Abstract
Cryptococcal meningitis affects normal hosts and immunocompromised patients exhibiting high mortality rates. The objective of this study was to design two molecular assays, visible microarray platforms and loop-mediated isothermal amplification (LAMP), to identify Cryptococcus spp. and the species neoformans and gattii from the cerebral spinal fluid (CSF). To identify Cryptococcus and the two species, we designed two microarrays DNA platforms based on the internal transcribed spacer (ITS) region and CAP59 gene and LAMP assays specific for Cryptococcus species. The assays were tested using CSF from patients with cryptococcal meningitis. CSF from patients with cryptococcal meningitis was cultured in Sabouraud culture medium, and the Cryptococcus spp. grown in the culture medium were also tested for LAMP and microarray platforms. The results were compared to DNA sequencing of the same genetic regions. A total of 133 CSF samples were studied. Eleven CSFs were positive for Cryptococcus (9 C. neoformans and 2 C. gattii), 15 were positive for bacteria, and 107 were negative. The CAP59 platform correctly identified 73% of the CSF samples, while the ITS platform identified 45.5%. CAP59 platform correctly identified 100% of the Cryptococcus isolates, and ITS platform identified 70%. The two sets of LAMP primers correctly identified 100% of the Cryptococcus isolates. However, for CSF samples, the amplification occurred only in 55.5% of C. neoformans. The methodologies were reliable in the identification of Cryptococcus species, mainly for isolates from culture medium, and they might be applied as adjunctive tests to identify Cryptococcus species.
Keywords: Cryptococcus, Microarray, LAMP, Molecular diagnosis, Cryptococcal meningitis
Introduction
Cryptococcosis is the leading systemic fungal infection worldwide with an estimated number of cases per year close to 1 million (1). In Brazil, cryptococcosis is associated with the AIDS epidemic when 40,000 new AIDS cases were being identified annually (2), coupled with 1,000 to 2,500 new cases of cryptococcal meningitis leading to an approximately 40% mortality rate (1,3). The cryptococcal meningitis caused by Cryptococcus neoformans and Cryptococcus gattii are clinically indistinguishable, however, some authors suggest that meningitis due to C. gattii appears to be more aggressive than that caused by C. neoformans, and may require extended periods of treatment (4,5).
The microbiological identification by automated systems and phenotypic methods applied in the differentiation of Cryptococcus species requires confirmation by molecular methods. Recently, matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) has identified correctly close to 100% of Cryptococcus species (6); however, this technology is available in few hospitals in Brazil. Immunoassays for the diagnosis of cryptococcosis have been applied in clinical practice to detect cryptococcal capsular antigen in body fluids, such as the Cryptococcus antigen lateral flow assay (CrAg LFA). The CrAg LFA assay identifies the four main serotypes of Cryptococcus (A, B, C, and D) (7), but not to species level. Cerebral lesions and hydrocephalus are more common in infections caused by C. gattii (8,9). In these cases, the accurate species identification would be useful in guiding the appropriate therapy scheme. The CrAg LFA assay is produced by only one biotech company (IMMY®, USA), and has not yet been approved by the Brazilian Health Regulatory Agency (Anvisa), which is an autarchy of the Ministry of Health that regulates health products in the country. Because the CrAg LFA assay is not available here, other techniques, including molecular ones, are necessary to improve the diagnosis of cryptococcosis.
A reliable identification of Cryptococcus species may facilitate the proper antifungal treatment and improve clinical and epidemiological surveillance strategies. Several studies have been published on the application of molecular techniques for the identification and diagnosis of Cryptococcus, but there is still a need for new studies and technologies to implement the diagnosis of this mycosis (10 –13).
Molecular techniques have contributed significantly to improve the diagnosis of fungal diseases. DNA sequencing has been an important complementary method to classical mycological identification of fungal pathogens, but it is time-consuming and expensive. Thus, there is a need for simpler molecular diagnostic methodologies with similar specificity. DNA microarray platforms have been developed to identify a diversity of microorganisms such as viruses (14), bacteria (15), and fungi (16,17), simultaneously or individually, with good sensitivity and specificity. Due to the low stability of most fluorescent dyes and the expensive scanning equipment used in this technique to visualize the result, our research group developed a more accessible DNA microarray system for identification of fungi at the genus and species level, the result of which is visualized with the naked eye (18).
Loop-mediated isothermal amplification (LAMP) has been used as a molecular diagnostic methodology based on the principle of isothermal amplification of simple nucleic acid. This technique has shown to be fast, specific, and economical. The assay has been validated by different clinical studies presenting a similar performance as the conventional PCR (19,20), and it has been applied for the detection of several pathogenic microorganisms, including viruses (20,21), bacteria (22), protozoa (23), and fungi (24 –29).
Therefore, the objective of this study was to develop two assays based on molecular techniques, visible DNA microarrays and LAMP, to identify the genus Cryptococcus and the species neoformans and gattii directly from clinical samples of cerebral spinal fluid (CSF) and from isolates of Cryptococcus grown on culture plates.
