SUMMARY
The Mediator complex relays regulatory signals from gene-specific transcription factors to the basal transcriptional machinery. However, the role of individual Mediator subunits in different tissues remains unclear. Here, we demonstrate that MED19 is essential for adipogenesis and maintenance of white adipose tissue (WAT) by mediating peroxisome proliferator-activated receptor gamma (PPARγ) transcriptional activity. MED19 knockdown blocks white adipogenesis, but not brown adipogenesis or C2C12 myoblast differentiation. Adipose-specific MED19 knockout (KO) in mice results in a striking loss of WAT, whitening of brown fat, hepatic steatosis, and insulin resistance. Inducible adipose-specific MED19 KO in adult animals also results in lipodystrophy, demonstrating its requirement for WAT maintenance. Global gene expression analysis reveals induction of genes involved in apoptosis and inflammation and impaired expression of adipose-specific genes, resulting from decreased PPARγ residency on adipocyte gene promoters and reduced association of PPARγ with RNA polymerase II. These results identify MED19 as a crucial facilitator of PPARγ-mediated gene expression in adipose tissue.
Graphical Abstract
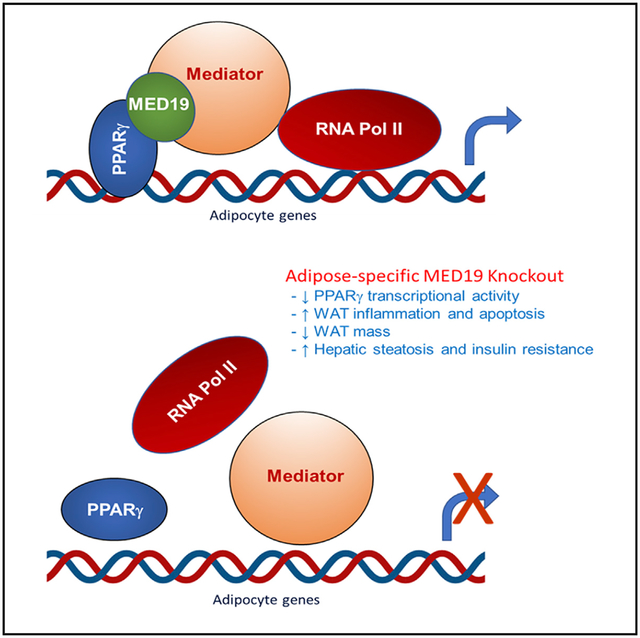
In Brief
The Mediator complex relays signals from upstream signaling pathways to the basal transcriptional machinery. Dean et al. show that the MED19 subunit is essential for white adipogenesis and maintenance of white adipose tissue mass by mediating PPARγ activity. Adipose-specific MED19 knockout promotes dramatic loss of WAT and whitening of BAT.
INTRODUCTION
Adipose tissue has emerged as a complex metabolic organ with a variety of functions. Two major types of adipose tissue are found in mammals: white adipose tissue (WAT) and brown adipose tissue (BAT). WAT primarily stores energy. In the context of obesity, WAT dramatically expands to store excess calories, contributing to metabolic consequences. Alternatively, BAT dissipates energy in the form of heat and is related to positive metabolic outcomes. A subtype of brown fat, called beige fat, can form in subcutaneous WAT in response to cold exposure and beta adrenergic signaling. Adipose tissue development is the result of a complex transcriptional cascade of adipocyte-specific factors. As the master regulator of adipogenesis, peroxisome proliferator-activated receptor gamma (PPARγ) plays a key role in this process, conducting the expression of various other transcription factors and proteins to ensure that adipocyte differentiation is completed (Rosen and Spiegelman, 2014). However, how PPARγ regulates the development of two functionally distinct types of adipose tissue as well as the browning of white fat remains unclear. Previous work in our lab contributed to this knowledge gap by showing that the peroxisomal lipid synthetic enzyme PexRAP acts as a transcriptional regulator of adipose tissue gene expression by preventing the interaction of PPARγ with PRDM16 and inhibiting PRDM16-mediated thermogenic gene expression (Lodhi et al., 2017). A global protein-protein interaction screen indicated that PexRAP (encoded by Dhrs7b) interacts with the Mediator complex protein MED19 (Stelzl et al., 2005), suggesting the possibility that MED19 could be involved in adipocyte gene expression.
The Mediator complex is composed of more than 30 subunits and acts as a bridge between gene-specific transcription factors and the basal transcription machinery. The subunit composition of the Mediator can vary, and different subunits interact with different transcription factors in a cell-context-dependent manner. Overall, very little is known about the regulation of this process or the specific function of the individual subunits (Allen and Taatjes, 2015). Several Mediator components have been implicated in adipogenesis and adipocyte gene expression (Zhang et al., 2013). In particular, MED1 is important for white adipogenesis through interactions with PPARγ (Ge et al., 2002, 2008), a process which is facilitated by MED14 (Grøntved et al., 2010). MED1 also plays a role in brown-fat-specific gene expression by facilitating recruitment of PRDM16 to the UCP1 enhancer (Chen et al., 2009; Iida et al., 2015). MED23 is necessary for hormone-induced adipogenesis by facilitating the expression of the early adipogenesis transcription factor Krox20 (Wang et al., 2009), and it plays a critical role in controlling the cell fate preference directing differentiation of smooth muscle cells or adipocytes (Yin et al., 2012). MED17 is required for the transcriptional activation of lipogenic genes in response to insulin (Viscarra et al., 2017). However, little is known about the function of MED19. In mammals, some studies have implicated MED19 as a facilitator of RE1 silencing transcription factor (REST)-imposed neuronal gene silencing (Ding et al., 2009), as well as roles in cancer metastasis (He et al., 2016; Liu et al., 2019; Wen et al., 2013). We now identify MED19 as a key player in adipogenesis and maintenance of WAT mass.
RESULTS
MED19 Is Required for White Adipogenesis
To study the role of MED19 in adipogenesis, we treated stromal vascular fraction (SVF) cells derived from inguinal WAT (iWAT) with MED19-targeting or scrambled lentiviral short hairpin RNA (shRNA) and stimulated the cells to differentiate into white adipocytes (Figure 1). MED19 knockdown almost completely blocked adipogenesis, leading to a striking decrease in lipid droplet accumulation (Figure 1A), as well as significant decreases in adipocyte-specific markers and transcription factors (Figure 1B). Surprisingly, Krüppel-like factor 5 (KLF5), a zinc-finger transcription factor that is involved early in the adipogenic transcription cascade and critical for WAT development (Oishi et al., 2005), was increased following MED19 knockdown, suggesting possible compensation for a downstream transcriptional block. In support of this, we performed a time course analysis of key transcriptional events during in vitro adipogenesis in MED19 inducible knockout (KO) iWAT SVF cells (Figure S1A–S1G). Despite blocking adipogenesis in these cells, MED19 KO had no effect on or increased the expression of the early adipogenic transcription factors, such as CCAAT-enhancer-binding protein beta (C/EBPβ), C/EBPδ, and KLF5 (Figures S1B–S1D). However, MED19 KO cells failed to induce PPARγ and C/EBPα (Figures S1E and S1F) or downstream transcriptional targets, such as CD36 (Figure S1G). These results suggest that MED19 is required for maintaining C/EBPα and PPARγ gene expression and inactivation of this Mediator protein results in a feedback induction of KLF5, a transcription factor known to link C/EBPβ and C/EBPδ to PPARγ and C/EBPα in the adipogenic transcription cascade (Oishi et al., 2005).
Figure 1. MED19 Is Required for White Adipogenesis, and Mice with Adipose-Specific Knockout (KO) of MED19 Exhibit a Severe Loss of WAT and Whitening of BAT.
