Abstract
Aberrant activation of Wnts is common in human cancers, including prostate. Hypermethylation associated transcriptional silencing of Wnt antagonist genes SFRPs (Secreted Frizzled-Related Proteins) is a frequent oncogenic event. The significance of this is not known in prostate cancer. The objectives of our study were to (i) profile Wnt signaling related gene expression and (ii) investigate methylation of Wnt antagonist genes in prostate cancer. Using TaqMan Low Density Arrays, we identified 15 Wnt signaling related genes with significantly altered expression in prostate cancer; the majority of which were upregulated in tumors. Notably, histologically benign tissue from men with prostate cancer appeared more similar to tumor (r = 0.76) than to benign prostatic hyperplasia (BPH; r = 0.57, p < 0.001). Overall, the expression profile was highly similar between tumors of high (≥ 7) and low (≤ 6) Gleason scores. Pharmacological demethylation of PC-3 cells with 5-Aza-CdR reactivated 39 genes (≥ 2-fold); 40% of which inhibit Wnt signaling. Methylation frequencies in prostate cancer were 10% (2/20) (SFRP1), 64.86% (48/74) (SFRP2), 0% (0/20) (SFRP4) and 60% (12/20) (SFRP5). SFRP2 methylation was detected at significantly lower frequencies in high-grade prostatic intraepithelial neoplasia (HGPIN; 30%, (6/20), p = 0.0096), tumor adjacent benign areas (8.82%, (7/69), p < 0.0001) and BPH (11.43% (4/35), p < 0.0001). The quantitative level of SFRP2 methylation (normalized index of methylation) was also significantly higher in tumors (116) than in the other samples (HGPIN = 7.45, HB = 0.47, and BPH = 0.12). We show that SFRP2 hypermethylation is a common event in prostate cancer. SFRP2 methylation in combination with other epigenetic markers may be a useful biomarker of prostate cancer.
Keywords: SFRP2, prostate cancer, hypermethylation, Wnt signaling
Prostate cancer is the most commonly diagnosed noncutaneous malignancy and third leading cause of cancer related deaths in men in the Western world.1 The heterogeneous nature of the disease results in a broad spectrum of clinical behavior from slow-growing indolent tumors to aggressive, metastatic disease. The Gleason grading system is a pathologic determinant of disease biology and prognosis, based on the glandular pattern of the tumor, serving as an independent prognostic factor. The predominant (primary) and next most prevalent (secondary) architectural patterns are graded from 1–5 (well differentiated–poorly differentiated) and summed, yielding an overall Gleason score, which is indicative of potential behavior, with Gleason score ≥7 predictive of a poor prognosis.2,3
Wnt (wingless-type)/β-catenin signaling is a major regulator of cell proliferation, migration and differentiation, controlling tissue homeostasis and tumor progression.4 The binding of a canonical Wnt ligand to its cell-surface receptor complex, consisting of Frizzled (FZD) and one of two low-density-lipoprotein-receptor-related proteins (LRP-5 and LRP-6), initiates a signaling cascade that activates disheveled (DVL), which releases β-catenin (CTTNB1) from an inhibitory complex consisting of Axin, APC and glycogen synthase kinase 3β (GSK3B). On dephosphorylation and release, β-catenin translocates to the nucleus, where it interacts with members of the T-cell factor/lymphoid enhancer factor (TCF/LEF) families of transcription factors to stimulate expression of genes involved in cell survival, proliferation and osteoblastic differentiation (e.g., MMPs, CCND1, PTGS2, MYC, JUN and VEGFR).5 Wnt signaling is regulated by several classes of negative regulators. The Secreted Frizzled-Related Protein (SFRP) class comprises SFRP1–SFRP5, Wnt inhibitory factor 1 (WIF1) and Cerberus. SFRPs are a family of soluble glycoproteins that possess a cysteine-rich domain (CRD) structurally similar to the extracellular Wnt-binding domain of the FZD receptors. SFRPs can thus modulate Wnt signaling by sequestering Wnts through their CRD or by acting as dominant-negative inhibitors, forming inactive complexes with the FZD receptors.6
Constitutive Wnt signaling is a feature of many human cancers.4 Several different mechanisms are involved, including loss of function mutations in the APC gene, stabilizing mutations or indirect activation of β-catenin and hypermethylation of Wnt antagonist genes.7,8 In prostate cancer, hyperactive Wnt signaling has been linked with androgen independent growth—through molecular-crosstalk with the Androgen Receptor,9 development of bone metastases10 and self-renewal of prostate cancer stem cells.11 However, the prevalence of APC and CTTNB1 mutations is low in prostate tumors, and the causes of hyperactive Wnt signaling remain poorly understood.
The heterogeneity of prostate cancer is reflected by a wide range of genetic and epigenetic abnormalities. Evidence indicates that an epigenetic “catastrophe” occurs during the earliest stages of prostate cancer development and is maintained in a clonal manner through metastatic progression.12 This catastrophe encompasses changes in DNA methylation through de novo promoter hypermethylation and concomitant silencing of tumor suppressor genes and genes with important regulatory functions; genome-wide hypomethylation resulting in genomic instability; and chromatin remodeling affecting the compaction of DNA into nucleosomes and its accessibility to the transcriptional apparatus.13 Promoter hypermethylation of GSTP1 (Glutathione S-transferase pi) is the most common somatically acquired genome alteration identified to date in prostate cancer. Glutathione S-transferase pi functions as an intracellular phase II drug metabolizing enzyme and is commonly overexpressed in human cancers. However, concomitant biallelic GSTP1 hypermethylation and loss of expression are widely reported in 75%–100% of prostate tumors and high-grade prostatic intraepithelial neoplasia (HGPIN) lesions.12,14,15
The primary objective of our study was to profile expression of canonical Wnt signaling genes in prostate cancer and evaluate a potential role for epigenetic gene silencing through promoter hypermethylation in hyperactivity of this pathway. A comprehensive expression analysis of Wnt signaling genes, including 19 Wnt ligands, 11 FZD and LRP receptors, ten soluble Wnt antagonists and four LEF/TCF transcription factors was performed in a panel of cell lines and tissue specimens. The effect of DNA methylation on gene expression was investigated by pharmacological demethylation. Tissue specimens from a range of disease states were used to represent the stepwise progression of prostate cancer, including benign prostatic hyperplasia (BPH), histologically benign epithelium adjacent to tumor (HB), preinvasive HGPIN and primary localized tumors categorized into low- and high-grade disease.
Material and Methods
Ethics statement
Ethical approval for our study was obtained from the ethics committee of St. James’s Hospital and The Adelaide and Meath incorporating the National Children’s Hospital. Ethical approval was granted for retrospective analysis of tissue specimens archived at the individual hospitals. Therefore, informed consent was not obtained from all participating patients.
