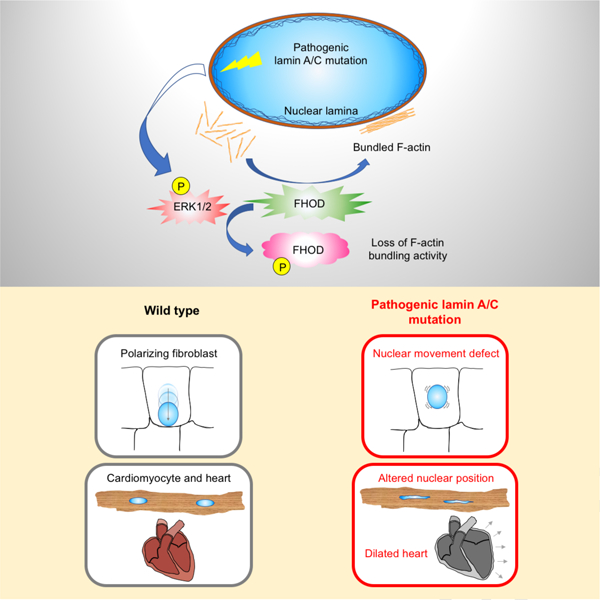
SUMMARY
Mutations in the lamin A/C gene (LMNA) cause cardiomyopathy and also disrupt nuclear positioning in fibroblasts. LMNA mutations causing cardiomyopathy elevate ERK1/2 activity in heart and its inhibition ameliorates pathology, but the downstream effectors remain largely unknown. We now show that cardiomyocytes from mice with a LMNA mutation and elevated cardiac ERK1/2 activity have altered nuclear positioning. In fibroblasts, ERK1/2 activation negatively regulated nuclear movement by phosphorylating S498 of FHOD1. Expression of an unphosphorylatable FHOD1 variant rescued the nuclear movement defect in fibroblasts expressing cardiomyopathy-causing lamin A mutant. In hearts of mice with LMNA mutation-induced cardiomyopathy, ERK1/2-mediated phosphorylation of FHOD3, an isoform highly-expressed in cardiac tissue. Phosphorylation of FHOD1 and FHOD3 inhibited their actin bundling activity. These results show that phosphorylation of FHOD proteins by ERK1/2 is a critical switch for nuclear positioning and may play a role in the pathogenesis of cardiomyopathy caused by LMNA mutations.
Graphical Abstract
eTOC blurb
Lamin A/C gene mutations cause dilated cardiomyopathy through elevated ERK1/2 activity. Antoku et al. find disrupted nuclear positioning in cardiomyocytes expressing pathogenic lamin A/C. ERK1/2 phosphorylation of FHOD inhibits its actin bundling to disrupt nuclear positioning. These results establish a link between ERK1/2 and nuclear positioning in disease pathogenesis.
INTRODUCTION
The nuclear lamina is a meshwork of intermediate filaments that lies beneath the inner nuclear membrane (Aebi et al., 1986; Turgay et al., 2017). One of its principal functions is controlling the physical properties of the nucleus (Lele et al., 2018). Expression of lamin A, one of the main somatic cell lamins, promotes nuclear stiffness (Lammerding et al., 2006) and is developmentally regulated such that its expression is proportional to tissue stiffness (Swift et al., 2013). The lamina also serves to anchor the linker of nucleoskeleton and cytoskeleton (LINC) complex, which couples the nucleus to the cytoskeleton to control nuclear position, shape and mechanochemical signaling (Chang et al., 2015b; Gundersen and Worman, 2013; Kirby and Lammerding, 2018). The lamina itself contributes to signaling by binding various factors and functions in organizing peripheral heterochromatin (Lammerding et al., 2006; Solovei et al., 2013). The extent to which these various functions are integrated is unknown.
A potentially powerful means to understand how lamina functions are integrated is to determine whether processes it regulates are coordinately affected by mutations in genes encoding its protein building blocks. Such a strategy is particularly relevant to the lamin A/C gene (LMNA). Hundreds of LMNA mutations have been described that cause diseases primary affecting striated muscle, adipose tissue, peripheral nerve or lead to multisystem conditions with features of accelerated aging (Dauer and Worman, 2009). Of these, the most prevalent are mutations that cause dilated cardiomyopathy frequently accompanied by muscular dystrophy, often in an Emery-Dreifuss type distribution (Bonne et al., 1999).
Altered ERK1/2 signaling is a characteristic phenotype associated with expression of striated disease variants of lamin A. ERK1/2 is hyperactivated in hearts from human subjects with cardiomyopathy caused by LMNA mutations and from LmnaH222P/H222P mice, which mimic the human disease (Muchir et al., 2007; Muchir et al., 2012). Treatment of LmnaH222P/H222P mice with inhibitors of the ERK1/2-activating kinase MEK1/2 delays the onset of cardiac pathology and restores normal nuclear morphology in cardiomyocytes (Muchir et al., 2012; Muchir et al., 2009).
Mis-positioning of nuclei in striated muscle may be another characteristic phenotype of the muscular dystrophy caused by LMNA mutations (Sewry et al., 2001). In skeletal muscle, nuclei are observed in the center of myofibers in mouse models expressing pathogenic lamin A variants (Muchir et al., 2013). Central nuclei are also observed during muscle regeneration (Folker and Baylies, 2013), so it is not yet clear whether this reflects pathological or physiological positioning of nuclei. Nonetheless, knockout of LMNA or expression of striated muscle disease-causing variants of lamin A in fibroblasts and myoblasts lead to nuclear positioning defects (Chang et al., 2015a; Folker et al., 2011). The effect of LMNA mutations on nuclear positioning likely involve the LINC complex as deletions of Syne1/2 in mice (Banerjee et al., 2014), and mutations in SUN1/2 (Meinke et al., 2014) and EMD (Bione et al., 1994) in humans, genes that encode LINC complex components and associated proteins, cause both striated muscle diseases and nuclear movement defects in fibroblasts (Chang et al., 2013; Luxton et al., 2010; Meinke et al., 2014). In cardiomyocytes where LMNA mutations have the most deleterious consequences, elongation of nuclei has been reported (Muchir et al., 2009), but nuclear positioning has not been examined. Whether there is a relationship between altered ERK1/2 signaling and nuclear mis-positioning is unknown.
We now identify and characterize nuclear mis-positioning in cardiomyocytes isolated from LmnaH222P/H222P mice with increased cardiac ERK1/2 activity. We further use a wounded fibroblast monolayer system to explore the relationship between ERK1/2 activity and nuclear positioning. Our findings indicate that ERK1/2 signaling and nuclear positioning deficits associated with striated muscle disease-causing lamin A variants are directly coupled through the FHOD family of formin proteins.
RESULTS
Lmna mutation causing dilated cardiomyopathy results in altered nuclear positioning in cardiomyocytes.
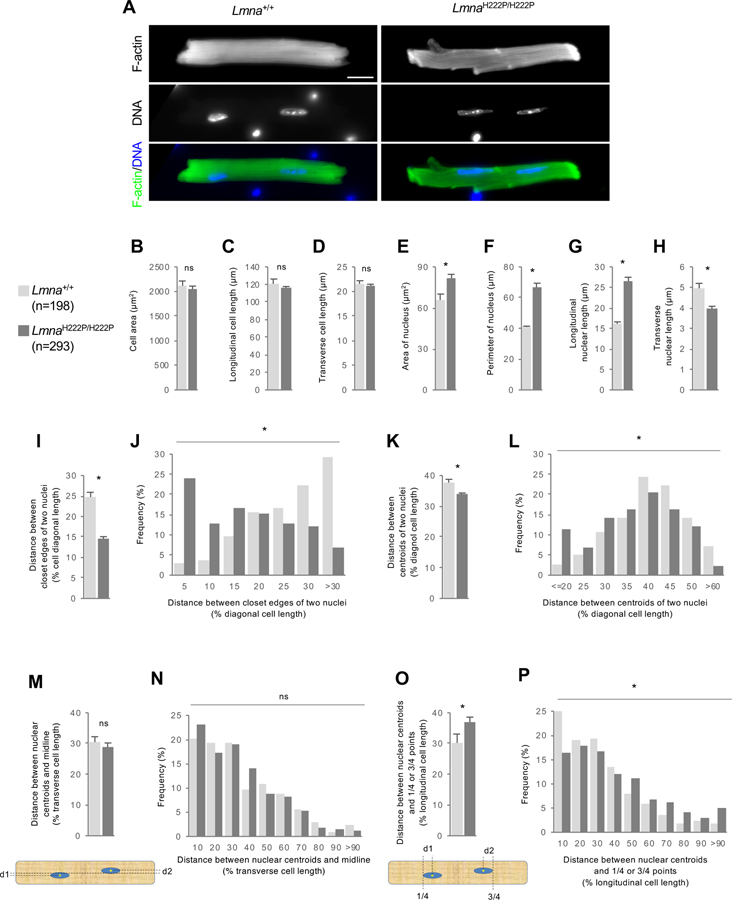
To examine if nuclear positioning was altered in cardiomyocytes expressing pathogenic lamin A/C variants, we isolated cardiomyocytes from male LmnaH222P/H222P mice (H222P cardiomyocytes) and wild type mice (WT cardiomyocytes) and stained nuclei and actin filaments to gauge cell size and shape. There was no significant difference in cell area or longitudinal and transverse cell lengths between WT and H222P cardiomyocytes (Figures 1A–D). Nuclei in H222P cardiomyocytes were considerably elongated compared to WT cardiomyocytes as previously reported (Muchir et al., 2009), with greater longitudinal and shorter transverse lengths and an increased perimeter (Figures 1A, and 1E–H).
Figure 1. Cardiomyocytes from LmnaH222P/H222P mice exhibit altered nuclear position.
(A) Images of isolated cardiomyocytes from 19-week-old Lmna+/+ or LmnaH222P/H222P mice stained for F-actin and DNA. Bar, 20 μm. (B-D) Mean cell area (B), longitudinal (C) and transverse (D) lengths of isolated cardiomyocyte from Lmna+/+ or LmnaH222P/H222P mice. (E-H) Mean nuclear area (E), perimeter (F), longitudinal length (G) and transverse length (H) of isolated cardiomyocyte from Lmna+/+ or LmnaH222P/H222P mice. (I,J) Means (I) and distribution (J) of closest distances between edges of two nuclei from isolated cardiomyocytes of Lmna+/+ or LmnaH222P/H222P mice. (K,L) Means (K) and distributions (L) of distances between centroids of the two nuclei in cardiomyocytes isolated from Lmna+/+ or LmnaH222P/H222P mice. Values were normalized to longitudinal cell length. (M,N) Means (M) and distributions (N) of distances between nuclear centroids and the midline (see diagram) of cardiomyocytes isolated from Lmna+/+ or LmnaH222P/H222P mice. Values were normalized to cell transverse cell length. (O,P) Means (O) and distributions (P) of distances of nuclear centroids from points ¼ and ¾ along the longitudinal axis (see diagram) of cardiomyocytes isolated from Lmna+/+ or LmnaH222P/H222P mice. Values were normalized to longitudinal cell length. In B-I, K, M, and O, values are means ± SEM; n, cells examined. *p < 0.05; ns, p > 0.05. In J, L, N and P, *p < 0.05; ns, p > 0.05.
More than 90 percent of adult mouse cardiomyocytes are binucleated (Soonpaa et al., 1996) and the distance between the two nuclei has been used as a measure of nuclear positioning (Banerjee et al., 2014). Compared to WT cardiomyocytes, the distance between the two nuclei was much shorter in H222P cardiomyocytes (Figures 1I and 1J). As the closer distance between the two nuclei may reflect the elongated nuclei of H222P cardiomyocytes, we also measured the distance between centroids of the nuclei. This also showed that the two nuclei were positioned more closely in H222P cardiomyocytes (Figures 1K and 1L).
We next determined if there were absolute changes in nuclear positioning relative to cell boundaries. There was no significant difference in how well the nuclear centroids aligned with respect to the midline bisecting the longitudinal axis of the cell (Figures 1M and 1N). We next assessed whether nuclei were positioned in such a way as to maximize the cytoplasmic space they occupied by determining how closely their centroids localized to points ¼ and ¾ of the longit udinal length of the cell (see Figure 1O). Nuclei in WT cardiomyocytes were closely clustered around these points, suggesting that they were spaced to minimize the distance from the nucleus to points in the cytoplasm (Figures 1O and 1P). In contrast, nuclei in H222P cardiomyocytes exhibited a skewed distribution relative to these points (Figures 1O and 1P). These data indicate that the H222P lamin A/C variants disrupt the equal positioning of nuclei in cardiomyocytes.
ERK1/2 inhibition rescues nuclear positioning in cells expressing striated muscle disease-associated lamin A variants.
To determine whether there might be a relationship between the elevated ERK1/2 activity and the nuclear positioning defect in cardiomyocytes, we turned to a simple model system where these have been well characterized. We used serum-starved, wounded monolayers of NIH3T3 fibroblasts in which the serum factor lysophosphatidic acid (LPA) triggers both ERK1/2 activation (Howe and Marshall, 1993) and actin-dependent nuclear movement (Gomes et al., 2005). The rearward movement of nuclei (measured relative to the cell centroid) coupled with the maintenance of the centrosome at the cell centroid, results in anterior orientation of the centrosome in fibroblasts at the wound edge (see (Chang et al., 2016) for method details).
