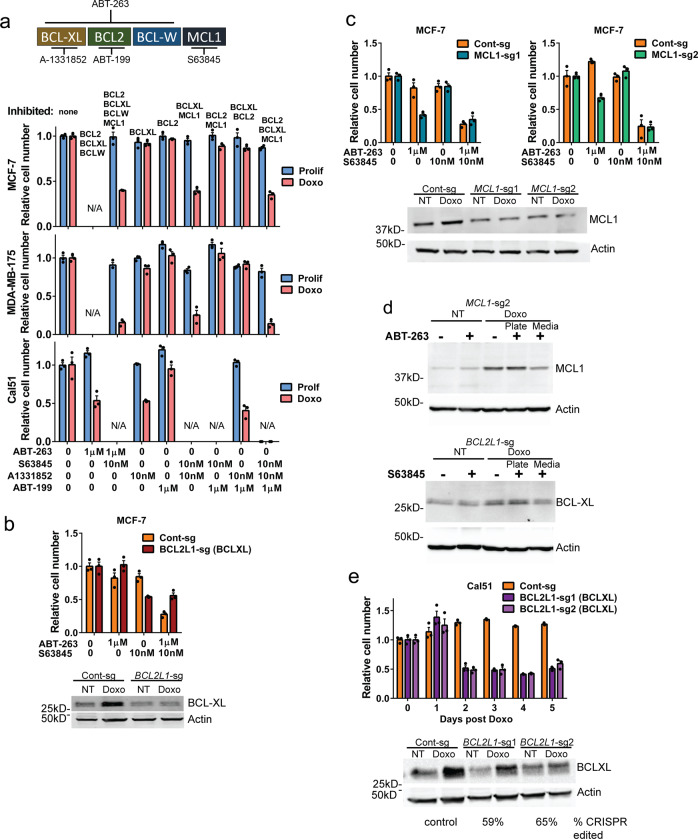
BCL-XL and MCL1 mediate survival of doxorubicin-induced senescent cells
Because ABT-263 targets BCL-XL, BCL2, and BCL-W, we investigated which specific BCL2 family member was responsible for survival of chemotherapy-treated cells in senescence. For A/S-sensitive cell lines, we used combinations of ABT-199, a BCL2 inhibitor, A-1331852, a BCL-XL inhibitor, and S63845, an MCL1 inhibitor (Fig. 6a), on doxorubicin-induced senescent MCF-7 and MDA-MB-175 cells. Cell lines sensitive to ABT-263 plus MCL1 inhibition were also sensitive to BCL-XL/MCL1 dual inhibition (Fig. 6a, upper and middle), suggesting activity of either BCL-XL or MCL1 is sufficient for survival of A/S-sensitive cells. ABT-263 sensitive senescent breast cancer cell line Cal51 died when treated with any drug combination that targeted BCL-XL, but did not respond to BCL2 inhibitor ABT-199, suggesting that BCL-XL is required for survival of senescent Cal51 (Fig. 6a, lower). To verify the role of BCL-XL and MCL1 in survival of senescent MCF-7 A/S-sensitive cells, we infected with CRISPR/Cas9 lentivirus and guide RNAs targeting either BCL2L1 (BCL-XL) or MCL1, and hypothesized edited cells would die when treated with only the MCL1 inhibitor S63845 or ABT-263, respectively. This hypothesis was supported as the number of senescent MCF-7 BCL2L1-sg cells was significantly reduced with S63845 alone (Fig. 6b), and senescent MCF-7 MCL1-sg cell number was reduced by treatment with ABT-263 alone (Fig. 6c). MCF-7 Control CRISPR cells (Cont-sg) required combined ABT-263 and S63845 treatment, consistent with the parental cells (Fig. 1). Immunoblotting showed reduction, but not complete loss, of BCL-XL and MCL1 proteins, consistent with a heterogeneous population of CRISPR edited cells that includes in-frame indels [62]. Further analysis by immunoblot showed that in the heterogeneous population of MCF-7 MCL1-sg edited cell line, cells that underwent apoptosis and lifted off the plate were more likely to be edited, as they expressed lower levels of MCL1 protein compared with cells that survived ABT-263 treatment (Fig. 6d, upper). Likewise, cells from the BC2L1-sg population that survived S63845 treatment had higher levels of BCL-XL compared with those that underwent apoptosis and lifted off the plate (Fig. 6d, lower). We similarly created a Cal51 BCL2L1 edited cell line, treated with doxorubicin, and determined cell number each day after treatment. We hypothesized that these cells would die over the days following doxorubicin treatment as senescence was induced. Indeed, 2 days after doxorubicin treatment, Cal51 BCL2L1-sg cell number was reduced (Fig. 6e). These results confirm that survival of senescent, chemotherapy-treated breast cancer cells is mediated in A-sensitive cells by BCL-XL, and in A/S-sensitive cells by BCL-XL or MCL1.
Fig. 6. Cells made senescent by doxorubicin are dependent on BCL-XL alone or can rely on either BCL-XL or MCL1 for survival.
a Schematic of BH3 mimetic drugs and targets. Indicated cell lines were plated, treated with doxorubicin for 24 h, and then 7 days later treated with various combinations of drugs as indicated below the graphs targeting BCL2 family members indicated above the graphs. MCF-7 and MDA-MB-175 were not treated with ABT-263 alone since they were shown to be resistant (Fig. 1b) and Cal51 was not treated with S63845 because they are sensitive to ABT-263 alone. b CRISPR guides targeting the functional domains of BCL2L1 (gene for BCL-XL) were cloned into a pLenti-CRISPR-mCherry vector and viral particles used to transduce MCF-7 cells. FACS sorted cells were plated and doxorubicin treated for 24 h, and then 7 days later ABT-263, S63845, or both ABT-263 + S63845 were applied as indicated and cell number determined by MTT assay 24 h later. Immunoblot for BCL-XL is shown below graph. c MCF-7 cells were transduced with pLenti-CRISPR-MCL1 vector and sorted as in (b). Indicated cells were plated and doxorubicin treated for 24 h, and then 7 days later ABT-263, S63845, or both ABT-263 + S63845 were applied as indicated and cell number determined by MTT assay 24 h later. Immunoblot for MCL1 is shown below graph. d MCF-7 MCL1-sg and BCL2L1-sg cells were plated and the next day doxorubicin treated for 24 h, and then 7 days later, ABT-263 or S63845 were applied as indicated. Media was removed from cells 24 h later and centrifuged to pellet nonadherent cells. Cells that remained adherent following treatment and nonadherent cells were lysed and immunoblots for MCL1 or BCL-XL and actin were performed as indicated. e Cal51 cells were transduced with pLenti-CRISPR-BCL2L1 vector and sorted as in (b). After plating and doxorubicin treatment, MTT assay was performed each day following for 5 days. Immunoblot for BCL-XL is shown below graph. Statistics (complete analysis available in Supplementary Table 4) used were as follows: (a) ANOVA with Tukey’s posttest comparing all treatment groups separately for Prolif and Doxo Rx. Control (indicated by “0” representing DMSO treatment) for Prolif and Doxo Rx were each normalized to 1. b–c ANOVA with Tukey’s posttest comparing all treatment groups separately for the Cont-sg and BCL2L1-sg cell lines. Control (indicated by “0” representing DMSO treatment) for each cell line was normalized to 1. e ANOVA with Tukey’s posttest comparing all days post doxorubicin treatment for the Cont-sg and BCL2L1-sg cell lines separately. Zero (“0”) days post doxorubicin for each cell line was normalized to 1. Each treatment group was in triplicate and individual data points are shown. Error bars indicate SEM. Nonnormalized data are shown in Supplementary Fig. 3c–f.