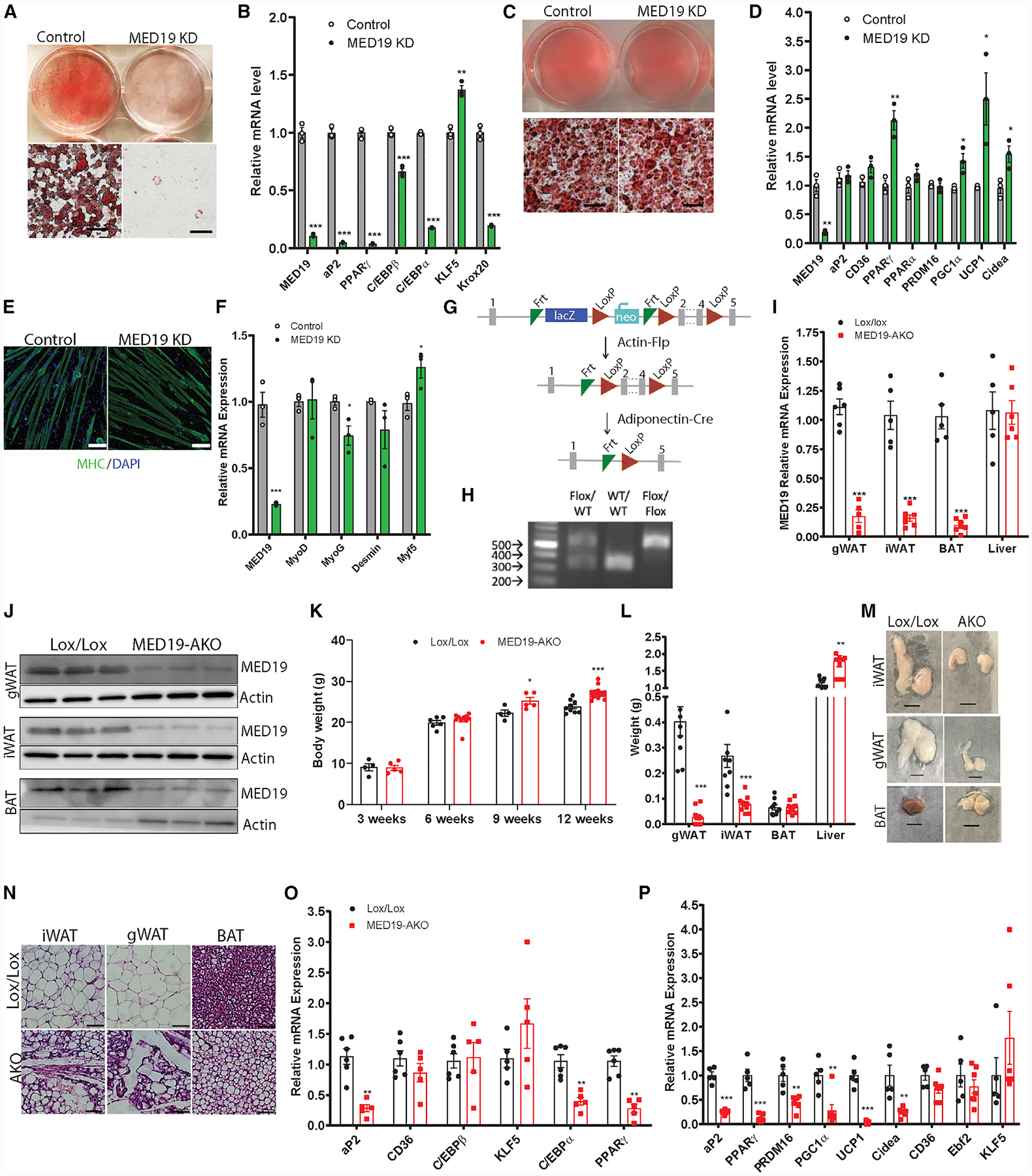
(A) Oil red O staining in differentiated iWAT SVF cells. Scale bar, 300 μm
(B) qPCR analysis of differentiated iWAT SVF cells; n = 3 each.
(C) Oil red O staining in differentiated BAT SVF cells. Scale bar, 300 μm
(D) qPCR analysis of differentiated BAT SVF cells; n = 3 each.
(E) Images of differentiated C2C12 cells stained with DAPI (blue) and myosin heavy chain (MHC) antibody (green) at 50×. Scale bar, 100 μm
(F) qPCR analysis of differentiated C2C12 cells; n = 3 each.
(G) Schematic of MED19 gene targeting strategy.
(H) Genotyping of MED19 floxed mice.
(I) qPCR analysis of MED19 expression in adipose tissue depots and liver of MED19-AKO and control animals; n = 5–7.
(J) Western blot analysis using MED19 and actin antibodies in adipose tissue depots.
(K) Body weight of control and MED19-AKO mice at 3 (n = 4 each), 6 (n = 7 Lox/Lox, 10 AKO), 9 (n = 4 Lox/Lox, 5 AKO), and 12 weeks of age (n = 9 Lox/Lox, 20 AKO).
(L) Tissue weight of gWAT, iWAT, BAT, and liver; n = 9 Lox/Lox and n = 10 MED19-AKO mice.
(M) Gross images of iWAT, gWAT, and BAT from Lox/Lox and MED19-AKO mice. Scale bar, 0.5 cm.
(N) H&E staining in iWAT, gWAT, and BAT. Scale bar, 75 μm.
(O) qPCR analysis of gWAT; n = 6 Lox/Lox and n = 5 MED19-AKO.
(P) qPCR analysis of BAT; n = 5 Lox/Lox and n = 7 MED19-AKO.
*p < 0.05, **p < 0.005, and ***p < 0.0005. For all panels, the error bars represent the SEM. See also Figure S1.
To determine if MED19 is a specific mediator of white adipogenesis, we also knocked down its expression in SVF cells derived from BAT of neonatal mice, followed by differentiation into brown adipocytes. Notably, MED19 knockdown did not inhibit adipogenesis in brown adipocytes and even led to a significant increase in expression of the brown adipocyte genes (Figures 1C and 1D). Moreover, MED19 knockdown in C2C12 myoblasts did not influence their ability to differentiate into myotubes (Figures 1E and 1F), suggesting that MED19 plays a specific role in adipogenesis rather than acting as a general regulator of transcription. Consistent with this notion, previous studies demonstrate that MED19 deficiency does not affect the structural integrity of the Mediator complex in yeast or mammalian cells or cause a gross transcriptional defect and cell death (Baidoobonso et al., 2007; Ding et al., 2009). Assessment of cell viability using the WST-1 reagent assay confirmed that MED19 knockdown does not affect the metabolic health of iWAT SVF cells and C2C12 myoblasts (Figures S1H and S1I).
Generation of Adipose-Specific MED19 KO Mice
To investigate the role of MED19 in adipose tissue, we obtained mice with a conditional-potential allele of MED19 from the European Conditional Mouse Mutagenesis Program (EUCOMM) repository, crossed them with actin-Flp transgenic mice to remove the targeting cassette, and then subsequently crossed the mice with adiponectin-Cre mice to generate adipose-specific MED19 KO (MED19-AKO) mice (Figures 1G and 1H). The mutant mice were overtly normal into the early adulthood (8 weeks) and fertile, at least until 12 weeks of age. Quantitative real-time PCR and western blot analyses showed that MED19 RNA and protein were significantly reduced in gonadal WAT (gWAT), iWAT, and BAT, but not the livers of MED19-AKO mice (Figures 1I and 1J).
MED19-AKO Mice Exhibit a Severe Atrophy of WAT and Whitening of Brown Fat
Although younger MED19-AKO mice weighed the same as their control littermates, older male MED19-AKO mice had increased body weight (Figure 1K), a trend that we saw in female mice as well (Figure S1J). Food intake was not significantly different between control and MED19-AKO mice (Figure S1K). Body composition analysis revealed reduced fat mass but slightly increased lean mass in both male and female mice (Figures S1L and S1M). Despite the increased body weight, weights of white fat depots (gWAT and iWAT) were dramatically reduced in MED19-AKO mice (Figure 1L), resembling mice with adipose-specific KO of PPARγ (Wang et al., 2013) or insulin receptor (Qiang et al., 2016), which weigh more than wild-type mice in the context of a dramatic reduction in adipose tissue mass. Other smaller WAT depots, such as anterior subcutaneous WAT (asWAT), retroperitoneal WAT (rWAT), and mesenteric WAT (mWAT), were also reduced in MED19-AKO mice (Figure S1N). Liver weight was markedly increased in MED19-AKO mice (Figure 1L), explaining their elevated body weight. In contrast to the striking loss of WAT, MED19-AKO mice had no change in BAT mass (Figure 1L). We also determined the effect of MED19 inactivation on adipose tissue of young mice at ~3 weeks of age, when they are undergoing a rapid growth of adipose tissue. Interestingly, the young MED19-AKO mice also showed a significant reduction in gWAT and iWAT mass but no change in BAT mass, suggesting that MED19 might play a role early on in WAT development (Figure S1O). In adult mice, gross examination revealed that the interscapular BAT of KO animals was much paler, resembling WAT (Figure 1M). The small amount of remaining white fat in MED19-AKO animals appeared unhealthy in histological sections, showing sparse, irregularly sized adipocytes among a fibrous extracellular matrix, and while brown fat contained many white-adipocyte-like unilocular cells (Figure 1N). Gene expression analysis of gWAT and iWAT from MED19-AKO animals showed decreased levels of the adipocyte genes aP2, C/EBPα, and PPARγ (Figures 1O and S1P). Consistent with the in vitro results, C/EBPβ and KLF5 expression was either unchanged or elevated in MED19-AKO animals, suggesting that MED19 may play a role downstream of these factors in the adipogenic transcription cascade. The expression of BAT genes, including UCP1, was also decreased in MED19-AKO mice (Figure 1P).
MED19-AKO Mice Exhibit Fatty Liver and Insulin Resistance
Given the decreased adiposity and increased liver weight, we next examined whether MED19-AKO mice exhibit hepatic steatosis and insulin resistance (Figure 2). Upon gross examination, livers of 12-week-old MED19-AKO animals were enlarged and pale (Figure 2A). Histological examination showed a massive accumulation of lipid droplets (Figure 2A) and 4-fold higher triglyceride content in the livers of MED19-AKO mice (Figure 2B). Assessment of glucose homeostasis revealed that both male and female MED19-AKO mice were hyperglycemic and glucose intolerant (Figures 2C and S2). Moreover, MED19-AKO mice displayed an impaired response to insulin in an insulin tolerance test (Figure 2D), as well as elevated fasting serum insulin levels (Figure 2E). KO mice had impaired hepatic insulin signaling, as assessed by phosphorylation of Akt in response to insulin (Figures 2F and 2G).