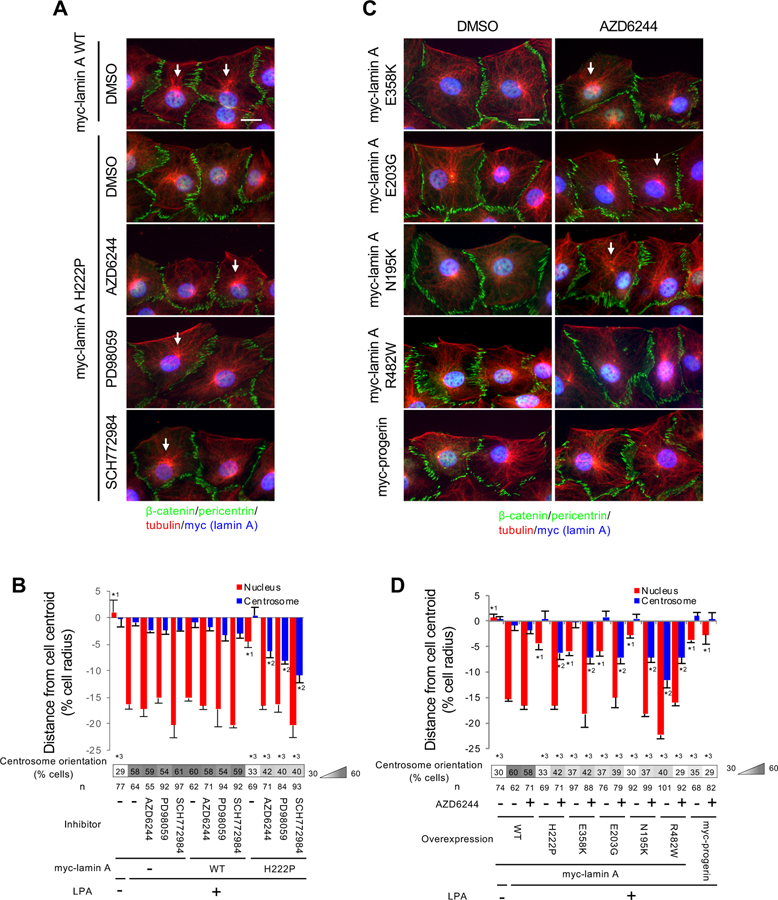
Expression of striated muscle disease-causing lamin A variants prevents nuclear movement and centrosome orientation in the NIH3T3 fibroblast system (Folker et al., 2011), but lamin A H222P was not previously tested. We confirmed that fibroblasts expressing lamin A H222P had defects in rearward nuclear movement and centrosome orientation (Figures 2A, 2B, and S1). Treatment of these cells with inhibitors of ERK1/2 (SCH772984) or MEK1/2 (AZD6244 and PD98059) (Dudley et al., 1995; Morris et al., 2013; Yeh et al., 2007) rescued the impaired nuclear positioning in cells expressing lamin A H222P (Figures 2A and 2B). The nuclear movement defect caused by expression of other muscle disease-associated variants of lamin A, E358K, E203G, and N195K (Folker et al., 2011), was also rescued by treatment with a MEK1/2 inhibitor (Figures 2C, 2D, and S1). Although nuclear movement was rescued by ERK1/2 inhibition, centrosome orientation was not rescued due to a failure to maintain the centrosome at the cell centroid (Figures 2, A–D). Consistent with this, the centrosome centration defect caused by expression of the lamin A R482W variant (Folker et al., 2011), which causes familial partial lipodystrophy, was not rescued by MEK1/2 inhibitor (Figures 2C, 2D, and S1). Cells expressing progerin, the truncated prelamin A variant in Hutchinson-Gilford progeria syndrome, also have a nuclear movement defect (Chang et al., 2019); however, this was not rescued by treatment with a MEK1/2 inhibitor (Figures 2C, 2D, and S1). These data indicate that inhibition of ERK1/2 specifically restores nuclear movement in fibroblasts expressing muscle disease-causing lamin A variants. Expression of these lamin A variants did not caused any apparent change in the expression of LINC complex components involved in nuclear movement in fibroblasts (Figure S1).
Figure 2. ERK1/2 inhibition rescues defective nuclear movement in NIH3T3 fibroblasts expressing muscle disease-causing lamin A variants.
(A) Images of wound-edge NIH3T3 fibroblasts expressing WT or H222P myc-lamin A stained for the indicated proteins after treatment with the indicated drugs (dimethyl sulfoxide [DMSO] is vehicle) and LPA stimulation for 2 hr. The wound edge is oriented toward the top of this and all subsequent panels of cells. White arrows, oriented centrosomes. Bar, 20 μm. (B) Centrosome and nuclear positions relative to the cell centroid defined as “0” (+ values toward the leading edge, - values toward the rear) for cells treated as in (A). Values are means ± SEM; n, cells examined. Centrosome orientation is shown in the heat map bar below the histogram. Random orientation is ~33%. n, cells examined. *1 and *2 indicate p < 0.05 compared to the rest of the samples for each category. *3 indicates p < 0.05 compared to LPA-stimulated myc-lamin A non-expressing control cells without drug treatment. (C) Images as in A for cell expressing the indicated lamin A variants: E358K, E203G and N195K (muscle disease), R482W (familial partial lipodystrophy), and progerin (Hutchinson-Gilford progeria syndrome). Bar, 20 μm. (D) Centrosome and nuclear positions (as described in B) for the indicted lamin A variants described in C. See also Figure S1.
ERK1/2 negatively regulates nuclear movement by preventing assembly of LINC complexes into transmembrane actin-associated nuclear (TAN) lines.
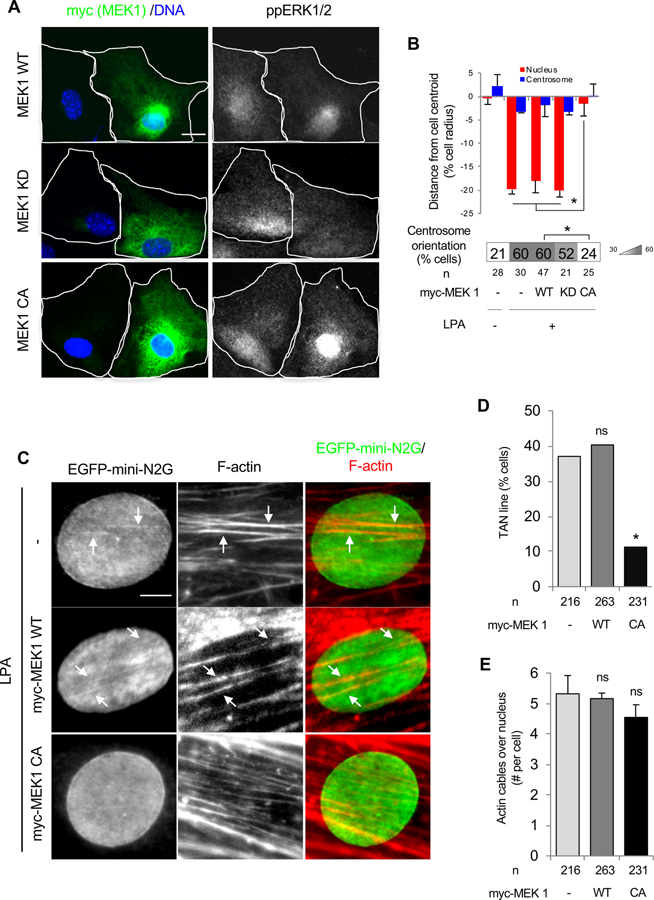
To understand how ERK1/2 signaling prevents nuclear movement in cells expressing muscle disease-causing lamin A variants, we first addressed the role of this kinase in nuclear positioning in WT cells. This question is relevant to LPA stimulation of nuclear movement, as this factor also stimulates a burst of ERK1/2 activity in serum-starved fibroblasts (Howe and Marshall, 1993). We first altered ERK1/2 activity in NIH3T3 fibroblasts by expressing active or inactive variants of MEK1. Cells expressing constitutively active MEK1 failed to displace their nuclei rearward, whereas cells expressing WT MEK1 or kinase defective MEK1 showed normal nuclear displacement (Figures 3A and 3B). In contrast, NIH3T3 fibroblasts treated with a MEK1/2 inhibitor did not show an apparent change in nuclear displacement or centrosome reorientation from controls at 2 hr after LPA stimulation, when nuclear displacement has reached its full extent (Figures S2A and S2B). However, at earlier time points, fibroblasts treated with the MEK1/2 inhibitor displaced their nuclei and oriented their centrosomes earlier than controls (Figures S2A–S2C). These observations indicate that activation of ERK1/2 negatively regulates nuclear movement at early times after LPA stimulation.
Figure 3. ERK1/2 negatively regulates nuclear movement in NIH3T3 fibroblasts.
(A) Images of LPA-stimulated wound-edge NIH3T3 fibroblasts expressing MEK1 constructs and stained for the indicated proteins and with DAPI. White outlines indicate cell borders. Bar, 20 μm. (B) Centrosome and nuclear position relative to the cell centroid in NIH3T3 fibroblasts expressing the indicated MEK1 constructs. Centrosome orientation is shown in the heat map bar below the histogram. Values are means ± SEM; n, cells examined. *p < 0.05. (C) Images of nuclei in LPA-stimulated wound-edge NIH3T3 fibroblasts expressing EGFP-mini-N2G and myc-MEK1 constructs and stained for the indicated proteins and DAPI. White arrows, colocalized EGFP-mini-N2G and F-actin indicating TAN lines. Bar, 5 μm. (D) Quantification of TAN lines 1 hr after LPA stimulation of NIH3T3 fibroblasts expressing the indicated MEK1 constructs. n, cells examined; ns, p > 0.05; *p < 0.05 compared to cells not microinjected with MEK1 constructs. (E) Quantification of actin cables over the nucleus 1 hr after LPA stimulation of NIH3T3 fibroblasts expressing the indicated MEK1 constructs. Values are means ± SEM; n, cells examined. ns, p > 0.05 compared to cells not microinjected with MEK1 constructs. See also Figure S2.
To understand how ERK1/2 inhibited nuclear movement, we examined the two structures necessary for nuclear movement: TAN lines and dorsal actin cables (Luxton et al., 2010; Luxton et al., 2011). TAN lines are linear assemblies of nesprin-2G-SUN2 LINC complexes and attach the nuclear envelope to dorsal actin cables that move retrogradely by actin flow. We visualized TAN lines by expressing enhanced green fluorescent protein (EGFP)-mini-nesprin-2G (mini-N2G) composed of the calponin homology (CH) and KASH domains of nesprin-2G (Luxton et al., 2010). The formation of TAN lines was severely impaired in NIH3T3 fibroblasts overexpressing constitutively active MEK1 compared to controls or those overexpressing WT MEK1 (Figures 3C and 3D). Conversely, the number of dorsal actin cables formed following LPA stimulation was not significantly different between controls and cells expressing WT or constitutively active MEK1 (Figures 3C and 3E). We further examined the kinetics of TAN line and actin cable formation in WT fibroblasts after LPA stimulation. Treatment of LPA-stimulated fibroblasts with a MEK1/2 inhibitor increased the formation of TAN lines at all time points examined coupled with more rapid formation of dorsal actin cables (Figures S2D–S2F). Although dorsal actin cables formed more rapidly, their rate of retrograde movement was unaffected by MEK1/2 inhibitor treatment (Figure S2G). Thus, ERK1/2 activation inhibits nuclear movement by targeting TAN line formation rather than disrupting the formation or movement of dorsal actin cables.
ERK1/2 phosphorylates the TAN line component FHOD1.
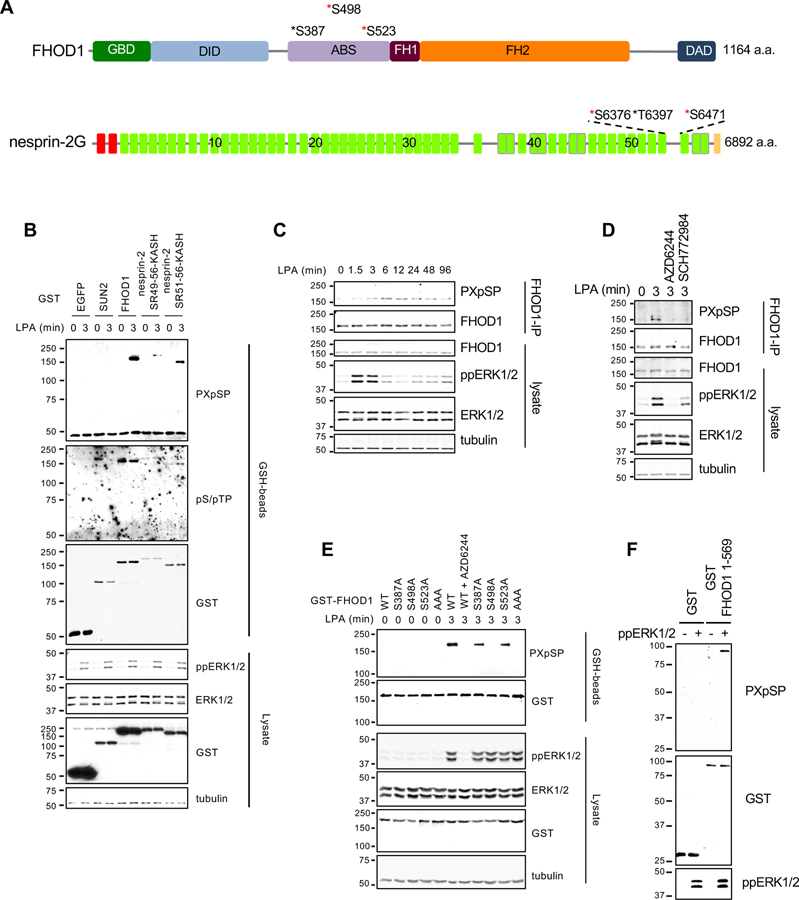
We next tested the possibility that ERK1/2 negatively regulates TAN line formation by directly phosphorylating TAN line components. We searched for consensus ERK1/2 phosphorylation sites among TAN line components FHOD1 (Kutscheidt et al., 2014), fascin1 (Jayo et al., 2016), nesprin-2G (Luxton et al., 2010), SUN2 (Luxton et al., 2010), Samp1 (Borrego-Pinto et al., 2012), and emerin (Chang et al., 2013) using PhosphositePlus (Hornbeck et al., 2012). We considered only consensus sites that were identified ≥5 times by mass spectroscopy and excluded sites that reside in the perinuclear space for the transmembrane proteins. With these criteria, we identified consensus ERK1/2 phosphorylation sites in FHOD1, nesprin-2G, and SUN2 (Figures 4A and S3A). To test whether these proteins were phosphorylated by ERK1/2 after LPA stimulation, we expressed glutathione-S-transferase (GST) fusions of FHOD1, SUN2, and the C-terminal portion of nesprin-2G (nesprin-2 SR51-56-KASH or SR49-56-KASH), which contains its consensus phosphorylation sites. We monitored phosphorylation with antibodies that recognize either specific PXpSP or broader pS/pTP consensus sites. Following LPA stimulation, FHOD1 and the nesprin-2G C-terminal polypeptides showed increased phosphorylation as detected by the anti-PXpSP antibody (Figure 4B). Nesprin-2 SR51-56-KASH also showed increased phosphorylation as revealed by the anti-pS/pTP antibody (Figure 4B). Phosphorylation of SUN2 was not detected by either antibody (Figure 4B).
Figure 4. ERK1/2 directly phosphorylates FHOD1.
(A) Predicted consensus ERK1/2 phosphorylation sites in FHOD1 and nesprin-2G. Red asterisks, predicted phosphorylation sites from Phosphosite Plus; black asterisks, possible phosphorylation sites deduced by visual inspection of the sequence. FHOD1 domains indicated are: GBD, GTPase binding domain; DID, DAD interaction domain; ABS, actin binding site, FH1 and FH2, formin homology domains; and DAD, Dia autoregulatory domain. Nesprin-2G, domains are color-coded: red, CH domains; green, predicted spectrin repeats (SRs); and yellow, KASH domain. The numbers on the SRs indicate SR number. (B) Western blots (antibodies indicated to the right) of lysates from LPA-stimulated NIH3T3 fibroblasts expressing the indicated GST constructs. GST-tagged proteins were captured on GSH-beads before western blotting. Tubulin is a loading control. (C) Western blots (antibodies indicated to the right) of lysates and endogenous FHOD1 immunoprecipitants from LPA-stimulated NIH3T3 fibroblasts. (D) Western blots (antibodies indicated to the right) of lysates and FHOD1 immunoprecipitants from LPA-stimulated NIH3T3 fibroblasts treated with or without AZD6244 or SCH772984. (E) Western blots (antibodies indicated to the right) of lysates and GSH-beads from LPA-stimulated NIH3T3 fibroblasts expressing the indicated GST-FHOD1 constructs. GST-FHOD1 AAA carries S387A, S498A, and S523A mutations. (F) Western blots (antibodies indicated to the right) of GST and GST-FHOD1 1-569 after in vitro phosphorylation by ppERK1/2. In B-F, migration of molecular mass standards (kDa) is indicated at the left of each blot. See also Figure S3.
