Abstract
The innate immune system acts as the first line of defense against pathogens, including coronaviruses (CoVs). Severe acute respiratory syndrome (SARS)-CoV and Middle East respiratory syndrome (MERS)-CoV are epidemic zoonotic CoVs that emerged at the beginning of the 21st century. The recently emerged virus SARS-CoV-2 is a novel strain of CoV that has caused the coronavirus 2019 (COVID-19) pandemic. Scientific advancements made by studying the SARS-CoV and MERS-CoV outbreaks have provided a foundation for understanding pathogenesis and innate immunity against SARS-CoV-2. In this review, we focus on our present understanding of innate immune responses, inflammasome activation, inflammatory cell death pathways, and cytokine secretion during SARS-CoV, MERS-CoV, and SARS-CoV-2 infection. We also discuss how the pathogenesis of these viruses influences these biological processes.
Keywords: RNA virus, coronavirus, SARS-CoV-2, COVID-19, SARS-CoV, MERS-CoV, MHV, innate immunity, inflammasome, cell death, PANoptosome, PANoptosis, cytokines, inflammation
Highlights
Toll-like receptor (TLR)- and retinoic acid-inducible gene I-like receptor (RLR)-mediated type I interferon (IFN) production is essential for providing protection against coronavirus (CoV) infection; the timing of the IFN response relative to CoV replication determines infection outcomes.
Optimal NLR family pyrin domain-containing 3 (NLRP3) inflammasome activation is beneficial for the host but aberrant activation may lead to detrimental CoV infection outcomes.
Specific CoV infections can activate inflammatory cell death (PANoptosis), thereby inducing cytokine release.
CoV disease tolerance occurs in age-, species-, and sex-dependent manners.
More studies are needed to define the innate immune response, specifically during severe acute respiratory syndrome (SARS)-CoV-2 (SARS-CoV-2) infection, and to inform the development of candidate therapeutics.
Innate Immunity and Coronaviruses
Innate Immunity and Inflammasomes
The innate immune system functions as the first line of host defense against microbial infections. During viral infections, the innate immune system is crucial for identifying and removing infected cells while also coordinating an adaptive immune response. To detect and defend rapidly against various microbes, mammalian hosts have evolved multiple pattern-recognition receptors (PRRs; see Glossary) including Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), the nucleotide-binding oligomerization domain (NOD)-like receptor family proteins (NLRs), and absent in melanoma 2 (AIM2). In response to specific pathogen-associated molecular patterns (PAMPs) and danger-associated molecular patterns (DAMPs), some PRRs, particularly members of the NLR family and AIM2, have the ability to assemble a large multiprotein complex called the inflammasome [1]. Upon assembly, the inflammasome induces membrane pore formation and proinflammatory cytokine processing, leading to a form of inflammatory cell death known as pyroptosis [1., 2., 3.].
Innate immune signaling and inflammasome activation are well-established key barriers during viral infection. However, activation of the innate immune system must be tightly regulated, because excessive activation can lead to systemic inflammation and tissue damage, which are detrimental to the host. Systemic hyperinflammation is common in a wide range of infectious diseases [4]. Therefore, the balance between the host innate immune response and viral intracellular replication has been considered for potential therapeutic approaches in viral infections; this balance must be finely controlled to reduce excessive inflammation while retaining antiviral functions.
Coronaviruses
Coronaviruses (CoVs) are positive-sense single-stranded (ss)RNA viruses and have an extensive range of natural hosts [5]. There are seven human-infecting CoVs identified to date: human coronavirus 229E (HCoV-229E), HCoV-OC43, severe acute respiratory syndrome (SARS)-CoV, HCoV-NL63, HCoV-HKU1, Middle East respiratory syndrome CoV (MERS)-CoV, and SARS-CoV-2 [6,7]. The endemic HCoV-229E and HCoV-OC43 were isolated >50 years ago and are responsible for about one-third of the common cold cases each year. SARS-CoV was isolated in 2003 in China [8], while HCoV-NL63 and HCoV-HKU1 were identified shortly following the SARS-CoV outbreak. Ten years after SARS-CoV, MERS-CoV emerged in Middle Eastern countries [9]. The most recently identified human-infecting CoV is SARS-CoV-2, the virus that causes coronavirus disease 2019 (COVID-19) [10,11], a respiratory illness in humans (Table 1 ). In addition to the human-infecting CoVs, there are also several other CoV strains that infect various animals. Among these, the most studied is murine hepatitis virus (MHV), which mimics many of the key aspects of human CoV biology [6]. Due to receptor specificity of human CoVs, MHV has been an ideal model for examining the pathogenesis and immune response to CoVs as well as for studying the basics of viral replication [6].
Table 1.
Systemic Hyperinflammation and Tissue Damage during SARS-CoV-2 Infection in Humansa
| Organ/system | Symptoms and clinical outcome |
|---|---|
| Circulation | Cytokine storm |
Lymphocyte changes
| |
| Thrombocytopenia | |
| Ferritin | |
| Blood vessels | Vasculitis and vascular dysfunction, endothelial cell inflammation and damage cause blood vessels to leak; vascular endothelial growth factor (VEGF) ↑, IL-10 ↑, IL-8 ↑ [11,121,129] |
| Lung | Pneumonia with ARDS and dyspnea [10,11,130] |
| Kidneys | Acute renal injury resulting in proteinuria and hematuria [131,132] |
| Brain | Neurological complications, headache, and ischemic stroke in patients with severe disease [11,133] |
| Heart | Acute myocardial injury and chronic damage to cardiovascular system [11,134] |
| Liver | Steatosis and abnormal liver function with aspartate transaminase (AST) ↑ and alanine aminotransferase (ALT) ↑ [130,135]; C-reactive protein (CRP) ↑ and albumin ↑ [121,136] |
| Intestine | Gastrointestinal symptoms (e.g., diarrhea and severe acute ulcerative colitis) [11,137,138] |
↑, increased concentrations or amounts; ↓ decreased concentrations or amounts.
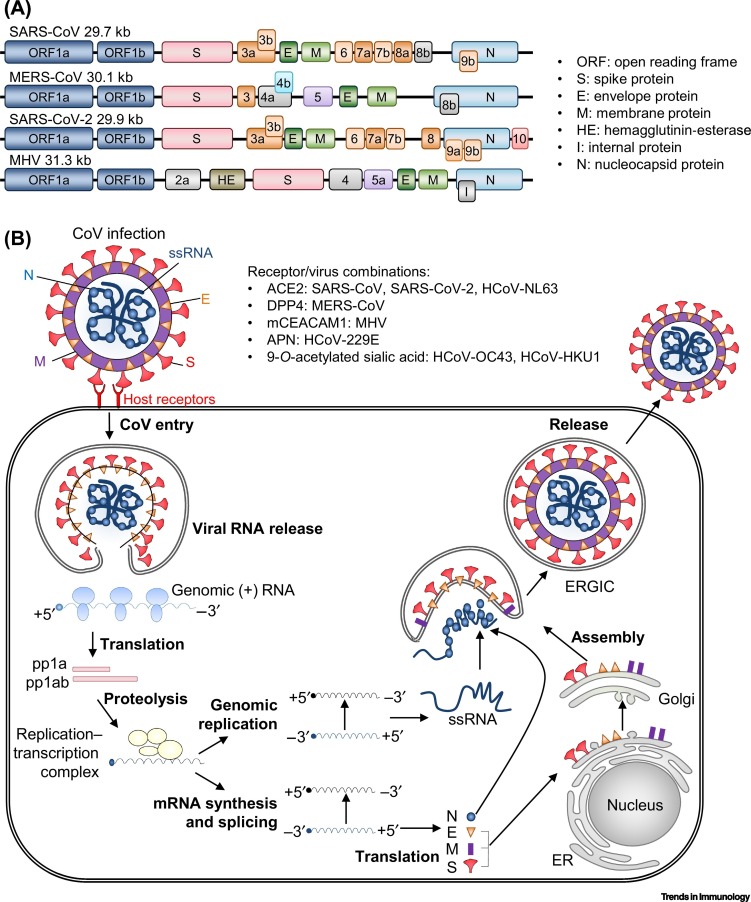
SARS-CoV, MERS-CoV, SARS-CoV-2, and MHV belong to the Betacoronavirus genus in the Coronaviridae family and have RNA genomes of 27.9 kb, 30.1 kb, 29.9 kb, and 31.3 kb, respectively [6,12]. Within the viral membrane, the genomic RNA is complexed with the nucleocapsid (N) protein to create a helical capsid. The viral proteins spike (S), envelope (E), and membrane (M) make up the membrane proteins of all CoVs. The S protein is a type I glycoprotein responsible for forming peplomers on the virion surface. The E protein is highly hydrophobic, and the M protein has a cytoplasmic tail that contains a short N-terminal ectodomain (Figure 1A) [6].
