Graphical abstract
Key Words: cardio-oncology, epigenomics, immunolomics, metabolomics, proteomics, transcriptomics
Abbreviations and Acronyms: BNP, B-type natriuretic peptides; CHIP, clonal hematopoiesis of indeterminate potential; cTn, cardiac troponin; CyTOF, mass cytometry by time-of-flight; DCM, dilated cardiomyopathy; GWAS, genome-wide association study; iPSC, induced pluripotent stem cells; lncRNA, long non-coding RNA; miRNA, microRNA; scRNA-seq, single-cell RNA-sequencing
Remarkable progress in cancer treatments has resulted in improved long-term survival. However, cardiovascular complications related to these therapies may result in treatment interruptions and worse oncologic and cardiovascular outcomes. Early diagnosis and management of cancer therapy cardiotoxicity are therefore critical for safe and effective cancer treatment and long-term survival. In this population, established clinical biomarkers such as cardiac troponin (cTn) and natriuretic peptides (B-type natriuretic peptide [BNP], and N-terminal pro–B-type natriuretic peptide [NT-proBNP]) have shown promise, but studies have been limited by variable sensitivity, relatively small study populations with low cardiotoxicity event rates, and inconsistencies in cardiotoxicity definitions and timing of biomarker ascertainment. Additionally, these biomarkers are not specific to drug-induced toxicity. Thus, there is a need to discover novel biomarkers to accurately identify at-risk populations, diagnose toxicity early, monitor disease course, and guide therapies, enabling the implementation of precision cardio-oncology. This paper provides a brief overview of the future of biomarker discovery in cardiotoxicity, translating novel omics technologies to the clinic.
Biomarker Sources and Their Comparative Properties
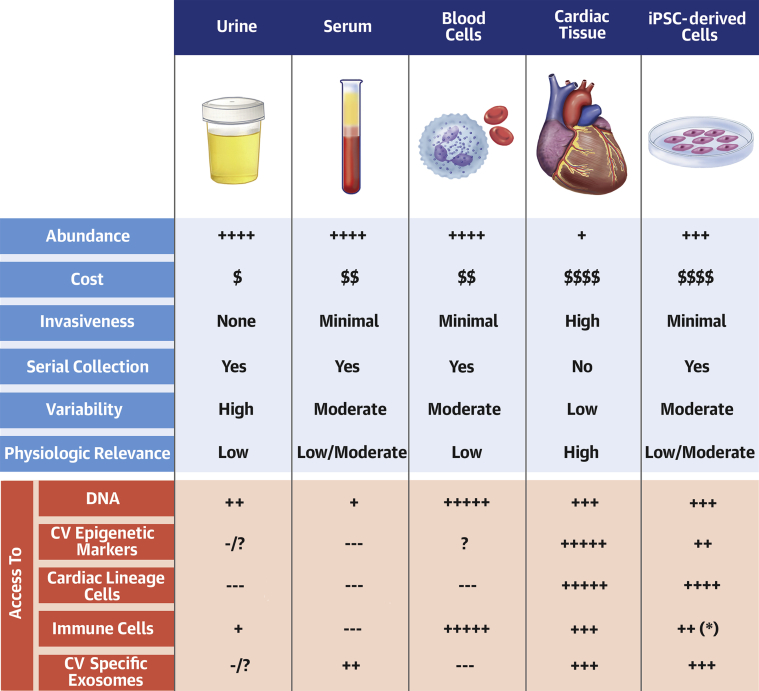
Biomarkers can be obtained from any products or parts of the body, including urine, blood, and tissues. Human-induced pluripotent stem cells (iPSCs) can be generated from patients’ somatic cells such as peripheral blood mononuclear cells and differentiated into relevant cardiovascular or immune cell types, providing a novel source of biomarkers. Figure 1 lists common sources of biomarkers and their properties. Briefly, urine and blood are abundant, noninvasive, and easy to collect. Blood cells provide an excellent source of genomic materials and immune phenotyping. However, biomarkers from those sources may lack specificity for cardiac pathology and suffer from wide variability and poor reproducibility due, in part, to the multiorgan effects of cancer treatments. Biomarkers obtained directly from cardiac tissue provide tissue-specific and physiologic information; however, those samples are often difficult to obtain due to procedural risk and cost. iPSC-derived cardiovascular cells are cardiac-specific, albeit immature. They can be cultured in a dish with a nearly limitless supply of cells and allow serial collection without requiring invasive procedures or additional clinical sampling. They retain patient-specific genetic information and enable personalized screening. However, reprogramming and maintenance of iPSCs are costly procedures and are labor intensive, and iPSC-derived cells alone lack environmental and physiologic relevance (1).
