Abstract
Data science is likely to lead to major changes in cardiovascular imaging. Problems with timing, efficiency, and missed diagnoses occur at all stages of the imaging chain. The application of artificial intelligence (AI) is dependent on robust data; the application of appropriate computational approaches and tools; and validation of its clinical application to image segmentation, automated measurements, and eventually, automated diagnosis. AI may reduce cost and improve value at the stages of image acquisition, interpretation, and decision-making. Moreover, the precision now possible with cardiovascular imaging, combined with "big data" from the electronic health record and pathology, is likely to better characterize disease and personalize therapy. This review summarizes recent promising applications of AI in cardiology and cardiac imaging, which potentially add value to patient care.
Keywords: artificial intelligence, cardiovascular imaging, deep learning, machine learning
To date, the application of data science to medicine has been less apparent than in other areas of daily life. However, growing interest in its application to clinical practice may portend an era of more dramatic change, and this may be particularly true in imaging. As in other data-heavy areas of biomedical science (such as proteomics, metabolomics, lipidomics, and genetics), the integration of imaging and clinical data, including the electronic health record (EHR) and pathology, may provide new discoveries. indeed, the precision now possible with cardiovascular imaging makes it an important potential contributor to “big data” approaches to sub-phenotype heterogeneous disease groups, such as heart failure and atrial fibrillation, which at present receive uniform rather than personalized treatments. Hypotheses generated from the associations identified from “big data” will inform traditional research designs that will explain these new observations. More importantly, the current model of image acquisition, interpretation, and decision-making presents problems with timing, efficiency, missed diagnoses, and false positive diagnoses. The growth of cardiovascular imaging has come at a significant financial cost, and by facilitating image acquisition, measurement, reporting, and subsequent clinical pathways, artificial intelligence (AI) may reduce cost and improve value. This review summarizes recent promising AI applications in cardiology and cardiac imaging that potentially add value to patient care.
INTRODUCTION TO AI
TERMINOLOGY AND TECHNIQUES.
AI describes a computational program that can perform tasks that are normally characteristic of human intelligence (such as pattern recognition and identification, planning, understanding language, recognizing objects and sounds, and problem solving). In practical terms, AI can be thought of as the ability of a machine, or device, to make autonomous decisions based on data it collects (1). In medicine, this typically involves data (health records or information extracted from images), being used to predict a likely diagnosis, identify a new disease, or select a best choice of treatment (2,3). Initial pioneers of AI emerged in the 1950s. However, tangible progress was not made until robust computational methods were developed to stratify and weight data (2,4), and datasets began to increase significantly in size over the last 25 years. Within the last few years, techniques for processing data through multilayered networks, the unprecedented increase in accessible datasets, and the emergence of user-friendly software packages to work with the data have meant an AI-driven revolution in health care is becoming realistic (3).
Machine learning (ML) is a technique used to give AI the ability to learn. Specifically, the technique can learn rules and identify patterns progressively from large datasets, without being explicitly programmed or any a priori assumptions. ML techniques have been effectively used for prediction and intelligent decision-making in many areas of everyday living, including internet search engines, customized advertising, filtering of spam emails, character recognition and language processing, finance trending, and robotics (5,6). Two key requirements for ML to function are: 1) data that are relevant and detailed enough to answer the question being asked; and 2) a computational ML technique appropriate for the type, amount, and complexity of the data available (1). Finally, what is generated from the machine learning needs to be validated and shown to be useful in clinical practice (Central Illustration).
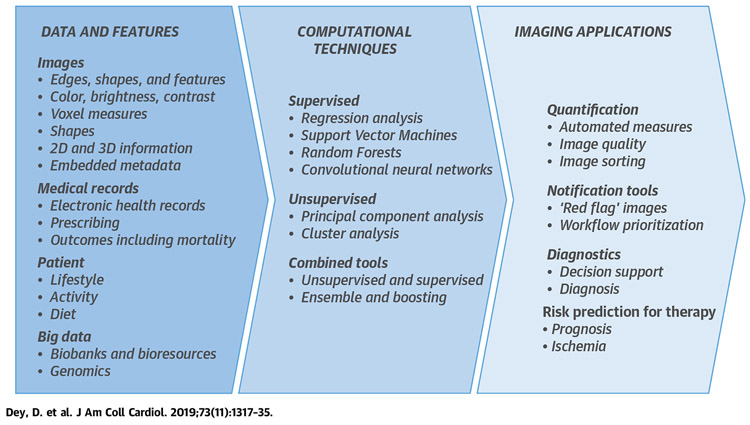
CENTRAL ILLUSTRATION. Steps to Use Artificial Intelligence in Imaging.
The starting point is high quality data–imaging findings are best contextualized with patient, medical records, and “big data.” Computational methods may be supervised, unsupervised, or combined. Imaging applications include quantification, notification tools, diagnostics, and risk prediction. 2D = 2-dimensional; 3D = 3-dimensional.
DATA AND INFORMATION.
“Big data” is used to describe large amounts of collected data. In health care, this includes medical health records, patient results, outcome data, genomic data, and importantly, image-derived information (7). “Biobanks” and “bioresources” are a particular form of organized “big data” collection usually as part of a formal research program with rigorous data collection and quality control (8,9). “Atlases” are big data collections in which data is merged to provide reference information on variation within an organized system, such as the structure of the heart (10,11).
Although “big data” is valuable for the application of AI, it is not essential, and simple ML applications can be applied to most datasets. However, for many datasets to be usable for ML, the original dataset is enriched by the addition of new information pertaining to data structure. This is achieved by breaking the data into individual pieces of information described as features. Each feature describes a particular characteristic or datapoint that can then be used within computational techniques. In imaging, this could be a pixel density or brightness, a vector of motion or a measurement from images, or a clinical report (12). The quality, accuracy, and richness of features in data will determine how effective computational techniques can deliver an AI. Provision of inappropriate or incorrectly categorized data effectively means that the dataset does not resemble the real world closely enough for ML to create a representative model. This may result in inappropriate decisions.
COMPUTATIONAL APPROACHES AND TOOLS.
There are, broadly, 2 possible ways for a machine to learn. Either unsupervised, in which pattern recognition is allowed to develop freely within the data supplied, or the program can be supervised to look for patterns of data that fit a particular outcome of interest (13) (Table 1).
TABLE 1.
Supervised and Unsupervised Approaches to Machine Learning
| Supervised (Ref. #) | |
|---|---|
| Regression analysis (13) | Uncomplicated form of supervised machine learning that generates an algorithm to describe a relationship between multiple variables and an outcome of interest. Stepwise models "automatically" add or remove variables based on the strength of their association with the outcome variable, until a significant model is developed or "learned." |
| Support vector machines (14,15) | Whereas regression analysis may identify linear associations, "support vector machines" provide nonlinear models by defining "planes" in higher-dimension that best separate out features into groups that predict certain outcomes. |
| Random forests (14,15) | Identify the best cutpoint values in different features of individual groups of related data to be able to separate them out to predict a particular outcome. |
| Neural networks | Features are fed through a nodal network of decision points, meant to mimic human neural processing. |
| Convoluted neural networks | A multilayered network, often applied to image processing, simulating some of the properties of the human visual cortex. A mathematical model is used to pass on results to successive layers. |
| Deep learning (DL) (12,16) | DL is defined as a class of artificial neural network algorithms, in which more internal layers are used than in traditional neural network approaches ("deep" merely describes a multilayered separation approach). Often described as convolutional neural networks. |
| Unsupervised (Ref. #) | |
| Principal component analysis (11) | Simple form of unsupervised learning in which the features that account for the most variation in a dataset can be identified. |
| Hierarchical clustering (e.g., agglomerative hierarchical clustering, divisive hierarchical clustering) | Creates a hierarchical decomposition of the data based on similarity with another cluster by aggregating them (an agglomerative approach) or dividing them as it moves down in hierarchy (a divisive approach). Strength–easy comprehension and illustration using dendrograms, insensitive to outliers. Difficulty–arbitrary metric and linkage criteria, does not work with missing data, may lead to misinterpretation of the dendrogram, difficulty finding optimal solution. |
| Partitioning algorithms (e.g., K-means clustering) | Form of cluster analysis that identifies degree of separation of different features within a dataset and tries to find groupings in which features are most differentiated. It does this by defining similarity on the basis of proximity to the centroid (mean, median, or medoid) of the cluster. The algorithm modulates the data to build the cluster by iteratively evaluating the distance from the centroid. Strengths–simple and easy to implement, easy to interpret, fast, and efficient. Has remained relatively underutilized in cardiology despite its simplicity in implementation and interpretation. Difficulty–uniform cluster size, may work poorly with different densities of clusters, difficult to find k, sensitive to outliers. |
| Model-based clustering (e.g., Expectation-Maximization Algorithm) | This clustering algorithm makes a general assumption that the data in each cluster is generated from probabilistic (primarily Gaussian) model. Strengths–distributional description for components, possible to assess clusters-within-clusters, can make inference about the number of clusters. Difficulties–computationally intensive, can be slow to converge. |
| Grid-based algorithms (e.g., Statistical Information Grid-Based Clustering [STING], OptiGrid, WaveCluster, GRIDCLUS, GDILC) | Strengths–can work on large multidimensional space, reduction in computational complexity, data space partitioned to finite cells to form grid structure. Difficulties–difficult to handle irregular data distributions; limited by predefined cell sizes, borders, and density threshold; difficult to cluster high-dimensional data. |
| Density-based spatial clustering of applications with noise | Strengths–does not require number of clusters, can find clusters of arbitrary shapes, robust to outliers. Difficulties–not deterministic, quality depends on the distance measure, cannot handle large differences in densities. |
In supervised learning, there is an iterative analysis of data, with individual features selected, processed, and weighted to identify the best combination to fit the outcome of interest. Regression analysis is an uncomplicated form of supervised ML has been in widespread use for many years. Although this is not typically thought of as AI, stepwise models are effectively learned (13). Newer statistical programs have now emerged to perform this associative analysis in a more sophisticated way, including support vector machines and random forests (14,15). Deep learning (DL) methods mimic human cognition by using multiple layers of convolutional neural networks. DL methods provide a means of informing associations based on previous experience, effectively training the process so that the probability of correct classification increases (12,16). These learning methods have been applied to image segmentation using techniques such as Marginal Space Learning, which undertakes parameter estimation in a series of approximations of object position, orientation, and anisotropy, reducing the number of testing hypotheses by about 6 orders of magnitude compared with full space learning.