We further confirmed the phosphorylation of endogenous FHOD1. FHOD1 phosphorylation increased shortly after ERK1/2 activation following LPA stimulation (Figure 4C) and was inhibited by ERK1/2 or MEK1/2 inhibitors (Figure 4D). To identify ERK1/2 phosphorylation sites on FHOD1, we mutated the three possible consensus phosphorylation sites in FHOD1 (S387, S498, and S523) to alanine either singly or in combination. Expression of these constructs in NIH3T3 fibroblasts followed by LPA stimulation revealed that the triple alanine mutant or the single S498A mutant eliminated the phosphorylation signal detected by the PXpSP antibody (Figure 4E), indicating that S498 was the sole ERK1/2 consensus phosphorylation site in FHOD1. Furthermore, an in vitro kinase assay showed that GST-FHOD1 N-terminus (1-569) containing S498 was directly phosphorylated by ERK1/2 (Figure 4F).
In addition to FHOD1, nesprin-2 was phosphorylated in an ERK1/2 activation-dependent manner in cells (Figure S3B). Using an in vitro kinase assay, we found that residues S6376, T6397, and S6471 of nesprin-2 were directly phosphorylated by ERK1/2 (Figures S3A and S3C).
Unphosphorylatable FHOD1 rescues nuclear movement in cells expressing a muscle disease-associated variant of lamin A
Nesprin-2 and FHOD1 knockdown or nesprin-2 knockout in fibroblasts inhibits nuclear movement (Kutscheidt et al., 2014; Luxton et al., 2010; Woychek and Jones, 2019). To test the impact of phosphorylation of nesprin-2 or FHOD1 on nuclear movement, phosphomimetic and unphosphorylatable forms of these proteins were expressed in fibroblasts depleted of these proteins (Figures S4A and S4B). To test nesprin-2, we prepared a chimeric construct containing the phosphorylation sites (SR51-56-KASH) and the N-terminal CH domains of nesprins-2, which are required for actin-dependent nuclear movement (Luxton et al., 2010). Expression of this construct (nesprin-2 ∆SR3-50) in its unphosphorylatable or phosphomimetic forms rescued the nuclear movement defect in nesprin-2 depleted cells (Figures S4C and S4D). This indicates that nesprin-2 phosphorylation by ERK1/2 is not responsible for inhibition of nuclear movement. Both WT nesprin-2 ∆SR3-50 and its phosphomimetic variant exhibited defects in centrosome centration and reorientation, suggesting that they may participate in these processes (Figures S4C and S4D).
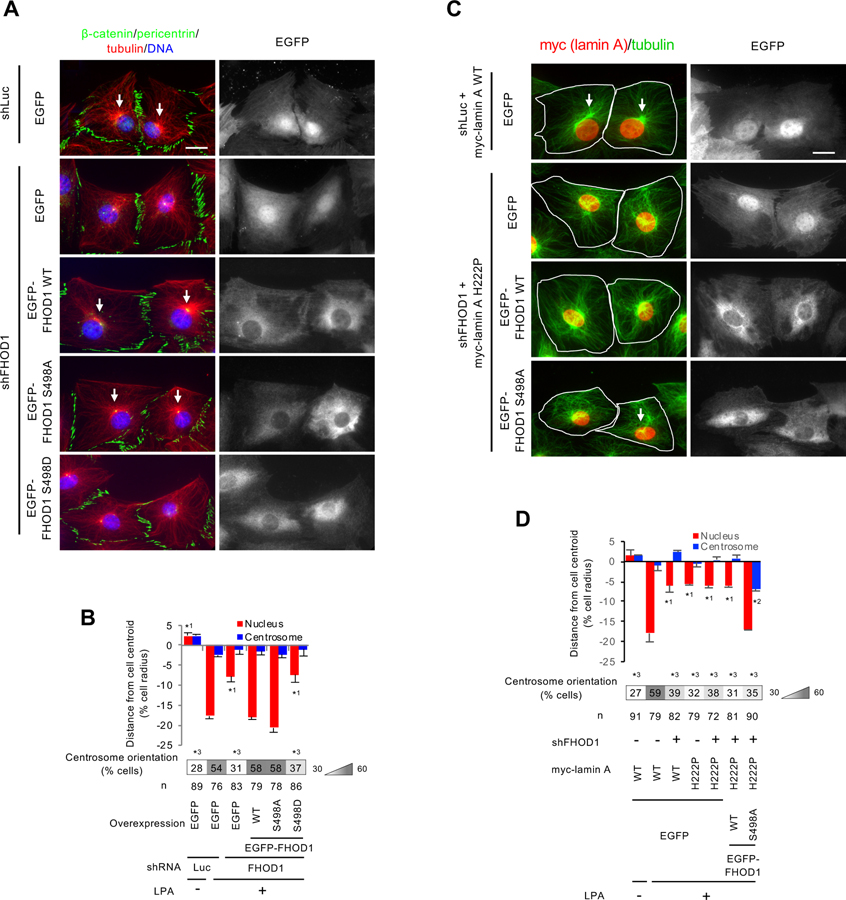
In contrast to phosphomimetic nesprin-2 ∆SR3-50, expression of phosphomimetic FHOD1 S498D in FHOD1-depleted cells did not rescue the nuclear movement defect (Figures 5A and 5B) (Kutscheidt et al., 2014). However, expression of FHOD1 WT or unphosphorylatable FHOD1 S498A rescued it (Figures 5A and 5B). These results support the idea that FHOD1 phosphorylation by ERK1/2 negatively regulates nuclear movement.
Figure 5. Phosphomimetic and unphosphorylatable FHOD1 mutants inhibit and promote, respectively, nuclear movement and centrosome orientation.
(A) Images of LPA-stimulated wound-edge NIH3T3 fibroblasts expressing EGFP or FHOD1 proteins after knockdown of FHOD1 and stained for the indicated proteins and DAPI. shLuc (shRNA against luciferase) is a negative control. White arrows, oriented centrosomes. Bar, 20 μm. (B) Centrosome and nuclear positions and centrosome orientation for the cells treated as in A. Values are means ± SEM; n, cells examined. Centrosome orientation (mean % of cells), is shown in the heat map below the histograms. *1 indicates p < 0.05 compared to the rest of the samples for each category. *3 indicates p < 0.05 compared to the LPA-stimulated control. (C) Images of lamin WT or lamin A H222P expressing wound-edge NIH3T3 fibroblasts expressing EGFP or FHOD1 proteins after knockdown of FHOD1 and stained for the indicated proteins and DAPI. shLuc is a negative control. White arrows, oriented centrosomes. White outlines, cell borders. Bar, 20 μm. (D) Centrosome and nuclear positions and centrosome orientation for cells treated as in B. *1 and *2 indicate p < 0.05 compared to the rest of the samples for each category. *3 indicates p < 0.05 compared to the LPA-stimulated control. See also Figure S4.
To test whether FHOD1 was the critical target of ERK1/2 in regulating nuclear movement in cells expressing muscle disease-causing lamin A variants, we tested whether expression of unphosphorylatable FHOD1 S498A might rescue defective nuclear movement. Thus, we knocked down FHOD1 in cells stably expressing lamin A H222P and expressed EGFP or EGFP-tagged FHOD1 S498A or FHOD1 WT (Figure S3E). Nuclear movement in cells expressing lamin A H222P was rescued upon expressing FHOD1 S498A, but not FHOD1 WT or EGFP (Figures 5C and 5D). These results indicate that FHOD1 is the principal substrate for ERK1/2’s inhibitory regulation of nuclear movement in cells expressing lamin A H222P. Although nuclear movement was rescued by FHOD1 S498A, as with the rescue of nuclear movement by ERK1/2 inhibition, lamin A H222P cells re-expressing FHOD1 S498A failed to orient their centrosomes due to a defect in centrosome centration (Figures 5C and 5D). Nonetheless, these results support the conclusion that muscle disease-causing lamin A variants exert their effects on actin-dependent nuclear movement through phosphorylation of FHOD1.
ERK1/2-dependent phosphorylation of FHOD3 is upregulated in hearts of LmnaH222P/H222P mice.
Results in the fibroblast model system indicate that defective nuclear movement is linked to ERK1/2 phosphorylation of FHOD1. We used LmnaH222P/H222P mice, which develop cardiomyopathy with elevated ERK1/2 activity and nuclear mis-positioning, to examine whether FHOD1 phosphorylation is relevant to pathology. We first examined FHOD1 expression in cardiac and skeletal muscle from WT and LmnaH222P/H222P adult mice. Consistent with other reports (Sanematsu et al., 2019; Tojo et al., 2003; Uhlen et al., 2015), we found that FHOD1 was expressed highly in skeletal muscle but only at very low levels in cardiac muscle (Figure S5A). Combined with the lack of cardiac defects in FHOD1 knockout mouse (Sanematsu et al., 2019), this suggests that FHOD1 is unlikely to contribute to the cardiac pathology in LmnaH222P/H222P mice.
FHOD3 is the second member of the FHOD family and highly homologous to FHOD1 (Figure S5B). It is essential for heart development and is required for normal cardiac function in the adult (Kan-o et al., 2012; Kan et al., 2012; Taniguchi et al., 2009). Mutations in FHOD3 are also associated with cardiomyopathy (Arimura et al., 2013; Ochoa et al., 2018). Thus, we hypothesized that FHOD3 instead of FHOD1 may be the relevant substrate of ERK1/2 in cardiac muscle. We confirmed earlier reports (Kan-o et al., 2012; Uhlen et al., 2015) that FHOD3 is highly expressed in cardiac tissue but not skeletal muscle (Figure S5A). Expression of the longer, cardiac-enriched FHOD3 isoform (FHOD3-1)(Kan-o et al., 2012) and the shorter isoform (FHOD3-2) in fibroblasts revealed that ERK1/2 specifically phosphorylated the longer isoform (Figure S5C).
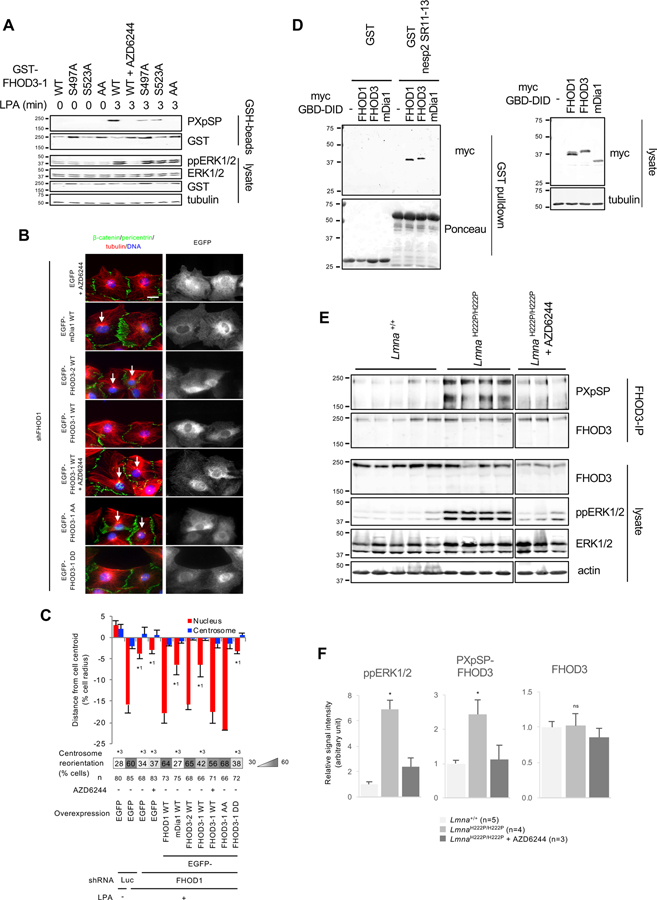
FHOD3-1 differs from FHOD3-2 by two inserts, one near the N-terminus and one near the C-terminus, but only the N-terminal insert has consensus ERK1/2 phosphorylation sites (Figure S5B). When either of the two PXpSP ERK1/2 sites in the N-terminal insert of FHOD3-1 was mutated to alanine, phosphorylation was substantially reduced and when both were mutated, phosphorylation was abolished (Figure 6A). Expression of FHOD3-2, but not the non-FHOD formin family member mDia1, rescued the nuclear movement defect in FHOD1 depleted fibroblasts (Figures 6B, 6C, and S5D). FHOD3-1, which is phosphorylated by ERK1/2, did not rescue the defect. However, when the ERK1/2 phosphorylation of FHOD3-1 was blocked by treatment with a MEK1/2 inhibitor, FHOD3-1 rescued the nuclear movement defect in fibroblasts lacking FHOD1 (Figures 6B and 6C). Furthermore, unphosphorylatable FHOD3-1 AA, but not phosphomimetic FHOD3-1 DD rescued the nuclear movement defect (Figures 6B and 6C). The interaction of FHOD1’s GBD-DID domain with spectrin repeats 11-13 of nesprin-2G is required for nuclear movement (Kutscheidt et al., 2014). We found that FHOD3’s GBD-DID domain also bound spectrin repeats 11-13 of nesprin-2G (Figure 6D). These results strongly suggest that the function of FHOD1 and FHOD3 and their regulation by ERK1/2 in nuclear movement are similar.
Figure 6. FHOD3 is inactivated by ERK1/2 phosphorylation and its phosphorylation is upregulated in hearts of LmnaH222P/H222P mice.
(A) Western blots (antibodies indicated to the right) of lysates from LPA-stimulated NIH3T3 fibroblasts expressing the indicated GST-FHOD3-1 constructs. GST proteins were capture on GSH-beads before western blotting. FHOD3-1 AA carries S497A and S523A mutations. (B) Images of LPA-stimulated wound-edge NIH3T3 fibroblasts stained for the indicated proteins and with DAPI. FHOD1 was depleted from cells using shRNA and the indicated constructs were expressed. FHOD3-1 DD carries S497D and S523D mutations. White arrows, oriented centrosomes. Bar, 20 μm. (C) Centrosome and nuclear positions and centrosome orientation in LPA-stimulated NIH3T3 fibroblasts knocked down for FHOD1 and expressing the indicated constructs as in B. Values are means ± SEM; n, cells examined. Centrosome orientation is shown in the heat map bars below the histograms. *1 and *2 indicate p < 0.05 compared to the rest of samples for each category. *3 indicates p < 0.05 compared to LPA-stimulated control. (D) Western blots (antibodies indicate on the right) of lysates from 293T cells transfected with the indicated constructs and the pulldown of GST or GST-nesprin2-SR11-13 against the lysates. Tubulin is a loading control. (E) Western blots (antibodies indicate on the right) of immunoprecipitated FHOD3 and lysates from heart tissue of 27-week-old Lmna+/+ mice, LmnaH222P/H222P mice and LmnaH222P/H222P mice treated with AZD6244. GAPDH is a loading control. (F) Quantification of western blot signals showing means ± SEM (n indicated in figure) from western blots as in E. *p < 0.05 compared to the rest of samples. ns indicates p > 0.05 compared to the rest of samples. In A, D, and E, migration of molecular mass standards (kDa) is indicated at the left of each blot. See also Figure S5.