Figure 1.
Common Sources of Biomarkers and Their Properties
Common sources of biomarkers include urine, blood (serum and blood cells), cardiac tissues, and iPSC-derived cardiac lineage cells (e.g., cardiomyocytes, endothelial cells, vascular smooth muscle cells, fibroblasts) and iPSC-derived immune cells such as T cells and lymphocytes (∗). Their comparative strengths and weaknesses to identify clinically useful biomarkers are summarized. CV = cardiovascular; iPSC = induced pluripotent stem cells.
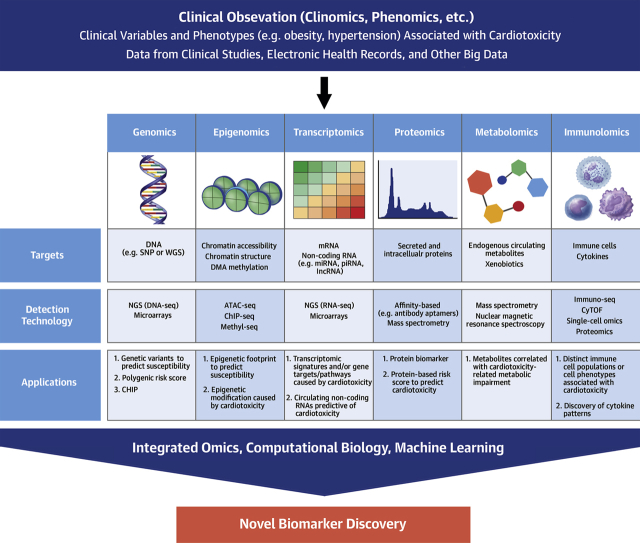
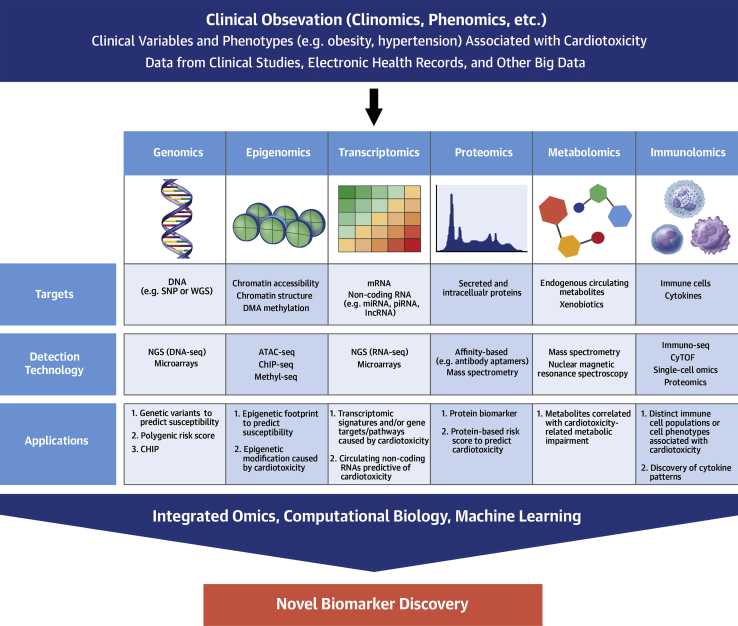
Omics Approaches For Novel Biomarker Discovery
Omics technologies allow high-throughput generation of large amounts of data for a specific molecular type such as DNA, proteins, and metabolites (Figure 2). By using bioinformatic tools combined with detailed clinical phenotyping, researchers can determine whether particular genetic or molecular patterns are associated with increased cardiotoxicity risk. This approach may enable novel biomarker discovery, provide mechanistic and therapeutic insights, and generate hypotheses for future investigations.
Figure 2.
Omics Strategies for Biomarker Discovery
Clinically informed, omics-based investigations using biological samples, including genomics, epigenomics, transcriptomics, proteomics, metabolomics, and immunolomics, combined with advances in bioinformatics and computational tools may provide an unparalleled opportunity for the discovery of novel, molecularly targeted biomarkers. ATAC-seq = assay for transposase-accessible chromatin sequencing; CHIP = clonal hematopoiesis of indeterminate potential; ChIP-seq = chromatin immunoprecipitation sequencing; CyTOF = cytometry by time-of-flight; lncRNA = long noncoding RNA; mRNA = messenger RNA; miRNA = microRNA; NGS = next-generation sequencing; piRNA = piwiRNA; seq = sequencing; SNP = single nucleotide polymorphism; WGS = whole-genome sequencing.