Material and Methods
Study design
The study was performed at the Clinical Hospital of the University of Campinas, Sao Paulo, Brazil, which is the referral hospital for tertiary care for more than 3 million inhabitants. All the laboratory assays were performed at the Molecular Epidemiology and Infectious Diseases Laboratory (LEMDI) of the Faculty of Medical Sciences, University of Campinas. This study was approved by the Institutional Ethical Committee (No. CAAE: 54943116000005404). We tested Cryptococcus isolates grown on culture plates after CSF cultivation and Cryptococcus spp. from the culture collection of Instituto Adolfo Lutz (IAL), which is the reference laboratory for infectious diseases for the State of São Paulo and Federal governments. LAMP and two visible DNA microarray platforms were tested.
Clinical samples
From August 2016 to October 2018, we studied samples of CSF from patients hospitalized at the Clinical Hospital. The microbiology laboratory of the Clinical Pathology Department previously analyzed the CSF samples. An aliquot was stored at 4°C as counterproof for further studies. For our study, we tested the CSFs that were positive for bacteria, Cryptococcus spp., and the ones that were negative for bacteria, mycobacteria, and fungi. The microbiological identification was performed by the automated system Vitek® 2 (Lab Equipment bioMérieux Inc., USA) or BD Phoenix (Becton Dickinson, USA). The results of fungal identification obtained with the automated systems were compared with the ones from DNA microarray, LAMP, and DNA sequencing. All CSF samples were submitted to DNA extraction (see methods) including the ones with a positive microbiological identification for fungi, bacteria or that cultured negative for pathogens. DNA sequencing, LAMP, and DNA microarray were performed for all CSF samples in which DNA extraction resulted in a positive amplification in the PCR reaction. The molecular tests were performed after all the CSF standard analyses and microbiological and biochemical tests were completed. Our results were not reported to the assistant physician and they were not used for therapeutic proposes.
For the purpose of this study, a loop of all CSF samples was cultivated in agar Sabouraud. The yeasts recovered from the culture plates were then stored in sterile water at 4°C and in 10% glycerol at −80°C.
Yeast strains
Eleven Cryptococcus spp. strains from CSF samples and 30 clinical and environmental isolates identified as C. gattii, generously donated by the Mycology Division of the IAL (Table 1), were included to develop the molecular assays of Cryptococcus species identification. The DNA samples of C. neoformans genotypes VNI to VNIII and C. gattii VGI to VGIII were kindly provided by the Oswaldo Cruz Foundation (FIOCRUZ) for LAMP assays.
Table 1. Cerebral spinal fluid (CSF) samples used in this study according to sample number and microbiological identification.
| CSF samples | Sample Number | Fungal identification in CSF by Vitek® 2 or BD Phoenix | Patient case number |
|---|---|---|---|
| Group A (n=11) | P232 | Cryptococcus spp. | Case 1 |
| P260 | Cryptococcus neoformans | Case 2 | |
| P261 | C. neoformans | Case 2 | |
| P272 | Negative | Case 3 | |
| P273 | C. neoformans | Case 4 | |
| P274 | C. neoformans | Case 4 | |
| P275 | C. neoformans | Case 5 | |
| P276 | Cryptococcus spp. | Case 6 | |
| P283 | C. neoformans | Case 7 | |
| P284 | C. neoformans | Case 8 | |
| P285 | Cryptococcus gattii | Case 9 | |
| Group B (n=15) | P63 | Staphylococcus haemolyticus and Staphylococcus epidermidis | |
| P113 | Pseudomonas aeruginosa | ||
| P228 | Enterobacter cloacae | ||
| P240 | Staphylococcus aureus | ||
| P257 | P. aeruginosa | ||
| P259 | P. aeruginosa | ||
| P262 | P. aeruginosa | ||
| P263 | Streptococcus pneumoniae | ||
| P264 | E. cloacae | ||
| P265 | Staphylococcus caprae | ||
| P266 | S. caprae | ||
| P267 | Haemophilus influenzae | ||
| P268 | S. pneumoniae | ||
| P269 | Pantoea aerogenes | ||
| P270 | S. pneumoniae | ||
| Group C (n=107) | Negative | ||
| IAL samples (n=30) | C. gattii |
Group A: 11 CSF samples from 9 patients that cultured positive for Cryptococcus; Group B: 15 CSF samples from patients with bacterial meningitis; Group C: 107 CSF samples that cultured negative for fungi, bacteria, and mycobacteria. IAL: Instituto Adolfo Lutz.
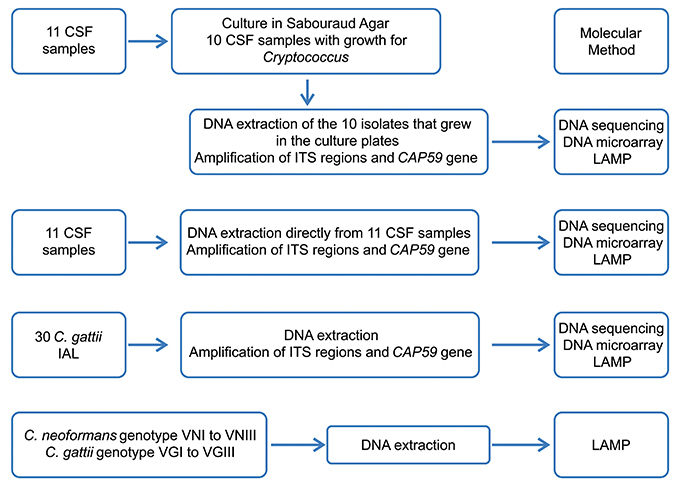
The flowchart of experiments is shown in Figure 1.