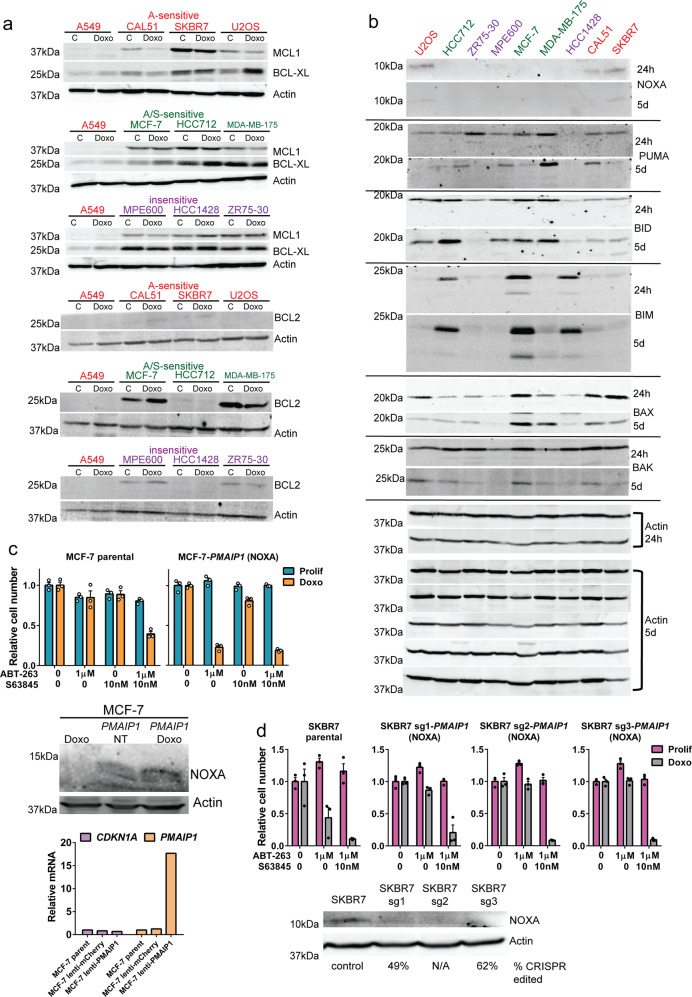
Expression of NOXA confers sensitivity to ABT-263 alone in doxorubicin-induced senescent cells
To address why cell lines might differ in sensitivity to BH3 mimetic drugs, we examined expression of pro- and antiapoptotic proteins. Surprisingly, some doxorubicin-induced senescent cells, such as SKBR7, that were sensitive to ABT-263 alone (not requiring inhibition of MCL1) expressed relatively high levels of MCL1. MCL1 levels have been shown to decline in some tumor cells after doxorubicin treatment [63], suggesting a mechanism to preclude use of an MCL1 inhibitor, but we observed MCL1 reductions in both A- and A/S-sensitive cell lines (Fig. 7a). Protein levels of PUMA, BID, BIM, BAK, and BAX did not show a clear trend among groups of A-sensitive, A/S-sensitive, and insensitive cell lines. However, doxorubicin-induced senescent A-sensitive cells expressed NOXA at both 24 h and 5 days following treatment, while A/S-sensitive and insensitive cells did not (Fig. 7b). Since NOXA is a BH3 mimetic that inhibits MCL1 [64, 65] to induce apoptosis [66, 67], we surmised that A/S-sensitive cells require S63845 in addition to ABT-263 because they do not express NOXA. To test this, we overexpressed PMAIP1, the gene for NOXA, in MCF-7 cells. When these cells were made senescent with doxorubicin, ABT-263 alone was sufficient to kill them (Fig. 7c). MCF-7 parental cells required ABT-263 and S63845 treatment to induce cell death (Fig. 7c), consistent with Fig. 1. Immunoblotting and qPCR of mRNA confirmed overexpression of NOXA in infected MCF-7 cells (Fig. 7c). Over expression of another proapoptotic BH3 only gene, BID, did not confer sensitivity to ABT-263 alone in MCF-7 cells, although this could be due to lack of activation by cleavage (Supplementary Fig. 7). To test if an A-sensitive cell line could be converted to A/S-sensitive, we generated cell lines infected with a CRISPR/Cas9 lentivirus and guide RNA targeting PMAIP1 and found that senescent SKBR7 PMAIP1-sg cells no longer died after ABT-263 alone, but now required additional treatment with S63845 to inhibit MCL1 (Fig. 7d). Senescent parental SKBR7 died from ABT-263 treatment alone (Fig. 7d), consistent with previous experiments (Fig. 1). Our results show that NOXA, by inhibiting MCL1, confers sensitivity to ABT-263 in doxorubicin-induced senescent cells, and that editing of PMAIP1 (NOXA) was sufficient to convert A-sensitive cells to A/S-sensitive.
Fig. 7. NOXA expression confers sensitivity to ABT-263.
a Indicated cell lines were untreated and harvested or treated with doxorubicin for 24 h and harvested 5 days later. Immunoblots for Actin, MCL1, BCL-XL, and BCL2 were performed. All blots were done in parallel, and A549 lysate is included in each gel. Red font denotes A-sensitive cell lines, green font denotes A/S-sensitive cell lines, and purple font denotes insensitive cell lines, as determined in Fig. 1. b Indicated cell lines were treated with doxorubicin for 24 h and harvested. Immunoblots for indicated proteins were performed. Actin blots at bottom are for the different membranes used to perform all of the 24 h and 5 day treated blots as indicated. c Lentivirus expressing PMAIP1 was used to transduce MCF-7 and cells were selected by FACS. MCF-7 parental and NOXA overexpressing cells were plated and treated with doxorubicin for 24 h, and then either ABT-263, S63845 or ABT-263 + S63845 were applied. Immunoblots for NOXA and actin, and quantification of PMAIP1 and CDKN1A mRNA levels are shown below graphs. d CRISPR guides targeting the functional domains of PMAIP1 (NOXA) were cloned into a pLenti-CRISPR-mCherry vector and virus generated. SKBR7 cells were transduced, selected by FACS. SKBR7 parental and PMAIP1-sg cells were plated and doxorubicin treated for 24 h, and then treated as indicated with ABT-263 or ABT-263 + S63845. MTT assay was performed 24 h after treatments. Immunoblots for NOXA and actin are shown beneath the graphs. Statistics (complete analysis available in Supplementary Table 5) used were as follows: (c) ANOVA with Tukey’s posttest comparing all treatment groups separately for Prolif and Doxo Rx, and separately for each of the four cell lines. Control (indicated by “0” representing DMSO treatment) for Prolif and Doxo Rx were each normalized to 1. d ANOVA with Tukey’s posttest comparing all treatment groups separately for Prolif and Doxo Rx, and separately for each of the two cell lines. Control (indicated by “0” representing DMSO treatment) for Prolif and Doxo Rx were each normalized to 1. Each treatment group was in triplicate and individual data points are shown. Error bars indicate SEM. Nonnormalized data for (c–d) are shown in Supplementary Fig. 3g–h.