Figure 1.
Coronavirus (CoV) RNA Genome and Replication.
(A) Schematic diagram of representative positive-sense RNA genomes of severe acute respiratory syndrome (SARS)-CoV (29.7 kB), Middle East respiratory syndrome (MERS)-CoV (30.1 kB), SARS-CoV-2 (29.9 kB), and murine hepatitis virus (MHV) (31.3 kB). The 5′-end encodes a polyprotein (ORF1a/b), and the 3′-end encodes four structural proteins [spike (S), membrane (M), envelope (E), nucleocapsid (N)], hemagglutinin-esterase (HE), internal (I) protein, and accessory proteins (ORF2a, 3, 3a, 3b, 4, 4a, 4b, 5a, 5, 6, 7a, 7b, 8a, 8b, 9a, 9b, and/or 10). The S, M, E, and N proteins are unique to each virus. (B) Representative diagram of CoV replication. After host receptor interaction with S viral protein [angiotensin-converting enzyme 2 (ACE2) for SARS-CoV, SARS-CoV-2, and HCoV-NL63; dipeptidyl peptidase 4 (DPP4) for MERS-CoV; murine carcinoembryonic antigen-related adhesion molecule 1 (mCEACAM1) for MHV; aminopeptidase N (APN) for HCoV-229E; 9-O-acetylated sialic acid for HCoV-OC43 and HCoV-HKU1], viral RNA and proteins are released to the cytoplasm, and open reading frame (ORF)-1a and ORF1ab are translated to produce pp1a and pp1ab. Next, pp1a and pp1ab form the replicase–transcriptase complex through cotranslational proteolytic processing. The complex produces an antisense negative-strand template, and the subgenomic mRNAs are synthesized. Subsequent translation of subgenomic mRNAs generates structural viral proteins, including the N, E, M, and S proteins, which are assembled at the endoplasmic reticulum (ER)–Golgi intermediate compartment (ERGIC). Newly synthesized virion-containing vesicles fuse with the plasma membrane to release the virus [15., 16., 17., 18.]. ACE2, angiotensin-converting enzyme 2; APN, aminopeptidase N; CoV, coronavirus; DPP4, dipeptidyl peptidase 4; mCEACAM1, murine carcinoembryonic antigen-related adhesion molecule 1; ORF, open reading frame.
To infect host cells, the S protein on the viral envelope binds to its cellular receptor, which is angiotensin-converting enzyme 2 (ACE2) for SARS-CoV, SARS-CoV-2, and HCoV-NL63; dipeptidyl peptidase 4 (DPP4) for MERS-CoV; murine carcinoembryonic antigen-related adhesion molecule 1 (mCEACAM1) for MHV; aminopeptidase N (APN) for HCoV-229E; and 9-O-acetylated sialic acid for HCoV-OC43 and HCoV-HKU1 [6,13,14]. S protein binding to these receptors promotes viral and host membrane fusion [6,13,14]. After membrane fusion, the viral RNA genome is released from the endosome into the cytoplasm (Figure 1B). CoVs have a unique coding strategy, and viral RNA is translated into two large polyproteins, pp1a and pp1ab, encoding 16 nonstructural proteins (nsp1–nsp16) that make up the viral replicase–transcriptase complex [15,16]. This complex generates an antisense negative-strand template; subgenomic negative-strand templates are predicted to enable subgenomic mRNAs to be produced. Membranes derived from the endoplasmic reticulum (ER) and Golgi are reorganized by the viral nonstructural protein (N) to form double-membrane vesicles, where viral replication and transcription occur [17]. The transcribed viral RNAs also encode S, E, M, and N proteins that are not involved in viral replication but mediate virus entry (S) and assembly and release (E and M). To create new virions, newly formed viral proteins traffic to the ER or Golgi membranes, while the genomic RNA and N proteins combine to form nucleocapsids. The full viral particle can then be assembled into the ER–Golgi intermediate compartment (ERGIC). This allows newly synthesized virion-containing vesicles to fuse with the plasma membrane and release the virus [18].
In this review, we discuss innate immune recognition, inflammasome activation, programmed cell death, and cytokine release in response to CoV infections, and describe how innate immune responses regulate disease to contribute to resistance to viral infection and disease tolerance.
Studying the Innate Immune Response to CoVs
Data from patients with CoVs have served as a key starting point for studying these viruses. However, mechanistic dissections of innate immune signaling pathways typically require the use of animal models. Due to species-specific CoV S protein binding to host cellular receptors, there is no single animal model for CoV infection that reproduces all aspects of the human disease. However, adaptation of SARS-CoV (Urbani strain) by serial passage in the lungs of BALB/c mice led to the creation of the MA15 virus, which is lethal following intranasal inoculation in mice [19]. MA15 is used as the mouse model for SARS-CoV, because infection with this virus recapitulates several aspects of the severe human disease, including pulmonary pathology, morbidity, and mortality [19]. For infections with MERS-CoV and SARS-CoV-2, human DPP4 transgenic mice with a mouse-adapted strain of MERS-CoV and human ACE2 (hACE2) transgenic mice with SARS-CoV-2 have been used to replicate several features of severe human disease, including pulmonary pathology and mortality [20,21]. Limitations with these systems have been noted. In human DPP4 transgenic mice, the lung disease that develops in response to the mouse-adapted strain of MERS-CoV infection may depend on species-specific mutations in the new host, which may not recapitulate all aspects of the human infection [20]. A potential disadvantage of studying SARS-CoV-2 in hACE2 transgenic mice is that the brain infection observed in mice that succumb to disease may not reflect the pathogenesis of SARS-CoV-2 in humans [21]. Additionally, hACE2 is expressed under the control of the HFH4/FOXJ1 promoter in these mice, which is largely specific to epithelial cells [21]. Consequently, hematopoietic cells from hACE2 transgenic mice cannot be infected with SARS-CoV-2 [21]. Despite these limitations, significant work has been done to molecularly characterize the innate immune pathways involved in detecting and controlling CoV infections.
Innate Immune Recognition of CoV
The innate immune response acts as a powerful barrier against CoV infection. CoVs infect the host through the oral or nasal cavities, where they first encounter the respiratory epithelium. After infecting respiratory epithelial cells, MHV, SARS-CoV, MERS-CoV, and SARS-CoV-2 can infect both non-immune and immune cells in the respiratory tract. During a typical viral infection, viral RNA can be recognized by various PRRs, including TLRs [22,23], RLRs [24,25], and NLRs [26., 27., 28., 29.] for the production of proinflammatory cytokines and the induction of an antiviral state (Figure 2 and Table 2 ). Prototypically, the proinflammatory cytokines are largely beneficial to the host and trigger the infiltration of immune cells to remove infectious viral agents. However, dysregulated proinflammatory cytokines drive detrimental systemic hyperinflammation and tissue damage [10].
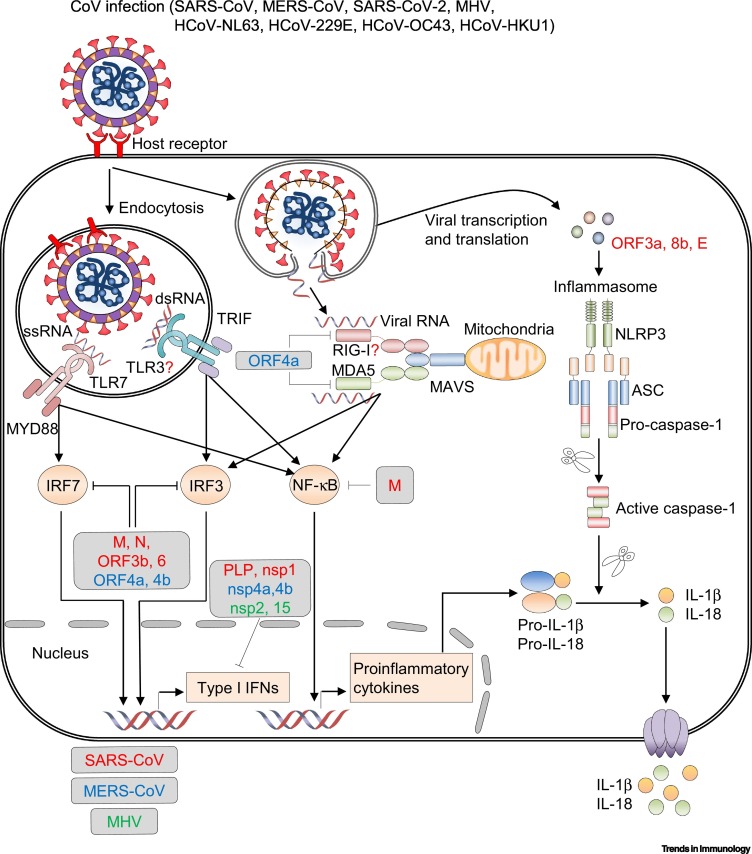
Figure 2.