Figure 1. Flowchart of the molecular experiments using cerebral spinal fluid from patients with cryptococcal meningitis, Cryptococcus gattii isolates, and DNA samples of Cryptococcus spp. ITS: internal transcribed spacer; IAL: Instituto Adolfo Lutz; CSF: cerebral spinal fluid; LAMP: loop-mediated isothermal amplification.
DNA extraction
Three protocols of CSF DNA extraction were previously tested and the protocol that used alkaline solution before the extraction by Qiagem DNA mini kit (Qiagen Sciences Inc., USA) had better performance (data not included). Two hundred microliters of CSF were added to 500 μL of alkaline solution (0.5 M NaOH + 0.05 M dehydrated trisodium citrate) and centrifuged at 16,000 g for 5 min at room temperature. The supernatant was removed and the precipitate was suspended in 500 μL of PBS followed by centrifugation at 16,000 g for 5 min at room temperature. The mixture was homogenized using MagNA Lyzer (Roche Applied Science, USA) at 5,000 g for 10 min. The precipitate was transferred to a 1.5-mL tube and proceeded with the DNA extraction using QIAmp DNA mini Kit, according to the manufacturer’s instructions. For DNA extraction from yeasts grown on culture plates, one loop from one pure colony was inserted in 500 μL of alkaline solution and followed the same protocol for CSF samples. The DNA extracted from CSF samples and from the grown yeasts on culture plates were quantified by NanoDrop™ 2000 Spectrophotometers (Thermo Fisher Scientific™, USA). The PCR products were amplified using specific primers for fungi targeting the internal transcribed spacer (ITS) region and CAP59 gene.
To determine the detection threshold of Cryptococcus DNA in CSF, 107 to 101 yeast cells/mL of C. neoformans and C. gattii were artificially inoculated in CSF samples. For this experiment, we used CSFs with biochemical parameters (protein and glucose) and cytology within the normal range, and that cultured negative for bacteria, mycobacteria, and fungi. The DNA detection threshold was used for LAMP and DNA microarray assays.
PCR reactions and DNA sequencing
PCR was performed using 5′-biotin-labeled fungus-specific universal primers; ITS1-bio (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4-bio (5′TCCTCCGCTTATTGATATGC-3′) (Sigma-Aldrich, USA) in order to amplify the entire ITS region and biotin labeling. For the gene amplification of CAP59, biotin-labeled specific primers CAP59-2R (5′-CRTTCATGAARACGACCG-3′) and CAP59-1F (5′-CKTGGTCCAACKCYGGCTG-3′) were used. The size of the amplified fragments varied from 426 to 930 bp depending on the fungal species. PCR was performed using PCR Master Mix (Promega®, USA). PCR reactions were incubated in Veriti 96-well Thermal Cycler (Applied Biosystems, USA) under the following conditions: 2 min for initial denaturation at 95°C, 40 cycles of DNA denaturation at 95°C for 30 s, primer annealing at 55°C for 30 s, elongation at 72°C for 1 min, and a final elongation step at 72°C for 5 min. For CAP59 PCR reaction, the annealing temperature was 52°C. PCR products were verified by electrophoresis in a 2% agarose gel, 100 v, for 30 min. For DNA sequencing the PCR products were performed with the universal fungus-specific primers: ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′TCCTCCGCTTATTGATATGC-3′), and CAP59 Cap59-1F(5′-CKTGGTCCAACKCYGGCT-3′) and Cap59-2R(5′- CRTTCATGAARACGACCG-3′) (Sigma-Aldrich) followed by purification with ExoSAP-IT for PCR Product Clean-up (Affymetrix USB, USA), and the products were sequenced with the BigDye Terminator regent kit (Applied Biosystems), according to the manufacturer's protocols on an ABI Prism 3500 genetic analyzer (Applied Biosystems). The sequenced data were assembled and analyzed using BLAST (Basic Local Alignment Search Tool; http://blast.ncbi.nlm.nih.gov). DNA sequences were edited and assembled by Sequencher version 5.2.4 (Gene Codes Corp., USA). For identification, a homology search for the sequences ITS region and CAP59 gene was done using the BLAST tool of the NCBI database (GenBank).
