Abstract
The causative agent of severe acute respiratory syndrome (SARS) reported by the Chinese Center for Disease Control (China CDC) has been identified as a novel Betacoronavirus (SARS-CoV-2). A computational approach was adopted to identify multiepitope vaccine candidates against SARS-CoV-2 based on S, N and M proteins being able to elicit both humoral and cellular immune responses. In this study, the sequence of the virus was obtained from NCBI database and analyzed with in silico tools such as NetMHCpan, IEDB, BepiPred, NetCTL, Tap transport/proteasomal cleavage, Pa3P, GalexyPepDock, I-TASSER, Ellipro and ClusPro. To identify the most immunodominant regions, after analysis of population coverage and epitope conservancy, we proposed three different constructs based on linear B-cell, CTL and HTL epitopes. The 3D structure of constructs was assessed to find discontinuous B-cell epitopes. Among CTL predicted epitopes, S257-265, S603-611 and S360-368, and among HTL predicted epitopes, N167-181, S313-330 and S1110-1126 had better MHC binding rank. We found one putative CTL epitope, S360-368 related to receptor-binding domain (RBD) region for S protein. The predicted epitopes were non-allergen and showed a high quality of proteasomal cleavage and Tap transport efficiency and 100% conservancy within four different clades of SARS-CoV-2. For CTL and HTL epitopes, the highest population coverage of the world’s population was calculated for S27-37 with 86.27% and for S196-231, S303-323, S313-330, S1009-1030 and N328-349 with 90.33%, respectively. We identified overall 10 discontinuous B-cell epitopes for three multiepitope constructs. All three constructs showed strong interactions with TLRs 2, 3 and 4 supporting the hypothesis of SARS-CoV-2 susceptibility to TLRs 2, 3 and 4 like other Coronaviridae families. These data demonstrated that the novel designed multiepitope constructs can contribute to develop SARS-CoV-2 peptide vaccine candidates. The in vivo studies are underway using several vaccination strategies.
Introduction
The causative agent of severe acute respiratory syndrome (SARS) reported by the Chinese Center for Disease Control (China CDC) has been identified as a novel Betacoronavirus (SARS-CoV-2) [1]. The genomic sequence of SARS-CoV-2 was similar but its composition was diverse as compared to SARS-CoV’s and MERS-CoV’s genome [2]. Accumulated clinical and experimental knowledge on these previous coronaviruses has led to an easier prediction of host immune responses against this particular virus. Genomic RNA of SARS-CoV-2 encodes non-structural replicase polyprotein and structural proteins including spike (S), envelope (E), membrane (M) and nucleocapsid (N). The entry of SARS-CoV-2 into host cells is mediated by attachment of S glycoprotein on the virion surface to the angiotensin-converting enzyme 2 (ACE2) receptor [3] mainly expressed in type 2 alveolar cells of lungs [4]. Enhanced binding affinity between SARS-CoV-2 and ACE2 receptor was proposed to correlate with increased virus transmissibility [5]. The trimeric S protein will be cleaved into two subunits of S1 and S2 during viral infection [6]. S1 and S2 subunits are responsible for binding to the ACE2 receptor and the fusion of the viral and cellular membranes, respectively [3]. Being the main antigenic component, S protein has been selected as an important target for vaccine development.
Anti-viral drugs, broad-spectrum antibiotics such as Remdesivir, Chloroquine, Ribavirin, Favipiravir or Baricitinib are potential therapeutic strategies used to reduce the viral load [7] by blocking the SARS-CoV-2 replication [8, 9]. Recently, the plasma exchange using convalescent sera of COVID-19 showed promising results [10, 11]. Also, the monoclonal antibody (CR3022) binding with the spike receptor-binding domain of SARS-CoV-2 had the potential to be developed as a therapeutic candidate [12]. Efforts toward developing an effective vaccine have been ignited in many countries. Actually, several projects have been reported by companies and researchers to start SARS-CoV-2 vaccine development. There are different kinds of novel vaccines including DNA-based, viral vector-based, recombinant S protein-based, adenovirus-based, mRNA-based and peptide-based vaccines. The mRNA-1273 candidate, an encapsulated mRNA vaccine encoding S protein developed by Moderna (NCT04283461), the Ad5-nCov candidate, an adenovirus type 5 vector expressing S protein developed by CanSino Biologicals (NTC04313127), the INO-4800 candidate, a DNA plasmid encoding S protein developed by Inovio Pharmaceuticals (NCT04336410), the LV-SMENP-DC candidate, dendritic cells modified with lentiviral vector (NCT04276896), and the pathogen-specific aAPC candidate, an aAPC modified with a lentiviral vector (NCT04299724) both developed by Shenzhen Geno-immune Medical Institute are few vaccines in phase I of the clinical trial against SARS-CoV-2 [13].
However, each type of vaccine has a number of advantages and disadvantages. Although platforms based on DNA or mRNA are flexible and effective for antigen manipulation, peptide-based vaccines are customizable multipurpose therapeutics which does not have the implication of stability or translation [14] and by the use of multiepitope approach, a single peptide-based vaccine can be designed to target different strains [15]. Despite safety and cost-effectiveness, peptide-based vaccines are difficult to design. The epitope-mapping is a crucial but time-consuming step in the design of a peptide-based vaccine. That is why no peptide-based vaccine for SARS-CoV-2 has reached phase I clinical trial to date. A successful peptide-based vaccine comprises immunodominant B-cell and T-cell being able to induce strong and long-lasting immunity against the desired pathogen [16]. Thus, the understanding of epitope interaction with major histocompatibility complex (MHC) is necessary. In the current study, a computational approach was adopted to identify multiepitope vaccine candidates against SARS-CoV-2 based on S, N and M proteins.
Materials and methods
Collection of targeted proteins sequences
The reference sequences of the targeted proteins including S, N and M proteins of SARS-CoV-2 were obtained from the NCBI database and used as an input for more bioinformatics analyses.
Linear B-cell epitope prediction
A successful vaccine must elicit strong cellular and humoral immune responses. Thus, it is important to show that the constructed immunogens are able to induce protective immunity. It should be considered that optimal peptide-based vaccines must be presented in a desired secondary structure of peptides in order to induce a specific humoral response. In this subsection, we used BepiPred-2.0 prediction module (http://www.cbs.dtu.dk/services/BepiPred-2.0/) for linear B-cell prediction of the conserved regions in S, N and M proteins of SARS-CoV-2 to produce the B-cell mediated immunity. In this study, epitope threshold value was set as 0.5 (the sensitivity and specificity of this method are 0.58 and 0.57, respectively) [17].
T-cell epitope identification
The initial step on applying bioinformatics to design synthetic peptide vaccines is to determine whether epitopes are potentially immunoprotective. T-cell epitopes presented by MHC are linear form containing 12 to 20 amino acids. This fact facilitates modeling for the interaction of ligands and T-cells with accuracy [18]. Binding of the MHC molecule is the most selective step in the presentation of antigenic peptide to T-cell receptor (TCR).
For MHC class I, we adapted Artificial Neural Networks (NetMHCpan4.1 server (http://www.cbs.dtu.dk/services/NetMHCpan/) to predict high-potential T-cell epitopes. This server is meant to predict MHC I binding with accuracy of 90–95% [19, 20]. Human alleles were used and the threshold for NetMHCpan was set at 0.5% for strong binders and 2% for weak binders.
For MHC class II, we used NetMHCIIpan 4.0 server (http://www.cbs.dtu.dk/services/NetMHCIIpan/) [21] to predict potential interaction of helper T-cell epitope peptides and MHC class II. Human alleles were used and the threshold for strong and weak binders was set at 2% and 10%, respectively.
Prediction of MHC class I peptide presentation pathway
Best ranked peptides extracted from NetMHCpan database were used in transporter associated with antigen presentation (TAP) transport efficacy and proteasomal cleavage analysis. In MHC class I presentation pathway, this section is as essential as binding affinity prediction. We employed NetCTL 1.2 server combined with Tap transport/proteasomal cleavage tools (http://www.cbs.dtu.dk/services/NetCTL) to assess the prediction of antigen processing through the MHC-I antigen presentation pathway. In this method, weight on C-terminal cleavage set on 0.15, and tap transport efficacy and epitope identification were set on 0.05 and 0.75, respectively.
Conservancy analysis
Up to now, more than 16667 full-sequences of SARS-CoV-2 have been registered globally in GISAID database classified into four clades of V, G, S and O. To calculate the degree of conservancy of each epitope, IEDB epitope conservancy tool (http://tools.immuneepitope.org/tools/conservancy/) was employed [22]. This tool computes the degree of conservancy of an epitope within a given protein sequence set at a given identity level. In this study, we determined epitope conservancy of each protein including S, N and M obtained from GISAID database.
Population coverage
Due to a phenomenon known as denominated MHC restriction of T-cell responses, selecting multiple epitopes with different HLA binding specificities will afford more increases in population coverage. Prediction based on HLA binding at population level in defined geographical regions where the peptide-based vaccine might be employed is essential. Since MHC polymorphisms are dramatically at different frequencies in different ethnicities, without careful consideration, a vaccine with ethnically biased population coverage will result. In this study, we used IEDB population coverage tool [23] (http://tools.iedb.org/population/) to assess the coverage rate of population for each epitope.
Antibody-specific epitopes prediction
IgPred module [24] (https://webs.iiitd.edu.in/raghava/igpred/index.html) was developed for predicting different types of B-cell epitopes inducing different classes of antibodies. We used this server to identify epitope tendency for inducing IgG and IgA antibodies.
Prediction of cytokine inducer peptides
It is important to understand that all MHC class II binders will not induce the same type of cytokines. Thus, we used IL-10 Pred [25] (http://crdd.osdd.net/raghava/IL-10pred/) and IFNepitope [26] webserver (http://crdd.osdd.net/raghava/ifnepitope/index.php) to predict Il-10 and Interferon-gamma inducing peptides, respectively. We used Support Vector Machine (SVM)-based model as prediction model in both servers. Other features including SVM threshold left at the default value. Through using these servers, we improved insight into the future in vivo studies.
Allergenicity and cross-reactivity assessment
The prediction of potential allergenicity is an important step in safety assessment as proteins and polypeptides have significant roles in inducing allergenic reactions. The allergenicity of the selected epitopes was calculated by PA3P server (http://lpa.saogabriel.unipampa.edu.br:8080/pa3p/pa3p/pa3p.jsp) using AFDS-motif, Allergen online (6aa and 80-word match) algorithms. The specificity of these methods is 95.43% (6aa), 92.88% (80aa), and 88.1% (ADFS) [27].
Peptide-protein flexible docking
To estimate the formation of MHC-peptide complex, we used GalexyPepDock peptide-protein flexible docking server [28] (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=PEPDOCK). This study presents an example of GalexyPepDock performed by each epitope and available PDB file of HLA alleles, separately.
Vaccine construction
To construct effectual vaccine components, we fused the antigenic epitopes with the help of specific peptide linkers. Three different constructs for each linear B lymphocyte (LBL), cytotoxic T lymphocyte (CTL) and helper T lymphocyte (HTL) were designed.