Innate Immune-Sensing Pathways in Coronavirus (CoV) Infection and Immune Evasion.
Upon CoV infection, incoming double-stranded RNA (dsRNA) and genomic single-stranded RNA (ssRNA) are recognized by Toll-like receptor 3 (TLR3) and TLR7, respectively. Signaling downstream of these TLRs induces activation of nuclear factor-κB (NF-κB) to produce proinflammatory cytokines and phosphorylation of interferon regulatory factor 3 (IRF3) and IRF7 to drive type I interferon (IFN) production. Viral RNA in the cytosol is recognized by retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5). The helicases of RIG-I and MDA5 associate with mitochondrial antiviral signaling protein (MAVS), leading to the activation of NF-κB and phosphorylation of IRF3. Severe acute respiratory syndrome (SARS)-CoV open reading frame (ORF)-3a, ORF8b, and envelope (E) protein stimulate the inflammasome, which results in caspase-1 activation and release of the cytokines interleukin-1β (IL-1β) and IL-18. Many CoV viral proteins [red: SARS-CoV, blue: Middle East respiratory syndrome (MERS)-CoV, green: murine hepatitis virus (MHV)] antagonize type I IFN signaling [85., 86., 87., 88., 89., 90., 91.]. Abbreviations: ASC, apoptosis-associated speck-like protein containing a caspase activation domain; M, membrane protein; N, nucleocapsid protein; NLRP3, nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing 3; nsp, nonstructural protein; PLP, papain-like protease.
Table 2.
Engaged Innate Immune Sensors, Subsequent Cell Death Pathways, and Notable Cytokines Released in Response to Specific CoVsa,b
| Virus | TLRs | RLRs | NLRs | Cell death | Notable cytokines |
|---|---|---|---|---|---|
| SARS-CoV | TLR7 [22] | N/A | NLRP3 [26,27,51] | Pyroptosis [26,27,51], apoptosis [60,61] | IL-1β [26,27,51], TNF [51], IL-6 [51] |
| MERS-CoV | TLR7 [22] | N/A | NLRP3 [28] | Pyroptosis [28], apoptosis [62,63] | IL-1β [28], TNF [28], IL-6 [28] |
| SARS-CoV-2 | N/A | N/A | NLRP3 [42] | Pyroptosis [42], apoptosis [65,66] | IL-1β [11,38,40., 41., 42.], IL-18 [42], TNF [11], IL-6 [11] |
| MHV | TLR7 [23] | MDA5 [24,25,33] | NLRP3 [29,52] | Pyroptosis [29,52], apoptosis [52,64,139], necroptosis [52] | IL-1β [29,52], IL-18 [52], TNF [52], IL-6 [52] |
Results are summarized from studies in both humans and mice.
Abbreviations: MDA5, melanoma differentiation-associated protein 5; N/A, not applicable (data for a particular virus and receptor combination have not been reported).
TLR- and RLR-Mediated CoV Recognition
TLR- and RLR-mediated signaling lead to the secretion of type I interferons (IFNs), which stimulate the expression of IFN-stimulated genes (ISGs) by infected and neighboring cells, inducing an antiviral state [22,23,25,30]. The TLR- and RLR-signaling cascades also lead to the secretion of proinflammatory cytokines, including interleukin-6 (IL-6) and tumor necrosis factor (TNF) [22,25].
TLR7 is a membrane-bound PRR on endosomes, where it can recognize ssRNA motifs from invading SARS-CoV, MERS-CoV, and MHV [22,23]. After recognizing the ssRNA, TLR7 dimerizes and recruits the myeloid differentiation primary response 88 (MYD88) adaptor protein [10,23,31]. MYD88 recruitment to the endosome results in signaling to trigger transcription factors, including interferon regulatory factor 7 (IRF7) and nuclear factor-κB (NF-κB), which stimulates the production of type I IFNs and proinflammatory cytokines, respectively, for host defense [22,23]. Indeed, Tlr7 -/- and Myd88 -/- mice have more severe disease following MERS-CoV or MHV infection compared with wild-type (WT) mice, with reduced survival and increased spread of the virus [22,23]. Another membrane-bound PRR, TLR3 has not been found to regulate the expression of type I IFNs and proinflammatory cytokines in response to SARS-CoV and MHV infection because Tlr3 -/- mice respond similarly to WT mice [23,32].
Melanoma differentiation-associated gene 5 (MDA5) is a cytosolic PRR in the RLR family that canonically senses double-stranded long RNAs. It plays a role in MHV detection and subsequent type I IFN production [24,25,33]. MDA5 contains two caspase recruitment domain (CARD)-like motifs near its N terminal and a downstream DExD/H-box helicase domain near its C terminal. Upon viral ligand binding in the C terminal domain, MDA5 facilitates conformational changes that enable CARD to bind the signaling adaptor mitochondrial antiviral signaling protein (MAVS), eventually leading to the production of type I IFNs and proinflammatory cytokines via the activation and nuclear translocation of IRF3 and NF-κB [24,25,34].
RIG-I is another cytosolic RLR, and it canonically senses 5′-triphosphate-double-stranded RNA. While RIG-I has been extensively characterized during infection with other respiratory RNA viruses, such as influenza A virus (IAV) [35], the MHV infection-mediated type I IFN response is RIG-I-independent in mice [24]. Further studies are required to determine the dominant innate sensor that drives type I IFN induction in vivo in response to infection with different CoVs (see Outstanding Questions).
Type I IFN Response during CoV Infection
Different studies have come to opposing conclusions regarding the involvement of the IFN response in CoV infection. In general, type I IFN production is thought to restrict SARS-CoV, MERS-CoV, SARS-CoV-2, and MHV infection in human and murine models [10,22,23,30]. Indeed, mice deficient in key mediators of type I IFN production (Tlr7 -/-, Myd88 -/-, or Mda5 -/- mice) show more severe disease following MERS-CoV or MHV infection compared with WT mice; this increased disease severity may be due to the reduced type I IFN production and elevated concentrations of proinflammatory cytokines [10,22,25,31,34]. During infection with SARS-CoV MA15, rapid viral replication in WT mice delays type I IFN signaling but orchestrates a robust proinflammatory cytokine response, including the induction of TNF and IL-6, which promotes lung immunopathology with diminished survival [30]. Ifnar -/- mice have reduced morbidity and mortality following lethal SARS-CoV (MA15) infection, suggesting a pathological role for IFN signaling; however, administration of recombinant IFN-β in WT mice early during the SARS-CoV infection reduces the viral titer in the lungs and improves animal survival [30]. Collectively, these results suggest that the timing of the IFN response is crucial and that delayed type I IFN causes lethal pneumonia during SARS-CoV infection [30]. A subsequent study also showed that type I IFN signaling is protective during MERS-CoV infection in the murine model; specifically, relative to control mice, blocking IFN signaling using an anti-IFNAR antibody via intraperitoneal injection led to delayed viral clearance, enhanced neutrophil infiltration, and impaired MERS-CoV-specific CD4+ and CD8+ T cell responses, causing fatal pneumonia [22].