Clinical specimens
Prostate tissue specimens (of tumor, HGPIN and histologically benign tissue) from men undergoing radical prostatectomy for primary prostate cancer were obtained through the histopathology archives dating from 1999–2008 at St. James’s Hospital and the Adelaide and Meath incorporating the National Children’s Hospital. Clinical and pathological data were obtained from the relevant patient records and are summarized in Table 1. For control purposes, BPH lesions were collected from 35 men with no diagnosis of prostate cancer who underwent transurethral resection of the prostate.
Table 1.
Clinicopathologic characteristics of the study population
| Prostate cancer cases | BPH cases | |
|---|---|---|
| Tissue specimens | 74 | 35 |
| Histologically benign | 69 | |
| HGPIN | 30 | |
| Age in years, mean (range) | 60.08 (44–79) | 75.41 (58–89) |
| Pre-op PSA ng/ml, mean (range) | 8.54 (0.7–37.3) | 6.39 (0.7–30) |
| <4 | 6 | 9 |
| 4–10 | 42 | 5 |
| >10 | 19 | 3 |
| Biochemical recurrence | ||
| No | 27 | |
| Yes | 2 | |
| Gleason score, n | ||
| ≤6 | 30 | |
| ≥7 | 44 | |
| TNM classification | ||
| pT2 | 54 | |
| pT3 | 19 | |
| pT4 | 2 |
PSA values were not available for all patients as there is no national PSA screening program in Ireland. Biochemical recurrence was defined as a single postprostatectomy PSA level > 0.2 ng/ml on two consecutive tests.
Suitable formalin fixed paraffin embedded (FFPE) blocks of each case representing as many Gleason grades as possible were selected. Corresponding Haematoxylin and Eosin (H&E) stained sections were reviewed and annotated by a pathologist to histologically identify areas of prostate cancer, HGPIN and HB tissue (not infiltrated with tumor or HGPIN).
Tissue microdissection
A total of 74 cases of prostate cancer and 35 cases of BPH were used for nucleic acid isolation. A series of 5 μm sections were cut from the FFPE blocks. The first and last sections were H&E stained and compared to the pathologically evaluated slides. Intervening sections were deparaffinized, and tissue was scraped from within the marked target areas. Genomic DNA and total RNA were extracted using a RecoverAll™ Total Nucleic Acid Isolation Kit (Ambion, Austin, TX).
Cell culture and pharmacological demethylation
Benign prostate cell lines PWR-1E and RWPE1 and four malignant prostate epithelial cell lines: primary androgen sensitive 22Rv1, metastatic androgen dependent LNCaP and metastatic androgen independent PC-3 and DU145 were obtained from the American Type Culture Collection and maintained under standard cell culture conditions. Human RC58/T prostate cancer cells were kindly provided by Prof. Rhim, Center for Prostate Disease Research, Uniformed Service University of Health Sciences, Bethesda, MD. PC-3 cells were treated with 1 μM 5-Aza-2′Deoxycytidine (5-Aza-CdR) at 48-hr intervals over a period of 1 week in induce global demethylation. Cells were harvested, and DNA and RNA were isolated using a QIAamp® DNA blood minikit and RNeasy kit, respectively (Qiagen, UK).
TaqMan low density array
The TaqMan low density array (TLDA) Human Wnt Gene set v1.0 microfluidic card (Applied Biosystems, Foster City, CA) was used to profile expression of Wnt signaling genes in cell lines and tissue specimens. The TLDA consisted of four identical 96-gene sets preconfigured in a 384-well format, including 19 Wnt ligands, 11 FZD and LRP receptors, ten soluble Wnt antagonists and four LEF/TCF transcription factors. The full list of genes configured on the customized TLDA is available through the Gene Expression Omnibus, accession number GSE33557 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33557).
To overcome the limited amount of RNA obtained from FFPE tissues, RNA samples were pooled. Four different pools were generated: prostate cancer, HGPIN, HB and BPH. Prostate cancer pools were further subdivided into Gleason score ≥ 7 and Gleason score ≤ 6. Each pool consisted of DNase-treated total RNA (100 ng), isolated from microdissected tissue from four individual cases, selected on the basis of similar histological and clinical features and previous epigenetic characterization in our laboratory.15 Biological replicates for each pool were prepared (Supporting Information Table 1). The high capacity cDNA Archive Kit (Applied Biosystems) was used to reverse transcribe pooled RNA (400 ng). TaqMan® reactions were performed in duplicate on a 7900HT Sequence Detection System.
Bisulfite modification and methylation specific PCR
Genomic DNA from cell lines (500 ng) and tissue specimens (50 ng) was bisulfite modified using the EZ DNA methylation™ kit (Zymo Research, Orange County, CA). The CpGenome Universal Methylated DNA and Unmethylated DNAs (Chemicon International, Temecula, CA) were used as positive controls. Promoter hypermethylation of the four SFRP genes was first analyzed by methylation specific PCR (MSP) in cell lines and in a subset of 20 tumors using the HotStar Taq PCR kit (Qiagen). PCR primer sets complementary to both modified, methylated DNA and modified, unmethylated DNA were designed for the four genes (Supporting Information Table 2).
Pyrosequencing
Bisulfite-treated DNA was amplified for 45 cycles with a biotinylated primer using the PyroMark PCR kit (Qiagen). PCR products were immobilized on Streptavidin Sepharose beads (GE Healthcare, UK) and pyrosequencing reactions were performed using the PyroMark Q24 System. Percentage methylation was calculated using the PSQ HS 96A 1.2 software, under the CpG mode. Primer sequences are listed in Supporting Information Table 2.
Quantitative methylation specific PCR
Quantitative methylation specific PCR (QMSP) was performed according to the method described by Eads et al.16 Bisulfite-treated DNA was amplified in parallel TaqMan® PCR reactions performed with oligonucleotides targeted to (i) endogenous control gene ACTB and (ii) target gene SFRP2. Samples were considered positively amplified when a comparative threshold cycle (CT) of < 50 was detected in all three replicates, with < 1 CT variance. A total of 10-fold serial dilutions of universal methylated DNA (Chemicon International, Temecula, CA) were used to generate a standard curve to quantify the amount of fully methylated SFRP2 in each reaction. A normalized index of methylation (NIM) was calculated, as previously described,12 to determine the ratio of the normalized amount of methylated SFRP2 to the normalized amount of ACTB in any given sample, by applying the formula:
where SFRP2sample is the quantity of fully methylated copies of SFRP2 in any individual sample, SFRP2MC is the quantity of fully methylated copies of SFRP2 in the methylated control DNA, ACTBsample is the quantity of bisulfite modified templates in any individual sample and ACTBMC is the quantity of bisulfite modified templates in the universally methylated control DNA.