Figure 2. MED19-AKO Mice Have Fatty Liver and Insulin Resistance.
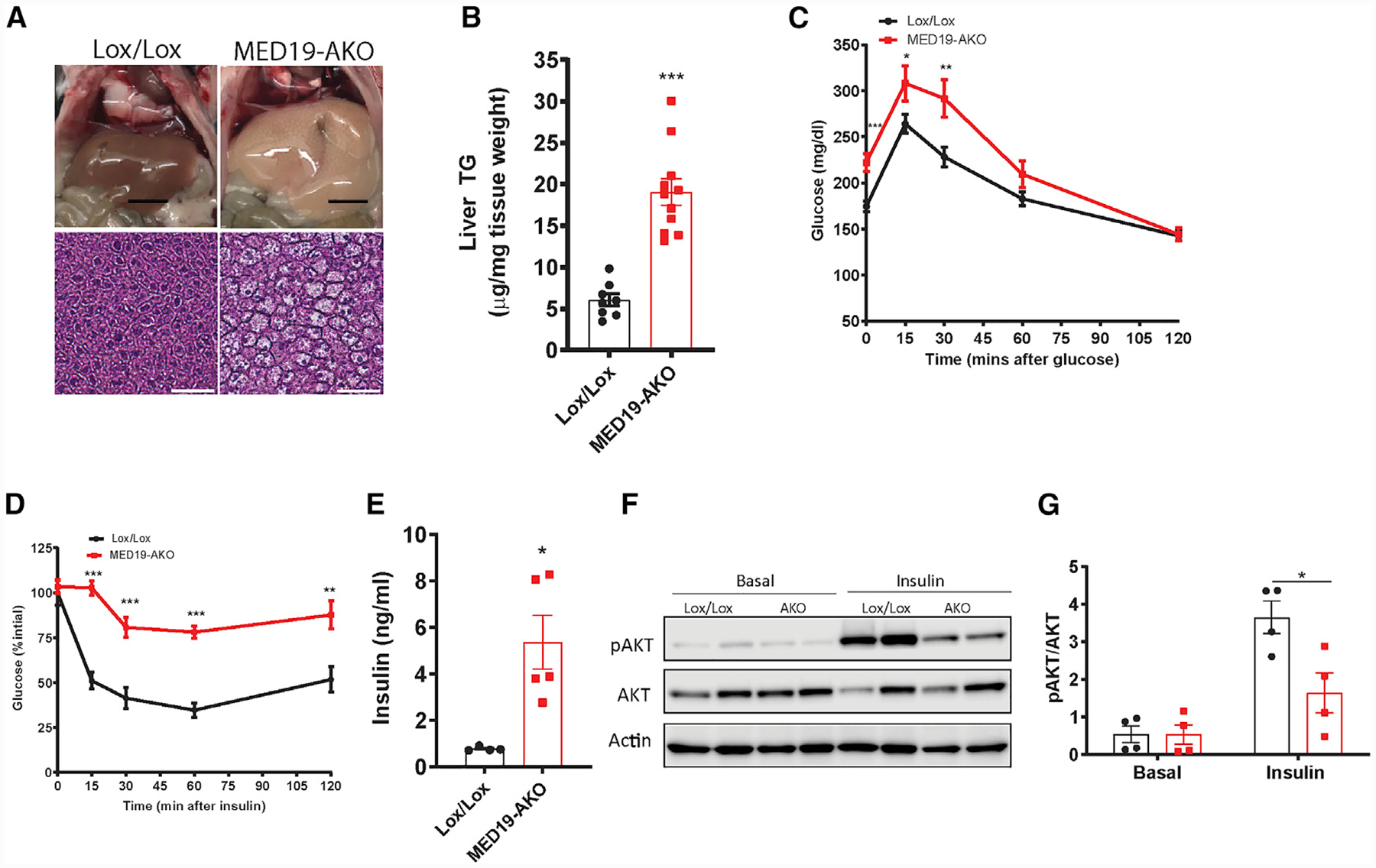
(A) Gross and histological images of H&E-stained liver. Scale bars represent 0.5 cm (gross liver images) and 75 μm (liver histology).
(B) Liver triglyceride content; n = 8 Lox/Lox mice and n = 11 MED19-AKO mice.
(C) Glucose tolerance test; n = 20 Lox/Lox mice and n = 13 MED19-AKO mice.
(D) Insulin tolerance test; n = 7 each.
(E) Serum insulin ELISA; n = 4 Lox/Lox mice and n = 5 MED19-AKO mice.
(F) Western blot analysis of Akt phosphorylation in livers of MED19-AKO and control mice at baseline and 10 min following insulin injection.
(G) Quantification of the results in (F); n = 4 each.
*p < 0.05, **p < 0.005, and ***p < 0.0005. For all panels, the error bars represent the SEM. See also Figure S2.
Deletion of MED19 in Mature Adipose Tissue Causes Lipodystrophy due to Increased Apoptosis and Macrophage Infiltration
To investigate the role of MED19 in mature adipose tissue, we created an adipose-specific, tamoxifen-inducible MED19 KO (MED19-iAKO) mouse line (Figure 3A). MED19-iAKO and Lox/Lox control mice were injected with tamoxifen for 5 consecutive days to induce the activity of the adiponectin-CreER fusion protein, resulting in KO of the gene in adipose tissue depots (Figure 3B). Twenty-one days later, MED19-iAKO mice had no change in body weight (Figure S3A) but showed a dramatic reduction in gWAT and iWAT mass (Figures 3C and 3D). Histologic analysis of the residual white fat in MED19-iAKO mice demonstrated a markedly decreased number of irregularly sized adipocytes, while BAT exhibited whitening, as indicated by accumulation of unilocular lipid droplets (Figure 3E). Oil red O staining confirmed that the pockets of white-adipocyte-like cells within BAT of MED19-iAKO were in fact adipocytes (Figure 3E).
Figure 3. Tamoxifen-Inducible KO of MED19 in Mature Adipose Tissue Causes Loss of WAT due to Increased Apoptosis and Macrophage Infiltration.
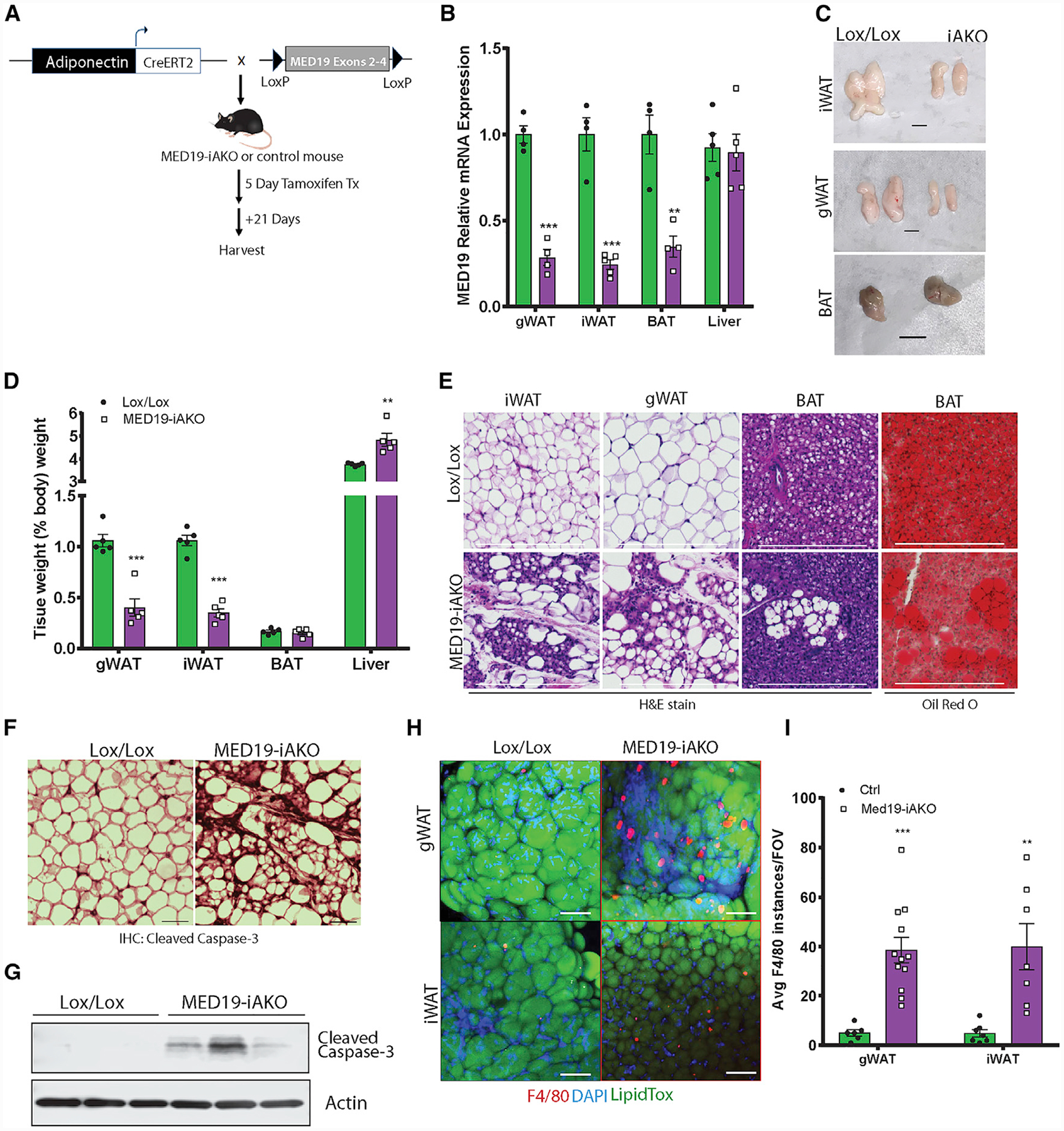
(A) Schematic of tamoxifen-inducible, adipose-specific MED19 KO.