Genomics
Genomic influence on the risk of cancer therapy-induced cardiotoxicity has been actively researched, given widely variable interindividual susceptibility. Hypothesizing shared genetic risk between dilated cardiomyopathy (DCM) and cancer-therapy induced cardiotoxicity, Garcia-Pavia et al. (2) sequenced putative DCM genes and found an increased prevalence of TTN-truncating variants in cardiotoxicity cases. In a separate study, Aminkeng et al. (3) conducted a genome-wide association study (GWAS) in 280 patients of European ancestry treated with anthracyclines (32 cases, 248 controls) and identified a protein-altering variant in RARG that was highly associated with cardiotoxicity; findings validated in other cohorts and independently supported by investigations using patient-specific and genome-edited iPSC-derived cardiomyocytes (4). Hence, genomics can be used to identify key genetic variants or develop polygenic risk scores predicting cancer therapy cardiotoxicity risk. However, for GWAS to yield meaningful discoveries, many factors need to be considered, including the overall prevalence of cardiotoxicity associated with a particular cancer drug, the number of cardiotoxicity cases and controls being studied, expected variant frequencies, and the extent to which cardiotoxicity risk is influenced by variants (5). Thus far, genomic studies in cardio-oncology have largely been limited by small numbers and relatively low cardiotoxicity event rates. This may be overcome by combining genomic data from various sources.
Another promising genetic biomarker is clonal hematopoiesis of indeterminate potential (CHIP), which has been linked to an increased risk of aging-related conditions including cardiovascular disease (6). Emerging data suggest that CHIP is more common in patients after cancer treatments such as bone marrow transplantation and is associated with higher risk of leukemia and all-cause mortality (7). Although CHIP is currently an area of active research, it may serve as an attractive biomarker to identify both cardiovascular and oncologic risk.
Epigenomics
Epigenomics are reversible genetic modifications that regulate gene expressions without altering the DNA sequence. Established epigenomic factors include: 1) DNA methylation and histone modifications altering chromatin accessibility and structure; and 2) expression of noncoding RNA, such as microRNA (miRNA) and long noncoding RNA (lncRNA), directly interacting with gene transcriptions. A patient-specific epigenomic footprint may influence the response to environmental insults or cancer therapies. Additionally, chemotherapies and radiation therapies may also alter the epigenome, further impacting gene expression and subsequent phenotypes.
Recent developments in epigenetic technologies to sequence accessible chromatin regions or quantitatively interrogate methylation sites across the genome have advanced the understanding of epigenetic regulations. With advances in technologies, investigations using miniscule amounts of genomic samples (<1 μg) can now provide comprehensive epigenomic information. Epigenomic profiling of the heart, however, has been hampered by difficulties in obtaining myocardial tissues from patients. Meder et al. (8) performed epigenome-wide mapping of DNA methylation in endomyocardial biopsies obtained from 41 DCM patients and 31 controls and compared the results with the methylation profiles of whole peripheral blood samples from the same patients. The authors observed distinct epigenetic patterns associated with DCM, identifying 27 epigenetic loci significantly enriched in the DCM cohorts, and also identified a minor subset of DCM-specific methylation sites conserved in cardiac and blood tissues. Further studies are needed to examine whether epigenetic signatures of the heart or other biosamples can serve as a useful biomarker of cardiotoxicity.
Transcriptomics
Next-generation sequencing (NGS) has enabled rapid and affordable sequencing of RNA, making it possible to quantitatively assess thousands of gene transcripts. Transcriptomic profiling of drug-treated iPSC-derived cardiac cells revealed distinct expression patterns and mechanistic insights for (9,10) tyrosine kinase inhibitor- (11), and trastuzumab-related cardiotoxicity (12). Together, these studies have also demonstrated interindividual variation correlating with cellular toxicity, suggesting underlying genetic contributions in modulating cellular response to various treatments. These findings exemplify potential utility of transcriptomic biomarkers to improve accurate diagnosis of cardiotoxicity. Additionally, the recent development of single-cell RNA sequencing (scRNA-seq) technologies allows cell-specific transcriptomic evaluation, enabling the discovery of new, relevant cell populations such as inflammatory cells and important genes and pathways that mediate cardiotoxicity (13).