Visual DNA microarray slides
The DNA microarray slide protocol used was as previously described by Sakai et al. (18) with modifications. Briefly, the oligonucleotide probes consisted of species/genus-specific nucleotide sequences with biotin-labeled poly(T) anchors at the end of each nucleotide (Sigma-Aldrich), which were designed based on the ITS sequences (ITS1 and ITS2) of the typed strains (GenBank database, American Type Culture Collection, CBS, and the Medical Mycology Research Center, Chiba University, Japan). This microarray platform covered 14 genera and 31 species of pathogenic fungi (Supplementary Figure S1A). A new platform, CAP59 platform, was designed to better differentiate the two Cryptococcus species using the probes for CAP59 gene regions (Supplementary Figure S1B). The previous platform was based on ITS regions resulting in cross-reaction between C. neoformans and C. gattii. The probe sequences were spotted onto polycarbonate slides (NGK Insulators LTD, Japan) with a KCS mini microarray printer (Kubota Comps Corporation, Japan). All slides had positive controls for fungi, where universal signals for fungi were enclosed in dotted line frame at the bottom right of each slide. After the hybridization was completed, the results were visible to the naked eye. This microarray assay could be done in up to 6 h.
Loop-mediated isothermal amplification
LAMP-method was performed as described elsewhere with modifications (24). A set of six primers were used for amplification of Cryptococcus neoformans: Set3_neoformans-(F3: GGTCGGTCTGAGGATCATCA; B3: TCTGTCTCCTACTCTGCCAA; FIP: TCGAACTTCGGCGAGGTATTCGGAACATCTATGCGTACCCGC; BIP: CGAGCTTCGTGACAATGACGGACAAGTCGTCCACGCAAGG; LF: GGTCTGTTCAACCATCGTAT; LB: GAAGTCTTCGACTCGGT) and for Cryptococcus gattii: Set1_gattii-(F3:CCAGACAAAGGCGCTCTTG; B3: ATCGTTCATGAAGACGACCG; FIP: ACGGGTACGCATGGATGTTCGGATTTTCGATGCCCTCGCG; BIP: TCGAATACCTTGCCGAAGTCCGCCGTCATTGTCACGGAGTTC; LF: GATGATGATCCTGAGACCGACG; LB: AAACGCCGCCATGCTGC). The primer sets were designed for the CAP59 gene using Primer Explorer V5 (https://primerexplorer.jp/e/). The amplification reaction of the assay was composed of 10 μL of each FIP and BIP primer at a concentration of 40 pmol, 10 μL of each F3 and B3 primer at 5 pmol concentration, 10 μL of LF and LB in a concentration of 20 pmol, 2 μL of extracted DNA, 100 μL of reaction mix (1 mL of 8 M betain, 500 μL of 165 mM MgCl2, 500 μL Tris-HCL, 500 μL KCL, 500 μL NHSO4, 500 μL 2% Tween 20, 350 μL 10 mM dNTP, and 100-300 μL dNTP 10 mM filtered water), 10 μL Bst DNA polymerase, and 30 μL filtered water. This mixture was incubated in a turbidimeter (Loopamp EXIA, Japan) at 63°C. Positive results were observed when the amplification curve appeared up to 60 min after starting the reaction. The turbidimeter of the LAMP reaction was analyzed in a real-time turbidimeter (Loopamp EXIA).
Results
A total of 133 CSF samples were included. Eleven CSFs were positive for Cryptococcus from nine patients, 15 samples were positive for bacteria, and 107 CSF samples were negative for fungi, bacteria, and mycobacteria (Table 1). Six patients had meningitis due to C. neoformans and two patients due to C. gattii. The CSF samples were named as followed: P232 (patient 1; C. neoformans), P260 and P261 (patient 2; C. neoformans), P272 (patient 3; C. gattii), P273 and P274 (patient 4; C. neoformans), P275 (patient 5; C. neoformans), P276 (patient 6; C. neoformans), P283 (patient 7; C. neoformans), P284 (patient 8; C. neoformans) and P285 (patient 9; C. gattii) (Table 1). P272 CSF sample did not grow in culture, and the diagnosis was made by microscopy that showed fungal elements suggestive of Cryptococcus and by DNA sequencing of the PCR product. The DNA quantification of the 11 CSF samples varied from 4.2 to 11.2 ng/μL whereas the DNA obtained from Cryptococcus isolates varied from 11.9 to 65.4 ng/μL. For the artificially inoculated CSF samples, the limit of detection of the PCR reactions targeting ITS regions of C. neoformans and C. gattii was 102 cells/mL in both species (data not shown).
DNA sequencing of ITS regions and </emph>CAP59 gene amplification
The DNA sequencing of ITS regions and CAP59 gene was made for all PCR amplification products to ensure that the resulting PCR amplification products were Cryptococcus and to confirm the species. The 15 CSFs positive for bacteria and the 107 negative CSF samples did not show any amplification for ITS regions and CAP59 gene. The 30 C. gattii strains from the IAL collection analyzed by DNA sequencing presented 99 to 100% homology with the NCBI database. The sequencing of the ITS regions and CAP59 gene of the DNA extracted directly from the 11 clinical samples and from their respective isolates grown on culture plates showed a percentage of homology ranging from 98 to 100% compared to the NCBI database. Our results suggested that CAP59 gene amplification was a suitable target for sequencing Cryptococcus (Supplementary Table S1). The sequences were deposited in the NCBI data bank accession numbers (MK369690-MK369710).