In unsupervised learning, the program is not trying to fit data to an outcome, but instead is just trying to identify any potential consistent patterns in the data (10). These approaches include principal component analysis (11) and cluster analysis approaches, which include a vast diversity of the algorithms, including “k-means” clustering (Table 1). The choice of cluster analysis not only depends on the data and the intended use of the results, but also on the domain knowledge of the researcher, which can introduce subjectivity. Newer techniques of clustering have recently evolved for understanding the similarities between patients (Table 2). For example, network graphs are structures that describe the relationship between objects and have played an important role in social networking, engineering, and molecular and population biology (17). A novel and improved technique to integrate and visualize cumulative associations in data, topological data analysis, has been recently adopted and may allow visualization of multidimensional imaging and patient parameters (18-21). Topological data analysis can cluster complex, high-dimensional data when most traditional data mining methods falter. For example, the connection between cardiac imaging utilization and its relationship to hospitalization outcomes can be displayed in a network for isolating patients with high-risk features (Figure 1). Once a model is developed, a new patient’s risk features may be recognized using similarity analysis, and some of these data could be useful for physicians for phenotypic differentiations of cardiovascular diseases. Indeed, combining these different statistical tools increases the discriminatory and learning power of AI. For instance, unsupervised learning can be applied to patients with known outcomes to identify novel features that could then be fed into supervised models (22). Data can also be tested in parallel or sequentially through different methods, such as a “random forest” and a “neural network,” to identify the best fit differentiation, a method known as “ensemble” machine learning. Once the statistical program has identified a consistent way of differentiating data, then the terms algorithm and model start to be used. These refer to the fact a particular pattern has now been identified, which can be used to predict the likely relevance or meaning of future data.
TABLE 2.
Comparison of Clustering With Other Analysis
| Clustering | Classification | Graph | Topology |
|---|---|---|---|
|
|
|
|
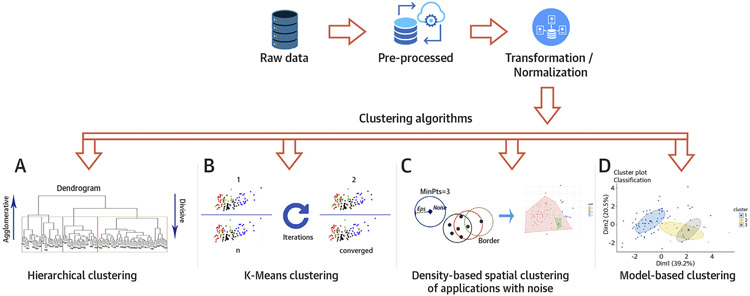
FIGURE 1. Steps in Performing Clustering Algorithm on the Data.
The raw data is initially pre-processed and transformed, if needed. (A) Dendrogram of hierarchical clustering where height is the distance and each leaf represent a patient. The colored boxes represent the patients within the cluster. The number of clusters depends on where the dendrogram is cut. Agglomerative clustering is the bottom-up approach, where the patients are grouped in the higher hierarchy. Divisive clustering is the top-down approach where a single cluster is divided as it moves down the hierarchy. (B) Panel showing k-means cluster at multiple iterations. The algorithm calculates the centroid at each iteration and modulates the cluster until it converges. (C) Eps is the minimum distance between 2 points and MinPts is the minimum number of points to form a dense region. The algorithm searches the points based on these parameters and creates the cluster if the criteria are met. (D) Cluster analysis using model-based algorithm which detects 3 clusters in the mixture using Gaussian probabilistic model.
APPLICATION AND VALIDATION.
After a model or algorithm has been defined, an important next step is to determine whether the model remains accurate if new data are fed into the model. Therefore, typically, data are needed for both “training” (i.e., developing the model) and for “testing” (i.e., seeing how well the defined algorithm continues to predict the same outcome if new data are supplied). For the validation regimen in machine learning, it is very important that the data used for training is not used for testing of ML models. Sometimes the same dataset is presented as being used for both training and testing, but this is only valid if the dataset has been subdivided and the testing performed iteratively on the data not used for training, referred to as “x-fold cross-validation.” For example, training can be performed on 90% of the data and testing on the remaining unseen data iteratively 10 times (23). This would be referred to as 10-fold cross-validation and has been shown to have smaller bias for discriminant analysis than a traditional split-sample approach (training and testing) (24).
With small datasets or datasets without much heterogeneity, there is a risk of overfitting–in effect, the model may only be suited to the dataset from which it was derived. If the dataset available is of sufficient size, then a portion of the data can be exclusively held back for this testing, or new data may be collected from external sources or from a different time period. These new data may have slightly different characteristics to explore how generalizable a model is to other data or to identify what degree of variation in features starts to make the model fail. As methods start to be tested in clinical practice, terminology is also developing around how AI can be used as a tool. Typically these uses include the use of a quantitative strategy to automatically generate measures (25), enabling notification devices to flag up particular problems, and as diagnostic support tools to generate recommendations and related information that can be used by a clinician to reach a conclusion. Fully automated diagnostic tools will ultimately provide medical opinions (12,26), but will also require extensive validation before regulatory approval.
APPLICATION OF AI FOR IMAGE AND DATA INTERPRETATION
DISEASE PHENOTYPING AND CLUSTER ANALYSIS.
Cluster analysis is an unsupervised ML technique that provides a process of creating homogenous, related groups from hidden patterns in data without a priori knowledge (i.e., oblivious to current classification of the data) (Table 1). Clustering can be an extremely effective tool for understanding the connecting links between clinical information from electronic medical records and clinical imaging for discovering relevant disease phenotypes and taxonomies (Table 3). Clustering proliferated in the past decades in various other fields, but its application in cardiology has remained relatively sparse (27-36).
TABLE 3.
Clustering in Cardiac Imaging
| First Author (Ref. #) | Assessment | Type of Clustering | Result |
|---|---|---|---|
| Ernande et al. (27) | Left ventricular function | Agglomerative Hierarchical Clustering | 3 clusters with significantly different echocardiographic phenotypes of type 2 diabetes mellitus. |
| Sanchez-Martinez et al. (28) | Heart failure with preserved ejection fraction | Agglomerative Hierarchical Clustering | 2 groups to capture healthy and heart failure patients with preserved ejection fraction. |
| Shah et al. (29) | Heart failure with preserved ejection fraction | Agglomerative Hierarchical clustering and model-based clustering | Hierarchical clustering to conceptualize similar and redundant features of phenotypic features. Model-based clustering with Gaussian distribution identified 3 clusters where cluster 1 had the least severe electric and myocardial remodeling while cluster 3 had the most severe. |
| Katz et al. (30) | Heart failure with preserved ejection fraction | Model-based clustering | 2 clusters with significantly different phenogroups of hypertensive patients. |
| Omar et al. (40) | Left ventricular diastolic dysfunction | Agglomerative Hierarchical Clustering | 3 groups corresponding to severity of diastolic dysfunction based on speckle-tracking echocardiography data. |
| Lancaster et al. (32) | Left ventricular diastolic dysfunction | Agglomerative Hierarchical Clustering | 2 groups with the severity of diastolic dysfunction. |
| Horiuchi et al. (33) | Acute heart failure | k-means clustering | 3 clusters with vascular failure, renal failure, and older patients with atrial fibrillation and preserved ejection fraction, respectively. |
| Bansod et al. (34) | Endocardial border estimation and ellipse fitting on more than 10 video sequences | Density-based spatial clustering of applications with noise | Endocardial border estimation using density-based spatial clustering of applications with noise. |
| Carbotta et al. (35) | Indication of coronary heart disease | Oblique principal components clustering procedure |
2 groups with elevated thyroid parameter with reduced cardiovascular parameter increased metabolic parameter to indicate coronary heart disease. |
| Peckova et al. (36) | Diastolic dysfunction | Hierarchical Clustering | 2 groups showed no association of deterioration of left ventricular relaxation with a mild-to-moderate decrease in eGFR. |
CLINICAL APPLICATIONS OF TYPES OF CLUSTERING.
Clustering may be used as a stand-alone tool to acquire insight into the distribution and complex summary of the cardiac imaging parameters or patient data, an exploratory or hypothesis generation and testing method, or a pre-processing tool for other ML algorithms (Table 1). Hierarchical clustering (Figure 2) is particularly relevant for cardiac diseases that are highly heterogenous. In hypertensive patients, model-based clustering has been used to show 2 phenogroups with distinct pathophysiological subtypes that may benefit from targeted therapies (30). Similarly, the k-means approach has been used to identify subgroups of patients with differing degrees of organ damage and the genetic patterns of renin-angiotensin-aldosterone system polymorphisms in hypertensive patients (37). Several other algorithms, including density models, have been used in breast cancer research (38), identifying protein complexes (39), and segmentation of lung nodule image sequences.
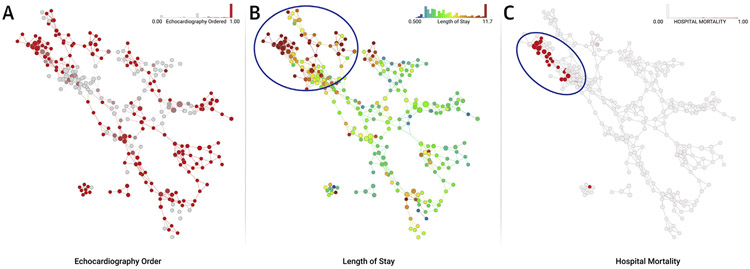
FIGURE 2. Topological Data Analysis for Isolating Patient Clusters.
An existing published dataset containing clinical features of hospitalized patients with inpatient echocardiography utilization was used to create a network where clinically similar hospitalized subjects clustered in nodes and connected with overlapping subjects to form the edges. The topological network allows rapid visualization and interpretation of outcomes of interest. The distribution and frequency of echocardiograms performed in hospitalized patient clusters is shown in shades of red (A). The nodes are subsequently color coded with outcomes of interest like the length of stay (in days) to reflect visually any relationships with performance of echocardiography (B). The upper left region of the map (circle) shows an area where there is high utilization of echocardiography. Interestingly, this region has patients who are sicker, have longer length of stay (B), and also higher mortality (C).