We next analyzed FHOD3-1 phosphorylation in hearts of male LmnaH222P/H222P mice, which develop progressive left ventricular dilatation and depressed left ventricular fractional shortening (Arimura et al., 2005). We examined FHOD3 in hearts of these mice at 27 weeks of age, when they have significant cardiac dysfunction and elevated ERK1/2 activity. FHOD3 levels in heart lysates were similar in WT and LmnaH222P/H222P mice, but phosphorylation at the ERK1/2 consensus PXpSP site was strongly elevated in the mutant mice (Figures 6E and 6F). Phosphorylation of this site was strongly suppressed in mice treated with the MEK1/2 inhibitor AZD6244 (Figures 6E and 6F). These results show that ERK1/2-dependent phosphorylation of FHOD3 is elevated in hearts of LmnaH222P/H222P mice.
ERK1/2 phosphorylation of FHOD proteins specifically impairs their F-actin bundling activity.
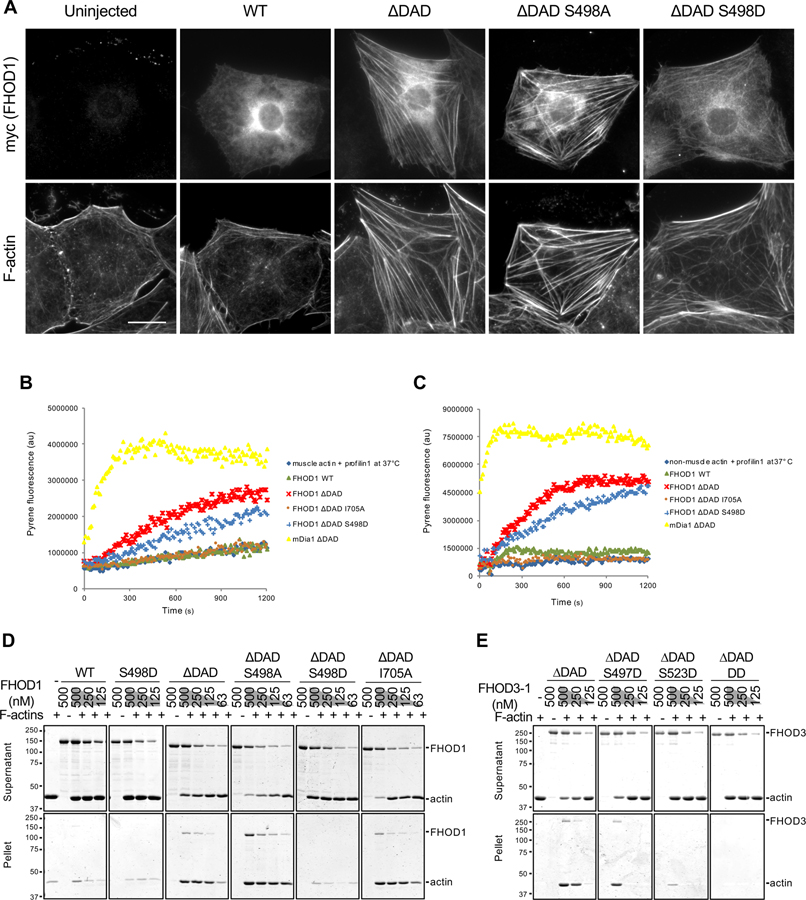
To understand how ERK1/2 phosphorylation of FHOD proteins affects their activity, we first examined formation of actin cables in wounded monolayers of serum-starved NIH3T3 fibroblasts overexpressing phosphorylation mutants of FHOD1. FHOD lacking the Dia autoregulatory domain (FHOD ∆DAD) is constitutively active and induces actin bundles in cells (Takeya and Sumimoto, 2003). We confirmed that expression of FHOD1 ∆DAD induced actin cable formation whereas WT FHOD1 did not (Figure 7A). FHOD1 ∆DAD S498A induced actin cables and, like FHOD1 ∆DAD, decorated them (Figure 7A). In contrast, FHOD1 ∆DAD S498D did not induce actin cables or decorate them. These results suggest that ERK1/2 phosphorylation of FHOD1 reduces its ability to induce actin cables and localize to them in cells.
Figure 7. Effect of FHOD phosphorylation site mutants on actin binding, polymerization and bundling activities.
(A) Images of serum-starved wound-edge NIH3T3 fibroblasts expressing the indicated myc-FHOD1 mutants and stained for F-actin and myc. Bar, 20 μm. (B) Actin polymerization activity of FHOD1 mutants examined using an actin pyrene-assay. Fluorescence of pyrene-rabbit skeletal muscle actin (2 μM, 10% pyrene-labeled) pre-incubated with 4 μM human profilin1 and then incubated with 200 nM of the indicated FHOD1 mutants or 20 nM mDia1 ∆DAD at 37°C. (C) Actin polymerization activity of FHOD1 mutants examined using an actin pyrene-assay. Fluorescence of pyrene-non-muscle actin (4 μM, 10% pyrene-labeled) pre-incubated with 8 μM profilin1 and then incubated with 400 nM of various FHOD1 proteins or 40 nM mDia1 ∆DAD at 37°C. (D) Actin bundling activity of FHOD1 mutants examined using a low-speed (16,000 x g) actin co-sedimentation assay. Coomassie blue-stained SDS-polyacrylamide gels of proteins in the supernatant and pellet are shown. (E) Actin bundling activity of FHOD3-1 mutants as in D. FHOD3-1 ∆DAD DD is FHOD3-1 ∆DAD S497D S523D. In D and E, migration of molecular mass standards (kDa) is indicated at the left of each gel or blot. See also Figures S6 and S7.
We next explored the impact of S498 phosphorylation on the ability of FHOD1 to bind, polymerize, and bundle actin filaments in vitro. After expression and purification FHOD1 proteins from 293T cells, FHOD1 was not detectably phosphorylated on the ERK1/2 phosphorylation site (Figure S6A). Using a high-speed F-actin co-sedimentation assay, we found no difference in the F-actin binding activity of FHOD1 ∆DAD, WT FHOD1 or FHOD1 S498D (Figure S6B).
Active forms of vertebrate FHOD1 inhibit polymerization of muscle actin in vitro (Patel et al., 2018; Schonichen et al., 2013) and we confirmed these results (Figure S6C). However, in the presence of mammalian profilin1 or profilin2a at 37°C, FHOD1 ∆DAD stimulated polymerization of mammalian muscle and non-muscle actin, although not to as great an extent as the ∆DAD construct of the related formin mDia1 (Figures 7B, 7C, and S6D–I). Under these conditions, FHOD1 ∆DAD S498D had slightly reduced actin polymerization activity compared to WT FHOD1 ∆DAD, but it still stimulated both muscle and non-muscle actin polymerization compared to actin alone (Figures 7B and 7C). FHOD1 ∆DAD I705A failed to stimulate actin polymerization, confirming the role of the FH2 domain in actin polymerization (Figures 7B and 7C) (Bartolini et al., 2008; Patel et al., 2018). These results suggest that ERK1/2 phosphorylation of FHOD1 does not affect its actin polymerization activity.
We used low-speed actin co-sedimentation to examine the bundling activity of FHOD1 mutants in vitro. As expected (Schonichen et al., 2013), WT FHOD1 weakly bundled F-actin, whereas FHOD1 ∆DAD robustly bundled F-actin (Figure 7D). The S498D phosphomimetic mutant strongly inhibited actin bundling by FHOD1 ∆DAD, whereas the unphosphorylatable S498A mutant had no effect (Figure 7D). As previously reported (Patel et al., 2018), the I705A FH2 mutation did not impair the F-actin bundling activity of FHOD1 ∆DAD (Figure 7D). The loss of bundling activity of the S498D mutant was also confirmed by visualizing actin bundles by total internal reflection fluorescence (TIRF) microscopy (Figure S7A). We also examined the F-actin bundling activity of phosphomimetic mutants of FHOD3-1 proteins in vitro. FHOD3-1 isolated from 293T cells was phosphorylated (Figure S7B) and had weak actin bundling activity that was enhanced by treatment with λ protein phosphatase (Figures S7B and S7C). Accordingly, we retreated all of the FHOD3-1 proteins with λ phosphatase before testing them for bundling activity (Figure S7D). The single mutants of FHOD3-1 (S497D and S523D) and the double mutant had reduced actin bundling activity compared to the WT protein (Figure 7E). Thus, similar to FHOD1, ERK1/2 phosphorylation of FHOD3-1 negatively regulates its F-actin bundling activity.
DISCUSSION
Our results show that ERK1/2 phosphorylation of FHOD proteins links two of the major phenotypes in cardiomyopathy caused by LMNA mutations: upregulated ERK1/2 signaling and nuclear mis-positioning. We describe the first evidence that a striated muscle disease variant of lamin A/C induces altered nuclear positioning in cardiomyocytes. Using a model system for nuclear positioning, we find that a single phosphorylation site in FHOD1 is the key target for ERK1/2 in its negative regulation of nuclear movement in fibroblasts. We suggest that the closely related FHOD3 performs a similar function in cardiomyocytes, given that its phosphorylation by ERK1/2 is upregulated in hearts from LmnaH222P/H222P mice, it functionally rescues nuclear movement in FHOD1 deficient fibroblasts, it interacts with nesprin-2 and ERK1/2 phosphorylation regulates the actin bundling activity of both FHOD proteins. ERK1/2 also phosphorylates cofilin-1 in hearts of LmnaH222P/H222P mice, which leads to actin filament disassembly (Chatzifrangkeskou et al., 2018). These processes together may contribute to cardiomyocyte dysfunction in the disease.
Our data support a new model for the role of FHOD1 in actin-dependent nuclear movement. Previously, FHOD1 binding to nesprin-2G was proposed to provide a second attachment point between the actin cable and the LINC complex (Kutscheidt et al., 2014). Our new data suggest that FHOD1 does not simply bind to the actin cables, but is required to reinforce them by enhancing actin bundling. This conclusion is supported by three pieces of evidence: 1) the inability of phosphomimetic FHOD1 to rescue nuclear movement in FHOD1 depleted cells, 2) the dramatic decrease in actin bundling activity of phosphomimetic FHOD1 and 3) the rescue of nuclear movement in lamin A H222P-expressing cells by unphosphorylatable FHOD1. Additionally, the ERK1/2 phosphorylation site in FHOD1 resides in the ABS of FHOD1 (Takeya and Sumimoto, 2003), which was previously shown to be important for its actin bundling activity (Schonichen et al., 2013). We do not believe that FHOD1 is absolutely required for the formation of the actin bundles, as we did not detect a difference in the number of dorsal actin cables localized over the nucleus or their rate of retrograde flow in cells with increased ERK1/2 activity or in FHOD1 depleted cells (Kutscheidt et al., 2014). A separate study reported a small effect on the formation of dorsal actin cables in FHOD1-depleted cells (Schulze et al., 2014). We cannot exclude the possibility that the bundling activity of FHOD1 increases the thickness of the dorsal actin cables. Based on these considerations, we propose that the actin bundling activity of FHOD1 mechanically reinforces actin cables so that they are able to resist the high forces needed to move the nucleus, which are estimated to be 50-100 nN (Lele et al., 2018).
FHOD1 has previously been shown to be phosphorylated by Src, Aurora-B, and RockI/II kinases (Floyd et al., 2013; Iskratsch et al., 2013; Takeya et al., 2008). These kinases phosphorylate sites distinct from that for ERK1/2 and have been proposed to activate FHOD1. In contrast to these, ERK1/2 phosphorylation of FHOD1 negatively regulates its actin bundling activity without affecting F-actin binding and polymerization activities. ERK1/2 phosphorylation blocked the bundling of active forms of FHOD1/3, indicating that this form of regulation can occur after the proteins have been released from autoinhibition. These results suggest that ERK1/2 phosphorylation of FHOD1/3 can tune the extent to which they reinforce actin bundle formation.
We speculate that FHOD3 in cardiomyocytes may play a similar role to FHOD1 in linking nuclei to actin filaments. FHOD3 is highly expressed in cardiomyocytes, interacts with the LINC complex component nesprin-2G and can substitute for FHOD1 in nesprin-2G- and actin-dependent nuclear movement in fibroblasts. Mice lacking both nesprin-1 and nesprin-2 in cardiomyocytes have abnormal heart development and cardiomyocyte nuclei show positioning and shape defects (Banerjee et al., 2014). Together these results suggest that FHOD3 may couple nuclei to actin filaments to position them in cardiomyocytes. Previous studies have revealed that FHOD3 localizes to cardiac sarcomeres and is required for their formation, indicating a role in sarcomere organization (Kan-o et al., 2012; Kan et al., 2012; Taniguchi et al., 2009). Whether FHOD3 links nuclei to forming sarcomeres in cardiomyocytes has not been tested.(Stewart et al., 2019).
During development of skeletal muscle, nuclear positioning is closely linked to the formation of sarcomeres (Roman et al., 2017). In Drosophila muscle development, loss of LINC complex proteins, including nesprin and SUN orthologs, disrupts nuclear positioning and results in disorganized sarcomere formation (Auld and Folker, 2016). Combined with our data, these results suggest that mis-positioning of nuclei in cardiomyocytes may contribute to cardiomyopathy by altering sarcomere formation and/or function.
Recent studies have focused on structural components required for actin-dependent nuclear movement (Borrego-Pinto et al., 2012; Folker et al., 2011; Kutscheidt et al., 2014; Luxton et al., 2010). We have identified ERK1/2 as the first negative regulator of this process. As nuclear movement and the resulting centrosome orientation contribute to cell polarity in migrating cells, ERK1/2 can potentially negatively regulate cell migration through this pathway. There are reports of both positive and negative effects of ERK1/2 on cell migration (Huang et al., 2004; Petrie et al., 2014). It may be that low ERK1/2 activity stimulates migration whereas high levels become inhibitory. We observe inhibition of nuclear movement in cells with highly activated ERK1/2, such as cells expressing constitutively active MEK1 and early times after LPA stimulation. Given the ubiquity of ERK1/2 signaling and the active positioning of nuclei in most cells, it is likely that this protein kinase plays an active role in fine-tuning nuclear positioning in many tissues.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Gregg G. Gundersen (ggg1@cumc.columbia.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture.
NIH3T3 fibroblasts were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Corning Inc.) containing 10 mM HEPES pH 7.4 and 10% (v/v) bovine calf serum (GE Health Life Science). 293T cells were maintained in DMEM containing 10 mM HEPES pH 7.4 and 5% (v/v) bovine calf serum and 5% (v/v) fetal bovine serum (Gemini Bio-Products).