The physicochemical parameters
The physicochemical properties of the designed LBL, CTL and HTL epitopes including molecular weight, theoretical PI, positive and negative charge residue, solubility and stability were evaluated by ProtParam online server (http://us.expasy.org/tools/protparam.html) [29].
3D structure prediction
I-TASSER server [30] (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) was used for modeling the 3D structure of designed constructs. This server is in active development with the goal to provide the most accurate protein structure and function predictions using state-of-the-art algorithms. After analysis, the models with the highest confidence score (C-score) were selected for refinement analysis.
Refinement and validation of tertiary structure
GalaxyRefine 2 Server [31] (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE2) was used to refine predicted tertiary structures. GalaxyRefine2 performs iterative optimization with several geometric operators to increase the accuracy of the initial models. Final Refined models were analyzed by SAVE5.0 (https://servicesn.mbi.ucla.edu/SAVES/) server to validated tertiary structures. SAVE server gives Ramachandran plot of the whole structure, determines the overall quality of tertiary structure, and calculates buried protein atoms, stereochemical quality and atomic interaction of predicted 3D structure.
Discontinuous B-cell epitope prediction
Prediction of discontinuous B-cell epitope needs tertiary structure of a protein or polypeptide since the interaction between antigen epitopes and antibodies is very important. As regards, after refinement and validation analysis, the 3D structure of constructs were assessed by the Ellipro server [32] (https://tools.iedb.org/ellipro/help/) to find discontinuous B-cell epitopes. ElliPro web-based server uses modified Thornton’s method along with residue clustering algorithms. In this study, epitope prediction parameters (minimum score and maximum distance) were set to default values (0.5 and 6).
Docking between vaccine constructs and Toll-Like Receptors (TLRs)
TLRs are sensors recognizing molecular patterns of pathogens to initiate innate immune system. It was demonstrated that TLRs 2, 3 and 4 are more susceptible to Coronaviridae family including SARS-CoV and MERS-CoV [33–35]. Thus, PDB files of TLRs 2, 3 and 4 were obtained from Protein Data Bank (http://www.rcsb.org/) and then protein-protein docking between three vaccine constructs and TLRs were performed by ClusPro server [36] (https://cluspro.bu.edu/). ClusPro uses three steps algorithms containing 1) Rigid-body docking, 2) Cluster retained conformations, and 3) Refine by CHARMM minimization.
Results
The sequences of the structural SARS-CoV-2 proteins
The reference sequences of the structural proteins (S, N and M, NC_045512.2) were obtained from NCBI. The sequence was downloaded in a FASTA format to carry out further analyses.
Prediction of linear B-cell epitopes
We obtained a total of 44 sequential linear B-cell epitopes with variable lengths from IEDB server within three main proteins of SARS-CoV-2 (i.e., S, N and M), and the ability of epitopes in inducing different classes of antibody in IgPred server were analyzed. In S protein, S1133-1172 (VNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGI), S440-501 (NLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTN), S59-81(FSNVTWFHAIHVSGTNGTKRFDN) and S304-322 (KSFTVEKGIYQTSNFRVQP), and in N protein, N232-269(SKMSGKGQQQQGQTVTKKSAAEASKKPRQKRTATKAYN), N164-216(GTTLPKGFYAEGSRGGSQASSRSSSRSRNSSRNSTPGSSRGTSPARMAGNGGD), N1-51(MSDNGPQNQRNAPRITFGGPSDSTGSNQNGERSGARSKQRRPQGLPNNTAS) and N361-390(KTFPPTEPKKDKKKKADETQALPQRQKKQQ) were chosen as they had the ability to induce antibody (Table 1). In case of M protein, we found three epitopes. However, we ruled out M epitope for potential B-cell epitope as they were unable to induce any class of the antibodies.
Table 1. The selected LBL* epitopes of SARS-CoV-2 based on binding affinity.
| Protein Name | Position | Epitope Sequence | Antibody Class Prediction |
|---|---|---|---|
| S | 1133–1172 | VNNTVYDPLQPELDSFKEELDKYFKNHTSPDVDLGDISGI | IgG |
| 440–501 | NLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTN | IgG | |
| 59–81 | FSNVTWFHAIHVSGTNGTKRFDN | IgG | |
| 304–322 | KSFTVEKGIYQTSNFRVQP | IgA | |
| N | 232–269 | SKMSGKGQQQQGQTVTKKSAAEASKKPRQKRTATKAYN | IgG |
| 164–216 | GTTLPKGFYAEGSRGGSQASSRSSSRSRNSSRNSTPGSSRGTSPARMAGNGGD | IgG | |
| 1–51 | MSDNGPQNQRNAPRITFGGPSDSTGSNQNGERSGARSKQRRPQGLPNNTAS | IgG | |
| 361–390 | KTFPPTEPKKDKKKKADETQALPQRQKKQQ | IgG |
*Linear B lymphocyte
Prediction of T-cell epitopes
Identification of CD8+ cytotoxic T lymphocyte (CTL) epitopes is a crucial step in epitope-driven vaccine design as MHC class I restricted CTL plays a critical role in controlling viral infections. In this study, we employed NetMHCpan and NetMHCIIpan as mentioned procedure in below.
MHC class I prediction
The SARS-CoV-2 protein sequences were analyzed by NetMHCpan 4.1 server to identify the most immunodominant regions. In each protein, peptides with the highest binding affinity scores were determined as high-potential CTL epitope candidates. In each protein, the best epitopes with higher binding affinity were selected as the putative CTL epitope based on calculated average immunogenicity scores. Chosen MHC-I epitopes were listed in Table 2 with encountering MHC alleles, average rank scores, conservancy prediction and allergenicity assessment. Also, all of the chosen sequences of epitopes were non-allergen and 100% conserved within four clades.
Table 2. The selected CTL* epitopes of SARS-CoV-2 based on binding affinity.
| Protein Name | Position | Epitope Sequence | No. of Alleles | NetMHCpan Average Rank Scores** | Conservancy | Allergenicity |
|---|---|---|---|---|---|---|
| S | 27–37 | YTNSFTRGVYY | 12 | 0.67 | S:100%*** V: 100% G: 100% O: 100% |
Non-allergen |
| 686–696 | VASQSIIAYTM | 12 | 0.69 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| 191–199 | FVFKNIDGY | 10 | 0.66 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| 1051–1061 | FPQSAPHGVVF | 10 | 1.08 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| 257–265 | WTAGAAAYY | 9 | 0.44 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| 603–611 | TSNQVAVLY | 9 | 0.44 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| 868–876 | MIAQYTSAL | 9 | 0.61 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| 161–169 | SANNCTFEY | 8 | 0.96 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| 360–368 | CVADYSVLY | 8 | 0.48 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| 1094–1102 | FVSNGTHWF | 8 | 0.69 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| N | 103–113 | LSPRWYFYYLG | 10 | 1.04 | S:100% V: 100% G: 100% O: 100% |
Non-allergen |
| 304–313 | AQFAPSASAF | 8 | 0.87 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| 47–55 | NTASWFTAL | 6 | 0.70 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| 264–273 | TKAYNVTQAF | 6 | 0.90 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| 52–60 | FTALTQHGK | 5 | 0.64 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| M | 36–46 | FAYANRNRFLY | 13 | 1.23 | S:100% V: 100% G: 100% O: 100% |
Non-allergen |
| 168–180 | TVATSRTLSYY | 9 | 0.89 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| 195–204 | YSRYRIGNYK | 9 | 0.31 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| 90–99 | MWLSYFIASF | 7 | 1.05 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | |
| 102–111 | FARTRSMWSF | 7 | 0.59 | S:100% V: 100% G: 100% O: 100% |
Non-allergen |
* Cytotoxic T lymphocyte
** Lower rates show better binding affinity
*** There are four clades of SARS-CoV-2 according to GISAID database
MHC class II prediction
The SARS-CoV-2 protein sequences were analyzed by NetMHCIIpan 4.0 server to identify MHC-II epitope. Epitopes with the maximum number of binding HLA-DR alleles were selected as putative HTL epitope candidate. Chosen MHC-II epitopes were listed in Table 3 with encountering MHC alleles, average rank scores, conservancy and antibody-specific epitopes prediction, and allergenicity assessment. Also, all of the chosen sequences of epitopes were non-allergen and 100% conserved within four clades.
Table 3. The selected HTL epitopes of SARS-CoV-2 based on binding affinity.
| Protein Name | Position | Epitope Sequence | No. of Alleles | NetMHCpan Average Rank Scores* | Conservancy | Allergenicity | IFN-gamma prediction | IL-10 prediction |
|---|---|---|---|---|---|---|---|---|
| S | 196–231 | NIDGYFKIYSKHTPINLVRDLPQGFS | 25 | 4.29 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Positive |
| 303–323 | LKSFTVEKGIYQTSNFRVQPT | 25 | 3.96 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Negative | |
| 313–330 | YQTSNFRVQPTESIVRFP | 25 | 3.17 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Positive | |
| 1009–1030 | TQQLIRAAEIRASANLAATKMS | 25 | 3.89 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Positive | |
| 32–53 | FTRGVYYPDKVFRSSVLHSTQD | 24 | 4.45 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Positive | |
| 1057–1074 | PHGVVFLHVTYVPAQEKN | 22 | 5.7 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Positive | |
| 689–704 | SQSIIAYTMSLGAENS | 21 | 5.2 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Positive | |
| 801–817 | NFSQILPDPSKPSKRSF | 20 | 3.51 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Negative | Positive | |
| 1110–1126 | YEPQIITTDNTFVSGNC | 19 | 3.19 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Negative | Positive | |
| 114–130 | TQSLLIVNNATNVVIKV | 19 | 5.37 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Positive | |
| N | 328–349 | GTWLTYTGAIKLDDKDPNFKDQ | 25 | 4.5 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Negative |
| 126–143 | NKDGIIWVATEGALNTPK | 24 | 3.85 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Negative | |
| 342–361 | KDPNFKDQVILLNKHIDAYK | 24 | 4.39 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Positive | |
| 48–63 | NTASWFTALTQHGKED | 23 | 3.24 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Negative | |
| 167–181 | LPKGFYAEGSRGGSQ | 23 | 2 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Negative | Negative | |
| 405–419 | KQLQQSMSSADSTQA | 22 | 3.58 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Negative | Negative | |
| M | 198–217 | RYRIGNYKLNTDHSSSSDNI | 22 | 4.76 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Negative |
| 173–191 | SRTLSYYKLGASQRVAGDS | 20 | 4.53 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Negative | |
| 107–128 | RSMWSFNPETNILLNVPLHGTI | 19 | 6.02 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Negative | |
| 163–181 | DLPKEITVATSRTLSYYKL | 17 | 3.57 | S:100% V: 100% G: 100% O: 100% |
Non-allergen | Positive | Negative |
*lower rates show better binding affinity
Tap transport/proteasomal cleavage
Tap transport and proteasomal cleavage are as important as binding affinity in antigen presentation pathway to CTLs. In this case, NetCTL1.2 server was used. All of the epitopes shown in Table 4 have upper cut off identification scores (> 0.75) which show a high quality of proteasomal cleavage and Tap transport efficiency. Among all epitopes, S257-265 and S603-611 have the highest epitope identification score of 3.14 and 3.07, respectively.