Real-time Reverse Transcription PCR (RT-PCR)
DNase-treated total RNA (30 ng) was reverse transcribed using the high capacity cDNA Archive Kit and subjected to TaqMan® preamplification. Expression of SFRP2 was quantified in samples of tumor (n = 57), HGPIN (n = 18), HB (n = 42) and BPH (n = 24) by QRT-PCR using the TaqMan® gene expression Assay ID: Hs00293258_m1. Human phosphoglycerate kinase 1 (PGK1) was used as an endogenous control. TaqMan® PCR reactions were performed in triplicate on an ABI Prism 7900 Sequence Detection System. SFRP2 expression was calculated relative to the BPH cohort, using SDS RQ Manager 1.2 software, which automatically determined relative quantities (RQ), by applying the arithmetic formula 2−ΔΔCT. All equipment and reagents were supplied by Applied Biosystems, Foster City, CA.
Statistical analysis
RQ data from TLDAs were analyzed using Real Time StatMiner® v3.1 (Integromics, Granada, Spain). The optimal endogenous control was selected using the minimal variance of median algorithm. Differences in gene expression were assessed for significance using the Limma test and adjusted for a false discovery rate using the Benjamini–Hochberg method. Hierarchical clustering was performed by complete linkage analysis with similarity measured by Euclidean distance. Linear correlation between biological replicates was calculated by Spearman correlation coefficient (r) on normalized data (2−ΔCt), where ΔCt = Ct_target gene Ct_endogenous control (PGK1). A heat map was generated using the R statistical software package with the enhanced heat map function (heatmap.2), as part of the gplots package.17
Other statistical analysis was performed using MINITAB v15 (Minitab, UK). An unpaired t-test was used to calculate the differences in age and PSA between patient groups. Differences in methylation frequencies were assessed using Fisher’s exact test. Differences in SFRP2 methylation levels and gene expression were analyzed by examining NIM and RQ values, respectively, between samples using the Kruskal–Wallis one-way analysis of variance (ANOVA) test and the Spearman Rank correlation. For all tests, significance was ascribed at p < 0.05.
Results
Expression profiling of Wnt signaling genes in prostate tissue specimens and cell lines
To identify Wnt signaling genes that are dysregulated in prostate cancer, RNA was isolated from tumors (both low and high grades), adjacent HB areas and HGPIN, obtained from radical prostatectomy specimens and compared to BPH, obtained from men with no evidence of cancer and analyzed on a human Wnt signaling TLDA. The mean age of the BPH patient pool (69 years) was significantly greater than the tumor patient pool (mean age 61.24 years), (mean difference = 7.063, p = 0.015, 95% CI = 1.73, 12.40) and HB patient pool (mean age 59.13 years; mean difference = 9.875, p = 0.003, 95% CI = 3.91, 15.84). The mean PSA level of tumor pools (6.97 ng/ml) was significantly higher than BPH pools (3.78 ng/ml; mean difference = 3.451, p = 0.026, 95% CI = 6.74, 0.56). There was no marked difference in age or PSA levels between the high Gleason score and low Gleason score tumor pools (Supporting Information Table 1).
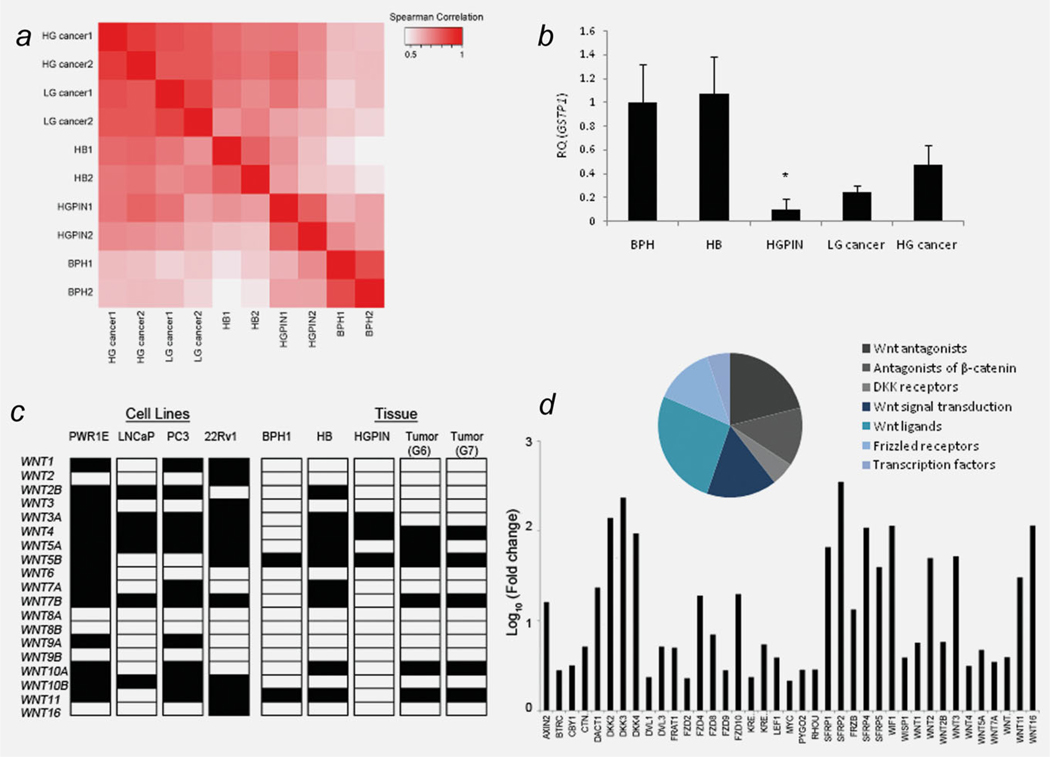
The performance of biological replicates of each pool was determined by measuring correlations in gene expression between the five sample types. Each pair of biological replicates yielded a correlation coefficient > 0.8 (based on normalized data (2−ΔCt) for all 96 genes), indicating similar patterns in gene expression for each biological group (Fig. 1a). High-grade (Gleason score ≥ 7) and low-grade (Gleason score ≤ 6) tumors appeared more similar to each other (r = 0.79) than to nonmalignant pools (r = 0.64; p = 0.007). Interestingly, HB pools from men with prostate cancer appeared more similar to the malignant pools (r = 0.72) than to BPH (r = 0.51; p = 0.004). The inclusion of GSTP1 on the TLDA served as an internal control to validate the purity of the biological groups. Both BPH and HB pools expressed GSTP1, whereas expression was lower, although not absent, in tumors and HGPIN (Fig. 1b). The TLDA data (normalized, non-normalized and fold-change) have been deposited in NCBI’s Gene Expression Omnibus18 and are accessible through GEO Series accession number GSE33557 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33557).