Circulating miRNAs are attractive biomarkers as they are readily detectable in serum and are stable against degradation with a long half-life. More than 1,900 human miRNAs have been annotated in the miRBase database (miRBase: MicroRNA, University of Manchester, Manchester, United Kingdom) and a number of miRNAs have already been shown to be associated with myocardial injury and cardiovascular death. Oatmen et al. (14) compared serum miRNA profiles in anthracycline-treated pediatric patients with age-matched controls. Using a customized microarray of 84 miRNAs associated with cardiovascular diseases, the authors observed significantly altered miRNA expression with anthracyclines and identified 8 miRNAs that correlated with cardiotoxicity. Although these findings suggest the potential utility of miRNAs as cardiotoxicity biomarkers, few markers have been validated, due in part to differences in isolation and measurement techniques. Future endeavors in optimized measurement and unbiased sequencing of miRNA may accelerate the discovery of novel miRNA cardiotoxicity biomarkers.
Proteomics
Although transcriptomic information provides insights into the proteome, significant discordance exists, due in part to complex protein regulation in cells. First, protein synthesis may be directly correlated with the abundance of RNA transcripts, but many factors contribute to protein degradation including ubiquitin-proteasome and lysosome-mediated proteolysis. Second, there are far more proteins in the body than there are protein-coding genes, further complicating proteomic evaluation. Third, although proteomic changes in cardiac tissues may reflect direct cardiac pathologies, the associated procedural risk has prevented its widespread use, and researchers have used plasma samples for proteomics analysis. Beer et al. (15) used mass spectrometry to comprehensively analyze plasma proteomic profiles of 3 cardiotoxicity cases versus 4 age- and cancer-matched controls without cardiomyopathy. Their results suggested that immunoglobulin E had the largest and most consistent differences in the levels between cases and controls (15). In another study in noncancer patients, researchers used aptamer-based proteomic technology to probe the plasma proteome in patients with coronary heart disease (16). The study’s 9-protein risk score outperformed the Framingham secondary event risk score in predicting cardiovascular events among patients with stable coronary heart disease. As such, proteomics-based studies may allow the discovery of novel protein biomarkers or the development of a risk score to predict those at risk for cardiotoxicity.
Metabolomics
Metabolomics is the analysis of metabolites in the body such as amino acids, lipids, and organic acids. According to the Human Metabolome Database, there are >100,000 metabolites, which include endogenous metabolites and metabolites from external sources such as food and environmental pollutants. Circulating metabolites not only reflect the end products of bodily processes but provide unique insights into the interplay between environmental exposure and development of cardiotoxicity. Although metabolomic profiling of plasma or urine samples can provide valuable insight into metabolic pathways critical to cardiotoxicity, cancer therapies typically have a broad impact in the metabolism of multiple organ systems, complicating interpretation. One way to circumvent this problem would be to use cardiac-specific tissues, such as iPSC-derived cardiovascular cells, to identify cardiac-specific metabolic perturbation by specific cancer therapies (1).
Immunolomics
With the introduction of immunotherapies, there have been increasing reports of immunity-related cardiovascular complications such as myocarditis. Omics-based immune profiling of whole blood or affected tissues, immunolomics, may provide insights into new mechanisms and biomarkers. The major approaches include: 1) genetic sequencing of the complementarity-determining regions and the antigen-binding portion of the T-cell–receptor beta chain, so-called immuno-seq; 2) scRNA-seq of immune cells; and 3) a single cell-level proteomic evaluation of immune cell surface receptors using a mass spectrometry-based approach called mass cytometry by time-of-flight (CyTOF), all of which are used to characterize cellular compositions and molecular characteristics of immune cells. Johnson et al. (17) used immuno-seq to identify selective clonal T-cell populations potentially involved in immune checkpoint inhibitor myocarditis. Although immunolomics are still at an early stage, they may provide a novel class of biomarkers with which to identify immune-related complications.
Integrated multi-omics and systems biology
With advances in bioinformatics and computational biology, the aforementioned multi-level omics data can be simultaneously and longitudinally studied, which may help narrow biomarker candidates and also reveal important interaction networks. Rose et al. (18) performed integrative multi-omics profiling from patient samples collected quarterly for up to 8 years. They constructed prediction models to identify patients at risk for developing type 2 diabetes mellitus and also reported >60 clinically actionable health discoveries to implement diet and exercise changes. With accumulating integrative and longitudinal multi-omics data, comprehensive molecular signatures specific to cardiotoxicity of cancer therapy may accelerate actionable health and biomarker discoveries.
Conclusions
Despite significant progress, there is an important need to continue to develop the necessary infrastructure and technologies to advance biomarker science. Emerging omics technologies and bioinformatics tools coupled with access to large patient populations may provide an unparalleled opportunity to discover novel, molecularly targeted biomarkers. This would facilitate better risk stratification, prevention, and treatment of cancer therapy-associated cardiotoxicity.