ITS and </emph>CAP59 DNA microarray platforms
The microarray platforms were tested for the identification of species/genus-specific hybridization patterns. The original microarray platform was designed to target the ITS region; however, the probes showed cross-hybridization within the Cryptococcus genus because of their highly conserved sequence (18). To improve the discrimination of Cryptococcus species, the CAP59 microarray platform was then developed. CAP59 microarray was firstly tested for the 30 C. gattii isolates from the IAL collection and showed 100% concordant results with DNA sequencing. For the experiments using DNA extracted from CSFs, CAP59 platform showed a better performance in the identification and discrimination of Cryptococcus species than the ITS platform. CAP59 platform identified the species of 73% (n=8/11) CSF samples whereas ITS platform identified 45.5% (n=5/11) (Figures 2 and 3). Better results were observed when the DNA microarray was performed using DNA extracted from cultivated isolates from CSFs samples (Figure 2B). CAP59 identified genus/species correctly in 100% and ITS in 70% of the Cryptococcus DNA.
Figure 2. Results of the identification of ITS and CAP59 DNA microarray platforms. Flowchart A shows the results of DNA extractions directly from clinical samples of cerebral spinal fluids (CSF). Flowchart B shows the results of DNA extractions obtained from Cryptococcus isolates grown in culture medium after CSF cultivation.
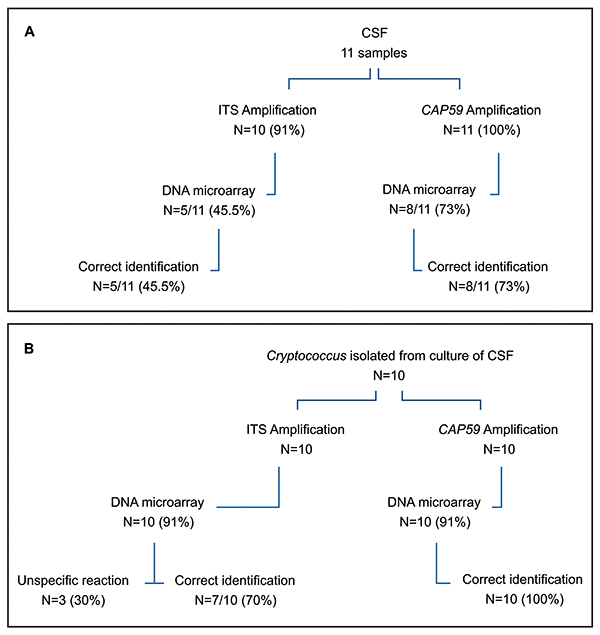
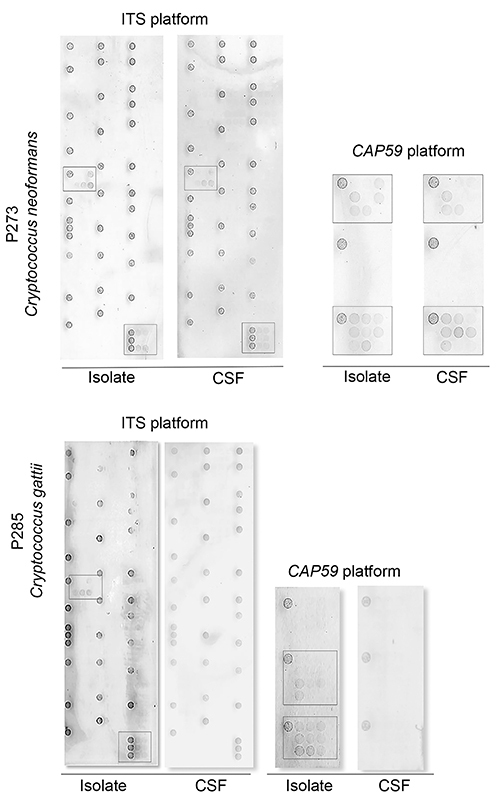
Figure 3. Representative results of Cryptococcus identification by DNA microarray platforms. P273 cerebral spinal fluid (CSF) sample and cultured isolate of C. neoformans tested by ITS and CAP59 platforms showing positive results, and P285 CSF sample and cultured isolated of C. gattii tested by ITS and CAP59 platforms showing positive results only for DNA extraction obtained from cultured isolates.