(B) qPCR analysis of MED19 expression in adipose and liver of MED19-iAKO and control mice; n = 4–5 each.
(C) Gross images of iWAT, gWAT, and BAT from MED19-iAKO and control mice. Scale bar, 0.5 cm.
(D) Tissue weights of MED19-iAKO and control mice normalized to body weight; n = 5 each.
(E) Histology images of iWAT, gWAT, and BAT from MED19-iAKO and control mice. Scale bar, 300 μm.
(F) Staining of iWAT tissue with cleaved caspase-3 antibody. Scale bar, 75 μm.
(G) Western blot analysis of cleaved caspace-3 in iWAT.
(H) Whole mounts of iWAT from control and MED19-iAKO mice 10 days after tamoxifen injections were stained with F4/80 (red), LipidTox (green), and DAPI (blue). Scale bar, 100 μm.
(I) Quantification of F4/80 staining from (H).
**p < 0.005 and ***p < 0.0005. For all panels, the error bars represent the SEM. See also Figure S3.
We next sought to understand the mechanism underlying the dramatic loss of WAT following acute inactivation of MED19 in adult animals. To determine if increased lipolysis was responsible, we measured glycerol release from adipose tissue explants in the presence or absence of isoproterenol. Surprisingly, iWAT from MED19-iAKO mice displayed impaired lipolytic activity (Figure S3B), likely due to decreased gene expression of factors involved in lipolysis, including adipose triglyceride lipase (ATGL), hormone-sensitive lipase (HSL), and Abhd5 (Figure S3C).
Next, we determined whether MED19 inactivation promotes apoptosis in WAT. Western blot analysis and immunohistochemistry staining of cleaved caspase-3 indicated an accumulation of apoptotic cells in the iWAT of MED19-iAKO animals (Figures 3F and 3G). Consistent with increased adipose tissue inflammation and death, we observed significant macrophage infiltration in the gWAT and iWAT of MED19-iAKO mice as measured by F4/80 immunofluorescence analysis (Figures 3H and 3I).
MED19 Maintains Adipocyte Gene Expression by Facilitating PPARγ-Mediated Gene Expression
To gain a better understanding of how MED19 regulates the transcriptional landscape in adipose tissue, we performed RNA sequencing (RNA-seq) on gWAT of MED19-iAKO mice 14 days after tamoxifen injection (Figure 4A), followed by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of genes that were significantly up- or downregulated in MED19-iAKO animals compared to controls (Figure 4B). Induced genes established a clear pattern of almost exclusively being involved in pathways associated with cell death, inflammation, and inflammatory cell signaling/recruitment (Figure 4B). This analysis supports the hypothesis that MED19 KO in WAT leads to lipoatrophy via cell death and inflammation. KEGG pathway enrichment analysis for downregulated genes identified pathways important in both adipocyte-specific metabolism and signaling (Figure 4B). In particular, genes involved in the PPAR signaling pathway were significantly downregulated in MED19-iAKO adipose tissue, suggesting that MED19 may be involved in facilitating PPARγ-mediated gene expression. In order to determine transcriptional circuits facilitated by MED19, we applied the dataset of downregulated genes identified in RNA-seq (false discovery rate [FDR] <0.05) to Hypergeometric Optimization of Motif EnRichment (HOMER) analysis (Heinz et al., 2010), which found an enrichment of DNA-binding motifs for PPARγ, C/EBP transcription factors, and KLF4 (Figure 4C).
Figure 4. MED19 Maintains Adipocyte Gene Expression by Facilitating PPARγ-Mediated Gene Expression.
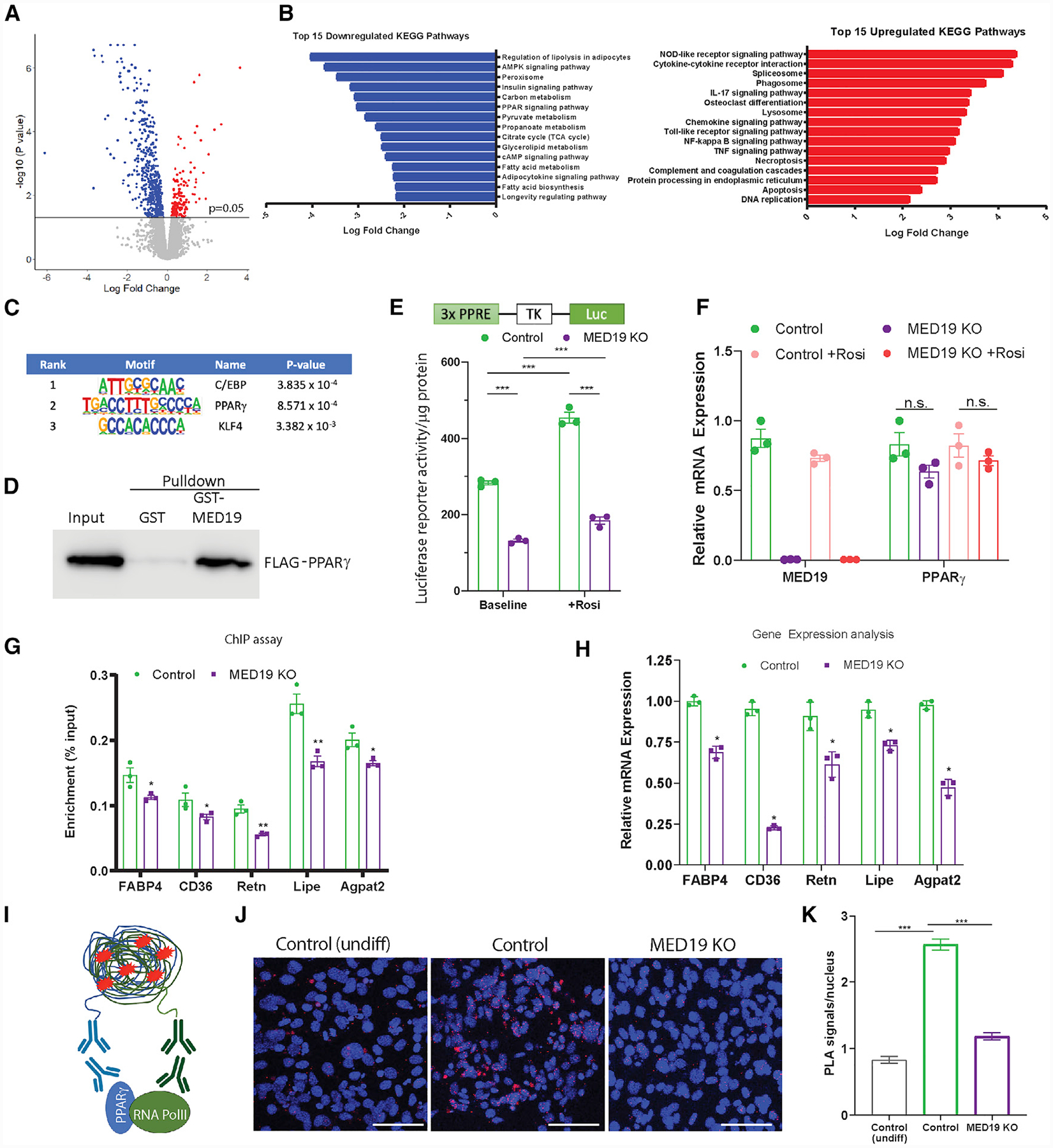
(A) Volcano plot of RNA-seq analysis in gWAT of 12-week-old MED19-iAKO and control mice 14 days after tamoxifen injections.
(B) Top 15 KEGG pathways identified in FDR-corrected, up-/downregulated gene expression subsets.
(C) HOMER analysis on FDR-corrected, downregulated genes. p values represent enrichment of indicated binding motif.
(D) FLAG-PPARγ2 pull-down using GST-tagged MED19.
(E) PPARγ-dependent luciferase reporter activity in MED19-iKO iWAT SVF cells transfected with pLJM1-PPRE-TK-Luc, pBABE-RXRα, and pLJM1-FLAGPPARγ2 and treated with vehicle or 1 μM 4-hydroxy-tamoxifen (4OH TAM) for 2 days; n = 3.
(F) Gene expression analysis of the cells from (E); n = 3 each.
(G) MED19-iAKO iWAT SVF cells were differentiated and treated with vehicle or 4-OH TAM before being harvested. ChIP was performed using an anti-PPARγ antibody, followed by qPCR using primers for the indicated genes. Results are background corrected to a normal immunoglobulin G (IgG) control and expressed as percent input; n = 3 each.