Acknowledgments
We thank Drs. Amanda Chase, Mark Chandy, and Edward Lau for their review of the manuscript. The figures were originally created using BioRender (Toronto, Ontario, Canada).
Footnotes
This paper has received funding support from National Institute of Health R01 HL113006, R01 Hl123968, and American Heart Association 17MERIT33610009 (to Dr. Wu). Dr. Wu is a cofounder of Khloris Biosciences but has no competing interests, as the work presented here is completely independent. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose. Aarti Asnani, MD, served as Guest Associate Editor for this paper. Anju Nohria, MD, served as Guest Editor-in-Chief for this paper.
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the JACC: CardioOncologyauthor instructions page.
References
- 1.Paik D.T., Chandy M., Wu J.C. Patient and disease-specific induced pluripotent stem cells for discovery of personalized cardiovascular drugs and therapeutics. Pharmacol Rev. 2020;72:320–342. doi: 10.1124/pr.116.013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia-Pavia P., Kim Y., Restrepo-Cordoba M.A. Genetic variants associated with cancer therapy-induced cardiomyopathy. Circulation. 2019;140:31–41. doi: 10.1161/CIRCULATIONAHA.118.037934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aminkeng F., Bhavsar A.P., Visscher H. A Coding variant in RARG confers susceptibility to anthracycline-induced cardiotoxicity in childhood cancer. Nat Genet. 2015;47:1079–1084. doi: 10.1038/ng.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christidi E., Huang H., Shafaattalab S. Variation in RARG increases susceptibility to doxorubicin-induced cardiotoxicity in patient specific induced pluripotent stem cell-derived cardiomyocytes. Sci Rep. 2020;10:10363. doi: 10.1038/s41598-020-65979-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purcell S., Cherny S.S., Sham P.C. Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 6.Jaiswal S., Natarajan P., Silver A.J. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377:111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson C.J., Lindsley R.C., Tchekmedyian V. Clonal hematopoiesis associated with adverse outcomes after autologous stem-cell transplantation for lymphoma. J Clin Oncol. 2017;35:1598–1605. doi: 10.1200/JCO.2016.71.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meder B., Haas J., Sedaghat-Hamedani F. Epigenome-wide association study identifies cardiac gene patterning and a novel class of biomarkers for heart failure. Circulation. 2017;136:1528–1544. doi: 10.1161/CIRCULATIONAHA.117.027355. [DOI] [PubMed] [Google Scholar]
- 9.Knowles D.A., Burrows C.K., Blischak J.D. Determining the genetic basis of anthracycline-cardiotoxicity by molecular response QTL mapping in induced cardiomyocytes. Elife. 2018 doi: 10.7554/eLife.33480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burridge P.W., Li Y.F., Matsa E. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med. 2016;22:547–556. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma A., Burridge P.W., McKeithan W.L. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aaf2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitani T., Ong S.-G., Lam C.K. Human-induced pluripotent stem cell model of trastuzumab-induced cardiac dysfunction in patients with breast cancer. Circulation. 2019;139:2451–2465. doi: 10.1161/CIRCULATIONAHA.118.037357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paik D.T., Cho S., Tian L., Chang H.Y., Wu J.C. Single-cell RNA sequencing in cardiovascular development, disease and medicine. Nat Rev Cardiol. 2020;17:457–473. doi: 10.1038/s41569-020-0359-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oatmen K.E., Toro-Salazar O.H., Hauser K. Identification of a novel microRNA profile in pediatric patients with cancer treated with anthracycline chemotherapy. Am J Physiol Heart Circ Physiol. 2018;315:H1443–H1452. doi: 10.1152/ajpheart.00252.2018. [DOI] [PubMed] [Google Scholar]
- 15.Beer L.A., Kossenkov A.V., Liu Q. Baseline immunoglobulin E levels as a marker of doxorubicin- and trastuzumab-associated cardiac dysfunction. Circ Res. 2016;119:1135–1144. doi: 10.1161/CIRCRESAHA.116.309004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganz P., Heidecker B., Hveem K. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. 2016;315:2532–2541. doi: 10.1001/jama.2016.5951. [DOI] [PubMed] [Google Scholar]
- 17.Johnson D.B., Balko J.M., Compton M.L. Fulminant myocarditis with combination immune checkpoint blockade. N Engl J Med. 2016;375:1749–1755. doi: 10.1056/NEJMoa1609214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schüssler-Fiorenza Rose S.M., Contrepois K., Moneghetti K.J. A longitudinal big data approach for precision health. Nat Med. 2019;25:792–804. doi: 10.1038/s41591-019-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]