Cluster analysis may provide robust opportunities for grouping relevant clinical and imaging information. Hierarchical clustering has been used to identify 3 clusters with different echocardiographic phenotypes that emphasize the prognostic advantage of left ventricular (LV) remodeling in patients with diabetes mellitus (27). Several other investigators have recently applied hierarchical clustering to understand phenotypic presentations in heart failure. Hierarchical clustering has been used to aggregate LV strain data to isolate 3 distinct clusters of patients with different severities of diastolic dysfunction and LV filling pressures (40). In another study, a similar agglomerative hierarchical clustering showed that 2 different clusters of patients with diastolic dysfunction showed differences in all-cause mortality, all-cause rehospitalization, cardiac mortality, and cardiac rehospitalization. A propensity score was generated to assess the likelihood of cluster membership and established that clustering was superior to expert consensus-based echocardiography guideline recommendations for classifying the severity of diastolic dysfunction. Similarly, other investigators have also used hierarchical clustering in heart failure patients with preserved ejection fraction (EF) to identify differences in clinical, cardiac structural, and invasive hemodynamics and associate these with clinical outcomes (32).
Clustering has been relatively little used in cardiovascular image processing, but may offer some value. An algorithm called density-based spatial clustering of applications with noise can help localizing the LV endocardial border in echocardiography for extracting cardiac measurements such as wall motion, area, and 3-dimensional (3D) visualization (34).
ROLE OF AI IN DIAGNOSTIC SUPPORT
Most of the diagnostic support for cardiovascular imaging assumes the segmentation of anatomical structures such as ventricles, valves, or coronaries, and the precise measurements of various parameters, including EF, perfusion defect, or the extent of coronary stenosis.
IMAGE SEGMENTATION.
Segmentation is the process of content extraction that takes as an input a medical image, volume, or sequence of images or volumes to produce associated shapes, such as 2D contours or 3D meshes. A 3D mesh represents the vertices of an anatomical 3D object, defined, for example, by the chambers of the heart or heart vasculature. Cardiovascular image segmentation is a developing field in which AI-based (especially DL) methods have recently shown substantial improvements in performance.
Multiple segmentation methods rely on landmarks or key points that anchor the contour to specific and well-defined anatomical points, such as the apex of the LV. One of the most recent methods for robust localization of anatomical landmarks relies on multiscale deep reinforcement learning (41,42). The modeling of anatomical appearance and the search for landmarks are coupled in a behavioral framework that exploits the strengths of deep reinforcement learning and multiscale representations. As a result, the method is trained not only to distinguish the target anatomical landmark, but also how to localize the landmark by learning and following an optimal navigation path through the volume. The reported method has been tested on 5,000 3D computed tomography (CT) volumes, totaling 2,500,000 slices. It achieves complete accuracy at detecting whether the landmarks are captured in the field-of-view of the scan, localizing multiple cardiovascular landmarks in <1 s.
Once specific landmarks in the data are known, various AI-powered segmentation methods can be called to delineate the heart and vascular structures. One of the earliest publications in this category relied on a database of annotated echocardiography images of the LV. It trains an adaptive boosting classifier to detect the localization, orientation, and scale of the LV and infer its shape, based on the joint distribution of appearances and shapes in the database. The method has been extended to 3D by formulating a learning process in marginal spaces of reduced dimensionality, and employing a probabilistic boosting tree as a classifier. The first application covered the segmentation of all 4 chambers of the heart from CT images (43). More recently, DL methods combined with level sets have been applied for the segmentation of LV from cardiac magnetic resonance (CMR) images (44). To evaluate myocardial motion, segmentation is typically followed by a temporal tracking algorithm that follows the myocardial border across frames (45,46).
Modeling and segmentation of the heart valves have received special interest due to the emergence of minimally invasive valve implant procedures, which require valve modeling and precise sizing of the implants. Marginal space learning has been used to model and segment all 4 heart valves (mitral, aortic, pulmonary, and tricuspid) from cardiac CT, together with the pulmonary trunk (47). In addition, this method used a constrained multilinear shape model to represent complex spatio-temporal variations of the heart valves. Modeling of the complex structures of mitral and aortic valves, including subvalvular apparatus, has been also proposed from transesophageal echocardiography data (48,49).
Interventional cardiology demands advanced imaging, modeling, segmentation of anatomical structures of the heart, and multimodality registration, all with a focus on real-time guidance. C-arm x-ray is one of the common modalities in the operating room due to its versatility to switch between 3D (CT) and real-time x-ray. As a result, multiple efficient methods for C-arm CT have been developed, including the segmentation of left atrium and pulmonary veins for image-guided ablation of atrial fibrillation (50) and aorta segmentation and valve landmark detection for transcatheter aortic valve implantation (51). The latest methods for registration are inspired by deep reinforcement learning and formulate image registration as strategy learning, with the goal of finding the best sequence of motion actions that result in image alignment (52).
Epicardial adipose tissue (EAT) is a metabolically active fat depot within the visceral pericardium that directly surrounds the coronary arteries. Several studies have shown that EAT exerts a local pathogenic effect on coronary vasculature and on the heart (53-57) and is related to adverse cardiovascular events (57,58). Deep learning has been used to perform automated quantification of EAT to measure EAT volume and density from calcium scoring CT scans in a study with 250 patients. In that study, fully automated quantification showed high agreement and correlation to expert manual measurement (59) (R = 0.924; p < 0.00001).
AUTOMATED MEASUREMENTS.
M-mode and Doppler echocardiography are common imaging modalities used for cardiac examination. Being based on 1 single interrogation beam, M-mode is capable of high temporal and spatial resolution along a single scan-line, and is thus effective in capturing subtle motion patterns. The Doppler signal is widely used to acquire a velocity-time image for assessing valvular regurgitation and stenosis. ML can be employed to automate (60) a number of measurements associated with M-mode (e.g., right ventricle internal dimension in diastole, interventricular septum thickness in diastole/systole, LV internal dimension in diastole/systole, LV posterior wall thickness in diastole/systole) and Doppler (e.g., mitral inflow, aortic regurgitation, tricuspid regurgitation). Research in B-mode echocardiography has been pursued to detect and estimate locally abnormal wall motion of the LV for early detection of coronary heart disease (61). Furthermore, the development of AI systems that directly evaluate medical images to derive a diagnosis (i.e., end-to-end learning) is an increasingly popular approach.
The detection and grading of coronary stenosis in computed tomography angiography (CTA) is an important field of cardiac measurements (62) based on coronary centerline tracing, lumen segmentation, and stenosis detection and classification. The hemodynamic importance of a given stenosis can be estimated through AI techniques, an efficient method with accuracy comparable to computationally expensive 3D flow simulations (63).
The development of personalized models from patient data that estimate not only the heart’s anatomy and dynamics, but also the hemodynamics (64), electrophysiology (65,66), and biomechanics (67) are capturing increasing attention. Such AI-based technologies, allowing comprehensive assessment of the heart, vasculature, and cardiac disease, will provide important steps toward precision medicine (68,69).
ROLE OF AI IN IMAGE INTERPRETATION
IMAGING DATABASES.
A critical aspect of ML for cardiac imaging applications is the availability of the data for learning and training. The quality and scope of the data will determine the applicability and accuracy of the algorithm, regardless of the ML approach. In cardiology, several efforts are underway to create large databases that allow development and application of ML systems. These databases can be created from the EHR, but imaging datasets are usually not integrated in these records and require separate time-consuming data collection and verification. Additionally, outcome data–key for training of ML systems–are not typically available in the EHR. Keeping in mind the goal of translating ML into wider clinical practices, it is of key importance that these databases include sufficient “real-world” heterogeneity, reflecting the spectrum of practice and imaging protocols in the field. Although there are large multicenter clinical databases containing cardiac imaging information, most often the imaging variables are limited to subjective visual scoring results. There are some recent initiatives, however, to develop imaging databases for ML that contain raw patient images. These will be invaluable in the development of ML methods for cardiac imaging.
An example of a cardiac imaging database that can be utilized for image-based ML is the CMR image database created for the 2016 Kaggle Data Science Bowl competition, in which >1,000 CMR datasets were provided by the National Institutes of Health. In this competition, the goal was to automatically compute LV end-systolic and -diastolic volumes from deidentified cine CMR. The LV volumes were also labeled by the experts for validation, and teams competed for the most accurate determination of LV volumes. However, currently, this database is limited to image analysis only, as it does not contain clinical data endpoints or outcomes.
Another large international imaging registry (>20,000 image cases from 9 centers), with associated clinical data and outcomes (revascularization, invasive coronary angiography, and major adverse cardiac events), was recently established with the goal of developing image-based ML for nuclear cardiology (70). The REFINE SPECT (REgistry of Fast Myocardial Perfusion Imaging with NExt generation SPECT) includes clinical data variables, stress test variables, and DICOM (Digital Imaging and Communications in Medicine) image datasets from single photon emission computed tomography (SPECT) myocardial perfusion image (MPI) scans (including gated, static, stress, and rest images) (71). Pre-processing by quantitative software allows derivation of individual imaging features including regional variables (Figure 3). Over 200 standard imaging features were automatically derived from images and thus were not influenced by the variability of clinical interpretation.
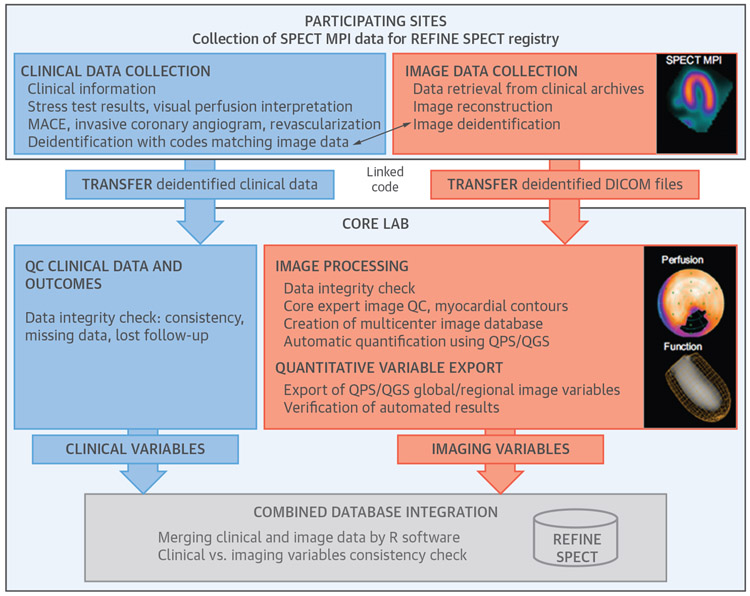
FIGURE 3. Organization and Content of the REFINE SPECT Registry for the Purposes of Machine Learning.