(H) Gene expression analysis by qPCR; n = 3 each.
(I) Schematic of the proximity ligation assay (PLA).
(J) Colocalization of PPARγ with RNA Pol II as determined by PLA in iWAT SVF cells after treatment with adipogenic cocktail. Red, PLA-positive signal; blue, DAPI. Scale bar, 75 μm.
(K) Quantification of the results from (J).
*p < 0.05, **p < 0.005, and ***p < 0.0005. For all panels, the error bars represent the SEM. See also Figure S4.
Given these findings and the similarities between our mice and those with adipose-specific deletion of PPARγ (Wang et al., 2013), we sought to determine whether MED19 interacts with PPARγ. Glutathione S-transferase (GST) pull-down assay indicated that MED19 interacts with FLAG-tagged PPARγ (Figure 4D). To investigate the effects of MED19 on PPARγ-mediated gene expression, we created a reporter assay by stably transfecting tamoxifen-inducible MED19 KO (MED19-iKO) iWAT SVF cell line with a PPARγ-dependent luciferase reporter along with PPARγ and RXRα. MED19 KO significantly blunted the ability of cells to activate the reporter at both baseline and in response to the PPARγ agonist rosiglitazone (Figure 4E), despite a lack of difference in PPARγ expression between control and MED19-KO cells (Figure 4F). To determine whether deletion of MED19 in adipocytes leads to decreased occupancy of PPARγ at PPARγ response elements (PPREs) of target genes, we performed chromatin immunoprecipitation (ChIP) assays using an antibody against PPARγ in adipocytes subjected to acute tamoxifen-inducible KO of MED19. Consistent with the result of the PPARγ reporter assay, ChIP-qPCR analysis of PPARγ binding to known PPREs (Lefterova et al., 2010; Rajakumari et al., 2013; Villanueva et al., 2011) was significantly reduced in MED19-KO cells (Figure 4G), resulting in decreased gene expression of PPARγ-dependent genes (Figure 4H).
Next, we determined whether overexpression of PPARγ alone or together with C/EBPα could rescue the effect of MED19 inactivation of adipogenesis. Interestingly, ectopic expression of PPARγ or PPARγ and C/EBPα in conjunction with rosiglitazone could not recue the adipogenesis defect in MED19-depleted cells (Figures S4A–S4E). Because the Mediator complex relays signals from upstream signaling pathways to the basal transcriptional machinery, we next determined whether MED19 links PPARγ to RNA polymerase II (RNA Pol II). To this end, we performed proximity ligation assay (PLA) (Figure 4I) under conditions identical to those used in the ChIP assay (Figure 4G). The results of the PLA assay indicate that KO of MED19 in iWAT SVF cells treated with adipogenic cocktail greatly reduces the association of PPARγ with RNA Pol II to levels similar to those in undifferentiated cells (Figures 4J and 4K), despite a lack of difference in PPARγ expression (Figure S4F). It is noteworthy that while sustained MED19 inactivation in mice or iWAT SVF cells results in decreased PPARγ gene expression, acute tamoxifen-inducible KO of the Mediator protein during adipogenesis in iWAT SVF cells as performed here did not impair PPARγ gene expression, thus allowing assessment of the effect on PPARγ transcriptional activity. Together, these results suggest that MED19 is necessary for the formation of an RNA Pol II/PPARγ adipogenic transcription complex and the maintenance of downstream PPARγ signaling required for adipocyte gene expression.
DISCUSSION
The Mediator is a multi-subunit complex that acts as a bridge between gene-specific transcription factors and the basal transcription machinery. MED19 is a poorly characterized Mediator subunit, but previous studies indicate that MED19 mutant yeast strains are viable, with no impairment of the global transcriptional program (Dettmann et al., 2010). Moreover, deletion of MED19 does not disrupt the structural integrity or function of the Mediator complex in both yeast (Baidoobonso et al., 2007) and HeLa cells (Ding et al., 2009). Given these findings, MED19 is a subunit that likely harbors more nuanced functions.
Here, we provide evidence suggesting that MED19 is essential for adipogenesis and maintenance of WAT mass. Knockdown of MED19 in iWAT SVF cells, but not C2C12 myoblasts, blocks differentiation, indicating a cell-specific role in adipogenesis. It is noteworthy that another Mediator subunit, MED23, is also dispensable for myoblast differentiation but important for hormone-induced adipogenesis in mouse embryonic fibroblasts by facilitating the activity of Elk1 early on in the adipogenic transcription cascade (Wang et al., 2009). As MED19 knockdown in iWAT SVF cells prevented induction of downstream adipogenic genes such as aP2 and C/EBPα but had a little to no effect on the expression of upstream genes such as C/EBPβ and KLF5, we hypothesized that MED19 is involved in the early stages of adipogenesis, similar to the role of MED23. However, unlike MED23, the block in adipogenesis due to MED19 inactivation could not be rescued by overexpression of C/EBPα and/or PPARγ (Figure S4). This suggests that MED19 plays a direct role in facilitating PPARγ activity rather than promoting PPARγ induction, since PPARγ overexpression and agonism has been shown to not only be sufficient for adipogenesis but also rescue upstream blocks in adipogenesis in vitro (Rosen et al., 2002; Tontonoz et al., 1994; Wang et al., 2009; Zuo et al., 2006).
In mice, adipose-specific deletion of MED19 resulted in near-ablation of WAT, leading to hepatic steatosis and insulin resistance. BAT mass was unaffected, but MED19-AKO mice manifested a significant whitening of the thermogenic fat. Although many genetic KO mouse models have been reported to have lipodystrophic phenotypes (Rochford, 2014; Savage, 2009), it is unusual to see such a severe defect in WAT that is not also seen in BAT. Of note, mice with a heterozygous KO for KLF5 (KLF+/−) show significant defects in neonatal WAT development but exhibit no difference in interscapular BAT mass compared to wild-type littermates (Oishi et al., 2005). Similarly, C/EBPα null mice rescued by transgenic hepatic expression have an absence of WAT, with no defects in BAT outside of mild adipocyte hypertrophy (Linhart et al., 2001). Thus, it is possible that MED19 is part of a transcription regulatory network that includes C/EBPα and KLF5.
To understand the cell-intrinsic role of MED19 in brown adipocytes, we knocked down its expression in SVF cells derived from BAT of neonatal mice. Surprisingly, the knockdown did not block brown adipogenesis and in fact significantly increased brown adipocyte gene expression. The inability of MED19 knockdown to block brown adipocyte differentiation supports the possibility that MED19 does not affect BAT development in mice. However, the effect of MED19 knockdown on gene expression in neonatal brown adipocytes is in contrast to the decreased PPARγ and UCP1 gene expression in BAT of adult MED19 KO mice. It is possible that MED19 is not necessary for BAT development and may even inhibit thermogenic gene expression in neonatal mice but is later required to maintain BAT gene expression as mice get older. Similarly, PRDM16 is dispensable for embryonic development of BAT but is required for maintenance of brown adipocyte identity and function in adult animals (Harms et al., 2014).
Adipose-specific MED19 KO in adult animals suggests that this factor is critical for maintenance of WAT mass. RNA-seq analysis of MED19-iAKO gWAT revealed upregulation of genes involved in apoptosis, necroptosis, and inflammation and downregulation of PPARγ-dependent genes. WAT in MED19-iAKO animals showed a marked increase in macrophage infiltration and accumulation of apoptotic cells, a phenotype very similar to that of mice with adipose-specific deletion of PPARγ (Wang et al., 2013). These data suggest that MED19 is necessary for mediating PPARγ transcriptional activity. HOMER analysis of our RNA-seq dataset revealed that downregulated genes in MED19-AKO adipose tissue had enrichment of PPARγ binding motifs, further implicating MED19 in PPARγ signaling. In support of this possibility, MED19-iKO iWAT SVF cells showed reduced residency of PPARγ on adipose-specific target genes and blunted PPARγ luciferase reporter activity.
In summary, our studies identify MED19 as a crucial facilitator of PPARγ-mediated gene expression that is required for adipogenesis and maintenance of WAT mass. Future studies aimed at understanding precisely how MED19 links signals from PPARγ to the basal transcriptional machinery to preferentially control white fat levels could identify a novel therapeutic strategy for treating obesity.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Irfan J. Lodhi (ilodhi@wustl.edu).
Materials Availability
The unique cell lines and mouse models generated in this study are available from the Lead Contact, Irfan J. Lodhi (ilodhi@wustl.edu) upon completion of a materials transfer agreement.
Data and Code Availability
The MED19-iAKO gWAT RNA-sequencing data have been deposited in the GEO repository and can be accessed with accession code GSE150250
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
To create MED19-AKO mice, we obtained mice with conditional-potential allele of MED19 (MED19tm1a(EUCOMM)Wtsi from the EUCOMM repository and crossed them with actin-Flp transgenic mice to remove the targeting cassette and then subsequently crossed the mice with adiponectin-Cre mice obtained from the Jackson Laboratory (028020) to generate adipose-specific MED19 knockout (MED19-AKO) mice. To generate adipose-specific, tamoxifen-inducible MED19 KO (MED19-iAKO) mice, MED19 floxed animals were crossed with Adiponectin-CreER mice obtained from the Jackson Laboratory (025124). To induce knockout, 12-week-od control (MED19Lox/Lox without Cre) and MED19-iAKO mice were treated daily with tamoxifen (100 μg/g body weight) for five consecutive days. All experiments used male mice unless otherwise stated. All mice were kept on a 12h light/dark cycle and provided ad libitum access to food (Purina 5053) and water. All animal experiments were performed in accordance with procedures approved by the Institutional Animal Care and Use Committee at Washington University School of Medicine.