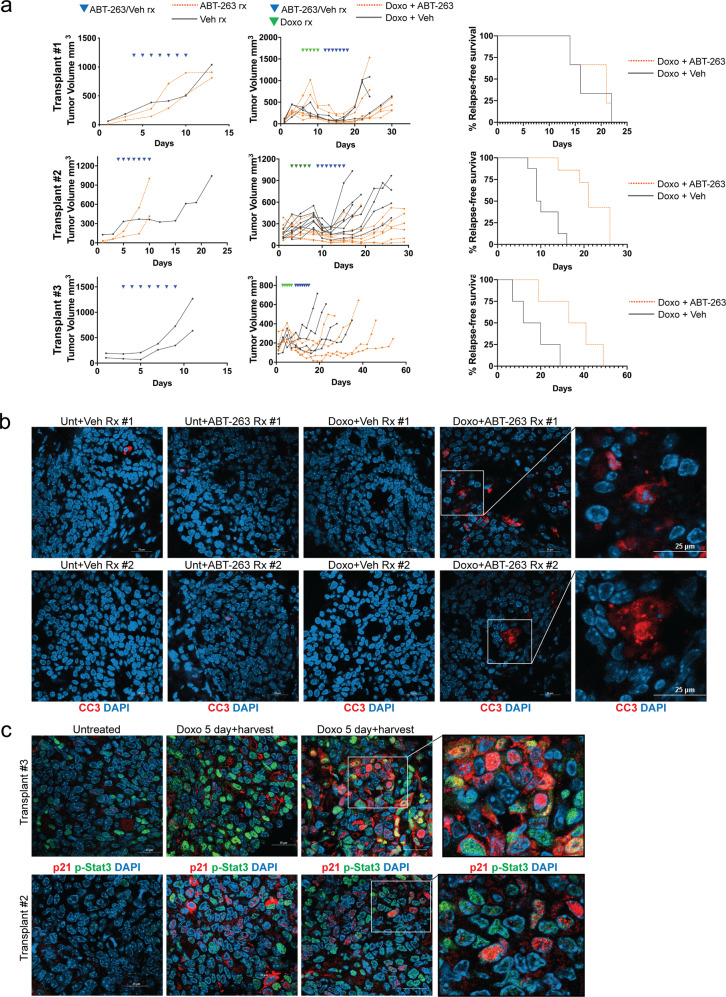
ABT-263 delays tumor relapse in vivo
Because p53 wild-type MMTV-Wnt1 tumors treated with doxorubicin undergo senescence, minimal tumor regression, and rapid relapse [12, 19], we determined if response in these tumors could be improved by treating mice with ABT-263 following chemotherapy treatment, when tumors are thoroughly senescent [12, 19]. Three different spontaneous tumors from MMTV-Wnt1 mice were orthotopically transplanted into the fat pad of C57BL/6j mice. After tumors formed, mice were treated with doxorubicin followed by either ABT-263 or vehicle treatment. Similar to our data from cell lines, we identified transplants that did respond to ABT-263 and those that did not. Transplant 1 tumors treated with doxorubicin then ABT-263 had similar tumor regression as mice treated with vehicle (p = 0.84) and also relapsed in approximately the same number of days (p = 0.26) (Fig. 8a, top). Doxorubicin-treated mice with tumor transplants 2 and 3, however, regressed more (p = 0.0003) (p = 0.04), took longer to relapse, and had longer survival when they were subsequently treated with ABT-263 than tumors treated with vehicle (p = 0.0009) (p = 0.04) (Fig. 8a, middle, bottom). In all experiments, tumors did not respond to vehicle or ABT-263 treatment alone. Next, to test if ABT-263 induced apoptosis in chemotherapy-induced senescent tumors in vivo, mice with tumors from transplant 2 were treated ± doxorubicin followed by ± ABT-263. Tumors were harvested 2 days following the final doxorubicin treatment, or after the 4th ABT-263 treatment. Consistent with previous reports [12], minimal apoptosis was detected in doxorubicin-treated tumors that arrest following treatment [12], or untreated tumors (Fig. 8b). Consistent with results in cell lines and tumor growth (Fig. 8a), ABT-263 treatment of mice with proliferating (not doxorubicin treated) tumors did not cause apoptosis, but treatment of mice that were previously doxorubicin treated resulted in striking examples of positive cleaved-caspase 3 staining and fragmented, condensed nuclei, similar to cell lines shown in Fig. 2 (Fig. 8b). Tumor transplants from doxorubicin-treated mice harvested 48 h following the final treatment stained intensely for senescence markers p-Stat3 (a SASP marker [12, 17, 19, 27, 68]) and p21 (a cell cycle arrest marker [12, 69]) compared with tumors from untreated mice (Fig. 8c). Because treatment with doxorubicin and/or ABT-263 can induce hematotoxicity [70, 71], we analyzed blood sampled from treated mice (Supplementary Fig. 8b). Consistent with previous reports, doxorubicin increased platelet count [71], and treatment with ABT-263 reduced platelet count [70]. The combination of doxorubicin followed by ABT-263 did not exacerbate or change the platelet number beyond treatment with ABT-263 alone (Supplementary Fig. 8a). Treatment with ABT-263 or doxorubicin + ABT-263 did not result in significant weight loss across the experiment (Supplementary Fig. 8b).
Fig. 8. ABT-263 improves response in chemotherapy-treated mice.
a Three separate MMTV-Wnt1 tumors were transplanted into C57BL/6j mice, and when a tumors reached ~300 mm3, mice were treated with 4 mg/kg IP doxorubicin on days indicated by green arrowheads above, followed by oral gavage with ABT-263 (black lines) or vehicle (red lines) on days indicated by blue arrowheads above. Measurements were taken with digital calipers every other day. In each transplant, mean tumor regression and days to relapse were calculated for doxorubicin followed by vehicle, vs. doxorubicin followed by ABT-263. Transplant 1: 24 days to relapse (Doxo + ABT + 263) vs. 20 (Doxo + Veh), 84% regression vs. 83%, both n.s.; transplant 2: 21 (Doxo + ABT-263) vs. 11 days (Doxo + Veh) to relapse, 48% vs. 23% tumor volume lost, both p < 0.05; transplant 3: 36 days (Doxo + ABT-263) to relapse vs. 16 (Doxo + Veh), 54% regression vs. 23%, both p < 0.05. Statistics used were two tailed, unpaired Student’s t test for comparisons of means in Doxo Rx followed by ABT-263 vs. Doxo Rx followed by vehicle. Kaplan–Meier survival analysis was performed to measure relapse-free survival of mice transplanted with MMTV-Wnt1 tumors (Transplants 1, 2, and 3), and treated with doxorubicin, and ABT-263 or vehicle (Transplant 1 p = 0.8658; Transplant 2 p = 0.0005; Transplant 3 p = 0.0510). Relapse-free survival was defined as the number of days between the largest tumor volume recorded before ending treatment, and when the tumor exceeded this volume following treatment. Differences in relapse-free survival were calculated by Log-rank (Mantel–Cox) test. b Mice bearing tumors (transplant 2) were treated as in (a), but harvested either 2 days following the final doxorubicin treatment or one day after the fourth ABT-263 treatment. FFPE sections were stained with cleaved casapse 3 and DAPI, and representative images were captured on the confocal microscope. Representative fields of view from each treatment group were quantified for cleaved-caspase 3 staining, and ANOVA was performed with Tukey’s posttest (Unt + Veh vs. Doxo + ABT-263 p < 0.0001; Unt + ABT-263 vs. Doxo + ABT-263 p < 0.0001; Doxo + Veh vs. Doxo + ABT-263 p < 0.0001). Scale bar represents 25 µm. c Mice bearing tumors (transplants 2 and 3) were treated with doxorubicin or vehicle. FFPE sections were stained with p21 (red) and p-STAT3 (green), and representative images were captured on the confocal microscope. Representative fields of view from each treatment group were quantified for p21 and p-STAT3 staining, and student’s t test for 2 way comparisons was performed (p21: Veh vs. Doxo p < 0.0001; p-STAT3: Veh vs. Doxo p < 0.0001).