(Blue) Clinical data collection and analysis; (orange) imaging data collection and analysis; and (gray) integration of clinical and imaging databases. Reproduced with permission from Slomka et al. (71). MACE = major adverse cardiovascular events; MPI = myocardial perfusion imaging; QC = quality control; QGS = quantitative gated SPECT; QPS = quantitative perfusion SPECT; SPECT = single photon emission computed tomography.
AI METHODS IN CARDIOVASCULAR IMAGING.
Two distinct approaches have been reported in the application of AI to cardiac imaging. Classical ML methods have been used with a multitude of clinical and/or pre-computed image features to predict diagnostic or prognostic outcomes from large datasets. More advanced AI methods, such as DL methods, have been applied to actual images to obtain diagnoses. Unlike conventional AI approaches, DL does not require so-called “feature engineering” (i.e., computation and extraction of “custom-tailored” imaging variables), but instead directly interrogates images for image segmentation or outcome prediction tasks. DL is particularly suited for large and complex datasets with many features–for example, genomics and imaging datasets.
Classical AI.
The underlying principle of this approach is that a set of weak base classifiers can be combined to create a single strong classifier by iteratively and automatically adjusting their appropriate weighting. A series of base classifier predictions and an updated weighting distribution are produced per iteration. These predictions are then combined by weighted majority voting to derive an overall classifier–the ML risk score ranging from 0 to 1–as a continuous estimate of the predicted risk.
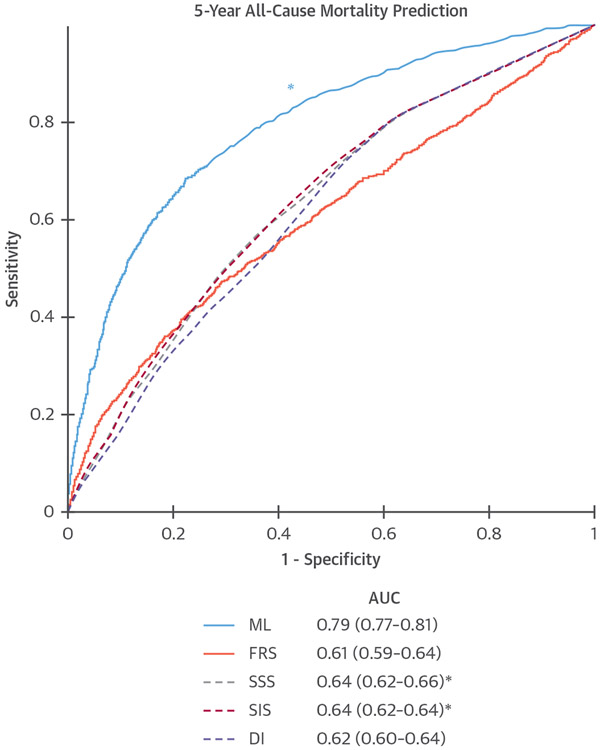
The accuracy of classical AI (LogitBoost) to predict all-cause mortality at 5-year follow-up was evaluated in the CONFIRM (Coronary CT Angiography EvaluatioN For clinical Outcomes: An InterRnational Multicenter registry) (N = 10,030). All available clinical (25 parameters) and visually assessed CTA parameters (44 parameters) were objectively evaluated. ML involved automated feature selection by information gain ranking, followed by model building with LogitBoost and 10-fold cross-validation. An ML risk score combining clinical and CTA data exhibited a significantly higher area under the curve (AUC) (ML AUC = 0.79) for the prediction of death compared with established risk indexes and visual CTA assessment (Figure 4) (23). After age, the number of segments with noncalcified and calcified plaque had the highest information gain for all-cause mortality (23).
FIGURE 4. ML to Predict All-Cause Mortality.
Receiver-operating characteristic curves for prediction of death with 5-year follow-up compared to the Framingham risk score (FRS) and computed tomography angiography (CTA) severity scores (Segment Stenosis Score [SSS], Segment Involvement Score [SIS], modified Duke Index [DI]). *ML had significantly higher AUC than all other scores (P < 0.001). Reproduced with permission from Motwani et al. (23). AUC = area under the curve; ML = machine learning.
In >6,800 asymptomatic patients undergoing coronary calcium scoring (CCS), in MESA (Multi-Ethnic Study of Atherosclerosis), AI demonstrated superior performance to CCS to predict adverse cardiovascular events (72). A composite risk score of coronary CTA stenosis and plaque measures has been shown to significantly improve identification of impaired myocardial flow reserve by 13N-ammonia positron emission tomography (ML AUC = 0.83 vs. CTA Stenosis = 0.66) (73).
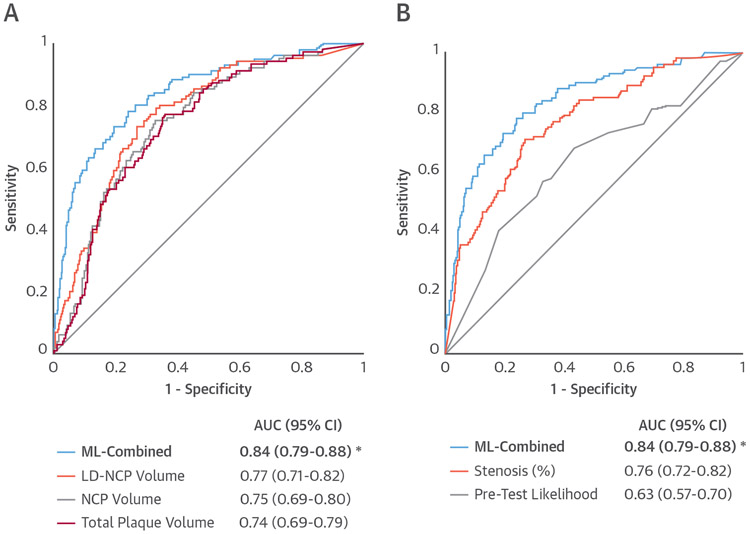
AI has also been applied to predict lesion-specific ischemia. In the NXT (Analysis of Coronary Blood Flow Using CT Angiography: Next Steps) trial, in which 254 patients underwent CTA prior to invasive coronary angiography with fractional flow reserve (FFR), an assessment of plaque characteristics was shown to improve the discrimination of lesion-specific ischemia compared with stenosis alone (74). A substudy explored whether clinical data, quantitative stenosis, and plaque metrics from CTA could be effectively combined with AI to predict lesion-specific ischemia (75). This combination provided a higher AUC for predicting ischemia than pre-test likelihood of coronary artery disease or quantitative CTA metrics (ML AUC = 0.84 vs. best clinical score = 0.63, CTA stenosis = 0.76, low-density noncalcified plaque volume = 0.77; p < 0.006) (75) (Figure 5). Such machine learning risk scores can be incorporated back into software tools and improve assessment of patient risk (Figure 6) (75). A random forest method to integrate several image-derived features has also been applied to estimate CTA image quality with equivalent results as expert visual assessment (76).
FIGURE 5. Prediction of Lesion-Specific Ischemia by the Integrated Ischemia Risk Score by ML-Combined.
(A) ML-combined versus quantitative plaque volumes (LD-NCP [low density noncalcified plaque], NCP, and total plaque volume).
(B) ML-combined versus quantitative stenosis and pre-test likelihood of coronary artery disease. ML-combined had a significantly higher AUC compared with individual quantitative CTA plaque measures or the pre-test likelihood. *indicates AUC significantly different (p < 0.05) than that from the other measures. Reproduced with permission from Lee et al. (12). Abbreviations as in Figure 4.
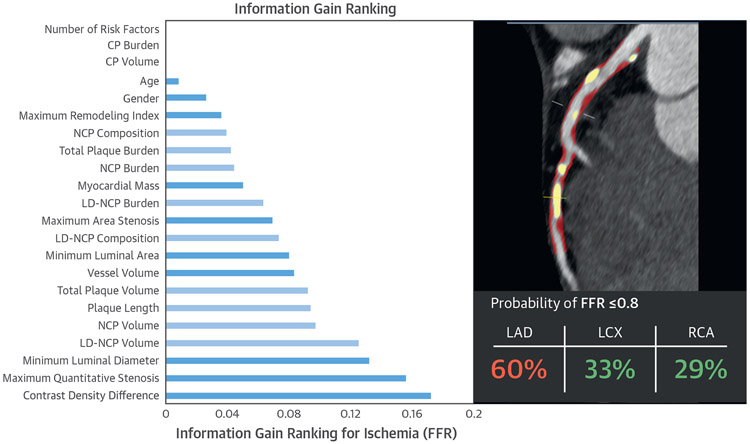
FIGURE 6. Information Gain for Age, Sex, and Quantitative CTA Measures for Lesion-Specific Ischemia.
In the left panel, measures directly related to plaque volumes are in light blue and the remaining measures are in dark blue. Variables with information gain >0.001 were used in machine learning. Contrast density difference had the highest information gain among quantitative CTA metrics. Reproduced with permission from Dey et al. (75). The right panel shows an example of the machine learning prediction of lesion-specific ischemia in a patient undergoing CTA. NCP and CP are shown in red and yellow image overlay in the left anterior descending (LAD) artery of a 67-year-old male symptomatic patient undergoing CTA, along with the integrated machine learning ischemia risk score (60% in the LAD). Invasive FFR measured in the LAD artery was 0.73. FFR = fractional flow reserve; LCX = left circumflex artery; RCA = right coronary artery; other abbreviations as in Figures 4 and 5.
Deep learning.
The seminal work by Krizhevsky and Hinton (77) applied DL to natural image classification of 1.3 million images, with over 1,000 identifiable objects on the images. The output of DL could provide a diagnosis, prediction, interpretation, or (more commonly) a transformed image–for example, anatomical labeling of the dataset, or an image with improved quality.