In the case of SARS-CoV-2 infection, data are just beginning to emerge. Single-cell RNA sequencing has shown that the type I IFN response coexists with TNF– and IL-1β–driven inflammation in peripheral blood mononuclear cells (PBMCs) from patients with severe COVID-19, indicating that type I IFN may play a role in exacerbating inflammation in severe COVID-19 [36]. Conversely, recent reports have observed that low amounts of type I and type III IFN proteins in the serum of patients with COVID-19 are accompanied by elevated protein amounts of chemokines and proinflammatory cytokines, including IL-6, CCL8, CXCL8, CXCL9, CXCL16, IL-1RA, CCL2, CXCL2, EN-RAGE, TNGSF14, and Oncostatin-M, resulting in severe or critical COVID-19 pathology [10,37]. A subsequent study monitoring protein amounts and viral titers in whole blood taken from patients with severe or critical COVID-19 also found that reduced amounts of type I IFNs in the blood during SARS-CoV-2 infection were associated with increased viral load in the blood, and exacerbation of the inflammatory response [38]. Furthermore, data from whole-genome sequencing of patients with life-threatening pneumonia from SARS-CoV-2 infection identified mutations in genes in the type I IFN pathway, such as IRF7, TLR3, and IFNAR1, along with autoantibodies against type I IFNs in the blood, among the patient cohort [39]. Together, these studies suggest that the location, timing, and duration of IFN exposure in both lungs and blood relative to CoV replication are important factors in driving or contributing to infection outcomes, and certainly warrant further investigation.
CoV Infection Stimulates NLRP3 Inflammasome Activation
Innate immune cells play a major role in antiviral immunity, inflammatory signaling, and cytokine production. Among the proinflammatory cytokines, IL-1β and IL-18 are key mediators of the inflammatory response, and increased amounts of IL-1β and IL-18 in plasma have been correlated with mortality or severity in patients with COVID-19 [38,40., 41., 42.]. The release of IL-1β and IL-18 requires proteolytic maturation of pro-IL-1β and pro-IL-18 mediated by inflammasomes and the activation of the protease caspase-1 [43]. The inflammasome sensors can recognize PAMPs and/or DAMPs produced during pathogen infection and cellular instability, respectively [43]. Among those sensors, the most well characterized is the NLR family pyrin domain-containing 3 (NLRP3), which has been implicated in many diseases, including pathogen-mediated disease, autoinflammatory diseases, colitis, and obesity [44., 45., 46., 47., 48., 49., 50.].
Studies have shown that SARS-CoV encodes at least three different proteins that can activate NLRP3 inflammasome assembly. First, SARS-CoV open reading frame (ORF) 3a protein-mediated K+ efflux and mitochondrial reactive oxygen species (ROS) induce NLRP3 inflammasome activation and IL-1β secretion in lipopolysaccharide (LPS)-primed murine bone marrow-derived macrophages (BMDMs) in vitro [27]. Second, the SARS-CoV ORF8b protein interacts with the LRR domain of NLRP3 and triggers NLRP3 inflammasome activation and IL-1β secretion in THP-1 macrophages [26]. Third, SARS-CoV E protein-mediated ion channel activity induces IL-1β secretion in the bronchoalveolar lavage fluid (BALF) from BALB/c mice [51]. Although the specific ligands from MHV that activate the NLRP3 inflammasome have not been characterized, a recent study demonstrated that MHV infection induces NLRP3 inflammasome activation and the subsequent release of IL-1β and IL-18 in murine BMDMs [52]. In the case of SARS-CoV-2, the specific ligands are also unknown, but infection with the virus can induce NLRP3 inflammasome activation, caspase-1 cleavage, and the release of IL-1β and IL-18 in PBMCs isolated from patients with COVID-19 [42].
Activation of the NLRP3 inflammasome is often vital in the host antiviral immune response [47,48,50]. The NLRP3 inflammasome is activated in response to several RNA viruses, including IAV and West Nile virus (WNV), leading to the inhibition of viral replication and reduced mortality in mouse models [50,53]. However, hyperactivation of the NLRP3 inflammasome can also lead to severe pathological injury. In a murine model of SARS-CoV infection, proinflammatory cytokines, including inflammasome-derived IL-1β, along with TNF and IL-6, promote lung damage and acute respiratory distress syndrome (ARDS) [51]. Additionally, MHV-3 infection induces dysregulated NLRP3 inflammasome activation and IL-1β secretion, resulting in severe liver disease and high mortality in mice [29]. While these studies indicate that optimal NLRP3 inflammasome activation may be beneficial for the host, but that aberrant activation could lead to detrimental outcomes, detailed molecular mechanisms describing how inflammasome-driven inflammation plays a role in ARDS remain largely unknown. Given that IL-1β has been associated with the severity of COVID-19 [38,40,42], future studies are needed to fully examine the factors required for SARS-CoV-2–induced NLRP3 inflammasome assembly and NLRP3 inflammasome-driven ARDS.
Programmed Cell Death and Proinflammatory Cytokines during CoV Infections
Programmed Cell Death
Cell death plays an important role in limiting infection with pathogens. However, inflammatory cell death also results in the release of proinflammatory cytokines and cellular contents, including alarmins, PAMPs, and DAMPs, which can cause severe inflammation [54,55]. Therefore, cell death has been considered a double-edged sword for the host during pathogen infection.
Pyroptosis, apoptosis, and necroptosis are mechanistically well-studied programmed cell death pathways (Figure 3 , Key Figure).
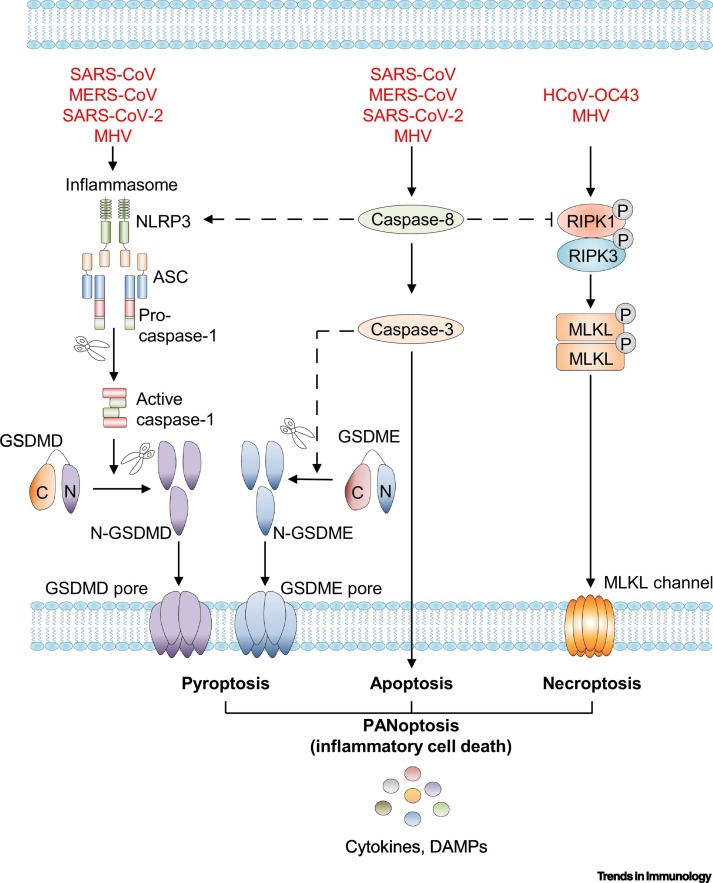
Figure 3.
Key Figure. Programmed Cell Death Pathways during Coronavirus (CoV) Infection.
Upon severe acute respiratory syndrome (SARS)-CoV, Middle East respiratory syndrome (MERS)-CoV, SARS-CoV-2, and murine hepatitis virus (MHV) infection, CoV viral proteins can function as cytosolic pathogen-associated molecular patterns (PAMPs) and stimulate inflammasome assembly, resulting in activation of caspase-1. Active caspase-1 then cleaves pro-interleukin (IL)-1β, pro-IL-18, and gasdermin D (GSDMD). The N-terminal fragment of GSDMD may then oligomerize within membranes to form membrane pores and execute pyroptosis [26., 27., 28., 29.,42,51,52]. SARS-CoV, MERS-CoV, SARS-CoV-2, and MHV infections also initiate a signaling cascade mediated by caspase-8 activation. Caspase-8 activates caspase-3 to drive apoptosis [52,60., 61., 62., 63., 64., 65., 66.]. Human (H)CoV-OC43 and MHV infection initiate necroptosis, a receptor-interacting serine/threonine-protein kinase 1 (RIPK1) and RIPK3 complex-dependent form of inflammatory cell death that depends on activation of the protein mixed lineage kinase domain-like pseudokinase (MLKL) to form channels in the membrane. PANoptosis, the coordinated regulation of pyroptosis, apoptosis, and necroptosis, is induced by MHV infection, and IL-1β, IL-18, and alarmins are released through GSDMD and MLKL membrane pores [52,57]. Broken lines represent connections that are known to exist in the context of inflammatory signaling but have not been shown for CoVs specifically. Abbreviations: DAMPs, danger-associated molecular patterns; GSDME, gasdermin E.