LAMP
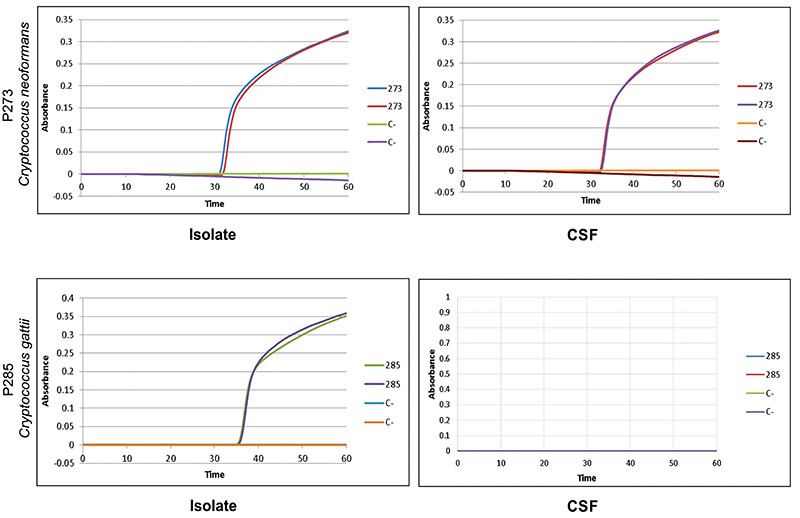
The primer set Set3_neoformans was firstly tested for the C. neoformans genotypes VNI, VNII, and VNIII, and the results are reported in Figure S2. The primer set Set1_gattii correctly identified the 30 isolates of C. gattii from IAL collection suggesting that this set of primer might be useful in the identification of cryptococcal DNA from CSF of patients with cryptococcal meningitis due to C. gattii. Set3_neoformans and Set1_gattii identified all C. neoformans and C. gattii isolates, respectively (Figure 4), when the DNAs used in the reaction were extracted from the isolates cultivated on culture plates. The two sets of primers identified the nine C. neoformans isolates (P232, P260, P261, P273, P274, P275, P276, P283, P284) and the P285 C. gattii isolate. However, when the same set of primers, Set3 neoformans and Set1 gattii, were applied to amplify the DNA extracted from CSF, exponential curves were seen in 55.5% (n=5/9) of C. neoformans and none for the two C. gattii samples. The threshold of detection test performed with C. neoformans ATCC90113 showed that 104 cells/mm3 were necessary to obtain a good exponential curve.
Figure 4. Representative loop-mediated isothermal amplification (LAMP) results of P273 cerebral spinal fluid clinical sample and culture (C). Isolate of Cryptococcus neoformans showed positive results after 30 min of reaction time. P285 showed a positive result after 35 min of reaction time only for DNA extracted from the clinical isolate of Cryptococcus gattii.
Discussion
In our study, two molecular methodologies, DNA microarray and LAMP, were tested in the identification and discrimination of Cryptococcus species from the CSF of patients with cryptococcal meningitis, and from the Cryptococcus isolates of CSF samples that were seeded in culture medium.
Several studies with array platforms were designed to identify one or more fungal species in the same assay (13,18,30,31). The insertion of oligonucleotide sequences for the detection of various genera and fungi species in a single platform reduces the operational cost and allows the identification of several species of fungi in the same reaction and the observation of cross reactions. In our study, ITS and CAP59 platforms identified and discriminated the genus Cryptococcus from both Cryptococci DNAs extracted from CSF and from the cultured isolates. ITS platform identified 45.5 and 70% and CAP59, 73 and 100% of the CSF samples and cultured isolates, respectively (Figure 2B). CAP59 platform seemed to be more discriminatory than ITS platform for the identification of the genus Cryptococcus and the species neoformans and gattii. Our microarray DNA platform developed to identify Cryptococcus to species level using probes designed based on CAP59 gene demonstrated 100% agreement in the identification of species of C. gattii when the DNA was extracted from clinical and environmental isolates, from the IAL collection. Although only 11 CSF samples were tested, these results might suggest that CAP59 could be included as a target gene in microarray platforms for further studies with Cryptococcus (Figure 2).
In our experiments, the amount of fungal DNA extracted from CSF was lower than the DNA obtained from the culture isolates, and the amount of DNA may had interfered with the microarray hybridization. For further conclusions, a larger number of clinical CSF samples from patients with cryptococcal meningitis will need to be tested in our platforms. O'Halloran et al. (32) failed to diagnose cryptococcal meningitis when CSF was studied by the approved commercialized FilmArray meningitis/encephalitis diagnostic panel (BioFire Diagnostic, USA). The authors suggested that patients with low fungal load in CSF were more likely to have a false negative test, emphasizing the importance of the amount of DNA in the molecular diagnostic tests.
LAMP tests were performed with two sets of primers that were designed based on the CAP59 gene: Set3_neoformans and Set1_gattii. These two sets were initially tested in DNA from clinical and environmental isolates of C. gattii from the IAL collection (genotypes VGI, VGII, VGIII, VNI, VNII, and VNIII) resulting in the correct identification of all the tested isolates (Figure S2). The primers underwent several tests with different samples, and we obtained 100% agreement with the results of DNA sequencing, absence of exponential curves for other microorganisms, such as Klebsiella sp., Candida albicans, Fusarium solani, Candida parapsilosis, Histoplasma capsulatum, and for negative controls. Only one previous study by Lucas et al. (33) had performed LAMP based on the CAP59 gene to identify the serotypes A, D, and B/C of Cryptococcus species complex, resulting in 83% agreement with a commercial serotyping kit.
Similarly to DNA microarray results, when testing CSF samples for LAMP assays, we had amplification in 55.5% of the CSF samples of patients with meningitis due to C. neoformans and no amplifications for the C. gattii CSF. Our results suggested that, for testing CSF samples, LAMP assay might need a higher number of Cryptococcus cells, for example, a larger volume of the clinical sample, to obtain a higher amount of DNA for the test. The mean amount of DNA obtained from the Cryptococcus grown in the culture media was significantly higher (P<0.001) (33.45 ng/μL) than the DNA obtained directly from CSF samples (6.85 ng/μL). However, the mean DNA quantification of CSF samples that resulted in a LAMP positive reaction (7.18 ng/μL) compared to the LAMP negative samples (6.46 ng/μL) was not significantly different (P=0.71) (data not shown). We observed that the specimen source played an important role in the results of DNA extraction. If the template was prepared from pure cultures, the amount of DNA was not a limiting factor. But working directly with clinical specimens such as CSF was challenging for many reasons. Low amounts of fungal cells can often be found in clinical specimens and host nucleic acids or proteins may occupy fungal targets, reducing the performance of the test. These problems might be overcome by optimizing the steps of precipitation and purification during DNA extraction to obtain sufficient amounts of DNA free of interferences.