Figure 1.
Wnt signaling expression profiling in prostate cancer. (a) Heatmap providing visual summary of the Spearman correlation matrix of biological groups, calculated on normalized RT-PCR data. Each sample type (HB, BPH, high grade cancer (HGcancer), low grade cancer (LGcancer)) and HGPIN has two biological replicates. (b) Fold change in GSTP1 expression relative to BPH. *p < 0.05. (c) Expression of 19 human WNT genes in prostate cell lines and tissue specimens by qRT-PCR. Filled square indicates expression, open square indicates no expression. (d) Genes upregulated in PC-3 cells by pharmacological demethylation.
A total of 15 genes showed a significant change in expression between cancer and BPH; the majority of which were upregulated in tumors (Table 2). Only 6/19 known human Wnt ligands were expressed in prostate cancer, and only WNT4 was significantly upregulated in tumors (Fig. 1c). All 12 genes that were upregulated in tumors were concurrently upregulated in HB samples, and 7/12 were also upregulated in HGPIN (results not shown). Overall, the expression profile was highly similar between tumors of high (≥ 7) and low (≤ 6) Gleason scores. However, significant differences were observed for three targets; DACT1, BTRC and AXIN1 were only expressed in high Gleason scoring tumors (p = 0.003).
Table 2.
Differentially expressed Wnt signaling genes between prostate cancer and BPH
| Gene | Description | Function | Fold change | p value* |
|---|---|---|---|---|
| Upregulated | ||||
| FZD4 | Frizzled homolog 4 | Wnt coreceptor | 54.68 | 0.0007 |
| WNT4 | Wingless-type MMTV integration site family, member 4 | Noncanonical WNT signaling; embryogenesis | 18.82 | 0.0092 |
| LEF1 | Lymphoid enhancer-binding factor 1 | Transcription factor in the Wnt pathway | 14.82 | 0.0071 |
| LRP5 | Low density lipoprotein receptor-related protein 5 | Wnt coreceptor | 11.85 | 0.0071 |
| NKD1 | Naked cuticle homolog 1 | Antagonist of Wnt signaling; binds DVL enabling GSK3B-dependent phosphorylation and ubiquitination of β-catenin | 11.79 | 0.0071 |
| CSNK2A2 | Casein kinase 2, alpha prime polypeptide | Cytoplasmic and nuclear stimulator of Wnt signaling; suppresses β-catenin degradation, enhances β-catenin:LEF1 transactivation of Wnt target genes | 11.20 | 0.0012 |
| SFRP4 | Secreted frizzled-related protein 4 | Secreted Wnt antagonist. Suppresses both canonical and noncanonical signaling | 9.28 | 0.0071 |
| FZD6 | Frizzled homolog 6 | Wnt coreceptor | 8.73 | 0.0305 |
| AXIN2 | Axin 2 | Antagonist of Wnt signaling; sequesters β-catenin in a multiprotein destruction complex | 6.69 | 0.0449 |
| DKK3 | Dikkopf homolog 3 | Secreted Wnt antagonist. Divergent member of DKK family; does not appear to function directly in Wnt signaling | 5.15 | 0.0508 |
| APC | Adenomatosis polyposis coli | Antagonist of Wnt signaling; sequesters β-catenin in a multiprotein complex | 4.76 | 0.0092 |
| GSK3β | Glycogen synthase kinase 3 beta | Antagonist of Wnt signaling; Phosphorylates β-catenin for proteasomal degradation | 3.77 | 0.0217 |
| DVL3 | Disheveled, dsh homolog 3 | Phosphoprotein, stimulates Wnt signaling; inhibits activity of GSK3β | 2.95 | 0.0217 |
| Downregulated | ||||
| FZD1 | Frizzled homolog 1 | Wnt receptor | 6.56 | 0.0449 |
| FZD8 | Frizzled homolog 8 | Wnt receptor | 4.15 | 0.0476 |
p value is adjusted for false discovery. Only those genes with a significant change in expression are listed in this table.
Wnt expression profiling was also performed in three prostate cancer cell lines, LNCaP, 22Rv1 and PC-3 and in normal prostate epithelial cell line PWR1E. Overall, the majority of targets showed downregulation relative to PWR1E. Pharmacological demethylation by 5-Aza-CdR reactivated 39 targets (≥ 2-fold) relative to untreated PC-3 cells, including eight soluble Wnt antagonists, five FZD receptors and ten Wnt ligands (Fig. 1d). Almost 40% of reactivated genes function as negative inhibitors of Wnt signaling and included all five genes.
Finally, we analyzed the expression profile of ten soluble Wnt antagonists. In the SFRP class, SFRP4 was significantly upregulated in both low and high Gleason scores and in all the tumor cell lines (Table 2). Interestingly, SFRP1 and WIF1 were significantly upregulated in HB samples (9-fold (p = 0.0427) and 15-fold (p = 0.0112), respectively), but with a moderate (< 3-fold), yet insignificant increase in tumor specimens, and expressed at very low levels or not detected in tumor cell lines. In the DKK class, the only significant change in expression was observed for DKK3, which was upregulated 32-fold in HB specimens (p < 0.0004) and 5-fold in tumors (p = 0.05) but was expressed at very low levels in all of the cell lines.