Discussion
This study demonstrates that ABT-263 and other BH3 mimetic drugs can selectively eliminate p53 wild-type breast and other cancer cells that have survived chemotherapy treatment by entering a state of senescence. Studies from our lab and others have shown that in chemotherapy-treated breast cancers: (1) TP53 wild-type tumors are much less likely to undergo a pCR [5–10]; (2) patients with TP53 wild-type tumors have a dismal prognosis [11]; (3) TP53 wild-type tumors undergo senescence and express SASP factors that can drive relapse [12, 13, 19]. Thus, eliminating senescent cells in residual disease stands to benefit those patients most in need of improved therapy options. Other studies have shown elimination of senescent normal cells in the stroma of tumors, using either genetic means or senolytic drugs, improved response to chemotherapy [72]. However, no studies have reported the use of senolytics to eradicate chemotherapy-induced senescent cancer cells to reduce residual disease burden. While BH3 mimetic sensitivity was not proven to be an integral part of senescence, we feel all data taken together support the conclusion that cells enter senescence to survive very different types of treatments (doxorubicin, IR, paclitaxel, nutlin), and in this senescent state they are sensitive to BH3 mimetics. Because senescent cells are the cells that survive treatment, it is important and clinically relevant that these cells are sensitive to BH3 mimetics.
We show some cell lines rely on BCL-XL for survival in the senescent state, while others can utilize either BCL-XL or MCL1. Interestingly, NOXA expression in senescent cells conferred sensitivity to inhibition of BCL-XL alone, and when not expressed, a second drug to inhibit MCL1 was required to induce apoptosis. These data support others’ findings that NOXA is a potent inhibitor of MCL1 [65, 73], and suggest that when treating senescent tumor cells with senolytics, NOXA expression should be used to direct co-administration of an MCL1 inhibitor with a BCL-XL inhibitor. Inhibition of multiple apoptotic proteins that include MCL1 is more likely to cause hematopoietic toxicities in vivo [74], but promising preclinical results in leukemia models suggest plausibility [75]. Interestingly, sensitivity to ABT-263 in nonsenescent cells can also be dependent on the ratio of MCL1 to BIM [44].
Cells from solid tumors, and particularly senescent cells, are notoriously difficult to kill [76, 77]. The extent and rapidity of apoptosis induced by ABT-263 in only the senescent cancer cells is notable. We observed little or no change in cell number or apoptosis in proliferating cells, or in cells 24 h following doxorubicin treatment. Our study, showing TP53 wild type tumor cells made senescent by chemotherapy can be eliminated in hours using senolytic drugs, is more likely to be far reaching due to prevalence of chemotherapy, which remains a first line therapy for many patients, and the only therapy for TNBC patients. Further, our study suggests a therapy that would benefit patients with TP53 wild-type breast tumors who have very poor response [5–10] and survival [11], and are thus in need of improved therapy options. Our results suggest that ABT-263 would be more effective in solid tumors if used in TP53 wild-type tumors and sequenced after chemotherapy treatment to allow for induction of senescent phenotypes.
Methods
Cell lines
4226 is a cell line made by plating and passage of an MMTV-Wnt1 tumor [50]; A549 was a gift from Joseph Lasky, Tulane School of Medicine; U2OS was a gift from Sean Lee, Tulane School of Medicine; MDA-MB-175 (University of Texas MD Anderson Characterized Cell Line Core); HCC712 was from University of Texas Southwestern Cell Line Core and Adi Gazdar, SKBR7 (gift of Jean-Paul Borg, Institut Universitaire de France); MPE-600, Cal51 (gift of Joe Gray, University of Oregon Health Science University); HCC1428, ZR75-30, MCF-7 were from ATCC; 293T cells used for virus production have been described [19]. 4226, A549, U2OS were grown in standard DMEM medium supplemented with 10% serum (Life Technologies). MDA-MB-175, MCF-7, and MPE-600 cells were grown in EMEM medium (ATCC) supplemented with 10% serum. HCC712, HCC1428, Cal51, SKBR7, and ZR75-30 cells were grown in RPMI medium (Life Technologies) supplemented with 10% serum. Cell lines were checked regularly for mycoplasma contamination by DAPI staining and microscopy. All cell lines were treated with 0.25 μM doxorubicin except for 4226 cells, which were treated with 1 μM, or as indicated in the figure. After 24 h on drug, cells were washed 3× in PBS and changed to complete media. SAβGal assay was performed as previously described [50, 59]. For paclitaxel experiments, cells were treated with 1 nM, 5 nM, or 10 nM paclitaxel for 24 h and then changed to complete media. For irradiation experiments, cells were exposed to 8 Gy gamma irradiation from a Cesium source. For Nutlin-3a experiments, cells were treated with Nutlin-3a (Sigma-Aldrich, 444152) every other day at concentrations of 2.5 μM (Cal51, A549, U2OS) or 15 μM (4226).
Cell number determination
For MTT assay, Thiazolyl Blue Tetrazolium Blue (MTT) (Sigma, M5655) was dissolved in PBS at 5 mg/ml. Cells were plated into 24 wells at ~80,000 cells per well (U2OS, A549, 4226) or 100,000 cells per well (MCF-7, MPE-600, MDA-MB-175, ZR-75). Cells were treated with ABT-263 at the time point indicated in the figure, and then MTT assay was performed 24 h later as described [78, 79]. Cell number of 4226-mCherry and MCF-7 GFP cells was determined on an IncuCyte S3 system at 37 °C and 5% CO2 in full growth medium at ×20 magnification and using 2018A software version. Cells were plated and treated as for MTT, and plates were scanned every 3 h. For senescent cells, total red object area was determined with the following parameters: segmentation: adaptive with a threshold of 0.2 RCU, Edge Split On at sensitivity of −45, minimum area 800 µm2. For proliferating cells, same settings but Edge Split sensitivity was −20, and minimum area 400 µm2.
Treatments
ABT-263, Q-VD-OPh, S63845, ABT-199, A-1331852, and paclitaxel were all purchased from Apexbio (Cat# A3007, A1901, A8737, A8194, and B6164). Drugs were resuspended in DMSO and used at indicated concentrations. Doxorubicin was purchased from Santa Cruz Biotech (SC-200923A), resuspended in water to a concentration of 10 mg/ml and then diluted with PBS to 50 µg/ml and used at indicated concentrations.
Western blot
Cells were lysed as described [51] and then Bicinchoninic Acid (BCA Protein Assay Kit, Pierce) was performed. Lysates (40–50 μg) were resolved on 10–13% SDS/PAGE gel and probed with antibodies for actin (MA5-15739, Invitrogen), BCL-XL (2764S, Cell signaling), BCL2 (D17C4, Cell signaling), MCL1 (sc-819, Santa Cruz), Bim (2819 S, Cell signaling), Bid (2002T, Cell Signaling), BAK (12105T, Cell Signaling), BAX (5023T, Cell Signaling), PUMA (12450T, Cell Signaling), NOXA (14766T, Cell Signaling), cleaved-caspase 3 (9661S, Cell Signaling), PARP (9542S, Cell Signaling). All antibodies were used at 1:1000 dilution.
Immunofluorescence staining
For cleaved-caspase 3 staining, cells were plated on glass coverslips (80,000 cells per well of a 24 well plate for Cal51, and 100,000 cells per well for MDA-MB-175), and treated the next day with doxorubicin for 24 h, followed by PBS washes and medium replenishment. On day 7, cells were treated with indicated senolytic for 2 h. Cells were then fixed with 4% formaldehyde, permeabilized in 1× PBS/0.5% Triton X-100, and then blocked in 5% BSA. Cleaved-caspase 3 primary antibody (Cell Signaling, Danvers, MA) was added at 1:400 concentration and incubated at 4 °C overnight. After washing, Alexa Fluor 594 Goat Anti-Rabbit secondary (ThermoFisher Scientific) at 1:500 concentration in 1% BSA was then added for 1 h at room temperature, followed by more washing. Cells were then DAPI stained and washed further.