Mice.
The Institutional Animal Care and Use Committee of Columbia University Irving Medical Center approved all protocols. Mice were kept at room temperature and fed normal chow. LmnaH222P/H222P mice have been described previously (Arimura et al., 2005).
METHOD DETAILS
Plasmids.
All constructs were confirmed by DNA sequencing. pGEX 6P-4 vector (GE Healthcare Life Science) was used to express GST-tagged proteins in bacteria. pMYC-C4 vector was derived from pEGFP-C4 (Clontech) vector by replacing EGFP with the myc tag. pMYC-C4 and pEGFP-C4 were used to express proteins in NIH3T3 fibroblasts by microinjection. pEF1a-GST-P-N4 was derived from the pEGFP-C4 vector by replacing the CMV promoter and EGFP sequence with an EF1a promoter and GST sequence from pEBG vector (Tanaka et al., 1995). pEBG and pEF1a-GST-P-N4 vectors were used to express GST-tagged proteins in 293T cells. pMSCV-GST-6P-4 was derived from pMSCV-puro (Clontech) by inserting GST into the multiple cloning site and deleting the PGK promoter and puro resistance gene; it was used to express GST-tagged proteins in NIH3T3 fibroblasts by retroviral infection. Similarly, pMSCV-myc, pMSCV-EGFP-C4, and pMSCV-puro EGFP-C4 were derived from pMSCV-puro and were used for myc- or EGFP-tagged protein expression in mammalian cells by retroviral infection. pLVX-EF1a-GST-6P-4 was derived from pLVX-puro vector (Clontech) by replacing the CMV promoter with an EF1a promoter and GST and deleting the PGK promoter and puro resistance gene. It was used for GST-tagged protein expression in mammalian cells by lentiviral infection. pSUPER-puro (Oligoengine) was used for expressing shRNA in NIH3T3 fibroblasts by retroviral infection.
cDNAs for human lamin A WT and variants were previously described (Folker et al., 2011; Muchir et al., 2007) and were inserted into a vector with BamHI and NotI restriction sites. Human MEK1 WT cDNA was obtained by PCR from HeLa cell mRNA. Constitutively active MEK1 was generated by introducing S218D (TCC to GAT) and S221D (TCC to GAC) point mutations. Kinase defective MEK1 was made by introducing a K97A (AAG to GCG) point mutation. These MEK1 cDNAs were inserted into a vector with BamHI and NotI restriction sites. Human FHOD1 WT, FHOD1 GBD-DID (1-339), FHOD1 1-569 and I705A mutant cDNAs were previously described (Kutscheidt et al., 2014). FHOD1 ∆DAD (1-1053) was generated by PCR. Various FHOD1 mutants were made by introducing S387A (TCA to GCC), S498A (AGC to GCC), S523A (AGC to GCC), AAA (S387A, S498A, and S523A) and S498D (AGC to GAC) point mutations. These FHOD1 cDNAs were inserted into a vector with BamHI and NotI restriction sites. Mouse FHOD3-2 WT and FHOD3-1 WT cDNAs were obtained by PCR from mouse E14.5 forebrain and mouse adult heart mRNA, respectively. FHOD3 GBD-DID (1-327), FHOD3-2 ∆DAD (1-1318) and FHOD3-1 ∆DAD (1-1477) were generated by PCR. Various FHOD3-1 mutants were made by introducing S497A (AGC to GCC); S523A (AGC to GCC); S497A and S523A; S497D (AGC to GAC); S523D (AGC to GAC); and S497D and S523D point mutations. These FHOD3 cDNAs were inserted into a vector with NotI restriction site. mDia1-2 WT cDNA was kindly provided by S. Narumiya (Kyoto University, Kyoto, Japan), and mDia1-2 GBD-DID (73-377) and mDia1-2 ∆DAD1-1182 was generated by PCR. These were inserted into a vector with NotI restriction site. EGFP-miniN2G was previously described (Luxton et al., 2010). Mouse nesprin-2 SR11-13 (1414-1736 aa), nesprin-2 SR49-56-KASH (5795-6892 aa), nesprin-2 SR51-56-KASH (6008-6892 aa), nesprin-2 SR51-54 (6008-6559 aa) and nesprin-2 ∆SR3-50 (3-484 aa + 6008-6892 aa) were generated from NIH3T3 fibroblast cDNA as a template. Nesprin-2 ∆SR3-50 AAA and DDD mutants were made by introducing AAA (S6376A, TCT to GCT, T6397A, ACC to GCC, and S6471A AGC to GCC) and DDD (S6376D, TCT to GAT, T6397D, ACC to GAC, and S6471D, GCC to GAC). All of these nesprin-2 cDNAs were inserted into a vector with NotI restriction site. Human profilin1 and 2a cDNAs were obtained by RT-PCR from Hela cell mRNA and inserted into a vector with BamHI and NotI restriction site. The shRNA sequences for Luc and Nesp2 were previously described (Chang et al., 2015a). The shRNA sequence for FHOD1 (5’-aggagccgaagaucacuagaag-3’) was obtained from a previously published sequence (Iskratsch et al., 2013).
Cardiomyocyte isolation.
Cardiomyocytes were isolated from adult mouse hearts using established protocols (Joseph et al., 2017). Briefly, the heart was removed and the aorta was cannulated. After calcium-free buffer was perfused for 2 min, 0.3 mg/ml collagenase solution was perfused through the coronary arteries for 7–8 min with calcium at 12.5 μM. Left ventricular tissue was teased apart and pipetted to release individual cells. Isolated cardiomyocytes were plated on 1 μg/ml laminin-coated coverslips. After 4 h incubation at room temperature, the cells were fixed for further processing.
Immunofluorescence microscopy.
For indirect immunofluorescence microscopy, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min, and permeabilized and blocked with PBS containing 0.1% Triton-X and 1% BSA for 30 min. The cells were labeled first with primary antibodies and then fluorescently-labeled secondary antibodies, phalloidin, and DAPI. Images were acquired with either a 60× PlanApo TIRF objective (NA 1.49) and an ORCA ERI CCD camera (Hamamatsu) or iXon X3 CCD camera (Andor) on a Nikon Eclipse Ti microscope controlled by Nikon’s NIS-Elements software. Some images were obtained with a 60× PlanApo objective (NA 1.4) and a CoolSNAP HQ CCD camera (Photometrics) on a Nikon TE300 inverted microscope controlled by MetaMorph.
Virus production and infection.
293T cells were transfected with retro or lentiviral vectors and ecotropic or pantropic packaging plasmids. Medium containing the produced virus was harvested 24 hr after transfection, added to the NIH3T3 fibroblasts in the presence of 2 μg/ml polybrene and incubated for one day.
Western blotting and interaction assays.
For western blotting, proteins suspended in SDS sample buffer were separated by SDS-PAGE. The proteins were transferred to nitrocellulose blots, probed with indicated antibodies and detected either by chemiluminescence with Odyssy Fc (LI-COR Inc.) or infra-red fluorescence with Odyssy CLx (LI-COR Inc.). For immunoprecipitation and GST-protein immobilization, antibody-bound Protein A/G Plus agarose beads or GSH-conjugated Sepharose beads were incubated overnight with lysates at 4 °C. The next day, the beads were washed with kinase lysis buffer (KLB: 25 mM Tris-HCl 7.4, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 10 mM sodium pyrophosphate, 10 mM β-glycerophosphate, 10 mM NaF, 1 mM sodium orthovanadate, 10% glycerol, and 1 mM phenylmethylsulfonyl fluoride) for 4 times. The beads were processed for further experiments or boiled with SDS sample buffer for western blotting. For GST-pulldown, lysates in KLB were prepared from transfected 293T cells, clarified by centrifugation and then incubated with GST-protein immobilized beads at 4 °C for 1.5 hr. The beads wer e then washed with KLB for 4 times and bound proteins eluted by boiling in SDS sample buffer.
LPA stimulation and drug treatment.
A day before serum-starvation, NIH3T3 fibroblasts were plated either directly on tissue-culture dishes for western blotting or on acid-washed coverslips for immunofluorescent staining. The next day, cells at about 40% confluency on coverslips or 80% confluency on plates were washed three times with DMEM and then DMEM containing 10 mM HEPES pH 7.4 and 0.1% (v/v) fatty acid free bovine serum albumin (BSA) was added. For western blotting, one day serum-starved cells were stimulated with 10 μM LPA and harvested after indicated time. For indirect immunofluorescent staining, cells were serum-starved for two days and then stimulated with 10 μM LPA and harvested after indicated time. For drug treatments, cells were incubated with final concentrations of 1 μM AZD6244, 50 μM PD98059 or 0.5 μM SCH772984 30 min before LPA treatments.
Microinjection.
For microinjection, each plasmid was suspended in 150 mM KCl and 10 mM HEPES pH 7.4 at 20 to 50 ng/μl and the plasmid injected in the nucleus of the cells. After 1.5 h, cells were either stimulated and fixed or fixed for analysis.
Sequence comparisons.
Sequence alignments were created by CLC Sequence Viewer software using the ClustalW algorithm.
Protein production and purification.
For production of proteins in mammalian cells, 293T cells on 150 mm plates were transfected with 30 μg of plasmid DNA with calcium phosphate precipitation for 6 hr. Two days after transfection, the cells were lysed with KLB. All other GST-tagged proteins were purified after expression in BL21(DE3). The bacteria were lysed by sonication with PBS containing 50 mM EDTA, 1% Triton-X, and 1 mM phenylmethylsulfonyl fluoride. GST-proteins were collected on GSH-Sepharose beads and then cleaved with Turbo3C protease. The released and cleaved proteins were run on a PD-10 column in PBS containing 1 mM DTT and 10% glycerol and concentrated by Amicon concentrator. Purified FHOD3 protein was dephosphorylated with ʎ protein phosphatase according to the manufacture’s protocol. The proteins were suspended in PBS containing 1 mM DTT and 10% glycerol and stored at −80 °C.
In vitro kinase assay.
Serum-starved NIH3T3 fibroblasts stimulated with 10% bovine calf serum for 3 min were lysed in kinase lysis buffer and phospho-ERK1/2 was immunoprecipitated by adding rabbit anti-phospho-ERK1/2 antibody and Protein A/G Plus agarose beads and incubating overnight at 4 °C. The immune-complex beads were washed once with KLB containing 500 mM NaCl and twice with KLB containing 150 mM NaCl. The phospho-ERK1/2 beads were suspended in kinase buffer (25 mM Tris-HCl pH 7.5, 10 mM MgCl2, and 5% glycerol), and mixed with the purified proteins. The reaction was pre-warmed at 24°C. The kinase reaction was initiated by adding ATP to 0.1 mM total concentration and stopped after 1 h by adding SDS sample buffer.
Actin assays.
For the actin binding assay (high speed pelleting assay), 16 μM of rabbit skeletal muscle actin was incubated in G-buffer (5 mM Tris-HCl pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP pH 7.4, and 0.5 mM DTT) at 4 °C for at l east 1 h. The G-actin was spun down at 100,000 x g for 20 min. The supernatant containing the G-actin was adjusted to F-buffer (50 mM KCl, 2 mM MgCl2, and 1 mM ATP pH 7.4) and incubated at 24°C for 2 h at 8 μM concentration. Proteins to be tested were preincubated in F-buffer at 4°C for 30 min. Insoluble material was removed by centrifugation at 100,000 x g for 20 min at 4°C and the proteins mixed with the F-actin solution at 1:1 ratio resulting in 4 μM F-actin. The solution was incubated at 24°C for 60 min and spun down at 100,000 x g for 20 min at 4°C. The supernatant was mixed with SDS sample buffer. The pellet was washed with F-buffer once and resuspended in SDS sample buffer. Proteins in these fractions were separated by SDS-PAGE and stained with SimplyBlue SafeStain.
For the actin bundling assay (low speed pelleting assay), G-actin was polymerized in KMEI buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole pH 7.0) at 24 °C for 2 h. After the incu bation, unlabeled phalloidin (actin:phalloidin = 1:2) was added and incubated for 5 min. Proteins to be tested were preincubated in KMEI buffer at 4°C for 30 min. The F-actin and the protein solutions were clarified by centrifugation at 16,000 x g at 4 °C for 5 min and the supernatants mixed at a 1:1 ratio resulting 2 μM F-actin. The solution was incubated at 24°C for 1 5 min and spun down at 16,000 x g for 5 min at 4°C. T he supernatant was mixed with SDS sample buffer. The pellet was washed with KMEI buffer once and resuspended in SDS sample buffer. Proteins in the samples were separated by SDS-PAGE and stained with SimplyBlue SafeStain.
For the fluorescent actin bundling assay, polymerized actin was incubated with Alexa Fluor 488 conjugated phalloidin (actin:phalloidin:488-phalloidin = 2:1:1) for 5 min and then incubated with proteins (as above) to test from bundling. After the incubation, the bundled F-actin solution was diluted in 20 times with KMEI buffer and applied to 0.01 % poly-L-lysine coated coverslips. The samples were imaged by TIRF microscopy with the microscope described above.
For the pyrene actin polymerization assay, proteins were incubated in 2X KMEI buffer at 37°C for 15 min. Pyrene labeled depolymer ized actin (4 μM rabbit skeletal muscle actin or 8 μM human non-muscle actin, 10% pyrene labeled) in G-buffer were incubated with the indicated profilin at 37°C for 5 min. The actin and test protein solution was mixed resulting in 2 μM muscle or 4 μM non-muscle actin. Pyrene actin fluorescent intensity was measured by SpectraMax i3x (Molecular Devices) every 10 s for 45 min.
Mice treatment.
Treatment of mice with AZD6244 has been described previously (Muchir et al., 2012); treatment was started at 26 weeks of age and continued for 9 days.
QUANTIFICATION AND STATISTICAL ANALYSIS
Cell and nuclear characteristics of isolated cardiomyocytes were quantified from images using ImageJ software. For the centrosome reorientation assays, the position of centrosome relative to the axis between the nuclei and the leading edge was analyzed from images of DAPI and tubulin and/or β-catenin/pericentrin antibody-labeled cells as previously described (Gomes et al., 2005; Palazzo et al., 2001). Nuclear and centrosomal positions of NIH3T3 fibroblasts were determined from images using Cell Plot software (Chang et al., 2016). The speed of actin retrograde flow was measured from movies of NIH3T3 fibroblasts stably expressing EGFP-LifeAct stimulated with LPA. TAN line formation and dorsal actin cable number were assessed from images of fixed and stained NIH3T3 fibroblasts as previously described (Kutscheidt et al., 2014; Luxton et al., 2010).