Promoter hypermethylation of SFRP2 and SFRP5 in prostate cancer
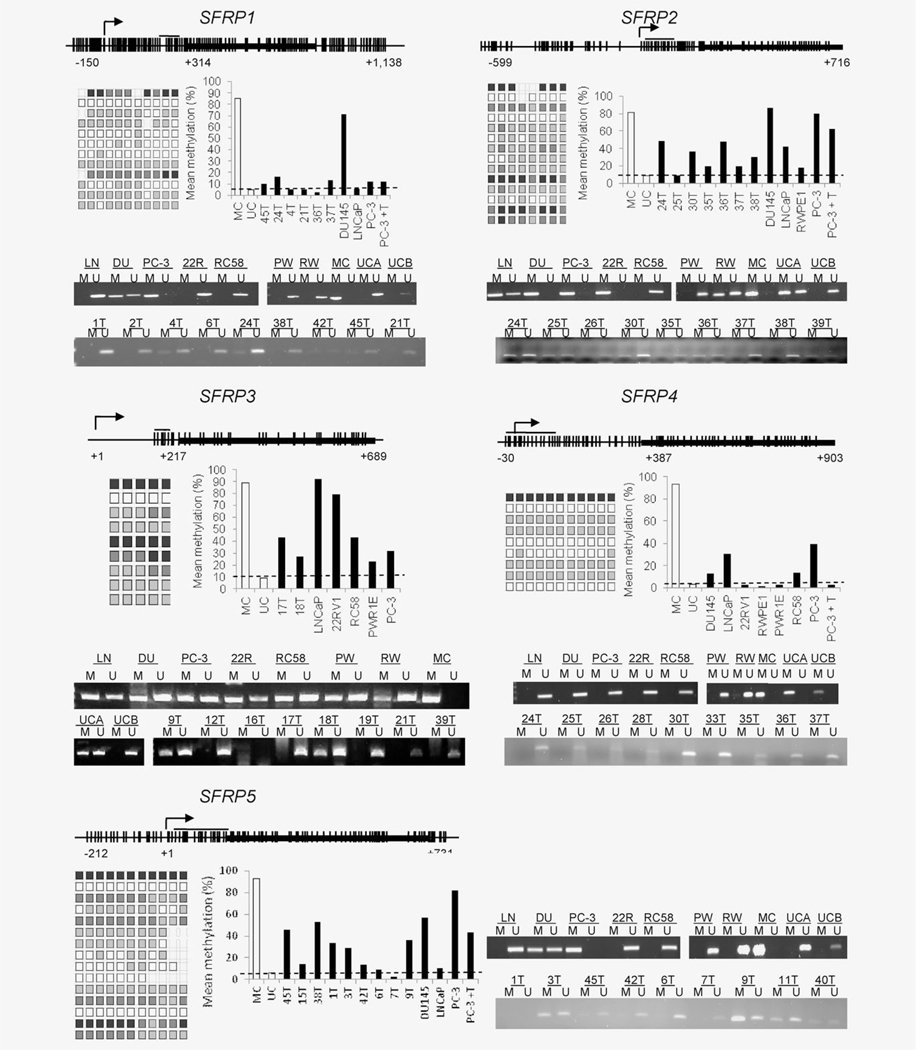
Based on the findings from the expression profiling and the evidence of tumor-associated hypermethylation of the SFRP genes in other cancers, we decided to test for hypermethylation of the SFRP genes in prostate cancer. MSP performed on a test set of 20 tumors and seven cell lines revealed frequent hypermethylation of SFRP2, SFRP3 and SFRP5. SFRP2 was methylated in 14/20 (70%) tumors, 4/5 prostate cancer cell lines (LNCaP, PC-3, DU145 and 22Rv1) and in one of the two benign cell lines (RWPE1). Similarly, SFRP5 appeared methylated in 12 (60%) tumors and in DU145 and PC-3 cell lines. Methylation of SFRP3 was detected in 6 (30%) tumors and in all the cell lines tested, including both benign and malignant. SFRP1 methylation was only detected in two tumors (20%) and in DU145 and PC-3 cells. All tumors and cell lines were negative for SFRP4 methylation. 5-Aza-CdR treatment of PC-3 cells induced partial demethylation of SFRP1, SFRP2 and SFRP5. Representative MSPs are shown in Figure 2. Pyrosequencing confirmed the MSP results, revealing moderate levels of methylation for SFRP2 (mean % methylation = 41.64), SFRP3 (mean % methylation = 48.42), SFRP5 (mean % methylation = 33) and lower levels for SFRP1 and SFRP4 (mean % methylation = 15 and 13.29, respectively; Fig. 2). Moderate levels of SFRP4 methylation were detected in LNCaP and PC3 cells, which were not apparent by MSP.
Figure 2.
Promoter hypermethylation of SFRP genes in prostate cancer. MSP and pyrosequencing of prostate cancer cell lines and tumors. Representative CpG islands are shown for SFRP1, SFRP2, SFRP3, SFRP4 and SFRP5. Individual vertical lines indicate the presence of a CpG dinucleotide and the location of sequenced CpG sites is indicated by a horizontal line. The transcriptional start site is shown by an arrow, and the first coding exon is illustrated by a thick black box. Each square represents the amount of methylation at a given CpG site, shaded from white (unmethylated) to black (fully methylated). Each row depicts the sequencing pattern for an individual sample, summarized in the bar charts. Dashed lines in the bar charts represent the mean level of methylation detected in an unmethylated sample. Abbreviations: PC-3 cells treated with 5AzaCdR are shown by PC-3+T, LNCaP: LN, DU145: DU, 22Rv1: 22R, PWR1E: PW, RWPE1: RW, MC: universal human methylated DNA, UCA and UCB: universal unmethylated DNA from human genomic DNA and a human fetal cell line, respectively.
Hypermethylation of SFRP2 occurs at significantly higher levels in prostate cancer than HB tissue
Based on the preliminary findings of SFRP2 methylation in prostate cancer by MSP and pyrosequencing, we next quantified methylation using QMSP in 74 cases of organ-confined prostate cancer, 69 samples of adjacent HB tissue, 20 HGPIN lesions and for control purposes, 35 men with BPH and no evidence of prostate cancer (Table 1). Notably, the mean PSA level of the BPH patients (6.39 ng/ml) was above 4.0 ng/ml, the widely accepted upper limit of normal and was only marginally less than that of the prostate cancer patients (8.54 ng/ml; mean difference = 2.156, p = 0.283, 95% CI = −1.93, 6.24). As expected, men with BPH were significantly older (mean age 75.41 years) than prostate cancer patients (mean age 60.08 years; mean difference 15.324, p < 0.0001, 95% CI = 18.54, 12.11), although there was considerable overlap in the age range.