In conclusion, DNA microarray and LAMP tested in this study based on ITS region and CAP59 gene could be applied in clinical and epidemiological studies for the diagnosis of cryptococcosis. CAP59 gene showed to be a promising target for future studies on molecular diagnostic tests. Moreover, they can be used as complementary tests to the microbiological diagnoses and antigen assays in the differentiation of Cryptococcus species in clinical practice.
Acknowledgments
We are very grateful to Prof. Akira Watanabe and Prof. Tohru Gonoi for providing part of ITS and CAP59 microarray slides. We thank the financial support of FAPESP (Fundação de Amparo a Pesquisa do Estado de São Paulo) for providing Grant #2016/12414-2 (P. Stivanelli scholarship) and Research Grant #2015/25035-7. We also thank the Brazilian National Council for Scientific and Technological Development (CNPq) for financial support (Grant #420163/2016-6).
References
- 1.Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, Chiller T, et al. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis. 2017;17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.United Nations AIDS . UNAIDS Website. 2018. http://www.unaids.org/en/resources/documents/2018/unaids-data-2018.Accessed December 23 2018. [Google Scholar]
- 3.Rajasingham R, Wake RM, Beyene T, Katende A, Letang E, Boulware DR. Cryptococcal meningitis diagnostics and screening in the era of Point-of-Care laboratory testing. J Clin Microbiol. 2019;57:e01238–e01218. doi: 10.1128/JCM.01238-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrnes EJ, Li W, Lewit Y, Ma H, Voelz K, Ren P, et al. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 2010;6:e1000850. doi: 10.1371/journal.ppat.1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, Falk R, et al. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc Natl Acad Sci USA. 2009; 4;106:12980–12985. doi: 10.1073/pnas.0902963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McTaggart LR, Lei E, Richardson SE, Hoang L, Fothergill A, Zhang SX. Rapid identification of Cryptococcus neoformans and Cryptococcus gattii by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011;49:3050–3053. doi: 10.1128/JCM.00651-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozel TR, Bauman SK. CrAg lateral flow assay for cryptococcosis. Expert Opin Med Diagn. 2012;6:245–251. doi: 10.1517/17530059.2012.681300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Severo LC, Oliveira FM, Londero AT. Cryptococcosis due to Cryptococcus neoformans var. gattii in Brazilian patients with AIDS. Rev Iberoam Micol. 1999;16:152–154. [PubMed] [Google Scholar]
- 9.Sorrell TC. Cryptococcus neoformans variety gattii. Med Mycol. 2001;39:155–168. [PubMed] [Google Scholar]
- 10.Arastehfar A, Wickes BL, Ilkit M, Pincus DH, Daneshnia F, Pan W, et al. Identification of mycoses in developing countries. J Fungi (Basel) 2019;5:90. doi: 10.3390/jof5040090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arastehfar A, Fang W, Pan W, Lackner M, Liao W, Badiee P, et al. YEAST PANEL multiplex PCR for identification of clinically important yeast species: stepwise diagnostic strategy, useful for developing countries. Diagn Microbiol Infect Dis. 2019;93:112–119. doi: 10.1016/j.diagmicrobio.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 12.WHO (World Health Organization) Guidelines on the diagnosis, prevention and management of cryptococcal disease in HIV-infected adults, adolescents and children: supplement to the 2016 consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. Geneva, Netherlands:: World Health Organization; 2018. https://www.who.int/hiv/pub/guidelines/cryptococcal-disease/en/.Accessed in November 2018. [PubMed] [Google Scholar]
- 13.Sturaro LL, Gonoi T, Lopes AF, Tararam CA, Levy CE, Lyra L, et al. Visible DNA microarray system as an adjunctive molecular test in identification of pathogenic fungi directly from a blood culture bottle. J Clin Microbiol. 2018;56:e01908–e01917. doi: 10.1128/JCM.01908-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou CC, Lee TT, Chen CH, Hsiao HY, Lin YL, Ho MS, et al. Design of microarray probes for virus identification and detection of emerging viruses at the genus level. BMC Bioinformatics. 2006;7:232. doi: 10.1186/1471-2105-7-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller MB, Tang YW. Basic concepts of microarrays and potential applications in clinical microbiology. Clin Microbiol. 2009;22:611–633. doi: 10.1128/CMR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang A, Li JW, Shen ZQ, Wang XW, Jin M. High-throughput identification of clinical pathogenic fungi by hybridization to an oligonucleotide microarray. J Clin Microbiol. 