For mitochondrial BAX staining [80], cells were plated and treated as above. On day 7, cells were incubated in 75 nM MitoTracker Red CMXRos (ThermoFisher Scientific, M7512) for 30 min. Cells were then treated with indicated senolytic for 2 h, fixed, permeabilized, and blocked as above. BAX primary antibody (Cell Signaling, Danvers, MA) was added at 1:200 concentration in 1% BSA and incubated at 4 °C overnight. After more washing, Alexa Fluor 488 Donkey Anti-Rabbit secondary (ThermoFisher Scientific, A21206) was then added at 1:500 concentration for 1 h at room temperature. Cells were then DAPI stained.
For cleaved-caspase 3 staining of MMTV-Wnt1 tumor sections, tumors were harvested, formalin fixed, paraffin embedded, and sectioned (5 μm). Sectioned tissue was deparaffinized and rehydrated by washing in the following solutions and order: (1) 3 × xylenes; (2) 2 × 100% EtOH; (3) 2 × 90% EtOH; (4) 2 × 70% EtOH; (5) 2 × H2O. Antigen retrieval was in citric acid buffer, pH6 in a steam bath. Sections were washed in TBS-T and blocked in 5% horse serum/0.1% Tween 20 in PBS. Cleaved-caspase 3 primary antibody was then added at 1:400 concentration and sections were incubated overnight at 4 °C. Alexa Fluor 594 Goat Anti-Rabbit secondary was then added at 1:500 concentration for 1 h at room temperature. Sections were then DAPI stained and coverslipped for imaging.
For p-STAT3 and p21 staining of MMTV-Wnt1 tumor sections, antigen retrieval was in Tris-EDTA buffer, pH9 in a steam bath. Sections were washed in TBS-T and blocked in 3% BSA/0.1% Tween 20 in PBS. p-STAT3 (Cell Signaling, 9145) (1:200) and p21 (Agilent, M720229-2) (1:50) primary antibodies were then added, and sections incubated overnight at 4 °C. Alexa Fluor 488 Donkey Anti-Rabbit and Alexa Fluor 555 Donkey Anti-Mouse secondary antibodies were then applied at 1:500 concentration for 1 h at room temperature. Sections were then DAPI stained and coverslipped for imaging.
For immunofluorescence quantification, at least three random, representative fields were analyzed for each experiment. Statistical differences were determined via ANOVA with Tukey’s posttest (MDA-MB-175, Cal51, and MMTV-Wnt1 cleaved-caspase 3; MDA-MB-175 and Cal51 MitoTracker/BAX) or two-tailed Student’s t test (MMTV-Wnt1 p21 and p-STAT3).
Microscopy
Confocal images of immunofluorescence staining were captured on a Nikon TiE-2 Inverted Research Microscope Nikon A1rSi Laser Point Scanning Confocal, with a Plan Apo λ 60× oil objective for both cell lines and tissue sections. Image brightness and background levels were adjusted uniformly and minimally for entire images using NIS-Elements software or Photoshop when necessary for visual clarity in publication.
Live-cell imaging
For cell engulfment experiments, GFP and mCherry labeled cells [50] were plated simultaneously in a six well plate. The following day, cells were treated with doxorubicin for 24 h (0.25 μM for MCF-7; 0.75 μM for 4226) followed by PBS washes and medium replenishment. On day 3, the plate was placed on an IncuCyte S3 system at 37 °C and 5% CO2 in full growth medium and imaged every 3 h at ×20 magnification and using 2018A software version. On day 7, cells were treated with indicated senolytic followed by imaging every hour for 24 h. Cells were identified as having engulfed or not by viewing the time-lapse images, and then scored for survival for the 24 h period following addition of senolytic. Survival of engulfing and never-engulfing cells was plotted on a Kaplan–Meier curve using GraphPad Prism software.
To determine senolytic drug sensitivity in senescence, cells were plated in 24 well plates (80,000 cells per well for Cal51, and 100,000 cells per well for MDA-MB-175). The following day, cells were treated or not with doxorubicin for 24 h (0.25 μM), followed by PBS washes and medium replenishment. On day 7, LysoTracker Red DND-99 (ThermoFisher Scientific; L7528) was added to media at concentration of 75 nM and incubated for 30 min. Cells were then treated with indicated senolytic or DMSO control and immediately imaged on IncuCyte for 4 h.
To determine selective induction of cell death in senescent but not proliferating cells, Cal51 cells were plated in six well plates (300,000 cells per well). The following day, cells were treated with doxorubicin for 24 h (0.25 μM), followed by PBS washes and medium replenishment. Cells were placed on IncuCyte and imaged every 3 h. On day 7, cells were treated with ABT-263 (1.0 μM), and cell death of senescent and proliferating cells was determined visually by live-cell imaging.
qPCR
RNA was extracted from MCF-7 parental and PMAIP1 overexpressing cells using Trizol reagent (Invitrogen, Carlsbad, CA), treated with DNAse (Roche, Indianapolis, IN), and then reverse transcribed using a kit (GE Healthcare, Piscataway, NJ). Real time PCR using SYBR green (BioRad, Valencia, CA) was performed using QuantStudio 6 (ThermoFisher Scientific). The following primers were used: PMAIP1 forward [1]: 5′-GAGCTGGAAGTCGAGTGTGCTA-3′; PMAIP1 reverse [1]: 5′-TGCCGGAAGTTCAGTTTGTCT-3′; PMAIP1 forward [2]: 5′-GCATTGTAATTGAGAGGAATGTGAA-3′; PMAIP1 reverse [2]: 5′-TGTATTCCATCTTCCGTTTCCA-3′. CDKN1A and RPLP0 primers are published [12].
CRISPR-Cas9 gene editing
All guides were designed to target functional domains. The BCL2L1 guide sequences in MCF-7 cells were BCL2L1 forward: 5′-GTTTGAACTGCGGTACCGGC-3′; BCL2L1 reverse: 5′-GCCGGTACCGCAGTTCAAAC-3′. The BCL2L1 sg1 guide sequences in Cal51 cells were BCL2L1 forward: 5′-TGCCCGGGAGGTGATCCCCA-3′; BCL2L1 reverse: 5′-TGGGGATCACCTCCCGGGCA-3′. BCL2L1 sg2 guide sequences in Cal51 cells were BCL2L1 forward: 5′-AGTTTGAACTGCGGTACCGG-3′; BCL2L1 reverse: 5′-CCGGTACCGCAGTTCAAACT-3′. The following primers were used: CRISPR BCL-XL forward: 5′-TTCTTACCCAGCCGCCGTTCTCCT-3′; CRISPR BCL-XL reverse: 5′-GCAACCCATCCTGGCACCTG-3.
MCL1 guides sequences in MCF-7 cells were MCL1-sg1 forward: 5′-GGAGACCTTACGACGGGTTG-3′; MCL1-sg1 reverse: 5′-CAACCCGTCGTAAGGTCTCC-3′. MCL1-sg2 forward: 5′-CAGCGACGGCGTAACAAACT-3′; MCL1-sg2 reverse: 5′-AGTTTGTTACGCCGTCGCTG-3′. The following primers were used to amplify the edited locus for sequencing: CRISPR MCL1 forward: 5′-CCTTACGAGAACGTCTGTGATAC-3′; CRISPR MCL1 reverse: 5′-TAATCGGACTCAACCTCTACTGT-3′.
The PMAIP1 (NOXA) guide sequences were PMAIP1sg1 forward: 5′-TCGAGTGTGCTACTCAACTC-3′; PMAIP1sg1 reverse: 5′-GAGTTGAGTAGCACACTCGA-3′. PMAIP1sg2 forward: 5′-ACGCTCAACCGAGCCCCGCG-3′; PMAIP1sg2 reverse: 5′-CGCGGGGCTCGGTTGAGCGT-3′. PMAIP1sg3 forward: 5′-TTCTTGCGCGCCTTCTTCCC-3′; PMAIP1sg3 reverse 5′-GGGAAGAAGGCGCGCAAGAA-3′. The following primers were used: CRISPR sg1 NOXA forward: 5′-GCTTTCGGTGAGTCACAATGTGTG-3′; CRISPR sg1 NOXA reverse: 5′-GGTTCCTGAGCAGAAGAGTTTGG-3. CRISPR sg2-3 NOXA forward: 5′-CACCCGATCCCAGCATCCCTGCC-3′; CRISPR sg2-3 NOXA reverse: 5′-CCAGCCGAGACCTCGACCC-3′.