Statistical analysis of data on nuclear positioning in cardiomyoctes (Fig. 1K–P), and centrosome reorientation (Fig. 2B,D; Fig. 3B; Fig. 5B,D; and Fig. 6D) and TAN line formation (Fig. 3D) was assessed by Chi-square test using GraphPad Software. Statistical analysis of cardiomyocte cell and nuclear parameters and two closest nuclear edges (Fig 1B–J), the number of actin cables over the nucleus (Fig. 3E), and the velocity of actin cable flow (Fig. S2G) were assessed by two-tailed unpaired student t-test using GraphPad Software. Statistical evaluation of the position of the nucleus and centrosome in NIH3T3 fibroblasts (Fig. 2B,D; Fig. 3B; Fig. 5B,D; and Fig. 6D) was by one-way ANOVA followed by Tukey’s multiple comparison test using SAS. All evaluated data were from at least N=3 experiments.
DATA AVAILABILITY
All raw data generated during this study are available from the corresponding author upon reasonable request. No new code was generated in this study
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit polyclonal myc | Santa Cruz Biotech | Cat# sc-789; RRID: AB_631274 |
| mouse monoclonal GFP | Santa Cruz Biotech | Cat# sc-9996; RRID: AB_627695 |
| mouse monoclonal GST | Santa Cruz Biotech | Cat# sc-138; RRID: AB_627677 |
| rabbit polyclonal ERK1/2 | Santa Cruz Biotech | Cat# sc-94; RRID: AB_2140110 |
| rabbit polyclonal emerin | Santa Cruz Biotech | cat# sc-15378, RRID: AB_2100047 |
| rabbit polyclonal FHOD1 | Santa Cruz Biotech | Cat# sc-99209, RRID: AB_2104511 |
| goat polyclonal FHOD1 | Santa Cruz Biotech | Cat# sc-46965, RRID: AB_2247011 |
| mouse monoclonal FHOD3 | Santa Cruz Biotech | Cat# sc-374601, RRID: AB_10987697 |
| HRP conjugated goat anti-mouse IgG | Santa Cruz Biotech | Cat# sc-2004, RRID: AB_631746 |
| HRP conjugated goat anti-rabbit IgG | Santa Cruz Biotech | Cat# sc-2005, RRID: AB_631736 |
| Alexa Fluor 488 conjugated donkey anti-mouse IgG | Jackson ImmunoResearch Laboratories | Cat# 715-545-150, RRID: AB_2340846 |
| Alexa Fluor 488 conjugated donkey anti-rabbit IgG | Jackson ImmunoResearch Laboratories | Cat# 711-545-152, RRID: AB_2313584 |
| Alexa Fluor 488 conjugated donkey anti-chicken IgY | Jackson ImmunoResearch Laboratories | Cat# 703-546-155, RRID: AB_2340376 |
| Rhodamine conjugated donkey anti-rat IgG | Jackson ImmunoResearch Laboratories | Cat# 712-025-153, RRID: AB_2340636 |
| Alexa Fluor 647 conjugated donkey anti-mouse IgG | Jackson ImmunoResearch Laboratories | Cat# 715-605-151, RRID: AB_2340863 |
| Alexa Fluor 647 conjugated donkey anti-rabbit IgG | Jackson ImmunoResearch Laboratories | Cat# 711-605-152, RRID: AB_2492288 |
| Alexa Fluor 647 conjugated donkey anti-rat IgG | Jackson ImmunoResearch Laboratories | Cat# 712-605-153, RRID: AB_2340694 |
| IRDye 680 conjugated goat anti-mouse IgG | LI-COR Biosciences | Cat# 926-32220, RRID: AB_621840 |
| IRDye 680 conjugatedgoat anti-rabbit IgG | LI-COR Biosciences | Cat# 926-32221, RRID:AB_621841 |
| IRDye 680 conjugated goat anti-rat IgG | LI-COR Biosciences | Cat# 926-32229, RRID: AB_1850020 |
| IRDye 800CW conjugated goat anti-mouse IgG | LI-COR Biosciences | Cat# 827-08364, RRID: AB_10793856 |
| IRDye 800CW conjugated goat anti-rabbit IgG | LI-COR Biosciences | Cat# 925-32219, RRID: AB_2721932 |
| IRDye 800CW conjugated goat anti-rat IgG | LI-COR Biosciences | Cat# 926-32219, RRID: AB_1850025 |
| mouse monoclonal β-catenin | Thermo Fisher Scientific | Cat# 71-2700, RRID: AB_2533982 |
| Alexa Fluor 568 conjugated donkey anti-mouse IgG | Thermo Fisher Scientific | Cat# A10037, RRID: AB_2534013 |
| Alexa Fluor 568 conjugated donkey anti-rabbit IgG | Thermo Fisher Scientific | Cat# A10042, RRID: AB_2534017 |
| rabbit polyclonal pS/pTP | EMD Millipore | Cat# 05-918, RRID: AB_441928 |
| chicken polyclonal GFP | EMD Millipore | Cat# AB16901, RRID: AB_90890 |
| rabbit polyclonal phospho-ERK1/2 | Cell Signaling Technology | Cat# 9101, RRID: AB_331646 |
| rabbit monoclonal PXpSP | Cell Signaling Technology | Cat# 2325, RRID: AB_331820 |
| rabbit polyclonal SUN2 | Abcam | Cat# ab87036, RRID: AB_1952674 |
| rabbit polyclonal FHOD3 | Abcam | Cat# ab224463 |
| mouse monoclonal pericentrin | BD Biosciences | Cat# 611814, RRID: AB_399294 |
| rat polyclonal a-tubulin | European Collection of Authenticated Cell Cultures | Cat# 92092402 |
| rabbit polyclonal nesprin-2G | (Luxton et al., 2010) | N/A |
| mouse monoclonal lamin A/C | Glen Morris, Wolfson Centre for Inherited Neuromuscular Disease, UK | N/A |
| rabbit polyclonal lamin B1 | (Cance et al., 1992) | N/A |
| Bacterial and Virus Strains | ||
| BL21(DE3) | NEB | Cat# C2527I |
| NEB 5-alpha | NEB | Cat# C2987I |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| lysophosphatidic acid (LPA) | Avanti Polar Lipids | Cat# 857130P |
| dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat# D8418 |
| bovine serum albumin (BSA) | Sigma-Aldrich | Cat# A7906 |
| fatty-acid free BSA | Sigma-Aldrich | Cat# A6003 |
| poly-L-lysine | Sigma-Aldrich | Cat# P8920 |
| laminin | Sigma-Aldrich | Cat# L2020 |
| collagenase (Liberase™ TM Research Grade) | Roche | Cat# 5401119001 |
| Alexa Fluor 488-phalloidin | Thermo Fisher Scientific | Cat# A12379 |
| Alexa Fluor 647-phalloidin | Thermo Fisher Scientific | Cat# A22287 |
| 4′,6-diamidino-2-phenylindole (DAPI) | Thermo Fisher Scientific | Cat# D3571 |
| Halt protease and phosphatase inhibitor cocktail | Thermo Fisher Scientific | Cat# 78440 |
| SimplyBlue SafeStain | Thermo Fisher Scientific | Cat# 465034 |
| PD98059 | Selleck Chemicals | Cat# S1177 |
| AZD6244 | Selleck Chemicals | Cat# S1008 |
| SCH772984 | MedChem Express | Cat# HY-50846 |
| phalloidin | Setareh Biotech | Cat# 6901 |
| rabbit skeletal muscle actin | Cytoskeleton | Cat# AKL99 |
| pyrene-labeled rabbit skeletal muscle actin | Cytoskeleton | Cat# AP05 |
| human non-muscle actin | Cytoskeleton | Cat# APHL99 |
| Turbo3C Protease | Accelagen | Cat# H0101S |
| polybrene | EMD Millipore | Cat# TR-1003 |
| Protein A/G PLUS-Agarose | Santa Cruz Biotech | Cat# sc-2003 |
| glutathione sepharose 4B | GE Healthcare Life Sciences | Cat# 17075601 |
| PD-10 column | GE Healthcare Life Sciences | Cat# 17085101 |
| Western Lightening Plus-ECL | PerkinElmer | Cat# NEL103001EA |
| ʎ protein phosphatase | NEB | Cat# P0753S |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| NIH3T3 | ATCC | Cat# CRL-1658, RRID: CVCL_0594 |
| HEK293T | ATCC | Cat# CRL-3216, RRID: CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| Male mus musculus 129S1/SvImJ | N/A | N/A |
| Oligonucleotides | ||
| 5’ primer for inserting a myc tag and making a pMSCV-puro-myc vector: GATCCACCATGGAGCAAAAGCTCATTTCTGAAGAGGACTTGAATGAAA |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a myc tag and making a pMSCV-puro-myc vector: CACCATGGAGCAAAAGCTCATTTCTGAAGAGGACTTGAATGAAAGATC |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a BamHI restriction site in 5’ end of lamin A cDNA: GTTAGGATCCACCATGGAGACCCCGTCCCAG | Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of lamin A cDNA: GTTAGCGGCCGCTTACATGATGCTGCAGTTC |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a myc tag and making a pMYC-C4 vector: CTAGCCACCATGGAGCAAAAGCTCATTTCTGAAGAGGACTTGAATGAAA |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a myc tag and making a pMYC-C4 vector: GATCTTTCATTCAAGTCCTCTTCAGAAATGAGCTTTTGCTCCATGGTGG |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a BamHI restriction site in 5’ end of MEK1 cDNA: GCCGCGGATCCACCATGCCCAAGAAGAAGCCG |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of MEK1 cDNA: GGCCCGCGGCCGCTTAGACGCCAGCAGCATG |
Integrated DNA Technologies | N/A |
| 3’ primer for introducing a MEK1 K97A (KD) point mutation: CGCTCTGGCCATGACCAG |
Integrated DNA Technologies | N/A |
| 5’ primer for introducing a MEK1 K97A (KD) point mutation: CTAATTCATCTGGAGATC |
Integrated DNA Technologies | N/A |
| 5’ primer for introducing MEK1 S218D S221D (CA) point mutations: GATATGGCCAACGACTTCGTGGGCACAAGG |
Integrated DNA Technologies | N/A |
| 3’ primer for introducing MEK1 S218D S221D (CA) point mutations: GTCGATGAGCTGCCCGCTG |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting GST cDNA and making a pMSCV-GST-6P-4 vector: GCCGGGATCCGCCACCATGTCCCCTATACTAGG |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting GST cDNA and making a pMSCV-GST-6P-4 vector: GAAAAGATCTCAGGGGCCCCTGGAACAG |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a BglIII restriction site in 5’ end of SUN2 cDNA: GATAAGATCTACCATGTCCCGAAGAAGCCAG |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of SUN2 cDNA: CATTGCGGCCGCCTAGTGGGCGGGCTCCCC |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a NotI restriction site in 5’ end of nesprin-2 SR49-56-KASH cDNA: GTACGCGGCCGCACCATGACTGCAGAGACCTGGGAC |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of nesprin-2 SR49-56-KASH cDNA: GTTAGCGGCCGCCTAGGTGGGAGGTGGCCC |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a NotI restriction site in 5’ end of nesprin-2 SR51-56-KASH cDNA: GTACGCGGCCGCACCATGTTTGCTTTCATTCAGCAG |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a BamHI restriction site in 5’ end of FHOD1 cDNA: GATAGGATCCACCATGGCGGGCGGGGAAGAC |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of FHOD1 cDNA: CATTGCGGCCGCTCACACCTCCAGGCCAGG |
Integrated DNA Technologies | N/A |
| 3’ primer for introducing a FHOD1 S387A point mutation: GCCTACCGGGGCGGCGGGGCCTGTGGGGCC |
Integrated DNA Technologies | N/A |
| 5’ primer for introducing a FHOD1 S387A point mutation: CCCGCCGCCCCGGTAGGCCCCACCTCTTC |
Integrated DNA Technologies | N/A |
| 3’ primer for introducing a FHOD1 S498A point mutation: GGGCAGGGGCCTGGGGTGTTCTGGCTGC |
Integrated DNA Technologies | N/A |
| 5’ primer for introducing a FHOD1 S498A point mutation: CACCCCAGGCCCCTGCCCCCTGTGTCCTG |
Integrated DNA Technologies | N/A |
| 3’ primer for introducing a FHOD1 S523A point mutation: CTTGGGGGCTGCTGGTATCAGTGGCTC |
Integrated DNA Technologies | N/A |
| 5’ primer for introducing a FHOD1 S523A point mutation: GATACCAGCAGCCCCCAAGGCTGAGCCCATC |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting EGFP cDNA and making a pMSCV-puro-EGFP-C4 vector: CAAAGGATCCATCGCCACCATGGTGAGCAAGGGCGAG |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting EGFP cDNA and making a pMSCV-puro-EGFP-C4 vector: CCCCCAGATCTCTTGTACAGCTCGTCCAT |
Integrated DNA Technologies | N/A |
| 3’ primer for introducing a FHOD1 S498D point mutation: GGGCAGGGTCCTGGGGTGTTCTGGCTGC |
Integrated DNA Technologies | N/A |
| 5’ primer for introducing a FHOD1 S498D point mutation: CACCCCAGGACCCTGCCCCCTGTGTCCTG |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting human EF1a promoter and making a pLVX-EF1a-GST-6P-4 vector: GTTTATCGATGGCTCCGGTGCCCGTCAG |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting human EF1a promoter and making a pLVX-EF1a-GST-6P-4 vector: GGTAGGATCCCTCACGACACCTGAAATG |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting GST cDNA and making a pLVX-EF1a-GST-6P-4 vector: GCCGAGATCTGCCACCATGTCCCCTATACTAGG |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting GST cDNA and making a pLVX-EF1a-GST-6P-4 vector: GAAATCTAGAACCGAATTCAGCGGATCCCAGGGGCCCCTGGAACAG |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a NotI restriction site in 5’ end of FHOD3-1 and −2 cDNAs: GAAAAGCGGCCGCGCCACCATGGCCACGCTGGCTTGTCG |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of FHOD3-1 and −2 cDNAs: GATATGCGGCCGCTCACAGTTGCAGTTCAGATG |
Integrated DNA Technologies | N/A |
| 3’ primer for introducing a FHOD3-1 S497A/D point mutation: GGGAGGTAGCAAGCGAGC |
Integrated DNA Technologies | N/A |
| 5’ primer for introducing a FHOD3-1 S497A point mutation: GCTCGCTTGCTACCTCCCGCCCCTGGCTTGGCCACTCG |