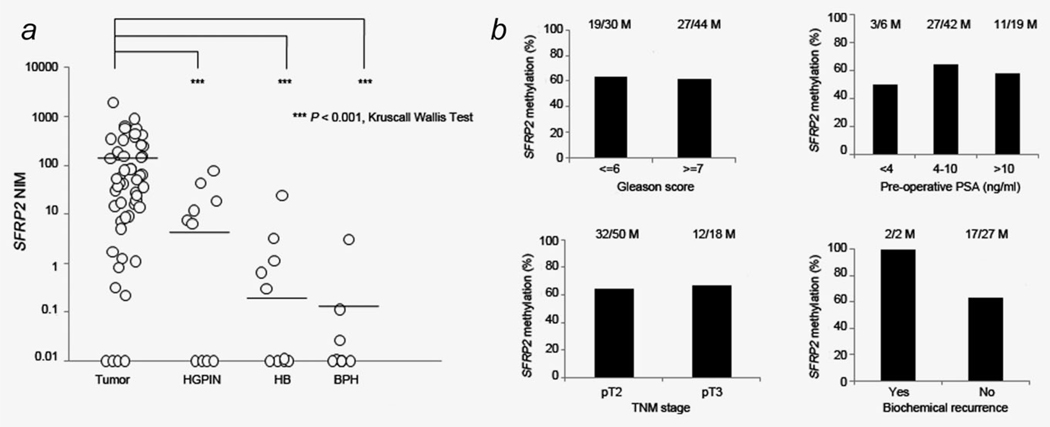
SFRP2 methylation was detected in 64.86% (48/74) of tumors, which was significantly higher than the frequency observed for HGPIN (30%, (6/20), p = 0.0096, 95% CI = 1.08, 4.31), tumor adjacent HB areas (8.82%, (7/69), p < 0.0001, 95% CI = 3.11, 13.16) and BPH (11.43%, (4/35), p < 0.0001, 95% CI = 2.22, 14.50). In addition, the quantitative level of methylation (NIM) was significantly higher in tumors (116) than in the other sample types (HGPIN = 7.45, HB = 0.47 and BPH = 0.12) (Fig. 3a).
Figure 3.
Hypermethylation of SFRP2 in prostate cancer. (a) Quantitative methylation analysis of SFRP2 in prostate. Values diagrammed at 0.01 represent zero methylation. (b) Association of SFRP2 methylation by clinicopathologic factors. M indicates number of samples that showed SFRP2 methylation within each clinicopathologic group.
Hypermethylation was not significantly associated with Gleason score ≥ 7, preoperative PSA level, TNM stage or biochemical recurrence (BCR) (Fig. 3b). Five-year postoperative follow-up data were available for 29 patients. Two patients died from metastatic prostate cancer, and one man died from another cause. Both patients who suffered BCR had SFRP methylation detected in their radical prostatectomy specimen.
Discussion
In our study, we performed gene expression analyses of Wnt signaling across a spectrum of prostate disease states and cell lines and identified key (epi)genetic aberrations involved in Wnt hyperactivity in prostate carcinogenesis.
Fifteen Wnt signaling genes were significantly altered in prostate cancer compared to BPH; the majority of which were upregulated. This is the first report of aberrant expression of FZD receptors FZD1 and FZD8 (elevated) and FZD4 and FZD6 (downregulated) along with coreceptor LRP5 (elevated). The WNT4 ligand showed an almost 19-fold induction. WNT4 activates noncanonical Wnt pathways, specifically in embryogenesis19 and is overexpressed in other human cancers,20 suggesting that upregulation of embryonic growth-promoting ligands may be an example of oncogenesis-recapitulating-ontogenesis to drive growth and proliferation. DVL3 (upregulated ~ 3-fold) is one of three mammalian DVL homologues and functions early in the cascade by inhibiting GSK3B activity, thus stimulating canonical Wnt signaling. Elevated levels of DVL3 have also been reported in nonsmall cell lung carcinoma.21 CSNK2A2 (overexpressed 11-fold) is a highly conserved and ubiquitous serine/threonine kinase, over-expressed in many tumors including prostate.22 Cytoplasmic CSNK2A2 opposes the inhibitory role of GSK3B by phosphorylating and activating DVL and β-catenin, while nuclear CSNK2A2 enhances β-catenin:LEF1 transactivation of Wnt target genes.23 Its nuclear localization was previously reported as a poor prognostic indicator in prostate cancer,24 potentially through its promotion of AR-dependent transcription.25 Downstream component of the pathway, transcription factor LEF1 was also significantly over-expressed and has recently shown to promote AR expression and consequently enhance growth and invasion ability of prostate cancer cells in vitro.26
Several components of the “destruction complex” consisting of GSK3B, APC, AXIN and casein kinase were also elevated in tumors and cell lines. GSK3B has also been shown to suppress AR-mediated transactivation and cell growth.27 AXIN2 is transcriptionally activated through Wnt signaling,28 thus forming a useful measure of pathway activity. It acts in a negative feedback loop by directing β-catenin for degradation through ubiquitin-dependent proteolysis and has previously been reported as upregulated in prostate cancer cell lines.29 Other Wnt signaling inhibitors were also overexpressed in tumors (SFRP4, DKK3 and NKD1). Overexpression of membranous SFRP4 in primary, androgen-dependent prostate cancer is reported as a good prognostic indicator. Functionally, it has been demonstrated to reduce proliferation, anchorage-dependent growth and invasiveness of prostate cancer cell lines.30 DKK3 is a proposed tumor suppressor and a divergent member of the dikkopf family, which does not appear to function directly in Wnt signaling and is frequently downregulated in human cancers.31 NKD1 binds to and inhibits the DVL family of scaffolding proteins, thus enabling GSK3B-dependent phosphorylation and ubiquitination of β-catenin.
Our finding that the expression profile of HB tissue was more similar to cancer than BPH strongly indicates that benign tissue adjacent/nearby tumor harbors molecular aberrations despite appearing histologically “normal.” In turn, this supports the theory of a “cancer field effect” in the prostate gland.32 Furthermore, our finding that expression of GSTP1 was high in HB tissue pools and low in tumor and HGPIN samples is in agreement with previous studies suggesting that that certain (epi)genetic targets may evade field cancerization.33,34 Alternatively, because we did not isolate pure epithelial populations through laser capture microdissection, the gene expression profiles of the biological pools may be influenced by fundamental differences between cells of epithelial versus mesenchymal lineage and represent differences in tumor-stroma content between benign and tumor tissue.
Among the genes that were differentially expressed, we selected the SFRP family for further analysis because they have previously been demonstrated to be hypermethylated in several cancer types, with only limited data available for prostate.7,35 Pyrosequencing and MSP data suggested that further investigation into methylation of SFRP3 and SFRP5 in prostate cancer was warranted. This is the first report of the methylation status and expression of the SFRP2 gene across a spectrum of prostate disease states. Promoter hypermethylation within the 5′UTR of SFRP2 was detected at a significantly higher frequency and quantitatively higher level in tumors than benign tissue.