Guides were cloned into pLenti-CRISPR-mCherry vector (Addgene, 75161) and transfected into 293T cells using Lipofectamine 2000 (ThermoFisher, 11668019) with packaging constructs, and then virus collected as described [19]. Infected cells were enriched by FACS (Louisiana Cancer Research Consortium). Editing was confirmed by PCR amplifying DNA with primers flanking the guide sequence followed by Sanger sequencing (Genewiz). Lenti-PMAIP1 overexpression vector pLV-CMV > EGFP-hPGK > hPMAIP1 was constructed by VectorBuilder and virus was generated as described [19]. Bid expression plasmid in pcDNA was a gift of Hua Lu, PhD.
To quantify editing in pooled populations, DNA extracts from cell lines were PCR amplified with primers flanking the sgRNA cut site, purified on columns (Qiagen cat# 28104), and then submitted for Genewiz AmpliconEZ sequencing using MiSeq 2 × 250 platform, yielding > 1000 2 × 250-nucleotide paired-end reads per sample. CRISPResso2 editing sequence analysis [81] was then performed under default parameters to quantify editing efficiency.
Mice
MMTV-Wnt1 tumors were injected into the mammary fat pads of 10 week old female C57BL/6j mice (1 × 106 tumor cells). Mice were monitored frequently for tumor formation. Once a palpable tumor was detected, tumors were measured every other day using digital calipers and tumor volume calculated as described [12]. When tumors reached a volume of ~300 mm3, they were treated with 4 mg/kg doxorubicin (Sigma-Aldrich) in PBS that was injected intraperitoneally (IP) once daily for 5 consecutive days [12, 19]. Two days after doxorubicin treatment, mice were treated with either vehicle or ABT-263 (in ethanol: polyethylene glycol 400: Phosal 50 PG) via oral gavage at 50 mg/kg body weight per day (mg/kg/d) for 7 consecutive days. All treatments were well tolerated with no observed weight loss or toxicity. Mice were sacrificed when tumors relapsed (reached a tumor volume greater than the largest volume observed before treatment). To capture tumors during response, mice with tumors formed from transplant 2 were treated with (1) doxorubicin followed by four doses of ABT-263, (2) doxorubicin alone, (3) four doses of ABT-263 alone, (4) vehicle alone. Two days following the final doxorubicin only treatment, or 1 day after the 4th ABT-263 treatment in doxorubicin or untreated mice, tumors were harvested, formalin fixed/paraffin embedded, and sectioned for staining as previously [12]. To determine potential drug toxicities in mice, blood was sampled either retro-orbitally or by terminal cardiac puncture from the following groups: (1) control/pretreatment; (2) 24 h following the 5th and final doxorubicin injection; (3) 24 h following the 7th and final ABT-263 gavage treatment; (4) 24 h following the 7th and final ABT-263 gavage treatment that followed five doxorubicin treatments. Complete blood count analysis was performed by Clinical Pathology Core, Division of Comparative Pathology, Tulane National Primate Research Center.
Statistics
All MTT assays and western blots are representative of at least two independent experiments. All MTT experiments were done in triplicate wells and normalized as indicated in the figure legends. Statistics used were as follows: Student’s t test for two way comparisons and ANOVA with Tukey’s posttest for comparisons of three or more groups. Variance between statistically compared groups was similar, and all statistical tests were performed with assumption of similar variance. For mouse experiments, days to relapse was expressed as the number of days between the largest tumor volume recorded before ending treatment, and when the tumor exceeded this volume following treatment. To determine the amount each tumor regressed following treatment, the smallest volume the tumor became after treatment was divided by the greatest tumor volume that was recorded prior to treatment. The mean of percent regression and days to relapse was calculated for Doxo + Vehicle and Doxo + ABT-263 tumors, and a two-tailed, unpaired t-test (GraphPad Prism software) was used to determine the p value. Mice numbers for in vivo efficacy experiments were calculated to detect tumor regression improvement from 35% (doxorubicin only), to 50% (doxorubicin followed by ABT-263), with 5% probability of incorrectly rejecting the null hypothesis using the standard deviation from response of spontaneous tumors in previous experiments [12]. Animals were randomly assigned to treatment groups, and investigator was blinded to group allocation. No animals or samples were excluded from analysis.
Supplementary information
Acknowledgements
The authors wish to acknowledge flow cytometry support from Dorota Wyczechowska, PhD.
Funding
This study was supported by the Department of Defense Breast Cancer Research Program (grant number W81XWH-14-1-0216 to JGJ) and The National Center for Advancing Translational Sciences (award TL1TR003106 to AS). Content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Advancing Translational Sciences. The Cellular Immunology and Immune Metabolism Core at the Louisiana Cancer Research Consortium is supported by a grant from the NIN/NIGMS (grant number 1P30GM114732-01).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All animal experiments were approved by the Tulane Institutional Animal Care and Use Committee, Protocol ID# 4381, and conformed to the guidelines of the United States Animal Welfare Act and the National Institutes of Health.
Footnotes
Edited by E. Baehrecke
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ashkan Shahbandi, Sonia G. Rao
Supplementary information
The online version of this article (10.1038/s41418-020-0564-6) contains supplementary material, which is available to authorized users.
References
- 1.Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, et al. TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum Mutat. 2016;37:865–76. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 2.Tonnessen-Murray C, Lozano G, Jackson JG. The p53 protein: from cell regulation to cancer. In: Lozano G, Levine AJ, editors. The p53 protein: from cell regulation to cancer. A cold spring harbor perspectives in medicine collection. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2016. p. 173–86.