Pyroptosis during CoV Infection
Pyroptosis is an inflammasome- or gasdermin-mediated form of inflammatory cell death [3]. Inflammasome assembly leads to the activation of inflammatory caspases [caspase-1 (human and mouse), caspase-4 (human), -5 (human), or -11 (mouse)], which proteolytically cleave and release the N-terminal fragment of gasdermin D (GSDMD) to form pores in the plasma membrane [1., 2., 3.]. Caspase-1-dependent GSDMD cleavage leads to the release of IL-1β and IL-18 [1]. SARS-CoV ORF3a, ORF8b, or E protein can induce NLRP3 inflammasome activation and IL-1β secretion, phenomena consistent with pyroptotic death in human THP-1 macrophages and HeLa cells [26,27,51], and pyroptotic death has been directly observed in THP-1 macrophages exposed to SARS-CoV ORF8b [26]. MERS-CoV can also cause pyroptosis and stimulate IL-1β release through the NLRP3 inflammasome in THP-1 cells and hDPP4 transgenic mice [28]. Furthermore, IL-1β release and caspase-1 activation have also been observed in murine peritoneal exudative macrophages (PEMs) and RAW264.7 cells [29]. A recent study revealed that MHV infection induces caspase-1 activation, GSDMD cleavage, IL-1β and IL-18 release, and pyroptosis in murine BMDMs in vitro [52]. Additionally, recent findings suggest that SARS-CoV-2 infection stimulates pyroptosis in PBMCs from patients with COVID-19, as evidenced by NLRP3 inflammasome activation, cleavage of caspase-1, and secretion of IL-1β and IL-18 [42] (Table 2).
Necroptosis during CoV Infection
Necroptosis is a mixed-lineage kinase domain-like pseudokinase (MLKL)-mediated form of inflammatory cell death, during which oligomerized MLKL is translocated and forms channels in the plasma membrane [56]. While evidence for necroptosis during CoV infection has been historically limited, new studies are beginning to identify this cell death pathway in response to CoVs. HCoV-OC43 induces necroptosis in human neural cells [57]. Additionally, a recent study reported that MHV infection induced phosphorylation of MLKL, causing necroptosis in murine BMDMs [52]. This study also found that Nlrp3 -/-, Casp1 -/- Casp11 -/-, or Gsdmd -/- BMDMs exhibited robust apoptosis and necroptosis, as evidenced from the cleavage of caspase-3, -7, and -8 and the phosphorylation of MLKL, driven by caspase-8 and receptor-interacting serine/threonine-protein kinase 3 (RIPK3) during MHV infection; indeed, Casp8 -/- Ripk3 -/- BMDMs were protected from MHV-induced death [52]. To date, there has been no clear evidence of necroptosis in the context of SARS-CoV-2 infection, or SARS-CoV and MERS-CoV (Table 2), and additional work is needed to determine whether this is occurring.
Apoptosis during CoV Infection
Pyroptosis and necroptosis are similar in that they are lytic forms of cell death driven by the GSDMD pore and MLKL channel, respectively, which release proinflammatory cytokines and other cellular factors to alert the surrounding cells of danger and to recruit innate and adaptive inflammatory cells [54,55]. Apoptosis was originally thought to be a less inflammatory form of cell death that disassembled the cell via membrane blebbing to avoid directly releasing cellular contents [58]. However, more recent studies have suggested that it is not always immunologically silent, because there is crosstalk between apoptotic proteins and lytic cell death executioners [58]. Apoptosis is driven by initiator caspase-8, -9, and -10 cleavage of executioner caspase-3 and -7. The apoptotic caspase-3 has been reported to activate gasdermin E (GSDME) to induce a lytic form of cell death [59]; caspase-8 has also been reported to activate GSDMD under specific conditions [58]. In the context of CoV infection, SARS-CoV infection and SARS-CoV ORF6 induce apoptosis in Vero E6 cells, indicated by the activation of caspase-3 [60,61]. Likewise, MERS-CoV infection can also activate apoptosis in renal proximal tubular cells (HK2) and human CD4+ and CD8+ T cells isolated from PBMCs, as evidenced by the activation of caspase-3 [62,63]. Additionally, infection with MHV induces apoptosis, as evidenced by cleavage of caspase-3, caspase-7, and caspase-8 in mouse BMDMs [52,64]. ORF3a from SARS-CoV-2 also induces cleavage of caspase-8 and -9 and causes apoptosis in HEK293T, HepG2, and Vero E6 cells [65]. A subsequent study of SARS-CoV-2 infection in human airway epithelial cells in vitro found an increase in cytopathic effects with apoptotic characteristics [66]. Collectively, these findings indicate that CoVs can induce apoptosis (Table 2).
Inflammatory Cell Death and Cytokines
Overt activation of inflammatory cell death pathways may lead to critical tissue damage as well as severe inflammation [54]. For instance, lactate dehydrogenase (LDH), a common marker for cell death, has now been linked to the pathogenesis of COVID-19; specifically, high LDH concentrations in the serum of patients are associated with tissue breakdown during COVID-19, and LDH amounts are considered a powerful predictive factor for the early recognition of lung injury in severe COVID-19 cases [67,68]. Indeed, LDH concentrations are upregulated in PBMCs from patients with COVID-19 [42]. Additionally, autopsy studies have observed lung damage in fatal cases of COVID-19 [69], suggesting that these proinflammatory cell death pathways are dysregulated in severe or critical cases. Because inflammatory cell death releases cytokines and chemokines, this process may contribute to the cytokine storm observed in patients with COVID-19. Patients with severe or critical COVID-19 cases have higher serum concentrations of proinflammatory cytokines and chemokines, such as granulocyte-colony stimulating factor (GCSF), monocyte chemoattractant protein 1 (MCP1), TNF, IL-6, and IL-1β, compared with healthy individuals [11,40]. Recent emerging studies examining the cytokine profiles from patients with COVID-19 have indicated a direct correlation between the cytokine storm and lung injury [70,71]. ARDS is another serious outcome of this cytokine storm [72]. Therefore, it is important to determine what innate immune processes contribute to cytokine release and identify what links may exist between cytokine signaling and inflammatory cell death in SARS-CoV-2 infections.
Additional studies are required to fully elucidate the inflammatory cell death pathways and functional consequences of these processes during CoV infections. Furthermore, based on the extensive co-regulation and crosstalk that has been identified between pyroptosis, apoptosis, and necroptosis [73., 74., 75., 76., 77., 78., 79., 80., 81., 82., 83.], recent work has introduced PANoptosis as a unique inflammatory programmed cell death regulated by the PANoptosome, which provides a molecular scaffold for interactions and activation of the machinery required for inflammasome/pyroptosis, apoptosis, and necroptosis [78,79,82,83]. MHV infection in mice induces PANoptosis in murine BMDMs, and inhibiting activation of the NLRP3 inflammasome pathway and pyroptosis leads to a robust increase in apoptosis and necroptosis, as evidenced by increased cleavage of the apoptotic caspase-3, -7, and -8 and the increased phosphorylation of the necroptotic executioner MLKL in Nlrp3 -/-, Casp1 -/- Casp11 -/-, or Gsdmd -/- BMDMs [52]. It will be important to determine if and how these findings translate to other CoVs, especially SARS-CoV-2, and what the implications of these findings in vivo will be.
CoV Immune Evasion
One of the goals of the innate immune response is to use PRRs to recognize pathogens (here, viral agents) and ultimately trigger an inflammatory response and programmed cell death to limit viral replication and promote clearance. In response, CoVs have undergone selective pressure to limit the innate immune response and permit their intracellular replication [84].