Cell Lines
Stromal vascular fraction cells from adult iWAT and postnatal day 1 interscapular BAT were isolated from wild-type C57 mice by collagenase and dispase digestion, immortalized, and differentiated into adipocytes as previously reported (Lodhi et al., 2017; Park et al., 2019). Inducible MED19 KO iWAT SV cells (MED19-iKO) were generated by treating iWAT SVF cells obtained from MED19Lox/Lox animals with MSCV CreERT2 puro retrovirus (Addgene #22776). C2C12 cells were maintained in DMEM supplemented with 10% FBS. To induce differentiation, C2C12 cells at ~80% confluence were switched to DMEM containing 2% horse serum. Human embryonic kidney 293T (HEK293T) cells were maintained in DMEM supplemented with 10% FBS. Cell viability was assessed by colorimetric assay using Cell Proliferation Reagent WST-1 (Roche).
METHOD DETAILS
Plasmid Constructs
Plasmid encoding lentivirus shRNA for mouse MED19 (TRCN000258189) was obtained from Sigma-Aldrich (St. Louis, MO). Packaging vector psPAX2 (12260), envelope vector pMD2.G (12259), and scrambled shRNA plasmid (1864) were obtained from Addgene. PPRE-TK-luc lentiviral reporter plasmid was generated by replacing the CMV-eGFP insert in pLJM1-eGFP with PPRE-TK-luciferase from the PPRE X3-TK-luc plasmid (Addgene #1015). Similarly, the pLJM1-FLAG-PPARγ was created by cloning FLAG-tagged PPARγ in place of eGFP in the pLJM1-eGFP vector.
Immunohistochemistry and Immunofluorescence Imaging
For C2C12 cell immunofluorescence imaging, cells were fixed using ice-cold methanol at −20°C for 10 min and permeabilized with 0.1% Triton X-100 in PBS (PBS-T). Cells were incubated with primary MYH1 anti-mouse (DSHB MF20) antibody at 1:100 dilution in PBS-T at 4°C overnight. Alexa Fluor conjugated secondary antibody (Invitrogen A-11008) was used at a dilution of 1:500 at room temperature for 1 hr. For IHC detection of cleaved caspase-3, formalin-fixed, paraffin-embedded adipose tissue sections were de-paraffinized and incubated with anti-cleaved caspase-3 antibody (Cell Signaling Technology D175) followed by detection using the Vectastain Universal Elite ABC kit (Vector Labs) before being mounted and imaged with a Leica DMI4000B fluorescence microscope. Immunofluorescent imaging of whole adipose tissue was done as previously described (Berry et al., 2014). Briefly, freshly harvested adipose tissue was washed with PBS before being incubated with Alexa fluor 647-conjugated rat anti-mouse F4/80 antibody (Biolegend 123121) at 1:100 overnight, HCS LipidTox (Invitrogen H34475) at 1:100 for 1hr, and DAPI (Invitrogen D1306) at 1:10,000 for 1hr. After antibody incubation, tissue was washed and mounted using Vectashield mounting medium (Vector Laboratories H-1000) and imaged Nikon A1Rsi Confocal Microscope. Quantification was performed using ImageJ software. For oil red O staining, tissue was harvested and transferred to a 4% paraformaldehyde solution overnight. The tissue was subsequently incubated overnight in a 15% sucrose solution and again in a 30% sucrose solution before being embedded in OCT compound and put in −20C overnight and sectioned. Slides are then dipped in 10% formalin for 10 mins, 60% isopropanol once, Oil Red O solution for 15mins, 60% isopropanol once, and then DI water 10x before mounting coverslips and imaging.
PPRE Luciferase Assay
MED19-iKO iWAT SVF cells were transduced with both pLJM1-FLAG-PPARγ, pBABE-RXR alpha, and pLJM1-PPRE-TK-Luciferase reporter plasmid lenti-/retroviruses. Cells were treated with media containing vehicle or 4-hydroxytamoxifen for 2 days. Cells were harvested and luminescence was measured using a Luciferase Assay System kit (Promega E1500). Luminescence was normalized to cellular protein as measured by Pierce BCA Assay kit (Thermo Fisher 23227).
RT-qPCR
Total RNA was isolated using TRIzol reagent (Invitrogen 15596026) or PureLink RNA Mini Kit (Invitrogen 12183018A). 2μg of isolated RNA was reverse transcribed using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems 4368814). qPCR was performed with a StepOnePlus Real-Time PCR System (Applied Biosystems) using PowerUP SYBR Green Master Mix (Applied Biosystems A25742). Assays were performed in duplicate and normalized to ribosomal protein L32.
Insulin ELISA and Insulin Signaling Assay
Serum was collected from 16-week old MED19-AKO males and insulin concentration was measured using an Ultra Sensitive Mouse Insulin ELISA kit (Crystal Chem 90080) according to the manufacturer’s suggested protocol. To measure insulin signaling in the liver, mice were fasted for 4–6 hr before tissue was collected and snap frozen in liquid nitrogen at the basal state or 10 mins after an IP injection of 5mU/g of insulin (Humulin R, Lilly). Tissue was placed in tissue lysis buffer (0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, 50mM Tris-HCL pH 7.6, 5mM EDTA, 150mM NaCl, 5mM NaF, and protease inhibitor) and homogenized (Glas-Col 099C-K54).
Ex vivo Lipolysis Assay
Ex vivo lipolysis assay was performed using the Lipolysis Colorimetric Assay Kit (Sigma, MAK211) according to the manufacturer’s instructions. Briefly, iWAT from control and MED19-iAKOmicewas harvested, minced, and ~50mg of tissue was cultured in 24-well plates overnight in DMEM F-12 containing insulin and dexamethasone. Tissue was washed 2x with adipocyte wash buffer and incubated in lipolysis buffer with or without 100 nM isoproterenol for 1.5 hr before measuring the amount of glycerol in the media using the assay kit.
Chromatin Immunoprecipitation
ChIP studies were conducted using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology) according to manufacturer’s instruction and as previously described (Lodhi et al., 2017). Briefly, MED19-iKO iWAT SVF cells were treated with differentiation media for 2 days before being switched to maintenance media with or without 4-hydroxy-tamoxifen for 2 days before being harvested. Cells were fixed with 1% formaldehyde for 15 min. Nuclei were harvested and chromatin digested by micrococcal nuclease before being lysed on ice by 3 sets of 10 s pulses from a Sonics VCX 500 probe sonicator at 30% amplitude. Chromatin was clarified by centrifugation before being incubated with normal rabbit IgG or rabbit anti-PPARγ (Cell Signaling Technology 81B8) antibodies at 4C overnight. ChIP-Grade protein G agarose beads were incubated for 4 h, spun, and washed. Crosslinks were reversed and DNA was purified by spin column. DNA was quantified by qPCR using PowerUp Sybr Green Master Mix (Thermo Fisher) in triplicate and data are shown as percentage of input. Primer sequences used in qPCR are as described previously (Lefterova et al., 2010; Rajakumari et al., 2013; Villanueva et al., 2011) and full sequences can be found in Table S1.
Proximity Ligation Assay
Proximity ligation assays were performed using Duolink reagents (Sigma) according to the manufacturer’s instructions with PPARγ (Cell Signaling 81B8) and RNA PolII (Sigma-Aldrich 05–623) primary antibodies. MED19-iKO iWAT SV cells were grown on chamber slides (Lab-Tek 177445) and differentiated into adipocytes by treating confluent cells with DMEM/F12 supplemented with 5μg/ml insulin, 0.25 μM Dexamethasone, 1μM rosiglitazone, and 0.5 μM isobutylmethylxanthine. After two days, the cells were switched to medium supplemented with only 1μM rosiglitazone and 5μg/ml insulin with or without 1μM 4-OHT for two days. Cells were then fixed with 4% paraformaldehyde (w/v) for 40 min and permeabilized with 0.5% (w/v) Triton X-100 in PBS for 15 min before being used in PLA assays.
GST Pull-down Assays
For GST pull-downs, cells were lysed using a buffer containing 50 mM HEPES (pH 7.4), 4 mM EDTA, 2 mM EGTA, 2 mM sodium pyrophosphate, 1% Triton X-100, 10 mM NaF, and protease inhibitors (P8340; Sigma-Aldrich) by rocking in the cold room for 15 mins followed by homogenization using a dounce homogenizer. Cell lysates were clarified by spinning down at 10,000 g for 10 mins and supernatant was mixed with an equal volume of the same buffer containing no Triton X-100, then 5 μg of GST alone beads or GST fusion protein beads and rocked at 4C for overnight. Samples were spun down at 2,500 g for 5 min and washed five times with lysis buffer containing 0.5% Triton X-100 before being resuspended in SDS sample buffer and subjected to SDS-PAGE.