Table 4. Proteasomal cleavage and TAP transport efficiency scores of MHC-I predicted epitopes.
| Protein Name | Position | Epitope Sequence | Proteasomal C -terminal cleavage Score* | TAP transport efficiency Score** | Epitope identification Score*** |
|---|---|---|---|---|---|
| S | 27–37 | YTNSFTRGVYY | 1.43 | 0.96 | 1.73 |
| 686–696 | VASQSIIAYTM | 1.49 | 0.93 | 1.73 | |
| 191–199 | FVFKNIDGY | 1.75 | 0.94 | 2.03 | |
| 1051–1061 | FPQSAPHGVVF | 0.98 | 0.93 | 1.18 | |
| 257–265 | WTAGAAAYY | 2.85 | 0.94 | 3.14 | |
| 603–611 | TSNQVAVLY | 2.87 | 0.95 | 3.07 | |
| 868–876 | MIAQYTSAL | 1.04 | 0.93 | 1.33 | |
| 161–169 | SANNCTFEY | 2.07 | 0.92 | 2.36 | |
| 360–368 | CVADYSVLY | 2.27 | 0.97 | 2.56 | |
| 1094–1102 | FVSNGTHWF | 1.46 | 0.86 | 1.72 | |
| N | 103–113 | LSPRWYFYYLG | 2.05 | 0.96 | 2.34 |
| 304–313 | AQFAPSASAF | 1.06 | 0.66 | 1.31 | |
| 47–55 | NTASWFTAL | 1.08 | 0.94 | 1.27 | |
| 264–273 | TKAYNVTQAF | 1.01 | 0.94 | 1.30 | |
| 52–60 | FTALTQHGK | 0.80 | 0.72 | 0.92 | |
| M | 36–46 | FAYANRNRFLY | 1.40 | 0.95 | 1.69 |
| 168–180 | TVATSRTLSYY | 2.31 | 0.96 | 2.61 | |
| 195–204 | YSRYRIGNYK | 1.36 | 0.84 | 1.64 | |
| 90–99 | MWLSYFIASF | 0.66 | 0.97 | 0.96 | |
| 102–111 | FARTRSMWSF | 1.10 | 0.95 | 1.38 |
*Higher rates show better quality of proteasomal cleavage
** Higher rates show better quality of tap transport efficiency
*** Higher rates show better quality of epitope identification
Population coverage
As mentioned above, MHC polymorphisms are dramatically at different frequencies in various ethnicities. Thus, careful consideration should be given to the way of effective vaccine development. In this study, population coverage was estimated separately for each putative epitope in different geographical regions (Tables 5 and 6). For CTL epitopes, the highest population coverage of the world’s population was calculated for S27-37 with 86.27%. For helper T-cell epitopes, the highest population coverage of the world’s population was calculated for S196-231, S303-323, S313-330, S1009-1030 and N328-349 with 90.33%.
Table 5. Population coverage of putative SARS-CoV-2 CTL epitopes.
| Area | S27-37 | S686-696 | S191-199 | S1051-1061 | S257-265 | S603-611 | S868-876 | S161-169 | S360-368 | S1094-1102 | N103-113 | N304-313 | N47-55 | N264-273 | N52-60 | M36-46 | M168-180 | M195-204 | M90-99 | M102-111 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Central Africa | 58.3% | 61.19% | 47.7% | 42.84% | 45.43% | 45.43% | 47.79% | 41.35% | 41.35% | 37.02% | 39.4% | 33.9% | 31.58% | 38.38% | 27.68% | 46.0% | 43.11% | 34.13% | 25.22% | 30.74% |
| East Africa | 69.68% | 67.18% | 54.97% | 51.17% | 50.59% | 50.59% | 54.01% | 44.19% | 44.19% | 48.33% | 47.25% | 27.24% | 27.26% | 29.76% | 31.08% | 53.0% | 48.86% | 41.54% | 28.22% | 28.53% |
| East Asia | 68.17% | 59.86% | 55.61% | 54.86% | 55.61% | 55.61% | 60.36% | 55.49% | 53.53% | 74.58% | 78.7% | 82.83% | 31.06% | 70.63% | 32.55% | 84.27% | 54.94% | 45.63% | 71.67% | 35.38% |
| Europe | 93.95% | 80.28% | 77.35% | 71.73% | 73.88% | 73.88% | 79.54% | 73.47% | 73.45% | 61.27% | 76.7% | 71.17% | 52.65% | 64.39% | 64.85% | 85.33% | 70.88% | 70.14% | 59.55% | 50.55% |
| North Africa | 76.03% | 73.45% | 57.9% | 50.94% | 55.03% | 55.03% | 59.15% | 52.23% | 52.0% | 52.87% | 55.62% | 44.7% | 30.4% | 34.85% | 36.01% | 57.8% | 50.85% | 46.2% | 37.72% | 29.94% |
| North America | 84.8% | 69.76% | 64.08% | 57.57% | 61.39% | 61.39% | 70.66% | 59.51% | 59.28% | 57.58% | 69.4% | 67.35% | 38.65% | 57.07% | 44.89% | 75.68% | 57.57% | 52.73% | 51.7% | 40.09% |
| Northeast Asia | 73.97% | 68.15% | 66.77% | 37.1% | 66.51% | 66.51% | 38.22% | 63.48% | 63.43% | 50.44% | 73.37% | 77.42% | 13.17% | 46.14% | 52.93% | 76.81% | 65.94% | 61.92% | 41.39% | 18.03% |
| Oceania | 63.86% | 58.32% | 54.78% | 34.54% | 54.21% | 54.21% | 31.12% | 52.96% | 52.96% | 66.01% | 84.26% | 83.22% | 21.79% | 59.46% | 50.2% | 86.64% | 52.61% | 51.88% | 60.32% | 17.61% |
| South Africa | 64.94% | 72.02% | 57.59% | 49.1% | 55.81% | 55.81% | 44.82% | 53.25% | 53.25% | 50.83% | 58.64% | 46.87% | 28.43% | 33.15% | 41.9% | 60.92% | 50.64% | 50.62% | 37.25% | 29.34% |
| South America | 56.97% | 45.93% | 40.8% | 25.13% | 37.66% | 37.66% | 36.75% | 36.75% | 36.75% | 39.03% | 44.64% | 52.03% | 16.62% | 37.72% | 31.08% | 57.06% | 27.16% | 34.24% | 36.7% | 14.41% |
| South Asia | 77.05% | 73.24% | 71.64% | 46.92% | 70.83% | 70.83% | 38.84% | 67.75% | 67.75% | 54.67% | 67.42% | 66.54% | 29.6% | 43.15% | 59.67% | 72.8% | 55.62% | 65.66% | 37.59% | 27.3% |
| Southeast Asia | 66.67% | 62.24% | 58.2% | 35.31% | 55.92% | 55.92% | 28.83% | 48.97% | 48.97% | 57.94% | 77.29% | 82.1% | 17.1% | 53.25% | 43.57% | 80.89% | 52.67% | 47.52% | 51.06% | 21.82% |
| Southwest Asia | 77.27% | 66.35% | 59.6% | 45.9% | 56.9% | 56.9% | 54.88% | 54.49% | 54.49% | 48.92% | 58.54% | 51.82% | 29.33% | 32.89% | 47.47% | 63.53% | 51.56% | 51.97% | 38.63% | 29.55% |
| West Africa | 70.28% | 74.43% | 58.96% | 53.85% | 56.47% | 56.47% | 57.29% | 51.92% | 51.92% | 48.96% | 55.22% | 47.07% | 38.36% | 43.67% | 33.19% | 61.05% | 55.9% | 41.03% | 41.74% | 42.38% |
| West Indies | 80.64% | 74.78% | 63.01% | 56.85% | 59.44% | 59.44% | 64.77% | 56.07% | 56.07% | 54.84% | 64.26% | 61.14% | 41.35% | 54.32% | 47.92% | 73.47% | 55.9% | 50.81% | 49.62% | 42.73% |
| World | 86.27% | 72.24% | 68.17% | 57.5% | 65.34% | 65.34% | 67.31% | 64.18% | 64.0% | 56.47% | 70.23% | 68.46% | 39.15% | 55.99% | 54.27% | 77.72% | 62.15% | 60.54% | 50.99% | 36.77% |
Table 6. Population coverage of putative SARS-CoV-2 HTL epitopes.