The most well-studied CoV immune evasion mechanism involves counteracting type I IFN innate immune signaling (Figure 2). SARS-CoV ORF3b, ORF6, and N protein can reduce type I IFN expression in A549 cells by inhibiting the activation of IRF3 and NF-κB nuclear translocation [85]. The M protein of SARS-CoV inhibits type I IFN production in HEK293T cells by delaying the formation of a functional TNF receptor-associated factor 3 (TRAF3)-containing complex, which is a key transcription factor that induces IFN production from cells through activation of TBK1 and IKKε upstream of IRF3 [86,87]. SARS-CoV papain-like proteases (PLP) also suppress type I IFN production in HEK293T cells mediated by stimulator of interferon genes (STING) [88]. SARS-CoV nsp1 suppresses type I IFN expression in HEK293T cells by promoting host mRNA degradation [89]. Additionally, MERS-CoV ORF4a suppresses the activation of RIG-I and MDA5 in HEK293T cells [90,91]. MERS-CoV nsp4a and nsp4b can also antagonize IFN expression in human A549 cells stably expressing the MERS-CoV receptor DPP4 [92]. Upon MHV infection, nsp15 and N protein suppress IFN production, as well as protein kinase R (PKR) and 2′,5′-oligoadenylate synthetase (OAS)/RNase L responses in murine 17Cl1 cells or BMDMs [64,93]. MHV nsp2 also antagonizes type I IFN production and blocks the 2′,5′-OAS/RNase L response in BMDMs [94]. Additionally, MHV infection induces high amounts of type I IFN secretion in plasmacytoid dendritic cells (pDCs), BMDMs, microglia, and oligodendrocytes [23,24,95], but relatively little production of type I IFNs in bone marrow-derived dendritic cells (BMDCs), hepatocytes, neurons, astrocytes, and murine 17CI-1 fibroblasts [24,96,97]; this suggests that MHV efficiently evades innate immune sensing in a cell type-dependent manner.
CoV Disease Tolerance versus Immunopathology
Innate host protection can play an important role in dictating CoV disease tolerance. First, SARS-CoV, MERS-CoV, and SARS-CoV-2 disease tolerance can occur in an age-dependent manner, because children with CoV-induced diseases, including COVID-19, have a less aggressive clinical course compared with adults [98., 99., 100.]. However, this may not be due to a virus-specific effect, because children have reduced expression of ACE2 in their nasal epithelium compared with expression in adults [101]; this might result in reduced viral infectivity in children compared with adults and contribute to the age-dependence of SARS- and SARS-CoV-2-induced disease. However, it is important to note that recent studies have found that SARS-CoV-2 infection is sometimes associated with multisystem inflammatory syndrome in children (MIS-C) [102], which is characterized by systemic inflammation, and may be caused by an aberrant immune response, although much remains unknown about this condition, and future robust studies are absolutely necessary.
Second, CoV disease tolerance is species specific. Bats are largely considered the original host of SARS-CoV, MERS-CoV, and SARS-CoV-2 [103., 104., 105.]. NLRP3 inflammasome-mediated inflammation in bat PBMCs is dampened in response to MERS-CoV infection compared with that observed in human PBMCs [106]. Another study also showed that bats have a mutation in an essential and highly conserved serine residue (S358) in STING that causes reduced IFN responses relative to controls in HEK293T cells [107]. These studies suggest that species-dependent host factors and molecular mechanisms regulate inflammatory responses during CoV infection.
Third, CoV disease tolerance is sex dependent. Specifically, men have had a higher case fatality rate than women during the SARS-CoV, MERS-CoV, and SARS-CoV-2 outbreaks [108., 109., 110., 111.]. This sex-dependent CoV disease tolerance in women is also more well defined with advancing age [108,110]. One recent study also found that lung infiltration of inflammatory cells and virus titers were further elevated in SARS-CoV-infected male mice compared with female mice and, furthermore, that estrogen receptor signaling induced by tamoxifen enhanced disease tolerance in female mice by moderating SARS-CoV-induced inflammation [112], suggesting that sex-based differences affect inflammation and susceptibility to CoV infection. These results suggest that innate host protection plays an important role in directing CoV disease resistance.
Concluding Remarks
We have highlighted the sensing of different CoVs by the innate immune system and the inflammatory cell death pathways activated in response to these CoV infections. Optimal cell death and inflammatory cytokine release are important for host defense. However, excessive host inflammatory responses and cell death can lead to cytokine storms and/or tissue damage, increasing morbidity and mortality in patients with CoV. Therefore, CoV-induced innate immunity and cell death must be finely controlled to reduce excessive inflammation while retaining antiviral functions.
In this regard, it will be important to mechanistically characterize the innate immune response to different CoV infections and evaluate the clinical efficacy of targeting innate immune pathways when designing evidence-based treatment strategies (see Outstanding Questions). For example, SARS-CoV-2 infection antagonizes type I and type III IFN production, but elevates the expression of chemokines and proinflammatory cytokines [10], which are associated with poor prognosis [10,11]. With other RNA viruses, IAV and WNV, inflammasome activation is required for host defense, but hyperactivation may lead to disease exacerbation [29,50,51,53]. It is possible that reduced innate antiviral defenses together with excessive inflammatory cytokine production are some of the driving features of CoV-mediated diseases.
The SARS-CoV-2 pandemic continues to spread globally, and we now desperately need effective therapies and vaccines to combat COVID-19. Given the urgency of the global health situation, researchers are exploring the possibility of repurposing existing therapeutic agents already approved, or which are under development/testing for other infections, including malaria (Plasmodium sp.), HIV, or other CoVs, or for blockade of the inflammasome (e.g., colchicine) or the proinflammatory cytokines IL-1 and IL-6 [113., 114., 115., 116., 117.]. The response to these therapies has been mixed. For example, a recent pilot study of COVID-19 in five patients who were already undergoing treatment with colchicine found that all five patients presented with mild/moderate disease and survived despite having multiple comorbidities [118]. However, this was a small observational study, preventing definitive conclusions. Despite promising early data from retrospective and single-center studies using IL-6-antibody blockade therapy [119], recent studies have found that blocking IL-6 can be associated with increased secondary infections [120], and the large Phase III trials using these inhibitors have had mixed results. The first two Phase III randomized double-blind, placebo-controlled trials that included anti-IL-6 antibody and placebo groups in patients with severe COVID-19, the COVACTA (NCT04320615)i and Kevzara trials (NCT04327388)ii, used improvement on the seven category ordinal scale as their primary outcomes. Both reported in company press releases that they failed to meet their primary endpoints. However, EMPACTA (NCT04372186)iii reportedly met its primary endpoint of a reduction in the cumulative proportion of participants requiring mechanical ventilation by day 28, although there was no statistical difference in 28-day mortality between the anti-IL-6 antibody and placebo groups, and the full data have not yet been made available. These results suggest that, while several potential therapies have been explored for COVID-19, balancing proinflammatory responses to clear the virus with preventing systemic inflammation remains challenging. An improved understanding of the mechanisms by which the innate immune system recognizes and responds to specific CoV infections will be key to informing the development of putative therapeutic strategies.
Outstanding Questions.
What is the dominant signaling pathway that controls the activation of type I IFNs and proinflammatory cytokine expression during CoV infections? What are the molecular mechanisms by which IFN-inducible genes drive robust inflammatory responses with delayed type I IFN production in these infections?
What are the ligands that activate the NLRP3 inflammasome, and what are the key host factors that regulate NLRP3 inflammasome activation in response to SARS-CoV-2 infection?
What types of programmed cell death are induced by CoV infection? Is there any redundancy among these types of cell death? What are the positive/negative regulators of cell death?
What makes older men more susceptible to CoV-related illness and fatality than other populations? What are the driving forces that lead to gain or loss of NLRP3 inflammasome functions in different species during CoV infections?
Alt-text: Outstanding Questions
Acknowledgments
We apologize to our colleagues in the field whose work could not be cited due to space limitations. We thank all the members of the Kanneganti laboratory for their comments and suggestions and Rebecca Tweedell for scientific editing and writing support. Work from our laboratory is supported by the US National Institutes of Health (AI101935, AI124346, AR056296, and CA253095 to T-D.K.) and the American Lebanese Syrian Associated Charities (to T-D.K.). Additional support was provided by the US National Institutes of Health (AG060222 to R.C.) The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Glossary
- Acute respiratory distress syndrome (ARDS)
type of respiratory failure characterized by the rapid onset of widespread inflammation in the lungs.
- Caspase-1
inflammatory caspase that is part of the inflammasome and cleaves IL-1β, IL-18, and GSDMD to drive pyroptosis and inflammatory cytokine release.
- Cytokine storm
over-reaction of the host immune response to release excess amounts of proinflammatory cytokines and chemokines.
- Gasdermin D (GSDMD)
executioner of pyroptosis that has pore-forming activity in the plasma membrane.
- Inflammasome
multiprotein intracellular complex consisting of caspase-1, ASC, and cytosolic PRRs.
- Interferon regulatory factor 7 (IRF7)
key transcriptional factor of type I IFN production against DNA and RNA viruses.