Following the spectacular success of DL in the computer vision field, numerous applications of DL to medical imaging have been proposed, with particular growth in the last 2 years. One of the contributing factors to this growth is the recent availability of relatively cheap computing graphic processing units (developed initially for the computer gaming industry), and multiple open software DL toolkits available to all researchers. DL algorithms have been applied to a fully automated organ or lesion segmentation, detection, and less commonly for classification, demonstrating large gains in performance compared with traditional methods. The most common application to date has been the analysis of pathology images, but several cardiac image analysis methods have recently been proposed.
In cardiology, DL has been applied to the segmentation of CMR, CT and ultrasound images of the LV, CCS (78), and coronary centerline extraction (79,80). The research has been enabled by the availability of publicly accessible training image datasets, especially for CMR, where a large publicly available repository has been created. In fact, the winning teams in the 2016 Kaggle CMR competition utilized a DL approach to estimate the LV volumes. In 1 published example of this work, researchers demonstrated successful fully automated measurement of LV volumes utilizing a total of 1,340 subjects for training and validation (81). DL approaches outperformed previous algorithms, which relied on painstaking feature engineering and image processing. The obtained DL results were also comparable to the reported inter-reader variability values for multiple independent expert readers. Considering that a skilled cardiologist must routinely analyze CMR scans to determine EF, which can often take up to 20 min to complete, such a fully automated approach may be of great clinical value, and may represent the first application of DL into clinical practice.
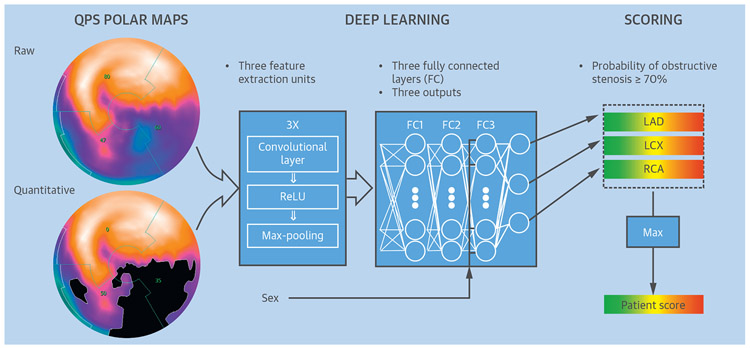
To date, to our knowledge, only 1 application in cardiology has attempted direct diagnosis by DL from the whole patient image. The challenge in this task is obtaining sufficient data, because the diagnosis is performed on a per-vessel or per-patient basis (utilizing the entire 3D or 4-dimensional image), in contrast to segmentation tasks, which are usually performed in 2D on a per-slice basis. Betancur et al. (82) used 1,638 SPECT MPIs from the REFINE SPECT registry, and obtained by the latest-generation SPECT cameras, in patients with suspected coronary artery disease (82) to train and validate a DL system for automatic detection of angiographically significant disease from SPECT MPI. To manage computational efficiency and memory, they utilized automatically derived compact 2D polar map displays as an initial input to the convolutional neural network, rather than entire 3D image datasets (Figure 7). The DL network was trained to detect obstructive disease in each vascular territory. With this approach (including cross-validation to separate training and testing data) they were able to outperform the current standard for quantification of these images on both per-patient and per-vessel basis. The time needed for the evaluation of a new patient with the pre-trained model was <1 s even without the dedicated graphics board used during the training of the system.
FIGURE 7. Training of a Deep Convolutional Neural Network.
Patterns of SPECT perfusion defects are identified by feature extraction (left) into a deep learning process (center) that combines parameters of location, shape, and density. This generates a probability of obstructive coronary artery disease in the left anterior descending artery (LAD), left circumflex artery (LCx), and right coronary artery (RCA) territories (right), which is trained by obstructive stenosis correlations by invasive coronary angiography. FC = fully connected layer; Max-pooling = filter that retains only the maximum value in a 2 × 2 patch; QPS = quantitative perfusion SPECT; ReLU = rectified linear unit (linear function mapping input to output values with a threshold). Adapted with permission from Betancur et al. (82).
DL in cardiac CT.
DL has been recently applied to cardiac CT–both for automated CCS from low-dose CT as well as CTA, showing good agreement with the expert reader (78,83).
DL can be used for image-based identification of disease or outcome prediction. In a recent study, DL was used for automated analysis of standard coronary CTA images to identify hemodynamically significant coronary stenosis (84). DL has also enabled faster onsite computation of noninvasive FFR. In recent studies, a DL method was trained on a large database of synthetically generated coronary models and was shown to be equivalent to an onsite computational fluid dynamics-based algorithm in a study of 85 patients with CTA followed by invasive FFR (AUC to predict invasive FFR: 0.91 for both), with the DL algorithm requiring significantly shorter execution times (about 2 s) (63,85). This method has been validated very recently in data from 5 centers, to compute onsite FFR from CTA in a consortium of 351 patients (with 525 vessels interrogated with invasive FFR) (86).
FULLY AUTOMATED DIAGNOSIS.
While most current applications of ML on cardiology image data were applied to segment the images or derive some quantitative parameters, researchers have also attempted to provide a classification of disease and diagnosis by ML. Ultimately, the physician’s final clinical diagnosis usually requires considering additional clinical information such as age, patient history, and symptoms, in addition to features extracted from the images. This complex task is currently performed “ad hoc” by physicians–often without clearly defined probabilistic algorithms. ML methods can potentially provide a rapid and precise computation of post-imaging disease or outcome probability, based on the integration of imaging and clinical variables. This approach was demonstrated in several recent studies, particularly in SPECT MPI, where the level of automation for image analysis is high compared to other modalities.
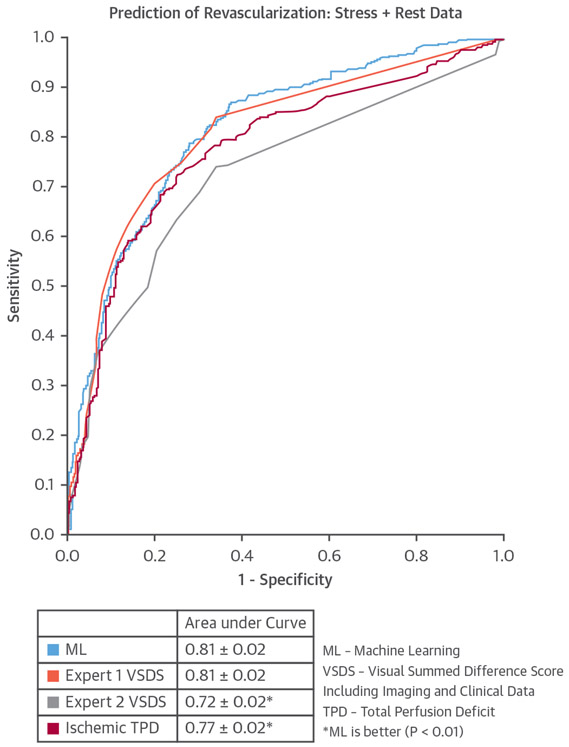
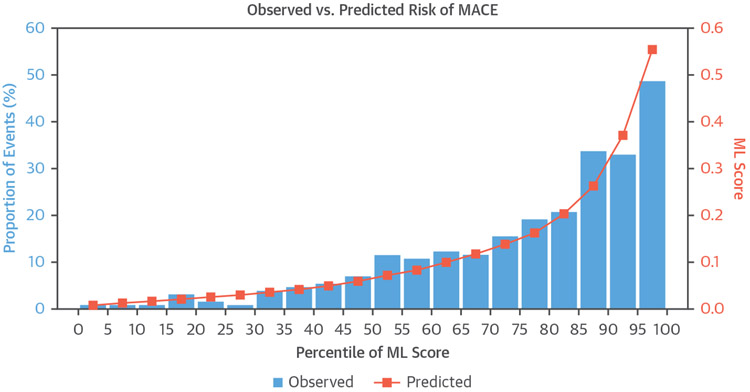
In a single-center study, the LogitBoost ML method was applied to integrate clinical and quantitative perfusion assessment (total perfusion deficit) from SPECT MPI images in 1,181 patients. When clinical information was provided to ML in addition to the imaging features, ML achieved higher AUC (0.94 ± 0.01) than total perfusion deficit (0.88 ± 0.01) or 2 visual readers (0.89, 0.85; p < 0.001), for the detection of coronary disease (22,87). In another study, ML was demonstrated to predict which patients would undergo early revascularization after SPECT MPI using 713 patient studies with available invasive angiography and SPECT MPI images, and compared with 2 experienced, board-certified clinicians (88). Quantitative SPECT MPI parameters were integrated with basic clinical parameters (patient sex, history of hypertension and diabetes, ST-segment depression on ECG, ECG and clinical response during stress, and post-ECG probability). The AUC for revascularization prediction by ML was similar to that for the visual scores of one reader and superior to that of the other reader (Figure 8). A similar LogitBoost approach was also shown to predict major adverse cardiac events in 2,619 patients with SPECT MPI more accurately than expert visual read or standard quantification (82), with good agreement for predicted and observed event rates (Figure 9). Unlike human readers, the ML algorithms provide a continuous probability for a given outcome, which can aid the clinicians in final diagnostic decision and treatment choice.
FIGURE 8. The ROC Curves Comparing the ML Algorithm Versus Ischemic TPD and Expert Visual SDS for Predicting Revascularization.
Reproduced with permission from Arsanjani et al. (88). ML = machine Learning; ROC = receiver-operating curve; TPD = total perfusion deficit; VSDS = visual summed difference score.
FIGURE 9. Observed Proportion of Events and Predicted ML Score Grouped by Every Fifth Percentile of Risk.
Blue bars indicate observed proportion of events, and orange points indicate predicted ML. Adapted with permission from Betancur et al. (70). MACE = major adverse cardiovascular events; ML = machine learning.
PITFALLS AND PROBLEMS
Although these initial results are exciting–potentially dramatically enhancing uniformity and objectivity of the patient diagnosis–many steps need to be undertaken for this approach to be translated into everyday clinical practice. Even if strict multiple split-sample regimens (10-fold cross validation) are used for ML validation, multicenter evaluations will be essential before clinical deployment (especially when clinical features are involved) to demonstrate the generalizability to various cohorts. A step further would be the external validation, where the developed models are applied to the population from centers that did not participate in model creation. When comparing ML techniques with currently established techniques, the latter is in some cases a visual assessment. Depending on the reference standard, ML might yield a continuous probability for a “normal” or “abnormal” scan diagnosis (e.g., positive or negative perfusion scan or stenosis severity), or as a means of quantification of an imaging biomarker (e.g., extent of perfusion abnormality or epicardial adipose tissue). Clearly, depending on the reference standard and validation, the utility of ML will be different for each of these tests.