| Area | S196-231 | S303-323 | S313-330 | S1009-1030 | S32-53 | S1057-1074 | S689-704 | S801-817 | S1110-1126 | S114-130 | N328-349 | N126-143 | N342-361 | N48-63 | N167-181 | N405-419 | M198-217 | M173-191 | M107-128 | M163-181 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Central Africa | 75.33% | 75.33% | 75.33% | 75.33% | 71.52% | 59.02% | 59.34% | 60.66% | 64.38% | 66.12% | 75.33% | 67.04% | 74.78% | 66.93% | 64.03% | 56.8% | 61.37% | 70.37% | 59.27% | 65.04% |
| East Africa | 81.34% | 81.34% | 81.34% | 81.34% | 76.57% | 65.54% | 58.27% | 68.08% | 62.42% | 65.07% | 81.34% | 73.27% | 80.55% | 74.6% | 69.31% | 56.04% | 65.89% | 74.4% | 62.42% | 64.24% |
| East Asia | 86.93% | 86.93% | 86.93% | 86.93% | 84.15% | 84.31% | 81.62% | 62.84% | 79.63% | 73.29% | 86.93% | 86.13% | 80.46% | 79.92% | 82.21% | 77.93% | 72.87% | 72.62% | 67.69% | 66.34% |
| Europe | 93.54% | 93.54% | 93.54% | 93.54% | 92.38% | 81.75% | 78.89% | 72.95% | 86.86% | 87.15% | 93.54% | 90.14% | 93.42% | 77.57% | 84.85% | 76.43% | 87.88% | 91.26% | 80.17% | 81.71% |
| North Africa | 88.26% | 88.26% | 88.26% | 88.26% | 87.65% | 74.7% | 72.59% | 73.18% | 80.37% | 82.82% | 88.26% | 85.14% | 87.95% | 71.69% | 78.01% | 75.59% | 83.78% | 83.49% | 68.5% | 76.74% |
| North America | 94.75% | 94.75% | 94.75% | 94.75% | 93.79% | 84.42% | 82.17% | 77.86% | 88.13% | 88.84% | 94.75% | 92.18% | 94.5% | 73.57% | 86.73% | 79.13% | 89.33% | 93.19% | 83.03% | 83.13% |
| Northeast Asia | 68.85% | 68.85% | 68.85% | 68.85% | 65.35% | 63.16% | 60.58% | 47.01% | 62.19% | 54.36% | 68.85% | 66.94% | 65.74% | 78.32% | 61.48% | 54.28% | 53.05% | 59.87% | 54.07% | 56.44% |
| Oceania | 74.82% | 74.82% | 74.82% | 74.82% | 69.27% | 73.93% | 62.41% | 62.75% | 61.98% | 58.06% | 74.82% | 74.55% | 68.85% | 56.81% | 68.59% | 54.01% | 64.28% | 53.78% | 52.62% | 43.72% |
| South Africa | 52.11% | 52.11% | 52.11% | 52.11% | 52.11% | 8.42% | 31.28% | 50.86% | 51.56% | 50.86% | 52.11% | 32.76% | 52.11% | 66.55% | 31.28% | 32.76% | 31.28% | 52.11% | 30.61% | 52.11% |
| South America | 76.19% | 76.19% | 76.19% | 76.19% | 75.35% | 69.22% | 68.04% | 62.65% | 58.82% | 68.62% | 76.19% | 73.99% | 67.94% | 31.28% | 70.7% | 68.41% | 68.85% | 71.95% | 65.09% | 54.89% |
| South Asia | 88.43% | 88.43% | 88.43% | 88.43% | 87.25% | 77.38% | 75.43% | 66.62% | 77.06% | 82.25% | 88.43% | 84.78% | 88.0% | 68.74% | 80.41% | 71.04% | 82.52% | 85.41% | 71.07% | 79.51% |
| Southeast Asia | 67.66% | 67.66% | 67.66% | 67.66% | 65.26% | 62.42% | 56.06% | 50.07% | 59.47% | 50.67% | 67.66% | 67.25% | 62.15% | 73.82% | 60.3% | 53.94% | 54.69% | 56.91% | 49.7% | 51.81% |
| Southwest Asia | 58.07% | 58.07% | 58.07% | 58.07% | 57.31% | 48.2% | 44.41% | 45.91% | 48.46% | 50.87% | 58.07% | 54.83% | 57.15% | 56.04% | 50.87% | 45.49% | 53.64% | 52.81% | 43.09% | 43.57% |
| West Africa | 84.27% | 84.27% | 84.27% | 84.27% | 83.28% | 75.29% | 72.9% | 71.93% | 64.44% | 72.66% | 84.27% | 79.83% | 83.27% | 48.87% | 79.11% | 70.05% | 73.8% | 78.54% | 60.66% | 69.82% |
| West Indies | 77.4% | 77.4% | 77.4% | 77.4% | 74.21% | 69.63% | 62.33% | 52.95% | 64.29% | 64.15% | 77.4% | 73.64% | 76.45% | 77.74% | 70.24% | 59.75% | 67.69% | 72.67% | 65.6% | 61.47% |
| World | 90.33% | 90.33% | 90.33% | 90.33% | 88.87% | 79.8% | 77.08% | 70.7% | 82.52% | 82.55% | 90.33% | 87.34% | 89.71% | 74.87% | 81.75% | 74.33% | 83.11% | 87.1% | 76.39% | 76.94% |
Peptide-protein flexible docking
At first, available structure data of MHC-I and MHC-II were downloaded from RCSB PDB server (https://www.rcsb.org/). All potential epitopes and MHC PDB files were submitted to the server separately. Then, top models with the highest interaction similarity score (similarity of the amino acids of the target complex aligned to the contacting residues in the template structure to the template amino acids, obtained from GalexyPepDock server) were selected for each peptide and its MHC. For CTL epitopes, N103-113, S868-876, M36-46, S1094-1102, M102-111, S1051-1061, S360-368, S191-199 and S686-696 had the highest average of interaction similarity score, respectively (Table 7). For HTL epitopes, M107-128, N48-63, S689-704, S1057-1074, S196-231, N328-349, S32-53, N126-143, M163-181 and S114-130 had the highest average of interaction similarity score, respectively (Table 8). Overall, CTL epitopes showed better quality of docking in comparison with HTL epitope.
Table 7. Interaction similarity scores of the selected CTL epitopes using GalexyPepDock flexible docking server.
| Protein | Epitope | HLA A0101 | HLA A0201 | HLA A0301 | HLA A2402 | HLA A1101 | HLA B0702 | HLA B0801 | HLA B2705 | HLA B3501 | HLA B5101 | Average* |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | YTNSFTRGVYY | 170 | 192 | 184 | 172 | 181 | 186 | 199 | 175 | 219 | 202 | 188.0 |
| SANNCTFEY | 195 | 170 | 153 | 160 | 158 | 171 | 167 | 168 | 213 | 185 | 174.0 | |
| FVFKNIDGY | 188 | 209 | 195 | 187 | 197 | 219 | 237 | 206 | 214 | 191 | 204.3 | |
| WTAGAAAYY | 151 | 181 | 165 | 178 | 159 | 170 | 189 | 176 | 201 | 190 | 176.0 | |
| CVADYSVLY | 203 | 231 | 223 | 190 | 216 | 201 | 192 | 207 | 232 | 212 | 210.7 | |
| TSNQVAVLY | 157 | 163 | 152 | 170 | 151 | 170 | 158 | 171 | 216 | 188 | 169.6 | |
| VASQSIIAYTM | 174 | 201 | 187 | 206 | 200 | 203 | 199 | 195 | 242 | 225 | 203.2 | |
| MIAQYTSAL | 205 | 266 | 239 | 237 | 220 | 219 | 227 | 219 | 229 | 225 | 228.6 | |
| FPQSAPHGVVF | 177 | 216 | 189 | 204 | 179 | 215 | 243 | 200 | 227 | 258 | 210.8 | |
| FVSNGTHWF | 199 | 207 | 189 | 217 | 208 | 235 | 256 | 221 | 220 | 201 | 215.3 | |
| M | FAYANRNRFLY | 210 | 232 | 202 | 268 | 213 | 213 | 236 | 211 | 253 | 226 | 226.4 |
| MWLSYFIASF | 171 | 231 | 204 | 219 | 185 | 197 | 214 | 187 | 191 | 195 | 199.4 | |
| FARTRSMWSF | 195 | 201 | 196 | 260 | 198 | 211 | 249 | 204 | 200 | 215 | 212.9 | |
| TVATSRTLSYY | 164 | 201 | 185 | 191 | 180 | 209 | 188 | 207 | 251 | 223 | 199.9 | |
| YSRYRIGNYK | 154 | 176 | 171 | 215 | 170 | 180 | 215 | 178 | 219 | 206 | 188.4 | |
| N | NTASWFTAL | 153 | 189 | 168 | 192 | 174 | 189 | 177 | 185 | 187 | 181 | 179.5 |
| FTALTQHGK | 167 | 186 | 166 | 170 | 176 | 206 | 232 | 191 | 188 | 176 | 185.8 | |
| LSPRWYFYYLG | 217 | 244 | 237 | 204 | 237 | 262 | 247 | 254 | 300 | 278 | 248 | |
| TKAYNVTQAF | 142 | 186 | 151 | 207 | 160 | 168 | 158 | 174 | 179 | 200 | 172.5 | |
| AQFAPSASAF | 172 | 201 | 192 | 203 | 188 | 195 | 192 | 191 | 213 | 215 | 196.2 |
* Higher rates show better quality of modeling
Table 8. Interaction similarity scores of the selected HTL epitopes using GalexyPepDock flexible docking server.
| Protein | Epitope | DRB1-0101 | DRB1-0301 | DRB1-0401 | DRB1-1101 | DRB1- 1501 | DRB5-0101 | Average* |
|---|---|---|---|---|---|---|---|---|
| S | NIDGYFKIYSKHTPINLVRDLPQGFS | 139 | 146 | 132 | 132 | 132 | 132 | 135.5 |
| LKSFTVEKGIYQTSNFRVQPT | 124 | 126 | 124 | 126 | 124 | 126 | 125.0 | |
| YQTSNFRVQPTESIVRFP | 118 | 105 | 118 | 118 | 108 | 118 | 114.1 | |
| TQQLIRAAEIRASANLAATKMS | 133 | 123 | 113 | 123 | 113 | 113 | 119.6 | |
| FTRGVYYPDKVFRSSVLHSTQD | 135 | 133 | 135 | 133 | 135 | 135 | 134.3 | |
| PHGVVFLHVTYVPAQEKN | 170 | 128 | 128 | 128 | 132 | 128 | 135.6 | |
| SQSIIAYTMSLGAENS | 141 | 129 | 141 | 141 | 141 | 141 | 139.0 | |
| NFSQILPDPSKPSKRSF | 148 | 126 | 126 | 126 | 126 | 126 | 129.6 | |
| YEPQIITTDNTFVSGNC | 123 | 116 | 120 | 116 | 123 | 120 | 119.6 | |
| TQSLLIVNNATNVVIKV | 131 | 131 | 131 | 130 | 130 | 131 | 130.6 | |
| M | RYRIGNYKLNTDHSSSSDNI | 117 | 113 | 117 | 117 | 117 | 117 | 116.3 |
| SRTLSYYKLGASQRVAGDS | 124 | 124 | 124 | 128 | 124 | 128 | 125.3 | |
| RSMWSFNPETNILLNVPLHGTI | 169 | 169 | 169 | 139 | 169 | 169 | 164.0 | |
| DLPKEITVATSRTLSYYKL | 137 | 124 | 125 | 137 | 137 | 137 | 132.8 | |
| N | GTWLTYTGAIKLDDKDPNFKDQ | 137 | 131 | 137 | 137 | 137 | 131 | 135 |
| NKDGIIWVATEGALNTPK | 136 | 133 | 131 | 136 | 131 | 136 | 133.8 | |
| KDPNFKDQVILLNKHIDAYK | 127 | 134 | 127 | 127 | 134 | 127 | 129.3 | |
| NTASWFTALTQHGKED | 137 | 137 | 137 | 137 | 149 | 137 | 139.0 | |
| LPKGFYAEGSRGGSQ | 107 | 107 | 107 | 105 | 105 | 107 | 106.3 | |
| KQLQQSMSSADSTQA | 142 | 118 | 118 | 118 | 118 | 118 | 122.0 |
* Higher rates show better quality of modeling
Construct design
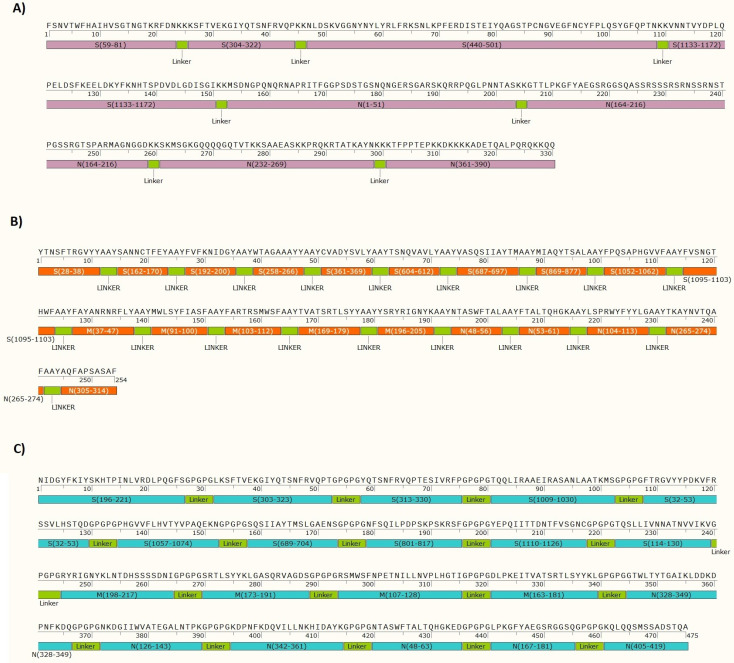
According to above-mentioned parameters including binding affinity between peptide and MHCs, epitope identification scores for T-cells, antibody-specific epitopes prediction for B-cells and T-cells, proteasomal cleavage and Tap transport scores, allergenicity, conservancy degree, population coverage and peptide-protein flexible docking scores, three different constructs were designed by top-ranked epitopes (Fig 1). For B-cell structure, S1133-1172, S440-501, S59-81, S304-322, N232-269, N164-216, N1-51 and N361-390 epitopes had the ability to induce antibodies. The B-cell epitopes were linked by KK linker [37]. For CTL structure, S27-37, S161-169, S191-199, S257-265, S360-368, S603-611, S686-696, S868-876, S1051-1061, S1094-1102, M36-46, M90-99, M102-111, M195-204, N47-55, N52-60, N103-113, N264-273 and N304-313 were selected. The CTL epitopes were linked by AAY linker [37], and finally, for Helper T-cell (HTL) structure, S32-53, S114-130, S196-231, S303-323, S313-330, S689-704, S801-817, S1009-1030, S1057-1074, S1110-1126, M107-128, M163-181, M173-191, M198-217, N48-63, N126-143, N167-181, N328-349, N342-361 and N405 were selected. The HTL epitopes were linked by GPGPG linker [37].