- Interferons (IFNs)
group of signaling cytokines released by host cells in response to several microbes for anti-microbial defense.
- Interleukin (IL)-1β
proinflammatory cytokine formed when the pro–IL-1β precursor is cleaved by active caspase-1.
- Interleukin (IL)-6
proinflammatory cytokine that is active in inflammation and B cell maturation.
- Interleukin (IL)-18
proinflammatory cytokine formed when the pro-IL-18 precursor is cleaved by active caspase-1.
- Lactate dehydrogenase (LDH)
a common marker of cell death.
- Mitochondrial antiviral signaling protein (MAVS)
signaling protein that binds RLRs during viral infection. Its activation eventually leads to the production of proinflammatory cytokines and type I IFNs.
- Mixed lineage kinase domain-like pseudokinase (MLKL)
pseudokinase that plays a key role in TNF-induced necroptosis, a programmed cell death process.
- Necroptosis
MLKL-mediated form of inflammatory cell death.
- Nuclear factor (NF)-κB
protein complex that regulates cytokine production, cell survival, and transcription of DNA.
- Oligoadenylate synthetase (OAS)/RNase L
IFN-induced ribonuclease that destroys both cellular and viral RNA within the cell upon activation.
- PANoptosis
integrated inflammatory cell death pathway that consists of components and machinery from three types of programmed cell death (pyroptosis, apoptosis, and necroptosis).
- PANoptosome
multiprotein complex containing components from multiple cell death pathways; initiates and coordinates pyroptosis, apoptosis, and necroptosis (PANoptosis).
- Pathogen-associated molecular patterns (PAMPs)
molecular signatures of infection.
- Pattern recognition receptors (PRRs)
group of host innate immune sensors that recognize PAMPs and DAMPs, thereby inducing inflammatory cytokine production.
- Pneumonia
respiratory lung infection that causes inflammation in one or both lungs.
- Polyprotein
protein cleaved to produce several polypeptides.
- Protein kinase R (PKR)
an interferon-induced, double-stranded RNA-dependent protein kinase; key host factor in the antiviral response.
- Pyroptosis
inflammasome- or gasdermin-mediated form of inflammatory cell death.
- Reactive oxygen species (ROS)
generally produced by mitochondria, resulting in oxidative damage of mitochondrial proteins, membranes, and DNA, impairing the function of mitochondria.
- Stimulator of interferon genes (STING)
downstream protein of cAMP-GMP synthase; plays an essential role in the innate immune response.
- T helper (Th) 17
subset of T helper cells induced by the production of IL-17.
- Tumor necrosis factor (TNF)
proinflammatory cytokine involved in systemic inflammation; plays a key role in the acute phase reaction.
Resources
ihttps://clinicaltrials.gov/ct2/show/NCT04320615iihttps://clinicaltrials.gov/ct2/show/NCT04327388iiihttps://clinicaltrials.gov/ct2/show/NCT04372186References
- 1.Man S.M. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol. Rev. 2017;277:61–75. doi: 10.1111/imr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He W.T. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res. 2015;25:1285–1298. doi: 10.1038/cr.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi J. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 4.Nathan C. Points of control in inflammation. Nature. 2002;420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 5.Hu B. Bat origin of human coronaviruses. Virol. J. 2015;12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong N.S. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaki A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 10.Blanco-Melo D. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim D. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann M. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawicki S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiel V. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 17.Knoops K. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Wit E. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts A. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3:e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li K. Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E3119–E3128. doi: 10.1073/pnas.1619109114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang R.D. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182:50–58. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Channappanavar R. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J. Clin. Invest. 2019;130:3625–3639. doi: 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cervantes-Barragan L. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roth-Cross J.K. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 2008;82:9829–9838. doi: 10.1128/JVI.01199-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zalinger Z.B. MDA5 is critical to host defense during infection with murine coronavirus. J. Virol. 2015;89:12330–12340. doi: 10.1128/JVI.01470-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi C.S. SARS-coronavirus open reading frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019;5:101. doi: 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen I.Y. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front. Microbiol. 2019;10:50. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y. Complement receptor C5aR1 inhibition reduces pyroptosis in hDPP4-transgenic mice infected with MERS-CoV. Viruses. 2019;11 doi: 10.3390/v11010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo S. The NLRP3 inflammasome and IL-1β accelerate immunologically mediated pathology in experimental viral fulminant hepatitis. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Channappanavar R. Dysregulated Type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheahan T. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Totura A.L. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. mBio. 2015;6 doi: 10.1128/mBio.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hackbart M. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. U. S. A. 2020;117:8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. U. S. A. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pichlmair A. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 36.Lee J.S. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arunachalam P.S. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hadjadj J. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Q. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King A. Anakinra in COVID-19: important considerations for clinical trials. Lancet Rheumatol. 2020;2:e379–e381. doi: 10.1016/S2665-9913(20)30160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cavalli G. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2020;2:e325–e331. doi: 10.1016/S2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodrigues T.S. Inflammasome activation in COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.08.05.20168872. Published online August 6, 2020. [DOI] [Google Scholar]
- 43.Kanneganti T.D. Central roles of NLRs and inflammasomes in viral infection. Nat. Rev. Immunol. 2010;10:688–698. doi: 10.1038/nri2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gurung P., Kanneganti T.D. Autoinflammatory skin disorders: the inflammasomme in focus. Trends Mol. Med. 2016;22:545–564. doi: 10.1016/j.molmed.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaki M.H. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stienstra R. Inflammasome is a central player in the induction of obesity and insulin resistance. Proc. Natl. Acad. Sci. U. S. A. 2011;108:15324–15329. doi: 10.1073/pnas.1100255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanneganti T.D. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 48.Kanneganti T.D. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J. Biol. Chem. 2006;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 49.Karki R. Concerted activation of the AIM2 and NLRP3 inflammasomes orchestrates host protection against Aspergillus infection. Cell Host Microbe. 2015;17:357–368. doi: 10.1016/j.chom.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas P.G. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nieto-Torres J.L. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng M. Impaired NLRP3 inflammasome activation/pyroptosis leads to robust inflammatory cell death via caspase-8/RIPK3 during coronavirus infection. J. Biol. Chem. 2020 doi: 10.1074/jbc.RA120.015036. Published online August 6, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramos H.J. IL-1β signaling promotes CNS-intrinsic immune control of West Nile virus infection. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bergsbaken T. Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pasparakis M., Vandenabeele P. Necroptosis and its role in inflammation. Nature. 2015;517:311–320. doi: 10.1038/nature14191. [DOI] [PubMed] [Google Scholar]
- 56.Dondelinger Y. MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep. 2014;7:971–981. doi: 10.1016/j.celrep.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 57.Meessen-Pinard M. Pivotal role of receptor-interacting protein kinase 1 and mixed lineage kinase domain-like in neuronal cell death induced by the human neuroinvasive coronavirus OC43. J. Virol. 2017;91 doi: 10.1128/JVI.01513-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Place D.E. PANoptosis in microbial infection. Curr. Opin. Microbiol. 2020;59:42–49. doi: 10.1016/j.mib.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rogers C. Gasdermin pores permeabilize mitochondria to augment caspase-3 activation during apoptosis and inflammasome activation. Nat. Commun. 2019;10:1689. doi: 10.1038/s41467-019-09397-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren L. Apoptosis induced by the SARS-associated coronavirus in Vero cells is replication-dependent and involves caspase. DNA Cell Biol. 2005;24:496–502. doi: 10.1089/dna.2005.24.496. [DOI] [PubMed] [Google Scholar]
- 61.Ye Z. A SARS-CoV protein, ORF-6, induces caspase-3 mediated, ER stress and JNK-dependent apoptosis. Biochim. Biophys. Acta. 2008;1780:1383–1387. doi: 10.1016/j.bbagen.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeung M.L. MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat. Microbiol. 2016;1:16004. doi: 10.1038/nmicrobiol.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu H. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J. Infect. Dis. 2016;213:904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deng X. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ren Y. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell. Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu N. Morphogenesis and cytopathic effect of SARS-CoV–2 infection in human airway epithelial cells. Nat. Commun. 2020;11:3910. doi: 10.1038/s41467-020-17796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yan L. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020;2:283–288. [Google Scholar]
- 68.Reeve J.L.V., Twomey P.J. Consider laboratory aspects in developing patient prediction models. Nat. Mach. Intell. 2020 doi: 10.1038/s42256-020-0221-2. Published online August 12, 2020. [DOI] [Google Scholar]