Ultimately, the most important ML application could be the quantitative ML-aided prediction of the potential benefit of available patient therapy, rather than only patient diagnosis. For example, it was previously established in a single-center study that a 10% threshold for ischemia after SPECT MPI represents a threshold for the benefit of revascularization versus therapy (89). In the era of patient-specific treatment prescriptions, it is conceivable that ML algorithms utilizing all available data could be more precise. Although such end-to-end ML application could be potentially seen as a threat by physicians, they and their patients will ultimately benefit from having the ability for individualized and objective treatment recommendations (5).
Even if an ML system is proven effective and thoroughly validated, there are practical difficulties in potential clinical deployment, such as data quality. Homogenization of clinical data recording and imaging protocols will be essential before data from different centers can be used as input by a standard ML model. Although the use of clinical data adds significant information to images, it will require seamless integration of EHR data with the image datasets.
The topic of cost for the development of ML has received scant attention. The admittedly early steps summarized in the previous text have been based on extensive software development and IT infrastructure, as well as large clinical datasets. Further advances to more complex ML will require significant investment.
Finally, there are legal issues. Typically, current diagnostic software tools are considered by the U.S. Food and Drug Administration in the United States as Class 2 (medium-risk) devices, with their role solely as a diagnostic aid for the physicians. If fully automated systems were to be used ultimately for the treatment prescription or diagnosis, they would need to be classified as Class 3 (high-risk) devices, with a much higher bar concerning performance and validation. Although overall performance of the ML maybe higher than that of the physician, there will always be individual cases where a physician could provide a better answer. Therefore, it is likely that physician control and override of these ML systems will always be required in the foreseeable future.
CURRENT APPLICATIONS
IMAGE ACQUISITION.
The broader medical applications of AI techniques include guiding how people perform procedures such as imaging scans, optimizing patient flows through departments, or automating image processing (3,90). The recognition of imaging planes may guide inexperienced clinicians in the acquisition of high-quality scans, and may also be used to resequence the display of studies so as to improve reading efficiency.
IMAGE ANALYSIS.
Medical imaging has been a vanguard for application of AI in medical practice due to the existing reliance on expert image interpretation. Radiology has been leading this field, and cardiovascular imaging is seeking to parallel its advances for optimizing the accuracy and quality of images (16,90,91). Medical imaging has provided some particularly interesting opportunities for application of AI from troves of image data (92). For example, features of each pixel can be considered in multiple ways: edges and patterns such as borders can be identified, movement of features can be characterized as vectors, colors can be extracted, and shapes with similar related characteristics such as image density can be identified. Data in images can also be enhanced through application of manual or automated techniques that define shapes within the image (for example, the LV border [43]), or the cardiac plane of interest (93). These techniques may identify a region of interest for study, such as a valve (48,94). Also, quantified metadata extracted from images such as EF, strain, or velocity are being usefully packaged with image data for subsequent analysis. Finally, data encoded within the stored image file, such as age, sex, or medical record number can be extracted. Thus, AI technologies are showing their potential to change how cardiovascular imaging is used and interpreted, improving quality control, test selection, quantification, reporting, diagnostics, ease of use, and workflow.
PATIENT OUTCOMES.
From published data to date, ML algorithms have already shown to improve accuracy of diagnostic tests (63,75,82,85,87) and prediction of disease (95). Concurrently, ML integration of clinical data and imaging measures has been shown to improve prediction of prognostic outcomes (23,70,96). Several of these studies have conclusively shown that for per-patient risk prediction, it is most effective to objectively rank and then integrate all available clinical and imaging measures with machine learning. ML algorithms also have the potential to find new insights in real data, particularly when objective feature ranking is performed (23,75). In a multicenter study, Dey et al. (75) found that contrast density difference had the highest information gain for lesion-specific ischemia by invasive FFR over quantitative stenosis and plaque metrics from CTA. A higher contrast density difference indicates a lower minimum luminal area and higher luminal attenuation gradient; thus, it includes the contribution of both quantitative measures. For plaque measures, low-density noncalcified plaque (97), which has been also shown to predict cardiac death, was the highest-ranked feature for lesion-specific ischemia.
INTEGRATION INTO CLINICAL ROUTINE.
In the current published data, development of ML models and their validation has been demonstrated in clinical studies. In the near future, it is easy to envisage ML working in the background of standard cardiac imaging reporting and quantitative analysis software, gathering the variables automatically and allowing on-the-fly risk score computation. This principle is already utilized daily by many applications that utilize ML “behind the scenes,” unknown to the user. For example, the personalized advertisements and browsing suggestions that appear in real-time during web-browsing are all based on the passive collection of variables and their seamless input into ML algorithms. With automated feature ranking, ML is almost fully automated, requiring only minimal input during model building. In the near future, as ML algorithms are incorporated into clinical routine for image acquisition, image analysis, and prediction of patient outcomes, we expect that such personalized medicine would help physicians find the right answers for their patients whose “lives and medical histories shape the algorithms” (5).
CONCLUSIONS
AI describes applications to identify patterns in data that are characteristic of human intelligence. It depends on appropriate data being supplied and its quality, and the right software being applied to identify the patterns. AI is likely to provide a process to improve speed and quality of acquisition, reduce measurement time, and allow prompt diagnoses, which in turn would improve workflow and patient care. The development of AI applications with big imaging registries will facilitate precision medicine and increase the utility of imaging tests for the patient. However, its introduction will bring a number of challenges. First, imaging specialists are mostly trained by manual analysis, and the “human neural network” is grown by practice. The replacement of the majority of this workload by AI will have training implications that warrant consideration in order to safeguard the “human neural network.” Second, consideration should be given by cardiologists as to the questions these techniques are applied to and determine what outcome needs to be achieved for patients. Finally, although AI may be used to predict likely outcomes, automatic generation of management decisions seems less desirable than a human neural network engaging with the clinical, personal, environmental, and social aspects of each individual patient. In the near future, therefore, the value-adding potential of AI is most likely to be as intelligent precision medicine tools for imaging specialists and clinicians.
HIGHLIGHTS.
Problems with timing, efficiency, and missed diagnoses occur at all stages of the imaging chain. The application of AI may reduce cost and improve value at all stages of image acquisition, interpretation, and decision-making.
The main fields of AI for imaging will pertain to disease phenotyping, diagnostic support, and image interpretation. Grouping of relevant clinical and imaging information with cluster analysis may provide opportunities to better characterize disease. Diagnostic support will be provided by automated image segmentation and automated measurements. The initial steps are being taken towards automated image acquisition and analysis.
"Big data" from imaging will interface with high volumes of data from the electronic health record and pathology to provide new insights and opportunities to personalize therapy.
Acknowledgments
This study was supported in part by a Partnership Grant from the National Health and Medical Research Council and from National Heart, Lung, and Blood Institute grant 1R01HL133616. Dr. Dey has received software royalties from Cedars-Sinai Medical Center and has a patent. Dr. Slomka has received a research grant from Siemens Medical Solutions; and has received software royalties from Cedars Sinai. Dr. Leeson is a founder, stockholder, and non-executive director of Ultromics Ltd.; and has received a research grant from Lantheus Medical Imaging. Dr. Comaniciu is a salaried employee of Siemens Healthineers. Dr. Marwick has received research grant support from GE Medical Systems for the SUCCOUR study. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose. Patrick W Serruys, M.D., Ph.D. served as Guest Associate Editor for this paper.