Fig 1. The suggested epitope constructs: a) the schematic diagram of the LBL epitope construct derived from the S and N proteins of the SARS-CoV-2 linked by KK linker; b) the schematic diagram of the CTL epitope construct derived from the S, M and N proteins of the SARS-CoV-2 linked by AAY linker; c) the schematic diagram of the HTL epitope construct derived from the S, M and N proteins of the SARS-CoV-2 linked by GPGPG linker.
The physicochemical parameters
Three constructs for each LBL, CTL and HTL epitope were analyzed by ProtParam server. Physicochemical properties of the constructed peptides were shown in Table 9. For LBL epitope, molecular weight (MW) was measured 36.5 kDa with theoretical isoelectric point (PI) of 10.24. For CTL and HTL epitopes, MWs were measured 28.6 and 49.5 kDa with PIs of 9.29 and 9.42, respectively. All constructs were soluble and stable.
Table 9. Physicochemical properties of the designed HTL and CTL and LBL epitopes.
| Construct | Molecular weight | Theoretical PI | Positive charge residue | Negative charge residue | Solubility | Stability |
|---|---|---|---|---|---|---|
| LBL-epitope | 36.5 kDa | 10.24 | 63 | 26 | Soluble | Stable |
| CTL-epitope | 28.6 kDa | 9.29 | 13 | 3 | Soluble | Stable |
| HTL-epitope | 49.5 kDa | 9.42 | 42 | 31 | Soluble | Stable |
3D structure prediction
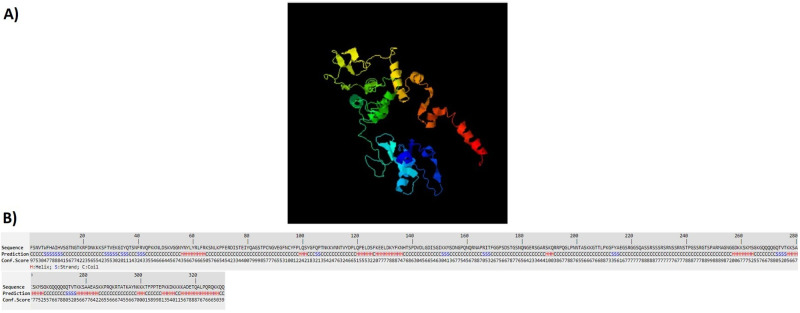
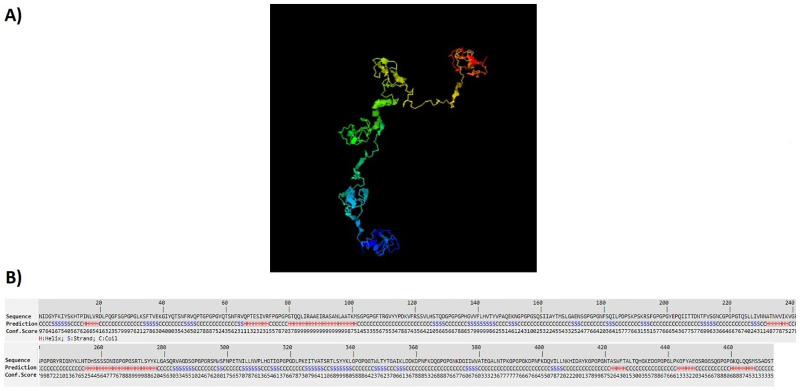
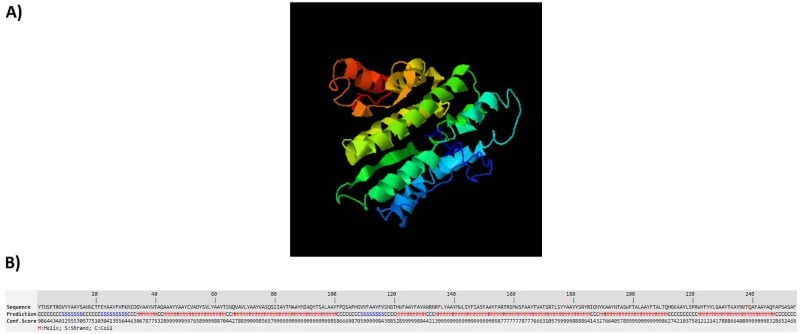
The designed structures were analyzed by I-TASSER server. This server generates some structural conformations, then uses SPICKER program to cluster all structures based on the pair-wise structure similarity. Finally, the top five models corresponding to the five largest clusters were reported by the server. The assurance of each model was calculated by C-score. The C-score values show the accuracy of the predicted model which usually is in the range of -5 to 2. Also, the higher value of the C-score signifies the better quality of prediction. The C-scores of the models for LBL, CTL and HTL polypeptide constructs were -2.39, -4.42 and -0.63, respectively. Figs 2–4 show tertiary structures of the predicted LBL, CTL and HTL structures.
Fig 2. The tertiary structure of the predicted LBL construct: a) 3D prediction of LBL multiepitope construct, b) sequence prediction and conformation scores of each amino acid in LBL multiepitope construct.
Fig 4. The tertiary structure of the predicted HTL construct: a) 3D prediction of HTL multiepitope construct, b) sequence prediction and conformation scores of each amino acid in HTL multiepitope construct.
Fig 3. The tertiary structure of the predicted CTL construct: a) 3D prediction of CTL multi-epitope construct, b) sequence prediction and conformation scores of each amino acid in CTL multiepitope construct.