- 69.Carsana L. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-centre descriptive study. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30434-5. Published online June 8, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. Med. 2020;8:e46–e47. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mehta P. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ragab D. The COVID-19 cytokine storm; what we know so far. Front. Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lamkanfi M. Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol. Cell. Proteomics. 2008;7:2350–2363. doi: 10.1074/mcp.M800132-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malireddi R.K. Cutting edge: proteolytic inactivation of poly(ADP-ribose) polymerase 1 by the Nlrp3 and Nlrc4 inflammasomes. J. Immunol. 2010;185:3127–3130. doi: 10.4049/jimmunol.1001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gurung P. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J. Immunol. 2014;192:1835–1846. doi: 10.4049/jimmunol.1302839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lukens J.R. Critical role for inflammasome-independent IL-1β production in osteomyelitis. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1066–1071. doi: 10.1073/pnas.1318688111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gurung P. NLRP3 inflammasome plays a redundant role with caspase 8 to promote IL-1β-mediated osteomyelitis. Proc. Natl. Acad. Sci. U. S. A. 2016;113:4452–4457. doi: 10.1073/pnas.1601636113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng M. Caspase-6 is a key regulator of innate immunity, inflammasome activation, and host defense. Cell. 2020;181:674–687.e613. doi: 10.1016/j.cell.2020.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Christgen S. Identification of the PANoptosome: a molecular platform triggering pyroptosis, apoptosis, and necroptosis (PANoptosis) Front. Cell. Infect. Microbiol. 2020;10:237. doi: 10.3389/fcimb.2020.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuriakose T. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 2016;1 doi: 10.1126/sciimmunol.aag2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kesavardhana S. ZBP1/DAI ubiquitination and sensing of influenza vRNPs activate programmed cell death. J. Exp. Med. 2017;214:2217–2229. doi: 10.1084/jem.20170550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Malireddi R.K.S. ZBP1 and TAK1: master regulators of NLRP3 inflammasome/pyroptosis, apoptosis, and necroptosis (PAN-optosis) Front. Cell. Infect. Microbiol. 2019;9:406. doi: 10.3389/fcimb.2019.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Samir P. The PANoptosome: a deadly protein complex driving pyroptosis, apoptosis, and necroptosis (PANoptosis) Front. Cell. Infect. Microbiol. 2020;10:238. doi: 10.3389/fcimb.2020.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kopecky-Bromberg S.A. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Siu K.L. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J. Biol. Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Häcker H. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439:204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- 88.Sun L. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Narayanan K. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008;82:4471–4479. doi: 10.1128/JVI.02472-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Siu K.L. Middle east respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J. Virol. 2014;88:4866–4876. doi: 10.1128/JVI.03649-13. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91.Niemeyer D. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J. Virol. 2013;87:12489–12495. doi: 10.1128/JVI.01845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Comar C.E. Antagonism of dsRNA-induced innate immune pathways by NS4a and NS4b accessory proteins during MERS coronavirus infection. mBio. 2019;10 doi: 10.1128/mBio.00319-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ye Y. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 2007;81:2554–2563. doi: 10.1128/JVI.01634-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao L. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe. 2012;11:607–616. doi: 10.1016/j.chom.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li J. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J. Virol. 2010;84:6472–6482. doi: 10.1128/JVI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou H., Perlman S. Preferential infection of mature dendritic cells by mouse hepatitis virus strain JHM. J. Virol. 2006;80:2506–2514. doi: 10.1128/JVI.80.5.2506-2514.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou H., Perlman S. Mouse hepatitis virus does not induce Beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J. Virol. 2007;81:568–574. doi: 10.1128/JVI.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qiu H. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect. Dis. 2020;20:689–696. doi: 10.1016/S1473-3099(20)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Memish Z.A. Middle East respiratory syndrome coronavirus disease in children. Pediatr. Infect. Dis. J. 2014;33:904–906. doi: 10.1097/INF.0000000000000325. [DOI] [PubMed] [Google Scholar]
- 100.Hon K.L. Clinical presentations and outcome of severe acute respiratory syndrome in children. Lancet. 2003;361:1701–1703. doi: 10.1016/S0140-6736(03)13364-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bunyavanich S. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA. 2020;323:2427–2429. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Verdoni L. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:1771–1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li W. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 104.Memish Z.A. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013;19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lu R. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ahn M. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat. Microbiol. 2019;4:789–799. doi: 10.1038/s41564-019-0371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie J. Dampened STING-dependent interferon activation in bats. Cell Host Microbe. 2018;23:297–301. doi: 10.1016/j.chom.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Karlberg J. Do men have a higher case fatality rate of severe acute respiratory syndrome than women do? Am. J. Epidemiol. 2004;159:229–231. doi: 10.1093/aje/kwh056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Leong H.N. SARS in Singapore--predictors of disease severity. Ann. Acad. Med. Singap. 2006;35:326–331. [PubMed] [Google Scholar]
- 110.Alghamdi I.G. The pattern of Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive epidemiological analysis of data from the Saudi Ministry of Health. Int. J. Gen. Med. 2014;7:417–423. doi: 10.2147/IJGM.S67061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Takahashi T. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020 doi: 10.1038/s41586-020-2700-3. Published online August 26, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Channappanavar R. Sex-based differences in susceptibility to severe acute respiratory syndrome coronavirus infection. J. Immunol. 2017;198:4046–4053. doi: 10.4049/jimmunol.1601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gao J. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 114.Cao B. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu X. Effective treatment of severe COVID-19 patients with tocilizumab. Proc. Natl. Acad. Sci. U. S. A. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Grein J. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huet T. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2020;2:e393–e400. doi: 10.1016/S2665-9913(20)30164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Montealegre-Gómez G. Colchicine: a potential therapeutic tool against COVID-19. Experience of 5 patients. Reumatol. Clin. 2020 doi: 10.1016/j.reuma.2020.05.001. Published online May 16, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Toniati P. Tocilizumab for the treatment of severe COVID-19 pneumonia with hyperinflammatory syndrome and acute respiratory failure: a single center study of 100 patients in Brescia, Italy. Autoimmun. Rev. 2020;19:102568. doi: 10.1016/j.autrev.2020.102568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kimmig L.M. IL6 inhibition in critically ill COVID-19 patients is associated with increased secondary infections. medRxiv. 2020 doi: 10.1101/2020.05.15.20103531. Published online September 12, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou F. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020;53:368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Qin C. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guo T. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020;5:811–818. doi: 10.1001/jamacardio.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu C. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao Q. Lymphopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a systemic review and meta-analysis. Int. J. Infect. Dis. 2020;96:131–135. doi: 10.1016/j.ijid.2020.04.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lippi G. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clin. Chim. Acta. 2020;506:145–148. doi: 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang X. Thrombocytopenia and its association with mortality in patients with COVID-19. J. Thromb. Haemost. 2020;18:1469–1472. doi: 10.1111/jth.14848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Becker R.C. COVID-19-associated vasculitis and vasculopathy. J. Thromb. Thrombolysis. 2020;50:499–511. doi: 10.1007/s11239-020-02230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xu Z. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cheng Y. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020;97:829–838. doi: 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Su H. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98:219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Beyrouti R. Characteristics of ischaemic stroke associated with COVID-19. J. Neurol. Neurosurg. Psychiatry. 2020;91:889–891. doi: 10.1136/jnnp-2020-323586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zheng Y.Y. COVID-19 and the cardiovascular system. Nat. Rev. Cardiol. 2020;17:259–260. doi: 10.1038/s41569-020-0360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang C. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol. Hepatol. 2020;5:428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang L. C-reactive protein levels in the early stage of COVID-19. Med. Mal. Infect. 2020;50:332–334. doi: 10.1016/j.medmal.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Carvalho A. SARS-CoV-2 gastrointestinal infection causing hemorrhagic colitis: implications for detection and transmission of COVID-19 disease. Am. J. Gastroenterol. 2020;115:942–946. doi: 10.14309/ajg.0000000000000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.D'Amico F. Diarrhea during COVID-19 infection: pathogenesis, epidemiology, prevention, and management. Clin. Gastroenterol. Hepatol. 2020;18:1663–1672. doi: 10.1016/j.cgh.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu Y. Role of the mitochondrial signaling pathway in murine coronavirus-induced oligodendrocyte apoptosis. J. Virol. 2006;80:395–403. doi: 10.1128/JVI.80.1.395-403.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]