Loss of SFRP2 in other human cancers has been largely attributed to promoter hypermethylation.36,37 However, TLDAs performed on pooled mRNAs and QRT-PCR on individual tissues (data not shown) both revealed widespread SFRP2 gene expression in benign, preinvasive and malignant glands. In contrast, SFRP2 mRNA transcripts were not detected in any of the prostate cancer cell lines, and treatment with 5-Aza-CdR had varying effects; only reactivating expression in androgen independent DU145 and PC-3 cells but not in androgen dependent LNCaP or androgen sensitive 22Rv1 cells (results not shown). We also performed immunohistochemical analysis of SFRP2 in tissue microarrays, which revealed strong cytoplasmic expression in benign prostate epithelia and negative/weak staining in the majority of prostate tumors.38
These findings suggest a complex mode of SFRP2 dysregulation during prostate carcinogenesis, in which loss of SFRP2 may largely occur through post-transcriptional mechanisms, underpinned by aberrant epigenetic modifications effective under certain environmental stimuli. Alternatively, it is possible that these results are influenced by aberrant SFRP2 expression in associated tumor-stroma. Immunohistochemical analysis of SFRP2 revealed no staining in prostate fibroblasts (both benign and malignant).38 However, this does not rule out the possibility of SFRP2 gene expression in these stomal cells. Whilst prostate adenocarcinomas are epithelial in origin, stromal cells adjacent to tumor epithelium are phenotypically different from stromal cells in benign glands.39 There is evidence that paracrine signals from prostatic tumor stroma can cause a phenotypic progression from a nontumorigenic to a tumorigenic state in adjacent epithelial cells.40 The molecular basis for the aberrant cell–cell communication between the tumor epithelium and tumor stroma is not well defined in prostate cancer. There is in fact precedent for involvement of the SFRP family in cellular crosstalk between the tumor epithelium and tumor mesenchyme. We and others have found that SFRP1 is expressed at low levels in the normal adult prostate.41 Ectopic expression of SFRP1 led to increased proliferation, reduced apoptosis and reduced Wnt/β-catenin signaling in a prostate cancer cell line model and increased proliferation in vivo. Elevated levels of SFRP1 are present in tumor stroma, which may provide a pro-proliferative paracrine signal to adjacent epithelial cells.41 In addition, SFRP3 has previously been shown to exhibit antitumor activity through the reversal of epithelial to mesenchymal transition and inhibition of matrix metalloproteinase activities in a subset of androgen independent prostate cancers.42
SFRPs have generally been thought to behave as tumor suppressors and are silenced by promoter hypermethylation in many cancers.43,44 SFRP2 has been shown to suppress transformation and invasion abilities of cervical cancer cells45 and has growth inhibitory effects in breast cancer.46 However, recent reports also indicate Wnt-activating properties for SFRPs in certain human tumors44 and Wnt-independent activities, including modulating the transcriptional activity of the androgen receptor.47,48 Our findings show that further investigation into the functional significance of SFRP2 promoter methylation on Wnt/β-catenin signaling in prostate cancer is warranted.
There is an urgent need for better prostate cancer-specific biomarkers that can distinguish between indolent and aggressive disease and avoid the over-treatment of clinically insignificant tumors. DNA methylation has several attributes, which make it an attractive avenue of biomarker research.49 We have shown that SFRP2 hypermethylation is a frequent aberration in prostate cancer, and its inclusion in an epigenetic biomarker panel for prostate cancer detection and prognosis is warranted.
Supplementary Material
What’s new?
Aberrant Wnt signaling is a characteristic of many cancers, but its causes are poorly understood. In a comprehensive gene expression and DNA methylation study, the authors identify a set of Wnt-related genes that are dysregulated in prostate cancer. They further demonstrate frequent hypermethylation of the SFRP family of Wnt antagonist genes, especially SFRP2, in tumors and pre-invasive lesions. These results bring new insight into the causes of dysregulated Wnt signaling and point toSFRP2 methylation as a novel biomarker in prostate cancer.
Acknowledgements
The authors thank Professor Ken Mills, Queen’s University Belfast and The Epigenetics Group, Department of Surgery and Cancer, Imperial College London for the use of their PyroMark Q24 systems.
Grant sponsors: the Irish Cancer Society, the Prostate Cancer Foundation
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin 2009; 59:225–49. [DOI] [PubMed] [Google Scholar]
- 2.Epstein JI, Partin AW, Sauvageot J, et al. Prediction of progression following radical prostatectomy. A multivariate analysis of 721 men with long-term follow-up. Am J Surg Pathol 1996; 20:286–92. [DOI] [PubMed] [Google Scholar]
- 3.Tabesh A, Teverovskiy M, Pang HY, et al. Multifeature prostate cancer diagnosis and Gleason grading of histological images. IEEE Trans Med Imaging 2007;26:1366–78. [DOI] [PubMed] [Google Scholar]
- 4.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer 2008;8:387–98. [DOI] [PubMed] [Google Scholar]
- 5.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature 2005;434:843–50. [DOI] [PubMed] [Google Scholar]
- 6.Lavergne E, Hendaoui I, Coulouarn C, et al. Blocking Wnt signaling by SFRP-like molecules inhibits in vivo cell proliferation and tumor growth in cells carrying active beta-catenin. Oncogene 2011;30:423–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki H, Toyota M, Carraway H, et al. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer 2008;98:1147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urakami S, Shiina H, Enokida H, et al. Combination analysis of hypermethylated Wnt-antagonist family genes as a novel epigenetic biomarker panel for bladder cancer detection. Clin Cancer Res 2006;12:2109–16. [DOI] [PubMed] [Google Scholar]
- 9.Wang G, Wang J, Sadar MD. Crosstalk between the androgen receptor and beta-catenin in castrate-resistant prostate cancer. Cancer Res 2008;68:9918–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai J, Hall CL, Escara-Wilke J, et al. Prostate cancer induces bone metastasis through Wnt-induced bone morphogenetic protein-dependent and independent mechanisms. Cancer Res 2008; 68:5785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bisson I, Prowse DM. WNT signaling regulates self-renewal and differentiation of prostate cancer cells with stem cell characteristics. Cell Res 2009; 19:683–97. [DOI] [PubMed] [Google Scholar]
- 12.Yegnasubramanian S, Kowalski J, Gonzalgo ML, et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res 2004;64:1975–86. [DOI] [PubMed] [Google Scholar]
- 13.Perry AS, Watson RW, Lawler M, et al. The epigenome as a therapeutic target in prostate cancer. Nat Rev Urol 2010;7:668–80. [DOI] [PubMed] [Google Scholar]