Refinement and validation of 3D structures
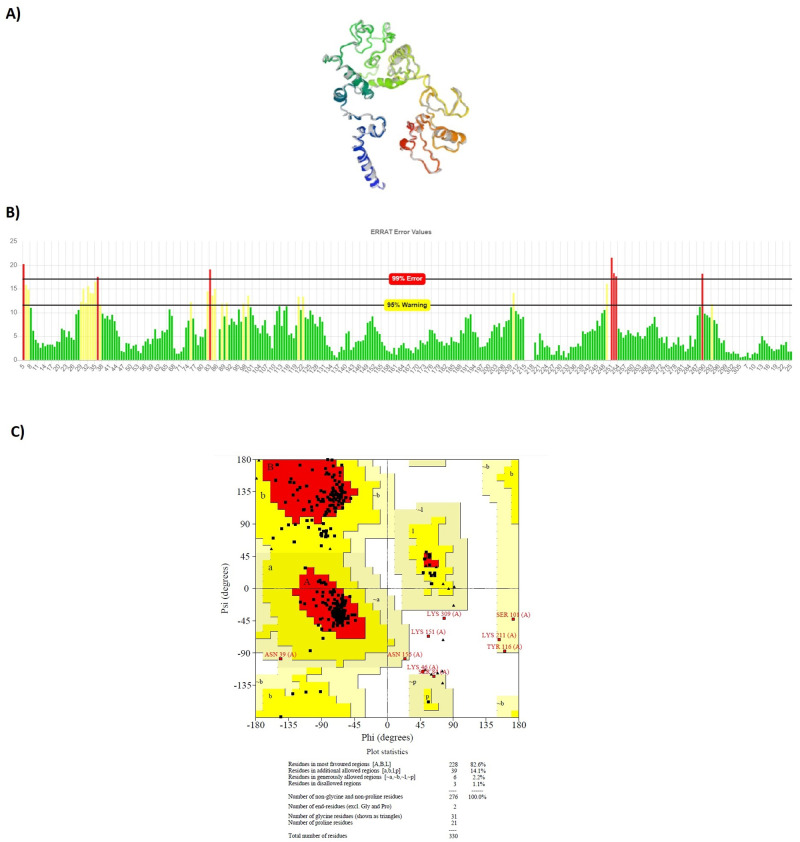
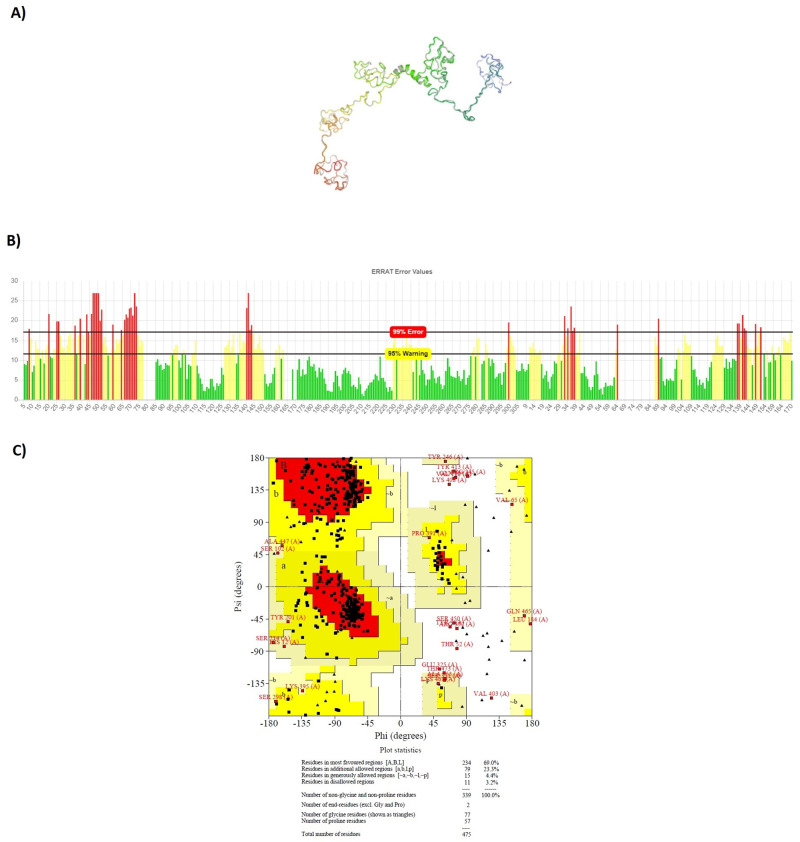
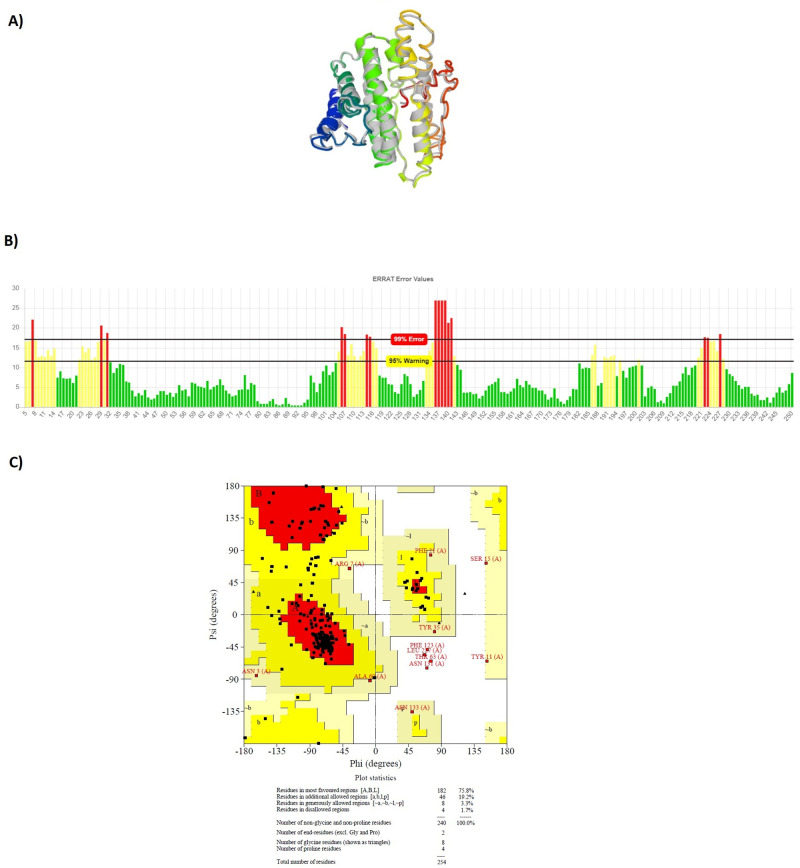
After tertiary structure prediction, the top model of each construct was submitted separately to GalaxyRefine 2 server. GalaxyRefine server rebuilds side-chain, and performs side-chain repacking and structure relaxation by molecular dynamic simulation. After refinement process, refined models were submitted to SAVE5.05 server for validation. The data indicated that the quality of tertiary structure was improved after refinement process. Most of the residues were found in favored and allowed regions: 98.9% for LBL, 98.3% for CTL and 96.8% for HTL constructs. Figs 5–7 show refined characteristics including secondary structures, overall quality and Ramachandran plots.
Fig 5. Refined characteristics of LBL multiepitope construct: a) Refined 3D prediction of LBL multiepitope construct, b) overall quality of refinement, c) Ramachandran plot.
Fig 7. Refined characteristics of HTL multiepitope construct: a) Refined 3D prediction of HTL multiepitope construct, b) overall quality of refinement, c) Ramachandran plot.
Fig 6. Refined characteristics of CTL multiepitope construct: a) Refined 3D prediction of CTL multiepitope construct, b) overall quality of refinement, c) Ramachandran plot.
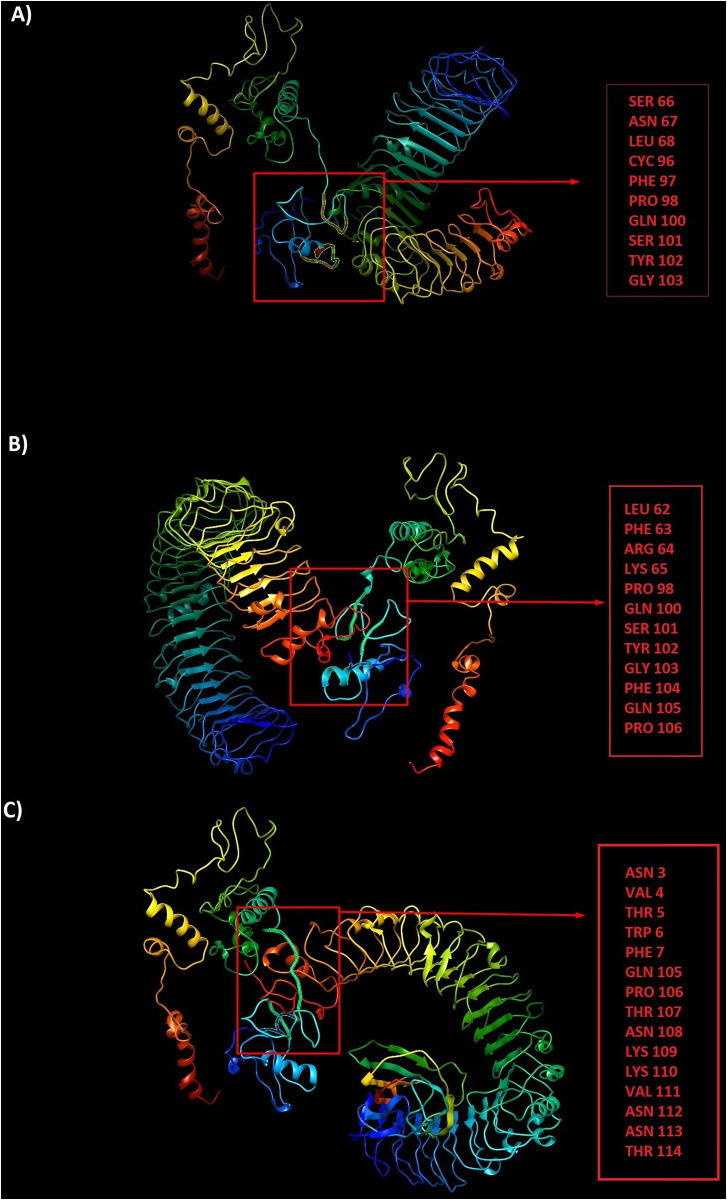
Prediction of discontinuous antibody epitopes
Linear antibody epitopes could be predicted through sequence-based algorithms. In contrast, prediction of discontinuous epitopes needs 3D structural information of the protein or polypeptide. Thus, the selected refined models were analyzed by Ellipro server to predict potential discontinuous B-cell epitopes. Ellipro servers identified 3 discontinuous B-cell epitopes for CTL, 4 for HTL and 3 for LBL. S1 Table indicates residues, number of residues and the 3D structure of putative B-cell epitopes in the designed constructs.
Docking between vaccine constructs and Toll-Like Receptors (TLRs)
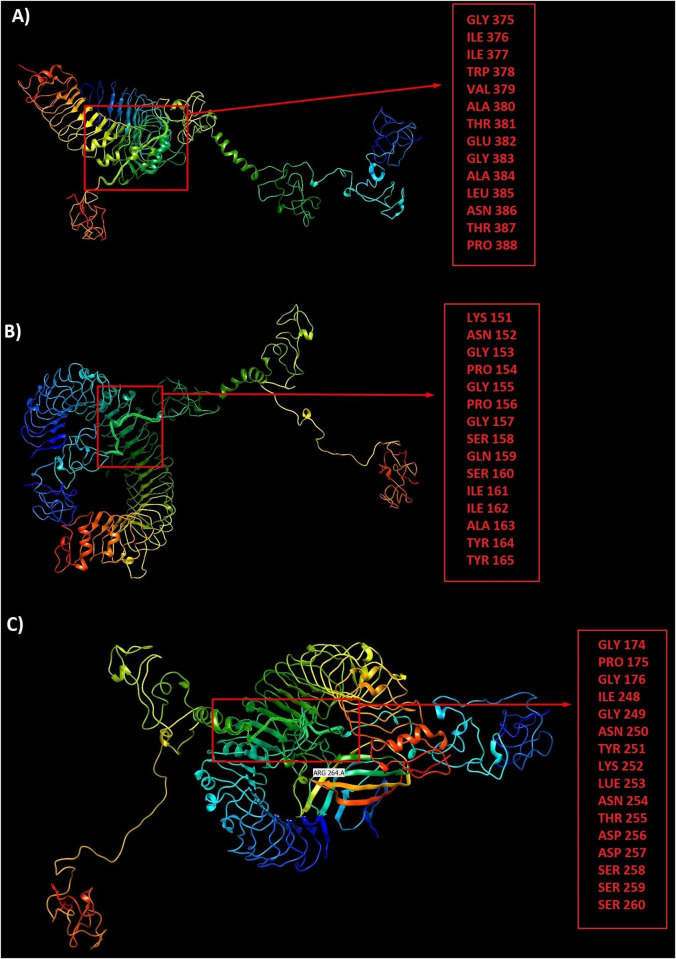
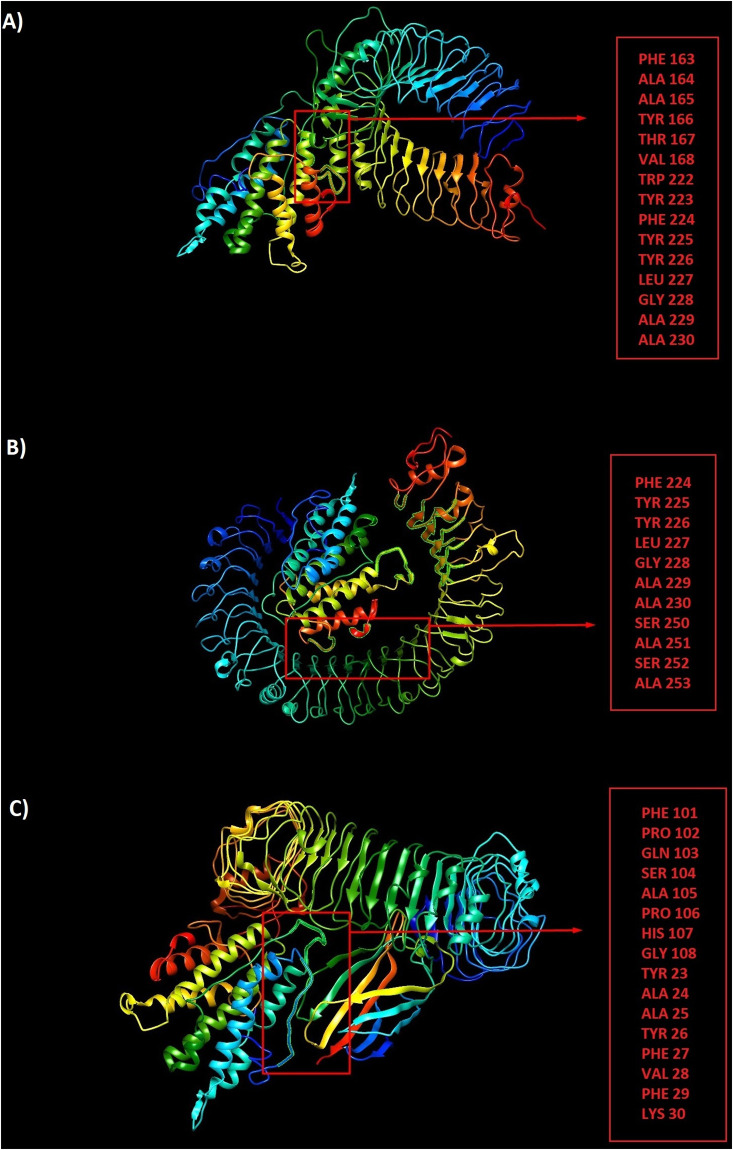
The peptide-protein docking between three vaccine constructs and TLRs 2, 3 and 4 were performed by ClusPro server. The lowest energy level was estimated for LBL-TLR2 complex-812.3, for LBL-TLR3 complex -894.8, for LBL-TLR4 complex -875.3, for CTL-TLR2 complex -1087.8, for CTL-TLR3 complex -1385.3, for CTL-TLR4 complex -1180.8, for HTL-TLR2 complex -1143.6, for HTL-TLR3 complex -1296.6 and for HTL-TLR4 complex -1119.6. Strong interactions between the designed constructs and TLRs 2, 3 and 4 supports the hypothesis of SARS-CoV-2 susceptibility to TLRs 2, 3 and 4 like other Coronaviridae families (Figs 8–10). All three constructs showed better interactions with TLR 3.