Integrated DNA Technologies | N/A |
| 3’ primer for introducing a FHOD3-1 S523A/D point mutation: GTGGGGCACGTAGGGCAGC |
Integrated DNA Technologies | N/A |
| 5’ primer for introducing a FHOD3-1 S523A point mutation: GCTGCCCTACGTGCCCCACGCCCCCTTCCACCTCTTCTC |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a NotI restriction site in 5’ end of mDia1-2 cDNA: GAAAAGCGGCCGCGCCACCATGGAGCCGTCCGGCGGGGG |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of mDia1-2 cDNA: GATTTGCGGCCGCTTAGCTTGCACGGCCAAC |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a NotI restriction site in 5’ end of nesprin-2 SR11-13 cDNA: GATAAGCGGCCGCACCATGCCTGAGGACAAGAAGTTAC |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of nesprin-2 SR11-13 cDNA: GATATGCGGCCGCCTACCCACTGGGCGCATAGCTG |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a EcoRI restriction site in 3’ end of FHOD1 GBD-DID cDNA: CATTGAATTCTCATCCATCCTCCAATTTC |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of FHOD3 GBD-DID cDNA: GAAAGCGGCCGCGCCACCATGTTTGAGCAGATGCTGGTG |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a NotI restriction site in 5’ end of mDia1-2 cDNA: GAAAGCGGCCGCGCCACCATGTTTGAGCAGATGCTGGTG |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of mDia1-2 cDNA: GAAAGCGGCCGCTCACTCCATGCGGATATCATC |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of FHOD1 ΔDAD cDNA: CATTGCGGCCGCTCACATACTAGCATGACTGTC |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a BamHI restriction site in 5’ end of profilin1 cDNA: GATAGGATCCGCCACCATGGCCGGGTGGAACGCC |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of profilin1 cDNA: GATTGCGGCCGCTCAGTACTGGGAACGCCG |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a BamHI restriction site in 5’ end of profilin2a cDNA: GATAGGATCCGCCACCATGGCCGGTTGGCAGAGCTACG |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of profilin2a cDNA: GATAGCGGCCGCCTAGAACCCAGAGTCTCTC |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of mDia1-2 ΔDAD cDNAs: GATTTGCGGCCGCTTACATCACACCTGTCTCATC |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of FHOD3-1 and −2 ΔDAD cDNAs: GATTTGCGGCCGCCTACATGTTCTCATGTTCGGC |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting human EF1a promoter and making a pEF1a-GST-P-N4 vector: GATTTATTAATGGCTCCGGTGCCCGTCAG |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting human EF1a promoter and making a pEF1a-GST-P-N4 vector: GGTAGGATCCCTCACGACACCTGAAATG |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting Precission cleavage site and making a pEF1a-GST-P-N4 vector: GGCCGCCGGCGAATTCCTGGAAGTTCTGTTCCAGGGGCCCCTGC |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting Precission cleavage site and making a pEF1a-GST-P-N4 vector: AATTGCAGGGGCCCCTGGAACAGAACTTCCAGGAATTCGCCGGC |
Integrated DNA Technologies | N/A |
| 5’ primer for inserting a GST cDNA and making a pEF1a-GST-P-N4 vector: GATTTGAATTCGCCACCATGTCCCCTATACTAGGTTATTG |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a GST cDNA and making a pEF1a-GST-P-N4 vector: GATTTCAATTGTTAATCCGATTTTGGAGGATG |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of FHOD3-1 and −2 ΔDAD cDNAs: GATTTGCGGCCGCCATGTTCTCATGTTCGGC |
Integrated DNA Technologies | N/A |
| 5’ primer for introducing a FHOD3-1 S497D point mutation: GCTCGCTTGCTACCTCCCGACCCTGGCTTGGCCACTCG |
Integrated DNA Technologies | N/A |
| 5’ primer for introducing a FHOD3-1 S523D point mutation: GCTGCCCTACGTGCCCCACGACCCCTTCCACCTCTTCTC |
Integrated DNA Technologies | N/A |
| 3’ primer for inserting a NotI restriction site in 3’ end of nesprin-2 SR51-54 cDNA: GTACGCGGCCGCTCACTGTTTGAGCCTGAGCTTG |
Integrated DNA Technologies | N/A |
| 3’ primer for introducing a nesprin-2 SR51-54 S6376A point mutation: CTGGGGAGCTGTGGGCTCCTCGCTCTC |
Integrated DNA Technologies | N/A |
| 5’ primer for introducing a nesprin-2 SR51-54 S6376A point mutation: GAGCCCACAGCTCCCCAGTCACTGTGTC |
Integrated DNA Technologies | N/A |
| 3’ primer for introducing a nesprin-2 SR51-54 S6471A point mutation: GGGAAGGGGCGTCAGGAACATGCCACGGG |
Integrated DNA Technologies | N/A |
| 5’ primer for introducing a nesprin-2 SR51-54 S6471A point mutation: GTTCCTGACGCCCCTTCCCATTCCAAGCATC |
Integrated DNA Technologies | N/A |
| 3’ primer for introducing a nesprin-2 SR51-54 T6397A point mutation: CTGACGGGGGCCTCACAGCCAGACCGCTC |
Integrated DNA Technologies | N/A |
| 5’ primer for introducing a nesprin-2 SR51-54 S6397A point mutation: GGCTGTGAGGCCCCCGTCAGTGTGGACTC |
Integrated DNA Technologies | N/A |
| Recombinant DNA | N/A | |
| pMSCV-puro-myc | This manuscript. This is derived from pMSCV-puro vector (Clontech) by inserting myc tag sequence in the 5’ side of MCS | N/A |
| pMSCV-puro-myc lamin A WT | This manuscript. (Muchir et al., 2009) | N/A |
| pMSCV-puro-myc lamin A H222P | This manuscript. (Muchir et al., 2009) | N/A |
| pMSCV-puro-myc lamin A E356K | This manuscript. (Folker et al., 2011) |
N/A |
| pMSCV-puro-myc lamin A E203G | This manuscript. (Folker et al., 2011) | N/A |
| pMSCV-puro-myc lamin A N195K | This manuscript. (Folker et al., 2011) | N/A |
| pMSCV-puro-myc lamin A R483W | This manuscript. (Folker et al., 2011) | N/A |
| pMSCV-puro-myc progerin | (Chang et al., 2019) | N/A |
| pMYC-C4 | This manuscript. This is derived from pEGFP-C1 vector (Clontech) by replacing EGFP sequence with myc tag sequence | N/A |
| pMYC-C4 MEK1 WT | This manuscript. Human Hela cDNA | N/A |
| pMYC-C4 MEK1 KD | This manuscript. Human MEK1 cDNA with a K97A (AAG to GCG) point mutation | N/A |
| pMYC-C4 MEK1 CA | This manuscript. Human MEK1 cDNA with S218D (TCC to GAT) and S221D (TCC to GAC) point mutations | N/A |
| pEGFP-C4 miniN2G | (Luxton et al., 2010) | N/A |
| pEGFP-C4 LifeAct | (Kutscheidt et al., 2014) | N/A |
| pMSCV-GST-6P-4 | This manuscript. This is derived from pMSCV-puro (Clontech) by inserting GST in the 5’ side of MCS and deleting the PGK promoter and puro resistance gene | N/A |
| pMSCV-GST-6P-4 EGFP | This manuscript | N/A |
| pMSCV-GST-6P-4 SUN2 WT | This manuscript. (Meinke et al., 2014) | N/A |
| pMSCV-GST-6P-4 nesprin-2 SR49-56-KASH | This manuscript. (Zhu et al., 2017) | N/A |
| pMSCV-GST-6P-4 nesprin-2 SR51-56-KASH | This manuscript. (Zhu et al., 2017) | N/A |
| pMSCV-GST-6P-4 FHOD1 WT | This manuscript. (Kutscheidt et al., 2014) | N/A |
| pMSCV-GST-6P-4 FHOD1 S387A | This manuscript. Human FHOD1 cDNA with a S387A (TCA to GCC) point mutation | N/A |
| pMSCV-GST-6P-4 FHOD1 S498A | This manuscript. Human FHOD1 cDNA with a S498A (AGC to GCC) point mutation | N/A |
| pMSCV-GST-6P-4 FHOD1 S523A | This manuscript. Human FHOD1 cDNA with a S523A (AGC to GCC) point mutation | N/A |
| pMSCV-GST-6P-4 FHOD1 S387A S498A S523A (AAA) | This manuscript. Human FHOD1 cDNA with S387A (TCA to GCC), S498A (AGC to GCC), and S523A (AGC to GCC) point mutations | N/A |
| pGEX-6P-1 | GE Healthcare Life Sciences | N/A |
| pGEX-6P-4 FHOD1 1-569 | (Kutscheidt et al., 2014) | N/A |
| pSUPER.retro.puro shLuc | (Zhu et al., 2017) | N/A |
| pSUPER.retro.puro shFHOD1 | (Zhu et al., 2017) | N/A |
| pMSCV-puro-EGFP-C4 | This manuscript. This is derived from pMSCV-puro (Clontech) by inserting EGFP sequence in the 5’ side of MCS | N/A |
| pMSCV-puro-EGFP-C4 FHOD1 WT | This manuscript. (Kutscheidt et al., 2014) | N/A |
| pMSCV-puro-EGFP-C4 FHOD1 S498A | This manuscript. Human FHOD1 cDNA with a S498A (AGC to GCC) point mutation | N/A |
| pMSCV-puro-EGFP-C4 FHOD1 S498D | This manuscript. Human FHOD1 cDNA with a S498D (AGC to GAC) point mutation | N/A |
| pLVX-EF1a-GST-6P-4 | This manuscript. This is derived from pLVX-puro vector (Clontech) by replacing CMV promoter with EF1a promoter and GST and deleting the PGK promoter and puro resistance gene. | N/A |
| pLVX-EF1a-GST-6P-4 FHOD3-2 WT | This manuscript. Mouse E12.5 forebrain cDNA | N/A |
| pLVX-EF1a-GST-6P-4 FHOD3-1 WT | This manuscript. Mouse adult heart cDNA | N/A |
| pLVX-EF1a-GST-6P-4 FHOD3-1 S497A | This manuscript. Mouse FHOD3-1 cDNA with a S497A (AGC to GCC) point mutation | N/A |
| pLVX-EF1a-GST-6P-4 FHOD3-1 S523A | This manuscript. Mouse FHOD3-1 cDNA with a S523A (AGC to GCC) point mutation | N/A |
| pLVX-EF1a-GST-6P-4 FHOD3-1 S497A S523A | This manuscript. Mouse FHOD3-1 cDNA with S497A (AGC to GCC) and S523A (AGC to GCC) mutations | N/A |
| pMSCV-puro-EGFP-C4 mDia1-2 WT | This manuscript. (Bartolini et al., 2008) | N/A |
| pLVX-EF1a-EGFP-C4 | This manuscript. This is derived from pLVX-puro vector (Clontech) by replacing CMV promoter with EF1a promoter and EGFP and deleting the PGK promoter and puro resistance gene. | N/A |
| pLVX-EF1a-EGFP-C4 FHOD3-2 WT | This manuscript. Mouse FHOD3-1 cDNA with S497A (AGC to GCC) and S523A (AGC to GCC) mutations | N/A |
| pLVX-EF1a-EGFP-C4 FHOD3-1 WT | This manuscript. Mouse FHOD3-1 cDNA with a S497D (AGC to GAC) and S523D (AGC to GAC) mutations | N/A |
| pGEX 6P-4 nesprin-2 SR11-13 | This manuscript. Mouse nesprin-2 1414-1736 aa | N/A |
| pMYC-C4 FHOD1 GBD-DID | This manuscript. Human FHOD1 1-339 aa | N/A |
| pMYC-C4 FHOD3 GBD-DID | This manuscript. Human FHOD3 1-327 aa | N/A |
| pMYC-C4 mDia1 GBD-DID | This manuscript. Mouse Dia1-2 73-377 aa | N/A |
| pGEX-6P-4 nesprin-2 SR51-54 | This manuscript. Mouse nesprin-2 6008-6559 aa | N/A |
| pGEX-6P-4 nesprin-2 SR51-54 S6376A | This manuscript. nesprin-2 SR51-54 cDNA with a S6376A (TCT to GCT) point mutation | N/A |
| pGEX-6P-4 nesprin-2 SR51-54 S6471A | This manuscript. nesprin-2 SR51-54 cDNA with a S6471A (AGC to GCC) point mutation | N/A |
| pGEX-6P-4 nesprin-2 SR51-54 SS->AA | This manuscript. nesprin-2 SR51-54 cDNA with S6376A S6471A point mutations | N/A |
| pGEX-6P-4 nesprin-2 SR51-54 T6397A | This manuscript. nesprin-2 SR51-54 cDNA with a T6397A (ACC to GCC) point mutation | N/A |
| pGEX-6P-4 nesprin-2 SR51-54 STS->AAA | This manuscript. nesprin-2 SR51-54 cDNA with S6376A T6397A S6471A point mutations | N/A |
| pEBG | (Tanaka et al., 1995) | N/A |
| pEBG FHOD1 WT | This manuscript. (Kutscheidt et al., 2014) | N/A |
| pEBG FHOD1 ΔDAD | This manuscript. Human FHOD1 1-1053 aa | N/A |
| pEBG FHOD1 ΔDAD S498D | This manuscript. Human FHOD1 ∆DAD cDNA with a S498D (AGC to GAC) point mutation | N/A |
| pGEX-6P-4 profilin1 | This manuscript. Human Hela cDNA | N/A |
| pGEX-6P-4 profilin2a | This manuscript. Human Hela cDNA | N/A |
| pEBG FHOD1 ΔDAD S498A | This manuscript. Human FHOD1 1-1053 aa cDNA with a S498A (AGC to GCC) point mutation | N/A |
| pEBG FHOD1 ΔDAD I705A | This manuscript. (Kutscheidt et al., 2014) | N/A |
| pEBG mDia1-2 ΔDAD | This manuscript. Mouse Dia1-2 1-1182 | N/A |
| pEF1a-GST-P-N4 | This manuscript. This is derived from pEGFP-C4 vector by replacing CMV promoter and EGFP sequence with EF1a promoter and GST sequence from pEBG vector | N/A |
| pEF1a-GST-P-N4 mFHOD3-1 ∆DAD | This manuscript. Mouse FHOD3-1 1-1477 aa | N/A |
| pEF1a-GST-P-N4 mFHOD3-1 ∆DAD S497D | This manuscript. Mouse FHOD3-1 ∆DAD cDNA with a S497D (AGC to GAC) mutation | N/A |
| pEF1a-GST-P-N4 mFHOD3-1 ∆DAD S523D | This manuscript. Mouse FHOD3-1 ∆DAD cDNA with a S523D (AGC to GAC) mutation | N/A |
| pEF1a-GST-P-N4 mFHOD3-1 ∆DAD S497D S523D | This manuscript. Mouse FHOD3-1 ∆DAD cDNA with S497D S523D mutations | N/A |
| pLP1 | ThermoFisher | Cat# 497500 |
| pLP2 | ThermoFisher | Cat# 497500 |
| pLP-VSV-G | ThermoFisher | Cat# 497500 |
| pHIT 60 | (Soneoka et al., 1995) | N/A |
| pHIT 123 | (Soneoka et al., 1995) | N/A |
| Software and Algorithms | ||
| MetaMorph | Molecular Devices | N/A |
| NIS-Element | Nikon | N/A |
| Fiji | ImageJ | N/A |
| Graphpad QuickCalcs | Prism | N/A |
| SAS | SAS Institute | N/A |
| Excel | Microsoft | N/A |
| Cell plot | (Chang et al., 2013) | N/A |
| CLC Sequence Viewer | Qiagen | N/A |
| Other | ||
Highlights.