- 14.Henrique R, Jeronimo C, Teixeira MR, et al. Epigenetic heterogeneity of high-grade prostatic intraepithelial neoplasia: clues for clonal progression in prostate carcinogenesis. Mol Cancer Res 2006;4:1–8. [DOI] [PubMed] [Google Scholar]
- 15.Perry AS, Loftus B, Moroose R, et al. In silico mining identifies IGFBP3 as a novel target of methylation in prostate cancer. Br J Cancer 2007; 96:1587–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 2000;28:E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Team RdC. R: A language and environment for statistical computing R Foundation for Statistical Computing, 2012. [Google Scholar]
- 18.Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res 2011;39:D1005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurus D, Heligon C, Burger-Schwarzler A, et al. Noncanonical Wnt-4 signaling and EAF2 are required for eye development in Xenopus laevis. EMBO J 2005;24:1181–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garnis C, Campbell J, Davies JJ, et al. Involvement of multiple developmental genes on chromosome 1p in lung tumorigenesis. Hum Mol Genet 2005;14:475–82. [DOI] [PubMed] [Google Scholar]
- 21.Wei Q, Zhao Y, Yang ZQ, et al. Dishevelled family proteins are expressed in non-small cell lung cancer and function differentially on tumor progression. Lung Cancer 2008;62:181–92. [DOI] [PubMed] [Google Scholar]
- 22.Wang G, Ahmad KA, Harris NH, et al. Impact of protein kinase CK2 on inhibitor of apoptosis proteins in prostate cancer cells. Mol Cell Biochem 2008;316:91–7. [DOI] [PubMed] [Google Scholar]
- 23.Song DH, Sussman DJ, Seldin DC. Endogenous protein kinase CK2 participates in Wnt signaling in mammary epithelial cells. J Biol Chem 2000; 275:23790–7. [DOI] [PubMed] [Google Scholar]
- 24.Laramas M, Pasquier D, Filhol O, et al. Nuclear localization of protein kinase CK2 catalytic subunit (CK2alpha) is associated with poor prognostic factors in human prostate cancer. Eur J Cancer 2007;43:928–34. [DOI] [PubMed] [Google Scholar]
- 25.Gotz C, Bachmann C, Montenarh M. Inhibition of protein kinase CK2 leads to a modulation of androgen receptor dependent transcription in prostate cancer cells. Prostate 2007;67: 125–34. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Wang L, Zhang M, et al. LEF1 in androgen-independent prostate cancer: regulation of androgen receptor expression, prostate cancer growth, and invasion. Cancer Res 2009;69:3332–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Lin HK, Hu YC, et al. Suppression of androgen receptor-mediated transactivation and cell growth by the glycogen synthase kinase 3 beta in prostate cells. J Biol Chem 2004;279: 32444–52. [DOI] [PubMed] [Google Scholar]
- 28.Jho EH, Zhang T, Domon C, et al. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol 2002;22: 1172–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang BE, Wang XD, Ernst JA, et al. Regulation of epithelial branching morphogenesis and cancer cell growth of the prostate by Wnt signaling. PLoS One 2008;3:e2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horvath LG, Lelliott JE, Kench JG, et al. Secreted frizzled-related protein 4 inhibits proliferation and metastatic potential in prostate cancer. Prostate 2007;67:1081–90. [DOI] [PubMed] [Google Scholar]
- 31.Niehrs C Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene 2006;25:7469–81. [DOI] [PubMed] [Google Scholar]
- 32.Nonn L, Ananthanarayanan V, Gann PH. Evidence for field cancerization of the prostate. Prostate 2009;69:1470–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cookson MS, Reuter VE, Linkov I, et al. Glutathione S-transferase PI (GST-pi) class expression by immunohistochemistry in benign and malignant prostate tissue. J Urol 1997;157:673–6. [PubMed] [Google Scholar]
- 34.Mehrotra J, Varde S, Wang H, et al. Quantitative, spatial resolution of the epigenetic field effect in prostate cancer. Prostate 2008;68:152–60. [DOI] [PubMed] [Google Scholar]
- 35.Lodygin D, Epanchintsev A, Menssen A, et al. Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res 2005;65:4218–27. [DOI] [PubMed] [Google Scholar]
- 36.Kawamoto K, Hirata H, Kikuno N, et al. DNA methylation and histone modifications cause silencing of Wnt antagonist gene in human renal cell carcinoma cell lines. Int J Cancer 2008;123: 535–42. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki H, Watkins DN, Jair KW, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet 2004;36:417–22. [DOI] [PubMed] [Google Scholar]
- 38.O’Hurley G, Perry AS, O’Grady A, et al. The role of secreted frizzled-related protein 2 expression in prostate cancer. Histopathology 2011;59:1240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayward SW, Rosen MA, Cunha GR. Stromal-epithelial interactions in the normal and neoplastic prostate. Br J Urol 1997;79(Suppl 2): 18–26. [DOI] [PubMed] [Google Scholar]
- 40.Olumi AF, Grossfeld GD, Hayward SW, et al. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res 1999;59:5002–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joesting MS, Perrin S, Elenbaas B, et al. Identification of SFRP1 as a candidate mediator of stromal-to-epithelial signaling in prostate cancer. Cancer Res 2005;65:10423–30. [DOI] [PubMed] [Google Scholar]
- 42.Zi X, Guo Y, Simoneau AR, et al. Expression of Frzb/secreted Frizzled-related protein 3, a secreted Wnt antagonist, in human androgen-independent prostate cancer PC-3 cells suppresses tumor growth and cellular invasiveness. Cancer Res 2005;65:9762–70. [DOI] [PubMed] [Google Scholar]
- 43.Kongkham PN, Northcott PA, Croul SE, et al. The SFRP family of WNT inhibitors function as novel tumor suppressor genes epigenetically silenced in medulloblastoma. Oncogene 2010;29: 3017–24. [DOI] [PubMed] [Google Scholar]
- 44.Kawakami K, Yamamura S, Hirata H, et al. Secreted frizzled-related protein-5 is epigenetically downregulated and functions as a tumor suppressor in kidney cancer. Int J Cancer 2011; 128:541–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung MT, Lai HC, Sytwu HK, et al. SFRP1 and SFRP2 suppress the transformation and invasion abilities of cervical cancer cells through Wnt signal pathway. Gynecol Oncol 2009;112: 646–53. [DOI] [PubMed] [Google Scholar]
- 46.Veeck J, Noetzel E, Bektas N, et al. Promoter hypermethylation of the SFRP2 gene is a high-frequent alteration and tumor-specific epigenetic marker in human breast cancer. Mol Cancer 2008;7:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bovolenta P, Esteve P, Ruiz JM, et al. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J Cell Sci 2008;121:737–46. [DOI] [PubMed] [Google Scholar]
- 48.Kawano Y, Diez S, Uysal-Onganer P, et al. Secreted Frizzled-related protein-1 is a negative regulator of androgen receptor activity in prostate cancer. Br J Cancer 2009;100:1165–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perry AS, Foley R, Woodson K, et al. The emerging roles of DNA methylation in the clinical management of prostate cancer. Endocr Relat Cancer 2006;13:357–77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.