ABBREVIATIONS AND ACRONYMS
- AI
artificial intelligence
- CMR
cardiac magnetic resonance
- CTA
computed tomography angiography
- DL
deep learning
- EAT
epicardial adipose tissue
- EF
ejection fraction
- EHR
electronic health record
- FFR
fractional flow reserve
- LV
left ventricular/ventricle
- ML
machine learning
- MPI
myocardial perfusion imaging/image
- SPECT
single photon emission computed tomography
REFERENCES
- 1.Russell S, Norvig P. Artificial Intelligence: A Modern Approach. 2nd edition. Upper Saddle River, New Jersey: Prentice Hall, 2003. [Google Scholar]
- 2.Szolovits P, Patil RS, Schwartz WB. Artificial intelligence in medical diagnosis. Ann Intern Med 1988;108:80–7. [DOI] [PubMed] [Google Scholar]
- 3.Darcy AM, Louie AK, Roberts LW. Machine learning and the profession of medicine. JAMA 2016;315:551–2. [DOI] [PubMed] [Google Scholar]
- 4.Nilsson N The Quest for Artificial Intelligence: A History of Ideas and Achievements. New York: Cambridge University Press, 2009. [Google Scholar]
- 5.Obermeyer Z, Emanuel EJ. Predicting the future - big data, machine learning, and clinical medicine. N Engl J Med 2016;375:1216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deo RC. Machine Learning in Medicine. Circulation 2015;132:1920–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrea DM, Marco G, Michele G. A formal definition of Big Data based on its essential features. Library Review 2016;65:122–35. [Google Scholar]
- 8.Petersen SE, Matthews PM, Bamberg F, et al. Imaging in population science: cardiovascular magnetic resonance in 100,000 participants of UK Biobank - rationale, challenges and approaches. J Cardiovasc Magn Reson 2013;15:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffey S, Lewandowski AJ, Garratt S, et al. Protocol and quality assurance for carotid imaging in 100,000 participants of UK Biobank: development and assessment. Eur J Prev Cardiol 2017;24:1799–806. [DOI] [PubMed] [Google Scholar]
- 10.Aye CYL, Lewandowski AJ, Lamata P, et al. Disproportionate cardiac hypertrophy during early postnatal development in infants born preterm. Pediatr Res 2017;82:36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewandowski AJ, Augustine D, Lamata P, et al. Preterm heart in adult life: cardiovascular magnetic resonance reveals distinct differences in left ventricular mass, geometry, and function. Circulation 2013;127:197–206. [DOI] [PubMed] [Google Scholar]
- 12.Lee JG, Jun S, Cho YW, et al. Deep learning in medical imaging: general overview. Korean J Radiol 2017;18:570–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayr A, Binder H, Gefeller O, Schmid M. The evolution of boosting algorithms. From machine learning to statistical modelling. Methods Inf Med 2014;53:419–27. [DOI] [PubMed] [Google Scholar]
- 14.Chykeyuk K, Clifton DA, Noble JA. Feature extraction and wall motion classification of 2D stress echocardiography with relevance vector machines. Proceedings - International Symposium on Biomedical Imaging 2011:677–80. [Google Scholar]
- 15.Domingos JS, Stebbing RV, Leeson P, Noble JA. Structured Random Forests for Myocardium Delineation in 3D Echocardiography. Cham: Springer International Publishing, 2014: 215–22. [Google Scholar]
- 16.Krittanawong C, Tunhasiriwet A, Zhang H, Wang Z, Aydar M, Kitai T. Deep Learning with unsupervised feature in echocardiographic imaging. J Am Coll of Cardiol 2017;69:2100–1. [DOI] [PubMed] [Google Scholar]
- 17.Pavlopoulos GA, Secrier M, Moschopoulos CN, et al. Using graph theory to analyze biological networks. BioData Min 2011;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres BY, Oliveira JH, Thomas Tate A, Rath P, Cumnock K, Schneider DS. Tracking resilience to infections by mapping disease space. PLoS Biol 2016;14:e1002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oxtoby NP, Garbarino S, Firth NC, et al. Data-driven sequence of changes to anatomical brain connectivity in sporadic Alzheimer's disease. Front Neurol 2017;8:580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruno JL, Romano D, Mazaika P, et al. Longitudinal identification of clinically distinct neurophenotypes in young children with fragile X syndrome. Proc Natl Acad Sci U S A 2017;114:10767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Cheng WY, Glicksberg BS, et al. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci Transl Med 2015;7:311ra174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arsanjani R, Xu Y, Hayes SW, et al. Comparison of fully automated computer analysis and visual scoring for detection of coronary artery disease from myocardial perfusion SPECT in a large population. J Nucl Med 2013;54:221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motwani M, Dey D, Berman DS, et al. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur Heart J 2017;38:500–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molinaro AM, Simon R, Pfeiffer RM. Prediction error estimation: a comparison of resampling methods. Bioinformatics 2005;21:3301–7. [DOI] [PubMed] [Google Scholar]
- 25.Stebbing RV, Namburete AI, Upton R, Leeson P, Noble JA. Data-driven shape parameterization for segmentation of the right ventricle from 3D+t echocardiography. Med Image Anal 2015;21:29–39. [DOI] [PubMed] [Google Scholar]
- 26.Fatima M, Pasha M. Survey of Machine learning algorithms for disease diagnostic. Journal of Intelligent Learning Systems and Applications 2017;9:1–16. [Google Scholar]
- 27.Ernande L, Audureau E, Jellis CL, et al. Clinical implications of echocardiographic phenotypes of patients with diabetes mellitus. J Am Coll Cardiol 2017;70:1704–16. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez-Martinez S, Duchateau N, Erdei T, et al. Machine learning analysis of left ventricular function to characterize heart failure with preserved ejection fraction. Circ Cardiovasc Imaging 2018;11:e007138. [DOI] [PubMed] [Google Scholar]
- 29.Shah SJ, Katz DH, Selvaraj S, et al. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation 2015;131:269–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katz DH, Deo RC, Aguilar FG, et al. Phenomapping for the identification of hypertensive patients with the myocardial substrate for heart failure with preserved ejection fraction. J Cardiovasc Transl Res 2017;10:275–84. [DOI] [PubMed] [Google Scholar]
- 31.Gan GCH, Ferkh A, Boyd A, Thomas L. Left atrial function: evaluation by strain analysis. Cardiovasc Diagn Ther 2018;8:29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lancaster MC, Salem Omar AM, Narula S, Kulkarni H, Narula J, Sengupta PP. Phenotypic clustering of left ventricular diastolic function parameters: patterns and prognostic relevance. J Am Coll Cardiol Img 2018. April 18 [E-pub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 33.Horiuchi Y, Tanimoto S, Latif A, et al. Identifying novel phenotypes of acute heart failure using cluster analysis of clinical variables. Int J Cardiol 2018;262:57–63. [DOI] [PubMed] [Google Scholar]
- 34.Bansod P, Desai UB, Burkule N. Endocardial segmentation in contrast echocardiography video with density based spatio-temporal clustering. Presented at: Proceedings of the First International Conference on Bio-inspired Systems and Signal Processing 2008 Funchal, Madeira, Portugal, January 28-31, 2008. [Google Scholar]
- 35.Carbotta G, Tartaglia F, Giuliani A, et al. Cardiovascular risk in chronic autoimmune thyroiditis and subclinical hypothyroidism patients. A cluster analysis. Int J Cardiol 2017;230:115–9. [DOI] [PubMed] [Google Scholar]
- 36.Peckova M, Charvat J, Schuck O, Hill M, Svab P, Horackova M. The association between left ventricular diastolic function and a mild-to-moderate decrease in glomerular filtration rate in patients with type 2 diabetes mellitus. J Int Med Res 2011;39:2178–86. [DOI] [PubMed] [Google Scholar]
- 37.Pontremoli R, Viazzi F, Nicolella C, et al. Genetic polymorphism of the renin-angiotensin-aldosterone system (RAAS) and organ damage (TOD) in essential hypertension (EH) (abstr). Am J Hypertension 1999;12:6A. [Google Scholar]
- 38.Shukla N, Hagenbuchner M, Win KT, Yang J. Breast cancer data analysis for survivability studies and prediction. Comput Methods Programs Biomed 2018;155:199–208. [DOI] [PubMed] [Google Scholar]
- 39.Lei X, Li H, Zhang A, Wu FX. iOPTICS-GSO for identifying protein complexes from dynamic PPI networks. BMC Med Genomics 2017;10:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Omar AMS, Narula S, Abdel Rahman MA, et al. Precision phenotyping in heart failure and pattern clustering of ultrasound data for the assessment of diastolic dysfunction. J Am Coll Cardiol Img 2017;10:1291–303. [DOI] [PubMed] [Google Scholar]
- 41.Ghesu FC, Georgescu B, Zheng Y, et al. Multiscale deep reinforcement learning for real-time 3D-landmark detection in CT scans. IEEE Trans Pattern Anal Mach Intell 2019;41:176–89. [DOI] [PubMed] [Google Scholar]
- 42.Ghesu FC, Georgescu B, Grbic S, Maier A, Hornegger J, Comaniciu D. Towards intelligent robust detection of anatomical structures in incomplete volumetric data. Med Image Anal 2018;48:203–13. [DOI] [PubMed] [Google Scholar]
- 43.Zheng Y, Barbu A, Georgescu B, Scheuering M, Comaniciu D. Four-chamber heart modeling and automatic segmentation for 3-D cardiac CT volumes using marginal space learning and steerable features. IEEE Trans Med Imaging 2008;27:1668–81. [DOI] [PubMed] [Google Scholar]
- 44.Ngo TA, Lu Z, Carneiro G. Combining deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance. Med Image Anal 2016;35:159–71. [DOI] [PubMed] [Google Scholar]
- 45.Comaniciu D, Zhou XS, Krishnan S. Robust real-time myocardial border tracking for echocardiography: an information fusion approach. IEEE Trans Med Imaging 2004;23:849–60. [DOI] [PubMed] [Google Scholar]
- 46.Yang L, Georgescu B, Zheng Y, Wang Y, Meer P, Comaniciu D. Prediction based collaborative trackers (PCT): a robust and accurate approach toward 3D medical object tracking. IEEE Trans Med Imaging 2011;30:1921–32. [DOI] [PubMed] [Google Scholar]
- 47.Grbic S, Ionasec R, Vitanovski D, et al. Complete valvular heart apparatus model from 4D cardiac CT. Med Image Comput Comput Assist Interv 2010;13:218–26. [DOI] [PubMed] [Google Scholar]
- 48.Ionasec RI, Voigt I, Georgescu B, et al. Patient-specific modeling and quantification of the aortic and mitral valves from 4-D cardiac CT and TEE. IEEE Trans Med Imaging 2010;29:1636–51. [DOI] [PubMed] [Google Scholar]
- 49.Voigt I, Mansi T, Ionasec RI, et al. Robust physically-constrained modeling of the mitral valve and subvalvular apparatus. Med Image Comput Comput Assist Interv 2011;14:504–11. [DOI] [PubMed] [Google Scholar]
- 50.Zheng Y, Wang T, John M, Zhou SK, Boese J, Comaniciu D. Multi-part left atrium modeling and segmentation in C-arm CT volumes for atrial fibrillation ablation. Med Image Comput Comput Assist Interv 2011;14:487–95. [DOI] [PubMed] [Google Scholar]
- 51.Zheng Y, John M, Liao R, et al. Automatic aorta segmentation and valve landmark detection in C-arm CT for transcatheter aortic valve implantation. IEEE Trans Med Imaging 2012;31:2307–21. [DOI] [PubMed] [Google Scholar]
- 52.Liao R, Miao S, Tournemire PD, et al. An artificial agent for robust image registrations. AAAI-Association for the Advancement of Artificial Intelligence 2017. Available at: https://www.groundai.com/project/an-artificial-agent-for-robust-image-registration. Accessed February 12, 2019.