Fig 8. The peptide-protein docking between LBL construct and TLRs 2, 3 and 4: a) LBL-TLR2 complex with participated residues in interaction, b) LBL-TLR3 complex with participated residues in interaction, c) LBL-TLR4 complex with participated residues in interaction.
Fig 10. The peptide-protein docking between HTL construct and TLRs 2, 3 and 4: a) HTL-TLR2 complex with participated residues in interaction, b) HTL-TLR3 complex with participated residues in interaction, c) HTL-TLR4 complex with participated residues in interaction.
Fig 9. The peptide-protein docking between CTL construct and TLRs 2, 3 and 4: a) CTL-TLR2 complex with participated residues in interaction, b) CTL-TLR3 complex with participated residues in interaction, c) CTL-TLR4 complex with participated residues in interaction.
Discussion
The SARS-CoV-2 has become a major global public health issue and scientists are struggling to find the best way to treat the disease and develop a vaccine against the virus. Numerous immune-bioinformatics methods have been developed in vaccine researches which can potentially save time and resources. These tools could help us to identify antigenic domains to design a multi-epitope vaccine. Since now we know enough information about SARS-CoV-2’s genomics and proteomics, we can design peptide vaccines based on a neutralizing epitope. These immunoinformatics methods have made a significant impact on the immunology researches and we can see many examples of in silico design of epitope-based vaccines against many viruses including human immunodeficiency virus (HIV) [16], human papillomavirus (HPV) [38, 39], SARS-CoV [40], rhinovirus [41].
SARS-CoV-2 is an RNA virus tending to mutate more frequently [42]. These mutations mostly occur at the surface of the protein like at S protein leaving the immune system in a blind spot. Being the main antigenic component, S protein of SARS-CoV-2 has been selected as an important target for vaccine development since it is a crucial factor modulating tropism and pathogenicity and has the ability to induce faster and longer-term immune response [43, 44]. Since the humoral response from memory B-cells can easily be overcome by the emergence of antigens, it is important to design constructs based on cell-mediated immunity leading to lifelong immunity. Thus, our in-silico approaches were intended to design a universal SARS-CoV-2 vaccine for induction of B- and T-cell immunity with efficient reactions to the virus and long-term immune responses based on the S protein of the virus and also two other structural proteins including N and M.
There are some efforts based on designing epitope-based vaccines against SARS-CoV-2 since the outbreak. In one study, Abdelmageed et al. suggested certain peptides in E protein of SARS-CoV-2 as promising epitope vaccine candidates against T-cell [45]. In another study, Singh et al. proposed one construct including E, S, N and M proteins as an epitope vaccine candidate [46]. On the other hand, Feng et al. suggested some putative B- and T-cell epitopes based on S, M and E proteins of SARS-CoV-2 [47]. Also, Enayatkhani et al. designed a multiepitope vaccine candidate based on N, M and open reading frame (ORF) 3a of SARS-CoV-2 [48], and Teimouri et al. tried to predict B- and T-cell epitopes of SARS-CoV-2 in comparison with SARS-CoV [49]. These papers contain some very worthwhile suggestions for ease of multi-epitope vaccine development and all of them demonstrate that a multi-epitope peptide vaccine targeting multiple antigens should be considered as an ideal approach for prevention and treatment of SARS-CoV-2.
According to the aforementioned finding, we tried to use computational and bioinformatics methods on the formulation of new SARS-CoV-2 vaccine against its structural proteins including S, N and M proteins in a more comprehensive way. In the beginning, the whole genome of SARS-CoV-2 was analyzed. Then, three major structural proteins including S, N and M were chosen for further analyses. We identified epitopes corresponding to B-cells and T-cells to design constructs being able to elicit both humoral and cellular immunity. We used BepiPred tool to predict putative B-cell epitopes and chose 8 putative epitopes of S and N protein being able to induce antibodies like IgG. In contrast, M protein of the virus could not induce any class of antibodies.
As CD8+ and CD4+ T-cells play a major role in antiviral immunity, we tried to evaluate the binding affinity to MHC class I and II molecules. Choosing S, N and M proteins of the virus as the antigenic site, we used NetMHCpan and NetMHCIIpan prediction tools to identify the most immunodominant regions. From all peptides predicted, we chose 20 putative epitopes for MHC class I and 20 putative epitopes for MHC class II. Since S protein is currently the most promising antigen formulation, we put the focus on the S protein epitopes and chose 10 epitopes of S protein and 10 epitopes of other proteins for each MHC class I and II. The IFN-γ and IL-10 cytokines were also measured as candidates in MHC class II epitopes as they promote the development of T-helper cells being required for B-cell, macrophage and cytotoxic T-cell activation. Among the CTL predicted epitopes, S257-265, S603-611 and S360-368 and among HTL predicted epitopes, N167-181, S313-330 and S1110-1126 had better MHC binding rank.
To predict antigen processing through the MHC class I antigen presentation pathway, we used NetCTL1.2 server. All of the predicted epitopes had upper cut off identification scores (> 0.75) showing a high quality of proteasomal cleavage and Tap transport efficiency. We also measured the epitopes for conservancy analysis. All predicted epitopes were 100% conserved within four different clades of SARS-CoV-2. In general, the selected epitopes had the potency to produce an immune response against S, V, G and O clades of SARS-CoV-2.
Population coverage is another important factor in vaccine design. We measured population coverage rate for CTL and HTL epitopes in 16 specified geographical regions. For CTL epitopes, and helper T-cell epitopes, the highest population coverage of the world’s population was calculated for S27-37 with 86.27%, and for S196-231, S303-323, S313-330, S1009-1030 and N328-349 with 90.33%, respectively. Overall, these results suggest a specific binding of CTL epitopes and HTL epitopes to the prevalent HLA molecules in the targeted populations. Another prominent obstacle in vaccine development is the probability of allergenicity since many vaccines stimulate the immune system into an allergenic reaction. In this study, we used PA3P to predict potential allergenicity and all of the epitopes were analyzed as non-allergen.
For CTL epitopes, N103-113, S868-876, M36-46, S1094-1102, M102-111, S1051-1061, S360-368, S191-199 and S686-696 had the highest average of interaction similarity score, respectively and For HTL epitopes, M107-128, N48-63, S689-704, S1057-1074, S196-231, N328-349, S32-53, N126-143, M163-181 and S114-130 had the highest average of interaction similarity score, respectively. Overall, CTL epitopes showed better quality of docking in comparison with HTL epitopes. Finally, the vaccine construction was completed after joining the LBL, CTL and HTL epitopes with KK, AAY and GPGPG linkers, respectively.
The molecular weights of the constructed LBL, CTL and HTL epitopes were obtained as 36.5, 28.6 and 49.5 kDa, respectively which were low molecular weights for a multiepitope vaccine. All constructs were soluble and stable indicating that the designed constructs had high solubility and stability for the initiation of an immunogenic reaction.
In the case of 3D modeling, we used I-TASSER server to predict the tertiary protein structure. The accuracy of the selected models was evaluated by C-score. The C-scores of the models for LBL, CTL and HTL polypeptide constructs were -2.39, -4.42 and -0.63, respectively. The higher value of the C-score is the better quality of prediction. Thus, HTL with the C-score of -0.63 showed higher accuracy of the predicted epitopes. Also, the quality of the predicted constructs was improved by refinement which leads to a higher quality of final models. Over the 96.8% of the residues were found in favored and allowed regions. At last, we used Ellipro server to predict potential discontinuous B-cell epitopes. Ellipro servers identified 3 discontinuous B-cell epitopes for CTL with 143 residues, 4 for HTL with 225 residues and 3 for LBL with 72 residues indicating the ability of the designed constructs for robust induction of humoral response. Also, peptide-protein docking between three vaccine constructs and TLRs 2, 3 and 4 were performed by ClusPro server, and all data showed strong interactions between the designed constructs and TLRs 2, 3 and 4 supporting the hypothesis of SARS-CoV-2 susceptibility to TLRs 2, 3 and 4 like other Coronaviridae family. All three constructs showed better interactions with TLR 3.