- 3.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062–78. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.TCGA-Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertheau P, Plassa F, Espie M, Turpin E, de Roquancourt A, Marty M, et al. Effect of mutated TP53 on response of advanced breast cancers to high-dose chemotherapy. Lancet. 2002;360:852–4. doi: 10.1016/S0140-6736(02)09969-5. [DOI] [PubMed] [Google Scholar]
- 6.Bertheau P, Turpin E, Rickman DS, Espie M, de Reynies A, Feugeas JP, et al. Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS Med. 2007;4:e90. doi: 10.1371/journal.pmed.0040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura Y, Oshima K, Naoi Y, Nakayama T, Kim SJ, Shimazu K, et al. 14-3-3sigma expression is associated with poor pathological complete response to neoadjuvant chemotherapy in human breast cancers. Breast Cancer Res Treat. 2012;134:229–36. doi: 10.1007/s10549-012-1976-x. [DOI] [PubMed] [Google Scholar]
- 8.Esserman LJ, Berry DA, Cheang MC, Yau C, Perou CM, Carey L, et al. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657) Breast Cancer Res Treat. 2012;132:1049–62. doi: 10.1007/s10549-011-1895-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen MB, Zhu YQ, Xu JY, Wang LQ, Liu CY, Ji ZY, et al. Value of TP53 status for predicting response to neoadjuvant chemotherapy in breast cancer: a meta-analysis. PloS One. 2012;7:e39655. doi: 10.1371/journal.pone.0039655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Xu Y, Chen J, Ouyang T, Li J, Wang T, et al. TP53 mutations are associated with higher rates of pathologic complete response to anthracycline/cyclophosphamide-based neoadjuvant chemotherapy in operable primary breast cancer. Int J Cancer. 2016;138:489–96. doi: 10.1002/ijc.29715. [DOI] [PubMed] [Google Scholar]
- 11.Ungerleider NA, Rao SG, Shahbandi A, Yee D, Niu T, Frey WD, et al. Breast cancer survival predicted by TP53 mutation status differs markedly depending on treatment. Breast Cancer Res. 2018;20:115. doi: 10.1186/s13058-018-1044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson JG, Pant V, Li Q, Chang LL, Quintas-Cardama A, Garza D, et al. p53-mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer Cell. 2012;21:793–806. doi: 10.1016/j.ccr.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.te Poele RH, Okorokov AL, Jardine L, Cummings J, Joel SP. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res. 2002;62:1876–83. [PubMed] [Google Scholar]
- 14.Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–78. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Novakova Z, Hubackova S, Kosar M, Janderova-Rossmeislova L, Dobrovolna J, Vasicova P, et al. Cytokine expression and signaling in drug-induced cellular senescence. Oncogene. 2010;29:273–84. doi: 10.1038/onc.2009.318. [DOI] [PubMed] [Google Scholar]
- 16.Cahu J, Bustany S, Sola B. Senescence-associated secretory phenotype favors the emergence of cancer stem-like cells. Cell Death Dis. 2012;3:e446. doi: 10.1038/cddis.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canino C, Mori F, Cambria A, Diamantini A, Germoni S, Alessandrini G, et al. SASP mediates chemoresistance and tumor-initiating-activity of mesothelioma cells. Oncogene. 2012;31:3148–63. doi: 10.1038/onc.2011.485. [DOI] [PubMed] [Google Scholar]
- 18.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–68. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tonnessen-Murray C, Ungerleider NA, Rao SG, Wasylishen AR, Frey WD, Jackson JG. p53 mediates vast gene expression changes that contribute to poor chemotherapeutic response in a mouse model of breast cancer. Transl Oncol. 2018;11:930–40. doi: 10.1016/j.tranon.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takamori H, Oades ZG, Hoch OC, Burger M, Schraufstatter IU. Autocrine growth effect of IL-8 and GROalpha on a human pancreatic cancer cell line, Capan-1. Pancreas. 2000;21:52–6. doi: 10.1097/00006676-200007000-00051. [DOI] [PubMed] [Google Scholar]
- 21.Dhawan P, Richmond A. Role of CXCL1 in tumorigenesis of melanoma. J Leukoc Biol. 2002;72:9–18. [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Lu P, Fujii C, Nakamoto Y, Gao JL, Kaneko S, et al. Essential contribution of a chemokine, CCL3, and its receptor, CCR1, to hepatocellular carcinoma progression. Int J Cancer. 2006;118:1869–76. doi: 10.1002/ijc.21596. [DOI] [PubMed] [Google Scholar]
- 23.Begley LA, Kasina S, Mehra R, Adsule S, Admon AJ, Lonigro RJ, et al. CXCL5 promotes prostate cancer progression. Neoplasia. 2008;10:244–54. doi: 10.1593/neo.07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karnoub AE, Weinberg RA. Chemokine networks and breast cancer metastasis. Breast Dis. 2006;26:75–85. doi: 10.3233/bd-2007-26107. [DOI] [PubMed] [Google Scholar]
- 25.Achuthan S, Santhoshkumar TR, Prabhakar J, Nair SA, Pillai MR. Drug-induced senescence generates chemoresistant stemlike cells with low reactive oxygen species. J Biol Chem. 2011;286:37813–29. doi: 10.1074/jbc.M110.200675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milanovic M, Fan DNY, Belenki D, Dabritz JHM, Zhao Z, Yu Y, et al. Senescence-associated reprogramming promotes cancer stemness. Nature. 2018;553:96–100. doi: 10.1038/nature25167. [DOI] [PubMed] [Google Scholar]
- 27.Toso A, Revandkar A, Di Mitri D, Guccini I, Proietti M, Sarti M, et al. Enhancing chemotherapy efficacy in Pten-deficient prostate tumors by activating the senescence-associated antitumor immunity. Cell Rep. 2014;9:75–89. doi: 10.1016/j.celrep.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 28.Rao SG, Jackson JG. SASP: tumor suppressor or promoter? Yes! Trends Cancer. 2016;2:676–87. doi: 10.1016/j.trecan.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Shahbandi A, Jackson JG. Analysis across multiple tumor types provides no evidence that mutant p53 exerts dominant negative activity. npj Precis Oncol. 2019;3:1. doi: 10.1038/s41698-018-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–81. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 31.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–5. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 32.Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 33.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lazzerini Denchi E, Attwooll C, Pasini D, Helin K. Deregulated E2F activity induces hyperplasia and senescence-like features in the mouse pituitary gland. Mol Cell Biol. 2005;25:2660–72. doi: 10.1128/MCB.25.7.2660-2672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–4. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 36.Freund A, Orjalo AV, Desprez PY, Campisi J. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol Med. 2010;16:238–46. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu Y, Tchkonia T, Pirtskhalava T, Gower AC, Ding H, Giorgadze N, et al. The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell. 2015;14:644–58. doi: 10.1111/acel.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dorr JR, Yu Y, Milanovic M, Beuster G, Zasada C, Dabritz JH, et al. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature. 2013;501:421–5. doi: 10.1038/nature12437. [DOI] [PubMed] [Google Scholar]
- 39.Merino D, Kelly GL, Lessene G, Wei AH, Roberts AW, Strasser A. BH3-mimetic drugs: blazing the trail for new cancer medicines. Cancer Cell. 2018;34:879–91. doi: 10.1016/j.ccell.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Yosef R, Pilpel N, Tokarsky-Amiel R, Biran A, Ovadya Y, Cohen S, et al. Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat Commun. 2016;7:11190. doi: 10.1038/ncomms11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu Y, Tchkonia T, Fuhrmann-Stroissnigg H, Dai HM, Ling YY, Stout MB, et al. Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell. 2016;15:428–35. doi: 10.1111/acel.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang J, Wang Y, Shao L, Laberge RM, Demaria M, Campisi J, et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat Med. 2016;22:78–83. doi: 10.1038/nm.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cory S, Roberts AW, Colman PM, Adams JM. Targeting BCL-2-like proteins to kill cancer cells. Trends Cancer. 2016;2:443–60. doi: 10.1016/j.trecan.