2006;44:3299–3305. doi: 10.1128/JCM.00417-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leinberger DM, Schumacher U, Autenrieth IB, Bachmann TT. Development of a DNA microarray for detection and identification of fungal pathogens involved in invasive mycoses. J Clin Microbiol. 2005;43:4943–4953. doi: 10.1128/JCM.43.10.4943-4953.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakai K, Trabasso P, Moretti ML, Mikami Y, Kamei K, Gonoi T. Identification of fungal pathogens by visible microarray system in combination with isothermal gene amplification. Mycopathologia. 2014;178:11–26. doi: 10.1007/s11046-014-9756-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Notomi T, Okayama H, Masubuchi H, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poon LL, Leung CS, Chan KH, Lee JH, Yuen KY, Guan Y, et al. Detection of human influenza A viruses by loop-mediated isothermal amplification. J ClinMicrobiol. 2005;43:427–430. doi: 10.1128/JCM.43.1.427-430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wamoto T, Sonobe T, Hayashi K. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J Clin Microbiol. 2003;41:2616–2622. doi: 10.1128/JCM.41.6.2616-2622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shahbazi E, Mollasalehi H, Minai-Tehrani D. Development and evaluation of an improved quantitative loop-mediated isothermal amplification method for rapid detection of Morganella morganii . Talanta. 2018;191:54–58. doi: 10.1016/j.talanta.2018.08.033. [DOI] [PubMed] [Google Scholar]
- 23.Poon LL, Wong BW, Ma EH, Chan KH, Chow LM, Abeyewickreme W, et al. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin Chem. 2006;52:303–306. doi: 10.1373/clinchem.2005.057901. [DOI] [PubMed] [Google Scholar]
- 24.de Souza M, Matsuzawa T, Sakai K, Muraosa Y, Muraosa Y, Lyra L, et al. Comparison of DNA microarray, loop-mediated isothermal amplification (LAMP) and real-time PCR with DNA sequencing for identification of Fusarium spp. obtained from patients with hematologic malignancies. Mycopathologia. 2017;182:625–632. doi: 10.1007/s11046-017-0129-5. [DOI] [PubMed] [Google Scholar]
- 25.Endo S, Komori T, Ricci G, Sano A, Yokoyama K, Ohori A, et al. Detection of gp43 of Paracoccidioides brasiliensis by the loop-mediated isothermal amplification (LAMP) method. FEMS Microbiol Lett. 2004;234:93–97. doi: 10.1111/j.1574-6968.2004.tb09518.x. [DOI] [PubMed] [Google Scholar]
- 26.Passos AIM, Dertkigil RP, Ramos MC, Busso-Lopes AF, Tararan C, Ribeiro EO, et al. Serum markers as an aid in the diagnosis of pulmonary fungal infections in AIDS patients. Braz J Infect Dis. 2017;21:606–612. doi: 10.1016/j.bjid.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheel CM, Zhou Y, Theodoro RC, Abrams B, Balajee SA, Litvintseva AP. Development of a loop-mediated isothermal amplification method for detection of Histoplasma capsulatum DNA in clinical samples. J Clin Microbiol. 2014;52:483–488. doi: 10.1128/JCM.02739-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inacio J, Flores O, Spencer-Martins I. Efficient identification of clinically relevant Candida yeast species by use of an assay combining panfungal loop-mediated isothermal DNA amplification with hybridization to species-specific oligonucleotide probes. J ClinMicrobiol. 2008;46:713–720. doi: 10.1128/JCM.00514-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trabasso P, Matsuzawa T, Fagnani R, Muraosa Y, Tominaga K, Resende MR, et al. Isolation and drug susceptibility of Candida parapsilosis sensu lato and other species of C. parapsilosis complex from patients with blood stream infections and proposal of a novel LAMP identification method for the species. Mycopathologia. 2015;179:53–62. doi: 10.1007/s11046-014-9830-9. [DOI] [PubMed] [Google Scholar]
- 30.Cao J, Gao S, Chen J, Zhu B, Min R, Wang P. The preparation and clinical application of diagnostic intracranial bacterial and fungal infections. Exp Ther Med. 2018;16:1304–1310. doi: 10.3892/etm.2018.6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campa D, Tavanti A, Gemignani F, Mogavero C, Bellini I, Bottariet F. DNA microarray based on arrayed-primer extension technique for identification of pathogenic fungi responsible for invasive and superficial mycoses. J Clin Microbiol. 2008;46:909–915. doi: 10.1128/JCM.01406-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Halloran JA, Franklin A, Lainhart W, Burnham CA, Powderly W, Dubberke E. Pitfalls associated with the use of molecular diagnostic panels in diagnosis of cryptococcal meningitis. Open Forum Infect Dis. 2017;4:oxf242. doi: 10.1093/ofid/ofx242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucas S, da Luz Martins M, Flores O, Meyer W, Spencer-Martins I, Inacio J. Differentiation of Cryptococcus neoformans varieties and Cryptococcus gattii using CAP59-based loop-mediated isothermal DNA amplification. Clin Microbiol Infect. 2010;16:711–714. doi: 10.1111/j.1469-0691.2009.02919.x. [DOI] [PubMed] [Google Scholar]