RNA-sequencing and Analysis
gWAT was collected from 12 to 13-week-old female control or Med19-iAKO mice 14 days post tamoxifen injections. RNA was harvested using a combination of TRIzol reagent (Invitrogen 15596026) and PureLink RNA Mini Kit (Invitrogen 12183018A) before being submitted to the Genome Technology Access Center (GTAC) at Washington University School of Medicine in St. Louis for RNA sequencing and analysis.
Samples were prepared according to library kit manufacturer’s protocol, indexed, pooled, and sequenced on an Illumina HiSeq. Basecalls and demultiplexing were performed with Illumina’s bcl2fastq software and a custom python demultiplexing program with a maximum of one mismatch in the indexing read. RNA-seq reads were then aligned to the Ensembl release 76 top-level assembly with STAR version 2.0.4b (Dobin et al., 2013). Gene counts were derived from the number of uniquely aligned unambiguous reads by Sub-read:featureCount version 1.4.5 (Liao et al., 2014). Isoform expression of known Ensembl transcripts were estimated with Sailfish version 0.6.13 (Patro et al., 2017). Sequencing performance was assessed for the total number of aligned reads, total number of uniquely aligned reads, and features detected. The ribosomal fraction, known junction saturation, and read distribution over known gene models were quantified with RSeQC version 2.3 (Wang et al., 2012).
All gene counts were then imported into the R/Bioconductor package EdgeR (Robinson et al., 2010) and TMM normalization size factors were calculated to adjust for samples for differences in library size. Ribosomal genes and genes not expressed in the smallest group size minus one samples greater than one count-per-million were excluded from further analysis. The TMM size factors and the matrix of counts were then imported into the R/Bioconductor package Limma (Ritchie et al., 2015). Weighted likelihoods based on the observed mean-variance relationship of every gene and sample were then calculated for all samples with the voomWithQualityWeights (Liu et al., 2015). The performance of all genes was assessed with plots of the residual standard deviation of every gene to their average log-count with a robustly fitted trend line of the residuals. Differential expression analysis was then performed to analyze for differences between conditions and the results were filtered for only those genes with Benjamini-Hochberg false-discovery rate adjusted p values less than or equal to 0.05.
For each contrast extracted with Limma, global perturbations in known Gene Ontology (GO) terms and KEGG pathways were detected using the R/Bioconductor package GAGE (Luo et al., 2009) to test for changes in expression of the reported log 2 fold-changes reported by Limma in each term versus the background log 2 fold-changes of all genes found outside the respective term. The R/Bioconductor package heatmap3 (Zhao et al., 2014) and Pathview (Luo and Brouwer, 2013) was used to display annotated KEGG graphs across groups of samples for each GO term or KEGG pathway (respectively) with a Benjamini-Hochberg false-discovery rate adjusted p value less than or equal to 0.05.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are expressed as mean ± SEM unless stated otherwise. Comparisons between two groups were performed using an unpaired, two-tailed t test. ANOVA was used for more than 2 groups. A p value less than 0.05 was considered significant. Statistical analysis and graphs were generated using GraphPad Prism software.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal Anti-FLAG M2 | MilliporeSigma | F1804; RRID: AB_262044 |
| Rabbit polyclonal anti-Actin | Santa Cruz Biotechnology | SC-1616-R; RRID: N/A |
| Mouse monoclonal anti-RNA polymerase II | MilliporeSigma | 05–623; RRID: AB_309852 |
| Rabbit polyclonal anti-HA | Santa Cruz Biotechnology | sc-805; RRID: AB_631618 |
| Rabbit polyclonal anti-Phospho-Akt (Ser473) | Cell Signaling Technology | 9271; RRID: AB_329825 |
| Rabbit polyclonal anti-AKT | Cell Signaling Technology | 9272; RRID: AB_329827 |
| Rabbit monoclonal anti-PPARγ (81B8) | Cell Signaling Technology | 2443S; RRID: AB_823598 |
| Rabbit polyclonal anti-MED19 | Invitrogen | PA5–44383; RRID: AB_2610610 |
| Rabbit polyclonal anti-Cleaved Caspase-3 (Asp175) | Cell Signaling Technology | 9661; RRID: AB_2341188 |
| Mouse monoclonal anti-MHC | Developmental Studies Hybridoma Bank | MF20; RRID: AB_1293549 |
| Alexa Fluor® 647 anti-mouse F4/80 Antibody | Biolegend | 123121; RRID: AB_893492 |
| Peroxidase IgG Fraction Monoclonal Mouse Anti-Rabbit IgG, light chain specific | Jackson Immunoresearch | 211-032-171; RRID: AB_2339149 |
| Peroxidase AffiniPure Goat Anti-Mouse IgG, light chain specific | Jackson Immunoresearch | 115-035-174; RRID: AB_2338512 |
| Mouse monoclonal anti-RNA Polymerase II | MilliporeSigma | 05-623; RRID: AB_309852 |
| Bacterial and Virus Strains | ||
| E. coli: Stable competent | NEB | C3040I |
| E.coli: DH5a competent cells | Thermo Fisher Scientific | 18265017 |
| Biological Samples | ||
| Fetal Bovine Serum | GIBCO | 26140–079 |
| Horse Serum | Thermo Fisher Scientific | 16050122 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cell Proliferation Reagent WST-1 | Roche | 5015944001 |
| Insulin (IP injections) | Lilly | Humalin, R |
| Vectashield mounting medium | Vector Labs | H-1000 |
| DMEM high glucose | MilliporeSigma | D6429 |
| DMEM F-12 Ham | MilliporeSigma | D6421 |
| Tamoxifen | MilliporeSigma | T5648 |
| Corn Oil | MilliporeSigma | C8267 |
| 4-hydroxy-tamoxifen | MilliporeSigma | H-6278 |
| Dexamethasone | MilliporeSigma | D-1756 |
| Insulin | MilliporeSigma | I-5523 |
| Rosiglitazone | Cayman Chemical | 71740 |
| 3-Isobutyl-1-methylxanthine | MilliporeSigma | I5879 |
| Collagenase D | Roche | 11088866001 |
| Dispase II | MilliporeSigma | D4693 |
| Ampicillin | GoldBio | A-301–5 |
| Penicillin-Streptomycin | GIBCO | 15140–122 |
| 0.05% Trypsin-EDTA | GIBCO | 25300–054 |
| Triton X-100 | MilliporeSigma | T8787 |
| Lipofectamine 2000 Reagent | Invitrogen | 11668–027 |
| HCS LipidTox | Invitrogen | H34475 |
| PowerUP SYBR Green Master Mix | Applied Biosystems | A25742 |
| TRIzol reagent | Invitrogen | 15596026 |
| SuperSignal West Femto Maximum Sensitivity Substrate | Thermo Fisher Scientific | 34096 |
| Protease inhibitor | MilliporeSigma | P8340 |
| DAPI | MilliporeSigma | 10236276001 |
| Precision Plus Protein Kaleidoscope Prestained Protein Standards | Bio-Rad | 1610375 |
| Protein A/G PLUS-Agarose beads | Santa Cruz | Sc-2003 |
| Critical Commercial Assays | ||
| Duolink In Situ Red Starter Kit Mouse/Rabbit | MilliporeSigma | DUO92101 |
| SimpleChIP® Plus Enzymatic Chromatin IP Kit | Cell Signaling Technology | 9004 |
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | 23225 |
| Ultra Sensitive Mouse Insulin ELISA kit | Crystal Chem | 90080 |
| Lipolysis Colorimetric Assay Kit | MilliporeSigma | MAK211 |
| Vectastain Universal Elite ABC kit | Vector Labs | PK-6200 |
| Luciferase Assay System kit | Promega | E1500 |
| High-Capacity cDNA Reverse Transcription kit | Applied Biosystems | 4368814 |
| PureLink RNA mini kit | Invitrogen | 12183018A |
| Deposited Data | ||
| MED19-iAKO gWAT RNA-seq data | This paper | GSE150250 |
| Experimental Models: Cell Lines | ||
| C2C12 | ATCC | CRL-1772 |
| HEK293T | ATCC | CRL3216 |
| iWAT SVF cells | Lodhi et al., 2017 | N/A |
| BAT SVF cells | Lodhi et al., 2017 | N/A |
| PPARγ-MEFs | Lodhi et al., 2017 | N/A |
| MED19 iWAT SVF cells | This paper | N/A |
| MED19-iKO | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: MED19-AKO | This paper | N/A |
| Mouse: MED19-iAKO | This paper | N/A |
| Mouse: Adiponectin Cre | The Jackson Laboratory | 028020 |
| Mouse: Adiponectin CreERT2 | The Jackson Laboratory | 025124 |
| Oligonucleotides | ||
| See Table S1 for oligonucleotide sequences | This paper | |
| Recombinant DNA | ||
| psPAX2 | Addgene | #12260 |
| pMD2.G | Addgene | #12259 |
| Scramble shRNA | Addgene | #1864 |
| pLJM1-EGFP | Addgene | #19319 |
| PPRE X3-TK-luc | Addgene | #1015 |
| pBABE puro PPAR gamma 2 | Addgene | #8859 |
| MED19 lentivirus shRNA | MilliporeSigma | TRCN000258189 |
| MSCV CreERT2 puro | Addgene | #22776 |
| pBABE-neo largeTcDNA | Addgene | #1780 |
| pLJM1-PPREX3-TK-luc | This paper | N/A |
| pLJM1-FLAG-PPARγ | This paper | N/A |
| pBABE puro human RXR alpha | Addgene | #11441 |
| pLJM1-FLAG C/EBP alpha | This paper | N/A |
| pcDNA-Myc PPARγ | This paper | N/A |
| pGEX-4T-1-MED19 | This paper | N/A |
| Software and Algorithms | ||
| Prism 8 | GraphPad | https://www.graphpad.com |
| ImageJ | ImageJ | https://imagej.nih.gov/ij/ |
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ngs/index.html |
| Image Studio | LI-COR Biosciences | https://www.licor.com/bio/image-studio/ |
Highlights.