Cardiomyocytes expressing pathogenic lamin A/C exhibit altered nuclear positioning
ERK1/2 phosphorylates FHOD1/3 and inhibits nuclear movement in fibroblast
ERK1/2 phosphorylated FHOD1/3 loses its F-actin bundling necessary for nuclear movement
FHOD3 phosphorylation by ERK1/2 is upregulated in dilated cardiomyopathy
ACKNOWLEDGEMENTS
Research reported in this publication was supported by NIH grants RO1 AR068636 to G.G.G. and H.J.W and R01 HL136758 to J.P.M. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTEREST
H.J.W. is on the scientific advisory board of and owns equity in Allomek Therapeutics, which is developing a MEK1/2 inhibitor to treat cardiomyopathy caused by LMNA mutation. The other authors declare no competing interests.
REFERENCES
- Aebi U, Cohn J, Buhle L, and Gerace L (1986). The nuclear lamina is a meshwork of intermediate-type filaments. Nature 323, 560–564. [DOI] [PubMed] [Google Scholar]
- Arimura T, Helbling-Leclerc A, Massart C, Varnous S, Niel F, Lacene E, Fromes Y, Toussaint M, Mura AM, Keller DI, et al. (2005). Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Hum Mol Genet 14, 155–169. [DOI] [PubMed] [Google Scholar]
- Arimura T, Takeya R, Ishikawa T, Yamano T, Matsuo A, Tatsumi T, Nomura T, Sumimoto H, and Kimura A (2013). Dilated cardiomyopathy-associated FHOD3 variant impairs the ability to induce activation of transcription factor serum response factor. Circ J 77, 2990–2996. [DOI] [PubMed] [Google Scholar]
- Auld AL, and Folker ES (2016). Nucleus-dependent sarcomere assembly is mediated by the LINC complex. Mol Biol Cell 27, 2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee I, Zhang J, Moore-Morris T, Pfeiffer E, Buchholz KS, Liu A, Ouyang K, Stroud MJ, Gerace L, Evans SM, et al. (2014). Targeted ablation of nesprin 1 and nesprin 2 from murine myocardium results in cardiomyopathy, altered nuclear morphology and inhibition of the biomechanical gene response. PLoS Genet 10, e1004114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F, Moseley JB, Schmoranzer J, Cassimeris L, Goode BL, and Gundersen GG (2008). The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J Cell Biol 181, 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bione S, Maestrini E, Rivella S, Mancini M, Regis S, Romeo G, and Toniolo D (1994). Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet 8, 323–327. [DOI] [PubMed] [Google Scholar]
- Bonne G, Di Barletta MR, Varnous S, Becane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, et al. (1999). Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat Genet 21, 285–288. [DOI] [PubMed] [Google Scholar]
- Borrego-Pinto J, Jegou T, Osorio DS, Aurade F, Gorjanacz M, Koch B, Mattaj IW, and Gomes ER (2012). Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci 125, 1099–1105. [DOI] [PubMed] [Google Scholar]
- Chang W, Antoku S, and Gundersen GG (2016). Wound-Healing Assays to Study Mechanisms of Nuclear Movement in Fibroblasts and Myoblasts. Methods Mol Biol 1411, 255–267. [DOI] [PubMed] [Google Scholar]
- Chang W, Antoku S, Ostlund C, Worman HJ, and Gundersen GG (2015a). Linker of nucleoskeleton and cytoskeleton (LINC) complex-mediated actin-dependent nuclear positioning orients centrosomes in migrating myoblasts. Nucleus 6, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Folker ES, Worman HJ, and Gundersen GG (2013). Emerin organizes actin flow for nuclear movement and centrosome orientation in migrating fibroblasts. Mol Biol Cell 24, 3869–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Wang Y, Luxton GWG, Ostlund C, Worman HJ, and Gundersen GG (2019). Imbalanced nucleocytoskeletal connections create common polarity defects in progeria and physiological aging. Proc Natl Acad Sci U S A 116, 3578–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Worman HJ, and Gundersen GG (2015b). Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol 208, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzifrangkeskou M, Yadin D, Marais T, Chardonnet S, Cohen-Tannoudji M, Mougenot N, Schmitt A, Crasto S, Di Pasquale E, Macquart C, et al. (2018). Cofilin-1 phosphorylation catalyzed by ERK1/2 alters cardiac actin dynamics in dilated cardiomyopathy caused by lamin A/C gene mutation. Hum Mol Genet 27, 3060–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer WT, and Worman HJ (2009). The nuclear envelope as a signaling node in development and disease. Dev Cell 17, 626–638. [DOI] [PubMed] [Google Scholar]
- Dudley DT, Pang L, Decker SJ, Bridges AJ, and Saltiel AR (1995). A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A 92, 7686–7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd S, Whiffin N, Gavilan MP, Kutscheidt S, De Luca M, Marcozzi C, Min M, Watkins J, Chung K, Fackler OT, et al. (2013). Spatiotemporal organization of Aurora-B by APC/CCdh1 after mitosis coordinates cell spreading through FHOD1. J Cell Sci 126, 2845–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker ES, and Baylies MK (2013). Nuclear positioning in muscle development and disease. Front Physiol 4, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker ES, Ostlund C, Luxton GW, Worman HJ, and Gundersen GG (2011). Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc Natl Acad Sci U S A 108, 131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes ER, Jani S, and Gundersen GG (2005). Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell 121, 451–463. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, and Worman HJ (2013). Nuclear positioning. Cell 152, 1376–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, and Sullivan M (2012). PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res 40, D261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe LR, and Marshall CJ (1993). Lysophosphatidic acid stimulates mitogen-activated protein kinase activation via a G-protein-coupled pathway requiring p21ras and p74raf-1. J Biol Chem 268, 20717–20720. [PubMed] [Google Scholar]
- Huang C, Jacobson K, and Schaller MD (2004). MAP kinases and cell migration. J Cell Sci 117, 4619–4628. [DOI] [PubMed] [Google Scholar]
- Iskratsch T, Yu CH, Mathur A, Liu S, Stevenin V, Dwyer J, Hone J, Ehler E, and Sheetz M (2013). FHOD1 is needed for directed forces and adhesion maturation during cell spreading and migration. Dev Cell 27, 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayo A, Malboubi M, Antoku S, Chang W, Ortiz-Zapater E, Groen C, Pfisterer K, Tootle T, Charras G, Gundersen GG, et al. (2016). Fascin Regulates Nuclear Movement and Deformation in Migrating Cells. Dev Cell 38, 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph LC, Kokkinaki D, Valenti MC, Kim GJ, Barca E, Tomar D, Hoffman NE, Subramanyam P, Colecraft HM, Hirano M, et al. (2017). Inhibition of NADPH oxidase 2 (NOX2) prevents sepsis-induced cardiomyopathy by improving calcium handling and mitochondrial function. JCI Insight 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan-o M, Takeya R, Taniguchi K, Tanoue Y, Tominaga R, and Sumimoto H (2012). Expression and subcellular localization of mammalian formin Fhod3 in the embryonic and adult heart. PLoS One 7, e34765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan-o M, Takeya R, Abe T, Kitajima N, Nishida M, Tominaga R, Kurose H, and Sumimoto H (2012). Mammalian formin Fhod3 plays an essential role in cardiogenesis by organizing myofibrillogenesis. Biol Open 1, 889–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby TJ, and Lammerding J (2018). Emerging views of the nucleus as a cellular mechanosensor. Nat Cell Biol 20, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutscheidt S, Zhu R, Antoku S, Luxton GW, Stagljar I, Fackler OT, and Gundersen GG (2014). FHOD1 interaction with nesprin-2G mediates TAN line formation and nuclear movement. Nat Cell Biol 16, 708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, and Lee RT (2006). Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem 281, 25768–25780. [DOI] [PubMed] [Google Scholar]
- Lele TP, Dickinson RB, and Gundersen GG (2018). Mechanical principles of nuclear shaping and positioning. J Cell Biol 217, 3330–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Gomes ER, Folker ES, Vintinner E, and Gundersen GG (2010). Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science 329, 956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Gomes ER, Folker ES, Worman HJ, and Gundersen GG (2011). TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus 2, 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke P, Mattioli E, Haque F, Antoku S, Columbaro M, Straatman KR, Worman HJ, Gundersen GG, Lattanzi G, Wehnert M, et al. (2014). Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet 10, e1004605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, Carr D, Deng Y, Jin W, Black S, et al. (2013). Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov 3, 742–750. [DOI] [PubMed] [Google Scholar]
- Muchir A, Kim YJ, Reilly SA, Wu W, Choi JC, and Worman HJ (2013). Inhibition of extracellular signal-regulated kinase 1/2 signaling has beneficial effects on skeletal muscle in a mouse model of Emery-Dreifuss muscular dystrophy caused by lamin A/C gene mutation. Skelet Muscle 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchir A, Pavlidis P, Decostre V, Herron AJ, Arimura T, Bonne G, and Worman HJ (2007). Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. J Clin Invest 117, 1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchir A, Reilly SA, Wu W, Iwata S, Homma S, Bonne G, and Worman HJ (2012). Treatment with selumetinib preserves cardiac function and improves survival in cardiomyopathy caused by mutation in the lamin A/C gene. Cardiovasc Res 93, 311–319. [DOI] [PubMed] [Google Scholar]
- Muchir A, Shan J, Bonne G, Lehnart SE, and Worman HJ (2009). Inhibition of extracellular signal-regulated kinase signaling to prevent cardiomyopathy caused by mutation in the gene encoding A-type lamins. Hum Mol Genet 18, 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa JP, Sabater-Molina M, Garcia-Pinilla JM, Mogensen J, Restrepo-Cordoba A, Palomino-Doza J, Villacorta E, Martinez-Moreno M, Ramos-Maqueda J, Zorio E, et al. (2018). Formin Homology 2 Domain Containing 3 (FHOD3) Is a Genetic Basis for Hypertrophic Cardiomyopathy. J Am Coll Cardiol 72, 2457–2467. [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Joseph HL, Chen YJ, Dujardin DL, Alberts AS, Pfister KK, Vallee RB, and Gundersen GG (2001). Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr Biol 11, 1536–1541. [DOI] [PubMed] [Google Scholar]
- Patel AA, Oztug Durer ZA, van Loon AP, Bremer KV, and Quinlan ME (2018). Drosophila and human FHOD family formin proteins nucleate actin filaments. J Biol Chem 293, 532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie RJ, Koo H, and Yamada KM (2014). Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science 345, 1062–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman W, Martins JP, Carvalho FA, Voituriez R, Abella JVG, Santos NC, Cadot B, Way M, and Gomes ER (2017). Myofibril contraction and crosslinking drive nuclear movement to the periphery of skeletal muscle. Nat Cell Biol 19, 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanematsu F, Kanai A, Ushijima T, Shiraishi A, Abe T, Kage Y, Sumimoto H, and Takeya R (2019). Fhod1, an actin-organizing formin family protein, is dispensable for cardiac development and function in mice. Cytoskeleton (Hoboken) 76, 219–229. [DOI] [PubMed] [Google Scholar]
- Schonichen A, Mannherz HG, Behrmann E, Mazur AJ, Kuhn S, Silvan U, Schoenenberger CA, Fackler OT, Raunser S, Dehmelt L, et al. (2013). FHOD1 is a combined actin filament capping and bundling factor that selectively associates with actin arcs and stress fibers. J Cell Sci 126, 1891–1901. [DOI] [PubMed] [Google Scholar]
- Schulze N, Graessl M, Blancke Soares A, Geyer M, Dehmelt L, and Nalbant P (2014). FHOD1 regulates stress fiber organization by controlling the dynamics of transverse arcs and dorsal fibers. J Cell Sci 127, 1379–1393. [DOI] [PubMed] [Google Scholar]
- Sewry CA, Brown SC, Mercuri E, Bonne G, Feng L, Camici G, Morris GE, and Muntoni F (2001). Skeletal muscle pathology in autosomal dominant Emery-Dreifuss muscular dystrophy with lamin A/C mutations. Neuropathol Appl Neurobiol 27, 281–290. [DOI] [PubMed] [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. (2013). LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152, 584–598. [DOI] [PubMed] [Google Scholar]
- Soneoka Y, Cannon PM, Ramsdale EE, Griffiths JC, Romano G, Kingsman SM, and Kingsman AJ (1995). A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23, 628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soonpaa MH, Kim KK, Pajak L, Franklin M, and Field LJ (1996). Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol 271, H2183–2189. [DOI] [PubMed] [Google Scholar]
- Stewart RM, Rodriguez EC, and King MC (2019). Ablation of SUN2-containing LINC complexes drives cardiac hypertrophy without interstitial fibrosis. Mol Biol Cell 30, 1664–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. (2013). Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341, 1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeya R, and Sumimoto H (2003). Fhos, a mammalian formin, directly binds to F-actin via a region N-terminal to the FH1 domain and forms a homotypic complex via the FH2 domain to promote actin fiber formation. J Cell Sci 116, 4567–4575. [DOI] [PubMed] [Google Scholar]
- Takeya R, Taniguchi K, Narumiya S, and Sumimoto H (2008). The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J 27, 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Gupta R, and Mayer BJ (1995). Differential inhibition of signaling pathways by dominant-negative SH2/SH3 adapter proteins. Mol Cell Biol 15, 6829–6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Takeya R, Suetsugu S, Kan OM, Narusawa M, Shiose A, Tominaga R, and Sumimoto H (2009). Mammalian formin fhod3 regulates actin assembly and sarcomere organization in striated muscles. J Biol Chem 284, 29873–29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojo H, Kaieda I, Hattori H, Katayama N, Yoshimura K, Kakimoto S, Fujisawa Y, Presman E, Brooks CC, and Pilch PF (2003). The Formin family protein, formin homolog overexpressed in spleen, interacts with the insulin-responsive aminopeptidase and profilin IIa. Mol Endocrinol 17, 1216–1229. [DOI] [PubMed] [Google Scholar]
- Turgay Y, Eibauer M, Goldman AE, Shimi T, Khayat M, Ben-Harush K, Dubrovsky-Gaupp A, Sapra KT, Goldman RD, and Medalia O (2017). The molecular architecture of lamins in somatic cells. Nature 543, 261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Woychek A, and Jones JCR (2019). Nesprin-2G knockout fibroblasts exhibit reduced migration, changes in focal adhesion composition, and reduced ability to generate traction forces. Cytoskeleton (Hoboken) 76, 200–208. [DOI] [PubMed] [Google Scholar]
- Yeh TC, Marsh V, Bernat BA, Ballard J, Colwell H, Evans RJ, Parry J, Smith D, Brandhuber BJ, Gross S, et al. (2007). Biological characterization of ARRY-142886 (AZD6244), a potent, highly selective mitogen-activated protein kinase kinase 1/2 inhibitor. Clin Cancer Res 13, 1576–1583. [DOI] [PubMed] [Google Scholar]
- Zhu R, Antoku S, and Gundersen GG (2017). Centrifugal Displacement of Nuclei Reveals Multiple LINC Complex Mechanisms for Homeostatic Nuclear Positioning. Curr. Biol. 27, 3097–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data generated during this study are available from the corresponding author upon reasonable request. No new code was generated in this study