- 53.Cheng VY, Dey D, Tamarappoo BK, et al. Pericardial fat burden on ECG-gated noncontrast CT in asymptomatic patients who subsequently experience adverse cardiovascular events on 4-year follow-up: a case-control study. J Am Coll Cardiol Img 2010:352–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahabadi AA, Massaro JM, Rosito GA, et al. Association of pericardial fat, intrathoracic fat, and visceral abdominal fat with cardiovascular disease burden: the Framingham Heart Study. Eur Heart J 2009;30:850–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamarappoo B, Dey D, Shmilovich H, et al. Increased pericardial fat volume measured from noncontrast CT predicts myocardial ischemia by. SPECT J Am Coll Cardiol Img 2010;3:1104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mazurek T, Zhang L, Zalewski A, et al. Human epicardial adipose tissue is a source of inflammatory mediators. Circulation 2003;108:2460–6. [DOI] [PubMed] [Google Scholar]
- 57.Mahabadi AA, Berg MH, Lehmann N, et al. Association of epicardial fat with cardiovascular risk factors and incident myocardial infarction in the general population: the Heinz Nixdorf Recall Study. J Am Coll Cardiol 2013;61:1388–95. [DOI] [PubMed] [Google Scholar]
- 58.Cheng VY, Dey D, Tamarappoo B, et al. Pericardial fat burden on ECG-gated noncontrast CT in asymptomatic patients who subsequently experience adverse cardiovascular events. J Am Coll Cardiol Img 2010;3:352–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Commandeur F, Goeller M, Betancur J, et al. Deep learning for quantification of epicardial and thoracic adipose tissue from non-contrast CT. IEEE Trans Med Imaging 2018;37:1835–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou SK, Guo F, Park J, et al. A probabilistic, hierarchical and discriminant framework for rapid and accurate detection of deformable anatomic structures. IEEE Int’l Conf Computer Vision 2007:1–8. Available at: https://ieeexpLore.ieee.org/document/4409045?arnumber=4409045. Accessed February 12, 2019. [Google Scholar]
- 61.Qazi M, Fung G, Krishnan S, et al. Automated heart wall motion abnormality detection from ultrasound images using Bayesian networks. International Joint Conferences on Artificial Intelligence, 2007. Available at: https://dl.acm.org/citation.cfm?id=1625358. Accessed March 6, 2019. [Google Scholar]
- 62.Kelm BM, Mittal S, Zheng Y, et al. Detection, grading and classification of coronary stenoses in computed tomography angiography. Med Image Comput Comput Assist Interv 2011;14:25–32. [DOI] [PubMed] [Google Scholar]
- 63.Itu L, Rapaka S, Passerini T, et al. A machine-Learning approach for computation of fractional flow reserve from coronary computed tomography. J Appl Physiol (1985) 2016;121:42–52. [DOI] [PubMed] [Google Scholar]
- 64.Mihalef V, Ionasec RI, Sharma P, et al. Patient-specific modelling of whole heart anatomy, dynamics and haemodynamics from four-dimensional cardiac CT images. Interface Focus 2011;1:286–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zettinig O, Mansi T, Neumann D, et al. Data-driven estimation of cardiac electrical diffusivity from 12-lead ECG signals. Med Image Anal 2014;18:1361–76. [DOI] [PubMed] [Google Scholar]
- 66.Neumann D, Mansi T, Itu L, et al. A self-taught artificial agent for multi-physics computational model personalization. Med Image Anal 2016;34:52–64. [DOI] [PubMed] [Google Scholar]
- 67.Mansi T, Voigt I, Georgescu B, et al. An integrated framework for finite-element modeling of mitral valve biomechanics from medical images: application to MitralCLip intervention planning. Med Image Anal 2012;16:1330–46. [DOI] [PubMed] [Google Scholar]
- 68.Kayvanpour E, Mansi T, Sedaghat-Hamedani F, et al. Towards personalized cardiology: multi-scale modeling of the failing heart. PLoS One 2015;10:e0134869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katus H, Ziegler A, Ekinci O, et al. Early diagnosis of acute coronary syndrome. Eur Heart J 2017;38:3049–55. [DOI] [PubMed] [Google Scholar]
- 70.Betancur J, Otaki Y, Motwani M, et al. Prognostic value of combined clinical and myocardial perfusion imaging data using machine learning. J Am Coll Cardiol Img 2017;16:30804–5. [Google Scholar]
- 71.Slomka PJ, Betancur J, Liang JX, et al. Rationale and design of the REgistry of Fast Myocardial Perfusion Imaging with NExt generation SPECT (REFINE SPECT). J Nucl Cardiol 2018. June 19 [E-pub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakanishi R, Dey D, Commandeur F, et al. Machine learning in predicting coronary heart disease and cardiovascular disease events: results from the Multi-Ethnic Study Of Atherosclerosis (MESA) (abstr). J Am Coll Cardiol 2018;71:A1483. [Google Scholar]
- 73.Dey D, Diaz Zamudio M, Schuhbaeck A, et al. Relationship between quantitative adverse plaque features from coronary computed tomography angiography and downstream impaired myocardial flow reserve by 13N-ammonia positron emission tomography: a pilot study. Circ Cardiovasc Imaging 2015;8:3255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gaur S, Øvrehus KA, Dey D, et al. Coronary plaque quantification and fractional flow reserve by coronary computed tomography angiography identify ischaemia-causing lesions. Eur Heart J 2016;37:1220–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dey D, Gaur S, Ovrehus KA, et al. Integrated prediction of lesion-specific ischaemia from quantitative coronary CT angiography using machine learning: a multicentre study. Eur Radiol 2018;28:2655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakanishi R, Sankaran S, Grady L, et al. Automated estimation of image quality for coronary computed tomographic angiography using machine learning. Eur Radiol 2018:1–9. [DOI] [PubMed] [Google Scholar]
- 77.Krizhevsky A, Sutskever I, Hinton GE. Image-Net classification with deep convolutional neural networks. Advances in neural information processing systems 2012:1097–105. [Google Scholar]
- 78.Wolterink JM, Leiner T, de Vos BD, van Hamersvelt RW, Viergever MA, Isgum I. Automatic coronary artery calcium scoring in cardiac CT angiography using paired convolutional neural networks. Med Image Anal 2016;21:30022–6. [DOI] [PubMed] [Google Scholar]
- 79.Chen HZY, Park J-H, Heng P-A, Zhou SK. Iterative multi-domain regularized deep learning for anatomical structure detection and segmentation from ultrasound images. Med Image Comput Comput Assist Interv 2016:487–95. [Google Scholar]
- 80.Yang H, Sun J, Li H, Wang L, Xu Z. Deep fusion net for multi-atlas segmentation: Application to cardiac MR images. Med Image Comput Comput Assist Interv 2016:521–8. [Google Scholar]
- 81.Tan LK, McLaughlin RA, Lim E, Abdul Aziz YF, Liew YM. Fully automated segmentation of the left ventricle in cine cardiac MRI using neural network regression. J Magn Reson Imaging 2018;48:140–52. [DOI] [PubMed] [Google Scholar]
- 82.Betancur JCF, Motlagh M, Sharir T, et al. Deep learning for prediction of obstructive disease from fast myocardial perfusion SPECT: a multicenter study. J Am Coll Cardiol Img 2018;11:1654–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lessmann N, van Ginneken B, Zreik M, et al. Automatic calcium scoring in low-dose chest CT using deep neural networks with dilated convolutions. IEEE Trans Med Imaging 2018;37:615–25. [DOI] [PubMed] [Google Scholar]
- 84.Zreik M, Lessmann N, van Hamersvelt RW, et al. Deep learning analysis of the myocardium in coronary CT angiography for identification of patients with functionally significant coronary artery stenosis. Med Image Anal 2018;44:72–85. [DOI] [PubMed] [Google Scholar]
- 85.Tesche C, De Cecco CN, Baumann S, et al. Coronary CT angiography-derived fractional flow reserve: machine learning algorithm versus computational fluid dynamics modeling. Radiology 2018;288:64–72. [DOI] [PubMed] [Google Scholar]
- 86.Coenen A, Kim YH, Kruk M, et al. Diagnostic accuracy of a machine-learning approach to coronary computed tomographic angiography-based fractional flow reserve: result from the MACHINE Consortium. Circ Cardiovasc Imaging 2018;11:e007217. [DOI] [PubMed] [Google Scholar]
- 87.Arsanjani R, Xu Y, Dey D, et al. Improved accuracy of myocardial perfusion SPECT for detection of coronary artery disease by machine learning in a large population. J Nucl Cardiol 2013;20:553–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Arsanjani R, Dey D, Khachatryan T, et al. Prediction of revascularization after myocardial perfusion SPECT by machine learning in a large population. J Nucl Cardiol 2015;22:877–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hachamovitch R, Rozanski A, Shaw LJ, et al. Impact of ischaemia and scar on the therapeutic benefit derived from myocardial revascularization vs. medical therapy among patients undergoing stress-rest myocardial perfusion scintigraphy. Eur Heart J 2011;32:1012–24. [DOI] [PubMed] [Google Scholar]
- 90.Slomka PJ, Dey D, Sitek A, Motwani M, Berman DS, Germano G. Cardiac imaging: working towards fully-automated machine analysis & interpretation. Expert Rev Med Devices 2017;14:197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Narula S, Shameer K, Salem Omar AM, Dudley JT, Sengupta PP. Machine-learning algorithms to automate morphological and functional assessments in 2D echocardiography. J Am Coll Cardiol 2016;68:2287–95. [DOI] [PubMed] [Google Scholar]
- 92.Douglas PS, Garcia MJ, Haines DE, et al. ACCF/ASE/AHA/ASNC/HFSA/HRS/SCAI/SCCM/SCCT/SCMR 2011 appropriate use criteria for echocardiography. A report of the American College of Cardiology Foundation Appropriate Use Criteria Task Force, American Society of Echocardiography, American Heart Association, American Society of Nuclear Cardiology, Heart Failure Society of America, Heart Rhythm Society, Society for Cardiovascular Angiography and Interventions, Society of Critical Care Medicine, Society of Cardiovascular Computed Tomography, Society for Cardiovascular Magnetic Resonance American College of Chest Physicians. J Am Coll Cardiol 2011;57:1126–66. [DOI] [PubMed] [Google Scholar]
- 93.Park J, Zhou S, Otsuki J, Simopoulos C, Comaniciu D. Automatic cardiac view classification of echocardiogram. In: IEEE, editor. International Conference on Computer Vision Rio de Janeiro: IEEE, 2007:1–8. [Google Scholar]
- 94.Jeganathan J, Knio Z, Amador Y, et al. Artificial intelligence in mitral valve analysis. Ann Card Anaesth 2017;20:129–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Miotto R, Li L, Kidd BA, Dudley JT. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci Rep 2016;6:26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digital Medicine 2018;1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hell MM, Motwani M, Otaki Y, et al. Quantitative global plaque characteristics from coronary computed tomography angiography for the prediction of future cardiac mortality during long-term follow-up. Eur Heart J Cardiovasc Imaging 2017;18:1331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]