MED19 knockdown blocks white adipogenesis, but not brown adipogenesis
Adipose-specific MED19 knockout promotes dramatic loss of WAT and whitening of BAT
MED19 KO decreases adipocyte genes and increases apoptosis and inflammation genes
MED19 interacts with PPARγ and links PPARγ to RNA Pol II
ACKNOWLEDGMENTS
This work was supported by NIH grants R01-DK115867 and R01-DK118333 to I.J.L. The core services of the Washington University Diabetes Research Center (DK020579) and the Nutrition Obesity Research Center (DK056341) also provided support for this work.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108228.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Allen BL, and Taatjes DJ (2015). The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol 16, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baidoobonso SM, Guidi BW, and Myers LC (2007). Med19(Rox3) regulates intermodule interactions in the Saccharomyces cerevisiae mediator complex. J. Biol. Chem 282, 5551–5559. [DOI] [PubMed] [Google Scholar]
- Berry R, Church CD, Gericke MT, Jeffery E, Colman L, and Rodeheffer MS (2014). Imaging of adipose tissue. Methods in Enzymology 537, 47–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Yang Q, and Roeder RG (2009). Dynamic interactions and cooperative functions of PGC-1alpha and MED1 in TRalpha-mediated activation of the brown-fat-specific UCP-1 gene. Mol. Cell 35, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmann A, Jäschke Y, Triebel I, Bogs J, Schröder I, and Schüller HJ (2010). Mediator subunits and histone methyltransferase Set2 contribute to Ino2-dependent transcriptional activation of phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Mol. Genet. Genomics 283, 211–221. [DOI] [PubMed] [Google Scholar]
- Ding N, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, and Boyer TG (2009). MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression. J. Biol. Chem 284, 2648–2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, and Roeder RG (2002). Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature 417, 563–567. [DOI] [PubMed] [Google Scholar]
- Ge K, Cho YW, Guo H, Hong TB, Guermah M, Ito M, Yu H, Kalkum M, and Roeder RG (2008). Alternative mechanisms by which mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor gamma-stimulated adipogenesis and target gene expression. Mol. Cell. Biol 28, 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grøntved L, Madsen MS, Boergesen M, Roeder RG, and Mandrup S (2010). MED14 tethers mediator to the N-terminal domain of peroxisome proliferator-activated receptor gamma and is required for full transcriptional activity and adipogenesis. Mol. Cell. Biol 30, 2155–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MJ, Ishibashi J, Wang W, Lim HW, Goyama S, Sato T, Kurokawa M, Won KJ, and Seale P (2014). Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab. 19, 593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He GY, Hu JL, Zhou L, Zhu XH, Xin SN, Zhang D, Lu GF, Liao WT, Ding YQ, and Liang L (2016). The FOXD3/miR-214/MED19 axis suppresses tumour growth and metastasis in human colorectal cancer. Br. J. Cancer 115, 1367–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida S, Chen W, Nakadai T, Ohkuma Y, and Roeder RG (2015). PRDM16 enhances nuclear receptor-dependent transcription of the brown fat-specific Ucp1 gene through interactions with Mediator subunit MED1. Genes Dev. 29, 308–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, Manduchi E, Grant GR, and Lazar MA (2010). Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol. Cell. Biol 30, 2078–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Linhart HG, Ishimura-Oka K, DeMayo F, Kibe T, Repka D, Poindexter B, Bick RJ, and Darlington GJ (2001). C/EBPalpha is required for differentiation of white, but not brown, adipose tissue. Proc. Natl. Acad. Sci. USA 98, 12532–12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Holik AZ, Su S, Jansz N, Chen K, Leong HS, Blewitt ME, Asselin-Labat ML, Smyth GK, and Ritchie ME (2015). Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 43, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Qi X, Zhang X, Gao D, Fang K, Guo Z, and Li L (2019). Med19 is involved in chemoresistance by mediating autophagy through HMGB1 in breast cancer. J. Cell. Biochem 120, 507–518. [DOI] [PubMed] [Google Scholar]
- Lodhi IJ, Dean JM, He A, Park H, Tan M, Feng C, Song H, Hsu FF, and Semenkovich CF (2017). PexRAP inhibits PRDM16-mediated thermogenic gene expression. Cell Rep. 20, 2766–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, and Brouwer C (2013). Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Friedman MS, Shedden K, Hankenson KD, and Woolf PJ (2009). GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi Y, Manabe I, Tobe K, Tsushima K, Shindo T, Fujiu K, Nishimura G, Maemura K, Yamauchi T, Kubota N, et al. (2005). Krüppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 1, 27–39. [DOI] [PubMed] [Google Scholar]
- Park H, He A, Tan M, Johnson JM, Dean JM, Pietka TA, Chen Y, Zhang X, Hsu FF, Razani B, et al. (2019). Peroxisome-derived lipids regulate adipose thermogenesis by mediating cold-induced mitochondrial fission. J. Clin. Invest 129, 694–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang G, Whang Kong H, Xu S, Pham HA, Parlee SD, Burr AA, Gil V, Pang J, Hughes A, Gu X, et al. (2016). Lipodystrophy and severe metabolic dysfunction in mice with adipose tissue-specific insulin receptor ablation. Mol. Metab 5, 480–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, Reed RR, and Seale P (2013). EBF2 determines and maintains brown adipocyte identity. Cell Metab. 17, 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochford JJ (2014). Mouse models of lipodystrophy and their significance in understanding fat regulation. Curr. Top. Dev. Biol 109, 53–96. [DOI] [PubMed] [Google Scholar]
- Rosen ED, and Spiegelman BM (2014). What we talk about when we talk about fat. Cell 156, 20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, and Spiegelman BM (2002). C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 16, 22–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DB (2009). Mouse models of inherited lipodystrophy. Dis. Model. Mech 2, 554–562. [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. (2005). A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, and Spiegelman BM (1994). Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 79, 1147–1156. [DOI] [PubMed] [Google Scholar]
- Villanueva CJ, Waki H, Godio C, Nielsen R, Chou WL, Vargas L, Wroblewski K, Schmedt C, Chao LC, Boyadjian R, et al. (2011). TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab. 13, 413–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscarra JA, Wang Y, Hong IH, and Sul HS (2017). Transcriptional activation of lipogenesis by insulin requires phosphorylation of MED17 by CK2. Sci. Signal 10, eaai8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Huang L, Huang Y, Yin JW, Berk AJ, Friedman JM, and Wang G (2009). Mediator MED23 links insulin signaling to the adipogenesis transcription cascade. Dev. Cell 16, 764–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, and Li W (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185. [DOI] [PubMed] [Google Scholar]
- Wang F, Mullican SE, DiSpirito JR, Peed LC, and Lazar MA (2013). Lipoatrophy and severe metabolic disturbance in mice with fat-specific deletion of PPARγ. Proc. Natl. Acad. Sci. USA 110, 18656–18661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Feng CC, Ding GX, Meng DL, Ding Q, Fang ZJ, Xia GW, Xu G, and Jiang HW (2013). Med19 promotes bone metastasis and invasiveness of bladder urothelial carcinoma via bone morphogenetic protein 2. Ann. Diagn. Pathol 17, 259–264. [DOI] [PubMed] [Google Scholar]
- Yin JW, Liang Y, Park JY, Chen D, Yao X, Xiao Q, Liu Z, Jiang B, Fu Y, Bao M, et al. (2012). Mediator MED23 plays opposing roles in directing smooth muscle cell and adipocyte differentiation. Genes Dev. 26, 2192–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiaoli, Zhao X, and Yang F (2013). The Mediator complex and lipid metabolism. J. Biochem. Pharmacol. Res 1, 51–55. [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Guo Y, Sheng Q, and Shyr Y (2014). Advanced heat map and clustering analysis using heatmap3. BioMed Res. Int 2014, 986048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Qiang L, and Farmer SR (2006). Activation of CCAAT/enhancer-binding protein (C/EBP) alpha expression by C/EBP beta during adipogenesis requires a peroxisome proliferator-activated receptor-gamma-associated repression of HDAC1 at the C/ebp alpha gene promoter. J. Biol. Chem 281, 7960–7967. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MED19-iAKO gWAT RNA-sequencing data have been deposited in the GEO repository and can be accessed with accession code GSE150250