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Faber AC, Farago AF, Costa C, Dastur A, Gomez-Caraballo M, Robbins R, et al. Assessment of ABT-263 activity across a cancer cell line collection leads to a potent combination therapy for small-cell lung cancer. Proc Natl Acad Sci USA. 2015;112:E1288–96. doi: 10.1073/pnas.1411848112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams MM, Lee L, Hicks DJ, Joly MM, Elion D, Rahman B, et al. Key survival factor, Mcl-1, correlates with sensitivity to combined Bcl-2/Bcl-xL blockade. Mol Cancer Res. 2017;15:259–68. doi: 10.1158/1541-7786.MCR-16-0280-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williams MM, Lee L, Werfel T, Joly MMM, Hicks DJ, Rahman B, et al. Intrinsic apoptotic pathway activation increases response to anti-estrogens in luminal breast cancers. Cell Death Dis. 2018;9:21. doi: 10.1038/s41419-017-0072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oakes SR, Vaillant F, Lim E, Lee L, Breslin K, Feleppa F, et al. Sensitization of BCL-2-expressing breast tumors to chemotherapy by the BH3 mimetic ABT-737. Proc Natl Acad Sci USA. 2012;109:2766–71. doi: 10.1073/pnas.1104778108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young AI, Law AM, Castillo L, Chong S, Cullen HD, Koehler M, et al. MCL-1 inhibition provides a new way to suppress breast cancer metastasis and increase sensitivity to dasatinib. Breast Cancer Res. 2016;18:125. doi: 10.1186/s13058-016-0781-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang BD, Broude EV, Dokmanovic M, Zhu H, Ruth A, Xuan Y, et al. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 1999;59:3761–7. [PubMed] [Google Scholar]
- 50.Tonnessen-Murray CA, Frey WD, Rao SG, Shahbandi A, Ungerleider NA, Olayiwola JO, et al. Chemotherapy-induced senescent cancer cells engulf other cells to enhance their survival. J Cell Biol. 2019;218:3827–44. doi: 10.1083/jcb.201904051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jackson JG, Pereira-Smith OM. Primary and compensatory roles for RB family members at cell cycle gene promoters that are deacetylated and downregulated in doxorubicin-induced senescence of breast cancer cells. Mol Cell Biol. 2006;26:2501–10. doi: 10.1128/MCB.26.7.2501-2510.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anwar T, Sen B, Aggarwal S, Nath R, Katoch A, Aiyaz M, et al. Differentially regulated gene expression in quiescence vs. senescence and identification of ARID5A as a quiescence associated marker. J Cell Physiol. 2018;233:3695–712. doi: 10.1002/jcp.26227. [DOI] [PubMed] [Google Scholar]
- 53.Litwiniec A, Grzanka A, Helmin-Basa A, Gackowska L, Grzanka D. Features of senescence and cell death induced by doxorubicin in A549 cells: organization and level of selected cytoskeletal proteins. J Cancer Res Clin Oncol. 2010;136:717–36. doi: 10.1007/s00432-009-0711-4. [DOI] [PubMed] [Google Scholar]
- 54.Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le Toumelin-Braizat G, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 2016;538:477–82. doi: 10.1038/nature19830. [DOI] [PubMed] [Google Scholar]
- 55.Rastogi S, Joshi B, Dasgupta P, Morris M, Wright K, Chellappan S. Prohibitin facilitates cellular senescence by recruiting specific corepressors to inhibit E2F target genes. Mol Cell Biol. 2006;26:4161–71. doi: 10.1128/MCB.02142-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 57.Arya AK, El-Fert A, Devling T, Eccles RM, Aslam MA, Rubbi CP, et al. Nutlin-3, the small-molecule inhibitor of MDM2, promotes senescence and radiosensitises laryngeal carcinoma cells harbouring wild-type p53. Br J Cancer. 2010;103:186–95. doi: 10.1038/sj.bjc.6605739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roninson IB, Broude EV, Chang BD. If not apoptosis, then what? Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resist Updates. 2001;4:303–13. doi: 10.1054/drup.2001.0213. [DOI] [PubMed] [Google Scholar]
- 59.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee BY, Han JA, Im JS, Morrone A, Johung K, Goodwin EC, et al. Senescence-associated beta-galactosidase is lysosomal beta-galactosidase. Aging Cell. 2006;5:187–95. doi: 10.1111/j.1474-9726.2006.00199.x. [DOI] [PubMed] [Google Scholar]
- 61.Kurz DJ, Decary S, Hong Y, Erusalimsky JD. Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J Cell Sci. 2000;113:3613–22. doi: 10.1242/jcs.113.20.3613. [DOI] [PubMed] [Google Scholar]
- 62.Aubrey BJ, Kelly GL, Kueh AJ, Brennan MS, O’Connor L, Milla L, et al. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10:1422–32. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 63.Wei G, Margolin AA, Haery L, Brown E, Cucolo L, Julian B, et al. Chemical genomics identifies small-molecule MCL1 repressors and BCL-xL as a predictor of MCL1 dependency. Cancer Cell. 2012;21:547–62. doi: 10.1016/j.ccr.2012.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 65.Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294–305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shibue T, Takeda K, Oda E, Tanaka H, Murasawa H, Takaoka A, et al. Integral role of Noxa in p53-mediated apoptotic response. Genes Dev. 2003;17:2233–8. doi: 10.1101/gad.1103603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, et al. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science. 2003;302:1036–8. doi: 10.1126/science.1090072. [DOI] [PubMed] [Google Scholar]
- 68.Ohanna M, Cheli Y, Bonet C, Bonazzi VF, Allegra M, Giuliano S, et al. Secretome from senescent melanoma engages the STAT3 pathway to favor reprogramming of naive melanoma towards a tumor-initiating cell phenotype. Oncotarget. 2013;4:2212–24. doi: 10.18632/oncotarget.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jackson JG, Pereira-Smith OM. p53 is preferentially recruited to the promoters of growth arrest genes p21 and GADD45 during replicative senescence of normal human fibroblasts. Cancer Res. 2006;66:8356–60. doi: 10.1158/0008-5472.CAN-06-1752. [DOI] [PubMed] [Google Scholar]
- 70.Wilson WH, O’Connor OA, Czuczman MS, LaCasce AS, Gerecitano JF, Leonard JP, et al. Navitoclax, a targeted high-affinity inhibitor of BCL-2, in lymphoid malignancies: a phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet Oncol. 2010;11:1149–59. doi: 10.1016/S1470-2045(10)70261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Molyneux G, Andrews M, Sones W, York M, Barnett A, Quirk E, et al. Haemotoxicity of busulphan, doxorubicin, cisplatin and cyclophosphamide in the female BALB/c mouse using a brief regimen of drug administration. Cell Biol Toxicol. 2011;27:13–40. doi: 10.1007/s10565-010-9167-1. [DOI] [PubMed] [Google Scholar]
- 72.Demaria M, O’Leary MN, Chang J, Shao L, Liu S, Alimirah F, et al. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 2017;7:165–76. doi: 10.1158/2159-8290.CD-16-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guikema JE, Amiot M, Eldering E. Exploiting the pro-apoptotic function of NOXA as a therapeutic modality in cancer. Expert Opin Ther Targets. 2017;21:767–79. doi: 10.1080/14728222.2017.1349754. [DOI] [PubMed] [Google Scholar]
- 74.Opferman JT, Iwasaki H, Ong CC, Suh H, Mizuno S, Akashi K, et al. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science. 2005;307:1101–4. doi: 10.1126/science.1106114. [DOI] [PubMed] [Google Scholar]
- 75.Teh TC, Nguyen NY, Moujalled DM, Segal D, Pomilio G, Rijal S, et al. Enhancing venetoclax activity in acute myeloid leukemia by co-targeting MCL1. Leukemia. 2018;32:303–12. doi: 10.1038/leu.2017.243. [DOI] [PubMed] [Google Scholar]
- 76.Brown JM, Wilson G. Apoptosis genes and resistance to cancer therapy: what does the experimental and clinical data tell us? Cancer Biol Ther. 2003;2:477–90. doi: 10.4161/cbt.2.5.450. [DOI] [PubMed] [Google Scholar]
- 77.Brown JM, Wouters BG. Apoptosis, p53, and tumor cell sensitivity to anticancer agents. Cancer Res. 1999;59:1391–9. [PubMed] [Google Scholar]
- 78.Jackson JG, White MF, Yee D. Insulin receptor substrate-1 is the predominant signaling molecule activated by insulin-like growth factor-I, insulin, and interleukin-4 in estrogen receptor-positive human breast cancer cells. J Biol Chem. 1998;273:9994–10003. doi: 10.1074/jbc.273.16.9994. [DOI] [PubMed] [Google Scholar]
- 79.Twentyman PR, Luscombe M. A study of some variables in a tetrazolium dye (MTT) based assay for cell growth and chemosensitivity. Br J Cancer. 1987;56:279–85. doi: 10.1038/bjc.1987.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang P, Wang P, Liu B, Zhao J, Pang Q, Agrawal SG, et al. Dynamin-related protein Drp1 is required for Bax translocation to mitochondria in response to irradiation-induced apoptosis. Oncotarget. 2015;6:22598–612. doi: 10.18632/oncotarget.4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol. 2019;37:224–6. doi: 10.1038/s41587-019-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.