Overall, we tried to consider three major structural proteins including S, N and M proteins of the virus and design three different constructs including LBL, CTL and HTL constructs to elicit more robust humoral and cellular immunity. Comparing our study with other studies in the field of multi-epitope vaccine design for SARS-CoV-2, all LBL epitopes obtained in Table 1 were reported in Bhattacharya et al. paper using the same server of Bepipred [50]. However, we chose the ones being able to induce different classes of antibodies including IgG and IgA. Among CTL epitopes obtained in Table 2, S686-696 (VASQSIIAYTM), S1051-1061 (FPQSAPHGVVF), N103-113 (LSPRWYFYYLG), N304-313 (AQFAPSASAF) and all of M epitope of CTL including M36-46 (FAYANRNRFLY), M168-180 (TVATSRTLSYY), M195-204 (YSRYRIGNYK), M90-99 (MWLSYFIASF) and M102-111 (FARTRSMWSF) have not been reported in any literature. Also, among HTL epitopes reported in Table 3, S303-323 (LKSFTVEKGIYQTSNFRVQPT), S1009-1030 (TQQLIRAAEIRASANLAATKMS), S801-817 (NFSQILPDPSKPSKRSF), N328-349(GTWLTYTGAIKLDDKDPNFKDQ), N126-143 (NKDGIIWVATEGALNTPK), N342-361 (KDPNFKDQVILLNKHIDAYK), N48-63 (NTASWFTALTQHGKED), N167-181 (LPKGFYAEGSRGGSQ), N405-419 (KQLQQSMSSADSTQA), M107-128 (RSMWSFNPETNILLNVPLHGTI) and M163-181 (DLPKEITVATSRTLSYYKL) have not been reported in any literature. The rest of the epitopes mentioned in Tables 2 and 3 were found in agreement with Teimouri et al. [49] for MHC class I and Feng et al. [47] for MHC class II, respectively. Also, we found one putative CTL epitope, S360-368 (CVADYSVLY) related to receptor-binding domain (RBD) region for S protein which was referred to the fragment of 347 to 520 amino acids [51]. We also identified overall 10 discontinuous B-cell epitopes for three multi-epitope constructs. Meanwhile, we investigated the interaction of three designed constructs with TLRs 2, 3 and 4 based on the previous studies on other Coronaviridae family such as SARS-CoV and MERS-CoV [33–35]. All three constructs showed strong interactions with TLRs 2, 3 and 4 supporting the hypothesis of SARS-CoV-2 susceptibility to TLRs 2, 3 and 4 like other Coronaviridae families. Albeit, SARS-CoV-2 was identified for only 5 month, but researches have recently begun to design a multiepitope vaccine. Thus, the collected data and information are very limited and need to be accumulated to improve existing processes and the designed multi-epitope vaccine needs to be tested clinically to validate vaccine safety.
Conclusion
In conclusion, we determined three vaccine constructs against three major structural proteins of SARS-CoV-2 designed based on robust vaccine design criteria including non-allergenicity, conservancy, affinity measurement to multiple alleles of MHC, worldwide population coverage, 3D prediction, refinement and validation, discontinuous B-cell epitope prediction, docking and effectiveness of molecular interaction with their respective HLA alleles and TLRs. These constructs require validation by in vivo and clinical experiments. Generally, with the help of in silico studies, experimental researches can march rapidly with higher probabilities of finding the desired solutions and controlling the current outbreak.
Supporting information
(DOCX)
Data Availability
All relevant data are completely within the manuscript.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Zhu N, Zhang D, Wang W. A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine. 2020; 382: 727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020; 38: 1–9 [DOI] [PubMed] [Google Scholar]
- 3.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020; 181: 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamming I, Timens W, Bulthuis M, Lely A, Navis Gv, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland. 2004; 203(2): 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W, Zhang C, Sui J. Receptor and viral determinants of SARS‐coronavirus adaptation to human ACE2. The EMBO Journal. 2005; 24: 1634–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020; 367: 1444–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang M, Cao R, Zhang L. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research. 2020; 30: 269–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richardson P, Griffin I, Tucker C. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020; 395: e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheahan TP, Sims AC, Leist SR. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nature Communications. 2020; 11: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derebail VK, Falk RJ. ANCA-associated vasculitis-refining therapy with plasma exchange and glucocorticoids. In: Mass Medical Soc. 2020; 382: 671–673 [DOI] [PubMed] [Google Scholar]
- 11.Keith P, Day M, Perkins L, Moyer L, Hewitt K, Wells A. A novel treatment approach to the novel coronavirus: an argument for the use of therapeutic plasma exchange for fulminant COVID-19. In: BioMed Central. 2020; 24: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian X, Li C, Huang A. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerging Microbes & Infections. 2020; 9: 382–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thanh LT, Andreadakis Z, Kumar A. The COVID-19 vaccine development landscape. Nature Reviews Drug Discovery. 2020; 1–20 [DOI] [PubMed] [Google Scholar]
- 14.Li J, Zhang C, Shan H. Advances in mRNA vaccines for infectious diseases. Frontiers in Immunology. 2019; 10: 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skwarczynski M, Toth I. Peptide-based synthetic vaccines. Chemical Science. 2016; 7: 842–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khairkhah N, Namvar A, Kardani K, Bolhassani A. Prediction of cross‐clade HIV‐1 T‐cell epitopes using immunoinformatics analysis. Proteins: Structure, Function, and Bioinformatics. 2018; 86: 1284–1293 [DOI] [PubMed] [Google Scholar]
- 17.Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Research. 2017; 45(W1): W24–W29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malherbe L. T-cell epitope mapping. Annals of Allergy, Asthma & Immunology. 2009; 103: 76–79 [DOI] [PubMed] [Google Scholar]
- 19.He Y, Rappuoli R, De Groot AS, Chen RT. Emerging vaccine informatics. BioMed Research International. 2011; 2010: 2011 [Google Scholar]
- 20.Hoof I, Peters B, Sidney J. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009; 61: 1 10.1007/s00251-008-0341-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reynisson B, Barra C, Kaabinejadian S, Hildebrand WH, Peters B, Nielsen M. Improved prediction of MHC-II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. Journal of Proteome Research. 2020; 19(6): 2304–2315. 10.1021/acs.jproteome.9b00874 [DOI] [PubMed] [Google Scholar]
- 22.Bui HH, Sidney J, Li W, Fusseder N, Sette A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinformatics. 2007; 8: 361 10.1186/1471-2105-8-361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bui HH, Sidney J, Dinh K, Southwood S, Newman MJ, Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics. 2006; 7: 153 10.1186/1471-2105-7-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta S, Ansari HR, Gautam A, Raghava GP, Consortium OSDD. Identification of B-cell epitopes in an antigen for inducing specific class of antibodies. Biology Direct. 2013; 8: 27 10.1186/1745-6150-8-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagpal G, Usmani SS, Dhanda SK. Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Scientific Reports. 2017; 7: 42851 10.1038/srep42851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhanda SK, Vir P, Raghava GP. Designing of interferon-gamma inducing MHC class-II binders. Biology Direct. 2013; 8: 30 10.1186/1745-6150-8-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chrysostomou C, Seker H. Prediction of protein allergenicity based on signal-processing bioinformatics approach. Paper presented at: 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. 2014; 2014 [DOI] [PubMed]
- 28.Lee H, Heo L, Lee MS, Seok C. GalaxyPepDock: a protein-peptide docking tool based on interaction similarity and energy optimization. Nucleic Acids Research. 2015; 43: W431–W435 10.1093/nar/gkv495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: The proteomics protocols handbook. 2005; 571–607 [Google Scholar]
- 30.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nature Methods. 2015; 12: 7 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee GR, Won J, Heo L, Seok C. GalaxyRefine2: simultaneous refinement of inaccurate local regions and overall protein structure. Nucleic Acids Research. 2019; 47: W451–W455 10.1093/nar/gkz288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ponomarenko J, Bui HH, Li W. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 2008; 9: 514 10.1186/1471-2105-9-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Totura AL, Whitmore A, Agnihothram S. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. M Bio. 2015; 6: e00638–00615 10.1128/mBio.00638-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olejnik J, Hume AJ, Mühlberger E. Toll-like receptor 4 in acute viral infection: Too much of a good thing. PLoS Pathogens. 2018; 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durai P, Batool M, Shah M, Choi S. Middle East respiratory syndrome coronavirus: transmission, virology and therapeutic targeting to aid in outbreak control. Experimental & Molecular Medicine. 2015; 47: e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozakov D, Hall DR, Xia B. The ClusPro web server for protein-protein docking. Nat Protoc. 2017; 12: 255–278 10.1038/nprot.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sayed SB, Nain Z, Abdullah F. Immunoinformatics-guided designing of peptide vaccine against Lassa virus with dynamic and immune simulation studies. Preprints. 2019; 2019 [Google Scholar]
- 38.Namvar A, Bolhassani A, Javadi G, Noormohammadi Z. In silico/In vivo analysis of high-risk papillomavirus L1 and L2 conserved sequences for development of cross-subtype prophylactic vaccine. Scientific Reports. 2019; 9: 1–22 10.1038/s41598-019-51679-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Namvar A, Panahi HA, Agi E, Bolhassani A. Development of HPV 16, 18, 31, 45 E5 and E7 peptides-based vaccines predicted by immunoinformatics tools. Biotechnology Letters. 2020; 1–16 10.1007/s10529-020-02792-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oany AR, Emran AA, Jyoti TP (2014) Design of an epitope-based peptide vaccine against spike protein of human coronavirus: an in silico approach. Drug Design, Development and Therapy. 2014; 8: 1139 10.2147/DDDT.S67861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lapelosa M, Gallicchio E, Arnold GF, Arnold E, Levy RM. In silico vaccine design based on molecular simulations of rhinovirus chimeras presenting HIV-1 gp41 epitopes. Journal of Molecular Biology. 2009; 385: 675–691 10.1016/j.jmb.2008.10.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanjuán R, Domingo-Calap P. Mechanisms of viral mutation. Cellular and Molecular Life Sciences. 2016; 73: 4433–4448 10.1007/s00018-016-2299-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Millet JK, Whittaker GR. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Research. 2015; 202: 120–134 10.1016/j.virusres.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma C, Li Y, Wang L. Intranasal vaccination with recombinant receptor-binding domain of MERS-CoV spike protein induces much stronger local mucosal immune responses than subcutaneous immunization: Implication for designing novel mucosal MERS vaccines. Vaccine. 2014; 32: 2100–2108 10.1016/j.vaccine.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdelmageed MI, Abdelmoneim AH, Mustafa MI. Design of multi epitope-based peptide vaccine against E protein of human 2019-nCoV: An immunoinformatics approach. BioRxiv. 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh A, Thakur M, Sharma LK, Chandra K. Designing a multiepitope peptide-based vaccine against SARS-CoV-2. BioRxiv. 2020; 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng Y, Qiu M, Zou S. Multi-epitope vaccine design using an immunoinformatics approach for 2019 novel coronavirus in China (SARS-CoV-2). BioRxiv. 2020; 2020 [Google Scholar]
- 48.Enayatkhani M, Hasaniazad M, Faezi S, Guklani H, Davoodian P, Ahmadi N, et al. Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in silico study. Journal of Biomolecular Structure and Dynamics 2020; 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teimouri H, Azad M. In-silico immunomodelling of 2019-nCoV. 2019
- 50.Bhattacharya M, Sharma AR, Patra P. Development of epitope‐based peptide vaccine against novel coronavirus 2019 (SARS‐COV‐2): Immunoinformatics approach. Journal of Medical Virology. 2020; 92: 618–631 10.1002/jmv.25736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu R, Zhao X, Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. 2020; 395: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are completely within the manuscript.